Abstract
Ebola virus (EBOV) hemorrhagic fever outbreaks have been challenging to deter due to the lack of health care infrastructure in disease-endemic countries and a corresponding inability to diagnose and contain the disease at an early stage. EBOV vaccines and therapies have improved disease outcomes, but the advent of an affordable, easily accessed, mass-produced rapid diagnostic test (RDT) that matches the performance of more resource-intensive polymerase chain reaction (PCR) assays would be invaluable in containing future outbreaks. Here, we developed and demonstrated the performance of a new ultrasensitive point-of-care immunoassay, the EBOV D4 assay, which targets the secreted glycoprotein of EBOV. The EBOV D4 assay is 1000-fold more sensitive than the U.S. Food and Drug Administration–approved RDTs and detected EBOV infection earlier than PCR in a standard nonhuman primate model. The EBOV D4 assay is suitable for low-resource settings and may facilitate earlier detection, containment, and treatment during outbreaks of the disease.
INTRODUCTION
Ebola virus (EBOV) outbreaks are a public health emergency due to the ease of contagion and high mortality rate associated with this hemorrhagic fever virus. The Centers for Disease Control and Prevention has determined that over 11,000 deaths occurred during the 2014–2016 West Africa outbreak, and more than 2200 deaths have occurred thus far (1) in two ongoing outbreaks in the North Kivu, Ituri, and Equateur provinces in the Democratic Republic of Congo (DRC) (2). Although EBOV vaccines and treatments provide hope for inhabitants of disease-endemic regions, there is a critical need for a low-cost and highly sensitive rapid diagnostic test (RDT) that could be deployed at the point of care (POC) to allow early-stage detection of EBOV infection, rapid triage, and effective contact tracing (3). One mathematical modeling analysis concluded that if available then, an RDT with the sensitivity and specificity of reverse transcription polymerase chain reaction (RT-PCR) would have reduced fatalities during the 2014 outbreak by ~40% (4). The need for early detection in the field was also highlighted by the 2019 Pamoja Tulinde Maisha (PALM) trial, in which survival rates for patients with Ebola treated with the neutralizing antibody (Ab) mAb114 at an early stage of infection (at low viral load) were 90%, whereas survival rates for patients treated at a later stage were 30% (5).
RT-PCR, the current gold standard for EBOV diagnosis, can diagnose EBOV 3 to 16 days earlier than serology or existing antigen (Ag) detection tests (3, 6, 7). Recent advances in RT-PCR have reduced assay complexity but the majority of molecular diagnostic tests for EBOV require laboratories and highly trained personnel (8). More recently, single-use cartridge systems containing all reagents needed for RT-PCR have expanded testing availability by reducing the need for skilled technicians and stringent biocontainment infrastructures (9), while delivering performance comparable to traditional RT-PCR (10, 11). In one example, the cartridge-based Xpert Ebola molecular test (Cepheid) was widely deployed in the recent DRC outbreaks. Although effective, this assay costs $22.50 USD (U.S. dollars) per test (3), has a run time of ~90 min per assay, and can only process one sample at a time per module. Thus, although cartridge-based RT-PCR systems represent an important advance for molecular testing, they do not yet conform to the desired target product profile for rapid EBOV diagnostic tests (3, 7) as defined by the World Health Organization (WHO) (12).
As an alternative to PCR, several lateral-flow assays have been developed and evaluated by the WHO and the U.S. Food and Drug Administration (FDA). Four LFA-based RDTs have received emergency use status or FDA approval: OraQuick Ebola (OraSure Technologies), ReEBOV (Corgenix), SD Q Line Ebola Zaire Ag (SD Biosensor), and the DPP Ebola Antigen System (Chembio). These LFA diagnostic tests are easy to use and yield results within 15 to 30 min but are limited by their lower sensitivity relative to RT-PCR, with analytical sensitivities of tens to hundreds of nanogram per milliliter for matrix protein (VP40) and/or nucleoprotein. Field trials of OraQuick Ebola (13), ReBOV (14), and QuickNavi Ebola (15) have shown clinical sensitivities as low as 60% for low–viral load samples and clinical specificities of ~90% (16, 17). Because of the potentially severe consequences of false-negative test results, the reduced sensitivity of LFAs has impeded their widespread adoption.
Here, we describe the development and preclinical testing of a highly sensitive POC rapid immunodiagnostic test for EBOV and other clinically relevant EBOV species including Sudan ebolavirus (SUDV) and Bundibugyo ebolavirus (BDBV) (18, 19). The development of this diagnostic assay involved five key strategies and technologies: (i) selection of a robust diagnostic target for early detection of EBOV infection; (ii) the use of single-chain Ab phage display to generate high-affinity Abs specific for EBOV; (iii) a high-throughput Ab-pairing strategy to identify the most sensitive capture Ab (cAb)–detection Ab (dAb) combination; (iv) the inkjet printing of cAbs and fluorescently labeled dAbs (FL-dAb) as microspots onto “D4 assay chips,” which consist of glass slides coated with a cell- and protein-resistant polymer brush coating (20–22), resulting in a single-step sandwich immunoassay with up to femtomolar sensitivity (23, 24); and (v) the development of a low-cost handheld wide-field fluorescence reader (the D4Scope), which quantifies the fluorescence output from the D4 chip with a performance similar to that of a fluorescence microarray scanner.
RESULTS
Target selection and Ab generation
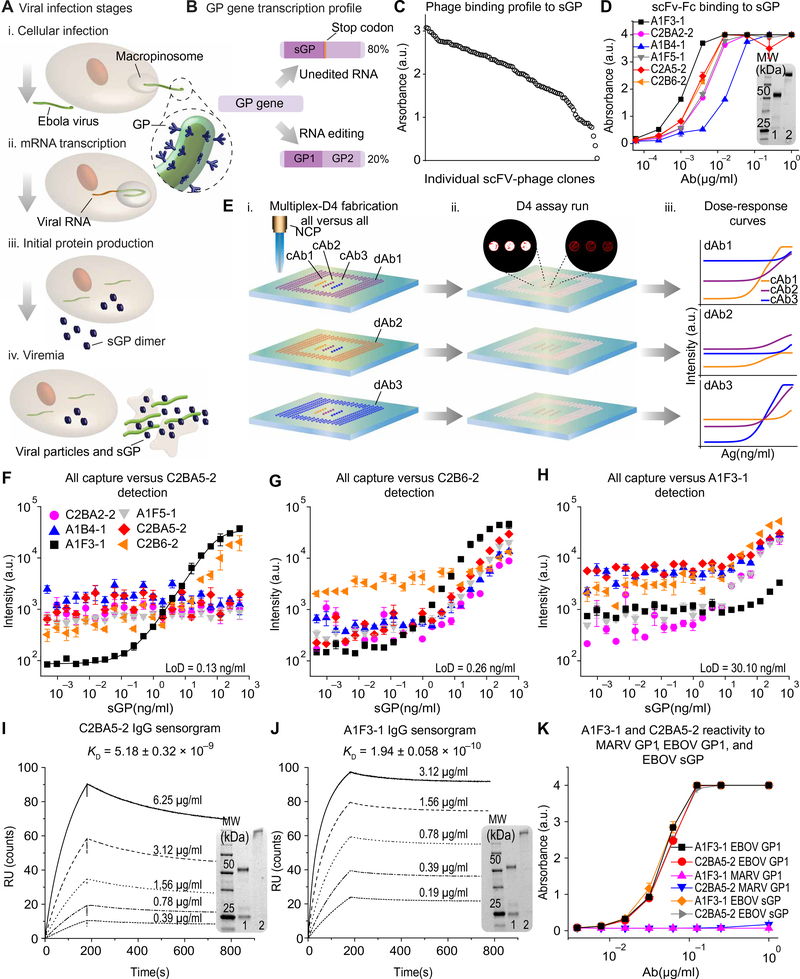
For our diagnostic target, we selected EBOV secreted glycoprotein (sGP), a truncated and actively secreted version of the GP1 glycoprotein, which acts as a decoy Ag that appears in serum earlier than other viral proteins. sGP is thought to subvert the immune response during EBOV infections and is produced in high amounts, accounting for ~80% of GP transcription (Fig. 1, A and B) (25, 26). Elevated concentrations of a circulating early-stage EBOV biomarker provide an opportunity to diagnose infection before contagion (27, 28). Motivated by this rationale, we selected sGP as the target Ag and incorporated it into the D4 assay (fig. S1) (24, 29, 30).
Fig. 1. sGP as a diagnostic target and selection of capture and detection Abs for the EBOV D4 assay.
(A) Schematic of EBOV infection. (B) Schematic of the GP gene transcription profile. (C) Binding of scFv-phage clones to EBOV sGP measured using Ag-down ELISA. Each data point is N = 1 technical replicate for scFv-phage clones with a distinct sequence. a.u., arbitrary units. (D) Binding of six scFv-Fc Abs to sGP, measured using Ag-down ELISA. Each data point represents the mean ± SEM of N = 2 replicates. (E) Schematic of Ab pairing using the D4 assay. (i) cAb and FL-dAb printed onto POEGMA-coated glass are exposed to sGP-spiked sample. (ii) Fluorescent spots are imaged; insets show spots with high (white) and low (red) fluorescence output. (iii) LoD and DR of each pair are calculated. (F to H) Dose-response curves using all six scFv-Fc Abs as cAb and C2BA5–2 (F), C2B6–2 (G), and A1F3–1 (H) as the dAb. Dose-response curves show the mean ± SEM of N = 4 test runs and were fitted using a five-parameter logistic fit. (I and J) SPR sensorgrams of C2BA5–2 and A1F3–1 binding to EBOV sGP. KD is the average ± SEM of N = 3 measurements. (K) Binding of A1F3–1 and C2BA5–2 binding to EBOV sGP, EBOV GP1, and MARV GP1. Each data point represents the average ± SEM of N = 2 replicates. Insets of (D), (I), and (J): SDS–polyacrylamide gel electrophoresis of purified scFv-Fc Abs under (1) reducing and (2) nonreducing conditions. NCP, noncontact printer; KD, equilibrium dissociation constant; MARV, Marburg virus; MW, molecular weight; kDa, kilodalton.
The D4 assay is a POC Ab microarray in a low-cost (~$0.25 USD per test), easy-to-fabricate format. Stable cAbs and soluble FL-dAbs are inkjet-printed onto a stealth, protein- and cell-resistant poly[oligo (ethylene glycol) methyl ether methacrylate] (POEGMA) brush surface grown in situ on glass slides by surface-initiated atom radical transfer polymerization (20–22, 31, 32). This polymer brush surface blocks nonspecific protein absorption and cell adhesion and, when combined with high affinity Abs, results in high signal-to-noise and femtomolar sensitivity (32). The dAbs printed on top of a soluble trehalose pad dissolve and bind to the target analyte when a liquid sample is added to the surface of a D4 chip. This dAb-analyte complex is then captured by the cAbs, and, after a rinse step (fig. S2), the fluorescence of the capture spots can be quantified by a fluorescence scanner. The resulting EBOV D4 assay is a user-friendly, accessible, highly portable, low-cost, mass-producible, ultrasensitive RDT that can be used to diagnose infection for clinically relevant species of EBOV.
To develop diagnostic Abs for the sGP D4 assay, M13 phage display (33, 34) was used to generate Abs that bind to sGP with high affinity. Mice were immunized with recombinant EBOV (Mayinga) sGP to elicit high-titer immunoglobulin G (IgG) responses (fig. S3A); mouse spleens were then harvested to obtain mRNA to generate an M13 phage library displaying single-chain variable fragment (scFv) Abs. Through iterative enrichment, phage clones with a high binding affinity for EBOV sGP and SUDV sGP were identified (Fig. 1C and fig. S3B). ScFv inserts from the phage clones with highest binding for sGP from EBOV, SUDV, and BDBV were subcloned and expressed on an Fc scaffold in Expi293 cells to obtain soluble scFv-Fc Abs (fig. S4A), which were purified using an IsoTag system (fig. S4, B to D) (35). ScFv-Fc Ab-sGP binding profiles were then evaluated using Ag-down enzyme-linked immunosorbent assay (ELISA) with EBOV sGP (Fig. 1D and fig. S4E).
ScFv-Fc Ab-sGP binding kinetics were determined using surface plasmon resonance (SPR) spectroscopy. Five of the six scFv-Fc Abs tested displayed an equilibrium dissociation constant (KD) of ~10−8 M−1 for sGP; one Ab, A1F3–1, displayed roughly 100-fold greater binding affinity with a KD of ~10−10 M−1 (fig. S5). We next identified the optimal cAb-dAb pair for the RDT application. Leveraging the multiplexing capability of the D4 assay, we generated dose-response curves for all six scFv-Fc Abs versus a single dAb in one experiment to characterize all 36 cAb-dAb combinations (Fig. 1E). Figure 1 (F to H) shows dose-response curves for all six scFv-Fc Abs as cAb, with C2BA5–2, C2B6–2, and A1F3–1 as the dAb. Dose-response curves for all 36 combinations are shown in fig. S6, as well as each pair’s limit of detection (LoD) and dynamic range (DR). The optimal pair—A1F3–1 as the cAb and C2BA5–2 as the dAb—showed an LoD of 0.13 ng/ml and a DR of 3.6 log10 for EBOV sGP.
After selecting A1F3–1 and C2BA5–2 as the optimal scFv-Fc cAb-dAb pair, their variable regions were subcloned into mouse γ2a and κ chain expression vectors to obtain full-length IgG2a Abs (fig. S7). The binding affinities of the full-length IgG2a Abs for EBOV sGP were three- to fivefold greater than those of their scFv-Fc counterparts (Fig. 1, I and J). The binding affinities of the IgG2a Abs for sGP from four other EBOV species—SUDV, BDBV, Reston ebolavirus (RESTV), and Taï Forest ebolavirus (TAFV)—were 3- to 1000-fold lower than for EBOV sGP (fig. S8), an expected result as the Abs were selected on the basis of their affinity for EBOV sGP. We also confirmed findings with Ag-down ELISA and evaluated if other Abs were specific to sGP from other EBOVs (fig. S9).
To further test the specificity of the IgG2a Abs, we assessed their reactivity to recombinant GP1 from Marburg virus (MARV), a closely related hemorrhagic virus, and to recombinant GP1 from EBOV (Fig. 1K). Neither Ab reacted with MARV GP1, whereas both Abs recognized EBOV GP1, indicating that their epitopes lie within a sequence common to sGP and GP1. This cross-reactivity between sGP and GP1 from EBOV may be beneficial in this RDT application because it should allow improved EBOV detection at later stages of the infection, when circulating sGP may decline and GP1 is elevated.
EBOV D4 assay development
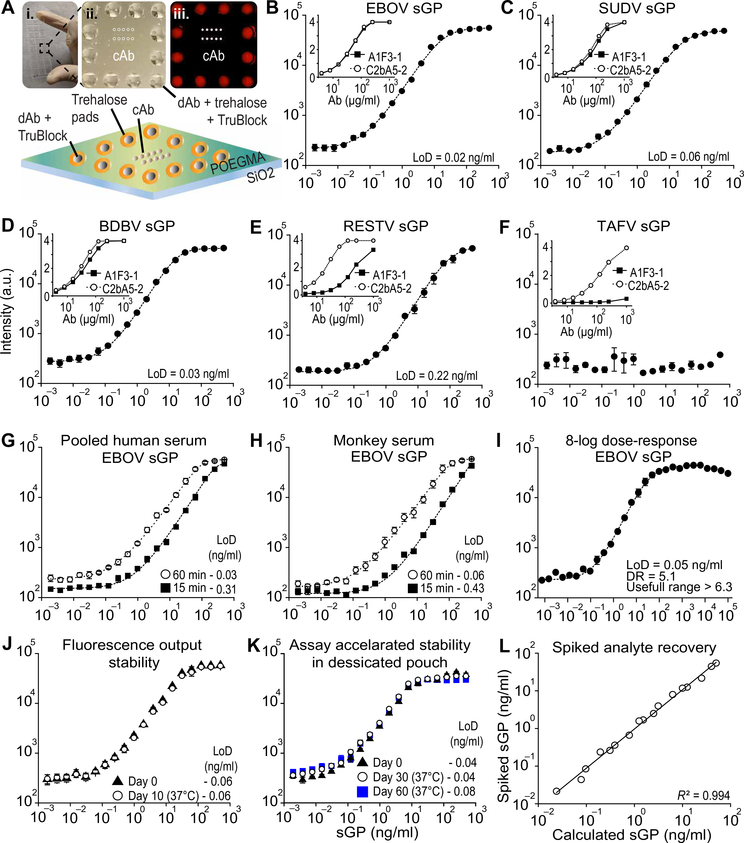
After their functional characterization, the A1F3–1 and C2BA5–2 IgG2a Abs were integrated into a new version of the D4 assay (Fig. 2A) in which the dAb is noncontact printed onto trehalose pads, reducing the fabrication time ~10-fold from that of the previous version in which dAbs were printed in 144 dissolvable detection spots forming multiple concentric rings around the cAbs. This reduction in fabrication time enables low-cost production of over 2000 tests per day in a research laboratory setting, an important decrease in fabrication time and increase in throughput that allows quicker turnaround for assay optimization and validation (3). Printing parameters, including the concentration of trehalose and Trublock—a reagent that minimizes cross-reactivity of mouse monoclonal Abs with human anti-mouse Abs (HAMA)—were optimized by varying their concentration and assessing background fluorescence, LoD, and DR of pooled human serum (HS) spiked with sGP. Twenty-four individual RDT chips, each of which consisted of 10 central cAb spots surrounded by a ring of 12 dAb spots, were fabricated on each POEGMA-coated glass slide, with a plexiglass gasket isolating each test (Fig. 2A, i and ii). Samples containing sGP at 125 ng/ml were detected by the fluorescence in the cAb spots after incubation with the analyte (Fig. 2Aiii).
Fig. 2. Performance of the EBOV D4 assay.
(A) (i) Photograph of a POEGMA-coated glass slide containing 24 EBOV D4 assay chips (one chip is indicated by the black box), with laser-cut acrylic and adhesive creating individual sample wells. (ii) Photograph of a single EBOV D4 assay. (iii) Fluorescence image of incubation with sample containing sGP, intense capture spots (white) and lower intensity detection spots (red) are visible. Bottom: Schematic of EBOV D4 assay. (B to F) Dose-response curves obtained using the EBOV D4 assay fabricated with A1F3–1 as cAb and C2BA5 as dAb, for EBOV sGP (B), SUDV sGP (C), BDBV sGP (D), RESTV sGP (E), and TAFV sGP (F) in FBS. Insets depict individual Ab-sGP binding profiles determined by Ag-down ELISA. (G and H) Dose-response curves using different incubation times (15 and 60 min) and sample types (pooled HS and rhesus MoS). (I) 8-log10 dose-response curve using sGP-spiked calf-serum. (J) Post-assay fluorescence stability: After running the assay, chips were incubated at 37°C and 50% humidity for 30 days. (K) Thermal stability of EBOV chips stored in pouches with desiccant at 37°C after 30 and 60 days. (L) Double-blinded spiked analyte recovery experiment in which sGP concentrations in pooled HS are determined by fluorescence intensity. In (B) to (H) and (J and K), each D4 assay data points represents the mean ± SEM of N = 4 assays; In (I), data points represent the mean of N = 3 ± SEM of N = 3 assays and in (L), data points represent the mean of N = 3 technical replicates. Ag-down ELISA data points represent the mean ± SEM of N = 2 assays. A five-parameter logistic fit (dashed line) was used to calculate LoD.
The EBOV D4 assay was initially tested using fetal bovine serum (FBS) spiked with recombinant sGP from EBOV, SUDV, BDBV, RESTV, and TAFV, using a 90-min incubation period (Fig. 2, B to F). The EBOV D4 assay achieved an LoD of 20 pg/ml when detecting EBOV sGP and LoDs of 60 pg/ml for SUDV sGP, 30 pg/ml for BDBV, and 220 pg/ml for RESTV sGP (Fig. 2, B to E) but was unable to detect TAFV sGP because the A1F3–1 Ab does not bind TAFV sGP (Fig. 2F).
After initial verification of the assay, we assessed the performance of the EBOV D4 assay on samples from different matrices with different incubation times. Dose-response curves were obtained using EBOV sGP spiked into FBS, pooled HS, and rhesus monkey serum (MoS), with incubation periods of 15, 60, and 90 min (Fig. 2, G and H, and fig. S10, A to D). An LoD of <100 pg/ml was observed for all sample matrices using a 60-min incubation. To assess the assay’s DR, defined as the range in which quantitation is accurate, and useful range, defined as the range in which a positive diagnosis can be obtained, we performed a hook-effect evaluation using an 8-log10 dose-response curve with a maximum sGP concentration of 0.1 mg/ml. The assay presented a DR range of 5.1 log10, and a useful range greater than 6.3 log10 (Fig. 2I). This large useful range reduces the risk of false negatives due to the hook effect at high concentrations of circulating sGP and GP1 in patients with high viremia.
We next assessed the stability of the assay in terms of its post-assay fluorescence when stored at 37°C at either 50% relative humidity or in an aluminum pouch with desiccant, as it would be advantageous if the chip could be read later if a fluorescence scanner was unavailable at the point of testing. Chips were processed using EBOV sGP–spiked samples at different sGP concentrations and were stored as described for up to 60 days before measuring their fluorescence. The fluorescence output was stable over 10 days when the chip was stored at 37°C with 50% relative humidity (Fig. 2J) and over 30 days when stored at 37°C in a dry pouch (fig. S11). We also evaluated the assay’s stability at warm temperatures. No loss of performance was observed for up to 60 days at 37°C when D4 assay chips were stored in aluminum pouches with desiccant (Fig. 2K), demonstrating that the assay does not require cold storage. These results demonstrate the excellent stability of the D4 assay under conditions likely to be encountered in the field. Next, we tested whether the fluorescence output could be used to quantify sGP concentration by performing a double-blinded analyte recovery experiment using different sGP concentrations in HS. The fluorescence output showed a strong linear correlation with sGP concentration spiked in the samples, (R2 = 0.994) (Fig. 2L).
EBOV D4 assay can detect sGP from clinically relevant EBOVs in whole human blood
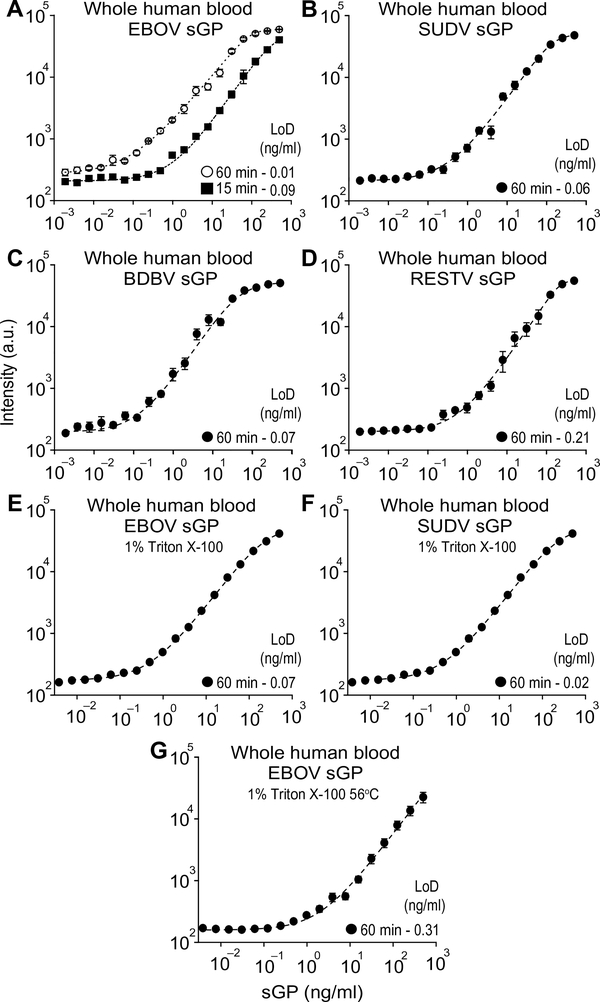
After validating the EBOV D4 assay in FBS, HS, and MoS, we sought to demonstrate that our device could detect sGP from all clinically relevant EBOVs in whole human blood (WHB), as this is the sample of choice for POC applications, especially when testing for high-risk pathogens where sample processing should be minimized. To assess the EBOV D4 assay performance, we generated dose-response curves for 15-, 60-, and 90-min incubation for single donor WHB spiked with EBOV sGP (Fig. 3A and fig. S12). Sixty- and 90-min incubations demonstrated optimal sensitivity for detecting EBOV sGP with LoDs of 0.01 and 0.02 ng/ml, respectively.
Fig. 3. EBOVs sGP detection in WHB.
(A) D4 assay dose-response curves using different incubation times (15 and 60 min) for EBOV sGP spiked in WHB. (B to D) D4 assay dose-response curves for SUDV sGP (B), BDBV sGP (C), and RESTV sGP (D) spiked in WHB and incubated on the chips for 60 min. (E and F) Dose-response curves for WHB samples spiked with EBOV sGP (E) and SUDV sGP (F) and treated for 60 min at room temperature (~23°C) with Triton X-100 to a final 1% (v/v) concentration before being added to D4 chips. Treated samples were incubated on the D4 chips for 60 min. (G) Dose-response curves for blood samples spiked with EBOV sGP and treated for 30 min at 56°C with Triton X-100 to a final 1% (v/v) concentration before being added to D4 chips. Treated samples were incubated on the D4 chips for 60 min. Each data point represents the mean ± SEM of N = 4 assays; a five-parameter logistic fit (dashed line) was used to calculate LoD.
Next, we evaluated whether sGP from other EBOVs could be detected in WHB such that the EBOV D4 assay could also be used during SUDV and BDBV outbreaks. Figure 3 (B to D) shows dose-response curves for WHB spiked with different concentration of sGPs from SUDV, BDBV, and RESTV. LoDs similar to those obtained in FBS after 90-min incubation were also achieved when detecting the different sGPs in WHB after incubation for 60 min.
We next assessed whether sGP could be detected in WHB after viral inactivation procedures. Exposure to detergents has been demonstrated to reduce infectivity by over 6 log10 when viral samples are treated with 0.1% (v/v) Triton X-100 for 1 hour (36). A 1% Triton X-100 (v/v) with 20-min contact time has also been shown to reduce EBOV infectivity in HS samples (37). To assess whether high detergent concentrations added to the WHB containing sGP could affect assay performance, we added Triton X-100 to a final 1% concentration in EBOV and SUDV sGP-spiked WHB. After 60 min of contact time at room temperature, we added the treated samples to EBOV D4 chips. Figure 3 (E and F) shows the dose-response curves generated with the samples treated with detergent, which had no effect on assay performance. We also evaluated whether heated samples treated with Triton X-100 would affect assay performance, as the combination of detergent and heat is recommended for inactivation of potentially infectious samples (38). Figure 3G shows the dose-response curve for WHB spiked with EBOV sGP that were treated with 1% (v/v) Triton X-100 for 30 min at 56°C. There was a ~10-fold loss in sensitivity when compared to untreated samples, although the assay still displayed a sub–nanogram-per-milliliter LoD.
D4 assay performance relative to lateral flow assays
To compare the performance of the EBOV D4 assay with that of a standard LFA, we fabricated LFA test strips using the same cAb-dAb pair (A1F3–1/C2BA5–2) and tested the resulting LFA in parallel with the EBOV D4 assay (fig. S13). The EBOV D4 assay was up to 600-fold more sensitive than an LFA fabricated using the same Ab reagents and displayed a sensitivity over 1000-fold greater than that reported for FDA-approved LFAs. The LoD of the LFA using our cAb-Ab pair (6 ng/ml) surpassed the performance of other LFA-based EBOV RDTs tested in the field (13, 14, 39, 40) (LoDs of 50 to 160 ng/ml for RDTs targeting VP40 Ag). It also matched the LoD of 7.8 ng/ml for an RDT targeting sGP that includes a silver-enhancing signal amplification scheme (41). These results demonstrate the effectiveness of the Ab selection methods used for the EBOV D4 assay and the greater sensitivity of the D4 assay compared to current LFAs.
A handheld detector to image the EBOV D4 assay
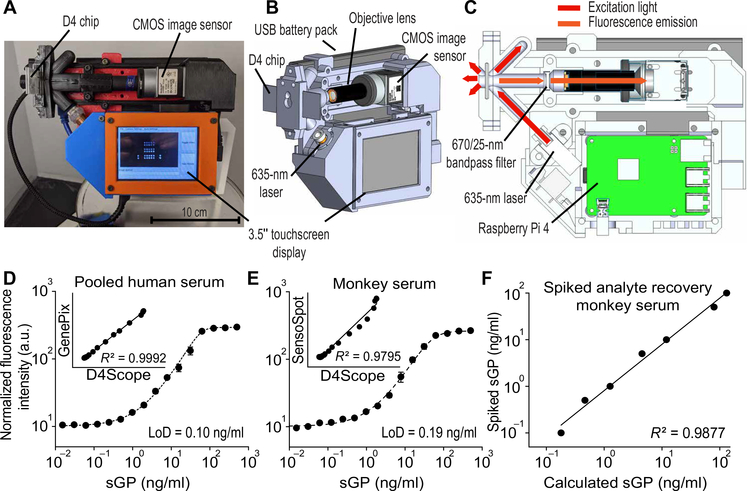
All EBOV D4 assay results described were obtained using a commercial table-top microarray scanner (GenePix), which is unsuitable for RDTs in low-resource settings. We initially attempted to use a cell phone image sensor as the detector; however, this method displayed a 20-fold decrease in sensitivity relative to the tabletop scanner (24). We therefore developed a low-cost, battery-powered, compact, wide-field fluorescence reader that can image microarray spots with high sensitivity, called the D4Scope (Fig. 4, A and B, and fig. S14). The D4Scope uses off-the-shelf components in a modular design, so that components can be easily replaced if a new wavelength is required for fluorescence imaging or if the reader is damaged in the field.
Fig. 4. The D4Scope, a handheld fluorescence detector.
(A) Photograph of the D4Scope. (B) Three-dimensional design, highlighting core components and computer-on-a-chip–based architecture, with an integrated touchscreen and CMOS camera. (C) D4Scope illumination scheme, depicting oblique angle laser excitation (at a 45° angle to the surface of the D4 chip), bandpass filter, and Raspberry Pi 4 processing unit. (D) Dose-response curve obtained by plotting fluorescence intensity measured by D4Scope versus concentration of EBOV sGP spiked into pooled HS. Inset: Comparison of fluorescence readout using a GenePix scanner and the D4Scope. (E) Dose-response curve for EBOV sGP spiked into rhesus MoS imaged by a technician using the D4Scope at the Galveston National Laboratory (GNL; BSL-4) after a 30-min training session. Inset: Comparison of fluorescence readouts from SensoSpot fluorescence scanner and D4Scope at GNL. (F) Double-blinded spiked analyte recovery experiment at GNL, with sGP concentrations in rhesus MoS determined by fluorescence intensity using D4Scope and SensoSpot scanner. D4 assay data point represents the mean ± SEM of N = 4 independent assays; blinded analyte recovery data points represent the mean of N = 3 technical replicates.
The fluorescence excitation in the D4Scope is set at an oblique angle (45°) relative to the assay chip and is achieved by collimating highly coherent red laser light onto the sample. The longer wavelength emission from the FL-dAb is filtered using a bandpass filter and is imaged onto a high-efficiency complementary metal-oxide semiconductor (CMOS) sensor for single snapshot capture without moving parts (Fig. 4C). The D4 scope has 21 cm by 16 cm by 9 cm dimensions, weighs 1.1 kg, and a single prototype costs less than $1000 USD to fabricate (table S1). In contrast, the commercial microarray scanner GenePix, a high-resolution nonconfocal tabletop fluorescence scanner, has dimensions of 43 cm by 65 cm by 34 cm, weighs 45 kg, and costs $150,000 USD. Despite its considerably smaller size and lower cost, the D4Scope displayed an LoD (0.10 ng/ml) (Fig. 4D) comparable to that of a commercial microarray scanner (GenePix; LoD, 0.07 ng/ml) (fig. S15A) and a near-perfect correlation (R2 = 0.9992) between its fluorescence readout and sGP concentration as measured by the GenePix when imaging a set of D4 assay chips tested with HS spiked with different concentrations of sGP.
We next evaluated the performance of the EBOV D4 assay at the Galveston National Laboratory (GNL) biosafety level 4 (BSL-4) facility. A GNL technician was instructed on the use of the assay in a 30-min training session. Figure 4E shows a dose-response curve generated with sGP-spiked rhesus MoS performed by the GNL technician. A double-blinded analyte recovery experiment was also performed in rhesus MoS by the GNL staff member, and sGP concentrations were determined on the basis of the measured fluorescence signal values, with R2 = 0.9877 (Fig. 4F). The D4Scope results (LoD, 0.19 ng/ml) were also compared on-site to results obtained using a commercial tabletop fluorescence imager (SensoSpot; LoD, 0.11 ng/ml) (fig. S15B). The SensoSpot and GenePix scanners were also compared by scanning the same set of D4 assay chips that were run with sGP-spiked FBS and showed excellence concordance in their fluorescence readouts with an R2 = 0.9977 (fig. S15, C to E). Collectively, these results demonstrate that the EBOV D4 assay can be performed using the D4Scope by a novice user after a brief training session.
sGP concentration throughout infection time course
We next validated the D4 assay for the early detection of EBOV infection using nonhuman primates challenged through intramuscular injection of EBOV (Makona), a well-established EBOV infection model (42, 43). Ten rhesus macaques were used. Baseline blood and serum samples were obtained from each animal before infection (day 0). Because of the need to fully anesthetize the animals to obtain samples and the strict constraint on blood volume drawn, only three samples were obtained from each animal during the course of the infection. The three collection time points were staggered to cover a 6-day period (Fig. 5A). For all time points, EBOV D4 assay fluorescence output was determined using the D4Scope and compared to plaque-forming units (PFU) obtained from plaque assays using Vero cells and viral genome copies, determined by RT-PCR with probes targeting the VP30 gene. Plaque assay and RT-PCR results for this set of samples are reported in (44) and further detailed in fig. S16 and table S2.
Fig. 5. Nonhuman primate model.
Healthy filovirus-negative rhesus macaques were challenged intramuscularly with 1000 PFU of EBOV Makona strain, n = 10. (A) Blood, plasma, and serum samples were collected at staggered time points during days 0 to 6 and tested using three assays: Serum samples were used on EBOV D4 assay, plasma samples were used for plaque assay with Vero cells, and blood samples were used in RT-PCR. The sample collection scheme and results are summarized in the panel. (B) Combined results for D4 assay, RT-PCR, and plaque assay. D4 assay: Normalized fluorescence intensity acquired with the D4Scope is reported for each time point. Each fluorescence value is the average of N = 3 technical replicates. Top: D4, PCR, and LFA results are summarized for each time point. (C) sGP concentration timeline determined using dose-response curve in Fig. 4E to convert fluorescence intensity to sGP concentration. Each data point shows the mean ± SD of sGP values for each time point. A statistically significant difference between sGP concentrations starts on day 3 (P < 0.0001; one-way ANOVA followed by Tukey’s post hoc test); values labeled with the same letter are significantly different. Inset: Western blot of serum samples from monkey 3 with sGP and GP as controls. (D) Correlation between sGP concentration and viremia determined by RT-PCR. Inset: Correlation of viral load and sGP concentration in the linear range of the EBOV D4 assay. (E) Correlation between fluorescence output measured by D4Scope and by SensoSpot scanner. Each data point is the mean ± SD of fluorescence values of each animal. RT-PCR LoD = ~103 viral genome copies/ml. Positive or negative results for PCR and D4 are color-coded. LFA results were estimated on the basis of infectious particle count results. “*” on day 4 (B) indicates one LFA with enough sensitivity to give positive results in 50% of the samples. PFU, plaque-forming units; UN, undetermined.
As the samples were previously characterized with an RT-PCR method developed in-house, we generated standard curves of cycle threshold (Ct) counts to assess the assay’s performance (fig. S17). The assay presented an LoD < 1000 viral genome copies/ml, a sensitivity that is comparable to the Xpert Ebola and other RT-PCR assays in the market (45).
All baseline (day 0) samples were negative by all assays. On day 1, one sample was positive by EBOV D4 assay and negative by the other two assays. On day 2, three samples were positive by EBOV D4 assay; two of these were negative by the other two assays, and one was negative by RT-PCR but presented a PFU of 8.3 by the plaque assay, which is above baseline albeit below the assay’s LoD of 25 PFU. All samples collected on day 3 or later were positive by all three assays, and fluorescence output intensities were statistically significant when compared to baseline values (P < 0.0001) (Fig. 5, A and B, fig. S16, and table S2). There was evidence of a hook effect at later time points in the EBOV D4 assay, as seen by the decreasing fluorescence intensity at days 5 and 6. Elevated PFU and genome copy counts indicate that this was due to high concentrations of sGP and GP1 associated with viremia in end-stage disease.
Samples tested positive by RT-PCR starting on day 3, 1 day later than by the EBOV D4 assay. The OraQuick Ebola and ReEBOV assays have sensitivities of ~1.0 × 106 PFU, based on WHO, FDA, and other reports (13, 39, 46, 47). In the EBOV (Makona) intramuscular model in our study, this PFU translates to day 5 of the infection, which is also when animals become symptomatic (44). The DPP Ebola Antigen System has a reported sensitivity of ~2.0 × 105 PFU (46), which translates to mixed results on day 4 in our study (one of the two samples had a PFU < 1.0 × 105 on day 4), but its evaluation with samples presenting low viremia indicated a sensitivity of only 56% (48). The LFA test using our A1F3–1 and C2BA5–2 Abs showed a sensitivity of ~1.0 × 106 viral genome copies, equivalent to ~3 × 104 PFU in the nonhuman primate model, a better sensitivity than that of current WHO- and FDA-approved RDTs.
sGP concentration in the rhesus macaques samples collected throughout the course of the infection were determined by converting D4 assay fluorescence intensity to analyte values using a dose-response curve with sGP-spiked rhesus MoS (Fig. 4E). sGP concentration increased on each day after infection, rising from picograms per milliliter on days 1 to 2, to nanograms per milliliter on Day 3, to micrograms per milliliter on day 4 (Fig. 5C). To confirm the presence of sGP in the samples, as cAb and dAb also bind GP1, serum samples from days 0, 3, and 6 of monkey 3 and sGP and GP controls were run on a Western blot (Fig. 5C and fig. S18). There was a strong linear correlation (R2 = 0.9921) between sGP concentration and viral load (determined by PCR) for sGP concentrations in the D4 assay’s linear range (Fig. 5D). The fluorescence signal measured by the D4Scope was in good agreement with fluorescence measurements using a tabletop fluorescence scanner (R2 = 0.9667) (Fig. 5E).
Following time-course studies in rhesus macaques, we also tested for sGP in a set of γ-irradiated samples from cynomolgus macaques. Three animals were intramuscularly challenged with 1000 PFU of Zaire EBOV. Blood and serum samples were collected at days 0 and 6 or 7 for each animal. Viral genome copy was determined by RT-PCR in blood samples and after viral neutralization by γ-irradiation (~5 mrad), serum samples were tested by the EBOV D4 assay, which indicated a statistically significant increase in fluorescence output (P < 0.0001) at later time points when compared to samples collected before viral challenge (fig. S19). sGP concentrations (table S3) were determined by interpolating a dose-response curve with spiked sGP in MoS (fig. S15B). sGP concentration in Makona and Zaire EBOV at similar time points were comparable.
DISCUSSION
Since the first major Ebola outbreak that devastated West Africa, public health experts have emphasized the need for rapid case ascertainment (49). The rapid diagnosis of Ebola infection facilitates triage and early treatment, reduces nosocomial transmission within health centers, and markedly improves the efficiency of contact tracing. In recent years, progress has been made on new diagnostics, therapies, and vaccines for EBOV (45, 50). Despite this, the speed at which infections can be identified and reported to health authorities remains suboptimal. In the 2018–2019 DRC epidemic, the median time for confirmation of infections during surveillance operations was 6 days, due, in large part, to the time required to get patients to testing sites, transport specimens to laboratories, and return results (51). Since 2014, the WHO has highlighted the urgent need for rapid POC EBOV diagnostic tests for use in decentralized health care facilities and has developed a target product profile for such assays (12). This need has been further underscored by the recent increase in size and frequency of EBOV outbreaks (52–54).
Here, we described the development of a rapid diagnostic immunoassay for EBOV that, in large measure, conforms to the desired WHO target product profile (12). The EBOV D4 assay is suitable for use in decentralized health care facilities where no laboratory infrastructure is available. Because the EBOV D4 displays a sensitivity comparable to and potentially better than that of RT-PCR, it should be capable of diagnosing acute EBOV infection in symptomatic patients without the need for confirmatory testing. Although the true clinical sensitivity of the EBOV D4 assay has not been determined, our studies in macaques suggest that it will be equal to or better than RT-PCR.
The EBOV D4 assay provides quantitative results and is designed to do so using capillary whole blood (WB) from a fingerstick. The EBOV D4 assay requires less than three steps to be performed by the operator and only one of these is timed. It requires no additional biosafety measures beyond personal protective equipment, although in its current embodiment, the assay requires a final wash step to be performed by the user. To address this limitation, D4 assay chips as self-contained passive microfluidic flow cells should be developed, which would require only the introduction of a drop of blood at one port and ~150 μl of wash buffer at a second port. In this design, all assay fluids could be collected by an internal wicking pad as the assay runs to completion. Microfluidic D4 assay chips are anticipated to provide improved biocontainment and biosafety. The EBOV D4 assay we describe here has no requirement for the transfer of precise volumes.
At 60 min, our current time to results does not meet the desired target time of less than 30 min but is well within the acceptable time of less than 3 hours. It is possible that shorter time to results can be achieved by incorporating cAbs and dAbs of higher affinity. D4 assay chips could also be constructed to include an internal control. Although, as we demonstrated here, samples can be treated with high concentrations of detergent and/or heat to allow viral inactivation, there is no need to process the sample before performing the test. In terms of operational characteristics, the EBOV D4 assay is heat stable, although it has not yet been tested against all WHO stability standards. It requires no reagent reconstitution, and even unskilled users can be trained in an hour. In terms of required equipment, the D4Scope, which is used to measure fluorescence output and interpret assay results, is a small portable handheld instrument weighing less than 2 kg that is capable of being powered by a rechargeable battery. Thus, although additional testing is required, the EBOV D4 assay appears capable of meeting almost all of the desired WHO benchmarks. It can also detect sGP from all clinically relevant EBOVs in human blood. This offers the advantage of using one assay to examine all potential cases of EBOV infection, irrespective of viral species, and could facilitate health care organizations’ responses to outbreaks (55).
As demonstrated, the current prototype EBOV D4 assay offers PCR-level sensitivity and specificity in a POC rapid diagnostic assay format. Hence, it has the potential to allow the accurate diagnosis EBOV infection in remote settings without the need for a confirmatory test. This could markedly improve the efficiency of surveillance and contact tracing operations by rapidly delivering clear disease diagnosis (56, 57). Its use could hence decrease the magnitude the future of EBOV outbreaks (4).
The EBOV D4 assay may also offer cost advantages over other diagnostic assay formats. The D4 required equipment cost is estimated to be less than $1000 USD for the reader, whereas the production costs of D4 assay chips at scale are expected to be around $0.25 USD (24). Both of these values are considerably less than those of RT-PCR–based diagnostics (3). Because LFAs require no equipment, their initial cost would be lower than that of the EBOV D4 assay; however, their production costs are higher, varying between $0.50 and $1 USD per unit (58, 59). Thus, the EBOV D4 assay would become more economical as the number of assays performed increases.
One characteristic of the D4 assay platform that we leveraged to conduct the functional Ab screen and pairing, but not in the final test, is the platform’s multiplexing capabilities. Several different Ab pairs can be printed on a single D4 assay chip, allowing the detection of multiple distinct Ags without loss of sensitivity (24). Thus, in principle, D4 chips could be produced to detect the presence of multiple pathogens in a single test. This, however, raises the question of whether the assay performance displayed by the EBOV D4 could be achieved in the case of the other infectious diseases. In our view, the sensitivity and specificity obtained with the EBOV D4 assay were due to four factors: the choice of sGP as a target, the affinity and specificity of the diagnostic Abs used, the optimization of cAb/dAb pairing, and the intrinsic sensitivity of the D4 assay platform. In many ways, EBOV sGP represents the ideal diagnostic target because it is secreted from infected cells in large amounts before viral replication occurs (60–63). Unfortunately, few viruses produce such targets. Lassa virus has been shown to produce sGP (64, 65), but other filoviruses, such as Marburgvirus, do not (66). D4 assays for other pathogens would therefore require the use of more traditional diagnostic targets. The clinical sensitivity for these targets that could be achieved in a D4 assay is not yet known but is expected to be better than that of LFAs.
Here, we used scFv phage display to generate our diagnostic Abs. This technology is widely applicable to other pathogens and is not unique in its ability to yield high-affinity Abs. Phage display offers two advantages in the production of diagnostic Abs. First, because scFv libraries can be subjected to positive selection for target Ags and negative selection against unwanted Ags, the specificities of the selected Abs can be tightly controlled. Second, because they are molecularly cloned, scFvs can be subjected to in vitro affinity maturation to increase Ab affinity by orders of magnitude (67, 68). Although the importance of cAb/dAb pairing is well recognized, identifying the optimal cAb/dAb pair can be extremely challenging and time-consuming when faced with a large number of candidate pairs. By inkjet printing Ab candidate pairs as microarrays, we were able to efficiently generate full dose-response curves for all possible Ab combinations. This allowed us to rapidly identify the optimum pair for use in the D4 format without the need for SPR analysis or epitope binning. The intrinsic sensitivity of the D4 assay platform has been described previously and is applicable to a wide variety of diagnostic applications (24, 69). Thus, we would expect that high-sensitivity D4 assays could be developed for a broad range of pathogens.
This study has limitations. In particular, the performance of the EBOV D4 assay in human samples is not yet determined, but the demonstration of sensitivity that rivals RT-PCR in nonhuman primates justifies further characterization with patient samples. Sample containment and the need for a user-assisted final rinse can also be problematic and may expose health care workers to biohazardous material, an important issue that may be overcome with improved biocontainment. The use of a VP30-targeting RT-PCR instead of the EBOV Xpert test is also a shortcoming of our study, as ideally, we would compare our test with the gold standard used in the field, even though both methods present similar analytical sensitivity (45). A direct comparison of the EBOV D4 and EBOV Xpert should be performed in future studies. sGP concentrations throughout infection time course have not been reported in humans although our study has demonstrated the temporal evolution of this biomarker in macaques. These results, however, must be confirmed with patient samples. Even with these caveats, however, this assay is promising for the POC diagnosis of EBOV disease.
MATERIALS AND METHODS
Study design
The goal of this study was to develop a new POC diagnostic system to detect EBOV sGP and demonstrate that EBOV nonstructural sGP protein is the ideal target for early disease diagnosis. Using phage display, we generated a new set of sGP-targeting Abs, which were later screened using the D4 assay as a selection and pairing tool. Fabrication parameters for the EBOV D4 assay using the ideal Ab pair were optimized, and the assay was evaluated with analyte spiked samples that simulate those generated in the field. The analytical performance of the test was evaluated with a tabletop and an in-house developed handheld fluorescence scanner. To demonstrate the presence of sGP and establish the evolution of this biomarker throughout the course of the infection, we selected samples previously collected and characterized in studies approved by the University of Texas Medical Branch (UTMB) at Galveston Institutional Animal Care and Use Committee (IACUC) and UTMB’s Institutional Biosafety Committee. A first set of samples from 10 male rhesus and cynomolgus macaques, 3 to 5 years of age, weighing between 4 and 8 kg that were intramuscularly challenged with EBOV isolate H.sapiens-tc/GIN/2014/Makona-Gueckedou-C07, accession number KJ660347.2. was used. This set had stacked samples that covered the initial stages of the disease when viremia is still undetected and the time point (day 3) where RT-PCR can detect the presence of viral genome in circulation. A second set of samples from three cynomolgus macaques, 3 to 5 years of age and weighing 4 to 8 kg challenged intramuscularly with Zaire EBOV from the 1995 outbreak (70, 71) was also used in this study. Samples were not blinded to investigators, unless specifically stated.
Generation of scFv-fc constructs
After immunizations, scFv library construction, and phage selection further detailed in the Supplementary Materials, eight scFv clones previously selected by Ag-down ELISA were subcloned into pcDNA5 (Thermo Fisher Scientific) fused to a mouse Fc gene for expression in a mammalian system. To this end, NEB 5-alpha competent Escherichia coli (New England Biolabs) with pcDNA5 plasmid was grown in ampicillin (Calbiochem) rich terrific broth (TB) media (Mo-Biolabs), and the plasmid was isolated with a QIAprep spin miniprep kit (QIAGEN), as recommended by the manufacturer. pcDNA5 plasmid was digested with Nhe I and Xho I (New England Biolabs) and gel purified. A gBlock DNA fragment (Integrated DNA Technology) encoding the mouse IgG2a Fc sequence, and appropriate overhangs was inserted in a pcDNA5 vector by Gibson Assembly (New England Biolabs), according to the manufacturer’s instructions. The resulting construct was then transformed into NEB 5-alpha competent cells (New England Biolabs), which were recovered with super optimal broth (SOC) media (MilliporeSigma) for 60 min at 37°C and later selected in TB (Mo-Biolabs) Agar (BD Biosciences) plates with ampicillin (50 μg/ml; Calbiochem). E. coli with pcDNA5 vector containing the Fc insert was grown and isolated, and the plasmid DNA was Sanger sequenced using appropriate primers (GENEWIZ). The plasmid was then restricted with Nhe I and Bam HI (New England Biolabs) and gel purified. gBlocks containing the eight scFv sequences (Integrated DNA Technology) were inserted in the Fc-pcDNA5 construct by Gibson Assembly. The resulting constructs were transformed into NEB 5-alpha competent cells and plated in ampicillin (Calbiochem) TB (Mo-Biolabs) Agar (BD Biosciences) plates. E. coli with pcDNA5 vector with scFv-Fc inserts were grown in rich TB media (Mo-Biolabs) with ampicillin (Calbiochem). The plasmids were isolated with QIAprep spin miniprep kits (QIAGEN) according to the manufacturer’s instructions, and the plasmid DNA was Sanger sequenced to ensure lack of point mutations and frame shifts.
Generation of IgG constructs
A1F3–1 and C2BA5–2 scFv-Fcs were transformed into full-length IgGs. gBlocks (Integrated DNA Technology) containing constant regions for mouse γ2a heavy chain and mouse k chains with appropriate overhangs were inserted in pcDNA5 vector through Gibson Assembly as recommended by the manufacturer instructions (Gibson Assembly Master Mix, New England Biolabs). Using the same procedure, gBlocks containing variable heavy and mouse γ2a immunoglobulin sequences for A1F3–1 and C2BA5–2 were also inserted in pcDNA5 (Thermo Fisher Scientific) vector through Gibson Assembly. The resulting constructs were then transformed into NEB 5-alpha competent cells (New England Biolabs), which were recovered with SOC media (MilliporeSigma) for 60 min at 37°C and later selected in TB (Mo-Biolabs) Agar (BD Biosciences) plates with ampicillin (50 μg/ml; Calbiochem). E. coli containing pcDNA5 vectors with inserts of interest were grown, isolated, and sequenced as previously described.
Expression of scFv-fc and IgG Abs
scFv-Fc and IgG Ab expression was performed with an Expi293 high-efficiency transient system (Thermo Fisher Scientific). Purified plasmids for each of the scFv-Fc and light and heavy chains of A1F3–1 and C2BA5–2 were retransformed into NEB 5-alpha competent cells, plated, and cultured overnight in ampicillin (Calbiochem) TB (Mo-Biolabs) Agar (BD Biosciences) plates. Isolated colonies were grown in ampicillin (Calbiochem) rich TB media (Mo-Biolabs) and DNA plasmid was purified using QIAprep spin miniprep plus kits (QIAGEN), as recommended by the manufacturer’s protocol. Purified DNA was Sanger sequenced (GENEWIZ) to guarantee correct expression products and transfected into Expi293 cells (Thermo Fisher Scientific), also following the manufacturer’s protocol. IgGs had plasmids with light and heavy chain cotransfected at 1:1.2 heavy to light chain molar ratios to a total of 1 μg of DNA/ml of culture. Culture supernatant was harvested 7 days after induction for Ab purification.
High-throughput Ab pairing
Following POEGMA brush synthesis described in the Supplementary Materials, D4 assays were fabricated as reported elsewhere in greater detail (24, 29), with minor modifications. Down-selected scFv-Fc Abs were directly conjugated to Alexa Fluor 647 (Thermo Fisher Scientific) following the manufacturers’ instructions and diluted with a trehalose (MilliporeSigma) stock solution in 1× phosphate-buffered saline (PBS) (MilliporeSigma) to a final 10% (w/v) trehalose (MilliporeSigma), Ab working concentration of 0.25 mg/ml. Aliquots of the scFv-Fcs to be tested as the cAb had trehalose (MilliporeSigma) added to a working concentration of 0.05% (w/v) in 1× PBS (MilliporeSigma). cAbs were inkjet printed as a vertical column of ~130-μm diameter spots in the central area of the chip with an interspot separation of 250 μm, and the FL-dAb with 10% (v/v) trehalose (MilliporeSigma) was also noncontact dispensed on a concentric pattern around the central area of the chip containing the cAbs with the sciFLEXARRAYER S11 spotter (Scienion). After printing, the chips were protected from light and left to dry in a vacuum chamber (-25 inch Hg) lined with desiccant material for 1 hour. After fabrication, dose-response curves were generated by incubating the D4 microarrays with FBS (Avantor) spiked with EBOV sGP at different concentrations for 90 min. After incubation, the chips were rinsed with 0.1% Tween 20 (v/v) (MilliporeSigma) in 1× PBS (MilliporeSigma) and spun dry with a C1303 slide spinner (Labnet International). Arrays were imaged and quantified with an Axon Genepix 4400 (Molecular Devices) with a photomultiplier gain of 750 and excitation power of 100. LoD was determined by using low-concentration samples (LCS) and the mathematical formula LoD = LoB + 1.645σLCS, where limit of blank (LoB) is determined by measuring the mean fluorescence intensity (μ) and SD (s) from 12 blank samples and using the definition LoB = μblank + 1.645δblank. DR was determined by the range of concentration from the LoD to the greatest concentration that had a fluorescent signal greater than 3σ of the next lowest concentration in the dilution series. Data were fitted with a five-parameter logistic curve using OriginPro 9.0 (OriginLab Corp).
Scalable D4 assay fabrication
To reduce D4 assay fabrication time, improve dissolution of the FL-dAb, and to minimize HAMA cross-reactivity, the manufacturing process used to fabricate the EBOV D4 assay chips was modified from that used for high-throughput Ab pairing in this study and reported in previous publications (24, 29). The first step in the fabrication of the D4 assay chip deviates significantly for the original protocol as follows. Before printing the cAb spots, trehalose pads are printed on the surface of the POEGMA substrate followed by printing the dAb and blocking reagents on top of the trehalose pads (Figs. 2 to 4 and figs. S10 to S12). The trehalose pads were inkjet printed from a 10% trehalose solution in deionized water as a 12-spot concentric pattern around the central region of the chip where the cAb is printed (Fig. 2A) with an AD1520 (BioDot) noncontact dispensing platform that allows nanoliter-sized drops to be dispensed—compared to the picoliter drop size of the Scienion printer—of highly viscous trehalose and dAb solutions. This reduced the number of dAb/blocking reagent drops and arm movements needed to fabricate a chip compared to the previous fabrication protocol of the D4 assay that solely used the Scienion sciEFLEXARRAYER S11 arrayer, resulting in a 10-fold increase in fabrication throughput (29). After printing, the trehalose pads were left to dry for 10 min at ambient conditions. Next, a solution of the FL-dAb at 0.05 mg/ml in a 10% trehalose (w/v) (MilliporeSigma) and 1× PBS (MilliporeSigma) solution was printed as nanoliter droplets using AD1520 (BioDot) arrayer and was then dried under ambient conditions for 10 min. Next, a solution of Trublock Ultra (Meridian Lifescience) at 12 mg/ml in 10% trehalose (w/v) (MilliporeSigma) and 1× PBS (MilliporeSigma) was also printed as nanoliter droplets on top of FL-dAbs also using the AD1520 (BioDot) arrayer. After the detection spots were printed, the protocol follows the previously reported process, where the cAb is inkjet printed as 350-pl droplets on the surface of a POEGMA-coated glass slide from a 1 mg/ml in 0.05% (w/v) trehalose in 1× PBS solution using a sciFLEXARRAYER S11 (Scienion). The capture spots are left to dry at ambient conditions for 10 min. Next, we assembled laser-cut 1-mm-thick Plexiglas (Astra Products) with double-sided 9474LE adhesive (3M) to form gaskets and create 24 separate D4 assay chips on a single POEGMA-coated glass slide. Chips are then stored in aluminum pouches with 0.5-g silica bags (EASE Medtrend) for later use.
Performance evaluation with multiple sGPs, matrix effect, hook effect, and thermal stability evaluation of EBOV D4 assay
The D4 assay chips were incubated with sGP from EBOV, SUDV, BDBV, RESTV, and TAFV spiked in FBS (Avantor) for 90 min to generate dose-response curves for each variant of sGP. For matrix effect assessment, D4 assay chips were incubated with different concentrations of EBOV sGP spiked in FBS (Avantor), pooled HS (Innovative Research), single donor human blood (Innovative Research), and rhesus MoS (Innovative Research) for 15, 60, and 90 min. sGP-spiked FBS was also incubated for 30 min. Hook effect evaluation was performed by generating an 8-log10 dose-response curve with a twofold dilution series from a stock concentration of EBOV sGP (0.1 mg/ml). D4 assay chips were also incubated for 60 min with sGP from SUDV, BDBV, and RESTV spiked into single donor human blood (Innovative Research). Single donor human blood (Innovative Research) spiked with different concentrations of EBOV and SUDV was treated with a 10% (v/v) Triton X-100 (MilliporeSigma) solution to a final concentration of 1% (v/v). Samples were incubated for 60 min at room temperature. Treated samples were then added to D4 assay chips. Single donor human blood (Innovative Research) spiked with different concentrations of EBOV was treated with a 10% (v/v) Triton X-100 (MilliporeSigma) solution to a final concentration of 1% (v/v). Samples were incubated for 30 min at 56°C. Treated samples were then added to D4 assay chips. After incubation, chips were rinsed, dried, and imaged on an Axon Genepix 4400 (Molecular Devices) with a photomultiplier gain of 750 and excitation power of 100. Figures of merit (FOMs)—the LoD and DR—were determined as previously described.
Accelerated stability testing was performed with chips that were individually packaged in heat-sealed aluminum pouches (EASE-Medtrend) with silica desiccant (EASE-Medtrend) and incubated at 37°C. Samples were tested at 30 and 60 days later by incubating chips with EBOV-spiked sGP in pooled HS for 90 min. After incubation, chips were rinsed, dried, and imaged on an Axon Genepix 4400 (Molecular Devices) with a photomultiplier gain of 750 and excitation power of 100. The LoD and DR were determined as previously described.
D4Scope fabrication and performance validation
The D4Scope’s fluorescence elements were mounted in an oblique angle laser excitation format in a three-dimensional (3D) printed body designed with SOLIDWORKS software (SOLIDWORKS) and fabricated with a Lulzbot Taz 6 (Lulzbot) 3D printer using 1.75-mm polylactic acid filament (HATCHBOX). The red 185-mW 635-nm laser diode (Sharp) with high optical coherence in the fluorophore’s excitation band (594 to 633 nm) is set at a 45° angle and excites the Alexa Fluor 647 fluorophore on the EBOV D4 assay surface placed in a chip holder that attaches to the D4Scope. The photons emitted by the fluorophore are filtered by a F01–676/37 25-mm bandpass optical filter (SHEMROCK) embedded in the camera’s field of view and are captured by a high-efficiency USB compatible AcA3088–57um CMOS camera (Basler) with a MC100X lens (Opto Engineering). To control the camera, and process and visualize the acquired images, a Raspberry Pi 4 (Raspberry) system-on-board computer with a 3.5-inch thin-film transistor touchscreen (UCTRONICST) was integrated into the detector. Custom software was written to control the camera’s exposure times, gain, and digital shift. Images are acquired by the user by a one-touch button, which activates the laser and records an image of the microspots, and the image and associated patient information are stored in the device and uploaded to a dedicated cloud Mango/DB server. The software also previews the images of the chips while they are being loaded on the device, thus ensuring proper use of the device. The entire system is powered by a 10,000-mAh power bank (Omars) mounted on the D4Scope.
To assess detector performance, optimized chips were exposed to EBOV sGP spiked in HS (Innovative Research) for 90 min, rinsed, dried, and scanned with an Axon Genepix with 750 gain and 100% power. The same chips were then loaded on the D4Scope and imaged with 1-s exposure, digital shift of 4- and 12-dB gain. Fluorescence values were normalized using the formula Fnorm. = ((Fm - Fblank)/F20 ng/ml) + cste where Fnorm. is the normalized fluorescence, Fm is the measured fluorescence, Fblank is the average fluorescence of blank samples, F20 ng/ml is the fluorescence of 20 ng/ml, and cste = 10. A set of chips was also transported to UTMB BSL-4 facility, where a technician was trained for about 30 min to run the EBOV D4 assay and operate the D4Scope. After training, EBOV D4 chips were exposed to EBOV sGP spiked in MoS (Innovative Research) for 90 min, rinsed, dried, and scanned with the D4Scope and a SensoSpot (Sensovation) fluorescence scanner with 1-s exposure.
Compatibility between the tabletop scanners—the Axon Genepix and the SensoSpot—and the D4Scope was ensured by scanning a set of EBOV D4 assay chips incubated with EBOV sGP in FBS (Avantor) for 90 min with the Genepix 4400 (Molecular Devices) and SensoSpot (Sensovation) fluorescence tabletop scanners. Dose-response curves were fit with a five-parameter logistic fit, and the output correlation among tabletop scanners and between tabletop scanners and D4Scope was determined by a linear fit. All fits and plots were performed using OriginPro 9.0 (OriginLab Corp).
Virus and challenge
The serum samples used in this work were previously collected and characterized in studies approved by the UTMB at Galveston IACUC and UTMB’s Institutional Biosafety Committee (44). In summary, a seed stock of EBOV Makona from a 2014 fatal human case that originated in Guékédou, Guinea was used. After passage in authenticated Vero E6 cells (American Type Culture Collection, CRL-1586), EBOV isolate H.sapiens-tc/GIN/2014/Makona-Gueckedou-C07, accession number KJ660347.2 was produced. This isolate was used to challenge 10 healthy, filovirus-negative, male rhesus and cynomolgus macaques of Chinese origin (PreLabs) that were 3 to 5 years of age and weighed between 4 and 8 kg with 1000 PFU intramuscularly. A smaller subset of three healthy, filovirus-seronegative adult cynomolgus macaques of Chinese origin (PreLabs) that were 3 to 5 years of age and weighed between 4 and 8 kg were challenged intramuscularly with 1000 PFU of EBOV (Zaire). The EBOV used in this study was obtained from a fatally infected human from the former Zaire in 1995 (70, 71). Animals were housed in the BSL-4 laboratory in the GNL and monitored postchallenge for clinical signs of disease.
Virus detection and quantification
For the 10 rhesus macaques challenged with Makona EBOV, viral titer was determined by plaque assay on Vero E6 cells from plasma samples, and RNA quantification was carried out by RT-PCR with probes targeting the VP30 gene (42) from blood samples. As reported elsewhere in greater detail (44), cells were plated and grown to confluency and virus was titrated in duplicate from 10−1 to 10−6 and counted with neutral red stain. RNA was isolated from WB using AVL buffer and Viral RNA Mini Kit (QIAGEN) using 100 ml of blood and 600 ml of AVL buffer. Primers targeting the EBOV VP30 gene were used in the RT-PCR assay (72) to detect EBOV RNA on a CFX96 (BioRad Laboratories) PCR instrument, using a One-Step Probe RT-PCR kit (QIAGEN) with a cycle of 50°C for 10 min, 95°C for 10 s, and 40 cycles of 95°C for 10 s, followed by 59°C for 30 s. Ct values representing EBOV genome equivalents (GEq) were analyzed with CFX Manager Software (BioRad Laboratories). A standard for the GEq was created from EBOV RNA stocks using the formula Ncopies (molecules) = (Namount × 6.0221 × 1023 (molecules/mole)/(Nlength × 1 × 109 (ng/g) × 340 (g/mole)), where Ncopies is the number of RNA copies, Namount is the length of RNA molecule (19 kb) (73), 340 g/mole = average mass of 1 bp of RNA, and the molecular weight of the EBOV genome (6,086,991.8 g/mol). For comparison purposes, we also evaluated the performance of RT-PCR targeting the GP gene with the AGA CAG CTG GCC AAC GAG AC forward and TCG CTG CAG CAA GAA ATC AA as reverse probes (fig. S17). Plaque assays (74) had an LoD of 25 PFU/ml and the LoD of RT-PCR was 1000 genome copies/ml. Three cynomolgus macaques challenged with Zaire EBOV had viral titers also quantified by RT-PCR from WB as previously described.
sGP detection in nonhuman primate models
Previously characterized samples from 10 rhesus macaques challenged with Makona EBOV were run on the EBOV D4 assay. Chips were imaged with the D4Scope and SensoSpot fluorescence scanners, as previously described. Each data point of fluorescence intensity reported is the average of three technical replicates. sGP levels were calculated from the dose-response curve in Fig. 4E. Samples from three cynomolgus macaques challenged with Zaire EBOV were run on the EBOV D4 assay after being irradiated with ~5 mrad. Each data point of fluorescence intensity reported results from a single measurement. sGP concentrations were calculated from the dose-response curve in fig. S15B.
Statistical analysis
Statistical analysis was performed with GraphPad Prism version 6.01 (GraphPad Software Inc). A one-way analysis of variance (ANOVA) followed by Tukey’s post hoc multiple comparisons test was used to evaluate the statistical significance of differences between groups. Raw data are provided in data file S1.
Supplementary Material
Fig. S1. Schematic depiction of EBOV D4 assay.
Fig. S2. D4 assay procedure.
Fig. S3. Mouse IgG titers for sGP and reactivity of individual scFv-phage clones for EBOV and SUDV sGP.
Fig. S4. scFv-Fc Ab expression, purification, and activity.
Fig. S5. Binding constants of scFv-Fc Abs for sGP.
Fig. S6. Ab pairing dose-response curves.
Fig. S7. Expression and purification of A1F3–1 and C2BA5–2 monoclonal IgG2a Abs.
Fig. S8. Binding constants of A1F3–1 (cAb) and C2BA5–2 (dAb) with multiple sGPs.
Fig. S9. Binding profiles of other scFv-Fc Abs with sGP.
Fig. S10. EBOV D4 assay performance.
Fig. S11. Post-assay fluorescence stability.
Fig. S12. EBOV D4 assay performance in human blood.
Fig. S13. EBOV sGP lateral flow assay fabrication.
Fig. S14. D4Scope design.
Fig. S15. Comparison of D4Scope, GenePix, and SensoSpot performance.
Fig. S16. RT-PCR and plaque assay.
Fig. S17. VP30- and GP-targeting RT-PCR.
Fig. S18. Western blot of MoS samples.
Fig. S19. Irradiated samples from EBOV-challenged nonhuman primates.
Table S1. D4Scope cost estimate.
Table S2. Summary of nonhuman primate model results.
Table S3. Summary of irradiated samples from EBOV-challenged nonhuman primates.
Acknowledgments:
We thank T. Hoenen at Rocky Mountain Laboratories for providing EBOV sGP (Mayinga) expression vector. We also thank B.8Watts at the Duke University Vaccine Institute for the assistance with SPR and R. Gordon for the GenePix usage.
Funding: The authors acknowledge funding from the NIH grant R01AI150888 to A.C. and U.S. Special Operations Command (contract number W81XWH-16-C-0219) to A.C. Partial support for some filovirus samples was provided by the Department of Health and Human Services, NIH grant U19AI142785 to T.W.G. and UC7AI094660 for BSL-4 operations support of the GNL. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the University of Texas Medical Branch or the NIH. C.M.F. also acknowledges financial support from the National Council for the Improvement of Higher Education (CAPES) and the Science without Borders project
Footnotes
Competing interests: The underlying technology of the D4 was developed by A.H. and A.C. and acquired by Immucor Inc. (Norcross, GA) in 2014. The other authors declare that they have no competing financial interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. Computer code that controls the D4Scope has been uploaded to Zenodo under DOI 10.5281/zenodo.4536655.
REFERENCE AND NOTES
- 1.Dyer O, Escalating Congo Ebola epidemic passes 2000 cases amid violence and suspicion. BMJ 365, l4062 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Maxmen A, Violence propels Ebola outbreak towards 1,000 cases. Nature 567, 153–154 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Cnops L, De Smet B, Mbala-Kingebeni P, van Griensven J, Ahuka-Mundeke S, Arien KK, Where are the Ebola diagnostics from last time? Nature 565, 419–421 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Nouvellet P, Garske T, Mills HL, Nedjati-Gilani G, Hinsley W, Blake IM, Van Kerkhove MD, Cori A, Dorigatti I, Jombart T, Riley S, Fraser C, Donnelly CA, Ferguson NM, The role of rapid diagnostics in managing Ebola epidemics. Nature 528, S109–S116 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulangu S, Dodd LE, Davey RT Jr., Tshiani Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D, Tshomba Oloma A, Ibanda A, Ali R, Coulibaly S, Levine AC, Grais R, Diaz J, Lane HC, Muyembe-Tamfum J-J; PALM Writing Group, Sivahera B, Camara M, Kojan R, Walker R, Dighero-Kemp B, Cao H, Mukumbayi P, Mbala-Kingebeni P, Ahuka S, Albert S, Bonnett T, Crozier I, Duvenhage M, Proffitt C, Teitelbaum M, Moench T, Aboulhab J, Barrett K, Cahill K, Cone K, Eckes R, Hensley L, Herpin B, Higgs E, Ledgerwood J, Pierson J, Smolskis M, Sow Y, Tierney J, Sivapalasingam S, Holman W, Gettinger N, Vallée D, Nordwal J; PALM Consortium Study Team, A randomized, controlled trial of Ebola virus disease therapeutics. N. Eng. J. Med. 381, 2293–2303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Towner JS, Rollin PE, Bausch DG, Sanchez A, Crary SM, Vincent M, Lee WF, Spiropoulou CF, Ksiazek TG, Lukwiya M, Kaducu F, Downing R, Nichol ST, Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J. Virol. 78, 4330–4341 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broadhurst MJ, Brooks TJG, Pollock NR, Diagnosis of Ebola virus disease: Past, present, and future. Clin. Microbiol. Rev. 29, 773–793 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler D, Speedy Ebola tests help contain Africa’s latest outbreak. Nature 558, 172 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Raftery P, Condell O, Wasunna C, Kpaka J, Zwizwai R, Nuha M, Fallah M, Freeman M, Harris V, Miller M, Baller A, Massaquoi M, Katawera V, Saindon J, Bemah P, Hamblion E, Castle E, Williams D, Gasasira A, Nyenswah T, Establishing Ebola virus disease (EVD) diagnostics using GeneXpert technology at a mobile laboratory in Liberia: Impact on outbreak response, case management and laboratory systems strengthening. PLOS Negl. Trop. Dis. 12, e0006135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen van Vuren P, Grobbelaar A, Storm N, Conteh O, Konneh K, Kamara A, Sanne I, Paweska JT, Comparative evaluation of the diagnostic performance of the prototype cepheid GeneXpert Ebola assay. J. Clin. Microbiol. 54, 359–367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinsky BA, Sahoo MK, Sandlund J, Kleman M, Kulkarni M, Grufman P, Nygren M, Kwiatkowski R, Baron EJ, Tenover F, Denison B, Higuchi R, Van Atta R, Beer NR, Carrillo AC, Naraghi-Arani P, Mire CE, Ranadheera C, Grolla A, Lagerqvist N, Persing DH, Analytical performance characteristics of the cepheid GeneXpert Ebola assay for the detection of Ebola virus. PLOS ONE 10, e0142216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization, Target Product Profile for Zaïre ebolavirus Rapid, Simple Test to Be Used in the Control of the Ebola Outbreak in West Africa (2014); https://www.who.int/medicines/publications/target-product-profile.pdf
- 13.VanSteelandt A, Aho J, Franklin K, Likofata J, Kamgang JB, Keita S, Koivogui L, Magassouba NF, Martel LD, Dahourou AG, Operational evaluation of rapid diagnostic testing for Ebola virus disease in Guinean laboratories. PLOS ONE 12, e0188047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boisen ML, Cross RW, Hartnett JN, Goba A, Momoh M, Fullah M, Gbakie M, Safa S, Fonnie M, Baimba F, Koroma VJ, Geisbert JB, McCormick S, Nelson DKS, Millett MM, Oottamasathien D, Jones AB, Pham H, Brown BL, Shaffer JG, Schieffelin JS, Kargbo B, Gbetuwa M, Gevao SM, Wilson RB, Pitts KR, Geisbert TW, Branco LM, Khan SH, Grant DS, Garry RF, Field validation of the ReEBOV antigen rapid test for point-of-care diagnosis of Ebola virus infection. J. Infect. Dis. 214, S203–S209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makiala S, Mukadi D, De Weggheleire A, Muramatsu S, Kato D, Inano K, Gondaira F, Kajihara M, Yoshida R, Changula K, Mweene A, Mbala-Kingebeni P, Muyembe-Tamfum JJ, Masumu J, Ahuka S, Takada A, Clinical evaluation of QuickNaviTM-Ebola in the 2018 outbreak of Ebola virus disease in the Democratic Republic of the Congo. Viruses 11, 589 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wonderly B, Jones S, Gatton ML, Barber J, Killip M, Hudson C, Carter L, Brooks T, Simpson AJH, Semper A, Urassa W, Chua A, Perkins M, Boehme C, Comparative performance of four rapid Ebola antigen-detection lateral flow immunoassays during the 2014–2016 Ebola epidemic in West Africa. PLOS ONE 14, e0212113 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy CN, Recent advances in the diagnosis and management of Ebola virus disease. Clin. Microbiol. Newslett. 41, 185–189 (2019). [Google Scholar]
- 18.Koch LK, Cunze S, Kochmann J, Klimpel S, Bats as putative Zaire ebolavirus reservoir hosts and their habitat suitability in Africa. Sci. Rep. 10, 14268 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob ST, Crozier I, Fischer II WA, Hewlett A, Kraft CS, de La Vega M-A, Soka MJ, Wahl V, Griffiths A, Bollinger L, Kuhn JH, Ebola virus disease. Nat. Rev. Dis. Primers. 6, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma H, Hyun J, Stiller P, Chilkoti A, “Non-fouling” oligo(ethylene glycol)-functionalized polymer brushes synthesized by surface-initiated atom transfer radical polymerization. Adv. Mater. 16, 338–341 (2004). [Google Scholar]
- 21.Ma H, Li D, Sheng X, Zhao B, Chilkoti A, Protein-resistant polymer coatings on silicon oxide by surface-initiated atom transfer radical polymerization. Langmuir 22, 3751–3756 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Ma H, Wells M, Beebe TP, Chilkoti A, Surface-initiated atom transfer radical polymerization of oligo(ethylene glycol) methyl methacrylate from a mixed selfassembled monolayer on gold. Adv. Funct. Mater. 16, 640–648 (2006). [Google Scholar]
- 23.Hucknall A, Kim D-H, Rangarajan S, Hill RT, Reichert WM, Chilkoti A, Simple fabrication of antibody microarrays on nonfouling polymer brushes with femtomolar sensitivity for protein analytes in serum and blood. Adv. Mater. 21, 1968–1971 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joh DY, Hucknall AM, Wei Q, Mason KA, Lund ML, Fontes CM, Hill RT, Blair R, Zimmers Z, Achar RK, Tseng D, Gordan R, Freemark M, Ozcan A, Chilkoti A, Inkjet-printed point-of-care immunoassay on a nanoscale polymer brush enables subpicomolar detection of analytes in blood. Proc. Natl. Acad. Sci. U.S.A. 114, E7054–E7062 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volchkov VE, Chepurnov AA, Volchkova VA, Ternovoj VA, Klenk HD, Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology 277, 147–155 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Misasi J, Sullivan NJ, Camouflage and misdirection: The full-on assault of Ebola virus disease. Cell 159, 477–486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST, The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. U.S.A. 93, 3602–3607 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez A, Ksiazek TG, Rollin PE, Miranda MEG, Trappier SG, Khan AS, Peters CJ, Nichol ST, Detection and molecular characterization of Ebola viruses causing disease in human and nonhuman primates. J Infect Dis 179, S164–S169 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Fontes CM, Achar RK, Joh DY, Ozer I, Bhattacharjee S, Hucknall A, Chilkoti A, Engineering the surface properties of a zwitterionic polymer brush to enable the simple fabrication of inkjet-printed point-of-care immunoassays. Langmuir 35, 1379–1390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hucknall A, Rangarajan S, Chilkoti A, In pursuit of zero: Polymer brushes that resist the adsorption of proteins. Adv. Mater. 21, 2441–2446 (2009). [Google Scholar]
- 31.Ma H, Hyun J, Zhang Z, Beebe TP, Chilkoti A, Fabrication of biofunctionalized quasi-three-dimensional microstructures of a nonfouling comb polymer using soft lithography. Adv. Funct. Mater. 15, 529–540 (2005). [Google Scholar]
- 32.Hucknall A, Simnick AJ, Hill RT, Chilkoti A, Garcia A, Johannes MS, Clark RL, Zauscher S, Ratner BD, Versatile synthesis and micropatterning of nonfouling polymer brushes on the wafer scale. Biointerphases 4, FA50–FA57 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Pande J, Szewczyk MM, Grover AK, Phage display: Concept, innovations, applications and future. Biotechnol. Adv. 28, 849–858 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Chen Y-H, Lipes BD, Kenan DJ, Staats HF, Gunn MD, Identification of recombinant antibodies against multiple distinct toll-like receptors by homolog mining a single immune scFv phage library. J. Immunol. Methods 340, 144–153 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madan B, Chaudhary G, Cramer SM, Chen W, ELP-z and ELP-zz capturing scaffolds for the purification of immunoglobulins by affinity precipitation. J. Biotechnol. 163, 10–16 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Colavita F, Quartu S, Lalle E, Bordi L, Lapa D, Meschi S, Vulcano A, Toffoletti A, Bordi E, Paglia MG, Di Caro A, Ippolito G, Capobianchi MR, Castilletti C, Evaluation of the inactivation effect of Triton X-100 on Ebola virus infectivity. J. Clinical Virol. 86, 27–30 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Burton JE, Easterbrook L, Pitman J, Anderson D, Roddy S, Bailey D, Vipond R, Bruce CB, Roberts AD, The effect of a non-denaturing detergent and a guanidiniumbased inactivation agent on the viability of Ebola virus in mock clinical serum samples. J. Virol. Methods 250, 34–40 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Haddock E, Feldmann F, Feldmann H, Effective chemical inactivation of Ebola virus. Emerg. Infect. Dis. 22, 1292–1294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cross RW, Boisen ML, Millett MM, Nelson DS, Oottamasathien D, Hartnett JN, Jones AB, Goba A, Momoh M, Fullah M, Bornholdt ZA, Fusco ML, Abelson DM, Oda S, Brown BL, Pham H, Rowland MM, Agans KN, Geisbert JB, Heinrich ML, Kulakosky PC, Shaffer JG, Schieffelin JS, Kargbo B, Gbetuwa M, Gevao SM, Wilson RB, Saphire EO, Pitts KR, Khan SH, Grant DS, Geisbert TW, Branco LM, Garry RF, Analytical validation of the ReEBOV antigen rapid test for point-of-care diagnosis of Ebola virus infection. J Infect Dis 214, S210–S217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tembo J, Simulundu E, Changula K, Handley D, Gilbert M, Chilufya M, Asogun D, Ansumana R, Kapata N, Ntoumi F, Ippolito G, Zumla A, Bates M, Recent advances in the development and evaluation of molecular diagnostics for Ebola virus disease. Expert Rev. Mol. Diagn. 19, 325–340 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Couturier C, Wada A, Louis K, Mistretta M, Beitz B, Povogui M, Ripaux M, Mignon C, Werle B, Lugari A, Pannetier D, Godard S, Bocquin A, Mely S, Béavogui I, Hébélamou J, Leuenberger D, Leissner P, Yamamoto T, Lécine P, Védrine C, Chaix J, Characterization and analytical validation of a new antigenic rapid diagnostic test for Ebola virus disease detection. PLOS Negl. Trop. Dis. 14, e0007965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mire CE, Geisbert JB, Agans KN, Deer DJ, Fenton KA, Geisbert TW, Oral and conjunctival exposure of nonhuman primates to low doses of Ebola Makona virus. J. Infect. Dis. 214, S263–S267 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geisbert TW, Strong JE, Feldmann H, Considerations in the use of nonhuman primate models of Ebola virus and marburg virus infection. J. Infect. Dis. 212 (Suppl 2), S91–S97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Versteeg K, Menicucci AR, Woolsey C, Mire CE, Geisbert JB, Cross RW, Agans KN, Jeske D, Messaoudi I, Geisbert TW, Infection with the Makona variant results in a delayed and distinct host immune response compared to previous Ebola virus variants. Sci. Rep. 7, 9730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loftis AJ, Quellie S, Chason K, Sumo E, Toukolon M, Otieno Y, Ellerbrok H, Hobbs MM, Hoover D, Dube K, Wohl DA, Fischer II WA, Validation of the Cepheid GeneXpert for detecting Ebola virus in semen. J. Infect. Dis. 215, 344–350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Food and Drug Administration, Emergency Use Authorizations for Medical Devices (2020); https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations-medical-devices
- 47.World Health Organization, Ebola Vaccines, Therapies, and Diagnostics (2015); https://www.who.int/medicines/emp_ebola_q_as/en/
- 48.Moran Z, Rodriguez W, Ahmadou D, Soropogui B, Magassouba NF, Kelly-Cirino C, Ben Amor Y, Comparative performance study of three Ebola rapid diagnostic tests in Guinea. BMC Infect. Dis. 20, 670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewnard JA, Ndeffo Mbah ML, Alfaro-Murillo JA, Altice FL, Bawo L, Nyenswah TG, Galvani AP, Dynamics and control of Ebola virus transmission in Montserrado, Liberia: A mathematical modelling analysis. Lancet Infect. Dis. 14, 1189–1195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feldmann H, Sprecher A, Geisbert TW, Ebola. N. Eng. J. Med. 382, 1832–1842 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Ilunga Kalenga O, Moeti M, Sparrow A, Nguyen V-K, Lucey D, Ghebreyesus TA, The ongoing Ebola epidemic in the Democratic Republic of Congo, 2018–2019. N. Eng. J. Med. 381, 373–383 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Munster VJ, Bausch DG, de Wit E, Fischer R, Kobinger G, Muñoz-Fontela C, Olson SH, Seifert SN, Sprecher A, Ntoumi F, Massaquoi M, Mombouli J-V, Outbreaks in a rapidly changing central Africa—Lessons from Ebola. N. Eng. J. Med. 379, 1198–1201 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Malvy D, McElroy AK, de Clerck H, Günther S, van Griensven J, Ebola virus disease. Lancet 393, 936–948 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Redding DW, Atkinson PM, Cunningham AA, Lo Iacono G, Moses LM, Wood JLN, Jones KE, Impacts of environmental and socio-economic factors on emergence and epidemic potential of Ebola in Africa. Nat. Commun. 10, 4531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shuchman M, Logistical challenges in the DR Congo Ebola virus response. Lancet 393, 117–118 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Building trust is essential to combat the Ebola outbreak. Nature 567, 433 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson A, Parker M, Martineau F, Leach M, Engaging ‘communities’: Anthropological insights from the West African Ebola epidemic. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yetisen AK, Akram MS, Lowe CR, Paper-based microfluidic point-of-care diagnostic devices. Lab Chip 13, 2210–2251 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Koczula KM, Gallotta A, Lateral flow assays. Essays Biochem. 60, 111–120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wahl-Jensen V, Kurz SK, Hazelton PR, Schnittler H-J, Ströher U, Burton DR, Feldmann H, Role of Ebola virus secreted glycoproteins and virus-like particles in activation of human macrophages. J. Virol. 79, 2413–2419 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mehedi M, Falzarano D, Seebach J, Hu X, Carpenter MS, Schnittler HJ, Feldmann H, A new Ebola virus nonstructural glycoprotein expressed through RNA editing. J. Virol. 85, 5406–5414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Volchkova VA, Klenk HD, Volchkov VE, Delta-peptide is the carboxy-terminal cleavage fragment of the nonstructural small glycoprotein sGP of Ebola virus. Virology 265, 164–171 (1999). [DOI] [PubMed] [Google Scholar]
- 63.Sanchez A, Yang Z-Y, Xu L, Nabel GJ, Crews T, Peters CJ, Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J. Virol. 72, 6442–6447 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Branco LM, Garry RF, Characterization of the Lassa virus GP1 ectodomain shedding: Implications for improved diagnostic platforms. Virol. J. 6, 147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Branco LM, Grove JN, Moses LM, Goba A, Fullah M, Momoh M, Schoepp RJ, Bausch DG, Garry RF, Shedding of soluble glycoprotein 1 detected during acute Lassa virus infection in human subjects. Virol. J. 7, 306 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manicassamy B, Wang J, Rumschlag E, Tymen S, Volchkova V, Volchkov V, Rong L, Characterization of Marburg virus glycoprotein in viral entry. Virology 358, 79–88 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Rathore AS, Sarker A, Gupta RD, Recent developments toward antibody engineering and affinity maturation. Protein Pept. Lett. 25, 886–896 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Lim CC, Choong YS, Lim TS, Cognizance of molecular methods for the generation of mutagenic phage display antibody libraries for affinity maturation. Int. J. Mol. Sci. 20, 1861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heggestad JT, Fontes CM, Joh DY, Hucknall AM, Chilkoti A, In Pursuit of Zero 2.0: Recent developments in nonfouling polymer brushes for immunoassays. Adv. Mater. 32, 1903285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jahrling PB, Geisbert J, Swearengen JR, Jaax GP, Lewis T, Huggins JW, Schmidt JJ, LeDuc JW, Peters CJ, Passive immunization of Ebola virus-infected cynomolgus monkeys with immunoglobulin from hyperimmune horses. Arch. Virol. Suppl. 11, 135–140 (1996). [DOI] [PubMed] [Google Scholar]
- 71.Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ, Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: Evidence that dendritic cells are early and sustained targets of infection. Am. J. Pathol. 163, 2347–2370 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thi EP, Mire CE, Lee ACH, Geisbert JB, Zhou JZ, Agans KN, Snead NM, Deer DJ, Barnard TR, Fenton KA, MacLachlan I, Geisbert TW, Lipid nanoparticle siRNA treatment of Ebola-virus-Makona-infected nonhuman primates. Nature 521, 362–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nanbo A, Watanabe S, Halfmann P, Kawaoka Y, The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci. Rep. 3, 1206–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shurtleff AC, Bloomfield HA, Mort S, Orr SA, Audet B, Whitaker T, Richards MJ, Bavari S, Validation of the filovirus plaque assay for use in preclinical studies. Viruses 8, 113–113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krebber A, Bornhauser S, Burmester J, Honegger A, Willuda J, Bosshard HR, Plückthun A, Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods 201, 35–55 (1997). [DOI] [PubMed] [Google Scholar]
- 76.Malone J, Sullivan MA, Analysis of antibody selection by phage display utilizing anti-phenobarbital antibodies. J. Mol. Recog. 9, 738–745 (1996). [DOI] [PubMed] [Google Scholar]
- 77.Lipes BD, Chen Y-H, Ma H, Staats HF, Kenan DJ, Gunn MD, An entirely cell-based system to generate single-chain antibodies against cell surface receptors. J. Mol. Biol. 379, 261–272 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meyer DE, Chilkoti A, Purification of recombinant proteins by fusion with thermallyresponsive polypeptides. Nat. Biotechnol. 17, 1112–1115 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Hill HD, Mirkin CA, The bio-barcode assay for the detection of protein and nucleic acid targets using DTT-induced ligand exchange. Nat. Protoc. 1, 324–336 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Levine CB, Bayle J, Biourge V, Wakshlag JJ, Cellular effects of a turmeric root and rosemary leaf extract on canine neoplastic cell lines. BMC Vet. Res. 13, 388–388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Schematic depiction of EBOV D4 assay.
Fig. S2. D4 assay procedure.
Fig. S3. Mouse IgG titers for sGP and reactivity of individual scFv-phage clones for EBOV and SUDV sGP.
Fig. S4. scFv-Fc Ab expression, purification, and activity.
Fig. S5. Binding constants of scFv-Fc Abs for sGP.
Fig. S6. Ab pairing dose-response curves.
Fig. S7. Expression and purification of A1F3–1 and C2BA5–2 monoclonal IgG2a Abs.
Fig. S8. Binding constants of A1F3–1 (cAb) and C2BA5–2 (dAb) with multiple sGPs.
Fig. S9. Binding profiles of other scFv-Fc Abs with sGP.
Fig. S10. EBOV D4 assay performance.
Fig. S11. Post-assay fluorescence stability.
Fig. S12. EBOV D4 assay performance in human blood.
Fig. S13. EBOV sGP lateral flow assay fabrication.
Fig. S14. D4Scope design.
Fig. S15. Comparison of D4Scope, GenePix, and SensoSpot performance.
Fig. S16. RT-PCR and plaque assay.
Fig. S17. VP30- and GP-targeting RT-PCR.
Fig. S18. Western blot of MoS samples.
Fig. S19. Irradiated samples from EBOV-challenged nonhuman primates.
Table S1. D4Scope cost estimate.
Table S2. Summary of nonhuman primate model results.
Table S3. Summary of irradiated samples from EBOV-challenged nonhuman primates.