Abstract
There are more than 55,000 variable number tandem repeats (VNTRs) in the human genome, notable for both their striking polymorphism and mutability. Despite their role in human evolution and genomic variation, they have yet to be studied collectively and in detail, partially owing to their large size, variability, and predominant location in noncoding regions. Here, we examine 467 VNTRs that are human-specific expansions, unique to one location in the genome, and not associated with retrotransposons. We leverage publicly available long-read genomes, including from the Human Genome Structural Variant Consortium, to ascertain the exact nucleotide composition of these VNTRs and compare their composition of alleles. We then confirm repeat unit composition in more than 3000 short-read samples from the 1000 Genomes Project. Our analysis reveals that these VNTRs contain highly structured repeat motif organization, modified by frequent deletion and duplication events. Although overall VNTR compositions tend to remain similar between 1000 Genomes Project superpopulations, we describe a notable exception with substantial differences in repeat composition (in PCBP3), as well as several VNTRs that are significantly different in length between superpopulations (in ART1, PROP1, DYNC2I1, and LOC102723906). We also observe that most of these VNTRs are expanded in archaic human genomes, yet remain stable in length between single generations. Collectively, our findings indicate that repeat motif variability, repeat composition, and repeat length are all informative modalities to consider when characterizing VNTRs and their contribution to genomic variation.
There are tens of thousands of variable number tandem repeats (VNTRs) in the human genome (Näslund et al. 2005), yet as a whole they remain uncharacterized. These VNTRs—that is, repeats with a repeat unit of seven base pairs (bp) or greater—are often too large or variable to be effectively captured using the short-read sequencing technologies typically used for whole-genome sequencing. In addition, they are frequently located in noncoding or intergenic regions, which until recently have garnered less attention than genomic variants in coding regions. VNTRs, however, are highly mutable, suggesting that they play influential roles in evolutionary biology, and along with short tandem repeats (STRs; repeats with a repeat unit of <7 bp), are a major source of human genetic diversity (Jeffreys et al. 1985; Berg et al. 2010; Hannan 2018). The recent advent of long-read sequencing has revealed that many VNTRs are much larger than the reference human genome suggests, and far more polymorphic. The few VNTRs that have been studied recently in more detail have provided insights into evolutionary history, replication mechanism, population structure, and disease. Characterizing more VNTRs at this higher resolution will continue to expand these insights.
Four VNTRs have recently been studied in detail, primarily owing to their involvement in disease. A 25-bp VNTR in the intron of ATP binding cassette subfamily A member 7 (ABCA7) influences alternative splicing and is associated with Alzheimer's disease (De Roeck et al. 2018). A 30-bp VNTR in calcium voltage-gated channel subunit alpha1 C (CACNA1C) shows varying repeat unit arrays correlated with “protective” or “risk” alleles in schizophrenia and bipolar disorder (Song et al. 2018). A 33-bp VNTR in the promoter of tribbles pseudokinase 3 (TRIB3) is associated with TRIB3 expression, and copy number of the repeat is correlated with certain disease-associated single-nucleotide polymorphisms (SNPs) (Örd et al. 2020). We also identified a 69-bp repeat in WD repeat domain 7 (WDR7) associated with ALS (Course et al. 2020). A closer look at long-read sequenced genomes from geographically diverse samples indicated that this particular repeat expands via duplication events and a replication error called template switching. Furthermore, a small number of repeat units were unique to certain superpopulations and were also found in short-read data sets of ancient genomes. While examining this VNTR along with several others, we recognized that performing a similar analysis on a larger number of VNTRs could illuminate how these repeats vary and the mutational processes that have shaped them.
As these examples show, the VNTRs studied in detail have so far been studied one at a time and for a particular reason, like association with disease. Instead of continuing to study these repeats one-by-one, we decided to study a subset of them methodically and as a group. Doing so could answer questions about these VNTRs as a category of genomic variant, like their general characteristics as well as timing and patterns of expansion. Here, we look at a set of 467 VNTRs chosen for the following characteristics: they show human-specific expansion, they are not associated with retrotransposons, and they are unique to one location in the genome. These parameters select the VNTRs most likely to have expanded recently—and that may still be expanding—so we can observe their changes in different populations more readily. Furthermore, expansion of the same genomic segment in multiple places in the genome would be unlikely unless there were a retrotransposon driving it, so these parameters select for VNTRs that have expanded via other mechanisms. We then assess these or a subset of these VNTRs in ancestral human genomes as well as modern human genomes from the 1000 Genomes Project. We observe the similarities and differences of VNTRs in various superpopulations and their timescale of expansion. Finally, by taking a closer look at these genomes in long-read sequenced samples, we define several modalities of internal nucleotide pattering, which provides a useful framework for future VNTR analysis.
Results
Identifying VNTRs that are unique, non-retrotransposon associated, human-specific expansions
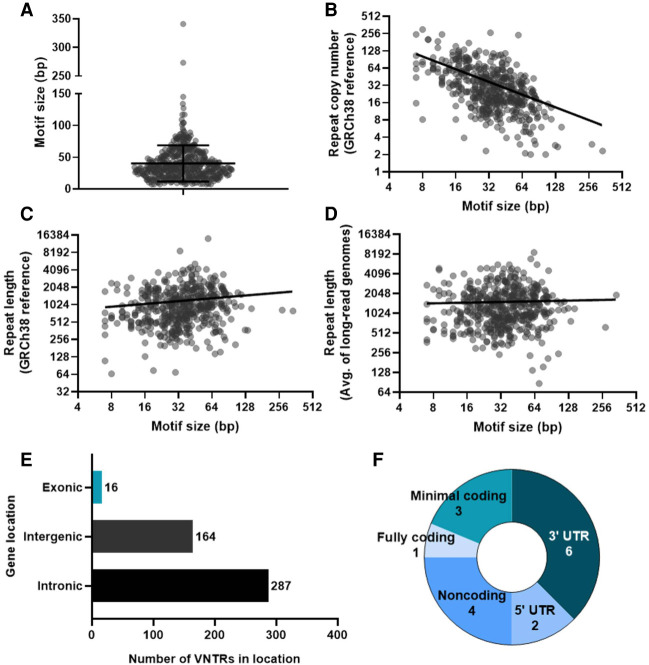
To generate a list of VNTRs of interest, we started with 1584 VNTRs that were recently described as having human-specific expansions (Sulovari et al. 2019). This list included both repeat expansions that arose ab initio (meaning they are only expanded in humans) and repeat expansions that are more expanded in humans than other primates. We then excluded VNTRs that were part of repetitive elements, like SINE-VNTR-Alu (SVA) repeats, and any repeats that were not unique to one location in the human genome (Supplemental Table S1). These parameters allowed us to generate a list of 467 unique VNTRs with human-specific expansions that were unlikely to have arisen because of a retrotransposon. These VNTRs ranged in repeat unit size from 7 to 341 bp (mean = 40.1 ± 28.6 bp; median = 34 bp) (Fig. 1A). Average repeat copy number in the GRCh38 human reference genome ranged from 2 to 300.5 copies (mean = 39.8 ± 40.4; median = 26.9). Comparing repeat motif size versus repeat copy number revealed an inverse correlation between motif size and copy number, fitting a log-log pattern of nonlinear regression (log-log slope = −0.74) (Fig. 1B). Accordingly, average total repeat length in the GRCh38 human reference genome, which ranged from 65 to 14,168 bp (mean = 1188 ± 1078 bp; median = 945 bp) was not correlated with motif size (log-log slope = 0.16) (Fig. 1C). This pattern was recapitulated in a separate data set covering eight geographically diverse genomes obtained through Pacific Biosciences (PacBio) small molecule, real-time (SMRT) long-read sequencing (Audano et al. 2019), in which the average total length of the repeats ranged from 86 to 8624 bp (mean = 1519 ± 1099 bp, median = 1230 bp) and showed no correlation between repeat motif size and total length (log-log slope = 0.034) (Fig. 1D). Although these lengths were obtained through disparate sequencing methods, their similar means and standard deviations, as well as their similar nonlinear regression statistics, are concordant and indicate that both types of sequencing are useful for analyzing VNTR expansions.
Figure 1.
Characterization of 467 human-specific VNTR expansions assessed. (A) Violin plot of motif size for each VNTR. Lines show mean (40.1 bp) and standard deviation (±28.6 bp). (B) Scatter plot of motif size versus repeat copy number in the GRCh38 human reference genome. Axes are log2 and line is log-log (best-fit slope = −0.74). (C) Scatter plot of motif size versus total repeat length in the GRCh38 human reference genome. Axes are log2 and line is log-log (best-fit slope = 0.16). (D) Scatter plot of motif size versus average total repeat length in eight SMRT long-read sequenced genomes representing different superpopulations. Axes are log2 and line is log-log (best-fit slope = 0.034). (E) Bar chart summarizing overall VNTR locations in the genome. (F) Pie chart breaking down specific locations of VNTRs in exons.
As for location in the genome, the vast majority of these VNTRs were in noncoding regions, with 287 intronic and 164 intergenic (Fig. 1E). Sixteen VNTRs were exonic, with four of these in noncoding exons, two in 5′ UTRs, and six in 3′ UTRs. Three overlapped coding regions by only 2, 5, or 15 bp out of a several hundred base pair sequence (termed “minimal coding”), leaving only one VNTR that was fully contained within a coding region (Fig. 1E,F). This 60-bp VNTR is divisible by three base pairs and resides in the gene MUC1, where it has previously been shown to play a role in medullary cystic kidney disease type 1 and other renal phenotypes (Kirby et al. 2013; Mukamel et al. 2021).
Most human-specific VNTR expansions are also expanded in ancient genomes
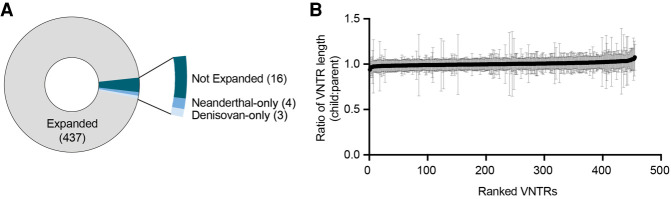
We assessed the lengths of these 467 VNTRs in the short-read sequenced genomes of ancestral humans—specifically, an Altai Neanderthal genome (Prüfer et al. 2014) and a Denisovan genome (Fig. 2A; Meyer et al. 2012). Of the 460 VNTRs that we successfully assessed, we found that only 16 VNTRs were not expanded in either genome, four were not expanded only in the Neanderthal genome, and three were not expanded only in the Denisovan genome (Fig. 2A). The number of expanded repeats may even be an underestimate because of low coverage (especially of AT-rich sequences) and short-read length in ancient genomes. Overall, we can conclude that the overwhelming majority of these 460 human-specific VNTR expansions are also expanded in ancient human genomes.
Figure 2.
Timing of expansion in 467 human-specific VNTRs. (A) Pie chart showing number of VNTRs expanded or not expanded in Neanderthal and/or Denisovan genomes. In total, 460 VNTRs were successfully assessed. (B) XY plot showing mean (black dots) and standard deviation (gray lines) ratio of child VNTR length to average parent VNTR length. Data are from 585 trios from the 1000 Genomes Project, and 455 VNTRs were successfully assessed and are ranked by mean ratio on the x-axis.
Human-specific VNTR expansions do not show intergenerational expansion
To estimate the rate of VNTR expansion in modern humans, we observed these VNTRs in short-read sequenced trio data sets from the 1000 Genomes Project (The 1000 Genomes Project Consortium 2015). Using 585 trios, we looked for events of expansion that occurred in one generation by comparing average repeat lengths in children versus parents. Ultimately, we observed that in virtually all of the 455 VNTRs successfully assessed, the average of the parents’ repeat lengths and the child's repeat length remained the same (meaning the ratio between the two stayed at or near one). The lack of any obvious change in copy number between generations indicates that expansion in a single generation is rare for these VNTRs and therefore they expand over a longer timescale (Fig. 2B).
Internal sequence variation of human-specific VNTR expansions can be divided into three categories
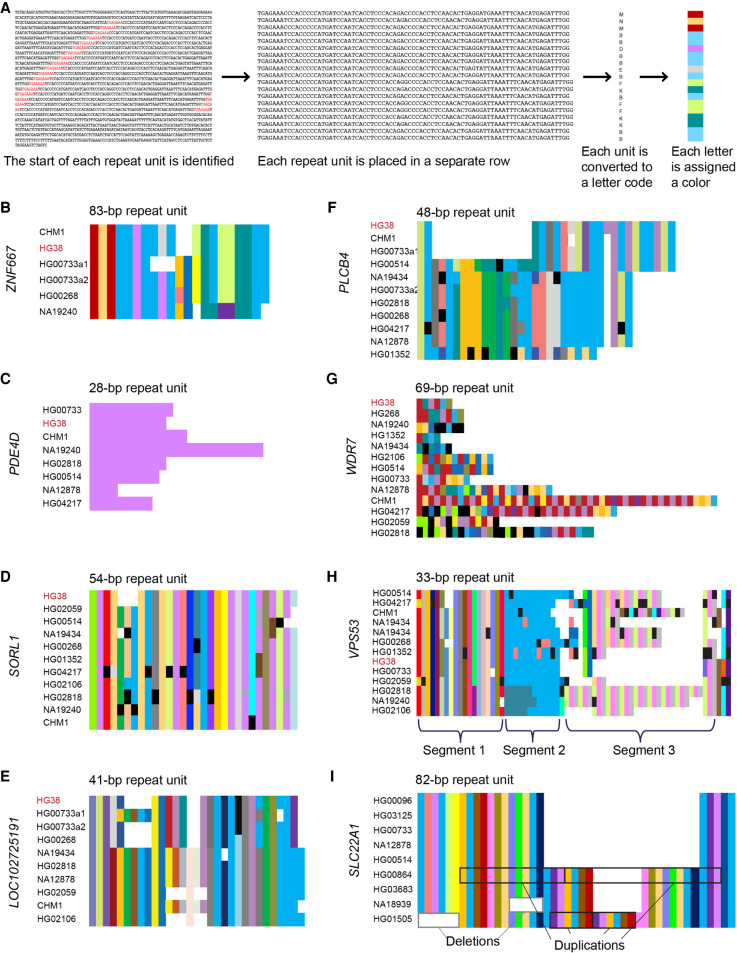
To assess the internal nucleotide patterns in these VNTRs, we evaluated their repeat unit composition using existing long-read SMRT-sequenced data sets in a subset of 53 VNTRs. These VNTRs were chosen for having the greatest standard deviation in length in the original data set (Sulovari et al. 2019), with the prediction that they would be the most likely to show a variety of different alleles across the genomes assessed. We extracted genomic sequences of the 53 VNTRs from 15 individuals who had undergone whole-genome long-read SMRT sequencing, representing the five superpopulations from the 1000 Genomes Project (African, Admixed American, East Asian, European, and South Asian) (Audano et al. 2019). Reads were divided into individual repeat units based on their nucleotide composition, beginning with the most common repeat, extracted from the simple tandem repeats finder track (Benson 1999) from the UCSC Genome Browser (Kent et al. 2002). Each repeat was assigned a single letter code and then aligned. We then color-coded the varying repeat units to visualize their overall repeat pattern (Fig. 3A).
Figure 3.
Composition plots illustrating modes of variability in VNTRs. (A) Schematic overview of how composition plots are generated from long-read sequencing. Example is from the CHM1 genome for the VNTR in ZNF667. (B–H) Composition plots showing varying patterns in example VNTRs: (B) ZNF667, (C) PDE4D, (D) SORL1, (E) LOC102725191, (F) PLCB4, (G) WDR7, (H) VPS53. At the left of each plot are listed the genomes from which the allele has been obtained, which were previously sequenced and published, and which represent geographically diverse populations (Audano et al. 2019). Different alleles from the same individual are denoted with “a1” and “a2.” At the top of each plot is the motif length given by Tandem Repeats Finder, which can vary in length by one or more bases depending on insertions or deletions in each motif. Black segments in the plots denote motifs with private variants. (I) Composition plot for the VNTR in SLC22A1, with examples of duplication and deletion boxed in black and gray, respectively. The genomes used for this plot were previously sequenced and published (Ebert et al. 2021).
From this visualization, we identified that there are three chief modes of variability to consider when categorizing VNTR internal nucleotide patterns. Defining a motif as the sequence of one repeat unit, alleles as a series of motifs, and allele groupings as groups of similarly patterned alleles, we observed the variability in length between alleles, variability in sequence between motifs, and variability in motif organization within groups of alleles. In the 53 VNTRs assessed, we saw that the majority contained several repeat motifs, some variability in length between alleles, and little variability in sequence between alleles. A good representation of this common pattern is found in a VNTR with an 84-bp repeat in ZNF667 (Fig. 3B). Some VNTRs showed far less variability in motifs, exemplified here by a 28-bp VNTR in PDE4D (Fig. 3C), which is “pure” or “uninterrupted,” although variable in allele length. Alternatively, in some VNTRs, most or even all repeat motifs were different from the one previous, such as in a 54-bp VNTR in SORL1 (Fig. 3D); yet, despite its varied motifs, this and many similar VNTRs show largely the same length and sequence between alleles. In another example, a 41-bp VNTR in LOC102725191 (Fig. 3E), we observed the same pattern of variability in repeat motifs, but this time observed two distinct groups of alleles. Taking this pattern a step further, a 48-bp VNTR in PLCB4 (Fig. 3F) shows about three distinct groups of alleles, which each vary in length, as well.
We also observed a few rare VNTRs that were highly variable in all three modalities: motif, length, and allele sequence, like the previously identified 69-bp repeat found in WDR7 (Fig. 3G; Course et al. 2020). For some VNTRs, we even observed multiple patterns of composition within alleles. This scenario is best exemplified by a 33-bp repeat in VPS53 (Fig. 3H) with a fixed length segment containing a variable internal sequence, followed by a variable length segment with fixed internal sequence, and finally a third variable length segment that repeats in groups of three motifs (Fig. 3H). Overall, we observed that most length and allelic differences were derived from duplications and deletions, as highlighted in an 82-bp repeat in SLC22A1 (long-read genomes used to analyze SLC22A1 were obtained from Ebert et al. 2021) (Fig. 3I), rather than unit-by-unit changes. Detailed motif information for each of these repeats is available in Supplemental Table S2.
VNTR motifs in PCBP3 are patterned across modern superpopulations
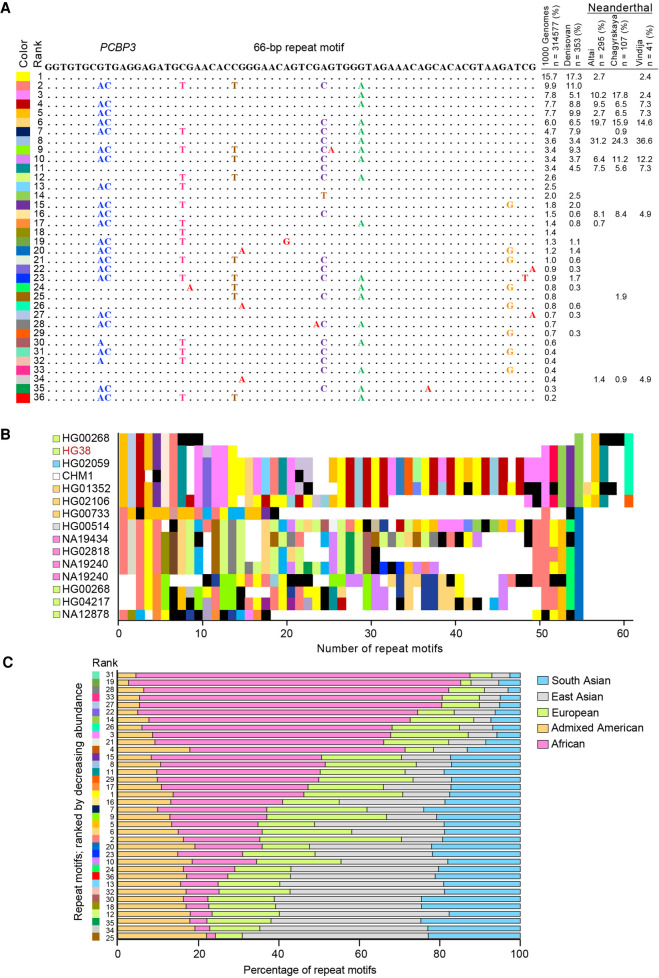
We also looked at the distribution of the repeat units in the most variable VNTRs using the 1000 Genomes Project to analyze population differences in repeat motif distribution in 25 geographically distinct modern populations (2504 individuals). For the most part, there were no notable differences in repeat motif distribution in these VNTRs. One exception, however, was a 66-bp repeat in intron 11 of poly(rC) binding protein 3 (PCBP3), which was variable in all three parameters: motifs, allele length, and allele sequence composition. A total of 8 bp of the 66 bp in the repeat unit were variable, and these variable positions result in 38 total primary repeat motifs (Fig. 4A). A number of these repeat motifs were also observed in one Denisovan (Meyer et al. 2012) and three Neanderthal genomes (Prüfer et al. 2014, 2017; Mafessoni et al. 2020). The repeat motif sequence varied between alleles, although what similarities existed partially clustered by superpopulation (Fig. 4B). The frequency of these repeat motifs differed across superpopulations, with a particularly noticeable inverse relationship between repeat motifs abundant in the African superpopulation versus the East Asian superpopulation (Fig. 4C).
Figure 4.
A VNTR in PCBP3 shows repeat motif differences in modern superpopulations. (A) PCBP3 repeat motifs with variable positions highlighted. At left is the assigned color code for each motif. At right is the relative abundance of each motif in the 1000 Genomes Project and in ancient genomes. (B) Composition plot of the PCBP3 VNTR in geographically diverse populations. At the left are listed the genomes from which the allele has been obtained, which were previously sequenced and published (Audano et al. 2019). Black segments in the plot denote motifs with private variants. (C) Cumulative frequency of repeat motifs in PCBP3 across superpopulations. Repeat motifs are ordered by decreasing abundance in the African superpopulation, and numbers on the y-axis correspond to their global ranked abundance.
Some human-specific VNTR expansion lengths are significantly different across modern superpopulations
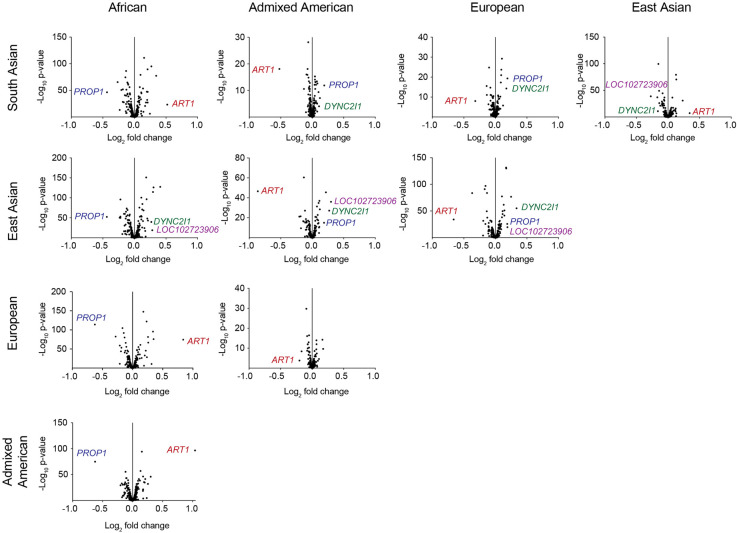
We then used the same 1000 Genomes Project data set to analyze the differences in length of all 467 VNTRs. We calculated the length of each repeat in these samples, merging combined reads for each of the five superpopulations. After estimating the average length of the VNTRs across these populations, we generated volcano plots to determine which VNTRs had significant length differences between superpopulations (Fig. 5). We observed that some superpopulations had more similar average VNTR lengths than others. For example, European and Admixed American populations showed strong concordance between repeat length, which corroborates previous population clustering determined by SNPs (The 1000 Genomes Project Consortium 2015; Glusman et al. 2017; Li et al. 2017), and South Asian and East Asian populations had only one VNTR that was a notable outlier. From these volcano plots, we chose the top four differentially expanded VNTRs (by both P-value and log2-fold change) to observe further, in: LOC102723906, PROP paired-like homeobox 1 (PROP1), ADP-ribosyltransferase 1 (ART1), and dynein 2 intermediate chain 1 (DYNC2I1).
Figure 5.
Comparing VNTR lengths across modern superpopulations. Volcano plots showing pair-wise comparisons of average VNTR lengths between superpopulations from the 1000 Genomes Project. The VNTRs with the greatest length differences (as determined by DESeq2) are labeled by the nearest gene, or gene in which they reside, and were determined based on both log2-fold change and P-value. Trial size for each superpopulation is 347–660 individuals.
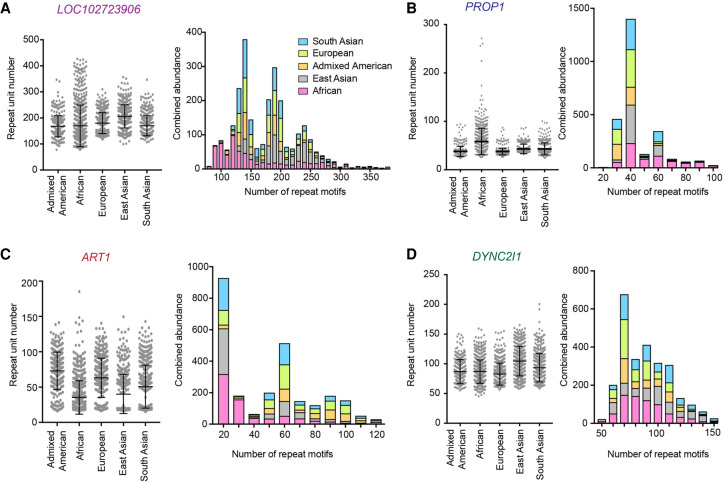
We compared both the pattern of individual data points as well as the cumulative abundance of the repeat lengths in all superpopulations (Fig. 6A−D). One-way ANOVAs to compare each population for each VNTR all gave P < 0.0001, and subsequent Tukey's multiple comparison tests corroborated the significant differences we observed in Figure 6 using DESeq2. Comparing these distributions, we saw that the VNTR in LOC102723906 is longer in East Asian than European and African populations, although samples of African ancestry show a larger range of allele lengths (Fig. 6A). The cumulative plot mirrors the right-tailed distribution observed in African samples, and reveals an unusual “trimodal distribution” in the remaining superpopulations (Fig. 6A). The VNTR in PROP1 shows longer repeats in the African superpopulation than in any other (Fig. 6B), whereas the VNTR in ART1 shows the opposite: fewer long repeats appearing in the African superpopulation compared to the other populations (Fig. 6C). The ART1 cumulative plot shows an overall right-tailed distribution of repeat copy number. In addition, the VNTR in ART1 shows a distinct gap in values in some of the superpopulations, akin to a bimodal distribution (Fig. 6C). Finally, the VNTR in DYNC2I1 shows a more normal distribution across the superpopulations, with longer repeats in the East Asian superpopulation (Fig. 6D).
Figure 6.
Length differences in the top four differentially expanded VNTRs in modern superpopulations. (A–D) Individual VNTR copy numbers plus mean and standard deviation for each superpopulation (left) and cumulative abundance binned into groups of 10 repeat motifs (right) for VNTRs in LOC102723906 (A), PROP1 (B), ART1 (C), and DYNC2I1 (D). Trial size for each superpopulation is 347–660. One-way ANOVAs gave P < 0.0001 for each comparison of superpopulations for each VNTR.
Differentially expanded human-specific VNTRs expanded at different time points
We also determined the evolutionary timing of these four VNTR expansions by observing them in the reference genomes of non-human primates. We found that they did not all initially expand around the same time. Instead, the VNTR in DYNC2I1 expanded between the branchpoint of New World monkeys and gibbons (the macaque genome has a portion of the sequence present), the VNTR in LOC102723906 expanded between gibbons and orangutans, and the VNTRs in PROP1 and ART1 expanded between the branching of chimpanzees and hominins. All of these VNTRs are expanded in the Denisovan and Neanderthal genomes.
Composition of differentially expanded human-specific VNTRs explains findings in short-read sequencing
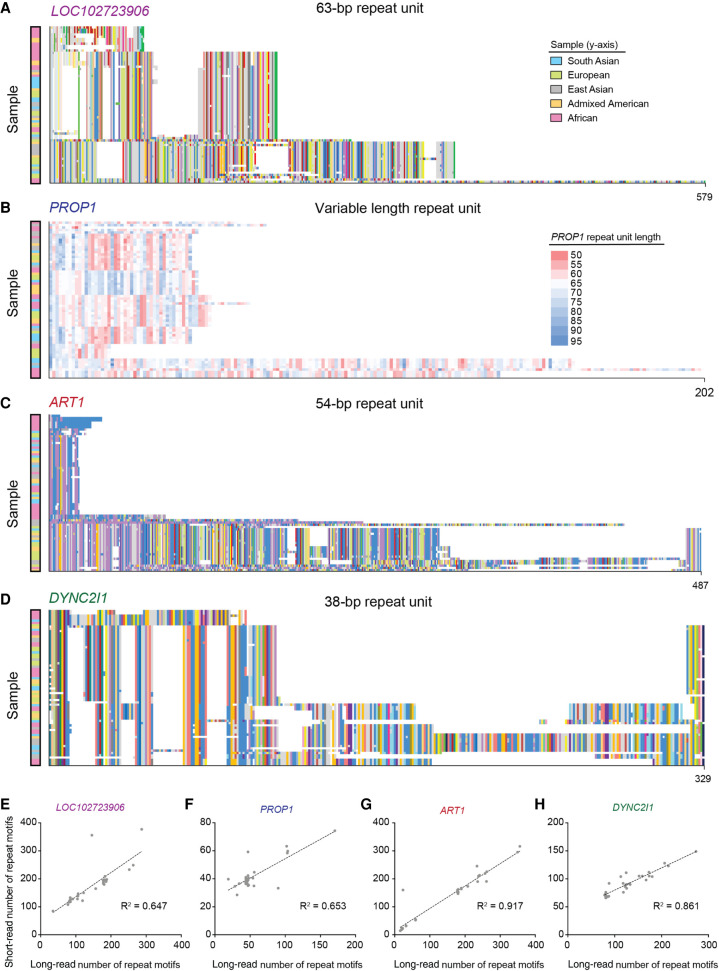
We then visualized the pattern of repeat motifs for these VNTRs as described in Figure 3A, this time using the 32 available PacBio HiFi SMRT-sequenced genomes available through the Human Genome Structural Variant Consortium (HGSVC) (Ebert et al. 2021), ultimately providing up to 64 alleles. This analysis revealed that the unusual trimodal distribution observed in the cumulative plot for the VNTR in LOC102723906 is explained by the three predominant groups of alleles of three differing sizes (averaging 58, 109, and 253 repeat units) found by SMRT sequencing (Fig. 7A). Long-read sequencing also revealed a highly unusual internal structure in PROP1, in which each repeat motif itself contains a TG-stretch that can vary widely in length (between 7 and 26 repeat copies) like an STR within a VNTR (Fig. 7B). This TG-stretch is likely the main cause of its length variability. Long-read sequencing also explained the distinct gap in values observed in ART1, showing that there are three predominant groups of alleles, two of which are similarly short (25 and 41 average repeat units), and one of which is quite long (381 average repeat units) (Fig. 7C). Larger expansions appear to occur as duplication events in blocks of 45–50 repeat units. Finally, the even distribution of lengths in DYNC2I1 is echoed by a varied distribution of alleles in long-read sequencing (Fig. 7D), not to mention more than 100 different motifs. All of these four VNTRs have markedly variable internal nucleotide sequence, a couple of major groups of alleles, and except for the VNTR in PROP1, their major length variation is caused by apparent deletion or duplication events. It is worth noting that these VNTRs tend to be longer and more variable than other VNTRs in this data set, which may be related to the fact that they are also the most different in length between superpopulations. Finally, comparison of the copy number of these VNTRs determined by short-read versus long-read sequencing reveals reasonable to strong concordance (Fig. 7E−H).
Figure 7.
Composition plots and copy number of the top four differentially expanded VNTRs in modern superpopulations. Composition plots for VNTRs in LOC102723906 (A), PROP1 (B), ART1 (C), and DYNC2I1 (D). The colors at the left of the plot denote the superpopulation from which the alleles were obtained (see key), which were previously sequenced and published (Ebert et al. 2021). Gray segments in the plot denote motifs that are rare or private. The y-axis shows the length of the repeat in number of repeat motifs. The heat map legend in B denotes the length of each repeat found in the PROP1 VNTR, which has been plotted based on this unique feature, instead of the motif structure used for the other VNTRs. (E–H) Plots comparing average number of repeat motifs estimated from short-read data and average number of repeat motifs (from both alleles per individual) from phased long-read genomes are given for the same VNTRs, along with their R2 values.
Discussion
In this study, we observed the patterns, timing of expansion, and geographical distribution of 467 VNTRs that are unique, non-retrotransposon-associated, and expanded specifically in humans. These particular characteristics were chosen to enrich for VNTRs that were likely to have expanded recently and via mechanisms other than association with repetitive elements. Even by generating this limited subset, we still observed a wide variability in repeat motif size and sequence, repeat length, internal sequence variability, and allele variability (Figs. 1, 3).
In considering the timing of expansion of these VNTRs, we first found that the overwhelming majority of VNTRs expanded in modern human genomes were already expanded in ancient human genomes (Fig. 2A). Human-specific expansion refers to both rare repeat expansions that arose ab initio (meaning they are only expanded in humans) as well as repeat expansions that are more expanded in humans than other primates. When we looked closer at the four VNTRs most different in length across superpopulations, we saw that they initially appeared at three different points in the primate phylogenetic tree, so there is no generalized pattern or moment that explains this initial expansion as a group.
Looking at modern humans, we found that none of the trios in the 1000 Genomes Project show obvious expansion over a single generation timescale. This intergenerational stability suggests that the VNTRs expand over a longer timescale, and therefore the events that lead to their expansion occur less frequently (Fig. 2B). This observation is consistent with what we previously observed using SMRT long-read sequencing in a very large pedigree for the WDR7 VNTR (Course et al. 2020). It is also consistent with the negative relationship we observed between motif size and copy number (Fig. 1B), which indicates that the longer a motif length is, the less likely it is to expand rapidly. That said, observing VNTRs in pedigrees has long been suggested as a way to differentiate between allelic variability attributed to homologous recombination versus unequal sister chromatid exchange, and looking for rare mutation events is still worthwhile (Jeffreys et al. 1985). Finally, a high percent of these VNTRs were intronic (61.5%, Fig. 1E) compared to the percentage of the human genome that is intronic (39.5%) (Piovesan et al. 2016). This discrepancy may be because many VNTRs are located in subtelomeric regions (Audano et al. 2019), which are known to be gene-rich (Calderón et al. 2014).
As for the geographical distribution of these VNTRs in modern humans, we see that some populations share more similar overall VNTR lengths than others, that is, European and Admixed American superpopulations share few VNTRs with a large log2-fold change, as is also observed for SNP data (The 1000 Genomes Project Consortium 2015; Glusman et al. 2017; Li et al. 2017), whereas there are many repeats whose lengths are significantly different between some populations (Fig. 5). The VNTRs with the greatest length differences were in LOC102723906, PROP1, ART1, and DYNC2I1. The function of these genes is disparate: LOC102723906 is an uncharacterized gene thought to produce noncoding RNA; PROP1 is a gene involved in pituitary development (Wu et al. 1998); ART1 encodes an enzyme that catalyzes the ADP-ribosylation of arginine residues (Paone et al. 2002); and DYNC2I1 may play a role in cilia development (McInerney-Leo et al. 2013; Hamada et al. 2018). Notably, DYNC2I1 is in a GWAS locus for height (Lettre et al. 2008), a trait that is under selection in humans (Turchin et al. 2012), highlighting how these and other VNTRs may contribute in part to the “missing heritability” of human disease at GWAS loci, as has been suggested (Supplemental Table S1; Hannan 2018). These four VNTRs were notably variable in internal nucleotide composition, but in most other ways they differed from one another, including in their length distribution patterns (Fig. 6) and their allele patterns revealed by long-read sequencing (Fig. 7). Long-read sequencing analysis of these VNTRs explained the patterns we observed from short-read data, with the primary groups of alleles accounting for the trimodal distribution pattern observed for the VNTR in LOC102723906 (Fig. 6A) and the gap in lengths observed for the VNTR in ART1 (Fig. 6C), for example. The reason why some populations have a higher frequency of certain expanded alleles is unclear, but it is consistent with population bottlenecks followed by drift of alleles that are possibly non- or weakly deleterious. This analysis also indicates that many VNTRs continue to expand in certain superpopulations. Although length may change noticeably between some superpopulations, the repeat motifs themselves generally do not, perhaps because the variation originated a long time ago. One exception to this observation is a VNTR in PCBP3, which shows an almost inverse correlation between repeat motifs common in the African superpopulation and those in the East Asian superpopulation (Fig. 4C).
We used existing SMRT long-read data sets to resolve the internal nucleotide patterns of 53 of these VNTRs, chosen for their high standard deviation in length (Sulovari et al. 2019), in addition to the four observed in the context of population structure. In doing so, we determined that the primary considerations for VNTR analysis are variability in motif organization within alleles, variability in motif sequence between alleles, and variability in length between alleles. Most VNTR analysis has revolved around assessing variability in length between alleles; however, we hope to illustrate here that the sequence compositions of VNTRs are a critical consideration, as well. Recent examples already indicate that sequence composition of VNTRs can influence disease state (Song et al. 2018), in addition to revealing population differences and expansion patterns (Course et al. 2020). Even in this subset of VNTRs, we observe a wide range of patterns. Some repeats are quite variable in allele length, whereas others seem to have only a few alleles fixed at certain lengths. Some repeats are not variable in sequence, whereas some are partially variable, and some others still are extremely variable, based on the number of different repeat units observed (Fig. 3). VNTRs can even have sections that represent more than one of these categories (e.g., Fig. 3H). We did observe that most of the VNTRs lacked high allele variance across superpopulations, which makes sense in light of the fact that most are already expanded in ancient genomes.
The primary force driving differences between alleles—both sequence and length—appears to be deletions and duplications (e.g., Fig. 3I), which could occur anywhere in the repeat, and are possibly the result of homologous recombination or unequal sister chromatid exchange. Compared to the VNTR in WDR7 that we had previously studied (Fig. 3G; Course et al. 2020), none of the VNTRs whose internal nucleotide patterns that we assessed showed as clear a directionality of expansion. This observation corroborates what has been seen for the VNTRs in ABCA7 (De Roeck et al. 2018, 2019), which is dynamically expanded in non-human primates and has much more internal variability, and in CACNA1A (Song et al. 2018), which has variability primarily in two regions in the middle of the repeat. Some of the earliest studies of VNTR evolution pointed toward both mitotic/meiotic recombination and replication slippage as influencing VNTR expansion and variability (Jeffreys et al. 1985). Repeat directionality (or polarity) has been observed in STRs, where the directionality played a role in instability of the repeat (Eichler et al. 1995). As more genomes undergo long-read sequencing, we may find additional examples of directionality in these longer VNTRs. In addition, recent efforts to long-read sequence model organisms, like that of the highly biodiverse 101 drosophilid genomes (Kim et al. 2020), will facilitate further exploration of the mechanisms responsible for VNTR expansion and contraction.
Sufficient sequence depth continues to be an important feature when measuring VNTR length. Short-read sequencing accuracy can be limited in situations with a low depth of coverage or techniques that require PCR amplification in library preparation, such as for Neanderthal and Denisovan genomes, both of which lead to lower reliability for GC-rich and GC-poor VNTRs. Here, we find that although there is reasonable to strong concordance between the copy numbers predicted by short-read sequencing versus those defined by long-read sequencing, the short-read sequencing generally appears to underestimate copy number. Although few long-read genomes have been publicly available until recently (Linthorst et al. 2020; Shumate et al. 2020), these can help resolve repeat length and copy number using methods like those used here. Fortunately, increasing numbers of high-quality phased long-read genomes are expected to be available starting with those from the HGSVC (Ebert et al. 2021). These genomes, along with improvements in long-read sequencing accuracy (Logsdon et al. 2020), will significantly aid in further defining the properties of VNTRs.
After observing the wide range of motif size in this set of VNTRs (7–341 bp), encompassing a mean size of 40 bp, together with the wide range of internal sequence variability and patterning, we submit that the current definition of a VNTR as a tandem repeat with a 7-bp motif or larger could benefit from more comprehensive categorization. We initially chose this set of VNTRs by selecting for parameters like human-specific expansion, but it may be more useful going forward to analyze VNTRs together in groups of similar physical characteristics, like motif size, number of variable motifs, and number of apparent groups of alleles. This kind of grouping is more likely to reveal common mechanisms of expansion and evolution.
Overall, these human-specific VNTR expansions are generally found in noncoding regions, are already expanded in ancient human genomes, and remain stable between single generations. They tend to have some variability in their motifs, but less variability between alleles, with their primary source of allelic variation driven by deletions and duplications. That said, there are exceptions to each of these generalizations, and what is clearly different between each VNTR is their motif size, which ranges widely and correlates inversely with copy number; their patterns of motif variability and sequence composition; and when they first arise in non-human primates. Subcategorizing VNTRs with these considerations in mind, and observing them in more detail with long-read sequencing, may lead to uncovering larger patterns that explain their expansion dynamics. The number of publicly available long-read genomes, both in humans and model organisms, is expected to increase significantly in the next several years, and this study provides a framework for conducting further analysis.
Methods
VNTR selection
We started with a list of 1584 tandem repeats categorized by their expansion specifically in humans (Sulovari et al. 2019). The motifs used in this list were previously identified using Tandem Repeats Finder (Benson 1999). We then excluded SVA retrotransposons and any repeats with a motif size <7 nt. These selection criteria yielded 467 VNTRs with unique matches in the human genome. In rare cases in which two motif lengths were possible and one was half the size of the other, we chose the smaller motif size; however, this did not lead to any situations in which the repeat dropped below the cutoff of >6 nt for VNTRs.
VNTR gene location
To determine the locations of the VNTRs, we started with previously published data (Sulovari et al. 2019). We then manually inspected VNTRs that were within or close to exons and compared VNTR location to GENCODE v32 transcripts in the UCSC Genome Browser (Kent et al. 2002).
Repeat length estimation
We adapted previous methods (Song et al. 2018; Course et al. 2020) to estimate read depth using short-read whole-genome sequencing data. Reads were counted that mapped to the repeat, compared to three sets of 100-kb windows of genomic sequence. The fraction of enrichment or depletion of reads was used to calculate the estimated length of each VNTR compared to the reference human genome (GRCh38).
Raw data for ancient DNA calculations were obtained from published genomes of Altai Neanderthal (Prüfer et al. 2014) and Denisovan (Meyer et al. 2012) samples. Data for Altai Neanderthal and Denisovan genomes can be accessed from the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena/browser/home) ERP002097 and ERP001519, respectively. Samples for Vindija and Chagyrskaya Neanderthals can be accessed from http://cdna.eva.mpg.de/neandertal/Vindija/ and http://cdna.eva.mpg.de/neandertal/Chagyrskaya/, respectively. We converted the coordinates of each VNTR from GRCh38 to hg19 using the liftOver tool in the UCSC Genome Browser, then queried the ancient genomes for the number of matches within those coordinates padded by 2 kb on either side of the VNTR repeat sequence as listed by Tandem Repeats Finder (Benson 1999). Of the 467 VNTRs, seven did not have corresponding matches in hg19. Regions corresponding to each VNTR were converted to SAM files and individual reads were queried for the presence of a complete repeat unit. For PCBP3, we also searched for repeat unit sequence matches from DNA reads for Chagyrskaya and Vindija Neanderthal Genomes (Prüfer et al. 2017; Mafessoni et al. 2020). Total counts for each repeat unit were aggregated across each superpopulation and used to identify their relative abundance.
Raw data for modern DNA calculations were obtained from the 1000 Genomes Project (The 1000 Genomes Project Consortium 2015), which is available from https://www.internationalgenome.org/data#download. Reads that mapped to each VNTR were extracted from BAM alignment files for each individual. The number of reads that corresponded to each repeat was counted across each sample and normalized first to the length of the repeat in the GRCh38 human genome and then to the read density across three separate 100-kb segments of DNA that did not contain a VNTR (which were confirmed to have consistent read density estimates across all three bins). This number of aligned reads from each sample was used as input for the DESeq2 program (Love et al. 2014) to identify differentially expanded repeats between superpopulations.
Long-read sequencing analysis
We obtained 15 PacBio SMRT-sequenced genomes from Audano et al. (2019) and 32 from the HGSVC (Ebert et al. 2021). We then aligned and visualized the VNTR alleles from these data sets as described previously (Fig. 3A; Course et al. 2020). Briefly, after extracting the repeat sequence along with flanking intronic sequence, each repeat unit per VNTR was assigned a single letter or number code. Each letter was then converted to its own unique color to improve visualization when aligning the series of repeat units for each sample. Motifs that appeared only once between all alleles were considered private variants, and we did not combine any of these motifs together. We then manually organized the repeat sequences based on their similarity, including creating gaps to shift units down to match the arrangement of neighboring alleles. Perhaps because of their repetitive nature, we did not always identify each repeat in every sequenced individual. Samples were not included if their repeat abutted the end of a sequence contig, which was a relatively common occurrence.
Whole-genome long-read sequence data used in this study were obtained from the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession numbers PRJNA246220 (CHM1), PRJNA300843 (HG00514), PRJNA300840 (HG00733), PRJNA288807 (NA19240), PRJNA339722 (HG02818), PRJNA385272 (NA19434), PRJNA339719 (HG01352), PRJNA339726 (HG02059), PRJNA323611 (NA12878), PRJNA481794 (HG04217), PRJNA480858 (HG02106), and PRJNA480712 (HG00268). Data from the HGSVC are available from https://www.internationalgenome.org/data-portal/data-collection/hgsvc2.
Phylogenetic analysis
Presence or absence of the VNTR repeat unit of interest was observed by extracting sequences in non-human primates in the UCSC Genome Browser. Using the “View in other genomes (convert)” function in the UCSC genome, we extracted sequences of 10 non-human primates to determine the presence of one or more copies of the repeat motif.
Statistical analyses
Statistical analyses were performed using Prism 8.0.1 (GraphPad Software). Log-log line slopes were determined by least squares regression. Volcano plots were generated by graphing log2 fold change against negative log10 P-value on an XY plot. A one-way analysis of variance (ANOVA) test was used to compare groups greater than two, followed by Tukey's multiple comparisons test when the ANOVA gave P < 0.05.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of General Medical Sciences (5T32GM007454-38 to M.M.C.), and the National Human Genome Research Institute (HG010169 to E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute. P.N.V. is supported in part by the Robert F. Schoeni Award for Research from the Ann Arbor Active Against ALS. We acknowledge the support of all members of the Valdmanis laboratory. Finally, we thank all individuals who donated biospecimens for their willingness to contribute to scientific research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275560.121.
Competing interest statement
The authors declare no competing interests.
References
- The 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526: 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah A, Dutcher SK, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176: 663–675.e19. 10.1016/j.cell.2018.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam KWG, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet 42: 859–863. 10.1038/ng.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón MDC, Rey MD, Cabrera A, Prieto P. 2014. The subtelomeric region is important for chromosome recognition and pairing during meiosis. Sci Rep 4: 6488. 10.1038/srep06488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Course MM, Gudsnuk K, Smukowski SN, Winston K, Desai N, Ross JP, Sulovari A, Bourassa CV, Spiegelman D, Couthouis J, et al. 2020. Evolution of a human-specific tandem repeat associated with ALS. Am J Hum Genet 107: 445–460. 10.1016/j.ajhg.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roeck A, Duchateau L, Van Dongen J, Cacace R, Bjerke M, Van Den Bossche T, Cras P, Vandenberghe R, De Deyn PP, Engelborghs S, et al. 2018. An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer's disease. Acta Neuropathol 135: 827–837. 10.1007/s00401-018-1841-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roeck A, De Coster W, Bossaerts L, Cacace R, De Pooter T, Van Dongen J, D'Hert S, De Rijk P, Strazisar M, Van Broeckhoven C, et al. 2019. NanoSatellite: accurate characterization of expanded tandem repeat length and sequence through whole genome long-read sequencing on PromethION. Genome Biol 20: 239. 10.1186/s13059-019-1856-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert P, Audano PA, Zhu Q, Rodriguez-Martin B, Porubsky D, Bonder MJ, Sulovari A, Ebler J, Zhou W, Serra Mari R, et al. 2021. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science 372: eabf7117. 10.1126/science.abf7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Hammond HA, Macpherson JN, Ward PA, Nelson DL. 1995. Population survey of the human FMR1 CGG repeat substructure suggests biased polarity for the loss of AGG interruptions. Hum Mol Genet 4: 2199–2208. 10.1093/hmg/4.12.2199 [DOI] [PubMed] [Google Scholar]
- Glusman G, Mauldin DE, Hood LE, Robinson M. 2017. Ultrafast comparison of personal genomes via precomputed genome fingerprints. Front Genet 8: 136. 10.3389/fgene.2017.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada Y, Tsurumi Y, Nozaki S, Katoh Y, Nakayama K. 2018. Interaction of WDR60 intermediate chain with TCTEX1D2 light chain of the dynein-2 complex is crucial for ciliary protein trafficking. Mol Biol Cell 29: 1628–1639. 10.1091/mbc.E18-03-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannan AJ. 2018. Tandem repeats mediating genetic plasticity in health and disease. Nat Rev Genet 19: 286–298. 10.1038/nrg.2017.115 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL. 1985. Hypervariable “minisatellite” regions in human DNA. Nature 314: 67–73. 10.1038/314067a0 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Wang JR, Miller DE, Barmina O, Delaney E, Thompson A, Comeault AA, Peede D, D'Agostino ERR, Pelaez J, et al. 2020. Highly contiguous assemblies of 101 drosophilid genomes. bioRxiv 10.1101/2020.12.14.422775 [DOI] [PMC free article] [PubMed]
- Kirby A, Gnirke A, Jaffe DB, Barešová V, Pochet N, Blumenstiel B, Ye C, Aird D, Stevens C, Robinson JT, et al. 2013. Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat Genet 45: 299–303. 10.1038/ng.2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, et al. 2008. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet 40: 584–591. 10.1038/ng.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cerise JE, Yang Y, Han H. 2017. Application of t-SNE to human genetic data. J Bioinform Comput Biol 15: 1750017. 10.1142/S0219720017500172 [DOI] [PubMed] [Google Scholar]
- Linthorst J, Meert W, Hestand MS, Korlach J, Vermeesch JR, Reinders MJT, Holstege H. 2020. Extreme enrichment of VNTR-associated polymorphicity in human subtelomeres: genes with most VNTRs are predominantly expressed in the brain. Transl Psychiatry 10: 369. 10.1038/s41398-020-01060-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Vollger MR, Eichler EE. 2020. Long-read human genome sequencing and its applications. Nat Rev Genet 21: 597–614. 10.1038/s41576-020-0236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafessoni F, Grote S, De Filippo C, Slon V, Kolobova KA, Viola B, Markin SV, Chintalapati M, Peyrégne S, Skov L, et al. 2020. A high-coverage neandertal genome from Chagyrskaya Cave. Proc Natl Acad Sci 117: 15132–15136. 10.1073/pnas.2004944117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney-Leo AM, Schmidts M, Cortés CR, Leo PJ, Gener B, Courtney AD, Gardiner B, Harris JA, Lu Y, Marshall M, et al. 2013. Short-rib polydactyly and Jeune syndromes are caused by mutations in WDR60. Am J Hum Genet 93: 515–523. 10.1016/j.ajhg.2013.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, De Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338: 222–226. 10.1126/science.1224344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel RE, Handsaker RE, Sherman MA, Barton AR, Zheng Y, McCarroll SA, Loh P-R. 2021. Protein-coding repeat polymorphisms strongly shape diverse human phenotypes. bioRxiv 10.1101/2021.01.19.427332 [DOI] [PMC free article] [PubMed]
- Näslund K, Saetre P, Von Salomé J, Bergström TF, Jareborg N, Jazin E. 2005. Genome-wide prediction of human VNTRs. Genomics 85: 24–35. 10.1016/j.ygeno.2004.10.009 [DOI] [PubMed] [Google Scholar]
- Örd T, Puurand T, Örd D, Annilo T, Möls M, Remm M, Örd T. 2020. A human-specific VNTR in the TRIB3 promoter causes gene expression variation between individuals. PLoS Genet 16: e1008981. 10.1371/journal.pgen.1008981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paone G, Wada A, Stevens LA, Matin A, Hirayama T, Levine RL, Moss J. 2002. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci 99: 8231–8235. 10.1073/pnas.122238899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan A, Caracausi M, Antonaros F, Pelleri MC, Vitale L. 2016. GeneBase 1.1: a tool to summarize data from NCBI gene datasets and its application to an update of human gene statistics. Database 2016: baw153. 10.1093/database/baw153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, De Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505: 43–49. 10.1038/nature12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, De Filippo C, Grote S, Mafessoni F, Korlević P, Hajdinjak M, Vernot B, Skov L, Hsieh P, Peyrégne S, et al. 2017. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358: 655–658. 10.1126/science.aao1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumate A, Zimin AV, Sherman RM, Puiu D, Wagner JM, Olson ND, Pertea M, Salit ML, Zook JM, Salzberg SL. 2020. Assembly and annotation of an Ashkenazi human reference genome. Genome Biol 21: 129. 10.1186/s13059-020-02047-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JHT, Lowe CB, Kingsley DM. 2018. Characterization of a human-specific tandem repeat associated with bipolar disorder and schizophrenia. Am J Hum Genet 103: 421–430. 10.1016/j.ajhg.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulovari A, Li R, Audano PA, Porubsky D, Vollger MR, Logsdon GA, Warren WC, Pollen AA, Chaisson MJP, Eichler EE. 2019. Human-specific tandem repeat expansion and differential gene expression during primate evolution. Proc Natl Acad Sci 116: 23243–23253. 10.1073/pnas.1912175116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchin MC, Chiang CWK, Palmer CD, Sankararaman S, Reich D, Hirschhorn JN. 2012. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nat Genet 44: 1015–1019. 10.1038/ng.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Cogan JD, Pfäffle RW, Dasen JS, Frisch H, O'Connell SM, Flynn SE, Brown MR, Mullis PE, Parks JS, et al. 1998. Mutations in PROP1 cause familial combined pituitary hormone deficiency. Nat Genet 18: 147–149. 10.1038/ng0298-147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.