Abstract
Hydroxycarbamide (HC, hydroxyurea) is a cytoreductive drug inducing cell cycle blockade. However, emerging evidence suggests that HC plays a role in the modulation of transcription through the activity of transcription factors and DNA methylation. Examining the global mechanism of action of HC in the context of myeloproliferative neoplasms (MPNs), for which HC is the first-line treatment, will provide a better understanding of its molecular effects. To explore the effects of HC genome-wide, transcriptomic analyses were performed on two clinically relevant cell types at different stages of differentiation treated with HC in a murine MPN model. This study was replicated in MPN patients by profiling genome-wide gene expression and DNA methylation using patient blood samples collected longitudinally, before and following HC exposure. The effects of HC on the transcriptome were not only associated with cell cycle interruption but also with hematopoietic functions. Moreover, a group of genes were restored to normal expression levels in murine hematopoietic stem cells (HSCs) following drug treatment, including the master regulator of hematopoiesis, RUNX1. In humans, HC significantly modifies DNA methylation levels in HSCs at several distal regulatory regions, which we show to be associated with SPI1 binding sites and at the SPI1 locus itself. We have identified novel targets of HC that include pivotal transcription factors involved in hematopoiesis, and for the first time we report abnormal methylation patterns in MPN patients at the master regulator gene SPI1 and its distal binding sites, which HC is able to restore to normal levels.
Myeloproliferative neoplasms (MPNs) are a heterogenous group of hematological disorders characterized by uncontrolled proliferation of hematopoietic stem and progenitor cells in the bone marrow and peripheral blood. The so-called “Philadelphia Chromosome negative” MPNs include polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis. MPN patients have a heterogeneous clinical phenotype with many developing disease-related symptoms, such as splenomegaly or constitutional symptoms, and have an increased risk of both thrombotic and hemorrhagic events (McMahon and Stein 2013). Collectively, mutations in the JAK2, MPL, or CALR genes are the most frequent disease-initiating driver mutations in these disorders. These mutations are normally mutually exclusive and result in constitutive activation of the JAK-STAT pathway that promotes cellular proliferation (Lu et al. 2008). A minority of patients with classical MPN do not present any detectable mutations in these genes, so-called “triple negative” (TN) MPN. Within MPNs, a range of concomitant somatic mutations have additionally been described including genes that regulate DNA methylation (e.g., TET2, DNMT3A) and chromatin modification (e.g., ASXL1, EZH2) (Tefferi et al. 2009; Stegelmann et al. 2011; Brecqueville et al. 2012).
Hydroxycarbamide (HC, also called hydroxyurea) remains a first-line therapy of choice for high-risk PV and ET patients, dependent on phenotype, age, and patient/physician choice. The primary mechanism is the inhibition of the ribonucleotide reductase (RNR) enzyme that catalyzes the reduction of ribonucleotide diphosphates into their corresponding deoxyribonucleotide triphosphates (dNTPs) used in DNA synthesis and repair. RNR inhibition depletes dNTPs during the synthesis phase causing cell cycle arrest (Young and Hodas 1964; Krakoff et al. 1968).
HC also induces transcription of fetal hemoglobin (HbF), which explains its therapeutic use in sickle cell anemia. Several mechanisms have been proposed to illuminate the clinical effect of HC, including the induction of DNA methylation changes (Walker et al. 2011) and influences on transcription factor (TF) regulation that ultimately increase HBG1 expression after HC exposure in sickle cell anemia patients (Chondrou et al. 2018).
Although this drug has been used for more than 30 years in the MPN clinical area, effects on DNA methylation have not been comprehensively studied genome-wide in relevant cell types, and currently there is a limited understanding about the global mechanistic effects of this agent. To gain insight into the effects of HC, gene expression analysis of hematopoietic stem and differentiated cells was performed in a well-characterized MPN mouse model carrying the Jak2V617F mutation (hereinafter referred to as Jak2VF) (Mullally et al. 2010; Chen et al. 2015) and treated with HC. This study was replicated in MPN patients, for which we conducted a longitudinal study over a period of 9 mo. DNA and RNA samples from patients’ hematopoietic stem and differentiated cells were taken before and at 3-mo intervals following HC treatment, and these were assayed for gene expression and DNA methylation. We present a comprehensive analysis of the effects of HC treatment genome-wide in both species and report novel effects on both gene expression and DNA methylation.
Results
HC significantly decreases blood cell counts and spleen size in Jak2VF mouse
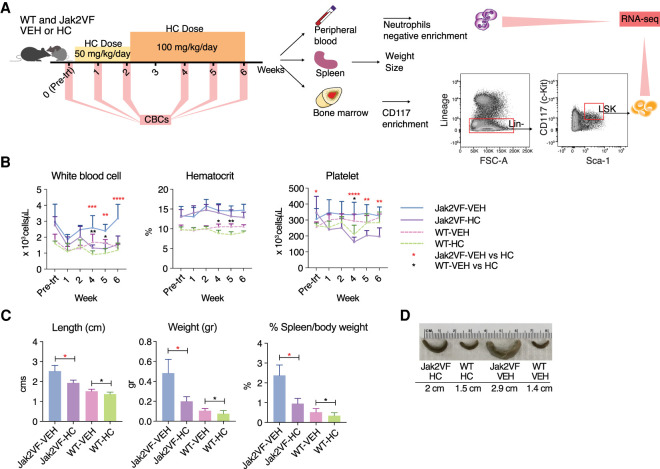
To investigate the effect of HC treatment on gene expression we treated a previously described heterozygous Jak2VF mouse that closely recapitulates the MPN phenotype found in patients (Mullally et al. 2010; Chen et al. 2015). Groups of Jak2VF and wild-type (WT) mice were treated with a starting dose of HC at 50 mg/kg/d or vehicle (Fig. 1A). After 2 wk, no noticeable changes in the hematocrit were observed in either of the mouse genotypes (Fig. 1B), so the dose was increased to 100 mg/kg/d for 4 wk. A significant reduction in white blood cells and platelets occurred at week 4 in both genotypes (Fig. 1B). The platelet compartment of Jak2VF mutant mice showed a greater sensitivity to the treatment compared to WT mice, showed by a severe reduction in platelet counts. Because Jak2VF mutant mice are known to develop splenomegaly, at the study end point spleens were isolated and we observed a significant decrease in size and weight in HC-treated mice compared to controls (Fig. 1C,D). These results show the efficacy of HC treatment in Jak2VF mutant mice and mimic the response seen in MPN patients treated with this drug.
Figure 1.
Treatment response to HC between Jak2VF mutant and wild-type mice. (A) Mouse study design: wild-type (WT) and JAK2VF mice were treated with HC or VEH for 6 wk, 5 d a week. Dose was maintained at 50 mg/kg/d for 2 wk and then increased to 100 mg/kg/d. After 6 wk, peripheral blood, spleen, and bone marrow were collected for further analyses. Spleens were measured for weight and size. Neutrophils were isolated from peripheral blood and LSK cells from bone marrow to perform RNA-sequencing on both cell types. (B) Mean blood cell counts per mouse group (Jak2VF-HC = 4, Jak2VF-VEH = 4, WT-HC = 7, WT-HC = 8), including white blood cells, platelets, and hematocrit. Data represent the mean ±SD. Statistical analysis performed in HC versus VEH. (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001 and (****) P ≤ 0.0001; analysis of variance (ANOVA). (C) Spleen weight and length following 6 wk of HC or VEH treatment. Nonparametric, unpaired Mann–Whitney U test. Error bars indicate mean with SD. (D) Representative examples of spleens (preserved in 10% formaldehyde) for each group. Length given corresponds to measurements on fresh tissue.
HC treatment perturbs gene expression more prominently in LSK cells compared to neutrophils, but immune system pathways are affected in both cell types
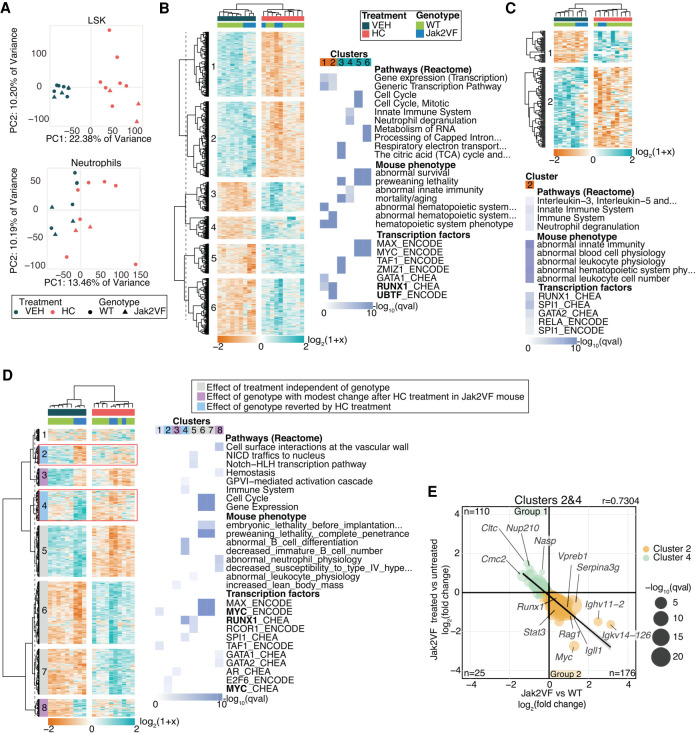
The effect of HC treatment on gene expression was assayed in Lin−Sca+cKit+ (LSK) cells, which are enriched for hematopoietic stem cells (HSCs) and in neutrophils using RNA-sequencing. The rationale underlying the choice of these two study populations is that (1) HSC are the disease-initating cell population in MPN, and (2) neutrophils are involved in the inflammatory response and have also been linked to thrombosis in MPN patients (Marin Oyarzún et al. 2016; Mesa et al. 2017; Barbui et al. 2018; Stone et al. 2018). Principal component analysis of the resulting expression profiles revealed a strong effect of HC in LSK cells (Fig. 2A), shown by two separated populations defined by the treatment (HC or vehicle).
Figure 2.
Gene expression analysis of LSK cells and neutrophils in mice. (A) Principal component analysis of gene expression profiles of LSK cells and neutrophil samples included in their analyses. Heatmap of DEGs and most significantly enriched terms for LSK cells (B) and neutrophils (C). (D) Heatmap plot of hierarchically clustered DEGs between WT-VEH versus Jak2VF-VEH and Jak2VF-HC versus Jak2VF-VEH and the top significant enriched terms of clusters identified. Clusters 2 and 4 (in the red square) correspond to genes that are differentially expressed between Jak2VF-VEH versus WT-VEH and that change their expression when treated with HC. TF name in bold indicates that its coding gene is differentially expressed. (E) Correlation plot of DEGs from Clusters 2 and 4 and their log2-fold change expression. Genes with the highest fold change are labeled. Linear regression.
In LSK cells, hierarchical clustering grouped samples from the same genotype, suggesting that the response to the treatment was influenced by the genotype (Fig. 2B). The differentially expresed genes (DEGs, Q-value < 0.05) were divided into six main clusters and enrichment analysis performed. Down-regulated genes (Fig. 2B, Clusters 1 and 2) were enriched for gene expression pathways associated with transcription regulation. Mouse phenotype enrichment analysis identified abnormal hematopoietic phenotypes revealing that a large proportion of down-regulated genes are involved in hematopoiesis. This was also shown in TF enrichment analysis, in which down-regulated genes were controlled mainly by RUNX1 and the Runx1 gene itself was differentially expressed and down-regulated (Supplemental Fig. S1). Conversely, up-regulated genes (Fig. 2B, Clusters 3, 4, 5, and 6) were enriched for four main pathways, including the citric acid cycle, the cell cycle, the immune system, and RNA metabolism.
In neutrophils, only down-regulated genes (Cluster 2) were significantly enriched (Fig. 2C) mainly in pathways involved in interleukin signaling and neutrophil degranulation. Comparative mouse phenotype enrichment analysis identified terms related to abnormal hematopoietic system physiology and TF enrichment analysis identified the hematopoietic regulators RUNX1 and SPI1.
Mouse LSK cells experienced more extensive changes with HC treatment than the more differentiated neutrophils, and treatment perturbs many pathways including the immune system and cell cycle. Thus, HC not only affects the cell cycle but it also influences hematopoietic functions along with master regulators of hematopoiesis, such as RUNX1 and SPI1.
HC partially reverts dysregulation of gene expression in Jak2VF mutant mice
The advantage of this MPN mouse model is that it carries the Jak2V617F mutation in every hematopoietic cell, whereas in human, MPN has a wide range of JAK2V617F allelic burden (Takahashi et al. 2013). Therefore, we examined the effect of the Jak2V617F mutation on gene expression in untreated mice to investigate the effect of HC on the genes affected by the mutation. To obtain a global view of how gene expression is affected by genotype and by treatment, all DEGs between Jak2VF mutant versus WT mice and between Jak2VF HC-treated versus untreated mice were analyzed (1596 genes). DEGs were hierarchically clustered according to their expression and enrichment analysis performed (Fig. 2D). Clusters 2 and 4 showed that the expression levels of misexpressed genes in Jak2VF mutant mice were partially restored to WT expression levels after treatment. These genes were enriched for immune system pathways and mouse phenotypes related to B cell development (Cluster 4). Furthermore, the TFs RUNX1, SPI1, and MYC were again identified as the main regulators of these genes (Supplemental Fig. S1).
The changes observed in Jak2VF compared to WT and in Jak2VF treated and nontreated (Fig. 2E) were examined and confirmed the negative correlation. One hundred ten genes were down-regulated in the Jak2VF mutant and up-regulated after treatment (Group 1), and 176 genes were up-regulated in Jak2VF compared to WT and down-regulated after treatment (Group 2). In Group 2, genes encoding immunoglobin proteins (Igll1, Ighv11-2, Igkv14-126, and Vpreb1) have the highest fold changes. Members of the JAK-STAT pathway, which is overactivated in MPN (Lu et al. 2008), were also down-regulated upon HC treatment (Jak3 and Stat3). These genes, which were identified as potential targets of RUNX1 (Fig. 2D), also included the Runx1 gene itself with an expression profile restored by HC treatment, highlighting a role for this transcription factor in HC response.
In neutrophils, a total of 497 genes were differentially expressed (Supplemental Fig. S2A,B). Hierarchical clustering confirmed that the main differences were at the genotype level rather than treatment level (Supplemental Fig. S2A). Nevertheless, Clusters 1 and 3 show that HC restored to WT expression levels, a subset of genes like in LSK cells (Supplemental Fig. S2B).
The effect of HC treatment is largely independent of the genotype. However, the restored expression to WT levels identified in Cluster 4 affecting the immune system reveals a novel aspect of HC in mutated cells that could contribute to changes in hematopoiesis through regulation of RUNX1. These observations may reflect the increasing awareness that inflammation may be as important in the pathogenesis and clinical features of MPN as JAK/STAT activation (Koschmieder et al. 2016; Lussana and Rambaldi 2017).
HC treatment effectively controls clinical parameters in MPN patients without affecting JAK2VF allele burden after 9 mo of treatment
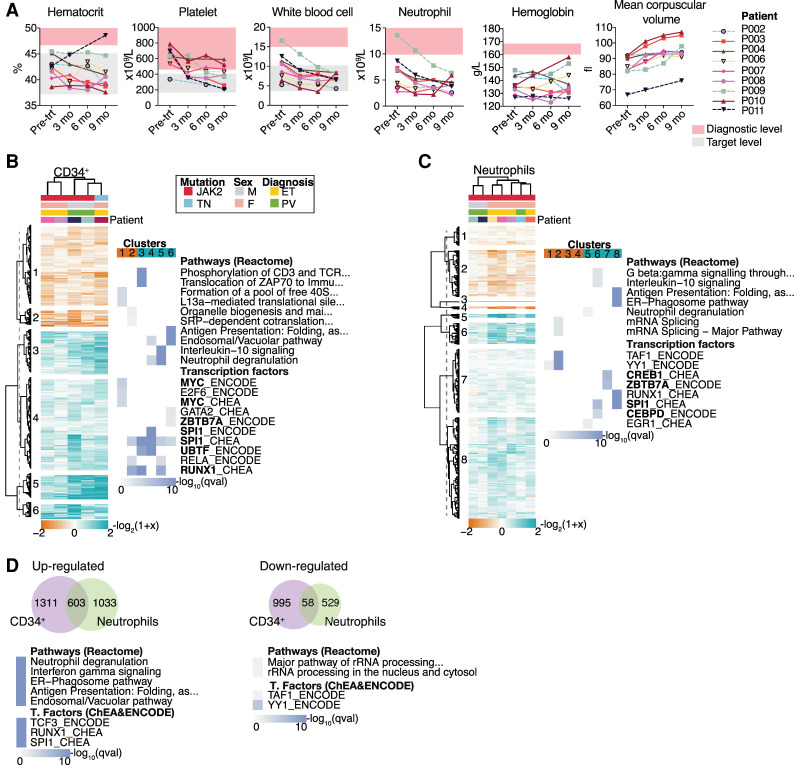
The effect of HC was assayed in MPN patients using a longitudinal study to minimize inter-subject variance. In this group of patients, no side effects from the HC treatment were documented, and blood cell counts decreased throughout the study period to within the desired ranges (Fig. 3A; Supplemental Table S1; Harrison et al. 2010; McMullin et al. 2019). In a single patient (P011) the hematocrit increased, which was confounded by the influence of on-going phlebotomy, whereas the remainder of the parameters decreased as expected.
Figure 3.
Blood cell counts through the course of the study and gene expression results of MPN patients. (A) Blood cell counts of clinical parameters monitored at four time points from each patient. The diagnostic criteria levels (Arber et al. 2016) and target level of the pharmacological treatment (Harrison et al. 2010; McMullin et al. 2019) are indicated. Hierarchical clustering of DEGs and the top significantly enriched terms for each cluster from CD34+ (B) and neutrophil cells (C). DEGs were clustered according to their log2-fold change expression over pretreatment samples (Euclidean correlation distance and ward.D2 agglomeration method). Clusters were divided at the same branch heights and numbered. Enrichment analyses for pathways and transcription factors were performed in each cluster. The top significantly enriched results from each cluster are represented as a heatmap plot in which color scale indicates significance. TF name in bold indicates that the gene is differentially expressed. (D) Venn diagram of common DEGs among CD34+ cells and neutrophils and results of enrichment analysis performed in the shared genes. Enrichment analysis was performed separately for up-regulated and down-regulated genes.
The JAK2V617F allele burden was quantified in neutrophils at two time points: pretreatment and at 9 mo of treatment. We observed a modest decrease of the JAK2V617F allele burden (1%–9%) after HC treatment, indicating that the changes occurring after treatment are not linked with a reduction in the proportion of mutant cells (Supplemental Fig. S3). Additionally, all patients were sequenced to identify common mutations using a “myeloid gene panel.” We identified two patients (P002 and P008) with mutations in the DNMT3A gene and one (P011) with mutations in the TET2 gene (Supplemental Table S1).
HC treatment of MPN patients affects the SPI1 and RUNX1 genes in the same way as in the mouse model
Human CD34+ cells, as a comparable population to LSK cells, and human neutrophils were isolated and RNA-sequenced before and following 9 mo of HC treatment.
In CD34+ cells, six main clusters were identified (Fig. 3B). Enrichment analysis showed that down-regulated genes were enriched for protein translation terms and up-regulated genes for immune system terms, including cytokine signaling and neutrophil degranulation. TF enrichment identified SPI1 and RUNX1 as the principal regulators of most clusters (Fig. 3B). SPI1 and RUNX1 were also up- and down-regulated, respectively (Supplemental Fig. S4), correlating with the pathways and TFs identified in mouse LSK cells.
In neutrophils, there was variability between PV and ET patients, whereby individuals with PV have fewer expression changes compared to ET patients (Fig. 3C). As seen in CD34+ cells, gene clustering analysis identified that down-regulated genes were enriched for protein translation terms, and up-regulated genes were enriched for immune system–related terms. Enrichment for potential TFs in up-regulated genes identified SPI1, ZBTB7A, and CEBPD, for which the encoding genes themselves were differentially expressed (Supplemental Fig. S5). Compared to mouse neutrophils, more genes were affected by HC in human neutrophils, but similar pathways involved in immune system functions were identified in both species. Thus, there are common pathways modulated by HC treatment in differentiated cells that are independent of cell cycle arrest, the main described mechanism of action of HC.
Enrichment analysis of up-regulated DEGs shared between CD34+ cells and neutrophils revealed the same terms as in mouse (Fig. 3D). TFs identified as regulators of these genes are RUNX1, SPI1, and TCF3. Other genes encoding TFs involved in hematopoiesis such as RARA, SPI1, CEBPB, and ZBTB7A were up-regulated in both cell types (Supplemental Figs. S4, S5). Significant terms in down-regulated DEGs shared between the two cell types included pathways related to protein translation (Fig. 3D). Thus HC regulates a subset of genes at two developmental stages, with a greater effect on up-regulation of genes mostly controlled by RUNX1 and/or SPI1.
Several TFs involved in hematopoiesis are commonly regulated in human and mouse, and RUNX1 is one of the major mediators of HC treatment in both species
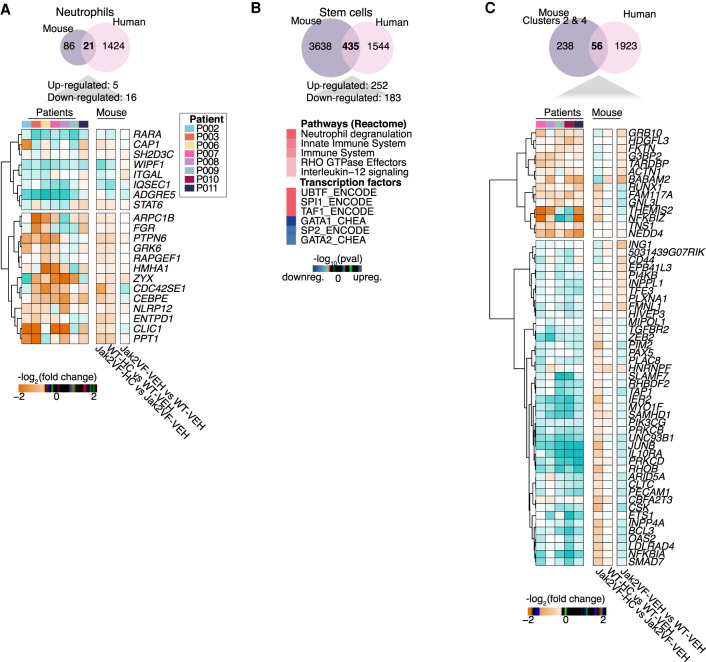
The extent of the conservation of the treatment response was investigated by comparing all conserved DEGs in the two species.
In neutrophils, only a few shared DEGs were identified (21 genes), so no enrichment analysis was performed. The log2-fold changes per patient (after vs. before HC treatment) and per mouse group comparison were plotted for these shared genes (Fig. 4A,C). Some DEGs in human have similar expression patterns to their counterparts in mouse treated with HC (ITGAL and CEBPE).
Figure 4.
Comparison of homologous differentially expressed genes between human and mouse. Venn diagrams show the total number of DEGs commonly regulated in human and mouse in neutrophils (A) and stem cells (CD34+ and LSK cells in human and mouse, respectively) (B). (C) Venn diagram shows the total number of genes identified in Clusters 2 and 4 from mouse LSK cells that overlap the DEGs from human CD34+ cells. (A,C) Heatmap plots represent the log2(fold change) of treated samples per patient versus pretreatment samples, along with pairwise comparison of each mouse condition (Jak2VF VEH vs. WT-VEH, WT-HC vs. WT-VEH, and Jak2VF-HC vs. Jak2VF-VEH). Bold is used to indicate the genes with similar expression between human and WT-HC or Jak2VF-HC mice.
At the stem cell level, 435 genes were commonly regulated in both species. Commonly up-regulated genes were enriched for neutrophil degranulation and innate immune system pathways (Fig. 4B), like individually observed in MPN patients and mice. SPI1 and GATA1 were shown by enrichment analysis to act as regulators for up- and down-regulated genes, respectively. No pathways were identified in down-regulated genes, but there was enrichment for “transcription regulation,” with several TF encoding genes, such as HOXB4 and BCL11A (Supplemental Table S2). Other DEGs encoding TFs such as RUNX1, ZBTB7A, ETS1, and ATF3 were identified as regulators of the common DEGs between human and mouse as well as genes involved in epigenetic regulation, KDM5B, a H3K4 demethylase, and TET1, which is involved in DNA demethylation (Iwase et al. 2007; Guo et al. 2011).
In mouse a subset of genes was partially restored back to WT expression levels by HC (Clusters 2 and 4) (Fig. 2). Homologous genes between human and mouse (Clusters 2 and 4) (Fig. 2) were compared to all DEGs from human CD34+ cells, and 56 genes were conserved (Fig. 4C). A subset of genes in human show similar expression patterns to WT or Jak2VF mice treated with HC (Fig. 4C, in bold), which included RUNX1 pointing to its important role in the therapeutic mechanism of HC.
In human neutrophils, DNA methylation levels are not affected by HC treatment
The effect of HC on DNA methylation has been documented in sickle cell anemia patients; however, the effects on MPN patients, in which abnormal gene expression and DNA methylation patterns can be expected as observed in other types of cancer (Hinoue et al. 2012; Sproul et al. 2012; Maupetit-Mehouas et al. 2018), have not been investigated. DNA methylation genome-wide was investigated in human CD34+ cells and neutrophils as a mediator of the HC effect. Differential methylation was measured at individual CpGs and at regions (1 kb) containing CpGs with consistent methylation changes, referred to as differentially methylated regions (DMRs).
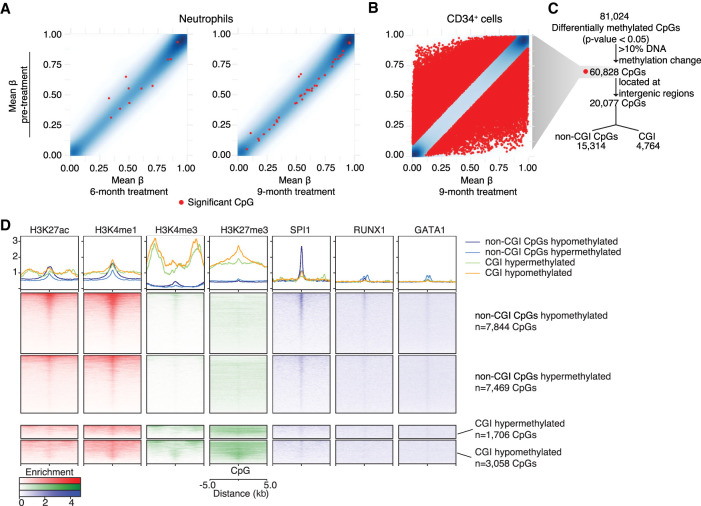
In neutrophils, DNA methylation was measured at four time points for eight patients. A relatively low number of differentially methylated CpGs were identified at 6 and 9 mo of treatment compared to pretreatment (Fig. 5A), and no DMRs were identified by any of the time point comparisons. DNA methylation in neutrophils is therefore highly stable throughout the course of HC treatment, and after 9 mo of treatment no significant changes were detected.
Figure 5.
DNA methylation analysis of neutrophils and CD34+ cells CpGs. (A,B) Scatter plots show the mean of DNA methylation level (β-value) from pretreated samples (y-axis) and after 6 and 9 mo of HC treatment in neutrophils and 9 mo of HC treatment in CD34+ cells (x-axis). Red dots correspond to significantly differentially methylated CpGs according to each time point comparison. (C) Workflow of differentially methylated CpGs analyzed in CD34+ cells. All differentially methylated CpGs identified were filtered by having >10% of DNA methylation change. From these, differentially methylated CpGs overlapping intergenic regions were further subdivided according to their position in the genome in respect to CpG islands (CGI), such as differentially methylated CpGs overlapping CGIs or non-CGI CpGs. (D) Heatmap plots that show enrichment of regulatory regions (H3K27ac, H3K4me1, H3K4me3, and H3K27me3) and transcription factor bindings sites (SPI1, RUNX1, and GATA1) at differentially methylated CpGs overlapping CGIs or non-CGI CpGs using ChIP-seq data from healthy CD34+ cells.
In CD34+ cells, HC modulates DNA methylation at regulatory regions of genes involved in hematopoiesis
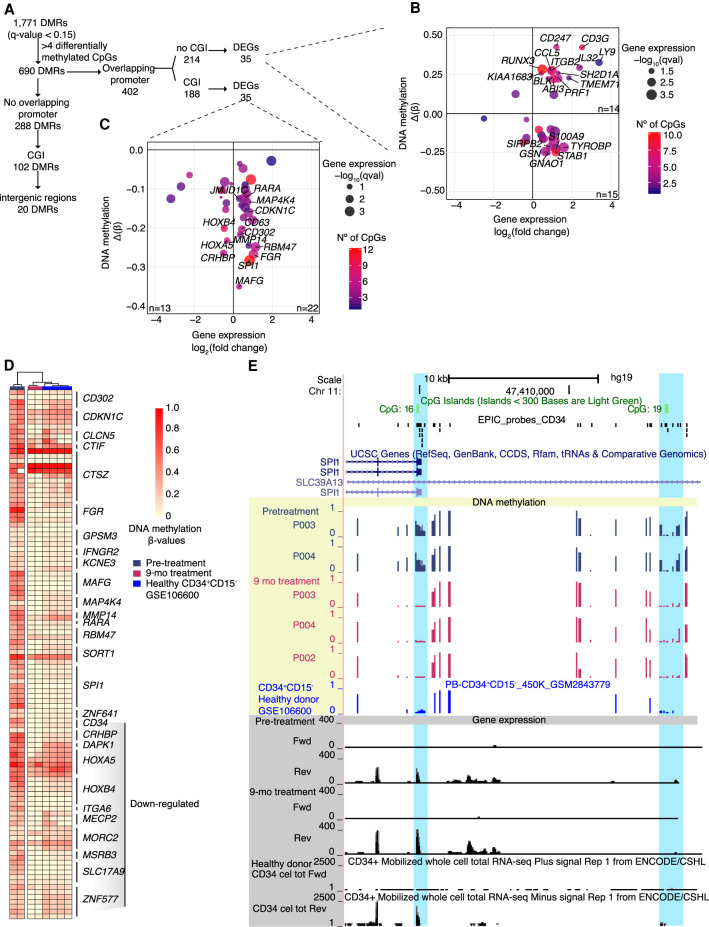
Owing to limited amounts of DNA from CD34+ cells, two of the nine patients (P003 and P004) were assayed at two time points: pretreatment and following 9 mo of treatment. Differential methylation testing identified 60,828 significant differentially methylated CpGs (P-value < 0.05) with >10% of DNA methylation change, and 690 DMRs (Q-value < 0.15) containing at least four differentially methylated CpGs (Figs. 5B,C, 6A).
Figure 6.
DNA methylation analysis of differentially methylated regions (DMRs) of human CD34+ cells. (A) Workflow of DMRs analyzed in CD34+ cells. DMRs with more than four differentially methylated CpGs were divided between DMRs overlapping promoters and no promoters. The DMRs overlapping promoters were subdivided between overlapping CpG islands (CGI) or not, and finally correlated to DEGs. The DMRs not overlapping promoters were filtered by overlapping CGI or intergenic regions. (B,C) Correlation plot between DNA methylation and gene expression changes between pretreatment and following 9 mo of HC treatment. DNA methylation of each region is expressed by the difference of the β-value (Δβ) before and following HC treatment (y-axis; n = 2). Gene expression change is expressed by the log2-fold change before and following HC treatment (x-axis; n = 5). Color scale represents the number of differentially methylated CpGs in the region. (B) DMRs that overlap gene promoters of DEGs with no CGIs described in the promoter region. (C) DMRs that overlap CGI at gene promoters of DEGs. (D) Heatmap of individual differentially methylated CpGs located in DMRs overlapping CGIs at pretreatment, 9 mo of HC treatment, and healthy CD34+ cells. Genes associated to the differentially methylated CpGs are indicated. (E) Locus of the promoter and regulatory region of SPI1 with DNA methylation and gene expression levels before and after 9 mo of HC treatment. Yellow shading indicates DNA methylation levels, blue shading indicates significant DMRs, and gray shading denotes gene expression levels.
Genomic context of DNA methylation changes is important and at distal regulatory regions some TFs can be affected by DNA methylation at one CpG in their binding site (Rahman et al. 2012; Mann et al. 2013). TF enrichment analysis at intergenic regions containing differentially methylated CpGs (±5 bp, Fig. 5C) was performed using the online resource i-cisTarget. The CpGs were divided into CpGs within CpG islands (CGIs) and non-CGI CpGs, which are outside of CGIs (Fig. 6B,C; Sandoval et al. 2011). Non-CGI CpGs were enriched for TFs binding sites, such as GATA1, GATA2, and SPI1 (Supplemental Table S3). The TFs associated with the highest number of DEGs, such as CEBPB, BCL11A, and SPI1, were themselves differentially expressed (Supplemental Table S3, in bold). CpGs overlapping CGIs were enriched for two members of the polycomb repressive complex 2 responsible for H3K27m3 deposition (SUZ12 and EZH2).
Because the analysis of DNA methylation at intergenic CpGs identified TF binding sites at distal regulatory regions, we investigated their function using publicly available ChIP-seq data from healthy control CD34+ cells (Huang et al. 2016). Peaks were identified around differentially methylated CpGs for H3K27ac and H3K4me1 (enhancer-associated histone modifications), H3K4me3 and H3K27me3 (CGI-associated histone modifications), and hematopoietic TFs (SPI1, RUNX1, and GATA1) (Fig. 5D). As expected, CGI CpGs were enriched for H3K4me3 and H3K27me3, confirming i-cisTarget prediction of polycomb repressive complex 2 proteins binding to these regions. Non-CGI CpGs were linked with enhancer-associated histone modifications (H3K27ac and H3K4me1) and, as predicted by i-cisTarget, these were highly enriched for SPI1 binding sites and to a lesser extent RUNX1 and GATA1 binding sites, confirming that non-CGI CpGs are likely to be found in enhancer regions in normal CD34+ cells, which are bound by SPI1 and reveal a role for them in hematopoiesis. To determine whether the differential methylation of non-CGI CpGs was correlated with changes in gene expression, genes near potential regulatory regions were assayed (Kaikkonen et al. 2013; Lam et al. 2013; Li et al. 2013). We identified 251 genes with nearby non-CGI CpGs that were differentially expressed (Supplemental Table S4) in CD34+ cells; in each case, differential expression could be linked to changes in DNA methylation at their enhancer regions (Supplemental Fig. S6).
Similarly, DMRs were investigated at intergenic CGIs, which can act as distal regulatory regions (Illingworth and Bird 2009). One intergenic hypermethylated CGI was identified as a binding site for RUNX3 (also differentially methylated and up-regulated) (Supplemental Tables S5, S6; Supplemental Fig. S7A). This region is linked to the IRF8 gene, which was up-regulated and differentially expressed (Supplemental Fig. S7B). Both of these genes are involved in hematopoiesis: Runx3 deletion leads to a myeloproliferative phenotype (Wang et al. 2013), and Irf8 deletion perturbs the self-renewal of HSCs in mouse (Qiu et al. 2015). This could be important for the regulation of the IRF8 gene and again shows that HC can regulate TFs involved in hematopoiesis, here linked to DNA methylation.
DNA hypermethylation on gene promoters returns to normal levels after HC treatment
At promoter regions, DNA methylation is associated with repression only if the promoter is associated with a CGI (Weber et al. 2007). Although rare in normal tissue (Auclair et al. 2014; Dahlet et al. 2020), repression of expression by DNA methylation at CGI-associated promoters is a hallmark of cancer (Esteller et al. 2000). DMRs overlapping with promoter regions were investigated (Fig. 6A). DMRs associated with non-CGI promoters or low CpG content promoters (214) were identified and among these, 35 DMRs were associated with DEGs. From these, 16 have a negative correlation with gene expression (hypomethylated and up-regulated), whereas 19 were positively correlated (hypermethylated and up-regulated) (Fig. 6B), confirming previous findings with low CpG content promoters, in which a negative correlation with gene expression is not observed but some active promoters may be methylated (Weber et al. 2007). On the other hand, 35 DMRs overlapping CGI promoters were associated with DEGs (Fig. 6C; Supplemental Table S7). Thirteen genes showed loss of DNA methylation and decreased gene expression including HOXB4 and HOXA5, explained by DNA methylation changes focused in small areas within the CGI, but not always localized into the promoter region (Supplemental Fig. S8A). Loss of DNA methylation in the promoter and increased gene expression occurred in 22 genes, which included kinases (FGR, CDKN1C, MAP4K4) and TFs (SPI1, RARA, JMJD1C). These regions were compared to publicly available DNA methylation data from healthy CD34+CD15− cells (Maupetit-Mehouas et al. 2018). Pretreatment samples were mostly hypermethylated compared to healthy samples, and during HC treatment, methylation was restored to healthy levels (Fig. 6D; Supplemental Fig. S8B). In addition, these promoters were enriched for permissive histone marks, such as H3K4me3 and H3K27ac, and depleted for the repressive mark H3K27me3 in normal CD34+ cells (Supplemental Fig. S8C), suggesting that these genes are normally expressed and that HC is able to restore their expression in association with loss of DNA methylation.
SPI1 was the most differentially expressed gene and had the highest number of differentially methylated CpGs at the promoter (Fig. 6C). Its regulatory region, located −17 kb away from the promoter (Okuno et al. 2005), was also differentially methylated (Fig. 6E). Comparison of these regions with healthy individual samples (Maupetit-Mehouas et al. 2018) revealed that they were aberrantly methylated in MPN patients. HC can restore the DNA methylation state to a level similar to healthy individuals. HC likely modulates DNA methylation and is correlated with gene expression changes at a subset of gene loci, although whether one is a downstream effect of the other is unclear. To investigate this, we used two hematopoietic cells lines (Jurkat and MT-4) that are methylated at the SPI1 promoter region. After optimization of the treatment conditions (Supplemental Fig. S9), we measured gene expression through RT-qPCR and DNA methylation using COBRA (Bilichak and Kovalchuk 2017). We observed that HC treatment increases gene expression of SPI1 across time (Supplemental Fig. S10A). We confirmed that SPI1 targets were also activated after HC treatment (CCR1 and PTGER4) (Supplemental Fig. S10B) and that RUNX1 was down-regulated (Supplemental Fig. S10C). The increase in SPI1 expression was not associated with changes in DNA methylation at its promoter region (Supplemental Fig. S10D), suggesting that the earliest consequence of the treatment is the activation of SPI1 expression. Of clinical relevance, and in line with a recent investigation that shows that SPI1 expression under inflammatory stress is required to restrict hematopoietic stem cell expansion (Chavez et al. 2021), our findings present SPI1 as a potential target for myeloproliferative diseases.
Discussion
HC is a frequently used cytoreductive therapy for high-risk PV and ET patients, and it inhibits the RNR enzyme leading to cell cycle arrest (Krakoff et al. 1968). However, this mechanism does not explain its therapeutic effects in sickle cell anemia (Platt et al. 1984), and the modulation of DNA methylation and TF expression following HC treatment in this disease has been investigated by others (Chondrou et al. 2018). Among MPN patients very limited evidence exists with regard to these potential effects. In granulocytes derived from chronic myeloid leukemia (CML) patients, the increased expression of RARA has been shown to reach healthy levels after 1 wk of HC treatment (Bruchova et al. 2002). However, a limited number of genes were differentially expressed because of the short duration of the study. Genome-wide DNA methylation in MPN patient neutrophils has been used to classify MPN patients, but the consequence of HC treatment has not been evaluated (Nischal et al. 2013). To understand the effects of HC treatment in MPN, we investigated for the first time two clinically relevant cell types from an epigenomic and transcriptomic point of view using an established MPN mouse model and MPN patients’ samples from a longitudinal study.
In mouse LSK cells, HC restores expression of a subset of genes abnormally expressed in the Jak2VF mutant mouse model. These genes are enriched for RUNX1 binding sites, which itself is among the HC down-regulated genes. These observations suggest a relationship between the expression profile linked with Jak2V617F mutation and Runx1 overexpression. High expression of RUNX1 in MPN patients compared to controls has been reported in granulocytes and erythroid precursor cells derived from CD34+ cells (Wang et al. 2010). Moreover, as in mouse, RUNX1 is down-regulated following HC treatment in human CD34+ cells, suggesting that it can contribute to the HC therapeutic effect in humans. In general, the effects of the Jak2V617F mutation on the transcriptome of LSK cells is small compared to the effects of HC treatment. LSK cells are highly affected by the treatment independently of the mutation, confirming previous findings that HC is not able to significantly reduce the JAK2V617F allele burden, but rather has a nonpreferential effect on mutant and non-mutant cells (Antonioli et al. 2010; Kubovcakova et al. 2013).
In mouse neutrophils, a strong effect of the Jak2V617F mutation is revealed in their transcriptome and is hardly modified by HC. Although gene expression in human neutrophils is affected by HC to a similar extent as in CD34+ cells, the lack of plasticity is manifested by DNA methylation levels whereby HC-mediated modulation of DNA methylation at any of the time points could not be detected. These findings need to be taken into account when selecting cell type populations for time-course experiments and additionally shows the importance of using paired samples from the same individual (Chatterjee et al. 2015). The striking differences in DNA methylation following HC treatment in human CD34+ cells support the importance of selecting the correct cell population for DNA methylation studies. These hematopoietic stem and progenitor cells are able to respond to physiological needs to maintain hemostasis (Ruf 2016). The DNA methylation machinery, including the DNMT enzymes, such as DNMT3A which is commonly mutated/comutated in MPN, play an important role during both development and differentiation. The lack of DNMT3B and DNMT3A, necessary for de novo methylation, increases self-renewal of HSCs and impairs differentiation (Challen et al. 2012, 2014; Jeong et al. 2018). These enzymes are highly expressed in early progenitor cells and their expression declines with differentiation (La Salle et al. 2004). Therefore, the ability of HC to affect DNA methylation in CD34+ cells and not in neutrophils is not surprising. The biological importance of these findings is shown by their correlation with gene expression. HC treatment modulates DNA methylation at non-CGI CpGs that are associated with DEGs and show characteristics of distal regulatory regions. Other studies have shown that demethylation at enhancers is associated with gene reactivation during differentiation (Mahé et al. 2017). However we do not know whether DNA methylation changes at enhacers is simply the readout of transcription factor availability or the mechanisms restricting transcription factor binding. Our results suggest that loss of DNA methylation upon HC treatment in distal regions, enriched for H3K27ac and H3K4me1 in healthy CD34+ cells, could be linked to gene reactivation. DNA methylation levels at these regions is dynamic during hematopoiesis and neoplastic transformation (Blattler et al. 2014; Rönnerblad et al. 2014; Agirre et al. 2015; Bell et al. 2016). Moreover, we identified enrichment for SPI1 in these regions, increasing the relevance of this TF. A dramatic loss of methylation occurred at the SPI1 promoter and its regulatory region following HC treatment and is negatively correlated with SPI1 expression. SPI1 expression is also impaired in chronic myeloid leukemia (CML), in which it can be recovered following drug treatment with interferon alpha and the tyrosine kinase inhibitor imatinib (Albajar et al. 2008). Aberrant methylation has also been identified at the SPI1 promoter in whole blood from CML patients (Baylin and Herman 2000). Altogether our study proposes a novel mechanism to understand MPN pathogenesis, in which RUNX1 down-regulation after HC treatment, which acts upstream of SPI1, could mediate the ability of HC to reduce myeloproliferation. Understanding these mechanisms can facilitate the development of more specific treatments to bypass undesired side effects of HC. Although several changes in DNA methylation were observed, not all the DEGs identified are associated with differentially methylated CpGs in the cell types studied. Therefore, other mechanisms are likely involved in the ability of HC to affect gene expression. The effects of HC at the SPI1 locus are proposed as a dynamic model showing that expression reactivation could precede DNA demethylation at CGIs promoters in hematopoietic malignancies. Further investigation in the SPI1 locus in MPN patients could lead to the use of DNA methylation and/or gene expression as markers of HC treatment efficiency.
Methods
Jak2V617F and control mice
Jak2V617F-knock-in mice were generated as previously described (Mullally et al. 2010; Chen et al. 2015). All mouse experiments were approved by the institutional ethic committee of Brigham and Women's Hospital, Boston (Protocol #2017N000025) and maintained in pathogen-free facilities at the Brigham and Women's Hospital, Boston.
Murine HC treatment
Hydroxycarbamide (HC, Sigma-Aldrich H8627) was diluted with injectable 0.9% NaCl solution (Baxter). HC solution was prepared fresh upon administration under sterile conditions. Intraperitoneal injections (IP) were administered to Jak2V617F and wild-type mouse groups 5 d a week for 6 wk. For the first 2 wk, 50 mg/kg/d was administered and for the following 4 wk, 100 mg/kg/d was used. Similarly, control groups per genotype received vehicle only on the same days.
Clinical parameters for murine study
Peripheral blood was sampled from the retro-orbital sinus using a micro-hematocrit capillary tube, sodium heparinized. Blood was collected in K2EDTA-coated tubes (BD Microtainer) to measure blood cell counts on an Advia 2120i Hematology System (Brigham and Women's Hospital). Blood cell counts and weight were controlled weekly for dose adjustment and monitoring toxicity.
LSK cell isolation from mouse
Whole bone marrow was obtained by crushing the femur, tibia, and spinal column. Erythocytes were removed using erythrocyte lysis buffer (10× BD-lysis-buffer, BD Bioscience). CD117 enrichment was performed using the Miltenyi CD117 microbeads (MACS Miltenyi Biotec). The CD117 enriched fraction was stained with CD5, Gr1, B220, CD3, CD11b, Ter119, CD117, and Sca-1 (Supplemental Table S8). LSK cells were sorted on a BD FACSAria cell sorter (BD Biosciences) and collected in RLT-plus buffer (Qiagen) with 1% of β-mercaptoethanol and stored at −80°C until use.
Neutrophil isolation from mouse
Peripheral blood was obtained from the retro-orbital venous plexus. Erythocytes were removed using 1× erythrocyte lysis buffer (BD Bioscience). The cells were stained with the Miltenyi Neutrophil Microbeads (MACS Miltenyi Biotec) following the manufacturer's instruction. The fraction containing the enriched neutrophils was washed and resuspended in 75 µL of RLT-plus buffer (Qiagen) with 1% of β-mercaptoethanol. The samples were stored at −80°C until use.
Human sample collection
This study was developed in collaboration with the Department of Hematology, Guy's and St Thomas’ NHS Foundation Trust. Ethical approval was obtained from the NRES Committee London—City Road and Hampstead, REC reference 15/LO/0265. Male and female patients (>18 yr old) with a confirmed diagnosis of MPN according to the World Health Organization 2016 criteria and who planned to start HC treatment as a first-line therapy were recruited to the study. Peripheral blood was collected at baseline and at 3, 6, and 9 mo after starting HC treatment. Clinical and hematological parameters were sequentialy collated. Patients’ characteristics are summarized in Supplemental Table S1.
Neutrophil isolation from human samples
Neutrophils were purified from 10 mL of peripheral blood as described previously (Heit et al. 2002). Neutrophil purity was assessed with flow cytometry by staining with CD16b, CD14, CD3, and CD19 (Supplemental Table S8).
CD34+ cell isolation from human samples
Peripheral blood was diluted in ice-cold PBS, and gradient centrifugation was performed using Histopaque-1077 (Sigma-Aldrich) to separate the peripheral blood mononuclear cells (PBMCs). Cells were resuspended with RPMI 1640 (Thermo Fisher Scientific) supplemented with 10% FBS and 5% Penicillin/Streptomycin. Cells were frozen with FBS+10%DMSO and stored in liquid nitrogen. Frozen PBMC aliquots, were thawed and resuspended in complete RPMI 1640 media. The cells were washed, resuspended as single cells, and stained with Miltenyi CD34 microbeads (MACS Miltenyi Biotec) following the manufacturer's instructions. Double column separation was performed to increase purity. DNA and RNA were isolated from the CD34+ fraction as described.
JAK2V617F allele burden
JAK2V617F allele burden was quantified in DNA isolated from neutrophils using the ipsogen JAK2 MutaQuant Kit (Qiagen) following the manufacturer's instructions. Samples from pretreatment and 9 mo of treatment were used to assess changes in the variant allelic frequency.
Myeloid gene panel
Neutrophil DNA samples were sequenced for 33 genes or gene mutation hotspots (Rio-Machin et al. 2020) on an Illumina MiSeq using TruSeq Custom Amplicon reagents at King's Haematological Malignancy Diagnostic Centre. Genes with mutations are reported in Supplemental Table S1.
DNA and RNA purification
DNA and RNA were simultaneously purified using AllPrep DNA/RNA Micro Kit (Qiagen) for mouse samples and AllPrep DNA/RNA Mini Kit (Qiagen) for human samples, according to the manufacturer's instructions. For RNA only extraction from CD34+ cells (patients P007, P008, P009, P010, and P011), the PicoPure RNA isolation kit (Thermo Fisher Scientific) was used.
Bisulfite conversion
Three hundred nanograms of DNA from human neutrophils or up to 50 ng of DNA from human CD34+ cells were processed using the EZ DNA Methylation Kit (Zymo Research), following the manufacturer's instructions for Infinium assays. Samples were stored at −20°C for up to 1 wk until use.
DNA methylation processing
DNA methylation was measured genome-wide with the Infinium Human MethylationEPIC BeadArray (Illumina). Raw files were processed in R (version 3.5.1) (R Core Team 2018). Quality control was performed using Minfi (version 1.28.4) (Aryee et al. 2014), and RnBeads's Greedy cut algorithm (version 2.0.1) (Assenov et al. 2014). Background correction was performed with the out-of-band model from Enmix (Xu et al. 2015). Quantile normalization was separately applied to each probe type and color channel to reduce systematic in-between array differences in the signal distributions. Beta mixture quantile normalization (BMIQ) was then used to correct for probe design type bias affecting Infinium II probes (Pidsley et al. 2013). Because multiple arrays were used to assess DNA methylation in neutrophils, the batch effect was removed using ComBat (Leek et al. 2012) from the sva package (version 3.30.1). Differential methylation testing was conducted with limma (version 3.38.3) (Ritchie et al. 2015) using a model including the time post-treatment factor (pretreatment vs. 3, 6, and 9 mo post-treatment) and patient as a covariant. Technical replicates were accounted for during differential methylation testing using the dupcorr function of the limma package (Ritchie et al. 2015).
For enrichment analysis, the coordinates of the DMRs and differentially methylated CpGs in BED file format were submitted to the online resource i-cisTarget (Imrichová et al. 2015) to identify potential regulatory regions. The enrichment analyses were performed separately for DMRs and differentially methylated CpGs divided in hypo- or hypermethylated sets following HC treatment. The configuration parameters minimum fraction of overlap, normalized enrichment score (NES) threshold, and AUC threshold were set to 0.4, 3.0, and 0.005, respectively.
The deepTools2 package (version 3.5.8) (Ramírez et al. 2016) was used to plot publicly available ChIP-seq data using differentially methylated CpGs at intergenic regions with the options ‐‐referencePoint center ‐‐beforeRegionStartLength 5000 ‐‐afterRegionStartLength 5000 ‐‐skipZeros. BEDTools “closest” function (Quinlan and Hall 2010) was used to identify the closest gene promoter to the differentially methylated CpG. The table with gene names and transcription start sites (TSS) used with BEDTools was retrieved from biomaRt (version 2.38.0) (Durinck et al. 2005) in R.
Processed ChIP-seq data from healthy CD34+ cells (A0) and DNA methylation data from healthy CD34+CD25− cells, Jurkat, and healthy CD34+ cells were obtained from the NCBI Gene Expression Omnibus database (GEO; https://www.ncbi.nlm.nih.gov/geo/) (accession numbers GSE70660, GSE106600, and GSE85525, respectively). For Jurkat and healthy CD34+ cells, M-values were converted to β-values using m2beta function of the lumi package (version 2.42) (Du et al. 2008) in R.
RNA library preparation
Between 1 and 5 ng of RNA was used to prepare libraries with the SMARTer Stranded Total RNA-Seqv2-Pico Input Mammalian kit (Clontech), according to the manufacturer's instructions. The same amount of RNA was used for paired samples. The libraries were diluted to equimolar concentrations, pooled, and sequenced in an Illumina HiSeq 4000 system.
Gene expression analysis
Raw sequencing reads were processed for quality control using FastQC (version 0.11.8). Samples with duplication rates >80% were removed from the analysis. Adapters were trimmed with BSTools (version 38.22) (Wang et al. 2015), and reads were pseudoaligned using kallisto (version 0.44) (Bray et al. 2016). Differential gene expression testing was performed with Sleuth (version 0.30) (Pimentel et al. 2017) using patient and time point as model factors in human, and genotype and treatment as model factors in mouse. DEGs (Q-value < 0.05) were hierarchically clustered using Euclidean distances and the ward.D2 agglomeration method. Gene set enrichment analyses were performed for each gene cluster using Enrichr (Chen et al. 2013; Kuleshov et al. 2016) database, through the R interface enrichR (https://cran.r-project.org/web/packages/enrichR/vignettes/enrichR.html, version 2.1), with gene sets defined by biological pathway, mouse phenotypes, or by their regulation via common TFs. To visualize the expression changes in patients, the log2-fold changes before and after treatment were calculated, and these values were hierarchically clustered.
Cell line treatment
Jurkat and MT-4 cell lines were treated with 0, 0.06, 0.125, 0.25, 0.5, and 1 mM of HC for 24 h to identify an optimal dose. Further experiments were performed using 0.125 and 0.5 mM of HC for Jurkat and MT-4, respectively. HC was added fresh every 24 h. Dead cell analysis was peformed using Annexin V (APC, Biolegend) and propidium iodide (Biolegend) as recommended by the manufacturer. Cells were resuspended with 400 µL of Annexin V binding buffer and analyzed using BD Accuri C6 Plus Flow Cytometry. Cell counts were simultaneusly obtained from BD Accuri, using the events/µL parameter.
RT-qPCR
RNA from each time point was isolated using TRIzol Reagent (Thermo Fisher Scientific) following the manufacturer's instructions. A DNase treatment step was performed using the Ambion DNA-free Kit (Thermo Fisher Scientific). cDNA was synthesized using the SuperScript VILO cDNA synthesis kit (Thermo Fisher Scientific). RT-qPCR was performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) in a 10 µL reaction including 1 µL cDNA, 5 µL Brilliant III Ultra-FAST SYBR Green QPCR Master Mix, and 0.5 µL of 0.1 mM primers (Supplemental Table S9).
DNA methylation analysis on cell lines
DNA methylation assay was performed using Bisulfite conversion, PCR amplification, and digestion for combined bisulfite restriction analysis (COBRA) (Bilichak and Kovalchuk 2017). Briefly, a PCR on SPI1 promoter (Supplemental Table S9) was performed in the bisulphite converted DNA. The PCR product was digested with TaqI enzyme (Thermo Fisher Scientific) for 2 h at 60°C. The digested product was loaded into a 3% agarose gel.
Data access
The raw RNA-sequencing and DNA Methylation data from human generated in this study have been submitted to the European Genome-Phenome Archive (EGA; https://ega-archive.org/) under accession number EGAS00001004583. The RNA-sequencing data from mouse generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA613592.
Supplementary Material
Acknowledgments
We thank the S.C.C. thesis committee, Cynthia Andoniadou, Deborah Cunninghame-Graham, and Donal McLornan, for their continuing advice. We thank the late David Grimwade for helping with the initial discussions about the project; Ismael Ranz Jimenez, Paul Lavender, and Matthew Shannon for helping with the set-up of experiments; Paul Lavender for providing some of the DNA methylation reagents; Adrian Signell and Michael Malim for providing the Jurkat and MT-4 cell lines; Catherine Sawai who kindly provided reagents and equipment; and Prodromos Chatzikyriakou and Marta Reyes-Corral for critical reading of the manuscript. The authors acknowledge the support from the Department of Health via the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy's and St Thomas’ National Health Service (NHS) Foundation Trust in partnership with King's College London for access to high-throughput sequencing and flow cytometry core facilities. This work was supported by Agencia Nacional de Investigación y Desarrollo, ANID DOCTORADO BECAS CHILE/2015—72160152 (PhD studentship to S.C.C.); European Hematology Association (S.C.C., A.M., R.J.O.); King's Health Partners R and D Challenge Fund (Medical Research Council), MC_PC_14105 (R.J.O., C.H.); King's Health Partners RandD Challenge (Medical Research Council) MC_PC_14105 (R.J.O., C.H.), NIHR Biomedical Research Centre at Guy's and St Thomas’ NHS Foundation Trust Genomic Research Award (S.C.C., C.H.); and Medical Research Council grant, MR/M019756/1 (R.S.).
Author contributions: S.C.C. performed the experiments, interpreted the data, and wrote the manuscript with B.M. and R.J.O. A.R. and W.D. supported the mouse experiments. B.M. interpreted the data and wrote the manuscript with S.C.C. and R.J.O. A.M. contributed to the experimental design of the mouse study and provided the mouse line. R.S. advised on the bioinformatics and data analysis. F.v.M. supported the genome-wide sequencing. C.H. provided advice with the clinical study design, facilitated access to the clinic and sample acquisition, provided clinical feedback, and participated in discussions. D.M. wrote the ethical approval and offered clinical advice and feedback. R.J.O. conceived the project, supervised the project, and wrote the manuscript with S.C.C. and B.M. All authors contributed to editing and reviewing the manuscript and approved its publication.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.270066.120.
Competing interest statement
A.M. has received honoraria from Blueprint Medicines, Roche, and Incyte and receives research support from Janssen. D.M. has received speaker fees and is on advisory boards for Jazz pharma, Celgene, Novartis, and Abbvie and has research support from Novartis. C.H. is on advisory boards and has received speaker fees from Novartis, Celgene, CTI, Gilead, Shire, Roche, Jannsen, Promedior, Geron, Galacteo, AOP, and Keros and has research support from Novartis, Celgene, and Constellation. The remaining authors have no conflicts of interests.
References
- Agirre X, Castellano G, Pascual M, Heath S, Kulis M, Segura V, Bergmann A, Esteve A, Merkel A, Raineri E, et al. 2015. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res 25: 478–487. 10.1101/gr.180240.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albajar M, Gutierrez P, Richard C, Rosa-Garrido M, Gómez-Casares MT, Steegmann JL, León J, Delgado MD. 2008. PU.1 expression is restored upon treatment of chronic myeloid leukemia patients. Cancer Lett 270: 328–336. 10.1016/j.canlet.2008.05.024 [DOI] [PubMed] [Google Scholar]
- Antonioli E, Carobbio A, Pieri L, Pancrazzi A, Guglielmelli P, Delaini F, Ponziani V, Bartalucci N, Tozzi L, Bosi A, et al. 2010. Hydroxyurea does not appreciably reduce JAK2 V617F allele burden in patients with polycythemia vera or essential thrombocythemia. Haematologica 95: 1435–1438. 10.3324/haematol.2009.021444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Beau Le MM, Bloomfield CD, Cazzola M, Vardiman JW. 2016. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127: 2391–2405. 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30: 1363–1369. 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. 2014. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods 11: 1138–1140. 10.1038/nmeth.3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair G, Guibert S, Bender A, Weber M. 2014. Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol 15: 545. 10.1186/s13059-014-0545-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbui T, Tefferi A, Vannucchi AM, Passamonti F, Silver RT, Hoffman R, Verstovsek S, Mesa R, Kiladjian JJ, Hehlmann R, et al. 2018. Philadelphia chromosome-negative classical myeloproliferative neoplasms: revised management recommendations from European LeukemiaNet. Leukemia 32: 1057–1069. 10.1038/s41375-018-0077-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Herman JG. 2000. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 16: 168–174. 10.1016/S0168-9525(99)01971-X [DOI] [PubMed] [Google Scholar]
- Bell RE, Golan T, Sheinboim D, Malcov H, Amar D, Salamon A, Liron T, Gelfman S, Gabet Y, Shamir R, et al. 2016. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res 26: 601–611. 10.1101/gr.197194.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilichak A, Kovalchuk I. 2017. The combined bisulfite restriction analysis (COBRA) assay for the analysis of locus-specific changes in methylation patterns. Methods Mol Biol 1456: 63–71. 10.1007/978-1-4899-7708-3_5 [DOI] [PubMed] [Google Scholar]
- Blattler A, Yao L, Witt H, Guo Y, Nicolet CM, Berman BP, Farnham PJ. 2014. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol 15: 469. 10.1186/s13059-014-0469-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotech 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brecqueville M, Rey J, Bertucci F, Coppin E, Finetti P, Carbuccia N, Cervera N, Gelsi-Boyer V, Arnoulet C, Gisserot O, et al. 2012. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer 51: 743–755. 10.1002/gcc.21960 [DOI] [PubMed] [Google Scholar]
- Bruchova H, Borovanova T, Klamova H, Brdicka R. 2002. Gene expression profiling in chronic myeloid leukemia patients treated with hydroxyurea. Leuk Lymphoma 43: 1289–1295. 10.1080/10428190290026358 [DOI] [PubMed] [Google Scholar]
- Challen Ga, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. 2012. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet 44: 23–31. 10.1038/ng.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, Mallaney C, Celik H, Yang L, Xia Z, et al. 2014. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell 15: 350–364. 10.1016/j.stem.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Stockwell PA, Rodger EJ, Duncan EJ, Parry MF, Weeks RJ, Morison IM. 2015. Genome-wide DNA methylation map of human neutrophils reveals widespread inter-individual epigenetic variation. Sci Rep 5: 17328. 10.1038/srep17328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez JS, Rabe JL, Loeffler D, Higa KC, Hernandez G, Mills TS, Ahmed N, Gessner RL, Ke Z, Idler BM, et al. 2021. PU.1 enforces quiescence and limits hematopoietic stem cell expansion during inflammatory stress. J Exp Med 218: e20201169. 10.1084/jem.20201169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A, Ma'ayan A, Ma A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Schneider RK, Breyfogle LJ, Rosen EA, Poveromo L, Elf S, Ko A, Brumme K, Levine R, Ebert BL, et al. 2015. Distinct effects of concomitant Jak2V617F expression and Tet2 loss in mice promote disease progression in myeloproliferative neoplasms. Blood 125: 327–335. 10.1182/blood-2014-04-567024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondrou V, Stavrou EF, Markopoulos G, Kouraklis-Symeonidis A, Fotopoulos V, Symeonidis A, Vlachaki E, Chalkia P, Patrinos GP, Papachatzopoulou A, et al. 2018. Impact of ZBTB7A hypomethylation and expression patterns on treatment response to hydroxyurea. Hum Genomics 12: 45. 10.1186/s40246-018-0177-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlet T, Argüeso Lleida A, Al Adhami H, Dumas M, Bender A, Ngondo RP, Tanguy M, Vallet J, Auclair G, Bardet AF, et al. 2020. Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity. Nat Commun 11: 3153. 10.1038/s41467-020-16919-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. 2008. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24: 1547–1548. 10.1093/bioinformatics/btn224 [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. 2005. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21: 3439–3440. 10.1093/bioinformatics/bti525 [DOI] [PubMed] [Google Scholar]
- Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, et al. 2000. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 92: 564–569. 10.1093/jnci/92.7.564 [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. 2011. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145: 423–434. 10.1016/j.cell.2011.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CN, Bareford D, Butt N, Campbell P, Conneally E, Drummond M, Erber W, Everington T, Green AR, Hall GW, et al. 2010. Guideline for investigation and management of adults and children presenting with a thrombocytosis. Br J Haematol 149: 352–375. 10.1111/j.1365-2141.2010.08122.x [DOI] [PubMed] [Google Scholar]
- Heit B, Tavener S, Raharjo E, Kubes P. 2002. An intracellular signaling hierarchy determines direction of migration in opposing chemotactic gradients. J Cell Biol 159: 91–102. 10.1083/jcb.200202114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CPE, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, Van Dijk CM, et al. 2012. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 22: 271–282. 10.1101/gr.117523.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC, et al. 2016. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev Cell 36: 9–23. 10.1016/j.devcel.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. 2009. Cpg islands—“A rough guide”. FEBS Lett 583: 1713–1720. 10.1016/j.febslet.2009.04.012 [DOI] [PubMed] [Google Scholar]
- Imrichová H, Hulselmans G, Atak ZK, Potier D, Aerts S. 2015. i-cisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res 43: W57–W64. 10.1093/nar/gkv395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JRR, Bonni A, Roberts TM, Shi Y. 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128: 1077–1088. 10.1016/j.cell.2007.02.017 [DOI] [PubMed] [Google Scholar]
- Jeong M, Park HJ, Celik H, Ostrander EL, Reyes JM, Guzman A, Rodriguez B, Lei Y, Lee Y, Ding L, et al. 2018. Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep 23: 1–10. 10.1016/j.celrep.2018.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. 2013. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51: 310–325. 10.1016/j.molcel.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmieder S, Mughal TI, Hasselbalch HC, Barosi G, Valent P, Kiladjian JJ, Jeryczynski G, Gisslinger H, Jutzi JS, Pahl HL, et al. 2016. Myeloproliferative neoplasms and inflammation: whether to target the malignant clone or the inflammatory process or both. Leukemia 30: 1018–1024. 10.1038/leu.2016.12 [DOI] [PubMed] [Google Scholar]
- Krakoff IH, Brown NC, Reichard P. 1968. Inhibition of ribonucleoside diphosphate reductase by hydroxyurea. Cancer Res 28: 1559–1565. [PubMed] [Google Scholar]
- Kubovcakova L, Lundberg P, Grisouard J, Hao-Shen H, Romanet V, Andraos R, Murakami M, Dirnhofer S, Wagner KUU, Radimerski T, et al. 2013. Differential effects of hydroxyurea and INC424 on mutant allele burden and myeloproliferative phenotype in a JAK2-V617F polycythemia vera mouse model. Blood 121: 1188–1199. 10.1182/blood-2012-03-415646 [DOI] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MTY, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al. 2013. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498: 511–515. 10.1038/nature12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM. 2004. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev Biol 268: 403–415. 10.1016/j.ydbio.2003.12.031 [DOI] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28: 882–883. 10.1093/bioinformatics/bts034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. 2013. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498: 516–520. 10.1038/nature12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Huang LJS, Lodish HF. 2008. Dimerization by a cytokine receptor is necessary for constitutive activation of JAK2V617F. J Biol Chem 283: 5258–5266. 10.1074/jbc.M707125200 [DOI] [PubMed] [Google Scholar]
- Lussana F, Rambaldi A. 2017. Inflammation and myeloproliferative neoplasms. J Autoimmun 85: 58–63. 10.1016/j.jaut.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Mahé EA, Madigou T, Sérandour AA, Bizot M, Avner S, Chalmel F, Palierne G, Métivier R, Salbert G. 2017. Cytosine modifications modulate the chromatin architecture of transcriptional enhancers. Genome Res 27: 947–958. 10.1101/gr.211466.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann IK, Chatterjee R, Zhao J, He X, Weirauch MT, Hughes TR, Vinson C. 2013. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res 23: 988–997. 10.1101/gr.146654.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin Oyarzún CP, Carestia A, Lev PR, Glembotsky AC, Castro Ríos MA, Moiraghi B, Molinas FC, Marta RF, Schattner M, Heller PG. 2016. Neutrophil extracellular trap formation and circulating nucleosomes in patients with chronic myeloproliferative neoplasms. Sci Rep 6: 38738. 10.1038/srep38738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupetit-Mehouas S, Court F, Bourgne C, Guerci-Bresler A, Cony-Makhoul P, Johnson H, Etienne G, Rousselot P, Guyotat D, Janel A, et al. 2018. DNA methylation profiling reveals a pathological signature that contributes to transcriptional defects of CD34+CD15− cells in early chronic-phase chronic myeloid leukemia. Mol Oncol 12: 814–829. 10.1002/1878-0261.12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon B, Stein BL. 2013. Thrombotic and bleeding complications in classical myeloproliferative neoplasms. Semin Thromb Hemost 39: 101–111. 10.1055/s-0032-1331153 [DOI] [PubMed] [Google Scholar]
- McMullin MF, Harrison CN, Ali S, Cargo C, Chen F, Ewing J, Garg M, Godfrey A, Steven Knapper S, McLornan DP, et al. 2019. A guideline for the diagnosis and management of polycythaemia vera. A British Society for Haematology Guideline. Br J Haematol 184: 176–191. 10.1111/bjh.15648 [DOI] [PubMed] [Google Scholar]
- Mesa RA, Jamieson C, Bhatia R, Deininger MW, Fletcher CD, Gerds AT, Gojo I, Gotlib J, Gundabolu K, Hobbs G, et al. 2017. Myeloproliferative neoplasms, version 2.2018 featured updates to the NCCN guidelines. JNCCN J Natl Compr Cancer Netw 15: 1193–1207. 10.6004/jnccn.2017.0157 [DOI] [PubMed] [Google Scholar]
- Mullally A, Lane SW, Ball B, Megerdichian C, Okabe R, Al-Shahrour F, Paktinat M, Haydu JE, Housman E, Lord AM, et al. 2010. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 17: 584–596. 10.1016/j.ccr.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nischal S, Bhattacharyya S, Christopeit M, Yu Y, Zhou L, Bhagat TD, Sohal D, Will B, Mo Y, Suzuki M, et al. 2013. Methylome profiling reveals distinct alterations in phenotypic and mutational subgroups of myeloproliferative neoplasms. Cancer Res 73: 1076–1085. 10.1158/0008-5472.CAN-12-0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H, Akashi K, Moreau-Gachelin F, Li Y, Zhang P, et al. 2005. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol 25: 2832–2845. 10.1128/MCB.25.7.2832-2845.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. 2013. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14: 293. 10.1186/1471-2164-14-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Platt OS, Orkin SH, Dover G, Beardsley GP, Miller B, Nathan DG. 1984. Hydroxyurea enhances fetal hemoglobin production in sickle cell anemia. J Clin Invest 74: 652–656. 10.1172/JCI111464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q, Liu P, Zhao X, Zhang C, Li D, Zhang R, Ren R. 2015. IRF8 regulates cell cycle of hematopoietic stem cells. Blood 126: 2353. 10.1182/blood.V126.23.2353.2353 [DOI] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman SM, Janssen RC, Choudhury M, Baquero KC, Aikens RM, De La Houssaye BA, Friedman JE. 2012. CCAAT/enhancer-binding protein β (C/EBPβ) expression regulates dietary-induced inflammation in macrophages and adipose tissue in mice. J Biol Chem 287: 34349–34360. 10.1074/jbc.M112.410613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Rio-Machin A, Vulliamy T, Hug N, Walne A, Tawana K, Cardoso S, Ellison A, Pontikos N, Wang J, Tummala H, et al. 2020. The complex genetic landscape of familial MDS and AML reveals pathogenic germline variants. Nat Commun 11: 1044. 10.1038/s41467-020-14829-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnerblad M, Andersson R, Olofsson T, Douagi I, Karimi M, Lehmann S, Hoof I, De Hoon M, Itoh M, Nagao-Sato S, et al. 2014. Analysis of the DNA methylome and transcriptome in granulopoiesis reveals timed changes and dynamic enhancer methylation. Blood 123: e79–e89. 10.1182/blood-2013-02-482893 [DOI] [PubMed] [Google Scholar]
- Ruf W. 2016. Hemostasis keeps the stem cell niche in order. Blood 128: 1027–1029. 10.1182/blood-2016-07-722421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J, Heyn HA, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. 2011. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6: 692–702. 10.4161/epi.6.6.16196 [DOI] [PubMed] [Google Scholar]
- Sproul D, Kitchen RR, Nestor CE, Dixon JM, Sims AH, Harrison DJ, Ramsahoye BH, Meehan RR. 2012. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome Biol 13: R84. 10.1186/gb-2012-13-10-r84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegelmann F, Bullinger L, Schlenk RF, Paschka P, Griesshammer M, Blersch C, Kuhn S, Schauer S, Döhner H, Döhner K. 2011. DNMT3A mutations in myeloproliferative neoplasms. Leukemia 25: 1217–1219. 10.1038/leu.2011.77 [DOI] [PubMed] [Google Scholar]
- Stone RM, Hultman C, Wolach O, Cherpokova D, Iliadou B, Ebert BL, Adams D, Wadleigh M, Galinsky I, Martinod K, et al. 2018. Increased neutrophil extracellular trap formation promotes thrombosis in myeloproliferative neoplasms. Sci Transl Med 10: eaan8292. 10.1126/scitranslmed.aan8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Patel KP, Kantarjian H, Luthra R, Pierce S, Cortes J, Verstovsek S. 2013. JAK2 p.V617F detection and allele burden measurement in peripheral blood and bone marrow aspirates in patients with myeloproliferative neoplasms. Blood 122: 3784–3786. 10.1182/blood-2013-07-515676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Pardanani A, Lim KHH, Abdel-Wahab O, Lasho TL, Patel J, Gangat N, Finke CM, Schwager S, Mullally A, et al. 2009. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia 23: 905–911. 10.1038/leu.2009.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AL, Steward S, Howard Ta, Mortier N, Smeltzer M, Wang Y, Ware RE. 2011. Epigenetic and molecular profiles of erythroid cells after hydroxyurea treatment in sickle cell anemia. Blood 118: 5664–5670. 10.1182/blood-2011-07-368746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Schwemmers S, Hexner EO, Pahl HL. 2010. AML1 is overexpressed in patients with myeloproliferative neoplasms and mediates JAK2V617F-independent overexpression of NF-E2. Blood 116: 254–266. 10.1182/blood-2009-11-254664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Motoda L, Satake M, Ito Y, Taniuchi I, Tergaonkar V, Osato M. 2013. Runx3 deficiency results in myeloproliferative disorder in aged mice. Blood 122: 562–566. 10.1182/blood-2012-10-460618 [DOI] [PubMed] [Google Scholar]
- Wang X, Yu X, Zhu W, Mccombie WR, Antoniou E, Powers RS, Davidson NO, Li E, Williams J. 2015. A trimming-and-retrieving alignment scheme for reduced representation bisulfite sequencing. Bioinformatics 31: 2040–2042. 10.1093/bioinformatics/btv089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466. 10.1038/ng1990 [DOI] [PubMed] [Google Scholar]
- Xu Z, Niu L, Li L, Taylor JA. 2015. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res 44: e20. 10.1093/nar/gkv907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young CW, Hodas S. 1964. Hydroxyurea: inhibitory effect on DNA metabolism. Science 146: 1172–1174. 10.1126/science.146.3648.1172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.