Abstract
RNA polymerase II (RNA Pol II) speed or elongation rate, i.e., the number of nucleotides synthesized per unit of time, is a major determinant of transcriptome composition. It controls co‐transcriptional processes such as splicing, polyadenylation, and transcription termination, thus regulating the production of alternative splice variants, circular RNAs, alternatively polyadenylated transcripts, or read‐through transcripts. RNA Pol II speed itself is regulated in response to intra‐ and extra‐cellular stimuli and can in turn affect the transcriptome composition in response to these stimuli. Evidence points to a potentially important role of transcriptome composition modification through RNA Pol II speed regulation for adaptation of cells to a changing environment, thus pointing to a function of RNA Pol II speed regulation in cellular physiology. Analyzing RNA Pol II speed dynamics may therefore be central to fully understand the regulation of physiological processes, such as the development of multicellular organisms. Recent findings also raise the possibility that RNA Pol II speed deregulation can be detrimental and participate in disease progression. Here, we review initial and current approaches to measure RNA Pol II speed, as well as providing an overview of the factors controlling speed and the co‐transcriptional processes which are affected. Finally, we discuss the role of RNA Pol II speed regulation in cell physiology.
Keywords: co‐transcriptional processes, RNA polymerase II, transcription speed
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics
This review discusses recent advances in understanding the regulation and functional effects of RNA polymerase II speed, as well as approaches for its experimental assessment.
Introduction
A cell’s response to changes in the environment or upon commitment to a specific developmental program is associated with major changes in genome expression. These changes involve quantitative alterations of messenger RNA (mRNA) or non‐coding RNA (ncRNA) expression, but also qualitative changes through differential RNA processing, such as alternative splicing. The correct regulation of genome expression is essential at all steps of an organism’s life. Among all the processes involved in genome expression control, transcription by RNA polymerase (pol) II is the first step and one of the most regulated. It has been extensively studied since it controls the production of all mRNAs in eukaryotic cells.
Transcription begins at the so‐called “Transcription Start Site” (TSS) immediately downstream of the promoter that contains specific DNA sequences providing stable binding sites for the RNA polymerase and transcription factors. RNA Pol II reaches the polyadenylation (poly(A)) site after transcribing through the whole transcription unit, which can span hundreds of kilobases. When transcribing a protein‐coding gene, RNA Pol II produces a pre‐mRNA that needs to undergo a maturation process before giving rise to a functional mRNA. This process involves capping of the 5′ end, removal of introns by splicing, and cleavage and polyadenylation at the 3′ end. All these steps stabilize mRNA and play important roles for its nuclear export and translation. In addition, these different pre‐mRNA processing steps are functionally coupled to transcription. Indeed, all these steps are initiated co‐transcriptionally, thereby allowing the connection of transcription with mRNA maturation in time and space (Bentley, 2014). Tight regulation of these various processing events allows cells to increase the diversity of the transcriptome, through alternative TSS selection, alternative splicing, back‐splicing, or alternative polyadenylation.
The various steps of transcription
RNA Pol II is a multiprotein complex composed of 12 subunits. RNA Pol II on its own cannot initiate transcription or transcribe long DNA sequences, but rather requires many additional protein complexes to perform these tasks. For example, topoisomerases which create transient DNA breaks to relieve DNA supercoiling play multiple roles during the transcription cycle (Pommier et al, 2016; Chen et al, 2018), including reduction of torsional stress to promote efficient transcription elongation (Baranello et al, 2016). Overall, transcription can be divided into four main steps: initiation, promoter‐proximal pausing, elongation, and termination (Fig 1) (for excellent reviews on these different steps, see (Porrua & Libri, 2015; Proudfoot, 2016; Core & Adelman, 2019; Schier & Taatjes, 2020)). The transitions between these steps often involve post‐translational modifications of RNA Pol II or associated factors.
Figure 1. Main regulated steps of the RNA Pol II transcription cycle.
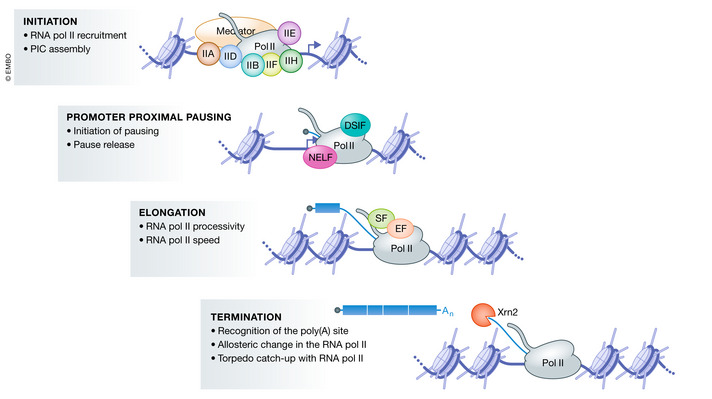
The RNA Pol II transcription cycle can be divided into four main regulated steps. First, transcription initiation starts with recruitment of RNA Pol II to the promoter and assembly of the pre‐initiation complex (PIC) which is composed of many factors including, but not restricted to, the general transcription factors TFIIA (IIA), TFIIB (IIB), TFIID (IID), TFIIE (IIE), TFIIF (IIF), TFIIH (IIH), RNA Pol II, and Mediator. The PIC opens the DNA and RNA Pol II starts transcription. Shortly after transcription initiation, RNA Pol II enters a paused state known as promoter‐proximal pausing. Promoter‐proximal pausing involves a conformational change in the RNA:DNA hybrid in the polymerase active site that prevents the addition of incoming NTPs by canonical base pairing. In many metazoans, the negative elongation factors NELF (Negative elongation factor) and DSIF (DRB‐sensitivity inducing factor) stabilize and extend the lifetime of the paused complex. After its release from promoter‐proximal pausing, RNA Pol II enters into productive elongation. During productive transcription elongation, many factors travel with RNA Pol II and help coordinate pre‐mRNA processing with transcription. These factors include elongation factors (EF, represented by an orange oval) and splicing factors (SF, represented by a green oval). The term transcription elongation includes at least two regulated processes: RNA Pol II processivity, i.e., the ability of RNA Pol II to travel the entire length of the gene and transcription speed, defined as the number of nucleotides synthesized per unit of time. Finally, transcription termination involves poly(A) site recognition by the cleavage and polyadenylation complex which cleaves the nascent pre‐mRNA that is then polyadenylated (An). Transcription of the poly(A) site induces an elongation slow‐down which promotes transcription termination upon catch‐up of RNA Pol II by the exonuclease Xrn2, which degrades the RNA synthesized beyond the poly(A) site from its 3′ end.
The first step of RNA Pol II transcription consists of the assembly of the pre‐initiation complex (PIC) on the promoter. PIC assembly often involves the recruitment of histone acetyltransferases and chromatin remodeling complexes that open chromatin to allow accessibility to promoters. The PIC is composed of many factors, including, but not restricted to, the general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, RNA Pol II, and Mediator (Schier & Taatjes, 2020). Once assembled on the promoter, the PIC opens the DNA double strand allowing transcription initiation (Dienemann et al, 2019). RNA Pol II thus starts transcribing and a short nascent RNA is produced.
Shortly after transcription initiation, RNA Pol II enters a paused state known as promoter‐proximal pausing. In mammals and many other metazoans, promoter‐proximal pausing occurs 25–50 base pairs downstream from the TSS and is a major rate‐limiting step of the transcription cycle (Jonkers et al, 2014; Mayer et al, 2015). It involves a conformational change in the RNA:DNA hybrid within the polymerase active site. This conformational change leads to the absence of a free DNA template base in the active site, which is required for the addition of incoming NTPs by canonical base pairing. As a consequence, the progression of RNA Pol II is prevented (Vos et al, 2018b). The negative elongation factors NELF (Negative elongation factor) and DSIF (DRB‐sensitivity inducing factor) stabilize and extend the lifetime of the paused complex. RNA Pol II is released from this pausing and allowed to resume transcription by the Cdk9 kinase component of the positive transcription elongation factor b (P‐TEFb) complex, which phosphorylates the Spt5 subunit of DSIF, NELF, and serines 2, 5, and 7 (Ser2, Ser5, and Ser7) of the RNA Pol II carboxy terminal domain (CTD) (Czudnochowski et al, 2012; Eick & Geyer, 2013; Core & Adelman, 2019). P‐TEFb is recruited to promoters through interactions with transcription factors, Mediator, and coactivators (Li et al, 2018). The P‐TEFb‐mediated phosphorylation events lead to both the dissociation of NELF from the RNA Pol II and the switch of Spt5 from a repressor to an activator of transcription (Ivanov et al, 2000; Yamada et al, 2006). Spt5 remains associated with RNA Pol II and serves as a recruitment platform for additional factors involved in RNA Pol II elongation and RNA processing.
Although RNA Pol II promoter‐proximal pausing also occurs in nematodes, fission yeast, and plants, NELF is absent in these species, suggesting that alternative modes of RNA Pol II promoter‐proximal pausing exist. Indeed, pausing at the +1 nucleosome, which does not involve DSIF or NELF, has been identified fission yeast (Booth et al, 2016) and plants (Kindgren et al, 2020) but also in mammals (Chiu et al, 2018). The mechanisms of this +1 nucleosome pausing are not yet fully understood, but regulation by the MYC transcription factor and sensitivity to flavopiridol (an inhibitor of Cdk9 and Cdk12) have been reported (Chiu et al, 2018). In nematodes, although promoter‐proximal pausing has been identified at stress response genes during starvation, its mechanism remains largely unknown (Kruesi et al, 2013; Maxwell et al, 2014). In contrast to these alternative mechanisms to pause RNA Pol II in promoter‐proximal regions in nematode, fission yeast, and plants, RNA Pol II promoter‐proximal pausing has, to date, not been reported in budding yeast (Booth et al, 2016).
After its release from this pause, RNA Pol II enters productive transcription elongation. The term transcription elongation refers to at least two regulated aspects of the transcript elongation process: RNA Pol II processivity and speed. RNA Pol II processivity is the ability of RNA Pol II to travel the entire length of the gene and thus controls the production of full‐length transcripts. Premature transcription termination reduces RNA Pol II processivity and can lead to the production of transcripts that depending on the context are either rapidly degraded or stabilized. Stabilized prematurely terminated transcripts can give rise to non‐coding RNAs or to proteins with different properties compared to those synthesized from full‐length transcripts (Kamieniarz‐Gdula & Proudfoot, 2019). The other transcription elongation parameter, transcription speed (or velocity), also referred to as the elongation rate, is defined as the number of nucleotides synthesized per unit of time and is the topic of this review. Note, however, that the term elongation rate is also sometimes used to describe other steps of the transcription cycle, such as RNA Pol II release from promoter‐proximal pausing or RNA Pol II processivity.
Finally, when RNA Pol II reaches the end of a gene, it terminates transcription. At the end of almost all protein‐coding genes (except replication‐dependent histone genes), the termination process involves poly(A) site recognition. The poly(A) site is recognized by the cleavage and polyadenylation complex composed of multi‐subunit factors including the cleavage and polyadenylation specificity factor (CPSF), the cleavage stimulatory factor (CstF), and the cleavage factors (CF) I and II. Upon recognition of the poly(A) site by the CPSF30 and WDR33 components of CPSF, the nascent pre‐mRNA is cleaved by the CPSF73 endonuclease. The cleaved 3′ end of the pre‐mRNA is then polyadenylated, while the unprotected 5′ end of the nascent RNA provides an entry site for the Xrn2 5′–3′ exonuclease, which co‐transcriptionally degrades the RNA synthesized beyond the poly(A) site (Eaton & West, 2020). In addition, transcription of the poly(A) site induces allosteric changes in the elongation complex (Logan et al, 1987) mediated by dephosphorylation of Spt5 by the PP1 phosphatase, which slow down elongation by at least two‐ to threefold (Cortazar et al, 2019). This reduction of elongation speed facilitates the catch‐up of RNA Pol II by the Xrn2 exonuclease, which dislodges RNA Pol II from the DNA template leading to transcription termination (Cortazar et al, 2019; Eaton et al, 2020).
Regulation of RNA Pol II speed
Initially, studies on transcriptional regulation focused on transcription initiation, demonstrating an essential role for this stage in the regulation of gene expression. However, over the years, it has become increasingly clear that other stages of transcription are critical to control gene expression. For example, RNA Pol II promoter‐proximal pausing, which has received increasing attention in the transcription field since the late 90s, was shown to be an early regulatory step that controls expression of most metazoan genes (for excellent reviews on this step, see (Jonkers & Lis, 2015; Chen et al, 2018)). Meanwhile, it has become clear that virtually all steps of the transcription cycle can be regulated by RNA Pol II‐associated factors and the epigenetic context of the locus.
Until recently however, transcription elongation by RNA Pol II was not considered as a major regulatory step in gene expression. However, studies published over the last 20 years have shown that transcription elongation speed is highly dynamic and extremely regulated within individual genes, as well as between genes, which is the topic of the first section of this review. The use of RNA Pol II mutants displaying accelerated or decelerated transcription rates has also allowed major advances in our understanding of the function of RNA Pol II speed, demonstrating an essential role in many co‐transcriptional processes and therefore in the repertoire of RNAs produced from a given locus as discussed in the second section of this review. We will also review the growing list of factors involved in the modulation of RNA Pol II speed highlighting that transcription velocity is a key regulatory step in the transcription cycle. Importantly, the introduction of a point mutation in RNA Pol II, which slows down transcription, was found to be embryonic lethal in mice (Maslon et al, 2019) and non‐viable in plants (Leng et al, 2020). These data demonstrate that correct RNA Pol II speed is essential for development (Maslon et al, 2019; Leng et al, 2020). Moreover, we and others have shown that transcription speed can be locally adapted in response to intrinsic or extrinsic stimuli, affecting the identity of RNAs produced in the cell (Schor et al, 2009; Schor et al, 2013; Sharma et al, 2014; Muniz et al, 2017).
In this review, we will therefore discuss the different techniques used to measure RNA Pol II velocity and the evidences indicating that transcription speed is not static, but highly variable between genes, as well as within individual genes. We will then describe the influence of RNA Pol II velocity on co‐transcriptional processes and thus on the identity of RNAs produced from a given gene, as well as the factors that regulate transcription speed. Finally, we will cover the dynamics of RNA Pol II speed regulation and the roles it may play in the response to environmental changes.
Measuring RNA Pol II speed
At which speed does RNA Pol II synthesize RNA? This a long‐standing question which has been investigated for over 40 years. Many different methods have been developed to estimate RNA Pol II speed on endogenous or integrated reporter genes in distinct species (see (Jonkers & Lis, 2015) for an excellent review describing some of these methods). These methods measure the distance traveled by RNA Pol II as a function of time. Importantly, we can only measure an average RNA Pol II speed on any given gene or part of gene, since it depends on both the instantaneous speed at which nucleotides are added by the polymerase to the nascent RNA, as well as polymerase pausing within the gene body. Indeed, transcription elongation is frequently interrupted by pauses induced by roadblocks such as nucleosomes, DNA‐binding proteins, or mis‐incorporated nucleotides (Mayer et al, 2017), which can thus affect the average speed of transcription. Most of the described methods developed to estimate RNA Pol II speed rely on inducible gene expression or treatments with small drugs globally inhibiting transcription either at the initiation step (with triptolide, for example) or, more commonly, at the promoter‐proximal pause release step with reversible P‐TEFb inhibitors (e.g., flavopiridol or DRB). Inhibition of P‐TEFb and thus repression of the release of RNA Pol II from promoter‐proximal pausing has been shown to block transcription of almost all genes (Henriques et al, 2013; Jonkers et al, 2014). In this scenario, RNA Pol II elongation complexes which are already in the productive elongation phase of the transcription cycle continue transcribing, while no new RNA Pol II enters productive elongation. As such, methods relying on the reversible P‐TEFb inhibitors theoretically allow measurement of RNA Pol II speed at most active genes. To calculate transcription speed, the disappearance or appearance of the RNA produced by transcription or RNA Pol II itself is then followed at various locations downstream of gene promoters in kinetics experiments. The distance traveled by RNA Pol II after transcription inhibition (also called the RNA Pol II “retreating wave”) can be followed in response to transcriptional switch‐off of inducible genes or after treatment with general transcription inhibitors. Conversely, the distance traveled by RNA Pol II after transcription induction (also called the RNA Pol II “emerging wave”) can be followed in response to transcriptional activation by different stimuli or after globally switching on transcription by removing a reversible inhibitor (Jonkers & Lis, 2015).
RNA Pol II speed measurements by biochemical approaches
Pioneering works have estimated average RNA Pol II speed only on a few genes. By monitoring the appearance of RNA by in situ hybridization, nuclear run‐on, slot blot, or RT–PCR after expression activation, RNA Pol II speed was estimated between 1.1 kb/min and 4.8 kb/min (Ucker & Yamamoto, 1984; Thummel et al, 1990; Shermoen & O'Farrell, 1991; O'Brien & Lis, 1993; Tennyson et al, 1995; Femino et al, 1998; Femino et al, 2003; Hanisch et al, 2013). Other studies have estimated average RNA Pol II speed between 2 and 6 kb/min by analyzing the sedimentation profile of RNA after [3H] uridine incorporation (Sehgal et al, 1976) or RNA Pol II levels by ChIP PCR at several locations in the gene (Mason & Struhl, 2005) after transcriptional repression.
By using quantitative methods to detect RNAs, these measures of RNA Pol II speed were then refined. For instance, the RNA Pol II “emerging wave” has been followed by monitoring the appearance of nascent RNAs by qRT–PCR on exon–intron junctions spanning the genes. This has been performed on a few long genes at different time points after stimulation by tumor necrosis factor‐α (TNFα) (Wada et al, 2009), interferon β, or DRB release (Singh & Padgett, 2009). In these studies, the average transcription speed was estimated between 3.1 kb/min and 3.8 kb/min with little variation between individual genes (Singh & Padgett, 2009; Wada et al, 2009).
In the late 2000s, many teams began developing methodologies to measure transcription speed genome wide (Table 1). A major improvement in the race for global measurement of RNA Pol II speed came from the demonstration that RNA Pol II “emerging” and “retreating” waves could be followed genome wide by global run‐on sequencing (GRO‐seq) (Hah et al, 2011). This technique has been applied in various human cell lines to follow the “emerging wave” of RNA Pol II after transcriptional activation by different cellular signaling pathways (Danko et al, 2013) or by DRB release (Saponaro et al, 2014). Other methods have been developed to specifically enrich nascent RNAs and have been applied to follow the RNA Pol II “emerging wave” after DRB or flavopiridol release. These include sequencing of nascent RNAs labeled with 4‐thiouridine (Fuchs et al, 2014b; Fuchs et al, 2015; Liang et al, 2018; Baluapuri et al, 2019; Gregersen et al, 2020) or bromouridine (Veloso et al, 2014). As for the RNA Pol II “retreating wave”, it has been followed genome wide by GRO‐seq after flavopiridol treatment (Jonkers et al, 2014). All these techniques showed in various human cell lines that the average RNA Pol II speed ranges between 1.25 kb/min and 3.5 kb/min (Danko et al, 2013; Fuchs et al, 2014b; Jonkers et al, 2014; Saponaro et al, 2014; Veloso et al, 2014; Liang et al, 2018; Baluapuri et al, 2019; Gregersen et al, 2020). Contrary to what had been initially shown for a few genes (Singh & Padgett, 2009; Wada et al, 2009), the measured transcription speed was found to be extremely variable between genes, with RNA Pol II velocity varying from 0.37 kb/min to 3.57 kb/min for individual genes (Danko et al, 2013). RNA Pol II speed was furthermore also shown to be highly variable within individual genes, as RNA Pol II accelerates while traveling throughout the gene body, starting at 0.5 kb/min in the first 10–15 kb and accelerating up to 2–4 kb/min downstream (Danko et al, 2013; Fuchs et al, 2014b; Jonkers et al, 2014; Saponaro et al, 2014). In addition, comparison of transcription speed between different cell lines showed that transcription speed on individual genes is overall well conserved between cell lines (Singh & Padgett, 2009; Veloso et al, 2014). However, Danko et al (2013) found that RNA Pol II velocity can differ by as much as 25–40% on individual genes in different cell lines and in response to diverse signaling pathways. Moreover, we and others have shown that RNA Pol II speed can vary on individual genes or downstream of genes in response to different stimuli (Schor et al, 2009; Schor et al, 2013; Sharma et al, 2014; Muniz et al, 2017). Thus, RNA pol II speed is highly regulated between as well as within genes and can change in response to different stimuli.
Table 1.
Average RNA pol II speed estimations on a genome‐wide basis.
| RNA pol II speed | Cell line, organism | Number of genes | Measurement method | Study |
|---|---|---|---|---|
|
2.1 kb/min 2.8 kb/min |
MCF‐7, Human AC16, Human |
140 genes 26 genes |
GRO‐seq (sequencing of nascent RNA extended in the presence of BrUTP in run‐on experiments) following transcriptional activation by estrogen signaling or cytokine treatment—follows the “emerging wave” of RNA pol II | Danko et al (2013) |
| 3.5 kb/min | HeLa, Human | 1,577 genes | 4sUDRB‐seq (sequencing of nascent RNA labeled with 4‐thiouridine following DRB release)—follows the “emerging wave” of RNA pol II | Fuchs et al (2014) |
|
1.25 kb/min 1.75 kb/min 1.25 kb/min 1.25 kb/min 1.75 kb/min |
HF1, Human TM, Human CS‐B, Human K562, Human MCF‐7, Human |
2,702 genes 2,469 genes 1,932 genes 2,270 genes 2,399 genes |
BruDRB‐seq (sequencing of nascent RNA labeled with bromouridine following DRB release)—follows the “emerging wave” of RNA pol II | Veloso et al (2014) |
| 3.13 kb/min | HEK293, Human | 237 genes | DRB/GRO‐seq (sequencing of nascent RNA extended in the presence of BrUTP in run‐on experiments following DRB release)—follows the “emerging wave” of RNA pol II | Saponaro et al (2014) |
| 2 kb/min | mESC, Mouse | More than 1,000 genes | GRO‐seq (sequencing of nascent RNA extended in the presence of BrUTP in run‐on experiments) following flavopiridol treatment to block RNA pol II promoter‐proximal pause release—follows the “retreating wave” of RNA pol II | Jonkers et al (2014) |
| 3.27 kb/min | U2OS, Human | 2,163 genes | 4sUDRB‐seq (sequencing of nascent RNA labeled with 4‐thiouridine following DRB release)—follows the “emerging wave” of RNA pol II | Baluapuri et al (2019) |
| Around 3.2 kb/min | HEK293, Human | 982 genes | 4sUFP‐seq (sequencing of nascent RNA labeled with 4‐thiouridine following flavopiridol release)—follows the “emerging wave” of RNA pol II | Liang et al (2018) |
| Around 1 kb/min | S2, Drosophila | Not mentioned | 4sUDRB‐seq (sequencing of nascent RNA labeled with 4‐thiouridine following DRB release)—follows the “emerging wave” of RNA pol II | Akhtar et al (2019) |
| 2.3 kb/min | HEK293, Human | 4,869 genes | DRB/TTchem‐seq (sequencing of nascent RNA labeled with 4‐thiouridine following DRB release)—follows the “emerging wave” of RNA pol II | Gregersen et al (2020) |
Finally, RNA Pol II speed has also been inferred from existing total RNA‐seq data (Ameur et al, 2011; Jonkers et al, 2014; Kawamura et al, 2019). Indeed, intronic reads form a decreasing gradient from the 5′ to the 3′ end of the intron. This gradient depends on the kinetics of RNA Pol II and co‐transcriptional splicing and leads to formation of a “saw‐tooth” pattern of read coverage over the length of the gene (Ameur et al, 2011). RNA Pol II speed can thus be inferred for long introns by calculating the 5′–3′ slope of intron reads coverage. Faster RNA Pol II will take less time to reach the downstream exon where co‐transcriptional splicing can take place, thus limiting RNA accumulation at the 5′ end of introns, thereby decreasing the 5′–3′ slope of intronic reads coverage. This technique is however far less accurate than the methods designed to follow the emerging or retreating waves of RNA Pol II. It may, nonetheless, give an opportunity to compare RNA Pol II speed in thousands of published datasets from different cell types and in response to many different stimuli.
RNA Pol II speed measurements by imaging technologies
Interestingly, studies performed in live cells came to very similar estimations of RNA Pol II speed. RNA Pol II speed in live cells was estimated to range from 0.6 kb/min to 4.3 kb/min by fluorescence recovery after photobleaching (FRAP) experiments using MS2 fluorescent proteins to label nascent MS2 repeat‐containing RNAs (Boireau et al, 2007; Darzacq et al, 2007; Ben‐Ari et al, 2010; Yunger et al, 2010; Brody et al, 2011; Maiuri et al, 2011; Muramoto et al, 2012; Corrigan et al, 2016). By using constructs carrying arrays of MS2 and PP7 repeats in the 5′ and 3′ regions of the same gene, RNA Pol II speed was estimated to range from 0.84 kb/min to 3.66 kb/min (Hocine et al, 2013; Fukaya et al, 2017). Other imaging techniques such as fluctuation analysis of fluorescently labeled nascent RNA or high‐speed 3D fluorescence nanoimaging techniques reported speed in the same range, albeit slightly higher between 0.6 and 12 kb/min (Larson et al, 2011; Martin et al, 2013; Annibale & Gratton, 2015). Measurement of RNA Pol II velocity on endogenous genes in living cells was also attempted by FRAP experiments using EGFP tagged RNA Pol II. The authors calculated an RNA Pol II speed between 1.1 to 1.5 kb/min when measured on the Hsp70 genes following heat shock in Drosophila (Yao et al, 2007; Ardehali et al, 2009). These studies also found that RNA Pol II speed was highly variable between cells (Hocine et al, 2013), as well as between identical copies of the same gene at different locations in a gene array, measured simultaneously in the same cell (Annibale & Gratton, 2015). Indeed, in this latter study, a fourfold variation in RNA Pol II velocity was measured between identical copies of the same gene and overall, the different RNA Pol II speeds measured in the gene array almost span the range of transcription speeds reported using the MS2 containing constructs, from below 0.6 kb/min to over 12 kb/min (Annibale & Gratton, 2015). Along the same line, Maiuri et al (2011) even reported an extremely high RNA Pol II speed (about 80 kb/min) on an HIV‐based vector integrated in one copy, whereas when integrated in 35 copies, RNA Pol II velocity was estimated at 1.6 kb/min, similar to earlier estimates from multicopy HIV‐derived vector insertions (Boireau et al, 2007). Nevertheless, single integration of the CCND1 gene under the control of an endogenous or viral promoter led to an RNA Pol II speed of 0.31–0.78 kb/min (Yunger et al, 2010) and the number of gene units in the tandem array had no effect on RNA Pol II speed in another study (Brody et al, 2011). In agreement with biochemical measurements of RNA Pol II speed, all these data show that RNA Pol II speed can be variable between genes but also within genes, highlighting the fact that RNA Pol II speed is highly regulated.
RNA Pol II speed measurements in organisms
As mentioned above, over the last 20 years, RNA Pol II speed has been estimated by several different methods. All these studies show that transcription speed is highly dynamic and highly regulated within, as well as between genes, suggesting that it might play an important role in the transcription cycle. The most accurate and least biased methods to measure RNA Pol II speed are based on activation or inhibition of transcription and following RNA Pol II progression throughout genes over time. This allows estimating the distance traveled by RNA Pol II during a given amount of time. However, most of these studies, in particular those analyzing RNA Pol II speed genome wide, used drugs that inhibit either transcription initiation or promoter‐proximal pause release. It is important to consider that these drugs may have important secondary effects, since most of them target kinases involved in cell‐cycle control, which may affect the measurements. Another issue with most of these studies is that they were performed using cell lines. Whether the observed features can be generalized or whether they are a peculiarity of the respective analyzed cell lines thus remains an open question. The use of drugs can be overcome by activating different signaling pathways to induce transcription of specific genes, hence limiting secondary effects. Although the use of these induction systems restricts the analysis to only a handful of genes, the method can be scaled up to whole organisms, as shown in a recent study measuring RNA Pol II speed in budding yeast and A. thaliana seedlings (Leng et al, 2020). Moreover, this study together with a recent study measuring RNA Pol II speed in mouse ESCs (Embryonic Stem Cells) and neurons showed that these methods are readily transferable to more complex systems and still give comparable results (Maslon et al, 2019; Leng et al, 2020). Clearly, further developing novel technologies allowing accurate measurement of RNA Pol II speed at genome‐wide level in physiological situations and in complex organisms will be a major advance in the field.
RNA Pol II speed controls many co‐transcriptional processes and thus affects the composition of the transcriptome
In the early 80s, a screen was performed to discover mutations rendering Drosophila RNA Pol II resistant to the transcription inhibitor α‐amanitin (Greenleaf et al, 1979). This screen led to the discovery of mutations affecting transcription speed. These mutations, mostly located in the largest RNA Pol II subunit RPB1 (Greenleaf, 1983), which either slow down or accelerate transcription, have allowed huge progress in the characterization of the role of RNA Pol II speed in the regulation of gene expression. At least two different mutations in human RNA Pol II are known to slow down transcription. The R749H substitution in the funnel domain is homologous to the C4 mutation in Drosophila (de la Mata et al, 2003) and likely affects translocation of the polymerase such that the forward and backward movement of the mutant elongation complex is slowed‐down (Chen et al, 1993; Chen et al, 1996). The H1108Y substitution is homologous to the H1085Y substitution in the yeast Rpb1 (Kaplan et al, 2008) and mutates a contact with the β‐phosphate of incoming NTPs (Fong et al, 2014) which also slows down transcription. Conversely, the E1126G substitution in human RPB1 (Fong et al, 2014) (homologous to the E1103G substitution in the yeast RPB1; Kaplan et al, 2008; Kireeva et al, 2008) accelerates transcription by stabilizing a triple alpha helix at the base of the trigger loop, concomitantly with a loss of fidelity that is reduced by α‐amanitin. Average speed for these RNA Pol II mutants is 0.5 Kb/min or even slower for the slow mutants (R749H and H1108Y, respectively) and 1.9 Kb/min for the fast mutant as compared to 1.7 Kb/min for the human α‐amanitin‐resistant WT pol II (Fong et al, 2014). Nevertheless, prolonged α‐amanitin treatment has been shown to induce accelerated degradation of several proteins including the elongation factor DSIF (Tsao et al, 2012). Measurements of RNA Pol II speed obtained using α‐amanitin‐resistant RNA Pol II mutants should thus be taken with care.
By analyzing these different mutants in different systems, RNA Pol II velocity has been shown to control many co‐transcriptional processes. Indeed, transcription speed determines the rate at which binding sites for RNA‐binding proteins are synthesized within the nascent RNA and is thus in kinetic competition with co‐transcriptional processes. Co‐transcriptional processes shown to be regulated by RNA Pol II speed include constitutive splicing, alternative splicing, back‐splicing, alternative polyadenylation, and transcription termination (Fig 2). All of these co‐transcriptional processes contribute to the generation of transcriptome diversity that is critical for proper cell functioning and will be discussed in further detail in the following sections.
Figure 2. Co‐transcriptional processes regulated by RNA Pol II speed.
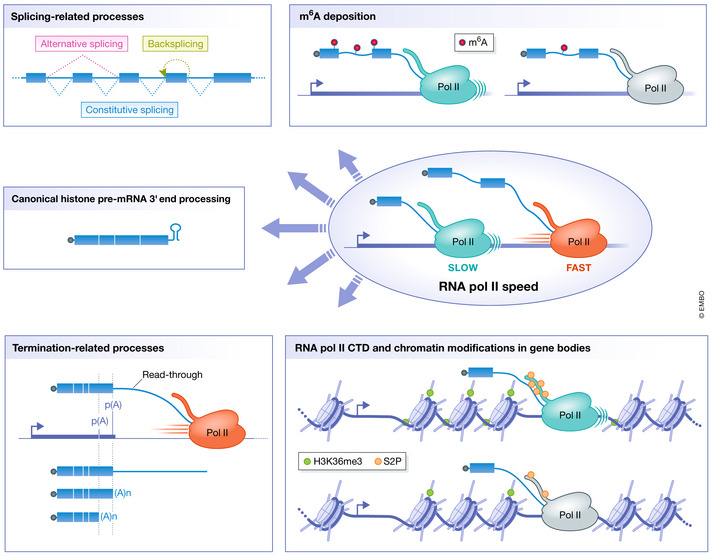
- Splicing‐related processes such as constitutive splicing, alternative splicing, and back‐splicing;
- The RNA methylome by controlling the deposition of N6‐methyladenosine in RNAs;
- Normal 3′ end processing of replication‐dependent core histone mRNAs by controlling the folding of nascent RNA at histone genes;
- Termination‐related processes such as alternative polyadenylation (represented by two poly(A) sites) and the distance traveled by RNA Pol II beyond the poly(A) site, i.e., read‐through transcription;
- Protein modifications on gene bodies: post‐translational modifications of RNA Pol II CTD such as serine 2 phosphorylation and chromatin modifications such as tri‐methylation of lysine 36 of histone H3.
Control of splicing‐dependent processes
Constitutive splicing is the joining of consecutive exons by removal of intervening introns. The majority of individual introns are constitutively spliced. However, splicing efficiency is not 100% and unspliced RNAs are predominantly retained in the nucleus and degraded through a quality‐control mechanism (Palazzo & Lee, 2018). Using microarrays designed to study splicing in budding yeast, slow RNA Pol II mutants were shown to enhance splicing, while fast mutants were shown to reduce constitutive splicing (Braberg et al, 2013). This anti‐correlation between transcription speed and splicing efficiency has been confirmed in budding yeast by RNA sequencing‐based methods (Aslanzadeh et al, 2018). Likewise, other studies using sequencing‐based methods showed that a slow mutant enhances constitutive splicing in Drosophila (Howe et al, 2003; Khodor et al, 2011; Braberg et al, 2013; Aslanzadeh et al, 2018), as does a fast mutant in plants (Leng et al, 2020). This discrepancy between plants on one hand and Drosophila and budding yeast on the other hand might seem surprising; however, both slow and fast RNA Pol II mutants inhibit constitutive splicing in human cells (Fong et al, 2014). These results show that while RNA Pol II speed differentially affects constitutive splicing in various species, proper control of intragenic RNA Pol II velocity is essential for proper regulation of constitutive splicing efficiency.
In contrast to constitutive splicing, alternative splicing contributes to the proteome diversity through the biogenesis of several mRNA splice variants from the same pre‐mRNA. Indeed, transcripts can undergo one or more alternative splicing events such as inclusion or skipping of an exon, use of alternative 5′ or 3′ splice sites, intron retention, or splicing of adjacent mutually exclusive exons, thus generating several different transcript isoforms from the same gene. More than 95% of human genes undergo alternative splicing in a developmental, tissue‐specific or signal transduction‐dependent manner (Nilsen & Graveley, 2010). Alternative splicing regulation is extremely complex, with dynamic alternative splicing networks identified in many different cell types and organs, as well as in different physiological conditions. Many types of RNA‐binding proteins can control alternative splicing through binding to cis elements (splicing enhancers or silencers), although the full set of factors involved is not known. Like constitutive splicing, alternative splicing is also sensitive to RNA Pol II speed. Indeed, the use of gel‐based methods in yeast, human, and mouse cells (de la Mata et al, 2003; Howe et al, 2003; Schor et al, 2009; Ip et al, 2011; Dujardin et al, 2014) or of RNA sequencing‐based methods in human cells (Fong et al, 2014) showed that slow and fast transcription either increased or decreased inclusion of alternative exons depending on the genes analyzed. Moreover, RNA sequencing analyses showed that slow transcription obtained using an RNA Pol II knock‐in mutant in mouse ESCs or fast transcription induced by an RNA Pol II point mutant in plants leads to changes in both exon inclusion and skipping (Maslon et al, 2019; Leng et al, 2020).
The effects of RNA Pol II velocity on splicing events can be explained by the fact that splicing is in kinetic competition with transcription elongation. According to the “window of opportunity” model, altering transcription speed may regulate splicing by modulating the presence of cis‐acting sequence elements on the nascent RNA and thus the recruitment of both positive and negative splicing factors. However, slow and fast RNA Pol II mutants often have the same effect on alternative exon inclusion or skipping (Fong et al, 2014; Aslanzadeh et al, 2018), which is not predicted by the “window of opportunity” model. These observations have thus led to the emergence of another model termed “Goldilocks” model in which an optimal RNA Pol II speed is required to allow proper nascent RNA folding and factors binding to splice sites, splicing enhancers and silencers (Fong et al, 2014). This model predicts that RNA Pol II speed has evolved under positive selection pressure to allow optimal co‐transcriptional splicing. However, it remains to be determined how RNA Pol II speed affects nascent RNA folding and/or binding of factors to influence co‐transcriptional splicing.
RNA Pol II speed, through controlling splicing efficiency, has also been implicated in the regulation of circular RNA (circRNA) production. CircRNAs belong to a newly described class of RNAs that also contribute to transcriptome diversity. CircRNAs are produced by back‐splicing of pre‐mRNAs, which corresponds to the joining of a downstream 5′ splice site to an upstream 3′ splice site in the reversed orientation, thus producing a circular RNA molecule (for excellent reviews on circRNA biogenesis and function, see (Chen, 2020; Xiao et al, 2020)). Tens of thousands of circRNAs have been identified in metazoans, some of which are evolutionarily conserved. While a majority of these circRNAs are expressed at low levels, some of them accumulate to levels exceeding those of their linear counterparts (Jeck et al, 2013; Salzman et al, 2013). Indeed, although back‐splicing is generally much less efficient than canonical splicing, owing to their circular structure, circRNAs are resistant to degradation by exonucleases, which allows them to accumulate to high levels. CircRNAs exhibit tissue and cell type‐specific expression patterns with a tendency to accumulate in the brain (Rybak‐Wolf et al, 2015). To date, only a small subset of all the identified circRNAs has been functionally studied, leaving a full repertoire of circRNAs of unknown function. Moreover, because of their low expression level, the biological significance of some circRNAs has been questioned; however, recent findings suggest that circRNAs can work as a group to achieve measurable effects. Furthermore, accumulating evidence indicates that circRNAs are tightly regulated transcripts that in some cases play essential biological functions. Indeed, a growing number of circRNAs have been implicated in the regulation of innate immunity, cell proliferation, transformation, and neuronal function (Chen et al, 2019; Liu et al, 2019; Hollensen et al, 2020). Misregulation of some circRNAs has also been implicated in multiple diseases among which cancer, some circRNAs playing important roles in cancer progression (Chen et al, 2019). CircRNAs can regulate transcription, splicing, and chromatin interactions. They can also act as microRNA decoys, function as protein scaffolds, sequester proteins, or be themselves translated (Kristensen et al, 2019). Moreover, because circRNAs are very stable and can be detected in body fluids (blood and urine, for example), they are now considered as promising disease biomarkers (Vo et al, 2019; Wang et al, 2021). Importantly, circRNA biogenesis is tightly regulated, although the mechanisms regulating their levels are not completely understood. Among key elements regulating their production, intronic complementary repeats in introns flanking the circularized exons have been shown to be important for the biogenesis of circRNAs (Liang & Wilusz, 2014). Base pairing between these repeats is required for the biogenesis of these circRNAs, most probably by bringing the splice sites into close proximity. Many proteins are also involved in the production of circRNAs, such as spliceosome components, RNA‐binding proteins, and RNA helicases (Patop et al, 2019). In addition, some recent studies point to RNA Pol II speed as another determinant of circRNA production. Indeed, the average RNA Pol II velocity is higher on genes producing circRNAs than on genes not producing circRNAs (Zhang et al, 2016) and especially on introns of these genes (Ragan et al, 2019). Moreover, the use of slow and fast RNA Pol II mutants has shown that transcription speed directly correlates with the circularization efficiency of circRNAs (Ashwal‐Fluss et al, 2014; Zhang et al, 2016).
Control of transcription termination‐related processes
RNA Pol II speed is also in competition with the rate of processes linked to transcription termination such as poly(A) site usage and transcription termination itself.
A gene can possess multiple poly(A) sites, and differential usage of these sites leads to the formation of distinct mRNA isoforms, a phenomenon termed alternative polyadenylation. Alternative polyadenylation is very common in eukaryotes, and around 70% of mammalian protein‐coding genes are subjected to alternative polyadenylation (Derti et al, 2012; Hoque et al, 2013). When an alternative poly(A) site is present in the coding region, the use of this alternative polyadenylation site can produce mRNA isoforms that differ in their coding capacity, resulting in different final protein products with specific cellular functions. In contrast, and most frequently, the alternative poly(A) sites are located downstream of the coding region and their differential usage generates mRNAs that share the same coding sequence, but have different 3′ untranslated region (UTR) lengths. The alternative 3′ UTR sequence may contain regulatory elements that interact with RNA‐binding proteins or miRNAs, modulating mRNA stability, translation, transport, and/or localization (Tian & Manley, 2017). On several genes possessing alternative poly(A) sites, a slow RNA Pol II mutant shows a switch to the use of upstream poly(A) sites (Pinto et al, 2011; Liu et al, 2017; Geisberg et al, 2020; Yague‐Sanz et al, 2020), while a fast RNA Pol II mutant shows a slight downstream switch in poly(A) site usage (Geisberg et al, 2020). In addition, a slow RNA Pol II mutant shows a tissue‐specific effect on alternative polyadenylation as it affects poly(A) site usage differentially in the Drosophila head or body (Liu et al, 2017).
Transcription termination is an essential step of the transcription cycle. It requires disassembly of the stable complex formed between the nascent RNA, RNA Pol II, and the DNA template. Transcription termination is important to recycle RNA Pol II and to prevent interference between adjacent transcription units. For most genes, transcription termination is triggered by recognition of the poly(A) site, but it does not occur at a conserved site or distance from the 3′ end of genes (Hofer & Darnell, 1981; Weintraub et al, 1981; Hagenbüchle et al, 1984). It rather occurs at diffuse sites, at variable distances, up to several kilobases, downstream of the poly(A) site (Fong et al, 2015). Transcription termination has been shown to be shifted closer to the poly(A) site by a slow RNA Pol II mutant (Fong et al, 2015), whereas a fast RNA Pol II mutant shifted transcription termination further downstream from the poly(A) site (Fong et al, 2015; Leng et al, 2020). Consistent with the results obtained with slow RNA Pol II mutants, slowing‐down transcription using mutants of the elongation factor TFIIS also shifts termination upstream (Sheridan et al, 2019; Zatreanu et al, 2019). RNA Pol II velocity can thus control the distance traveled by RNA Pol II downstream of genes, in other words, the extent of transcriptional read‐through. These results suggest that a kinetic competition between the elongating RNA Pol II and the degradation of the nascent RNA by Xrn2 is controlling termination (Fong et al, 2015). Interestingly, we recently showed that transcription speed increases downstream of two genes in response to activation of an oncogene and that this increase correlates with an increase in transcriptional read‐though downstream of these genes (Muniz et al, 2017). This implies that transcriptional read‐through might be regulated by RNA Pol II speed in response to intra‐cellular stimuli.
Control of mRNA modifications
N6‐methyladenosine (m6A) is the most abundant modification in mRNA, detected in thousands of human transcripts (about 25% of mRNAs contain at least one m6A) (Dominissini et al, 2012; Meyer et al, 2012). More than 99% of total m6A in polyadenylated mRNA is deposited by a heterodimeric complex composed of METTL3‐METTL14 in a highly specific manner (Shi et al, 2019; Zaccara et al, 2019). m6A influences splicing, degradation, and translation of mRNA (Knuckles & Bühler, 2018; He & He, 2021). A slow RNA Pol II mutant induces an increase in the level of m6A deposition on mRNAs, and this has been linked to a reduced efficiency of mRNA translation (Slobodin et al, 2017). Thus, transcription speed, by affecting m6A deposition on mRNAs, may influence mRNA stability as well as translation efficiency and therefore proteome composition.
Control of histone pre‐mRNA processing through alteration of nascent RNA structure
mRNAs encoding replication‐dependent core histones (H2A, H2B, H3, and H4) produced during S‐phase lack the normal poly(A) tail and instead present a 3′ stem‐loop (Marzluff & Koreski, 2017). They undergo endonucleolytic cleavage at the 3ʹ side of this stem‐loop, and correct folding of this structure is absolutely essential for normal 3′ processing of histone mRNAs (Williams et al, 1994; Battle & Doudna, 2001; Dominski et al, 2003). Slow RNA Pol II mutants reduce histone pre‐mRNA processing (Saldi et al, 2018), the stem‐loop failing to properly fold in transcripts synthesized by these mutants. This is likely due to a competition mechanism between the rate of RNA synthesis and the rate of nascent RNA folding, thus explaining the failure to process 3′ ends. These results show that regulation of transcription speed can also control pre‐mRNA processing by changing nascent RNA structure.
Control of protein modifications on gene bodies
The CTD of RNA Pol II, which consists of tandem repeats of the heptapeptide YSPTSPS, plays a central role in the coordination of co‐transcriptional processing through its association with a large number of enzymes and protein/RNA‐binding factors. The CTD is the target of many post‐translational modifications, yielding specific patterns (referred to as “the CTD code”), which are recognized by factors playing essential roles throughout the transcription cycle (Harlen & Churchman, 2017). Importantly, these RNA Pol II CTD modifications are dynamic, allowing spatial and temporal regulation of processing factors binding. Eukaryotic transcription units are, for instance, characterized by conserved 5′ to 3′ profiles of specific pol II CTD phosphorylation events deposited co‐transcriptionally. Phosphorylation on Ser5 is normally high at the 5′ ends of genes where it facilitates mRNA capping, whereas Ser2 phosphorylation is high at 3′ ends where it is important for mRNA 3′ end formation (Buratowski, 2009; Heidemann et al, 2013; Bentley, 2014). Interestingly, in vitro transcription analyses have shown that the RNA Pol II CTD phosphorylation patterns and the factors recognizing them change as a function of post‐initiation time rather than distance elongated (Joo et al, 2019). Moreover, slow RNA Pol II mutants shift RNA Pol II Ser2 hyperphosphorylation to the 5′ end of genes (Fong et al, 2017). These results suggest that the dwell time spent by RNA Pol II within the “target zone” where it associates with a modifier can modulate the deposition or removal of post‐translational modifications (Fong et al, 2017). Altogether, these studies indicate that CTD phosphorylation is controlled by RNA Pol II speed.
Eukaryotic transcription units are also characterized by conserved 5′ to 3′ profiles of specific histone modifications deposited co‐transcriptionally. These histone modifications profiles are sensitive to RNA Pol II speed. Indeed, fast transcription correlates with distal localization of H3K36me3 (Jonkers et al, 2014). Moreover, a fast RNA Pol II mutant shifts H3K4me2/me3 in the 3′ direction (Soares et al, 2017), while a slow RNA Pol II mutant shifts these marks toward the 5′ end of genes (Fong et al, 2017; Soares et al, 2017). In addition, H2Bub1 levels are highly correlated with RNA Pol II velocity and inhibition of transcription elongation strongly affects global H2Bub1 levels (Fuchs et al, 2014a). These results suggest that transcription speed can modulate co‐transcriptional chromatin modification patterns.
Control of TSS selection
Most eukaryotic promoters are known to utilize several different TSSs; nevertheless, how the TSSs are chosen remains an open question. Transcripts with different start sites can differ in their stability or translational efficiency (Rojas‐Duran & Gilbert, 2012) and give rise to transcriptome variations during development and cell differentiation (Hu et al, 2020). In budding yeast, RNA Pol II speed controls the use of differential TSSs, with fast RNA Pol II mutants shifting to upstream TSSs, while slow RNA Pol II mutants shift to downstream TSSs (Kaplan et al, 2012; Braberg et al, 2013; Qiu et al, 2020).
Control of pausing
Following transcription initiation and promoter clearance, RNA Pol II stalls and accumulates at high levels in the promoter‐proximal region. Promoter‐proximal pausing is a key rate‐limiting step of the transcription cycle that RNA Pol II experiences at almost all genes in metazoans (Henriques et al, 2013; Jonkers et al, 2014), at around 20% of genes in plants (Kindgren et al, 2020) and at about 30% of genes in fission yeast (Booth et al, 2016). In Drosophila, a slow RNA Pol II mutant has been shown to shift pausing, which is stabilized by NELF in this species, closer to the TSS (Li et al, 2013), while in plants, a fast RNA Pol II mutant decreases the extent of +1 nucleosome promoter‐proximal pausing (Leng et al, 2020). As such, RNA Pol II velocity can control both the location relative to the TSS and the extent of promoter‐proximal pausing.
Control of gene expression
Finally, RNA Pol II speed has been linked to regulation of gene expression levels in several studies. Indeed, genes with the highest measured RNA Pol II velocity have been shown to generate more mRNAs (Danko et al, 2013; Jonkers et al, 2014; Cohen et al, 2018). Moreover, transcription by a fast RNA Pol II mutant in A. thaliana increased production of nascent RNAs (Leng et al, 2020), suggesting that transcription speed can, to some extent, affect gene expression levels. However, other studies have found no relationship between RNA Pol II speed and expression levels (Singh & Padgett, 2009; Fukaya et al, 2017). Nevertheless, RNA Pol II velocity has also been suggested to affect the timing of mRNA accumulation and could thus play a role in the timing of some developmental regulatory processes (Thummel et al, 1990; Fukaya et al, 2017; Maslon et al, 2019). Indeed, early stages of embryonic development display great variations in cell‐cycle duration with some extremely fast cell divisions (Ciemerych & Sicinski, 2005; Ferree et al, 2016). It is thus possible that during these short cell divisions, RNA Pol II speed plays a crucial role in maintaining the required level of some essential mRNAs that need to be expressed at given developmental stages.
Factors controlling RNA Pol II speed
As described above, dynamic regulation of transcription speed within gene bodies is essential for controlling co‐transcriptional processes, which determine the composition of the transcriptome. Factors regulating RNA Pol II velocity therefore play an essential role in cells and are the topic of several recent studies using genome‐wide RNA Pol II speed measurement methods (Table 2) (Saponaro et al, 2014; Liang et al, 2018; Baluapuri et al, 2019; Cortazar et al, 2019; Gregersen et al, 2019; Hou et al, 2019; Sheridan et al, 2019; Fan et al, 2020). RNA Pol II speed can be controlled by different factors including, but not restricted to, DNA sequence, gene structure, histone modifications, chromatin remodelers, and RNA Pol II‐associated factors and modifications.
Table 2.
List of factors controlling transcription speed
| Factor | Study | Effect on RNA pol II speed | Organism | Number of genes analyzed and measurement method |
|---|---|---|---|---|
| RNA pol II‐associated factors and modifications | ||||
| Ccr4‐Not | Kruk et al (2011) | Increases | Yeast | Measured on 1 gene (GAL1‐YLR454W) by RNA pol II ChIP qPCR at +2 Kb into the gene body at several time points after transcription inhibition by switching from galactose to dextrose containing growth medium |
| CDK12 and CDK13 | Fan et al (2020) | Increases | Human | Measured on 368 genes longer than 100 Kb by PRO‐seq at several time points following DRB release |
| DBIRD | Close et al (2012) | Increases locally at AT‐rich exon–intron junctions | Human | Measured on 2 genes by RNA pol II ChIP qPCR and nascent RNA analysis at several locations into the gene |
| MYC | Liang et al (2018) | Increases | Human | Measured on 1,021 genes by 4sU‐seq 15 min after flavopiridol release |
| Baluapuri et al (2019) | Increases | Human | Measured on 2,163 genes by 4sU‐seq 10 min after DRB release | |
| Caggiano et al (2019) | Increases | Human | Measured on 1 gene (Sam68) by RT–qPCR with primers designed in two regions of the gene and by calculating the abundance of distal versus proximal pre‐mRNA 20 min following DRB release | |
| Paf1C | Hou et al (2019) | Increases | Mouse | Measured on 1,708 genes longer than 70 Kb by PRO‐seq at several time points following DRB treatment |
| Mason and Struhl (2005) | No effect | Yeast | Measured on 1 gene (GAL1‐YLR454W) by RNA pol II ChIP PCR at several locations into the gene 4 min after transcription inhibition by switching from galactose to glucose containing growth medium | |
| Rtf1 subunit | Vos et al (2020) | Increases | Sus scrofa RNA pol II | Measured in vitro using reconstituted elongation complexes |
| PNUTS‐PP1 | Cortazar et al (2019) | Decreases | Human | Measured on 4,324 non‐overlapping genes of more than 60 Kb by RNA pol II ChIP‐seq at several time points following DRB release |
| RECQL5 | Saponaro et al (2014) | Decreases | Human | Measured on 237 long genes by GRO‐seq at several time points following DRB release |
| SCAF8 | Gregersen et al (2019) | Increases | Human | Measured on 4,869 genes between 60 and 300 Kb by DRB/TTchem‐seq at several time points following DRB release |
| Super elongation complex | Liang et al (2018) | Increases | Human | Measured on 982 genes by 4sU‐seq 15 min after flavopiridol release |
| Spt5 | Quan and Hartzog (2010) | Increases | Yeast | Measured on 1 gene (GAL1‐YLR454W) by RNA pol II ChIP qPCR at several locations into the gene 5 min after transcription inhibition by switching from galactose to glucose containing growth medium |
| Fitz et al (2018) | No effect | Mouse | Measured on 8 long and 2 short genes by RT–qPCR on exon‐intron junctions at several time points following flavopiridol release | |
| Spt6 | Ardehali et al (2009) | Increases | Drosophila | Measured on 1 gene (Hsp70) by RNA pol II ChIP qPCR at several locations into the gene and at several time points following activation by heat shock |
| Sub1 | Garcia et al (2012) | Increases | Yeast | Measured on 1 gene (GAL1‐YLR454W) by RNA pol II ChIP PCR at several locations into the gene 4 min after transcription inhibition by switching from galactose to glucose containing growth medium |
| TFIIS | Mason and Struhl (2005) | No effect | Yeast | Measured on 1 gene (GAL1‐YLR454W) by RNA pol II ChIP PCR at several locations into the gene 4 min after transcription inhibition by switching from galactose to glucose containing growth medium |
| Sheridan et al (2019) | Increases | Human | Measured on 600–1,200 genes of more than 75 Kb by RNA pol II ChIP‐seq or Bru‐seq at several time points following DRB release | |
| Zatreanu et al (2019) | Increases | Human | Measured on 1 long gene by RT–qPCR at several locations on the nascent RNA (labeled with 4SU) 40 min following DRB release | |
| Factors affecting the chromatin landscape | ||||
| Brm (Swi‐Snf subunit) | Batsché et al (2006) | Decreases locally at alternative exon junctions | Human | Measured on 1 gene (CD44) by RNA pol II ChIP qPCR at several locations into the gene |
| H3K9me2 and H3K27me3 | Schor et al (2013) | Decreases | Mouse | Measured on 1 gene (NCAM) by RT–qPCR at several locations into the gene and at several time points following DRB release |
| Histone acetylation | Schor et al (2009) | Increases | Mouse | Measured on 1gene (NCAM) by RT–qPCR with primers designed in different introns and by calculating the abundance of distal versus proximal introns in a given region |
| Sharma et al (2014) | Increases | Mouse | Measured on 3 genes by RT–qPCR on the nascent RNA (labeled with BrU) with primers located in different exons spanning the entire genes at several time points following DRB release | |
| HP1 proteins | Saint‐André et al (2011) | Decreases | Human | Measured on 1 gene (CD44) by RNA pol II ChIP qPCR at several locations into the gene |
| Allo et al (2009) | Decreases | Human | Measured on 1 reporter gene by RT–qPCR with primers designed in different regions of the gene and by calculating the abundance of distal versus proximal pre‐mRNA in a given region | |
DNA sequence and nascent RNA folding
During transcription elongation, RNA Pol II encounters roadblocks that can cause it to enter a paused state and backtrack, sliding backwards on the DNA template. Backtracking thus causes a misalignment of the nascent RNA 3′ end with its active site, which needs to be resolved before transcription can restart. RNA poll II pausing and backtracking lead to an overall decrease in RNA Pol II average speed. The DNA sequence in the transcription bubble and the secondary structure adopted by the nascent RNA can influence RNA Pol II speed through controlling pausing and backtracking. Indeed, in vitro transcription assays showed that RNA Pol II pauses are shorter and less frequent on GC‐rich templates. This is probably due to co‐transcriptional folding of nascent RNA which, at GC‐rich sequences, may impose an energy barrier to pausing by impeding backtracking along the DNA (Zamft et al, 2012). Along the same line, in vivo experiments in budding yeast showed that fast transcription elongation by RNA Pol II is associated with strong nascent RNA structures which would prevent RNA Pol II from backtracking thus pushing it forward (Turowski et al, 2020). Short DNA sub‐sequences have also been shown to influence the frequency and duration of pauses in budding yeast. However, here, no correlation has been found between the GC content of these short sequences and RNA Pol II speed (Cohen et al, 2018). Moreover, the GC content within gene bodies has been shown to correlate negatively with RNA Pol II speed (Jonkers et al, 2014; Veloso et al, 2014). As GC rich templates have been shown to increase RNA pol II speed in vitro (Zamft et al, 2012), the role played by the GC content in RNA pol II speed in cellulo might be more complex than anticipated. For example, while high GC content in the transcription bubble slows down RNA polymerase I transcription, strong nascent RNA structures formed close to the polymerase promote forward movement (Turowski et al, 2020). Finally, in budding yeast, RNA Pol II pauses were found to correlate with the stability of the RNA:DNA hybrid, rather than the stability of the DNA:DNA duplex (Lukačišin et al, 2017).
Gene structure
Interestingly, the gene structure itself, and in particular its intron and exon composition, can also play a role in regulating RNA Pol II velocity. Indeed, a positive correlation has been identified between intron 1 size and RNA Pol II speed (Jonkers et al, 2014) and the presence of introns increases transcription speed (Fukaya et al, 2017). Conversely, exon density correlates negatively with RNA Pol II velocity (Jonkers et al, 2014; Veloso et al, 2014), which may be the result of several contributing factors.
First, increased nucleosome occupancy occurs on exons with respect to introns (Andersson et al, 2009; Nahkuri et al, 2009; Schwartz et al, 2009; Spies et al, 2009; Tilgner et al, 2009; Chodavarapu et al, 2010). Nucleosomes can induce RNA Pol II pausing in vitro and in vivo and may even be the major source of pausing (Churchman & Weissman, 2011; Bintu et al, 2012). Consistent with the idea that nucleosomes represent a transcriptional barrier, histone depletion from human cells increases transcription speed (Jimeno‐González et al, 2015). Although no general correlation has been found between nucleosome occupancy and RNA Pol II speed (Jonkers et al, 2014), nucleosomes bound within exons could nonetheless slow down RNA Pol II transcription.
Second, pre‐mRNA splicing may be involved in the reduction of RNA Pol II velocity over exons, which could, in turn, affect co‐transcriptional splicing. Indeed, RNA Pol II ChIP on chip and ChIP‐seq data showed that RNA Pol II accumulates on exons in yeast and human, suggesting that RNA Pol II pauses over exons (Brodsky et al, 2005; Schwartz et al, 2009; Alexander et al, 2010; Carrillo Oesterreich et al, 2010; Kwak et al, 2013; Mayer et al, 2015; Nojima et al, 2015; Harlen et al, 2016). Furthermore, in budding yeast, RNA Pol II ChIP qPCR allowed observation of RNA Pol II pausing at the 3′ splice site (Alexander et al, 2010). This transcriptional pausing over the 3′ splice site is splicing‐dependent, since it was lost upon introduction of a mutation in the 5′ or 3′ splice sites (Alexander et al, 2010; Chathoth et al, 2014). A mutation in the branch site of the intron also inhibited this RNA Pol II pausing, which was restored by introducing a complementary mutation in the U2 snRNA (Alexander et al, 2010). Finally, this pausing is enforced by ubiquitylation of the catalytic subunit of RNA Pol II (Milligan et al, 2017).
More recently, PRO‐seq (precision run‐on sequencing) and NET‐seq (native elongating transcript sequencing), methods offering global and strand‐specific mapping of RNA Pol II density at nucleotide resolution, were used to investigate RNA Pol II pausing throughout genes. These methods confirmed the existence of a pause at different positions over exons and intron‐exon boundaries in several different species. These methods also confirmed the existence of a pause at the 3′ splice site in budding yeast (Harlen et al, 2016) and Drosophila (Kwak et al, 2013), at the 3′ and 5′ splice sites in human cells (Mayer et al, 2015; Nojima et al, 2015) and at the 5′ splice site in plants (Kindgren et al, 2020). Moreover, higher RNA Pol II accumulation has been reported at spliced exons, as compared to skipped exons (Kwak et al, 2013; Mayer et al, 2015; Nojima et al, 2015). All these studies show that exons and intron–exon boundaries are regions of RNA Pol II transcription slow‐down and this slow‐down seems to be induced by the splicing process itself and could, in turn, affect co‐transcriptional splicing.
RNA Pol II‐associated factors and modifications
Many transcription factors play a role in transcription elongation; however, few of them have been shown to directly influence the speed of transcription by RNA Pol II (Table 2). Backtracked RNA Pol II can be rescued by the elongation factor TFIIS which stimulates RNA Pol II intrinsic cleavage activity (Cheung & Cramer, 2011). RNA Pol II cleaves the nascent RNA 3′ end, releasing an RNA fragment of 2–14 bases long (Izban & Luse, 1993) and producing a new 3′ end perfectly aligned with its active site. RNA Pol II can then resume transcription. TFIIS thus reduces the duration of pausing and has been shown to accelerate RNA Pol II transcription in vitro (Ishibashi et al, 2014; Schweikhard et al, 2014). Accordingly, TFIIS increases RNA Pol II speed in human cells (Sheridan et al, 2019; Zatreanu et al, 2019), while its deletion had no effect on RNA Pol II velocity in budding yeast (Mason & Struhl, 2005). Another factor which can accelerate transcription through reactivation of backtracked RNA Pol II is the Ccr4‐Not complex (Kruk et al, 2011), which is considered a master regulator of eukaryotic gene expression. Reactivation of backtracked RNA Pol II by the Ccr4‐Not complex is different from reactivation by TFIIS. In vitro transcription assays showed that in contrast to TFIIS, this complex is unable to reactivate RNA Pol II which has backtracked after incorporation of the chain terminator O‐me‐GTP, indicating that its activity does not involve stimulation of the intrinsic RNA Pol II cleavage activity (Kruk et al, 2011). The Ccr4‐Not complex is instead thought to push RNA Pol II forward through binding to the nascent RNA until realignment of the 3′ end of the nascent transcript in the active site (Kruk et al, 2011). Finally, interplays between these two complexes have been described: While Ccr4‐Not can rescue backtracked RNA Pol II in the absence of TFIIS, further in vitro characterization showed that Ccr4‐Not also interacts with TFIIS and recruits it to the arrested elongation complex resulting in enhanced transcript cleavage (Dutta et al, 2015).
Besides factors stimulating the progression of backtracked RNA Pol II, other general transcription factors are involved in controlling transcription speed by mechanisms that are for the most unclear. These factors, which we introduce in the following section, are mostly proteins or complexes associated with RNA Pol II which are also involved in other transcription steps. Sub1, which is a functional component of the pre‐initiation complex playing a role in TSS selection is also involved in transcription elongation and has been shown to increase RNA Pol II velocity in budding yeast (García et al, 2012). Another factor reported to increase RNA Pol II speed is the transcription elongation factor Spt6, which physically interacts with histone H3 (Bortvin & Winston, 1996; Winkler et al, 2000) and can facilitate nucleosome reassembly (Adkins & Tyler, 2006). Spt6 binds to the phosphorylated RNA Pol II CTD and facilitates RNA Pol II promoter‐proximal pause release (Vos et al, 2018a). In Drosophila, Spt6 has also been shown to increase RNA Pol II speed (Ardehali et al, 2009).
The super elongation complex (SEC) is a large protein complex with compositional and functional diversity and is one of the most active P‐TEFb containing complexes (Luo et al, 2012). In addition to its role in promoter‐proximal pause release and elongation processivity, the SEC facilitates RNA Pol II transcription, as disruption of this complex slows down RNA Pol II (Liang et al, 2018). RNA Pol II CTD heptapeptide residues Tyr1, Ser2, Thr4, Ser5, and Ser7 are dynamically phosphorylated and dephosphorylated by different CTD kinases and phosphatases throughout the transcription cycle (Harlen & Churchman, 2017). CDK12 and CDK13 are multitask RNA Pol II CTD kinases playing important roles for proper gene expression and genome stability (Greenleaf, 2019). Dual inhibition of both kinases significantly decreases RNA Pol II CTD Ser2 and Thr4 phosphorylation, as well as RNA Pol II processivity and speed (Fan et al, 2020).
Spt5 (suppressor of Ty 5) is a highly conserved RNA Pol II‐associated factor, which, in eukaryotes, is part of the DSIF complex together with Spt4. In budding yeast, Spt5 increases transcription speed (Quan & Hartzog, 2010). However, in mouse embryonic fibroblasts it controls RNA Pol II processivity, but not RNA Pol II velocity (Fitz et al, 2018). Nevertheless, consistent with a role of Spt5 in the regulation of RNA Pol II speed, the PP1 phosphatase together with its nuclear targeting subunit (PNUTS) can dephosphorylate Spt5, which slows down RNA Pol II transcription in human cells (Cortazar et al, 2019). Moreover, the transcription factor MYC, which directly binds to Spt5 and recruits it to promoters, allowing Spt5 transfer onto RNA Pol II (Baluapuri et al, 2019), has been shown to be required for fast transcription elongation (Liang et al, 2018; Baluapuri et al, 2019; Caggiano et al, 2019).
Furthermore, involved in regulating RNA Pol II speed is the conserved polymerase‐associated factor 1 complex (Paf1C), which plays multiple roles in chromatin transcription and genomic regulation (Van Oss et al, 2017). Paf1C specifically increases RNA Pol II velocity within the first part of gene bodies in mice (Hou et al, 2019), although no effect of the loss of this factor could be measured in budding yeast (Mason & Struhl, 2005). On the other hand, consistent with a role of the Paf1C complex in RNA Pol II speed regulation, recent data show that the dissociable Paf1C subunit Rtf1 increases RNA Pol II speed in in vitro transcription assays (Vos et al, 2020).
Two more components affecting RNA Pol II speed are SCAF8 and the DBIRD complex. SCAF8, an RNA Pol II CTD‐interacting protein, which plays a role in suppressing the use of proximal poly(A) sites, is a positive RNA Pol II elongation factor that increases RNA Pol II speed (Gregersen et al, 2019). Depletion of DBIRD, a protein complex formed by DBC1 and ZNF326, induces an accumulation of RNA Pol II on A/T‐rich alternatively spliced internal exons in human cells, suggesting a role for DBIRD in the control of RNA Pol II speed over exons (Close et al, 2012).
Finally, RECQL5, a member of the conserved family of RECQ DNA helicases, decreases RNA Pol II speed and suppresses genome instability (Saponaro et al, 2014). The hypothesis behind these two observations is that decreasing RNA Pol II speed smoothens transcription elongation and thus renders RNA Pol II less prone to pausing and arrest which can induce transcription stress (Saponaro et al, 2014).
As exemplified above, some factors such as Paf1C, TFIIS, or Spt5 have been reported to be important for transcription speed in certain species, but not in others. This may represent a real biological difference between species or could be due to the fact that these factors are involved only for some genes or under particular conditions. However, this difference could also be caused by technical issues: In yeast, genes are quite short (around 1.5–2 kb long on average, although the gene YLR454W, used for most of the experiments measuring RNA Pol II speed in budding yeast, is exceptionally large with a size of around 8 kb (Table 2) (Mason & Struhl, 2005; Quan & Hartzog, 2010; Kruk et al, 2011; García et al, 2012)). It might thus be difficult to assess the effect of a factor on transcription speed because the methods used to measure RNA Pol II speed offer poor resolution on short genes. Indeed, in mammalian cells most studies only consider very long genes to accurately decipher an elongation speed (Saponaro et al, 2014; Cortazar et al, 2019; Gregersen et al, 2019; Hou et al, 2019; Sheridan et al, 2019; Fan et al, 2020).
Chromatin landscape
Chromatin modifications can also regulate RNA Pol II velocity by facilitating or hindering RNA Pol II progression through nucleosomes (Table 2). Genome‐wide studies have shown that H3K79me2, H2B mono‐ubiquitylation, and H4K20me1 positively correlate with RNA Pol II speed in the gene body (Fuchs et al, 2014a; Jonkers et al, 2014; Veloso et al, 2014). Besides these global correlations, other histone modifications locally influence RNA Pol II velocity: modifications triggering chromatin relaxation increase RNA Pol II speed, while modifications triggering chromatin compaction slow down RNA Pol II. For instance, a reduction in the level of class II HDACs on specific genes and increased H3 and H4 acetylation correlate with faster RNA Pol II transcription on these genes (Sharma et al, 2014). Similarly, H3K9 hyperacetylation correlates with a change in the ratio of distal versus proximal pre‐mRNA levels suggesting a local increase in RNA Pol II speed (Schor et al, 2009), whereas an increase in H3K9 di‐methylation and H3K27 tri‐methylation correlates with a slow‐down of RNA Pol II transcription (Schor et al, 2013).
The evolutionary conserved family of non‐histone chromosomal proteins HP1 (heterochromatin protein 1) consists of three isoforms in human. HP1 proteins bind to di‐ and trimethylated H3K9, as well as to other proteins involved in various epigenetic functions, such as histone methyltransferases (HMTs) (Zeng et al, 2010). HP1α and HP1β isoforms are mainly heterochromatic, while HP1γ is present in both heterochromatin and euchromatin (Zeng et al, 2010). Through RNA Pol II ChIP measurements, HP1γ has been suggested to locally increase RNA Pol II pausing on alternatively spliced exons enriched in H3K9me3 marks (Saint‐André et al, 2011). The mechanism is thought to involve the RNAi machinery (Saint‐André et al, 2011; Ameyar‐Zazoua et al, 2012). Similarly, HP1α recruitment to local H3K9me2 enrichment induced by siRNA targeting correlates with a change in the ratio of distal versus proximal pre‐mRNA levels suggesting a decreased local RNA Pol II speed (Alló et al, 2009).
ATP‐dependent chromatin remodeling complexes regulate the access of different factors to genomic DNA during transcription, replication, and repair by altering the contacts between histones and DNA within nucleosomes. The SWI/SNF family of ATP‐dependent chromatin remodeling complexes controls the expression of many genes by sliding or evicting nucleosomes positioned on binding sites for transcription activators or repressors (Clapier et al, 2017). SWI/SNF complexes contain more than 10 protein subunits, including a single ATPase, either SMARCA4 (BRG1) or SMARCA2 (BRM), that utilizes the energy of ATP hydrolysis to alter nucleosome positioning (Euskirchen et al, 2012). The SWI/SNF complex subunit BRM has been suggested to locally decrease RNA Pol II speed in response to activation of the MAP kinase pathway. Indeed, BRM knockdown has been shown to decrease RNA Pol II accumulation on alternatively spliced exons as measured by RNA Pol II ChIP (Batsché et al, 2006).
Adapting RNA Pol II speed to the cellular environment
Over the last 20 years, the use of kinetic RNA Pol II mutants has shown that RNA Pol II speed regulation plays an essential role in many co‐transcriptional processes thus controlling the composition of the transcriptome. The use of such mutants has also shown that RNA Pol II speed regulation is essential for development (Maslon et al, 2019; Leng et al, 2020). However, RNA Pol II mutants globally affect RNA Pol II velocity and thus may have side effects on the expression of transcription factors involved in these co‐transcriptional events or in physiological processes such as development. These studies thus left the question of the physiological relevance of transcription speed regulation open. Recent studies have however given a hint of an answer to this question by revealing that RNA Pol II velocity can be adapted to respond to intra‐ and extra‐cellular stimuli (Fig 3). First, RNA Pol II velocity differs by as much as 25–40% on individual genes stimulated by diverse signaling pathways in different cell lines (Danko et al, 2013). Second, RNA Pol II velocity changes around alternatively spliced exons in specific genes, in response to cell depolarization and cellular differentiation (Schor et al, 2009; Schor et al, 2013; Sharma et al, 2014). Along the same lines, RNA Pol II accumulates on alternatively spliced exons of specific genes in response to activation of the protein kinase C or MAP kinase pathways, suggesting a decrease in RNA Pol II speed (Batsché et al, 2006; Saint‐André et al, 2011; Ameyar‐Zazoua et al, 2012). Third, transcription speed increases downstream of some protein‐coding genes in response to an oncogenic signal (Muniz et al, 2017). Finally, UV‐induced DNA damage has been proposed to induce a reduction in RNA Pol II speed (Muñoz et al, 2009; Lavigne et al, 2017; Williamson et al, 2017). Although this last result can be explained by transcription‐blocking DNA lesions accumulating along gene bodies, it remains plausible that DNA damage signaling also directly triggers RNA Pol II transcription slow‐down.
Figure 3. RNA Pol II speed is regulated in order to adapt the transcriptome composition in response to intra‐ or extra‐cellular stimuli.
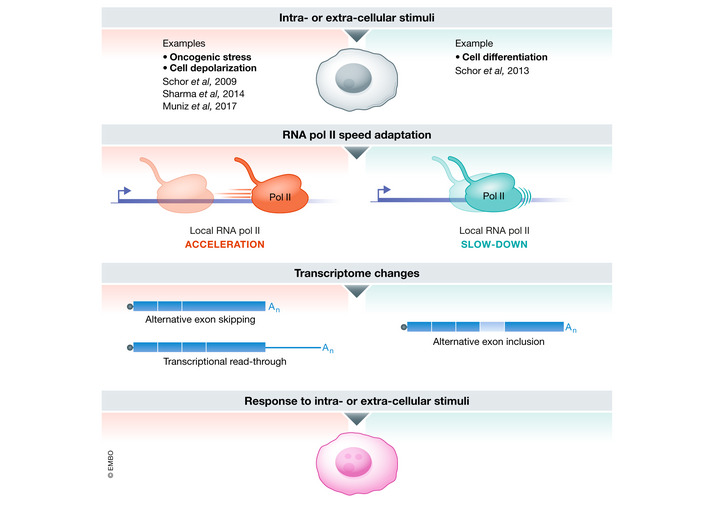
In response to intra‐ or extra‐cellular stimuli such as oncogenic stress, cell depolarization, or cell differentiation, RNA Pol II can either accelerate or slow down locally, inducing a change in alternative splicing or the extent of read‐through which could play a role in the response to stimuli.
These variations in transcription speed in response to changes in the cellular environment may have important consequences on transcriptome composition. Indeed, changes in RNA Pol II velocity measured in response to cell depolarization and cell differentiation regulate alternative splicing and thus RNA identity (Schor et al, 2009; Schor et al, 2013; Sharma et al, 2014). Moreover, we have shown that the increase in RNA Pol II velocity downstream of genes is accompanied by an increase in transcriptional read‐through which interferes with and thus decreases the expression of downstream convergent protein‐coding genes (Muniz et al, 2017). Increased transcriptional read‐through is known to interfere with the expression of downstream genes also in other contexts such as cancer or HSV‐1 infection in human cells (Grosso et al, 2015; Rutkowski et al, 2015; Hennig et al, 2018), as well as cold acclimation in plants (Kindgren et al, 2018). Transcriptional read‐through can also give rise to a chimeric transcript which can be spliced and translated into an aberrant fusion protein, if it invades the downstream gene in a pair of tandem genes (Grosso et al, 2015; Hennig et al, 2018). Therefore, by controlling the extent of transcriptional read‐through, RNA Pol II speed can have dramatic consequences on the expression of surrounding genes.
Importantly, transcriptome modifications induced by RNA Pol II speed changes can be extremely fast, since they do not require new transcription initiation events. Furthermore, these transcriptome modifications could participate in the response to intra‐ or extra‐cellular stimuli. Indeed, we have shown that those convergent genes, antisense to the transcriptional read‐through induced in response to oncogene‐induced senescence, are enriched in pro‐proliferative genes (Muniz et al, 2017). Thus, by repressing those genes, the transcriptional read‐through may participate in the control of the genetic program of senescence. Moreover, transcriptional read‐through is induced downstream of thousands of genes in response to different kinds of stresses (Rutkowski et al, 2015; Vilborg et al, 2015; Vilborg et al, 2017; Cardiello et al, 2018; Hennig et al, 2018) and has been proposed to play an important role in the stress response by reinforcing the nuclear scaffold integrity after nuclear stress (Vilborg et al, 2015). Finally, read‐through transcription can also affect the genome 3D structure: In read‐through regions, elongating RNA Pol II displaces cohesin from CTCF sites thus eliminating chromatin loops and locally decompacting chromatin (Heinz et al, 2018). Therefore, by controlling the extent of transcriptional read‐through, RNA Pol II speed could affect both gene expression and genome 3D organization. More generally speaking, regulation of RNA Pol II speed might thus be important for the cell response to intra‐ and extra‐cellular stimuli by affecting the whole transcriptome, from splicing‐dependent processes to gene expression and chromatin organization.
Conclusion and perspectives
More and more studies point to the importance of RNA Pol II speed in determining the identity of RNAs produced from a specific locus. Indeed, changes in transcription speed induce changes in the splicing repertoire, with modifications of alternative splicing and of the production of circular RNAs by back‐splicing (see section on co‐transcriptional processes regulated by RNA Pol II speed). These changes also affect transcription termination‐related processes such as poly(A) site usage and the extent of transcriptional read‐through. In summary, changes in RNA Pol II speed modify the whole spectrum of coding and non‐coding RNAs produced from the genome. By changing the spectrum of RNAs produced by transcribed genes, RNA Pol II speed can thus provide another layer of complexity in genome expression control.
An important issue is to which extent this speed can be regulated upon changes in the cellular environment. As exemplified above, a number of factors are known to participate in controlling transcription speed. Some of them are general transcription factors probably affecting all transcribing RNA polymerases. Thus, any modification of the function of these factors presumably affects RNA Pol II velocity genome wide, leading to major changes in the repertoire of coding and non‐coding RNAs. However, some features of the chromatin landscape, such as histone modifications or chromatin bound factors, can locally control RNA Pol II speed. Chromatin is a highly dynamic structure, which is known to harbor local changes depending on the cellular environment. Such local changes within a transcribed region are thus susceptible to lead to local changes in RNA Pol II velocity and thus to modifications in the identity of the RNAs produced by this locus. We recently exemplified such a local modification of chromatin and RNA Pol II speed in cells undergoing oncogene‐induced senescence downstream of two genes, leading to extended transcriptional read‐through (Muniz et al, 2017). Along the same lines, local changes of histone modifications such as acetylation or methylation and RNA Pol II speed occur on alternative spliced exons during cell response to depolarization and differentiation (Schor et al, 2009; Schor et al, 2013; Sharma et al, 2014). Whether such observations can be generalized to various environmental changes is clearly an open issue which merits further investigation. Deciphering the mechanisms underlying such changes, as well as the signaling pathways involved, will also clearly be a major step toward our understanding of the genome response to environmental changes. Similarly, the local regulation of RNA Pol II velocity upon activation of specific signaling pathways may be essential during the development of multicellular organisms and therefore represents a challenging line of research in the field of developmental biology.
It would also be important to investigate whether deficient regulations of RNA Pol II speed participate in disease progression, again through the resulting consequences on local or global genome expression. Cancer cells have probably exploited such mechanisms given the variety of circular and read‐through RNAs whose expression is affected during cancer progression (Grosso et al, 2015; Smid et al, 2019). Most importantly, recent results point to a causative role of circular RNAs on cancer progression (Chen et al, 2019). Investigating the developmental‐programmed and cancer‐induced modifications in RNA Pol II velocity and the complexity of the RNAs affected thus clearly opens a new avenue of research in the fields of physiology and physiopathology.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
The authors would like to thank the funders that support their research: the Ligue Nationale Contre le Cancer (équipe labellisée) (EL‐190350) and the Fondation Toulouse Cancer Sante (DRUGSPEED 2018‐2) to DT; and the Fondation de France (185742_ 00087515) and the Ligue Contre le Cancer 31 to LM. LM was also supported by a fellowship from the Fondation de France.
The EMBO Journal (2021) 40: e105740.
References
- Adkins MW, Tyler JK (2006) Transcriptional activators are dispensable for transcription in the absence of Spt6‐mediated chromatin reassembly of promoter regions. Mol Cell 21: 405–416 [DOI] [PubMed] [Google Scholar]
- Akhtar J, Kreim N, Marini F, Mohana G, Brüne D, Binder H, Roignant J‐Y (2019) Promoter‐proximal pausing mediated by the exon junction complex regulates splicing. Nat Commun 10: 521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD (2010) Splicing‐dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M , Agirre E, Plass M, Eyras E, Elela SA et al (2009) Control of alternative splicing through siRNA‐mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 [DOI] [PubMed] [Google Scholar]
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L (2011) Total RNA sequencing reveals nascent transcription and widespread co‐transcriptional splicing in the human brain. Nat Struct Mol Biol 18: 1435–1440 [DOI] [PubMed] [Google Scholar]
- Ameyar‐Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC et al (2012) Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 19: 998–1004 [DOI] [PubMed] [Google Scholar]
- Andersson R, Enroth S, Rada‐Iglesias A, Wadelius C, Komorowski J (2009) Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res 19: 1732–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annibale P, Gratton E (2015) Single cell visualization of transcription kinetics variance of highly mobile identical genes using 3D nanoimaging. Sci Rep 5: 9258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT (2009) Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J 28: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal‐Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell 56: 55–66 [DOI] [PubMed] [Google Scholar]
- Aslanzadeh V, Huang Y, Sanguinetti G, Beggs JD (2018) Transcription rate strongly affects splicing fidelity and cotranscriptionality in budding yeast. Genome Res 28: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluapuri A, Hofstetter J, Dudvarski Stankovic N, Endres T, Bhandare P, Vos SM, Adhikari B, Schwarz JD, Narain A, Vogt M et al (2019) MYC recruits SPT5 to RNA polymerase II to promote processive transcription elongation. Mol Cell 74: 674–687.e611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranello L, Wojtowicz D, Cui K, Devaiah BN, Chung HJ, Chan‐Salis KY, Guha R, Wilson K, Zhang X, Zhang H et al (2016) RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell 165: 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsché E, Yaniv M, Muchardt C (2006) The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 13: 22–29 [DOI] [PubMed] [Google Scholar]
- Battle DJ, Doudna JA (2001) The stem‐loop binding protein forms a highly stable and specific complex with the 3' stem‐loop of histone mRNAs. RNA 7: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Ari Y, Brody Y, Kinor N, Mor A, Tsukamoto T, Spector DL, Singer RH, Shav‐Tal Y (2010) The life of an mRNA in space and time. J Cell Sci 123: 1761–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2014) Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C (2012) Nucleosomal elements that control the topography of the barrier to transcription. Cell 151: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M , Knezevich A, Pradet‐Balade B, Bäcker V, Kornblihtt A, Marcello A, Bertrand E (2007) The transcriptional cycle of HIV‐1 in real‐time and live cells. J Cell Biol 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth GT, Wang IX, Cheung VG, Lis JT (2016) Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res 26: 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F (1996) Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272: 1473–1476 [DOI] [PubMed] [Google Scholar]
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F et al (2013) From structure to systems: high‐resolution, quantitative genetic analysis of RNA polymerase II. Cell 154: 775–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, Meyer CA, Swinburne IA, Hall G, Keenan BJ, Liu XS, Fox EA, Silver PA (2005) Genomic mapping of RNA polymerase II reveals sites of co‐transcriptional regulation in human cells. Genome Biol 6: R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Böhnlein EM, Neugebauer KM, Darzacq X, Shav‐Tal Y (2011) The in vivo kinetics of RNA polymerase II elongation during co‐transcriptional splicing. PLoS Biol 9: e1000573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S (2009) Progression through the RNA polymerase II CTD cycle. Mol Cell 36: 541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano C, Pieraccioli M, Panzeri V, Sette C, Bielli P (2019) c‐MYC empowers transcription and productive splicing of the oncogenic splicing factor Sam68 in cancer. Nucleic Acids Res 47: 6160–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiello JF, Goodrich JA, Kugel JF (2018) Heat shock causes a reversible increase in RNA polymerase II occupancy downstream of mRNA genes, consistent with a global loss in transcriptional termination. Mol Cell Biol 38: e00181–e218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM (2010) Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell 40: 571–581 [DOI] [PubMed] [Google Scholar]
- Chathoth KT, Barrass JD, Webb S, Beggs JD (2014) A splicing‐dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell 53: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Smith ER, Shilatifard A (2018) Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 19: 464–478 [DOI] [PubMed] [Google Scholar]
- Chen LL (2020) The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 21: 475–490 [DOI] [PubMed] [Google Scholar]
- Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, Zeng Y, Hua JT, Petricca J, Guo H et al (2019) Widespread and functional RNA circularization in localized prostate cancer. Cell 176: 831–843.e822 [DOI] [PubMed] [Google Scholar]
- Chen Y, Chafin D, Price DH, Greenleaf AL (1996) Drosophila RNA polymerase II mutants that affect transcription elongation. J Biol Chem 271: 5993–5999 [PubMed] [Google Scholar]
- Chen Y, Weeks J, Mortin MA, Greenleaf AL (1993) Mapping mutations in genes encoding the two large subunits of Drosophila RNA polymerase II defines domains essential for basic transcription functions and for proper expression of developmental genes. Mol Cell Biol 13: 4214–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AC, Cramer P (2011) Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature 471: 249–253 [DOI] [PubMed] [Google Scholar]
- Chiu AC, Suzuki HI, Wu X, Mahat DB, Kriz AJ, Sharp PA (2018) Transcriptional pause sites delineate stable nucleosome‐associated premature polyadenylation suppressed by U1 snRNP. Mol Cell 69: 648–663.e647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ et al (2010) Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS (2011) Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciemerych MA, Sicinski P (2005) Cell cycle in mouse development. Oncogene 24: 2877–2898 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL (2017) Mechanisms of action and regulation of ATP‐dependent chromatin‐remodelling complexes. Nat Rev Mol Cell Biol 18: 407–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close P, East P, Dirac‐Svejstrup AB, Hartmann H, Heron M, Maslen S, Chariot A, Söding J, Skehel M, Svejstrup JQ (2012) DBIRD complex integrates alternative mRNA splicing with RNA polymerase II transcript elongation. Nature 484: 386–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Zafrir Z, Tuller T (2018) A code for transcription elongation speed. RNA Biol 15: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L, Adelman K (2019) Promoter‐proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33: 960–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan AM, Tunnacliffe E, Cannon D, Chubb JR (2016) A continuum model of transcriptional bursting. Elife 5: e13051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortazar MA, Sheridan RM, Erickson B, Fong N, Glover‐Cutter K, Brannan K, Bentley DL (2019) Control of RNA Pol II speed by PNUTS‐PP1 and Spt5 dephosphorylation facilitates termination by a "sitting duck torpedo" mechanism. Mol Cell 76: 896–908.e894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czudnochowski N, Bösken CA, Geyer M (2012) Serine‐7 but not serine‐5 phosphorylation primes RNA polymerase II CTD for P‐TEFb recognition. Nat Commun 3: 842 [DOI] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL (2013) Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 50: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav‐Tal Y, de Turris V , Brody Y, Shenoy SM, Phair RD, Singer RH (2007) In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett‐Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T (2012) A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienemann C, Schwalb B, Schilbach S, Cramer P (2019) Promoter distortion and opening in the RNA polymerase II cleft. Mol Cell 73: 97–106.e104 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch‐Moshkovitz S, Schwartz S, Salmon‐Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob‐Hirsch J, Amariglio N, Kupiec M et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A‐seq. Nature 485: 201–206 [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Kaygun H, Dadlez M, Marzluff WF (2003) A 3' exonuclease that specifically interacts with the 3' end of histone mRNA. Mol Cell 12: 295–305 [DOI] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, de la Mata M , Marasco LE, Muñoz MJ, Le Jossic‐Corcos C, Corcos L, Kornblihtt AR (2014) How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell 54: 683–690 [DOI] [PubMed] [Google Scholar]
- Dutta A, Babbarwal V, Fu J, Brunke‐Reese D, Libert DM, Willis J, Reese JC (2015) Ccr4‐not and TFIIS function cooperatively to rescue arrested RNA polymerase II. Mol Cell Biol 35: 1915–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton JD, Francis L, Davidson L, West S (2020) A unified allosteric/torpedo mechanism for transcriptional termination on human protein‐coding genes. Genes Dev 34: 132–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton JD, West S (2020) Termination of transcription by RNA polymerase II: BOOM!. Trends Genet 36: 664–675 [DOI] [PubMed] [Google Scholar]
- Eick D, Geyer M (2013) The RNA polymerase II carboxy‐terminal domain (CTD) code. Chem Rev 113: 8456–8490 [DOI] [PubMed] [Google Scholar]
- Euskirchen G, Auerbach RK, Snyder M (2012) SWI/SNF chromatin‐remodeling factors: multiscale analyses and diverse functions. J Biol Chem 287: 30897–30905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Devlin JR, Hogg SJ, Doyle MA, Harrison PF, Todorovski I, Cluse LA, Knight DA, Sandow JJ, Gregory G et al (2020) CDK13 cooperates with CDK12 to control global RNA polymerase II processivity. Sci Adv 6: eaaz5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH (1998) Visualization of single RNA transcripts in situ. Science 280: 585–590 [DOI] [PubMed] [Google Scholar]
- Femino AM, Fogarty K, Lifshitz LM, Carrington W, Singer RH (2003) Visualization of single molecules of mRNA in situ. Methods Enzymol 361: 245–304 [DOI] [PubMed] [Google Scholar]
- Ferree PL, Deneke VE, Di Talia S (2016) Measuring time during early embryonic development. Semin Cell Dev Biol 55: 80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz J, Neumann T, Pavri R (2018) Regulation of RNA polymerase II processivity by Spt5 is restricted to a narrow window during elongation. EMBO J 37: e97965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, Bentley DL (2015) Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol Cell 60: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu XD, Bentley DL (2014) Pre‐mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev 28: 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Saldi T, Sheridan RM, Cortazar MA, Bentley DL (2017) RNA Pol II dynamics modulate co‐transcriptional chromatin modification, CTD phosphorylation, and transcriptional direction. Mol Cell 66: 546–557.e543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Hollander D, Voichek Y, Ast G, Oren M (2014a) Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res 24: 1572–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Voichek Y, Benjamin S, Gilad S, Amit I, Oren M (2014b) 4sUDRB‐seq: measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biol 15: R69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Voichek Y, Rabani M, Benjamin S, Gilad S, Amit I, Oren M (2015) Simultaneous measurement of genome‐wide transcription elongation speeds and rates of RNA polymerase II transition into active elongation with 4sUDRB‐seq. Nat Protoc 10: 605–618 [DOI] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M (2017) Rapid rates of Pol II elongation in the Drosophila embryo. Curr Biol 27: 1387–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A, Collin A, Calvo O (2012) Sub1 associates with Spt5 and influences RNA polymerase II transcription elongation rate. Mol Biol Cell 23: 4297–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Struhl K (2020) The transcriptional elongation rate regulates alternative polyadenylation in yeast. Elife 9: e59810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf AL (1983) Amanitin‐resistant RNA polymerase II mutations are in the enzyme's largest subunit. J Biol Chem 258: 13403–13406 [PubMed] [Google Scholar]
- Greenleaf AL (2019) Human CDK12 and CDK13, multi‐tasking CTD kinases for the new millenium. Transcription 10: 91–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf AL, Borsett LM, Jiamachello PF, Coulter DE (1979) Alpha‐amanitin‐resistant D. melanogaster with an altered RNA polymerase II. Cell 18: 613–622 [DOI] [PubMed] [Google Scholar]
- Gregersen LH, Mitter R, Ugalde AP, Nojima T, Proudfoot NJ, Agami R, Stewart A, Svejstrup JQ (2019) SCAF4 and SCAF8, mRNA anti‐terminator proteins. Cell 177: 1797–1813.e1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Mitter R, Svejstrup JQ (2020) Using TT chem‐seq for profiling nascent transcription and measuring transcript elongation. Nat Protoc 15: 604–627 [DOI] [PubMed] [Google Scholar]
- Grosso AR, Leite AP, Carvalho S, Matos MR, Martins FB, Vítor AC, Desterro JM, Carmo‐Fonseca M, de Almeida SF (2015) Pervasive transcription read‐through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife 4: e09214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbüchle O, Wellauer PK, Cribbs DL, Schibler U (1984) Termination of transcription in the mouse alpha‐amylase gene Amy‐2a occurs at multiple sites downstream of the polyadenylation site. Cell 38: 737–744 [DOI] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL (2011) A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Holder MV, Choorapoikayil S, Gajewski M, Özbudak EM, Lewis J (2013) The elongation rate of RNA polymerase II in zebrafish and its significance in the somite segmentation clock. Development 140: 444–453 [DOI] [PubMed] [Google Scholar]
- Harlen KM, Churchman LS (2017) The code and beyond: transcription regulation by the RNA polymerase II carboxy‐terminal domain. Nat Rev Mol Cell Biol 18: 263–273 [DOI] [PubMed] [Google Scholar]
- Harlen KM, Trotta KL, Smith EE, Mosaheb MM, Fuchs SM, Churchman LS (2016) Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho‐CTD residue. Cell Rep 15: 2147–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He PC, He C (2021) m6 A RNA methylation: from mechanisms to therapeutic potential. EMBO J 40: e105977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann M, Hintermair C, Voß K, Eick D (2013) Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta 1829: 55–62 [DOI] [PubMed] [Google Scholar]
- Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L et al (2018) Transcription elongation can affect genome 3D structure. Cell 174: 1522–1536.e1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig T, Michalski M, Rutkowski AJ, Djakovic L, Whisnant AW, Friedl MS, Jha BA, Baptista MAP, L'Hernault A, Erhard F et al (2018) HSV‐1‐induced disruption of transcription termination resembles a cellular stress response but selectively increases chromatin accessibility downstream of genes. PLoS Pathog 14: e1006954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K (2013) Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 52: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocine S, Raymond P, Zenklusen D, Chao JA, Singer RH (2013) Single‐molecule analysis of gene expression using two‐color RNA labeling in live yeast. Nat Methods 10: 119–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer E, Darnell JE (1981) The primary transcription unit of the mouse beta‐major globin gene. Cell 23: 585–593 [DOI] [PubMed] [Google Scholar]
- Hollensen AK, Thomsen HS, Lloret‐Llinares M, Kamstrup AB, Jensen JM, Luckmann M, Birkmose N, Palmfeldt J, Jensen TH, Hansen TB et al (2020) circZNF827 nucleates a transcription inhibitory complex to balance neuronal differentiation. Elife 9: e58478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B (2013) Analysis of alternative cleavage and polyadenylation by 3' region extraction and deep sequencing. Nat Methods 10: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Wang Y, Liu Y, Zhang N, Shamovsky I, Nudler E, Tian B, Dynlacht BD (2019) Paf1C regulates RNA polymerase II progression by modulating elongation rate. Proc Natl Acad Sci USA 116: 14583–14592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KJ, Kane CM, Ares M (2003) Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA 9: 993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhong J, Xiao Y, Xing Z, Sheu K, Fan S, An Q, Qiu Y, Zheng Y, Liu X et al (2020) Single‐cell RNA cap and tail sequencing (scRCAT‐seq) reveals subtype‐specific isoforms differing in transcript demarcation. Nat Commun 11: 5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ (2011) Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 21: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Dangkulwanich M, Coello Y, Lionberger TA, Lubkowska L, Ponticelli AS, Kashlev M, Bustamante C (2014) Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc Natl Acad Sci USA 111: 3419–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB (2000) Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol 20: 2970–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS (1993) The increment of SII‐facilitated transcript cleavage varies dramatically between elongation competent and incompetent RNA polymerase II ternary complexes. J Biol Chem 268: 12874–12885 [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno‐González S, Payán‐Bravo L, Muñoz‐Cabello AM, Guijo M, Gutierrez G, Prado F, Reyes JC (2015) Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre‐mRNA splicing. Proc Natl Acad Sci USA 112: 14840–14845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT (2014) Genome‐wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife 3: e02407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Lis JT (2015) Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 16: 167–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo YJ, Ficarro SB, Chun Y, Marto JA, Buratowski S (2019) In vitro analysis of RNA polymerase II elongation complex dynamics. Genes Dev 33: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamieniarz‐Gdula K, Proudfoot NJ (2019) Transcriptional control by premature termination: a forgotten mechanism. Trends Genet 35: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Jin H, Zhang IL, Belyanin A (2012) Dissection of Pol II trigger loop function and Pol II activity‐dependent control of start site selection in vivo. PLoS Genet 8: e1002627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Larsson KM, Kornberg RD (2008) The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha‐amanitin. Mol Cell 30: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Koyama S, Yoshida R (2019) Statistical inference of the rate of RNA polymerase II elongation by total RNA sequencing. Bioinformatics 35: 1877–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT, Rosbash M (2011) Nascent‐seq indicates widespread cotranscriptional pre‐mRNA splicing in Drosophila . Genes Dev 25: 2502–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Ard R, Ivanov M, Marquardt S (2018) Transcriptional read‐through of the long non‐coding RNA SVALKA governs plant cold acclimation. Nat Commun 9: 4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Ivanov M, Marquardt S (2020) Native elongation transcript sequencing reveals temperature dependent dynamics of nascent RNAPII transcription in Arabidopsis . Nucleic Acids Res 48: 2332–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M (2008) Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell 30: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P, Bühler M (2018) Adenosine methylation as a molecular imprint defining the fate of RNA. FEBS Lett 592: 2845–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20: 675–691 [DOI] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ (2013) Condensin controls recruitment of RNA polymerase II to achieve nematode X‐chromosome dosage compensation. Elife 2: e00808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruk JA, Dutta A, Fu J, Gilmour DS, Reese JC (2011) The multifunctional Ccr4‐Not complex directly promotes transcription elongation. Genes Dev 25: 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT (2013) Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH (2011) Real‐time observation of transcription initiation and elongation on an endogenous yeast gene. Science 332: 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne MD, Konstantopoulos D, Ntakou‐Zamplara KZ, Liakos A, Fousteri M (2017) Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat Commun 8: 2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng X, Ivanov M, Kindgren P, Malik I, Thieffry A, Brodersen P, Sandelin A, Kaplan CD, Marquardt S (2020) Organismal benefits of transcription speed control at gene boundaries. EMBO Rep 21: e49315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Rhee HS, Ghosh SK, Bai L, Pugh BF, Gilmour DS (2013) Kinetic competition between elongation rate and binding of NELF controls promoter‐proximal pausing. Mol Cell 50: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu M, Chen LF, Chen R (2018) P‐TEFb: Finding its ways to release promoter‐proximally paused RNA polymerase II. Transcription 9: 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28: 2233–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Smith ER, Aoi Y, Stoltz KL, Katagi H, Woodfin AR, Rendleman EJ, Marshall SA, Murray DC, Wang L et al (2018) Targeting processive transcription elongation via SEC disruption for MYC‐induced cancer therapy. Cell 175: 766–779.e717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H et al (2019) Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177: 865–880.e821 [DOI] [PubMed] [Google Scholar]
- Liu X, Freitas J, Zheng D, Oliveira MS, Hoque M, Martins T, Henriques T, Tian B, Moreira A (2017) Transcription elongation rate has a tissue‐specific impact on alternative cleavage and polyadenylation in Drosophila melanogaster . RNA 23: 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J, Falck‐Pedersen E, Darnell JE, Shenk T (1987) A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj‐globin gene. Proc Natl Acad Sci USA 84: 8306–8310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukačišin M, Landon M, Jajoo R (2017) Sequence‐specific thermodynamic properties of nucleic acids influence both transcriptional pausing and backtracking in yeast. PLoS One 12: e0174066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Shilatifard A (2012) The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13: 543–547 [DOI] [PubMed] [Google Scholar]
- Maiuri P, Knezevich A, De Marco A, Mazza D, Kula A, McNally JG, Marcello A (2011) Fast transcription rates of RNA polymerase II in human cells. EMBO Rep 12: 1280–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RM, Rino J, Carvalho C, Kirchhausen T, Carmo‐Fonseca M (2013) Live‐cell visualization of pre‐mRNA splicing with single‐molecule sensitivity. Cell Rep 4: 1144–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Koreski KP (2017) Birth and death of histone mRNAs. Trends Genet 33: 745–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslon MM, Braunschweig U, Aitken S, Mann AR, Kilanowski F, Hunter CJ, Blencowe BJ, Kornblihtt AR, Adams IR, Cáceres JF (2019) A slow transcription rate causes embryonic lethality and perturbs kinetic coupling of neuronal genes. EMBO J 38: e101244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K (2005) Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840 [DOI] [PubMed] [Google Scholar]
- de la Mata M , Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR (2003) A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- Maxwell CS, Kruesi WS, Core LJ, Kurhanewicz N, Waters CT, Lewarch CL, Antoshechkin I, Lis JT, Meyer BJ, Baugh LR (2014) Pol II docking and pausing at growth and stress genes in C. elegans . Cell Rep 6: 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, di Iulio J , Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS (2015) Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161: 541–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Landry HM, Churchman LS (2017) Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr Opin Cell Biol 46: 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149: 1635–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan L, Sayou C, Tuck A, Auchynnikava T, Reid JE, Alexander R, Alves FL, Allshire R, Spanos C, Rappsilber J et al (2017) RNA polymerase II stalling at pre‐mRNA splice sites is enforced by ubiquitination of the catalytic subunit. Elife 6: e27082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz L, Deb MK, Aguirrebengoa M, Lazorthes S, Trouche D, Nicolas E (2017) Control of gene expression in senescence through transcriptional read‐through of convergent protein‐coding genes. Cell Rep 21: 2433–2446 [DOI] [PubMed] [Google Scholar]
- Muñoz MJ, Pérez Santangelo MS, Paronetto MP, de la Mata M , Pelisch F, Boireau S, Glover‐Cutter K, Ben‐Dov C, Blaustein M, Lozano JJ et al (2009) DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137: 708–720 [DOI] [PubMed] [Google Scholar]
- Muramoto T, Cannon D, Gierlinski M, Corrigan A, Barton GJ, Chubb JR (2012) Live imaging of nascent RNA dynamics reveals distinct types of transcriptional pulse regulation. Proc Natl Acad Sci USA 109: 7350–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahkuri S, Taft RJ, Mattick JS (2009) Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle 8: 3420–3424 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, Carmo‐Fonseca M, Proudfoot NJ (2015) Mammalian NET‐Seq reveals genome‐wide nascent transcription coupled to RNA processing. Cell 161: 526–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Lis JT (1993) Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol Cell Biol 13: 3456–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES (2018) Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front Genet 9: 440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patop IL, Wüst S, Kadener S (2019) Past, present, and future of circRNAs. EMBO J 38: e100836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, Coelho PA, Carmo AM, Sunkel CE, Proudfoot NJ et al (2011) RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J 30: 2431–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, Nitiss JL (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17: 703–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Libri D (2015) Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol 16: 190–202 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ (2016) Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Jin H, Vvedenskaya I, Llenas JA, Zhao T, Malik I, Visbisky AM, Schwartz SL, Cui P, Čabart P et al (2020) Universal promoter scanning by Pol II during transcription initiation in Saccharomyces cerevisiae. Genome Biol 21: 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan TK, Hartzog GA (2010) Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4‐Spt5 in transcription. Genetics 184: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan C, Goodall GJ, Shirokikh NE, Preiss T (2019) Insights into the biogenesis and potential functions of exonic circular RNA. Sci Rep 9: 2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas‐Duran MF, Gilbert WV (2012) Alternative transcription start site selection leads to large differences in translation activity in yeast. RNA 18: 2299–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski AJ, Erhard F, L'Hernault A, Bonfert T, Schilhabel M, Crump C, Rosenstiel P, Efstathiou S, Zimmer R, Friedel CC et al (2015) Widespread disruption of host transcription termination in HSV‐1 infection. Nat Commun 6: 7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak‐Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal‐Fluss R et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58: 870–885 [DOI] [PubMed] [Google Scholar]
- Saint‐André V, Batsché E, Rachez C, Muchardt C (2011) Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat Struct Mol Biol 18: 337–344 [DOI] [PubMed] [Google Scholar]
- Saldi T, Fong N, Bentley DL (2018) Transcription elongation rate affects nascent histone pre‐mRNA folding and 3' end processing. Genes Dev 32: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell‐type specific features of circular RNA expression. PLoS Genet 9: e1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, Söding J, Stewart A, Svejstrup JQ (2014) RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell 157: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AC, Taatjes DJ (2020) Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 34: 465–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Fiszbein A, Petrillo E, Kornblihtt AR (2013) Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J 32: 2264–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Rascovan N, Pelisch F, Alló M, Kornblihtt AR (2009) Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci USA 106: 4325–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G (2009) Chromatin organization marks exon‐intron structure. Nat Struct Mol Biol 16: 990–995 [DOI] [PubMed] [Google Scholar]
- Schweikhard V, Meng C, Murakami K, Kaplan CD, Kornberg RD, Block SM (2014) Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc Natl Acad Sci USA 111: 6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal PB, Derman E, Molloy GR, Tamm I, Darnell JE (1976) 5,6‐Dichloro‐1‐Beta‐D‐ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science 194: 431–433 [DOI] [PubMed] [Google Scholar]
- Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H (2014) Calcium‐mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci USA 111: E4920–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan RM, Fong N, D'Alessandro A, Bentley DL (2019) Widespread backtracking by RNA Pol II is a major effector of gene activation, 5' pause release, termination, and transcription elongation rate. Mol Cell 73: 107–118.e104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen AW, O'Farrell PH (1991) Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell 67: 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wei J, He C (2019) Where, when, and how: context‐dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74: 640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA (2009) Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Han R, Calderone V, Vrielink JAFO, Loayza‐Puch F, Elkon R, Agami R (2017) Transcription impacts the efficiency of mRNA translation via co‐transcriptional n6‐adenosine methylation. Cell 169: 326–337.e312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smid M, Wilting SM, Uhr K, Rodríguez‐González FG, de Weerd V , Prager‐Van der Smissen WJC, van der Vlugt‐Daane M , van Galen A , Nik‐Zainal S, Butler A et al (2019) The circular RNome of primary breast cancer. Genome Res 29: 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, He PC, Chun Y, Suh H, Kim T, Buratowski S (2017) Determinants of histone H3K4 methylation patterns. Mol Cell 68: 773–785.e776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB (2009) Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson CN, Klamut HJ, Worton RG (1995) The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet 9: 184–190 [DOI] [PubMed] [Google Scholar]
- Thummel CS, Burtis KC, Hogness DS (1990) Spatial and temporal patterns of E74 transcription during Drosophila development. Cell 61: 101–111 [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL (2017) Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, Guigó R (2009) Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001 [DOI] [PubMed] [Google Scholar]
- Tsao DC, Park NJ, Nag A, Martinson HG (2012) Prolonged α‐amanitin treatment of cells for studying mutated polymerases causes degradation of DSIF160 and other proteins. RNA 18: 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski TW, Petfalski E, Goddard BD, French SL, Helwak A, Tollervey D (2020) Nascent transcript folding plays a major role in determining RNA polymerase elongation rates. Mol Cell 79: 488–503.e411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucker DS, Yamamoto KR (1984) Early events in the stimulation of mammary tumor virus RNA synthesis by glucocorticoids. Novel assays of transcription rates. J Biol Chem 259: 7416–7420 [PubMed] [Google Scholar]
- Van Oss SB, Cucinotta CE, Arndt KM (2017) Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem Sci 42: 788–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M (2014) Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 24: 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA (2015) Widespread inducible transcription downstream of human genes. Mol Cell 59: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Sabath N, Wiesel Y, Nathans J, Levy‐Adam F, Yario TA, Steitz JA, Shalgi R (2017) Comparative analysis reveals genomic features of stress‐induced transcriptional readthrough. Proc Natl Acad Sci USA 114: E8362–E8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR et al (2019) The Landscape of Circular RNA in Cancer. Cell 176: 869–881.e813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, Cramer P (2018a) Structure of activated transcription complex Pol II‐DSIF‐PAF‐SPT6. Nature 560: 607–612 [DOI] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Urlaub H, Cramer P (2018b) Structure of paused transcription complex Pol II‐DSIF‐NELF. Nature 560: 601–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Linden A, Urlaub H, Cramer P (2020) Structure of complete Pol II‐DSIF‐PAF‐SPT6 transcription complex reveals RTF1 allosteric activation. Nat Struct Mol Biol 27: 668–677 [DOI] [PubMed] [Google Scholar]
- Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A et al (2009) A wave of nascent transcription on activated human genes. Proc Natl Acad Sci USA 106: 18357–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, Xu X, Liang Q, Christiani DC, Wang M et al (2021) Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer 20: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Larsen A, Groudine M (1981) Alpha‐Globin‐gene switching during the development of chicken embryos: expression and chromosome structure. Cell 24: 333–344 [DOI] [PubMed] [Google Scholar]
- Williams AS, Ingledue TC, Kay BK, Marzluff WF (1994) Changes in the stem‐loop at the 3' terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res 22: 4660–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson L, Saponaro M, Boeing S, East P, Mitter R, Kantidakis T, Kelly GP, Lobley A, Walker J, Spencer‐Dene B et al (2017) UV irradiation induces a non‐coding RNA that functionally opposes the protein encoded by the same gene. Cell 168: 843–855.e813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M, Aus Dem Siepen T, Stamminger T (2000) Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J Virol 74: 8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao MS, Ai Y, Wilusz JE (2020) Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol 30: 226–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yague‐Sanz C, Vanrobaeys Y, Fernandez R, Duval M, Larochelle M, Beaudoin J, Berro J, Labbé S, Jacques P, Bachand F (2020) Nutrient‐dependent control of RNA polymerase II elongation rate regulates specific gene expression programs by alternative polyadenylation. Genes Dev 34: 883–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H (2006) P‐TEFb‐mediated phosphorylation of hSpt5 C‐terminal repeats is critical for processive transcription elongation. Mol Cell 21: 227–237 [DOI] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT (2007) Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell 28: 978–990 [DOI] [PubMed] [Google Scholar]
- Yunger S, Rosenfeld L, Garini Y, Shav‐Tal Y (2010) Single‐allele analysis of transcription kinetics in living mammalian cells. Nat Methods 7: 631–633 [DOI] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR (2019) Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20: 608–624 [DOI] [PubMed] [Google Scholar]
- Zamft B, Bintu L, Ishibashi T, Bustamante C (2012) Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc Natl Acad Sci USA 109: 8948–8953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatreanu D, Han Z, Mitter R, Tumini E, Williams H, Gregersen L, Dirac‐Svejstrup AB, Roma S, Stewart A, Aguilera A et al (2019) Elongation factor TFIIS prevents transcription stress and R‐loop accumulation to maintain genome stability. Mol Cell 76: 57–69.e59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Ball AR, Yokomori K (2010) HP1: heterochromatin binding proteins working the genome. Epigenetics 5: 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL (2016) The biogenesis of nascent circular RNAs. Cell Rep 15: 611–624 [DOI] [PubMed] [Google Scholar]


