Abstract
A wealth of genetic information is available describing single-nucleotide variants in the human population that appear to be well-tolerated and in and of themselves do not confer disease. These variant data sets contain signatures about the protein structure-function relationships and provide an unbiased view of various protein functions in the context of human health. This information can be used to determine regional intolerance to variation, defined as the missense tolerance ratio (MTR), which is an indicator of stretches of the polypeptide chain that can tolerate changes without compromising protein function in a manner that impacts human health. This approach circumvents the lack of comprehensive data by averaging the data from adjacent residues on the polypeptide chain. We reasoned that many motifs in proteins consist of nonadjacent residues, but together function as a unit. We therefore developed an approach to analyze nearest neighbors in three-dimensional space as determined by crystallography rather than on the polypeptide chain. We used members of the GRIN gene family that encode subunits of NMDA-type ionotropic glutamate receptors (iGluRs) to exemplify the differences between these methods. Our method, 3DMTR, provides new information about regions of intolerance within iGluRs, allows consideration of protein–protein interfaces in multimeric proteins, and moves this important research tool from one-dimensional analysis to a structurally relevant tool. We validate the improved 3DMTR score by showing that it more accurately classifies the functional consequences of a set of newly measured and published point mutations of Grin family genes than existing methods.
The field of genetics has had a transformative effect on biology and clinical medicine. For example, advances in genetics can provide definitive clinical diagnoses for multiple disorders, breaking the cycle of specialist referrals that fail to yield actionable diagnoses or effective therapeutics for many patients (Heinzen et al. 2015; Stefanski et al. 2021). It has allowed assessment of risk factors that can be used to inform clinical care and preventive medicine (Müller-Nedebock et al. 2021), illuminated networks of biological processes through gene regulation, and allowed modeling of human disease in vivo (Shore et al. 2020). Understanding the genetic code has enabled manipulation of protein structure via site-directed mutagenesis to further understand protein function (Myers et al. 2019).
The field is currently in the midst of transformation brought about by low-cost sequencing, which allows large amounts of genetic information to be obtained from both patients and healthy populations (Landrum et al. 2016; Lek et al. 2016; Karczewski et al. 2020; Wang et al. 2020). This information provides insight into protein function and structure by highlighting the regions that are critical to human health (Petrovski et al. 2013; Traynelis et al. 2017). Specifically, whole-exome sequences are now available in numbers that allow the analysis of single-nucleotide variation (SNV) within the coding regions of any given gene for human disease. Recent efforts to condense large SNV data sets into a score for each residue of a given protein have resulted in development of the missense tolerance ratio (MTR). The MTR was defined as a running average over some number (typically 31) of adjacent residues in the polypeptide chain of the ratio of observed to expected variants (Traynelis et al. 2017). The intended function of the MTR is readily observed in the GRIN gene family, which encodes subunits comprising the NMDA receptor (NMDAR) (Ogden et al. 2017; Li et al. 2019). The NMDAR mediates a slow component of excitatory synaptic transmission and is linked to critical brain functions such as development, learning, and memory. Furthermore, NMDARs are implicated in multiple neurological diseases including schizophrenia, Alzheimer's disease, and neuronal death following acute injury (Traynelis et al. 2010; Paoletti et al. 2013; Hansen et al. 2017; Hansen et al. 2021). NMDARs are heterotetramers consisting of two GRIN1 and two GRIN2 protein subunits (GluN1 and GluN2, respectively, according to IUPHAR nomenclature), which are coded by the GRIN1 and GRIN2 genes. MTR evaluation of the relative frequency of GRIN variants in the healthy population reveals regions of the NMDAR subunits that are tolerant to change, that is, where there are many variants in healthy population genomic databases. It also highlights regions of the receptor that are intolerant to change, such as the parts of the polypeptide chain that contribute to the receptor channel gate (Ogden et al. 2017). This information has provided an unbiased view that highlights structural features not previously recognized as critical for function (Amin et al. 2020; Perszyk et al. 2020), such as the gating triad comprising three short stretches of the polypeptide chain, each a considerable distance from one another in their position in the open reading frame. Specifically, this gating triad is virtually devoid of variants in the healthy population and is a hotspot for disease-associated variants.
Despite providing insight into critical regions of a protein that are tolerant or intolerant to change, the MTR algorithm could still become a more accurate in silico predictor for pathogenicity of variants identified in patients. By averaging residues along the polypeptide chain, information contained in the secondary and tertiary structure of the protein is lost (Lal et al. 2020), which might have improved variant classification (Iqbal et al. 2020). We reasoned that a better indicator of residues that define an intolerant region would be distance within the protein structure. We thus created an algorithm that calculated the MTR for neighbors over a three-dimensional (3D) distance from crystallographic and cryo-EM data. We implemented this new approach (entitled 3DMTR) using GRIN variants to illustrate its ability to provide an accurate diagnostic tool. Moreover, the 3DMTR approach can operate on multimeric proteins to reveal information about protein interfaces, such as the tetrameric glutamate receptors, which are potential hotspots for disease-causing SNVs. Furthermore, we use a validation data set to specifically compare the performance of the 3DMTR approach to the one-dimensional MTR (1DMTR).
Results
The 1DMTR score identifies only some of the critical elements of the GRIN2A subunit of NMDARs
The recently published MTR algorithm (1DMTR) converts population genomic variant data into a single metric by incorporating data from neighboring residues (typically 31 residues) because the variant data sets are currently sparse. For NMDARs, the 1DMTR does a good job identifying stretches of GRIN2A that are well-described as being critical for function (Fig. 1A). For example, the 1DMTR method readily identifies a stretch of completely intolerant residues (MTR = 0) that corresponds to the SYTANLAAF motif, which is conserved in all glutamate receptors (Traynelis et al. 2010). To understand what this score captures, one can observe the occurrence of the gnomAD variants (version v2.1.1) (Karczewski et al. 2020) throughout the gene sequence (Fig. 1B) or by viewing their location on the subunit structure (Fig. 1C). The 1DMTR score decreases as there are fewer missense variants than synonymous variants.
Figure 1.
The sequential MTR score (1DMTR) of GRIN2A and structural mapping of GRIN2A variants. (A) The 1DMTR score of GRIN2A, calculated using gnomAD v2.1.1. (B) Linear domain map of the GRIN2A gene semiautonomous domains of the receptor. The linear map matches the x-axis of the graph in A. A raster plot of the GRIN2A variants found in gnomAD is plotted below the linear map: (orange) synonymous variants; (cyan) missense variants. (C) View of the GRIN2A structure (homology model) (Supplemental File 1) depicting the GRIN2A variants shown in B. (D) View of the GRIN2A subunit illustrating the extracellular and transmembrane domains. The NTD is depicted in cyan, the first ABD segment (ABD-S1) in magenta, the second ABD segment (ABD-S2) in purple, the TMD in green, and the linkers in gray. (E) View of the GRIN2A subunit colored to depict its position along the polypeptide chain, represented by the gradient shown below the structure. The gradient is shown next to the linear domain map depicting the main receptor domains. Some residues in the M1-M2 linker are missing from the structure, shown by the black region in the linear scale. (F) A closer view of the GRIN2A ABD highlighting the two portions of the polypeptide chain (ABD-S1, ABD-S2) that form the domain.
The 1DMTR method, however, has a deficiency in that it disregards interactions between residues located in distal parts of the polypeptide chain that interact to form functional domains (Fig. 1B,D–F), which may be more important than the influence of neighboring residues. In NMDARs, the agonist binding domains (ABDs) of the receptor each consists of two noncontiguous portions of the polypeptide chain (ABD-S1, ABD-S2) and form a bilobed clamshell-like domain, which flanks the first three intervening transmembrane helices (Fig. 1B,E,F). Residues in both lobes participate in agonist binding and facilitate clamshell closure, which is an initial necessary step in receptor activation. This provides a clear example that the most relevant residues are those that are nearby in 3D space and not those residues that are sequentially closest (Fig. 1E,F). For this reason, we developed a method to calculate the MTR score based on the location of each residue in space (3D) instead of its location on the polypeptide chain (1D).
Calculation of the three-dimensional MTR score
Advances in methods for determining protein structure have provided high-resolution crystallographic and cryo-EM structures for integral membrane protein complexes (such as iGluRs). It is conceptually straightforward, using these structures, to calculate the average location for all residues of the protein and then determine the inter-residue distances for all residues in the structure (Supplemental Fig. S1A). In the resulting matrix of residue distances for GRIN2A, there is a clear signature of the multiple subunit domains that have been studied (Meguro et al. 1992; Monyer et al. 1992; Ishii et al. 1993). For instance, we can see that the two halves of the bilobed ABD are very close in space (within ∼40–50 Å) but that the TMD is very distal to the NTD (>50 Å). We can impose simple logical criteria based on these distances to determine which subset of residues to use to calculate the MTR score, for example, to include the 31 spatially closest residues. As a result, the closest 31 residues in 3D space are distinct from the closest 31 residues in the polypeptide chain (Supplemental Fig. S1A,B).
Analysis of Pro686 in the ABD-S2 segment clearly shows how these two methods differ (Fig. 2A–C). Pro686 is proximal to the agonist binding site (Fig. 2D; Jespersen et al. 2014; Yu and Lau 2018). The closest 31 residues in terms of gene sequence (1D) (Fig. 1B) included residues on the outer surface of the ABD. The mean distance of these residues is 14.6 Å away from Pro686. The closest residues in 3D space are clustered around Pro686 (mean distance is 9.1 Å), in the middle of the ABD, and including residues from both ABD segments (five from ABD-S1, 14 from ABD-S2) (Fig. 2C).
Figure 2.
GRIN2A comparison of the closest residues as determined by sequence (1D) or inter-residue distances (3D). (A) Residue distances from Pro686. (A1) A linear heatmap raster plot and heatmap showing the distance of all residues in the GRIN2A subunit to residue Pro686. (A2) A closer view of the ABD in A1. (B) Closest 31 residues (1D) to Pro686. (B1) A linear raster plot and structural view of the 31 sequentially closest (1D) residues (blue) to Pro686 (red), with the side chains depicted by sticks. (B2) A closer view of the ABD in B1. (C) Closest 31 residues (3D) to Pro686. (C1) A linear raster plot and structural view of the 31 closest (3D) residues (blue) to Pro686 (red), side chains depicted by sticks. (C2) A closer view of the ABD in C1.
With this spatially derived set of residues, the MTR can be simply recalculated using the same equation (Methods; Fig. 3A,B; Supplemental Data Set 1). Overall, the 1DMTR and 3DMTR produce similar MTR score profiles but with several notable differences within the various receptor domains. In general, the NTD tends to have a higher MTR score in both methods, and the ABD and TMD domains tend to have lower (more intolerant) scores (Fig. 3A,B). Most residues have different scores (Supplemental Fig. S2A) with an average difference in MTR score of 0.157 (either positive or negative). More specifically, the majority of the NTD residue scores are above average using both methods (Mean1D = 0.612, Mean3D = 0.584). For the TMD, several residues of the SYTANLAAF motif have an MTR score of 0 for both the 1DMTR and 3DMTR method (five of nine residues). There is rearrangement of the most intolerant TMD residues, with a portion of the residues deemed highly intolerant (defined as outside the two standard deviation range) in either the 1DMTR or the 3DMTR analysis; however, there are more highly intolerant residues resulting from the 3DMTR method (11 with 1DMTR vs. 19 with 3DMTR, total residues 89, Fisher's exact test P = 0.1602). The majority of ABD residues have MTR scores that are more intolerant than the average in either method, but only the 3DMTR method identifies highly intolerant residues. A trend similar to TMD occurs in the linker residues; however, the residues deemed highly intolerant are nonoverlapping with slightly more residues in the 3DMTR analysis (10 vs. 7, 66 total residues, Fisher's exact test P = 0.6044). In particular the subset of ABD-TMD linkers are those with more highly intolerant residues in the 3DMTR analysis compared to the 1DMTR (10 vs. 1, 37 total residues, Fisher's exact test P = 0.0066).
Figure 3.
Comparison of the GRIN2A 1DMTR and 3DMTR intra-subunit scores. (A) The linear (1D) MTR score of GRIN2A. (A1) The 1DMTR score shown via a scatter plot and raster plot (see color bar for score representation; intolerant scores are blue, tolerant scores are red). (A2) View of the 1DMTR score heatmap applied to the GRIN2A structure. (A3) A closer view of the ABD in A2. (A4) A closer view of the TMD in A2. (B) The 3DMTR score of GRIN2A, illustrated similarly as in A. (C) The difference in the 3DMTR and the 1DMTR score of GRIN2A. The color bar for the raster plots and the structure heatmaps is shown above C2.
When the MTR scores from both methods are viewed as a scatter plot (score vs. residue), the 3DMTR scores appears more variable than 1DMTR (Fig. 3A1,B1). However, when mapped onto the GRIN2A structure, the scores result in similar patterns (Fig. 3A2–4,B2–4). The overall variance of the 3DMTR scores in this data set is slightly lower than the 1DMTR scores for GRIN2A (variance3D = 0.049, variance1D = 0.052) (Supplemental Fig. S2C), and the average 3DMTR for GRIN2A is actually lower than the 1DMTR (Mean3D = 0.584, Mean1D = 0.612, P = 0.014, unpaired t-test). The differences between the 1DMTR and 3DMTR scores can be extreme (Fig. 3C). The largest decrease in the MTR score occurs for Pro686 (difference3D-1D = −0.78, 3DMTR = 0.11, 1DMTR = 0.89) and the largest increase is seen at Tyr700 (difference3D-1D = +0.70, 3DMTR = 1.05, 1DMTR = 0.35). Because Pro686 is proximal to the agonist binding site (Jespersen et al. 2014; Yu and Lau 2018), the 3DMTR score is more appropriate than the 1DMTR score. Most differences are not as extreme (mean amplitude, 0.16; median amplitude, 0.13). We observe modest but consistent decreases in the MTR score across the ABD-TMD linkers and the agonist binding pocket and modest increases in some stretches of the TMD (Fig. 3C3,4). These differences result in the 3DMTR assessing the agonist binding pocket (central portions of the ABD), the channel selectivity filter (M2), the channel gate (extracellular portion of M3), and the ABD-TMD linkers as the most intolerant positions (Fig. 3B).
Calculation of the 3DMTR score of an entire protein complex
NMDARs are heterotetrameric assemblies of two GRIN2 subunits in complex with two GRIN1 subunits. The 3DMTR methodology used on the isolated GRIN2A subunit can be applied to the entire GRIN1/2A receptor complex (Fig. 4A; Supplemental Fig. S3; Supplemental Data Set 1). We performed the 3DMTR analysis on a GRIN1/2A homology model based on the highest resolution NMDAR structure available (Chou et al. 2020). This model contains two GRIN1 (Chains A and C) and two GRIN2A subunits (Chains B and D) (Supplemental Fig. S3). Whereas the majority of the closest residues are from within the same chain, there are important instances in which there is close contact to other polypeptide chains at protein–protein interfaces (Supplemental Fig. S3; Supplemental Fig. S4). In particular, the TMDs show considerable cross talk. For example, GRIN2A-Phe652 is in the SYTANLAAF motif conserved in all iGluRs and faces away from the pore but has important cross-subunit interactions that are essential for receptor function (Fig. 4B). The closest 31 residues to Phe652 as determined by the 1D, 3D (intra-subunit), and 3D (intra-receptor) are partially overlapping but distinct. In the 1DMTR method, we see the included residues span the M3 helix and the TMD-ABD-S2-linker (Fig. 4B1). However, the closest residues determined using the 3DMTR method include those from the Pre-M1 and the Pre-M4 linkers (Fig. 4B2). Moreover, the 31 closest residues determined by the 3DMTR method are closer than the 31 determined by the 1DMTR method (the mean distance 3DMTRreceptor is 8.3 Å, compared to 15.2 Å for the 1DMTR and 9.3 Å in the 3DMTRsubunit methods). In the context of the receptor, residues in the M3 of both chains of GRIN1 (A and C), the Pre-M4 of GRIN1 (Chain C), and the Pre-M1 of the same chain are included in the closest 31 residues to Phe652 (Fig. 4B3, Chain B). The receptor elements identified by this method have recently been identified as a “gating triad” that has a critical role in channel function (Chen et al. 2017; Gibb et al. 2018; Amin et al. 2020; Perszyk et al. 2020).
Figure 4.
Comparison of the included residues in the 1DMTR, 3DMTR intra-subunit, and 3DMTR intra-receptor calculation. (A) Structural views of the GRIN1/2A receptor. The GRIN1 subunits are colored in pale yellow and the GRIN2A subunits are colored pale green. The full receptor is shown in A1, a top down view of the four isolated NTDs of the receptor in A2, and a side view of one NTD dimer in A3. B) The closest 31 residues as determined using the 1D polypeptide sequence (B1), the inter-residue distances within just the GRIN2A subunit (B2), and the inter-residue distances within the GRIN1/2A receptor structure (B3). This viewpoint is depicted by the cartoon eye labeled (B) in A1. Of the closest 31 residues in the receptor structure to GRIN2A-Chain B-Phe652, there are three residues from Chain A, nine residues from Chain C, and 19 residues from Chain B. A view of the receptor structure and a raster plot of the closest residues are shown. In the structure views, the closest 31 residue side chains are depicted by sticks and colored based on the subunit coloring in A: (yellow) GRIN1; (green) GRIN2; (red) Phe652.
Using the GRIN1/2A receptor structure, we can calculate the 3DMTR (intra-receptor, or 3DMTRreceptor) for the entire protein complex (Fig. 5). In general, we see a similar MTR profile for both GRIN1 (Fig. 5A1) and GRIN2A (Fig. 5A2), which both have similarities to their 1DMTR profile. On average we see a modest increase (more tolerant) in the mean 3DMTRreceptor score for GRIN1 compared to 1DMTR (Mean3D-rec-chainC = 0.499, Mean1D = 0.473, P = 0.024, unpaired t-test) and a decrease in the GRIN2A score (Mean3D-rec-chainB = 0.566, Mean1D = 0.612, P < 0.0001, unpaired t-test). Similar to the previous 3DMTR calculation of the GRIN2A subunit, scatter plots of MTR score versus residue have higher apparent variability; however, the variance of these data sets is similar (GRIN1 variance3D-rec-chainC = 0.056, variance1D = 0.056; GRIN2A variance3D-rec-chainB = 0.055, variance1D = 0.052). The differences between the 1DMTR and the 3DMTR methods are influenced by the fact that for the spatially determined inclusion method used by 3DMTR, residues can have a variable number of times they are selected (Supplemental Fig. S5). Typically, residues within the interior of the protein tend to be included more often, and peripheral residues are included less often.
Figure 5.
3DMTR intra-receptor score of GRIN1/2A. (A) Scatter plots of the GRIN1 (A1) and GRIN2A (A2) 3DMTR (intra-receptor) score. (Below) Raster plots of each subunit's 1DMTR, 3DMTR intra-subunit, and the 3DMTR intra-receptor scores. (*) Primary differences between the intra-subunit and intra-receptor 3DMTR scores. (B) Structural heatmap views showing the 3DMTR (intra-receptor) score on the full receptor (B1), a top down view of the four isolated NTDs of the receptor (B2), a side view of one NTD dimer (B3); one of the GRIN1 (B4) and GRIN2A (B5) subunits are shown in isolation. All raster plots and structural heatmaps use the same color bar, shown at the top right of B5.
There are several changes to the 3DMTR (intra-receptor) profile that arise from the calculation using the entire NMDAR. In general, the ABD-TMD linkers tend to be more intolerant, and there is a rearrangement of the MTR score for residues in the ABD (Fig. 5). Overall, the 3DMTR results for the GRIN1/2A NMDAR show the clearest hotspots of genetic intolerance are located within the agonist binding pockets of the ABD and the channel pore (Fig. 5B1). The NTD is the most tolerant portion of the receptor (Fig. 5B1). However, when the NTD layer is viewed in isolation, the NTD heterodimer contains some intolerant regions (e.g., the NTD dimer interfaces) when compared with the rest of the domain (Fig. 5B2-3; Supplemental Fig. S6). Similar to the 3DMTR analysis of the isolated GRIN2A subunit, the channel selectivity filter (M2), the channel gate (M3), the ABD-TMD-linkers, and the agonist binding pocket are the most intolerant portions of the GRIN2A subunit when calculated with the entire GRIN1/2A NMDAR (Fig. 5B4; Supplemental Fig. S6). Similarly, the GRIN1 3DMTR (intra-receptor) identifies many of the same portions of the subunit as the most intolerant (the M2 channel selectivity filter, the M3 channel gate, and the ABD-TMD-linkers). More of the GRIN1 ABD has highly intolerant scores (as compared to GRIN2A), with many intolerant residues concentrated at the ABD-dimer interface (Fig. 5B4).
Permutation analysis identifies the most intolerant and tolerant residues
Because structural data is not available for all proteins, the 3DMTR method cannot be applied to the entire genome. It is thus impossible to calculate a universal likelihood of any given 3DMTR score among all proteins; therefore a global occurrence and relevance of a specific 3DMTR score cannot be determined. An alternative means of assessing relevance is to use permutation analysis to estimate the likelihood of a specific score in a given data set (protein complex and gnomAD variant data). In other words, permutation analysis provides a distribution of the possible scores that each residue can have. The randomization distribution for each residue provides a background level that is expected simply by chance and can be used to determine if the actual 3DMTR score (signal) is highly unlikely. We performed two different permutation analyses, one in which the residue locations were randomized (Fig. 6; Supplemental Fig. S7) and one in which the gnomAD data were randomized (Supplemental Fig. S8; Supplemental Fig. S9).
Figure 6.
Permutation analysis (residue randomization) identifies the 3DMTR scores that are unlikely given random chance. (A1) Scatter plot of the GRIN1 (Chain C) 3DMTR score (magenta line, black dots), the permutation 3DMTR score mean (black line), and ±1 and ±2 standard deviations of the permutation 3DMTR score (gray areas). The mean and standard deviation are calculated for each residue based on the permutation results for that particular residue (for the distributions of several example data sets, see Supplemental Fig. S10). (A2) Raster plots of the GRIN1 3DMTR score and the calculated significance via permutation analysis. A residue is deemed highly intolerant (blue, less than the permutation analysis mean minus 2 standard deviations) and highly tolerant (red, more than the permutation analysis mean plus 2 standard deviations). (B) Permutation analysis for GRIN2A (Chain B) as shown by similar plots in A. (C) The structural map of calculated significance from permutation analysis (same as the raster plots above), mapped onto a GRIN1 subunit and GRIN2A subunit (C1). Closer view of the ABDs: (C2) GRIN1; (C3) GRIN2A. Closer view of the NTDs, shown with side chains (sticks) of the identified significant residues: (C4) top down; (C5) side view depicted in C4.
In the residue permutation analysis, all residue locations are randomly swapped with one another resulting in a new orientation relationship among all residues (Supplemental Fig. S7). This approach has one important implication: because just the location was swapped, each residue retains its original gnomAD data. By randomizing the surrounding residues, this permutation method probes how neighboring residues influence a residue's 3DMTR score. In the permutation analysis of the gnomAD data, each residue location is fixed, but the variant data used for the 3DMTR calculation is randomized (Supplemental Fig. S8). The main consequence of this is that a residue will have the same number of extra-chain contacts. Fluctuations in the 3DMTR permutation mean score can suggest if there is a bias for certain residues to have a different 3DMTR score based on how the amino acid chains in the structure are paired.
We performed both permutation analyses on the GRIN1/2A NMDAR. In each analysis we generated 1000 randomization instances and quantified the 3DMTR results of each permutation (Fig. 6; Supplemental Fig. S7; Supplemental Data Set 1). The resulting permutation MTR score for a given residue will be normally distributed (Supplemental Fig. S10), which typically occurs during randomization even if the sampled distribution is not normally distributed (Supplemental Figs. S2, S6). We found more residues with scores significantly different (defined outside two times standard deviation range from each residue's mean) than their permutation mean than would have been expected by random chance (Fig. 6A,B). A large number of residues were significant (135–169 residues) (Supplemental Table S1), many more than expected to fall beyond two times the standard deviation range based on random chance (about 40 residues). These residues were usually the most extreme scores (Fig. 6A,B). Furthermore, the excess number of significant residues appear to be ones with more intolerant scores (Supplemental Table S1). The number of significant residues with more tolerant scores were about as many as expected from random chance. Overall, we see that residue permutation analysis results, which identifies residues that have scores that are highly unlikely given random chance, corresponds with the key functional elements of the NMDAR (Fig. 6C–E). The results show that highly intolerant residues occur throughout the channel selectivity filter, channel gate, glutamate binding site, ABD-dimer interface, and ABD-TMD-linkers.
We found similar results from the gnomAD data permutation analysis (1000 randomization instances) (Supplemental Figs. S8, S9; Supplemental Data Set 1), in which there were many more residues with extreme scores than expected from random chance (138–175 residues) (Supplemental Table S1). As above, the extra number of significant residues appear to be ones with more intolerant scores. In general, the same key elements of the receptor were identified (Supplemental Fig. S9C). Stretches of residues with permutation means diverted from a general trend were mainly the portions with extensive contact with other polypeptide chains (Supplemental Fig. S4). The diversions in GRIN1 and GRIN2A were in opposite directions, suggesting that for these residues, there may be some influence on their 3DMTR (intra-receptor) score caused by the pairing of these subunits’ variant data sets. In general, both 3DMTR permutation analyses results were similar, which suggests that the specific residues at their location in the structure drive the highly intolerant (or tolerant) scores without significant influence of the pairing of GRIN1 and GRIN2A.
Examining a subdomain in isolation and altering the number of included residues may refine 3DMTR analysis
The resolution and conformation of the protein structure is an important consideration when using the 3DMTR method. High-resolution structures are required for accurate placement of the side chains (<∼4 Å). Furthermore, multiple structures have been described for some proteins in different conformations. Given that the 3DMTR permutation analysis did not show clear differences in the NTD when using the GRIN1/2A homology model based on the nonactive GRIN1/2B structure (Chou et al. 2020), we analyzed an isolated NTD in a crystal structure with Zn2+ bound (Romero-Hernandez et al. 2016). Zn2+ ions are an important endogenous modulator of GRIN2A-containing receptors (Paoletti et al. 1997; Traynelis et al. 1998). We performed the 3DMTR method on this structure using the closest 31 residues and performed residue permutation analysis (Supplemental Fig. S11; Supplemental Data Set 1). As expected, the NTD in the full-length structure (Full) compared to the isolated Zn2+-NTD structures had similar 3DMTR profiles with similar standard deviation (collective statistics for both GRIN1 and GRIN2A NTDs; meanFull = 0.630, STDFull = 0.165; meanZn2+ = 0.626, STDZn2+ = 0.167). However, the permutation analysis of the isolated NTD structure identified more highly intolerant residues in the Zn2+ structure (Supplemental Table S2). That is, 46 residues were highly tolerant and eight residues were highly intolerant in the NTD dimer in the full-length structure. In contrast, only 23 residues were highly tolerant and 25 residues were highly intolerant in Zn2+-NTD structure. The highly intolerant residues identified were distributed along the dimer interface between GRIN1 and GRIN2A.
It could be argued that using the closest 31 residues potentially includes irrelevant residues. Therefore, we calculated the 3DMTR using fewer residues (closest 11 residues, c11; closest 21 residues, c21). We observed in the 3DMTR profiles calculated with fewer than 31 included residues had more highly intolerant residues (number of 3DMTR scores of less than 0.3, 5 residues in c31, 25 residues in c21, and 68 residues in c11) (Supplemental Figs. S11B–D, S12). Overall, the mean 3DMTR scores were similar for these different analyses (0.626 in c31, 0.609 in c21, 0.600 in c11). However, the standard deviation increased by almost twofold (0.167 in c31, 0.204 in c21, 0.303 in c11). The pattern of significant residues in the c21 analysis is similar to c31, although fewer significant residues were found (Supplemental Table S2) and many of the identified residues were different. For c11, more residues were identified as significantly different than the average, however a clear pattern was absent. It appears that with the current level of genomic data, the 3DMTR method is capable of producing similar results using 21 residues or more, but reducing the number of residues to less than 21 risks producing a 3DMTR profile that is too variable to reveal differences (Supplemental Fig. S12).
The 3DMTR score better classifies the functional outcome of mutations than the 1DMTR score
We quantified the 1DMTR and 3DMTR methods’ ability to predict important residues by testing for functional consequences when a residue is mutated. We selected several published studies in which point mutations were generated and the glutamate EC50 was determined. We collected 67 Grin1 mutants (GRIN1/2A), 128 Grin2A mutants, 104 Grin1 mutants (GRIN1/2B), and 60 Grin2B mutants from 31 sources (Supplemental Data Set 2). We also determined glutamate EC50 values for 40 new Grin2A mutants (Supplemental Tables S3, S4; Supplemental Data Set 2). In addition to these mutant data, wild-type glutamate EC50 values were collected from the same publications and from our own testing (26 GRIN1/2A and 17 GRIN1/2B determinations) (Supplemental Data Set 2). We used the mean and standard deviation of all wild-type EC50 determinations (Supplemental Table S5) to determine the Z-score for all mutant determinations based on the wild-type distribution parameters.
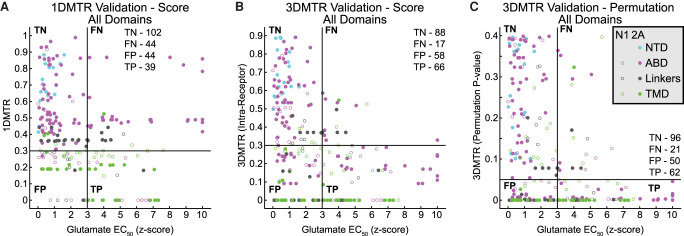
We selected MTR and Z-score cutoffs to create a binary classification to see if the MTR can detect mutants that have functional consequences (Fig. 7). The result of the 1DMTR analysis showed 39 of the 84 mutants with Z-scores suggestive of altered functional phenotype (Z-score > 3) had 1DMTR scores less than 0.3. In addition, 101 of 145 mutant determinations with Z-scores suggestive of wild-type function (Z-score < 3) had 1DMTR scores greater than 0.3 (Fig. 7A). In comparison, the 3DMTR analysis was better able to identify mutants that were functionally different than wild type. With the 3DMTR, 66 of the 84 residues where mutations were shown experimentally to influence glutamate EC50 were correctly identified by having an MTR score less than 0.3, whereas 87 of the 145 mutants with a wild-type EC50 phenotype were identified as tolerant to mutation (Fig. 7B). Additionally, with the 3DMTR permutation analysis, 62 of the 84 mutant determinations that were different than wild type were correctly identified, and 95 of the 145 mutant determinations that were like wild type were correctly identified (Fig. 7C). Fisher's exact test suggests the binary classification of all GRIN1/2A point mutations with the 1DMTR analysis failed to reach a high significance level (Table 1), whereas both of the 3DMTR methods achieved high significance (Table 1). The distribution of the glutamate EC50 Z-scores changes from the more tolerant MTR scores to the more intolerant MTR scores, especially the 3DMTR (Supplemental Fig. S13). In other words, the more tolerant residues (≥0.6) have tight distributions with Z-scores less than 3, whereas the more intolerant residues (<0.3) have broad Z-score distributions.
Figure 7.
The 3DMTR is a better predictor of functional consequences of GRIN1/2A point mutations. (A–C) Scatter plots of various residues’ glutamate EC50 (represented as a Z-score based on intra-study wild-type distribution) and MTR score: (A) 1DMTR; (B) 3DMTR; (C) 3DMTR permutation analysis. Thresholds for the MTR score, <0.3 (A), <0.3 (B), <0.05 (C), and for the glutamate EC50 Z-score (>3) were determined from the distribution of WT EC50 values across the studies included. Implementation of threshold criteria creates a binary classification to determine the portions of the residues which are deemed highly intolerant and have differences in receptor function when mutated. Thus, each residue can be classified as true negative (TN), false negative (FN), false positive (FP), and true positive (TP), and the various MTR methods can be compared.
Table 1.
Comparison of MTR method validation with GRIN1/2A glutamate EC50 data
Much of the improvement in the ability to predict functional consequences of point mutations of the 3DMTR score is attributable to the differences in the MTR score of the ABD and the domain linkers (Table 1; Supplemental Fig. S14). For the 1DMTR analysis of the ABD and linker mutants, 14 of the 45 mutant determinations that were different than wild type were correctly identified, and 64 of the 79 mutant determinations that were like wild type were correctly identified (Supplemental Fig. S14B). With the 3DMTR analysis and permutation analysis, 75% of the residues below their respective cutoffs were identified as functionally different (Supplemental Fig. S14B,C). Fisher's exact test suggests the binary classification of ABD and linker point mutations with the 1DMTR failed to reach significance (Table 1) but were highly significant with either the straight 3DMTR or the permutation analysis binary classifications (Table 1).
Additionally, we observed similar trends with the GRIN1/2B data (Supplemental Figs. S15, S16; Supplemental File 2). The 3DMTR method showed improved sensitivity than over the 1DMTR or the 3DMTR permutation analysis, in addition to classifying the most true positives correctly (Supplemental Table S6). The 3DMTR permutation method resulted in high specificity but had many more false negative residues than the other methods. Fisher's exact test suggests the binary classification of all GRIN1/2B point mutations with the 1DMTR failed to reach high significance (Supplemental Table S6), but achieved high significance levels with the binary classification of either 3DMTR method (Supplemental Table S6).
Discussion
The introduction of the MTR method (Traynelis et al. 2017) provided the biomedical research community with an important analytical tool that allowed regional intolerance to be assessed. Individuals working in all fields of biology can make use of this tool to better understand their protein of interest. However, the concept was limited by application to the polypeptide chain, which neglects tertiary structure of proteins that often juxtapose two residues that reside at considerable distance from one another on the polypeptide chain. 3DMTR analysis is an important refinement of this approach. For example, the results provided here for the GRIN family of genes yields several important differences when compared to 1DMTR analysis.
3DMTR analysis improves the application of population-level variant data
The 3DMTR method did qualitatively better in identifying residues in protein domains known to be critical for receptor function. These mainly include residues in the ABD, the domain linkers, and the channel gate. For example, mutations of residues in the inner cleft of the ABD that harbor the agonist binding pocket can alter glutamate potency up to 3000-fold, either by changing agonist affinity or by modifying the domain closure, movement that drives activation of the receptor. The TMD has key residues that enable opening of the channel as well as additional residues that control Mg2+ block and Ca2+ permeability. Also, the ABD-TMD linkers have been suggested to be important not only for translating conformational changes from the ABD to the TMD, but also for key interactions that maintain the closed state of the receptor as well as allow for the responsive activation required for rapid synaptic transmission (Amin et al. 2020; Perszyk et al. 2020). In the GRIN1/2A receptor 3DMTR analysis, these key portions of the receptor were labeled as highly intolerant.
The improvement provided by the 3DMTR method may reflect selection of residues that are closer and therefore more relevant to the central residue. The average of the mean residue distances of the selected 31 residues for the entire GRIN1/2A structure is 9.1 Å for the 3DMTRreceptor, 9.5 Å for the 3DMTRsubunit, and 14.4 Å for the 1DMTR. For the 3DMTRreceptor method, reducing the number of selected residues to 21 or 11 reduces the mean residue distances (7.8 Å and 6.0 Å, respectively). Using 21 or 31 residues in the 3DMTR calculation, the residues that directly interact with the central residue are included as well as those that might interact or may influence indirectly the central residue. In contrast, using a fewer number of residues (e.g., 11) selects only residues that are positioned more closely to the central residue. Owing to the lack of full coverage of the genomic database, a minimum number of residues is required to avoid errors in the MTR calculation (typically more than 11 residues) and using more residues (21 or 31 residues) provides a more consistent MTR score with lower overall variability.
The main drawback of the 3DMTR method is that it can only be used for proteins for which structural data is available. Thus, the method cannot be used for proteins or protein regions that lack crystallographic or cryo-EM structural data. In such cases, the only opportunity is to use homology models, the original 1DMTR analysis, or other methods to consolidate population genomic data into simple metrics or scores (Traynelis et al. 2017; Pérez-Palma et al. 2020). For example, MTR analysis of the intracellular C-terminal domain of NMDAR subunits is not possible because there are no structural data for this domain, which is critical for the NMDAR regulation and localization. Potentially new innovations in predicting protein structure (AlphaFold) could be implemented to fill in these gaps in structural information that lack experimental data (Senior et al. 2020).
Permutation analysis and the accuracy of the 3DMTR
The 3DMTR permutation analysis provides reassurance that the 3DMTR profile identifies real differences between the residues in a given protein data set. By calculating numerous permutations, the likelihood of each residue's score can be determined, which suggests whether the most extreme values in the data set result from random coincidence. In instances for which the overall 3DMTR score is moderately high (suggesting tolerance of variation), as was observed in the isolated NTD domains of the GRIN1 and GRIN2A, permutation analysis can detect which are the most intolerant residues. Additionally, as we saw in the evaluation of the isolated GRIN1/2A NTD, reducing the number of residues included can alter the MTR profile and increase the number of highly intolerant residue scores (<0.3). However, reducing the number of included residues (from 21 to 11) increases the permutation variability and thus suggests that these highly intolerant scores are more likely caused by random coincidences, resulting from the less than optimal level of SNV data set coverage. Thus, permutation analysis can be used to ensure that the 3DMTR analysis is producing reliable results.
3DMTR shows an improved ability to classify the functional outcomes for NMDAR mutations
An important component of this study was the statistical validation of the results we obtained. We found that the majority of the mutations produced glutamate EC50 values that were different from wild type. Comparing these experimental data sets with the results from the analysis by 3DMTR and 1DMTR methods showed that the 3DMTR score was better at predicting the functionally different mutation than the 1DMTR score. The 3DMTR permutation analysis also correctly classified the functional result of point mutations, similar to the 3DMTR score for GRIN1/2A. However, for GRIN1/2B, the permutation analysis had lower sensitivity in the binary classification. This may suggest that the raw 3DMTR score may be a better predictor of the functional outcome of mutations when analyzing data sets with numerous intolerant 3DMTR scores. As GRIN2B is on average more intolerant than GRIN2A (mean intra-receptor 3DMTR score 0.567 for GRIN2A, 0.389 for GRIN2B), the permutation analysis may exclude some potentially relevant residues that have highly intolerant scores (<0.3) because the method identifies only the most extreme cases in a data set.
The 3DMTR score was capable of detecting functionally relevant consequences of mutation with high sensitivity and a high negative predictive value; however, the 3DMTR score had lower positive predictive value, which was still comparable or better than 1DMTR score. The low positive predictive values are not problematic for several reasons. First, glutamate EC50 captures only one aspect of receptor function, with other important features being represented by glycine EC50, receptor deactivation time course, open probability, Mg2+ sensitivity, proton sensitivity, Zn2+ sensitivity, cell surface expression, and other descriptive parameters. The reason for a low positive predictive value might be that the false positives (e.g., low 3DMTR, but no change in glutamate EC50) capture differences in other functional properties that result in tolerance selection. An additional caveat is that all point mutants in the validation were assayed in NMDARs, where both copies of the subunit in the tetrameric receptor contains the mutation. In contrast, tolerance selection occurs in individuals that most often are heterozygous carriers of the mutation; therefore, the majority of the receptor population will contain only a single subunit with the mutation, along with a wild-type subunit. Next, because these were a random sampling of the mutations that we found in the literature, not all of them are likely to occur in individuals because some require multiple base pair changes to produce the specific codon change (they are not SNVs). The predictability of any MTR score might therefore be different for those changes that can be produced by one base pair change versus those that require multiple changes. Additionally, this might also contribute to false negative data points, in that if the specific point mutation tested is not a SNV, the functional consequences of that mutation might not be predicted by the MTR score. However, given that the 3DMTR is calculated by averaging SNV counts of multiple residues, this may result in a more generalized score of tolerance. In other words, a low 31-residue averaged 3DMTR score suggests that any perturbation in that 3D space is likely to alter function.
A more wholistic approach may be more appropriate than our binary classification of just a single functional parameter. The measurement of many parameters describing NMDAR function for a large number of NMDAR mutations could be determined and used to determine a single aggregate score representing functional alterations (Swanger et al. 2016). In addition, assessment of multiple parameters could allow a multivariable nonlinear regression to be performed to determine the optimal combination of functional parameters that contribute to MTR tolerance. Furthermore, there may be cellular or network scale end points (charge transfer, contribution to neuronal bursting behavior, changes in neuronal plasticity, etc.) that are altered by NMDAR mutations that may be a better predictor than basic receptor properties.
Future applications of the 3DMTR method
The 3DMTR method can be used in other protein analysis applications, including molecular dynamic (MD) simulations. Because the 3DMTR method is heavily based on the structure or model that is used, these data sets may provide further insight into the variability of the 3DMTR with small conformational changes. For NMDARs, important conformational changes occur during receptor activation, and thus the 3DMTR-MD evaluation may be able to detect important transient interactions that occur or alternative receptor conformations with local minima or highly variable 3DMTR scores. Additionally, many proteins interact as parts of multimeric complexes, sometimes with multiple analogs potentially interacting with the same site, such as the different GRIN2 subunits occupying the same position in the NMDAR complex or different G-proteins interacting with G-protein coupled receptors. Here, the 3DMTR method could be used to compare different analogs to reveal differences or similarities in protein interactions. Differences in the 3DMTR score will occur at interface positions based on the juxtaposition of different analog data sets, as well as potential alterations in protein complex conformation with different partners.
Alternative 3DMTR scores could be developed using inclusion metrics other than Euclidean distance, focusing on which residues we know interact with each other. Some examples of alternative inclusion criteria could be using residue distance with weighed factors such as residues of a similar class (polar, charges, etc.), residues that have correlated movements in MD simulations, or residue interactions (salt-bridge, hydrogen bond, or aromatic interaction networks). Additionally, a related method could be implemented incorporating weighing based on a variable mutation rate that may influence the 3DMTR score (Samocha et al. 2017).
Looking forward, as the precision of the MTR will improve with a larger variant data set, eventually providing enough synonymous and missense variants to produce a residue-specific MTR. It would appear at this point in time there might be a redundancy to the 3DMTR method, but there may be benefits to averaging data over local regions to provide a more generalizable assessment of tolerance to perturbation. Furthermore, because the 3DMTR is more effective at using this genomic data it may be useful in addressing concepts of polygenic diseases (where multiple gene variants contribute to a complex disease), ancestral/lineage-specific 3DMTR, or in analyzing other subsets of the complete databases. One could imagine that having a de novo variant in the GRIN2A gene might confer susceptibility to epilepsy but may require another variant in a second gene (such as SCN1A, also known as Nav1.1). Analyzing a subset variant genome data set, with or without GRIN2A, any mildly intolerant variants could be constructed and the 3DMTR evaluated for SCN1A (or vice versa). The 3DMTR profile might be different for these subsets, suggesting that mildly intolerant variants in either GRIN2A or SCN1A may be tolerated but mildly intolerant variants in both are removed from the gene pool. Thus, 3DMTR analysis is a useful tool with which to guide the study of the structure-function relationship of proteins and protein complexes.
Methods
Three-dimensional MTR calculation
The missense tolerance ratio (MTR) was calculated from population-level SNV data sets, similar to previous reports (Traynelis et al. 2017), by the following equation:
where MTRn is the missense tolerance ratio for residue n, incln is the set of residues to be included for each residue n, MSobs is the observed missense variant counts for a particular residue, SSobs is the observed synonymous or same sense variant counts for a particular residue, MSexp is the expected missense variant counts for a particular residue as determined by the codon, and SSexp is the expected synonymous variant counts for a particular residue as determined by the codon. The expected number of MS and SS variants assume an equal theoretical mutation rate at every base pair and were determined from the number of potential missense amino acid variants based on redundancy in the genetic code. That is, we determined the number of SNVs within a codon that would produce change in the amino acid represented by the codon, or alternatively yield the same amino acid.
For the 1DMTR, the 15 residues before and after residue n, in terms of the amino acid sequence, were included in the calculation. For the 3DMTR, the closest 31 residues in space are included, according to a given protein structure. To determine which set of neighboring residues to include for each individual residue, the locations of each residue in 3D space, as given by coordinates in the PDB file, was defined as the average location (X, Y, Z coordinates) of all atoms (present in the given model) comprising the residue (peptide backbone plus side chain). The Euclidean distance between residues was calculated using the 3D version of the Pythagorean theorem. Alternative 3DMTR scores can be calculated by simply using the closest N (11, 21) residues for each residue. gnomAD v2.1.1 was used for all analyses.
MTR permutation analysis
To assess the likelihood of the calculated 3DMTR scores from a given data set (gnomAD variant data set, protein structure), two permutation analysis methods were developed: residue and factor. For the residue permutation analysis, the residue locations were randomized with one another within the selected set, and the distances between each were recalculated. For the factor permutation analysis, a randomized look-up table was used when selecting the residues to be included in the 3DMTR calculation. For both methods, randomization was constrained to be within each polypeptide chain, if there were multiple polypeptide chains included in the analysis. The mean permutated score and standard deviation were determined for each residue. In all cases presented here, 1000 permutations were calculated.
Agonist concentration-response curves and two-point agonist EC50 estimation
Rat wild-type NMDARs were produced by coexpression cDNAs encoding the GRIN1-1A (NCBI Genbank [https://www.ncbi.nlm.nih.gov/genbank/] accession numbers U11418, U08261; hereafter GRIN1) and GRIN2A (D13211) subunits in Xenopus laevis oocytes as previously described (Erreger et al. 2007). X. laevis stage VI oocytes were isolated from X. laevis ovaries as previously described (Hansen et al. 2013). The Hill equation was fitted to glutamate concentration-response data as previously described (Hansen et al. 2013; Supplemental Methods).
For the two-point agonist EC50 estimation, current response to application of a submaximal and a saturating of concentration of glutamate (2 and 100 μM, respectively) were measured, and the following equation was used to estimate the EC50:
where EC50_estimate is the estimated half-maximally effective concentration of agonist, [Asub] is the submaximal concentration of the agonist used, Imax is the maximal current response, Isub is the submaximal current response, and h is the Hill slope. The Hill slope was assumed to be 1.2 for all mutants tested with this assay.
Preparation of the validation data set
Previously published NMDAR wild-type and point mutation construct glutamate EC50 values were obtained from the literature (Laube et al. 1993; Kuryatov et al. 1994; Wafford et al. 1995; Williams et al. 1995, 1998; Hirai et al. 1996; Kashiwagi et al. 1996, 1997; Anson et al. 1998; Fayyazuddin et al. 2000; Low et al. 2000; Wittekindt et al. 2001; Laube et al. 2004; Chen et al. 2005, 2017; Furukawa et al. 2005; Inanobe et al. 2005; Erreger et al. 2007; Maier et al. 2007; Ren et al. 2013; Yuan et al. 2014; Serraz et al. 2016; Swanger et al. 2016; Addis et al. 2017; Bledsoe et al. 2017; Ogden et al. 2017; Wang et al. 2017; Li et al. 2019; Hrcka Krausova et al. 2020; McDaniel et al. 2020; Skrenkova et al. 2020). The residue numbers were modified so that the initiating methionine was always one. All values were converted to the log EC50. The wild-type data points were treated as if each data point was a sample from the normal distribution of the mean wild-type parameter, despite being collected via slightly varying assays. The mean and standard deviation of the underlying distribution were estimated from the sample mean (meanwt) and standard deviation (STDwt). An EC50 Z-score for each point mutation was calculated using the wild-type normal distribution parameters:
Binary classification
MTR score and Z-score threshold were determined empirically. 1DMTR scores below 0.3 are considered extremely rare and meaningful (exome-wide MTR fifth percentile = 0.5462, MTR-viewer, http://mtr-viewer.mdhs.unimelb.edu.au/). For the permutation analysis, a threshold of 0.05 was chosen to align with the rest of the results presented by this method, which identifies the residues for which the 3DMTR score is more extreme than would be expected by chance (95%). The Z-score cutoff was chosen as 3 to select for those data points that are most certainly different than the wild type, especially given that many of the data points collected are reported specifically because of their functional consequences, which might result in inflated numbers of mutants or variants that have higher Z-scores. The data points were classified as either true negative (TN, MTR score >0.3, 0.05 and Z-score<3), false negative (FN, MTR score>0.3, 0.05 and Z-score>3), false positive (FP, MTR score<0.3, 0.05 and Z-score< 3), or true positive (TP, MTR score<0.3, 0.05 and Z-score>3). After classifying the data points, standard binary classification metrics were calculated (Supplemental Methods). Fisher's exact test was used to evaluate the effectiveness of the classification, using an α = 0.01 owing to the relatively large sample sizes we collected.
Software availability
All MTR calculations were performed using a MATLAB (Mathworks, version R2019b) encapsulated application (for source files, see Supplemental Code File) or other custom scripts (for details, see Supplemental Methods). Additionally, all code, the executable 3DMTR application, and future updates are available on GitHub (https://github.com/riley-perszyk-PhD/3DMTR, current version v1.008) along with an operation manual explaining how to use the application on other proteins.
Supplementary Material
Acknowledgments
We thank Pieter Burger for assistance with modeling of GRIN1/2A and GRIN1/2B. This work was supported by the National Institutes of Health (R35 NS111619 to S.F.T.) and Austin's Purpose to S.F.T. The opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the funding agencies.
Author contributions: R.E.P., A.S.K., and S.F.T. participated in research design. R.E.P., A.S.K., and P.L. conducted experiments. R.E.P. contributed new reagents or analytic tools. R.E.P., A.S.K., and S.F.T. performed data analysis. All authors wrote or contributed to the writing of the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275528.121.
Competing interest statement
S.F.T. is a PI on research grants from Biogen and Janssen to Emory University; is a paid consultant for Janssen; is a member of the Scientific Advisory Board for Eumentis, Sage Therapeutics, GRIN2B Foundation, and CureGRIN Foundation; is cofounder of NeurOp, Inc., and Agrithera; and has received licensing fees and royalties from Emory. S.F.T. is a coinventor on Emory University–owned Intellectual Property that includes allosteric modulators of glutamate receptor function.
References
- Addis L, Virdee JK, Vidler LR, Collier DA, Pal DK, Ursu D. 2017. Epilepsy-associated GRIN2A mutations reduce NMDA receptor trafficking and agonist potency-molecular profiling and functional rescue. Sci Rep 7: 66. 10.1038/s41598-017-00115-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin JB, Moody GR, Wollmuth LP. 2020. From bedside-to-bench: what disease-associated variants are teaching us about the NMDA receptor. J Physiol 599: 397–416. 10.1113/JP278705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anson LC, Chen PE, Wyllie DJA, Colquhoun D, Schoepfer R. 1998. Identification of amino acid residues of the NR2A subunit that control glutamate potency in recombinant NR1/NR2A NMDA receptors. J Neurosci 18: 581–589. 10.1523/JNEUROSCI.18-02-00581.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bledsoe D, Tamer C, Mesic I, Madry C, Klein BG, Laube B, Costa BM. 2017. Positive modulatory interactions of NMDA receptor GluN1/2B ligand binding domains attenuate antagonists activity. Front Pharmacol 8: 229. 10.3389/fphar.2017.00229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PE, Geballe MT, Stansfeld PJ, Johnston AR, Yuan H, Jacob AL, Snyder JP, Traynelis SF, Wyllie DJA. 2005. Structural features of the glutamate binding site in recombinant NR1/NR2A N-methyl-D-aspartate receptors determined by site-directed mutagenesis and molecular modeling. Mol Pharmacol 67: 1470–1484. 10.1124/mol.104.008185 [DOI] [PubMed] [Google Scholar]
- Chen W, Tankovic A, Burger PB, Kusumoto H, Traynelis SF, Yuan H. 2017. Functional evaluation of a de novo GRIN2A mutation identified in a patient with profound global developmental delay and refractory epilepsy. Mol Pharmacol 91: 317–330. 10.1124/mol.116.106781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TH, Tajima N, Romero-Hernandez A, Furukawa H. 2020. Structural basis of functional transitions in mammalian NMDA receptors. Cell 182: 357–371.e13. 10.1016/j.cell.2020.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Geballe MT, Kristensen A, Chen PE, Hansen KB, Lee CJ, Yuan H, Le P, Lyuboslavsky PN, Micale N, et al. 2007. Subunit-Specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Mol Pharmacol 72: 907–920. 10.1124/mol.107.037333 [DOI] [PubMed] [Google Scholar]
- Fayyazuddin A, Villarroel A, Le Goff A, Lerma J, Neyton J. 2000. Four residues of the extracellular N-terminal domain of the NR2A subunit control high-affinity Zn2+ binding to NMDA receptors. Neuron 25: 683–694. 10.1016/S0896-6273(00)81070-3 [DOI] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. 2005. Subunit arrangement and function in NMDA receptors. Nature 438: 185–192. 10.1038/nature04089 [DOI] [PubMed] [Google Scholar]
- Gibb AJ, Ogden KK, McDaniel MJ, Vance KM, Kell SA, Butch C, Burger P, Liotta DC, Traynelis SF. 2018. A structurally derived model of subunit-dependent NMDA receptor function. J Physiol 596: 4057–4089. 10.1113/JP276093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jørgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF. 2013. Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol Pharmacol 84: 114–127. 10.1124/mol.113.085803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Yi F, Perszyk RE, Menniti FS, Traynelis SF. 2017. NMDA receptors in the central nervous system. In NMDA Receptors. Methods in Molecular Biology, Vol. 1677 (ed. Burnashev N, Szepetowski P). Humana Press, New York. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Wollmuth LP, Bowie D, Furukawa H, Menniti FS, Sobolevsky AI, Swanson GT, Swanger SA, Greger IH, Nakagawa T, et al. 2021. Structure, function, and pharmacology of glutamate receptor Ion channels. Pharmacol Rev 10.1124/pharmrev.120.000131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Neale BM, Traynelis SF, Allen AS, Goldstein DB. 2015. The genetics of neuropsychiatric diseases: looking in and beyond the exome. Annu Rev Neurosci 38: 47–68. 10.1146/annurev-neuro-071714-034136 [DOI] [PubMed] [Google Scholar]
- Hirai H, Kirsch J, Laube B, Betz H, Kuhse J. 1996. The glycine binding site of the N-methyl-D-aspartate receptor subunit NR1: identification of novel determinants of co-agonist potentiation in the extracellular M3-M4 loop region. Proc Natl Acad Sci 93: 6031–6036. 10.1073/pnas.93.12.6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrcka Krausova B, Kysilov B, Cerny J, Vyklicky V, Smejkalova T, Ladislav M, Balik A, Korinek M, Chodounska H, Kudova E, et al. 2020. Site of action of brain neurosteroid pregnenolone sulfate at the N-methyl-D-aspartate receptor. J Neurosci 40: 5922–5936. 10.1523/JNEUROSCI.3010-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanobe A, Furukawa H, Gouaux E. 2005. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron 47: 71–84. 10.1016/j.neuron.2005.05.022 [DOI] [PubMed] [Google Scholar]
- Iqbal S, Pérez-Palma E, Jespersen JB, May P, Hoksza D, Heyne HO, Ahmed SS, Rifat ZT, Sohel Rahman M, Lage K, et al. 2020. Comprehensive characterization of amino acid positions in protein structures reveals molecular effect of missense variants. Proc Natl Acad Sci 117: 28201–28211. 10.1073/pnas.2002660117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Moriyoshi K, Sugihara H, Sakurada K, Kadotani H, Yokoi M, Akazawa C, Shigemoto R, Mizuno N, Masu M, et al. 1993. Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J Biol Chem 268: 2836–2843. 10.1016/S0021-9258(18)53849-7 [DOI] [PubMed] [Google Scholar]
- Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H. 2014. Structural insights into competitive antagonism in NMDA receptors. Neuron 81: 366–378. 10.1016/j.neuron.2013.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581: 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K, Fukuchi J, Chao J, Igarashi K, Williams K. 1996. An aspartate residue in the extracellular loop of the N-methyl-D-aspartate receptor controls sensitivity to spermine and protons. Mol Pharmacol 49: 1131–1141. [PubMed] [Google Scholar]
- Kashiwagi K, Pahk AJ, Masuko T, Igarashi K, Williams K. 1997. Block and modulation of N-methyl-D-aspartate receptors by polyamines and protons: role of amino acid residues in the transmembrane and pore-forming regions of NR1 and NR2 subunits. Mol Pharmacol 52: 701–713. 10.1124/mol.52.4.701 [DOI] [PubMed] [Google Scholar]
- Kuryatov A, Laube B, Betz H, Kuhse J. 1994. Mutational analysis of the glycine-binding site of the NMDA receptor: structural similarity with bacterial amino acid-binding proteins. Neuron 12: 1291–1300. 10.1016/0896-6273(94)90445-6 [DOI] [PubMed] [Google Scholar]
- Lal D, May P, Perez-Palma E, Samocha KE, Kosmicki JA, Robinson EB, Møller RS, Krause R, Nürnberg P, Weckhuysen S, et al. 2020. Gene family information facilitates variant interpretation and identification of disease-associated genes in neurodevelopmental disorders. Genome Med 12: 28. 10.1186/s13073-020-00725-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. 2016. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44: D862–D868. 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube B, Kuryatov A, Kuhse J, Betz H. 1993. Glycine-glutamate interactions at the NMDA receptor: role of cysteine residues. FEBS Lett 335: 331–334. 10.1016/0014-5793(93)80412-N [DOI] [PubMed] [Google Scholar]
- Laube B, Schemm R, Betz H. 2004. Molecular determinants of ligand discrimination in the glutamate-binding pocket of the NMDA receptor. Neuropharmacology 47: 994–1007. 10.1016/j.neuropharm.2004.07.041 [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang J, Tang W, Mizu RK, Kusumoto H, XiangWei W, Xu Y, Chen W, Amin JB, Hu C, et al. 2019. De novo GRIN variants in NMDA receptor M2 channel pore-forming loop are associated with neurological diseases. Hum Mutat 40: 2393–2413. 10.1002/humu.23895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low CM, Zheng F, Lyuboslavsky P, Traynelis SF. 2000. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-D-aspartate NR1/NR2A receptors. Proc Natl Acad Sci 97: 11062–11067. 10.1073/pnas.180307497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Schemm R, Grewer C, Laube B. 2007. Disruption of interdomain interactions in the glutamate binding pocket affects differentially agonist affinity and efficacy of N-methyl-D-aspartate receptor activation. J Biol Chem 282: 1863–1872. 10.1074/jbc.M608156200 [DOI] [PubMed] [Google Scholar]
- McDaniel MJ, Ogden KK, Kell SA, Burger PB, Liotta DC, Traynelis SF. 2020. NMDA receptor channel gating control by the pre-M1 helix. J Gen Physiol 152. 10.1085/jgp.201912362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguro H, Mori H, Araki K, Kushiya E, Kutsuwada T, Yamazaki M, Kumanishi T, Arakawa M, Sakimura K, Mishina M. 1992. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature 357: 70–74. 10.1038/357070a0 [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. 1992. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256: 1217–1221. 10.1126/science.256.5060.1217 [DOI] [PubMed] [Google Scholar]
- Müller-Nedebock AC, van der Westhuizen FH, Kõks S, Bardien S. 2021. Nuclear genes associated with mitochondrial DNA processes as contributors to Parkinson's disease risk. Mov Disord 36: 815–831. 10.1002/mds.28475 [DOI] [PubMed] [Google Scholar]
- Myers SJ, Yuan H, Kang JQ, Tan FCK, Traynelis SF, Low C-M. 2019. Distinct roles of GRIN2A and GRIN2B variants in neurological conditions. F1000Res 8: 1940. 10.12688/f1000research.18949.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KK, Chen W, Swanger SA, McDaniel MJ, Fan LZ, Hu C, Tankovic A, Kusumoto H, Kosobucki GJ, Schulien AJ, et al. 2017. Molecular mechanism of disease-associated mutations in the pre-M1 helix of NMDA receptors and potential rescue pharmacology. PLoS Genet 13: e1006536. 10.1371/journal.pgen.1006536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Ascher P, Neyton J. 1997. High-affinity zinc inhibition of NMDA NR1–NR2A receptors. J Neurosci 17: 5711–5725. 10.1523/JNEUROSCI.17-15-05711.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. 2013. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci 14: 383–400. 10.1038/nrn3504 [DOI] [PubMed] [Google Scholar]
- Pérez-Palma E, May P, Iqbal S, Niestroj LM, Du J, Heyne HO, Castrillon JA, O'Donnell-Luria A, Nürnberg P, Palotie A, et al. 2020. Identification of pathogenic variant enriched regions across genes and gene families. Genome Res 30: 62–71. 10.1101/gr.252601.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perszyk RE, Myers SJ, Yuan H, Gibb AJ, Furukawa H, Sobolevsky AI, Traynelis SF. 2020. Hodgkin–Huxley–Katz Prize Lecture: genetic and pharmacological control of glutamate receptor channel through a highly conserved gating motif. J Physiol 598: 3071–3083. 10.1113/JP278086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. 2013. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 9: e1003709. 10.1371/journal.pgen.1003709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Zhao Y, Wu M, Peoples RW. 2013. A novel alcohol-sensitive position in the N-methyl-D-aspartate receptor GluN2A subunit M3 domain regulates agonist affinity and ion channel gating. Mol Pharmacol 84: 501–510. 10.1124/mol.113.085993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Hernandez A, Simorowski N, Karakas E, Furukawa H. 2016. Molecular basis for subtype specificity and high-affinity zinc inhibition in the GluN1-GluN2A NMDA receptor amino-terminal domain. Neuron 92: 1324–1336. 10.1016/j.neuron.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Kosmicki JA, Karczewski KJ, O'Donnell-Luria AH, Pierce-Hoffman E, MacArthur DG, Neale BM, Daly MJ. 2017. Regional missense constraint improves variant deleteriousness prediction. bioRxiv 10.1101/148353 [DOI]
- Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Žídek A, Nelson AWR, Bridgland A, et al. 2020. Improved protein structure prediction using potentials from deep learning. Nature 577: 706–710. 10.1038/s41586-019-1923-7 [DOI] [PubMed] [Google Scholar]
- Serraz B, Grand T, Paoletti P. 2016. Altered zinc sensitivity of NMDA receptors harboring clinically-relevant mutations. Neuropharmacology 109: 196–204. 10.1016/j.neuropharm.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Shore AN, Colombo S, Tobin WF, Petri S, Cullen ER, Dominguez S, Bostick CD, Beaumont MA, Williams D, Khodagholy D, et al. 2020. Reduced GABAergic neuron excitability, altered synaptic connectivity, and seizures in a KCNT1 gain-of-function mouse model of childhood epilepsy. Cell Rep 33: 108303. 10.1016/j.celrep.2020.108303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrenkova K, Song J, Kortus S, Kolcheva M, Netolicky J, Hemelikova K, Kaniakova M, Krausova BH, Kucera T, Korabecny J, et al. 2020. The pathogenic S688Y mutation in the ligand-binding domain of the GluN1 subunit regulates the properties of NMDA receptors. Sci Rep 10: 18576. 10.1038/s41598-020-75646-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanski A, Calle-López Y, Leu C, Pérez-Palma E, Pestana-Knight E, Lal D. 2021. Clinical sequencing yield in epilepsy, autism spectrum disorder, and intellectual disability: a systematic review and meta-analysis. Epilepsia 62: 143–151. 10.1111/epi.16755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger SAA, Chen W, Wells G, Burger PBB, Tankovic A, Bhattacharya S, Strong KLL, Hu C, Kusumoto H, Zhang J, et al. 2016. Mechanistic insight into NMDA receptor dysregulation by rare variants in the GluN2A and GluN2B agonist binding domains. Am J Hum Genet 99: 1261–1280. 10.1016/j.ajhg.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. 1998. Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. J Neurosci 18: 6163–6175. 10.1523/JNEUROSCI.18-16-06163.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. 2010. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62: 405–496. 10.1124/pr.109.002451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis J, Silk M, Wang Q, Berkovic SF, Liu L, Ascher DB, Balding DJ, Petrovski S. 2017. Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res 27: 1715–1729. 10.1101/gr.226589.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wafford KA, Kathoria M, Bain CJ, Marshall G, Le Bourdelles B, Kemp JA, Whiting PJ. 1995. Identification of amino acids in the N-methyl-D-aspartate receptor NR1 subunit that contribute to the glycine binding site. Mol Pharmacol 47: 374–380. [PubMed] [Google Scholar]
- Wang TM, Brown BM, Deng L, Sellers BD, Lupardus PJ, Wallweber HJAA, Gustafson A, Wong E, Volgraf M, Schwarz JB, et al. 2017. A novel NMDA receptor positive allosteric modulator that acts via the transmembrane domain. Neuropharmacology 121: 204–218. 10.1016/j.neuropharm.2017.04.041 [DOI] [PubMed] [Google Scholar]
- Wang Q, Pierce-Hoffman E, Cummings BB, Alföldi J, Francioli LC, Gauthier LD, Hill AJ, O'Donnell-Luria AH, Armean IM, Banks E, et al. 2020. Landscape of multi-nucleotide variants in 125,748 human exomes and 15,708 genomes. Nat Commun 11: 2539. 10.1038/s41467-019-12438-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Kashiwagi K, Fukuchi JI, Igarashi K. 1995. An acidic amino acid in the N-methyl-D-aspartate receptor that is important for spermine stimulation. Mol Pharmacol 48: 1087–1098. [PubMed] [Google Scholar]
- Williams K, Pahk AJ, Kashiwagi K, Masuko T, Nguyen ND, Igarashi K. 1998. The selectivity filter of the N-methyl-D-aspartate receptor: a tryptophan residue controls block and permeation of Mg2+. Mol Pharmacol 53: 933–941. [PubMed] [Google Scholar]
- Wittekindt B, Malany S, Schemm R, Otvos L, Maccecchini ML, Laube B, Betz H. 2001. Point mutations identify the glutamate binding pocket of the N-methyl-D-aspartate receptor as major site of conantokin-G inhibition. Neuropharmacology 41: 753–761. 10.1016/S0028-3908(01)00112-5 [DOI] [PubMed] [Google Scholar]
- Yu A, Lau AY. 2018. Glutamate and glycine binding to the NMDA receptor. Structure 26: 1035–1043.e2. 10.1016/j.str.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Zhang J, Pierson TM, Markello TC, Fajardo KV, Holloman CM, Golas G, Adams DR, Boerkoel CF, et al. 2014. Functional analysis of a de novo GRIN2A missense mutation associated with early-onset epileptic encephalopathy. Nat Commun 5: 3251. 10.1038/ncomms4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.