Abstract
How organisms integrate metabolism with the external environment is a central question in biology. Here, we describe a novel regulatory small molecule, a proteogenic dipeptide Tyr‐Asp, which improves plant tolerance to oxidative stress by directly interfering with glucose metabolism. Specifically, Tyr‐Asp inhibits the activity of a key glycolytic enzyme, glyceraldehyde 3‐phosphate dehydrogenase (GAPC), and redirects glucose toward pentose phosphate pathway (PPP) and NADPH production. In line with the metabolic data, Tyr‐Asp supplementation improved the growth performance of both Arabidopsis and tobacco seedlings subjected to oxidative stress conditions. Moreover, inhibition of Arabidopsis phosphoenolpyruvate carboxykinase (PEPCK) activity by a group of branched‐chain amino acid‐containing dipeptides, but not by Tyr‐Asp, points to a multisite regulation of glycolytic/gluconeogenic pathway by dipeptides. In summary, our results open the intriguing possibility that proteogenic dipeptides act as evolutionarily conserved small‐molecule regulators at the nexus of stress, protein degradation, and metabolism.
Keywords: Arabidopsis, central carbon metabolism, dipeptides, GAPDH, NADPH
Subject Categories: Metabolism, Plant Biology
The dipeptide Tyr‐Asp redirects glycolytic flux, increases NADPH/NADP+ ratio, and improves stress resistance in plants.

Introduction
Glucose is a critical carbohydrate in living organisms serving both as a cellular fuel and as a precursor for other metabolites. While glucose is broken down in the process of glycolysis, it can also conversely be produced from specific non‐carbohydrate substrates in the process of gluconeogenesis. Glycolysis and gluconeogenesis share most, but not all, enzymatic reactions that are subject to multiple levels of control (Plaxton, 1996). Regulation of the specific and irreversible steps enables cells to quickly adjust glucose metabolism to both developmental and environmental cues by shifting the balance between glucose consumption and re‐synthesis (Plaxton, 1996). Moreover, regulation of the reversible steps is important to redirect the glycolytic intermediates into the pathways branching out of glycolysis (Ralser et al, 2007). In addition to their metabolic functions, glycolytic enzymes are also involved in a multitude of diverse non‐metabolic processes, referred to as moonlighting functions (Scheibe, 2019), including the control of gene expression, the bundling of actin fibers, and the process of apoptosis (Zaffagnini et al, 2013; Jung et al, 2014; Hildebrandt et al, 2015; Schneider et al, 2018).
Post‐translational modifications and non‐covalent binding of small‐molecule ligands enable cells to rapidly regulate both enzymatic and non‐enzymatic activities of the glycolytic enzymes and, thus, to integrate metabolism with developmental and environmental responses (Plaxton & Podesta, 2006; Zaffagnini et al, 2013; Hildebrandt et al, 2015). The best characterized allosteric regulation of the irreversible steps of glycolysis, that involve signaling metabolites such as fructose 2,6‐bisphosphate (Stitt & Sonnewald, 1995; Fernie et al, 2001), has been widely proven (Buchanan et al, 2015). However, there are many other examples. For instance, phosphatidic acid binding to the Arabidopsis glyceraldehyde 3‐phosphate dehydrogenase (GAPC) promotes its proteolytic degradation, effectively reducing GAPC protein levels in phosphatidic acid grown seedlings (Kim et al, 2013).
Considering the many and essential roles of the glycolytic enzymes, we aimed to explore the function of the recently reported in vitro interaction between a proteogenic dipeptide Tyr‐Asp and Arabidopsis GAPC (Veyel et al, 2018). GAPDH catalyzes the reversible oxidation of glyceraldehyde 3‐phosphate (G3P) to produce 1,3‐bisphosphoglycerate and NADH, using NAD+ and inorganic orthophosphate as co‐substrates. In contrast to animals, which have only one GAPDH isoform, plants are characterized by the presence of multiple GAPDH isoforms, which differ not only in their catalytic and regulatory properties, but also in their subcellular localization (Iglesias, 1990; Rius et al, 2008; Zaffagnini et al, 2013; Hildebrandt et al, 2015). In the cytosol, two different GAPDHs are involved in glycolysis, the phosphorylating NAD+‐dependent GAPDH (GAPC1 and GAPC2; EC 1.2.1.12) and the non‐phosphorylating, NADP+‐dependent GAPDH (GAPN; EC 1.2.1.9). GAPN irreversibly oxidizes G3P to 3‐phosphoglycerate (3PGA) and has no homology to GAPC (Habenicht et al, 1994; Michels et al, 1994). In the chloroplasts, a phosphorylating NADP+‐dependent GAPDH (GAPA/B; EC 1.2.1.13) is involved in carbon fixation by the Calvin‐Benson‐Bassham cycle (Wolosiuk & Buchanan, 1978). The coordinated activity of cytosolic and chloroplastic GAPDHs constitutes a shuttle system that transports energy and reducing power from the chloroplasts to the cytosol of photosynthetic cells (Kelly & Gibbs, 1973; Iglesias, 1990). Additionally, an NAD+‐dependent GAPDH (GAPCp), closely related to GAPC, operates in the plastidial glycolytic pathway (Petersen et al, 2003; Zaffagnini et al, 2013). Besides their role in carbon assimilation and partitioning, phosphorylating GAPDHs (particularly, GAPC1 and GAPA1) have additional moonlighting functionalities (Henry et al, 2015; Scheibe, 2019).
Relatively little is currently known concerning the function of proteogenic dipeptides. Dipeptides are short‐lived intermediates of proteolysis. However, this simple view has been challenged by recent studies reporting regulatory roles for various proteogenic dipeptides across different organisms. Prominent examples include a neurotransmitter and a potent analgesic, Tyr‐Arg, also known as kyotorphin (Takagi et al, 1979), and an anxiolytic dipeptide Tyr‐Leu (Kanegawa et al, 2010; Mizushige et al, 2013). Notably, in the case of Tyr‐Leu, neither the retro‐sequence peptide Leu‐Tyr nor the mixture of Tyr and Leu harbored anxiolytic activity, arguing for the dipeptide function being independent of the degradation to the constituent amino acids (Kanegawa et al, 2010). In other words, the function of dipeptides is more than a sum of the amino acids and their sequential order matters. Similarly, a single dipeptide Tyr‐Ala was shown to enhance both the lifespan and the healthspan of the model nematode Caenorhabditis elegans, with a particularly pronounced effect under heat and oxidative stress conditions (Zhang et al, 2016). Further evidence linking dipeptides with stress responses comes from metabolite profiling experiments in Arabidopsis. Dipeptide levels are affected by both abiotic and biotic conditions, with the exact response varying for the different dipeptides (Doppler et al, 2019; Thirumalaikumar et al, 2020). Moreover, dipeptide accumulation depends on autophagy, at least under heat stress (Thirumalaikumar et al, 2020). Dipeptides were also reproducibly measured in Arabidopsis root exudates, downstream of MPK3/6 signaling, suggesting a role in plant–microbe communication and plant–plant communication (Strehmel et al, 2017).
To exert a regulatory function, small molecules require a macromolecular partner, as a rule, a protein. Therefore, we were intrigued to find nearly 100 different dipeptides in protein complexes isolated from Arabidopsis cell cultures (Veyel et al, 2017; Veyel et al, 2018). By focusing on one selected dipeptide, Tyr‐Asp, we could narrow down the list of Tyr‐Asp protein interactors to GAPC. By testing multiple dipeptides and multiple GAPDHs, we showed that Tyr‐Asp, but none of the other dipeptides, binds to the plant and mammalian GAPDHs (Veyel et al, 2018). Furthermore, neither Tyr nor Asp was shown to interact with GAPDH in E. coli (Diether et al, 2019). Here, we demonstrate that Tyr‐Asp inhibits the enzymatic activity of GAPC. Tyr‐Asp feeding induced a shift of glucose 6‐phosphate (G6P) utilization from glycolysis to the pentose phosphate pathway (PPP), thereby altering redox equilibrium of the NADP(H) pool and improving tolerance to oxidative stress.
A regulatory function of dipeptides with connection to the stress response has been shown before; however, a detailed mechanistic insight, as provided by our study, has not been undertaken yet. Because Tyr‐Asp is a small‐molecule compound that can mitigate oxidative stress, it bears potential to be used to enhance crop performance under environmental stress.
Results
Tyr‐Asp inhibition of GAPC activity is associated with the shift of the glycolytic flux toward the PPP and increased NADPH/NADP+ ratio
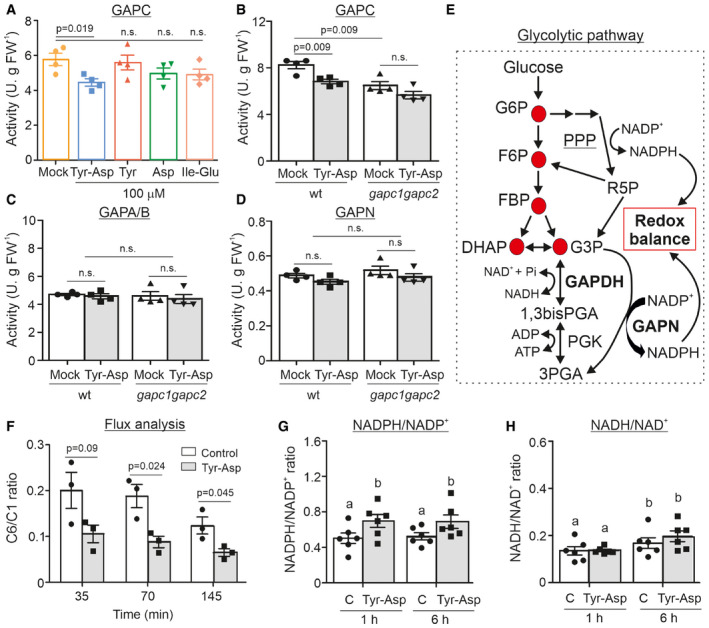
The main aim of our work was to understand the biological significance of our recently reported in vitro interaction between the dipeptide Tyr‐Asp and GAPC (Veyel et al, 2018). We began with the most straightforward hypothesis and tested GAPC activity in the absence and presence of Tyr‐Asp. Indeed, Tyr‐Asp application (100 µM) inhibited GAPC enzymatic activity, unlike treatment with single amino acids (Tyr and Asp) or chemically unrelated dipeptide (Ile‐Glu) (Fig 1A). The reduction of 23% may appear modest, but it is important to note that GAPC activity assay accounts not only for cytosolic GAPC but also for GAPCp, and moreover GAPA/B, which has substantial activity with NAD+ (Falini et al, 2003), whereas GAPCp activity is negligible in crude extracts from Arabidopsis rosettes harvested in the light, that is not the case for GAPA/B (Munoz‐Bertomeu et al, 2009). To differentiate between GAPC and GAPA/B activities, we introduced gapc1 gapc2 double mutant, which is entirely devoid of the cytosolic GAPC activity (Guo et al, 2012). By doing so, we could demonstrate that the reduction in GAPC activity produced by the addition of Tyr‐Asp is similar to that observed in the gapc1 gapc2 double mutant in the absence of the dipeptide (Fig 1B). Moreover, gapc1 gapc2 double mutant (Guo et al, 2012) was insensitive to Tyr‐Asp inhibition of GAPC activity. Based on the obtained results, we conclude that GAPC1 and GAPC2 are primary targets of Tyr‐Asp action and that 100 µM concentration of Tyr‐Asp is sufficient to completely inhibit the activity of the glycolytic GAPC. To further substantiate our results, we tested the activity of GAPA/B and GAPN in crude extracts from wild‐type plants and the gapc1 gapc2 double mutant (Guo et al, 2012). Fig 1C and D shows that neither GAPA/B nor GAPN activity was affected by Tyr‐Asp. Tyr‐Asp (100 µM) was sufficient to inhibit GAPC activity. In comparison, Tyr‐Asp amount measured in the Arabidopsis seedlings (control conditions) varied from 0.23 to 2.7 nmol g−1 FW−1 with an average of 0.62 nmol g−1 FW−1 (n = 20; Appendix Fig S1). We then used data from Koffler et al (2013) to estimate the concentration of Tyr‐Asp in planta. If we assume equal distribution of Tyr‐Asp in all compartments, the intracellular concentration of Tyr‐Asp would be approximately 1 µM, whereas it would rise to 26.5 µM if all Tyr‐Asp were exclusively located in the cytosol.
Figure 1. Tyr‐Asp inhibition of GAPC activity is associated with the shift of the glycolytic flux toward the PPP and increased NADPH/NADP+ ratio.
-
AGAPC enzymatic activity was measured in wild‐type crude extracts treated with H2O (mock), and 100 µM of Tyr‐Asp, Tyr, Asp, and Ile‐Glu.
-
B–DThe enzymatic activities of GAPC (B), GAPA/B (C), and GAPN (D) were measured in the wild‐type and double k.o. mutant gapc1 gapc2 treated with H2O (mock), and 100 µM of Tyr‐Asp.
-
ESchematic representation of glycolysis and pentose phosphate pathways.
-
FGlucose‐labeled C6 and C1 flux experiment in 10‐day‐old Arabidopsis seedlings. C6/C1 ratio for control (water treated) and Tyr‐Asp (100 µM)‐treated wild‐type seedlings after 35, 70, and 145 min is shown.
-
GNADPH/NADP+ ratio.
-
HNADH/NAD+ ratio. Ratios were calculated from the NAD+, NADH, NADP+, and NADPH measurements.
Data information: Data are mean ± SEM of n = 4 (A–D; four technical replicates), n = 3 (F; three independent flasks), and n = 5–6 (G–H; five to six independent seedling flasks). For (A–D, F), significance was assessed using unpaired two‐tailed Student’s t‐test. For (G–H), significance was assessed using two‐way ANOVA (P ≤ 0.05; letters show significance associated with the Tyr‐Asp treatment). G6P: glucose 6‐phosphate; F6P: fructose 6‐phosphate; FBP: fructose 1,6‐bisphosphate; DHAP: dihydroxyacetone‐phosphate; G3P: glyceraldehyde 3‐phosphate; 1,3bisPGA: 1,3bis‐phosphoglycerate; 3PGA: 3‐phosphoglycerate; PPP: pentose phosphate pathway; R5P: ribose 5‐phosphate. In (G) and (H), (C) stands for control; n.s.: not significant.
In animal and yeast cells, oxidative inactivation of the glycolytic GAPDH by redox modification of the catalytic cysteine was shown to increase the NADPH/NADP+ ratio by redirecting the glycolytic flux to the PPP (Fig 1E) (Ralser et al, 2007). Moreover, Arabidopsis double gapc1 gapc2 knock‐out mutant is characterized by an increase in the NADPH/NADP+ ratio (Guo et al, 2014). To test whether Tyr‐Asp‐treated Arabidopsis plants would have similar effects and shift glycolytic intermediates toward the PPP due to inhibition of GAPC, we exploited the proven ability of Arabidopsis roots to take up dipeptides from the growth media (Komarova et al, 2008). Seedlings were fed with 14C glucose labeled on position C1 or C6 in the absence or presence of 100 μM Tyr‐Asp (Nunes‐Nesi et al, 2005). We compared the rates at which 14CO2 was released from carbons 1 (C1) and 6 (C6) at 35, 70, and 145 min following the addition of the 14C glucose (Fig 1F; Appendix Fig S2A). 14CO2 release from C1 is related to the activity of both PPP and glycolysis, while from C6 only of glycolysis/TCA, and as consequence lower C6‐to‐C1 ratio is indicative of the more active PPP (Appendix Fig S2A). A significant decrease in the C6‐to‐C1 ratio measured in the Tyr‐Asp‐treated versus control seedlings argues for the G6P being redirected from glycolysis toward PPP (Fig 1F). To investigate whether observed changes in the PPP activity result in an altered NADPH/NADP+ ratio, NAD+, NADH, NADP+, and NADPH cellular concentrations were measured in mock (H2O‐treated control) and Tyr‐Asp‐treated 10‐day‐old Arabidopsis seedlings grown in liquid culture at 1 and 6 h using a targeted enzymatic assay (Appendix Fig S2B–F). Tyr‐Asp supplementation produced a significant reduction in NAD+, NADH, and NADP+ levels (Appendix Fig S2B–F), resulting in a significant increase in the deduced NADPH/NADP+ ratio (Fig 1G), but an unchanged NADH/NAD+ ratio (Fig 1H).
Finally, we used YFP and GFP marker lines of GAPC1 and GAPCp1/2 (Munoz‐Bertomeu et al, 2009; Guo et al, 2012) to determine GAPDH subcellular localization upon Tyr‐Asp treatment, and therefore its moonlight activities (Zaffagnini et al, 2013; Schneider et al, 2018). However, and at least in our experimental conditions, Tyr‐Asp treatment did not change GAPDH subcellular localization (Appendix Figs S3 and S4).
Tyr‐Asp supplementation confers resistance to oxidative stress in Arabidopsis and tobacco seedlings
Inactivation of GAPDH improves oxidative stress tolerance in both yeast and animal cells by supplying NADPH (Ralser et al, 2007). Here, we examined whether the reduction in GAPDH activity measured in the Tyr‐Asp‐treated plants would also confer an advantage under oxidative stress conditions. To test our hypothesis, we used two agents known to induce an oxidative stress response: hydrogen peroxide (H2O2:50 mM) and catechin (0.175 mM) (Scarpeci et al, 2008; Kaushik et al, 2010). To be consistent with the Tyr‐Asp feeding experiments, we used plants grown on synthetic MS media, and then, we transferred them to liquid MS media for oxidative stress and Tyr‐Asp treatments (see Methods section). Moreover, we tested both Arabidopsis and tobacco seedlings (in several experiments; Dataset EV1), the latter characterized by a more homogeneous growth under normal conditions. Plants were germinated on solid MS media and transferred to a 24‐well plate at 12 days after stratification (DAS). First, seedlings were incubated with either 100 µM Tyr‐Asp or mock (H2O) for 1 h before applying the stress. Fresh weight, as a proxy for growth, was measured after two and four days of catechin treatment in Arabidopsis and tobacco, respectively, and after 36 h and 2 days of H2O2 treatment in Arabidopsis and tobacco, respectively. While Tyr‐Asp supplementation did not affect the overall plant growth under control condition, it did lead to increased fresh weight under both oxidative stress regimes in both Arabidopsis and tobacco seedlings (Fig 2A–C and Appendix Fig S5A–D). The increase in fresh weight was further corroborated by the measurements of leaf area in tobacco plants treated with catechin (Fig 2C). Importantly, neither the combination of Tyr and Asp nor the two other tested dipeptides, Ser‐Leu and Gly‐Pro, exhibited the bioactivity of Tyr‐Asp (Fig 2A and Appendix Fig S5A–D).
Figure 2. Tyr‐Asp treatment improves growth performance of Arabidopsis and tobacco plants exposed to oxidative stress.
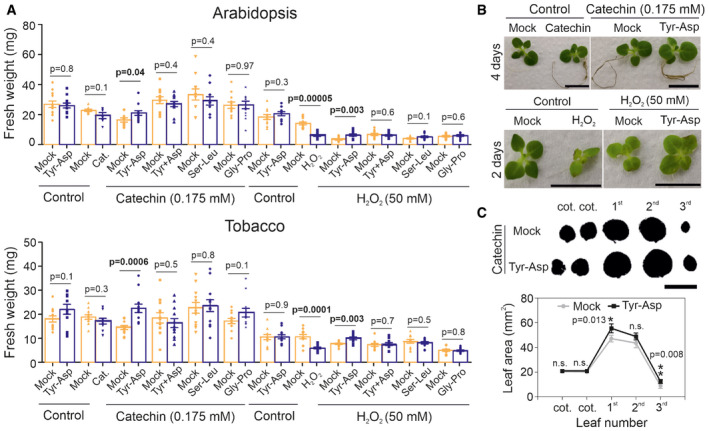
- Fresh weight quantification of Arabidopsis and tobacco seedlings. Seedlings were pretreated with mock (water), Tyr‐Asp (100 μM), Tyr and Asp (100 μM), Ser‐Leu (100 μM), or Gly‐Pro (100 μM) for 1 h prior an oxidative stress (catechin or H2O2). Each treatment was compared with its respective control, corresponding to plants grown in one 24‐well plate (represented by the adjoined bars).
- Catechin‐ (left upper panel) and (catechin‐) Tyr‐Asp‐treated tobacco plants (right upper panel) grown for four days in liquid media. H2O2‐ (left upper panel) and (H2O2‐) Tyr‐Asp‐treated tobacco plants (right upper panel) grown for two days in liquid media. All plants were 2 weeks old before starting the stress treatment.
- Leaf series prepared from tobacco seedlings pretreated (1 h) with either mock or Tyr‐Asp and exposed to catechin for 4 days.
Data information: Data are mean ± SEM of n = 10–12 (seedlings). Unpaired two‐tailed Student’s t‐test was performed to compare treatments with control. Scale bar: 10 cm.
To complement oxidative stress regimes, we decided to test the effect of Tyr‐Asp supplementation on plant performance under salt stress. As described above, 12‐day‐old tobacco seedlings were incubated with either 100 µM Tyr‐Asp or mock (H2O) for 1 h before applying the salt stress (50 mM NaCl). Plant performance was measured after 6 days of salt treatment by assessing the fresh weight of the plants. Again Tyr‐Asp treatment, but neither the combination of amino acids nor the two other tested dipeptides did improve plant performance under stress conditions (Appendix Fig S5E and F).
To test whether improved stress tolerance is restricted to the use of liquid media and thus dipeptide uptake via both roots and shoots, we performed catechin and salt experiments using tobacco plants grown on nylon mesh overlaying solid MS media (see Methods section). Two‐week‐old plants were transferred first to plates containing Tyr‐Asp and afterward to plates containing a combination of Tyr‐Asp, salt and/or catechin. Fresh weight measurements were taken after one week of treatment (Appendix Fig S6A). The addition of Tyr‐Asp had no effect on plant growth measured under control conditions, but it resulted in the overall higher biomass in plants subjected to the catechin and salt stresses (Appendix Fig S6B).
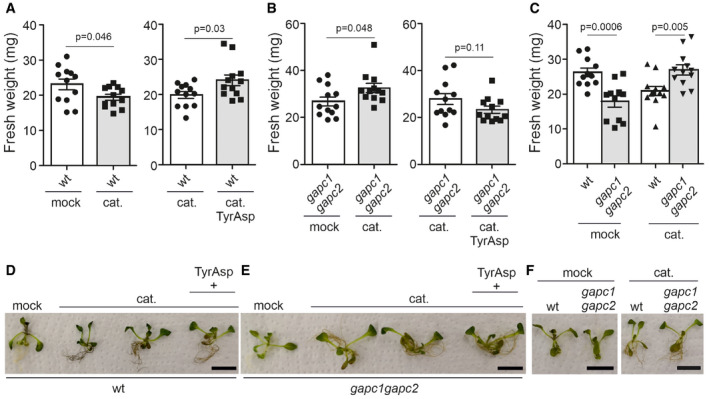
Finally, and to assess the contribution of the Tyr‐Asp inhibition of GAPC activity to the improved stress tolerance associated with the Tyr‐Asp treatment, we used the double gapc1 gapc2 double knock‐out mutant (Guo et al, 2012), which showed a similar effect as the observed for Tyr‐Asp treatment, in the reduction in the total GAPC activity (Fig 1B). Arabidopsis wild‐type and gapc1 gapc2 plants were subjected to catechin stress as described above. As observed previously, Tyr‐Asp supplementation increased biomass of catechin‐treated wild‐type seedlings (Fig 3A and D; Appendix Fig S7A). By contrast, no such improvement was observed for the gapc1 gapc2 mutant line (Fig 3B and E, and Appendix Fig S7B), arguing for the Tyr‐Asp‐associated stress tolerance being dependent on the inhibition of the GAPC1 and GAPC2 activities. Moreover, whereas under control conditions, gapc1 gapc2 mutant line was significantly smaller compared with the wild type, the opposite was true under treatment with catechin (Fig 3C and F; Appendix Fig S7C). Our results are in line with the previously reported improved drought tolerance of the gapc1 gapc2 mutant (Guo et al, 2012).
Figure 3. Tyr‐Asp improvement of Arabidopsis stress tolerance is associated with the GAPC1/2 inhibition.
-
A–CFresh weight measurements of Arabidopsis wild‐type and gapc1 gapc2 double k.o. plants subjected to the oxidative stress (e.g., catechin) and Tyr‐Asp treatments. Each treatment was compared with its respective control, corresponding to plants grown in one 24‐well plate (represented by the adjoined bars).
-
D–FRepresentative Arabidopsis wild‐type and gapc1 gapc2 double k.o. seedlings from the different treatments. All plants were 10 days old at the stress onset. The seedlings were exposed to catechin for 3 days.
Data information: Data are mean ± SEM of n = 10–12 (seedlings). Unpaired two‐tailed Student’s t‐test was performed to compare treatments with control. Scale bar: 10 cm. Data from an independent experiment are shown in (Appendix Fig S7).
In silico prediction of Tyr‐Asp binding to the Arabidopsis GAPC1 reveals two spatially close sites
Taking advantage of the available crystal structure for the Arabidopsis GAPC1‐NAD+ protein (PDB‐ID 4z0h) (Zaffagnini et al, 2016), we performed an in silico docking analysis. Tyr‐Asp was predicted to bind to two spatially close sites near the NAD+: at the catalytic site surrounded by amino acid residues “SCT”, “H”, “TAT”, and “R” (positions 148‐150, 176, 179‐181, and 231 of chain R), and close to the adenosine‐moiety of NAD+. In the first pocket, Tyr‐Asp binds with an associated Kd of 28.6 µM (Fig 4A–C, pocket 1). In the second pocket, lined by amino acid residues “KTVDGP” (sequence position 183‐188 of chain O) and “F”, “A”, and “Q” (positions 34, 179, and 181 of chain R), in close proximity to the ribonucleotide and nicotinamide moieties of NAD+, Tyr‐Asp binds with an associated Kd of 23.3 µM (Fig 4A–C, pocket 2). In addition, we found an alternative binding pocket with lower, but still appreciably, binding efficiency (Kd of 82.5 µM), relatively distant from the previous two pockets and lined by the amino acid residues “VGD” (sequence positions 284‐286 of chain O) and “R”, “HGQ”, “K”, and “W” (positions 17, 49‐51, 53, and 315 of chain R’) (Fig 4A, pocket 3). We subsequently removed the NAD+ molecule from the crystal structure (PDB‐ID 4z0h), resulting in Tyr‐Asp binding at the position of the removed adenosine of NAD+, binding to the amino acid residues “I”, “SAP”, “ASC”, “T”, “R”, “NE”, “Y” (positions 11, 119‐121, 147‐149, 179, 231, 313‐314, and 317, with a Kd of 5.1 µM; Fig 4B). Docking Tyr‐Asp to the S‐glutathionylated GAPC1‐NAD+ structure (PDB‐ID 6quq) (Berman et al, 2000; Zaffagnini et al, 2019) resulted in no binding pocket being detected at the catalytic site, and hence no predicted binding. The oxidized form of GAPC1‐NAD+ (PDB‐ID 6qun) (Zaffagnini et al, 2019) showed a 13 times decreased affinity for the binding to the catalytic pocket, with a Kd of 304.3 µM. Finally, we compared the predicted Tyr‐Asp binding sites with the published structure of the Arabidopsis GAPA protein with a small chloroplast protein Cp12‐2 (PDB‐ID 3qv1; Fig 4C) (Marri et al, 2008; Fermani et al, 2012). The Tyr‐Asp motif present at the C‐terminus of the Cp12‐2 was previously reported to be involved in the stabilization of GAPA/Cp12 interaction with Tyr76 forming a hydrogen bond with phosphate group of NAD+. The C‐terminal Tyr‐Asp motif of Cp12 overlaps with the computationally docked Tyr‐Asp at the catalytic site (Fig 4C, pocket 1). The predicted binding position of Tyr‐Asp denoted as the second pocket (Fig 4A and C, pocket 2) overlaps with glutamic acid (sequence position 12) of Cp12‐2. Hence, Tyr‐Asp may interfere with Cp12 binding, posing an interesting hypothesis to be tested in the future.
Figure 4. In silico prediction of Tyr‐Asp binding to GAPC.
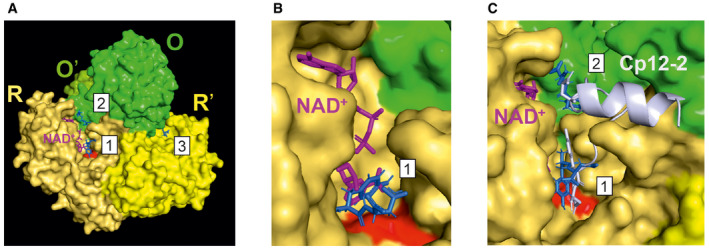
- Surface representation of the GAPC1 tetramer (dimer of O‐R‐dimer) with colors indicating the different chains and respective sequence identity (O = O' and R = R') and highlighting the conformations of Tyr‐Asp (blue) docked to the identified pockets, labeled 1‐3, as well as the catalytic site (Cys 149 and His 176 in red), and NAD+ (pink).
- Predicted binding conformation of Tyr‐Asp docked to GAPC1 without NAD+ (but shown for reference and taken from 4z0h, purple).
- Zoom‐in pockets 1 and 2 with overlaid Cp12‐2 (taken from PDB‐ID 3qv1, cartoon gray) with highlighted glutamic acid in sequence position 12 and C‐terminal Tyr‐Asp motif (both in stick representation), which sterically interfere with Tyr‐Asp binding to pockets 1 and 2.
Proteome characterization of the Tyr‐Asp feeding experiment revealed changes in protein and redox metabolism consistent with the Tyr‐Asp protein interactions beyond that with GAPC
Tyr‐Asp inhibition of the GAPC activity and the associated change in the NADPH/NADP+ ratio provides an explanation for the improved stress resistance of the Tyr‐Asp‐supplemented seedlings. To explore alternative mechanisms, we decided to follow two additional experimental strategies. First, we characterized proteome and metabolome changes associated with the Tyr‐Asp treatment. Specifically, 10‐day‐old Arabidopsis seedlings grown in liquid culture were supplemented with 100 µM Tyr‐Asp and harvested at five different time‐points (15, 30 min, 1, 6, and 24 h) prior to untargeted mass spectrometry‐based metabolomic and proteomic analyses. As before, mock (water‐)‐treated seedlings were used as control. Statistical analysis of the 5257 proteins and 201 metabolites revealed a total of 212 proteins, but only three metabolites (Tyr‐Asp, Tyr, and Asp‐Pro) were significantly affected by Tyr‐Asp (two‐way ANOVA, FDR corrected P ≤ 0.05) (Fig 5A; Dataset EV2). The Tyr‐Asp content measured in the treated Arabidopsis seedlings was elevated within 15 min of Tyr‐Asp supplementation, peaking at 1 h, followed by a decrease, and reduced accumulation at 24 h (Dataset EV2). Not surprisingly, considering the rapid turnover of the supplemented Tyr‐Asp, tyrosine levels also increased upon Tyr‐Asp feeding (Dataset EV2). Altogether, 164 of the 212 proteins affected by Tyr‐Asp treatment (38 up‐ and 126 down‐regulated) to 29 functional MapMan bins (Fig 5B; Dataset EV3), revealing changes in protein and redox metabolism. While ribosomal subunits were enriched among up‐regulated proteins, chaperones (e.g., HSP70, HSP90.7, HSP90.1, ROF1, and CPN10) and proteases (e.g., cysteine proteases RD21 and cathepsin) were down‐regulated by the Tyr‐Asp treatment. Decreased abundance was also measured for enzymes involved in redox metabolism (e.g., thioredoxin H1 and H3, glutathione reductase, ferredoxin‐thioredoxin reductase, and ascorbate oxidase). In fact, 22 of the differential proteins bind NADP(H), including glutathione reductase, which is involved in replenishing the pool of reduced glutathione, a small‐molecule indispensable for the oxidative stress response (Noctor et al, 2018). Further down‐regulated proteins included enzymes from amino acid and carbon metabolism, notably plastidial G6PDH, abscisic acid receptors PYR1 and PYL1, and cell wall‐associated proteins.
Figure 5. Tyr‐Asp affects redox and protein metabolism.
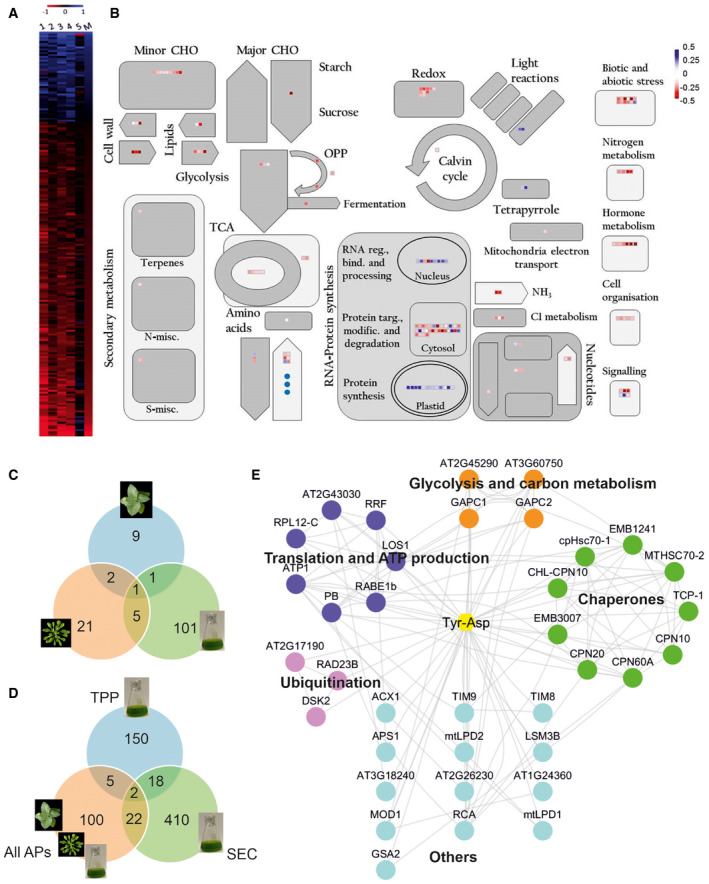
- Proteins and metabolites differentially accumulating in response to the Tyr‐Asp feeding were delineated using two‐way ANOVA embedded in MeV software (Howe et al, 2011), followed by false discovery rate (FDR) correction. Heat map representation of the, respectively, 212 and three differential (two‐way ANOVA; FDR corrected P ≤ 0.05) proteins and metabolites across five time‐points. Data are presented as log2 fold change between control and treatment. Red indicates down‐ while blue up‐regulation. 1: log2 fold change (FC) 15 min; 2: log2 FC 30 min; 3: log2 FC 1 h; 4: log2 FC 6 h; 5: log2 FC 24 h; M: median log2 FC of all time‐points.
- Median of the log2 fold change between control and treatment calculated from the five time‐points was used for MapMan visualization (Thimm et al, 2004) of the differential proteins (squares) and metabolites (circles). Blue indicates up‐ while red down‐regulation. MapMan automatically assigned bincodes and proteins were classified into different cellular functions/processes. Bind.: binding; CHO: carbohydrates; Misc.: miscellaneous; Modif.: modification; N: nitrogen; OPP: oxidative pentose phosphate pathway; Reg.: regulation; S: sulfur; Targ.: targeting; TCA: tricarboxylic acid cycle.
- Venn diagram comparison of putative Tyr‐Asp interactors identified in the AP experiments using different sources of starting material.
- Venn diagram comparison of putative Tyr‐Asp interactors identified in the AP, TPP, and PROMIS experiments.
- Cytoscape (Shannon et al, 2003) visualization of the Tyr‐Asp interactome. Nodes represent Tyr‐Asp interactors, and edges were imported from the STRING database (Szklarczyk et al, 2017) using experimental, database, and literature evidence (score set at 0.4). Functionality was assigned based on the UniProt (Apweiler et al, 2004). Disconnected nodes were removed from the network.
In parallel to the omics analysis of the Tyr‐Asp feeding experiment, we revisited the Tyr‐Asp protein interactome. Previously, agarose beads coupled to Tyr‐Asp were used to capture protein binders from the native cellular lysate prepared from Arabidopsis cell cultures (Veyel et al, 2018). Here, we performed two additional affinity purification (AP) experiments, this time using Arabidopsis and tobacco leaves. A total of 13 and 29 proteins were identified in tobacco and Arabidopsis AP experiments, respectively, in comparison with the 108 previously identified using cell cultures (Fig 5C; Dataset EV4 and EV5). A Venn diagram overlap of the three lists contained just one protein, cytosolic GAPC1 (Fig 5C), supporting a conserved role of Tyr‐Asp in the regulation of the NAD+‐dependent GAPDH activity.
To complement the AP experiments, we decided to investigate the Tyr‐Asp interactome by an independent small‐molecule‐centered approach, namely thermal proteome profiling (TPP) (Savitski et al, 2014). In the TPP experiment, putative protein targets are delineated by monitoring changes in protein thermal stability caused by small‐molecule binding. Arabidopsis native cellular lysate (prepared from the cell suspension culture grown in the light) was treated with either mock or 10 µM Tyr‐Asp, followed by temperature gradient and proteomic analysis. Tyr‐Asp treatment altered thermal stability of 177 out of 3092 quantified proteins (Dataset EV6). The list of putative Tyr‐Asp interactors was enriched in proteins associated with protein metabolism and abiotic stress response: chaperones (e.g., CPN10, CPN20, CPN60A), ribosomal proteins (e.g., RPL12‐C, RPL10, RPS24/35), proteases and peptidases (e.g., CLPR3, peptidase C15, DEG1, peptidase M20/M25/M40), and enzymes involved in oxidative stress defense (glutaredoxins and thioredoxins; Appendix Fig S8).
In a final step, we compared the lists of putative Tyr‐Asp interactors from the TPP and AP experiments. We also included a third list obtained from the published PROMIS experiment and comprising proteins co‐separating with Tyr‐Asp in size‐based fractionation (Veyel et al, 2018). We refer to 47 proteins identified by at least two independent approaches as high confidence putative Tyr‐Asp interactors (Fig 5D; Dataset EV7). By querying the STRING database for reported protein–protein interactions (Szklarczyk et al, 2017) and using Cytoscape as a visualization tool (Shannon et al, 2003), we built the final Tyr‐Asp interactome (network) comprising 36 interconnected proteins. Proteins were grouped into four functional classes (i) carbon metabolism (GAPC1, GAPC2, and TKL1, TKL2), (ii) chaperones (e.g., CPN10, CPN20), (iii) translation and ATP production (e.g., RRF, RPL12‐C), and (iv) protein degradation (e.g., RAD23B, DSK2; Fig 5E).
AthPEPCK1 activity is inhibited by the branched‐chain and polar amino acid dipeptides
Next, we wondered whether GAPC is the only glycolytic/gluconeogenic enzyme targeted by proteogenic dipeptides. The PROMIS dataset (Veyel et al, 2018), used in the identification of the Tyr‐Asp–GAPDH interaction, contains 92 additional proteogenic dipeptides, spanning the whole protein separation range (Dataset EV8). To test the likelihood of proteogenic dipeptides binding to other glycolytic/gluconeogenic enzymes, we analyzed co‐elution of all dipeptides and all glycolytic/gluconeogenic enzymes present in the PROMIS dataset (Veyel et al, 2018). Our analysis identified numerous putative interactions (Appendix Fig S9; Dataset EV8). We focused our attention on one of the co‐elution clusters containing the gluconeogenic enzyme phosphoenolpyruvate carboxykinase 1 (AthPEPCK1) and a group of six dipeptides characterized by the presence of either polar or branched‐chain amino acids (Ile‐Gln, Ile‐Ala, Phe‐Gln, Leu‐Thr, Ser‐Tyr, and Ser‐Val). The activity of recombinant AthPEPCK1 (Rojas et al, 2019) was analyzed at increasing concentrations of the selected dipeptides (Fig 6A). Remarkably, all six tested dipeptides inhibited AthPEPCK1 activity with I 0.5 values ranging from mid to high micromolar, while no effect was observed for Tyr‐Asp (Fig 6B). The Ala‐Ile dipeptide was the most potent inhibitor, followed by Ile‐Gln, Ser‐Tyr, Phe‐Gln, Ser‐Val, and Leu‐Thr. Contrarily, none of the tested amino acids affected AthPEPCK1 activity (Appendix Fig S10). To validate the inhibition of recombinant AthPEPCK1, we measured PEPCK activity in a protein lysate from Arabidopsis rosettes, using a fluorometric assay optimized ad hoc (Appendix Fig S11). In line with the results obtained for the recombinant enzyme, all six dipeptides (Ile‐Gln, Ile‐Ala, Phe‐Gln, Leu‐Thr, Ser‐Tyr, and Ser‐Val) inhibited PEPCK activity in crude extracts, with Ala‐Ile showing the most potent inhibitory effect, whereas no effect was observed with Tyr‐Asp (Fig 6C).
Figure 6. AthPEPCK1 activity is inhibited by dipeptides.
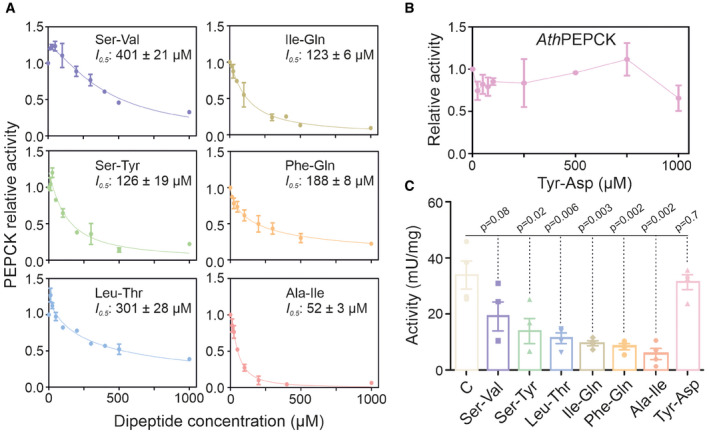
-
A, BRecombinant AthPEPCK1 activity was measured at increasing concentrations of six different H–P dipeptides (A) or Tyr‐Asp (B). I 0.5 values are indicated on each graph. Measurements were performed in the decarboxylation direction, as described in the Methods section. Data were adjusted to a modified Hill equation. The value of 1 corresponds to the activity of 4.3 ± 0.2 U mg−1. Data are mean ± SEM of 4 independent measurements.
-
CPEPCK activity was measured in a protein lysate prepared from 3‐week‐old Arabidopsis leaves in the absence (control, C) or presence of 500 µM of the different dipeptides. Measurements were performed in the carboxylation direction, as described in the Methods section. Data are mean ± SE of 4 independent lysate preparations. Unpaired two‐tailed Student’s t‐tests were performed to compare dipeptide treatments with the control.
Discussion
Production of NADPH is among the fastest known cellular responses to oxidative stress (Dick & Ralser, 2015). NADPH is at the heart of the cellular antioxidative system acting as a co‐factor of the enzymes involved in ROS scavenging and is required to replenish the pool of reduced glutathione (GSH) (Ying, 2008). In animals, NADPH is produced predominantly by the activity of oxidative PPP, which branches out from upper glycolysis, and uses G6P as a substrate (Buchanan et al, 2015). Reversible oxidative inactivation of the glycolytic enzymes downstream of G6P allowed to rapidly redirect the glycolytic flux to PPP and boost NADPH production (Ralser et al, 2007; Ralser et al, 2009). Similarly to animals, plant GAPC also undergoes oxidative inhibition, which not only affects the enzymatic activity but also affects the subcellular localization, and thus its moonlighting function(s) (Hildebrandt et al, 2015; Schneider et al, 2018; Scheibe, 2019). Arabidopsis GAPC1 protein has two cysteines. Catalytic cysteine 149 is particularly prone to oxidation via H2O2 followed by reversible S‐glutathionylation (Bedhomme et al, 2012); recovery of the glutathionylated enzyme is mediated by glutaredoxins and thioredoxins. Interestingly, it has been recently shown that, at least in vitro, persistent glutathionylation destabilizes AtGAPC1 conformation and promotes the formation of insoluble aggregates (Zaffagnini et al, 2019), possibly explaining the existence of an additional regulatory mechanism, such as the one described here, to decrease GAPDH activity under prolonged stress conditions.
In contrast to animal cells, the PPP is not the sole source of cytosolic NADPH, as the latter can also be produced by GAPN. Notably, GAPN is less sensitive than GAPC to oxidation and becomes an important source of NADPH under oxidative stress conditions (Rius et al, 2006; Piattoni et al, 2013). Moreover, during the day, NADPH is produced in chloroplasts via the light‐dependent reactions of photosynthesis, and reducing equivalents are transported into the cytosol by the activities of the "malate shuttle" (Selinski & Scheibe, 2019) and the “triose‐P shuttle” (Kelly & Gibbs, 1973; Iglesias, 1990). Tyr‐Asp inhibition of GAPC reported here provides an alternative mechanism to boost NADPH production redirecting carbon flux from glycolysis into PPP (Fig 7B). In line with this, Asp and/or Glu‐containing dipeptides (later referred to as acidic dipeptides) were shown to respond to multiple stress conditions. Specifically, levels of the acidic dipeptides increased under heat (Fig 7A) and dark but decreased under cold‐treatment (Thirumalaikumar et al, 2020). While oxidative inactivation of GAPDH happens within seconds after stress exposure (Ralser et al, 2009), accumulation of acidic dipeptides occurs over a longer time‐scale. For instance, in the above‐mentioned time‐course experiment, the increase in acidic dipeptides was first apparent 1 h after the onset of stress (Thirumalaikumar et al, 2020). We thus speculate that Tyr‐Asp accumulation may serve to support GAPC inhibition under persistent stress conditions. To address the above hypothesis, it will be crucial to understand how Tyr‐Asp inhibits GAPC activity, which entails experimental validation of the Tyr‐Asp binding site. Furthermore, however, 100 µM Tyr‐Asp is sufficient to inhibit GAPC activity in the cellular lysate; we miss information on the Tyr‐Asp binding affinity and its relation to the Tyr‐Asp cellular concentrations.
Figure 7. Working model of the Tyr‐Asp action.
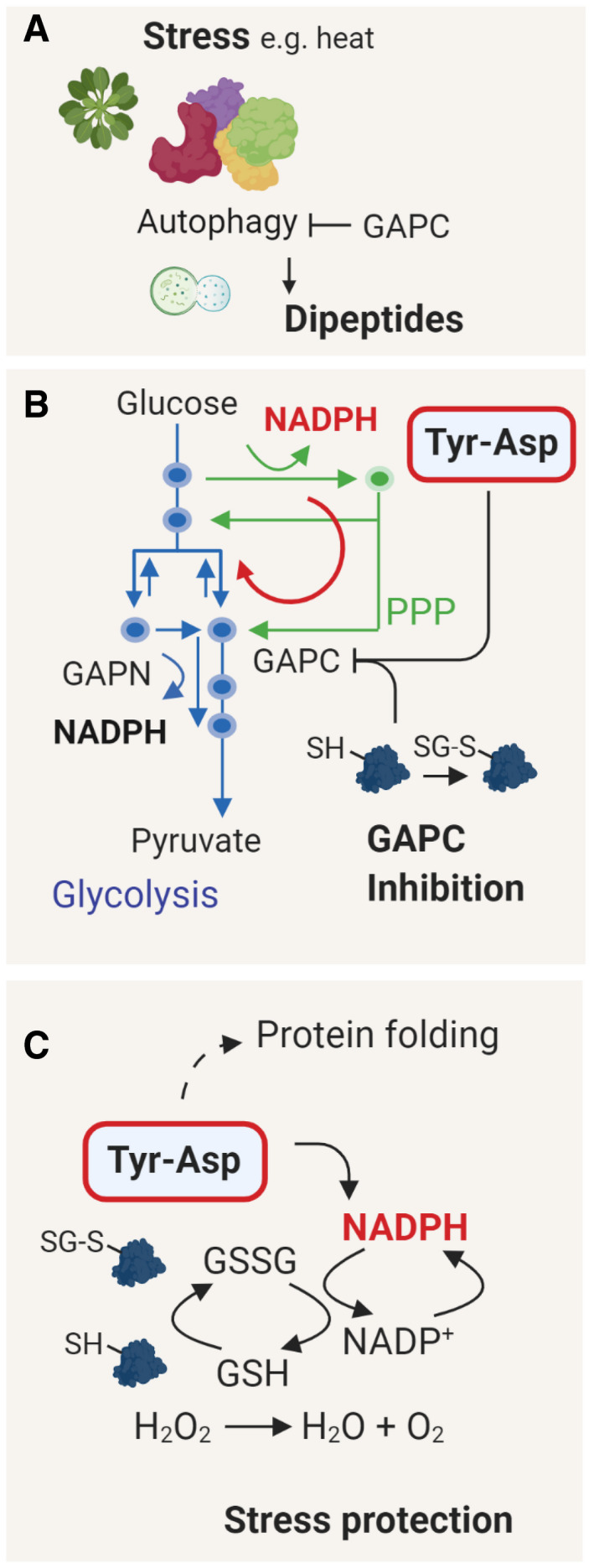
- Dipeptide accumulation under stress conditions depends on the autophagy and protein recycling (Thirumalaikumar et al, 2020). Interestingly, GAPC1 was reported to inhibit autophagy by an interaction with the ATG3 protein (Han et al, 2015; Henry et al, 2015). ROS‐mediated disruption of the GAPC1‐ATG3 complex promotes autophagy.
- Inhibition of the GAPC activity redirects the glycolytic pathway toward PPP, resulting in the production of NADPH(Ralser et al, 2007). While oxidative inactivation of GAPC is a fast event (Ralser et al, 2009; Zaffagnini et al, 2019), we speculate that Tyr‐Asp inhibition becomes critical under prolonged stress to maintain high levels of NADPH production. In contrast to GAPC, GAPN is unaffected by Tyr‐Asp and less sensitive to oxidation.
- NADPH is used to replenish the pool of reduced glutathione (GSH), needed to mitigate oxidative stress damage. Tyr‐Asp binding to chaperones suggests its role in protein folding, possibly further contributing to the Tyr‐Asp‐related stress protection. Figure was prepared using Biorender.com
In addition to its effect on GAPC activity, Tyr‐Asp is likely involved in other aspects of stress tolerance, such as protein folding (Fig 7C). The Tyr‐Asp interactome comprises multiple chaperones including heat‐shock proteins and the chloroplast chaperonin complex (CPN60/CPN20/CPN10). Notably, it has been previously demonstrated that dipeptides can support efficient folding of MHC class I molecules, transmembrane proteins important to the mammalian antiviral and antitumor immune response (Saini et al, 2013). By analogy, in vivo bioactivity could further contribute to the improved oxidative stress performance of the Tyr‐Asp‐supplemented seedlings. Multiple chaperones were also among the proteins affected by Tyr‐Asp feeding, but somewhat counterintuitively, their levels were down‐regulated. In addition, other proteins associated with stress such as enzymes involved in redox control and abscisic acid receptors were down‐regulated. The increase in NADPH level without the concomitant ROS accumulation was shown before to lead to reductive stress (Xiao & Loscalzo, 2019). Therefore, we postulate that the overall dampening of the redox stress machinery may be an adaptive response. In the future, it would be interesting to perform omics‐wide characterization of the Tyr‐Asp treatment in the context of the oxidative stress conditions, relevant for the Tyr‐Asp action.
Importantly, our results support previous studies (see Introduction) demonstrating that, in terms of function, dipeptides differ from their constituent amino acids. Tyr‐Asp supplementation leads to the rapid increase in the tyrosine level. However, neither tyrosine nor aspartic acid inhibited GAPC activity, and unlike Tyr‐Asp, tyrosine and aspartic acid supplementation had no effect on the oxidative stress tolerance.
While our work predominantly focused on Tyr‐Asp, we also questioned the broader role of dipeptides for the regulation of carbon metabolism. We showed that a group of branched‐chain and polar amino acid dipeptides, but neither their constituent amino acids nor Tyr‐Asp, inhibits the activity of AthPEPCK1, which controls the rate of gluconeogenesis (Rylott et al, 2003). PEPCK converts oxaloacetic acid to phosphoenolpyruvate, and its activity is crucial during germination and early seedling establishment. It has been demonstrated that, while PEPCK is of key importance for lipid mobilization, an alternative route operating via pyruvate orthophosphate dikinase (PPDK) enables sugar synthesis from pyruvate coming from protein breakdown (Eastmond et al, 2015). Notably, protein mobilization during germination (Avin‐Wittenberg et al, 2015), analogously to dipeptide accumulation (Thirumalaikumar et al, 2020), is autophagy dependent. We thus speculate that dipeptide inhibition of PEPCK activity could serve to promote the PPDK route. Further, circumstantial evidence linking protein degradation with PEPCK activity comes from the work of Raineri et al (2016), who showed that transgenic Arabidopsis lines with impaired protein degradation display higher rates of lipid consumption, with a concomitant increase in PEPCK activity.
Considering the evolutionary conservation of dipeptides, we anticipate that our findings will be of general relevance. Indeed, we could recently demonstrate that dipeptides accumulate in buddying yeast in response to glucose depletion before the diauxic shift (Luzarowski et al, 2021). The binding of a representative dipeptide Ser‐Leu, but not Tyr‐Asp, activates a different glycolytic enzyme, phosphoglycerate kinase 1 (PGK1). Consistent with the binding data, Ser‐Leu feeding results in metabolic changes and delays a diauxic shift's timing. Moreover, and analogously to Tyr‐Asp, Ser‐Leu interactome comprises metabolic enzymes and proteins associated with different facets of proteostasis, such as proteasomal subunits, chaperones, and ribosomal subunits. In summary, our results support the notion of dipeptides acting as metabolic regulators at the nexus of protein degradation and central carbon metabolism, and dipeptide role in the coordination of metabolism with the environment.
Materials and Methods
Reagents
Unless otherwise stated reagents were purchased from Sigma‐Aldrich. Tyr‐Asp, His‐Tyr, Leu‐Phe, and Ser‐Leu were purchased from Biomatik (Wilmington, Delaware, USA). Solid‐phase synthesis of the dipeptides Ser‐Val, Ser‐Tyr, Leu‐Thr, Ile‐Gln, Phe‐Gln, and Ala‐Ile was performed at the Bioactive Peptides Laboratory (FBCB, UNL, Argentina). The control of the final product was carried out by thin‐layer chromatography using UV, iodine, and ninhydrin. Note that Tyr‐Asp (10 mM) requires both heating (˜ 45°C) and time (several hours) to fully dissolve in water.
Plant growth
Cell culture (for TPP)
Cell suspension culture PSB‐L of A. thaliana, derived from MM2d cells, was grown in MSMO medium, in a continuous photoperiod, at 21°C, on an orbital shaker (110–120 rpm). Cells were harvested at the logarithmic growth phase (7 days after last passage) by filtration and immediately frozen in liquid nitrogen.
Arabidopsis plants (for AP)
Arabidopsis plants (Col‐0) were grown in a long‐day photoperiod (16‐h day/8‐h night), at 20/18°C, under an irradiance of 150 µmol m−2 s−1.
Arabidopsis plants (for enzymatic assays)
Arabidopsis plants (Col‐0 and the gapc1 gapc2 mutant) were grown in a chamber at 22–24°C, under an irradiance of 120 µmol m−2 s−1 with a long‐day photoperiod. Rosette leaves from three‐week‐old plants were harvested in the light, immediately frozen in liquid nitrogen, and pulverized to a fine powder.
Tobacco plants (for AP)
Tobacco plants were grown for four weeks in fully controlled plant growth chambers with constant light intensity and long‐day photoperiod (16‐h day length, 250 µE m−2 s−1, 22°/18°C—day/night) and then transferred to the greenhouse. Tobacco leaves were harvested from 6‐week‐old tobacco plants, immediately frozen in liquid nitrogen, and ground to a fine powder.
Tyr‐Asp feeding experiments
Arabidopsis Col‐0 plants were grown in liquid Murashige and Skoog medium (Murashige & Skoog, 1962) supplemented with 1% sucrose as described in Kosmacz et al (2019). Each culture corresponds to approximately 100 plants (1.5 mg of seeds). After 10 days, either Tyr‐Asp (100 μM) or water (mock control) was supplemented to the media. Seedlings were harvested into liquid nitrogen at different time‐points ranging from 15 min to 24 h.
Stress experiments
Arabidopsis and tobacco seeds were sterilized and germinated on solid MS medium supplemented with 3% sucrose. 12‐day‐old seedlings were transferred to a 24‐well plate filled with liquid MS medium as described in Moreno et al (2021) with some modifications. Briefly, seedlings were incubated in an orbital shaker (120 rpm) with either 100 µM Tyr‐Asp, Ser‐Leu, Gly‐Pro, combination of Tyr and Asp, or mock (water) for one h before applying the oxidative agents: H2O2 (50 mM) and catechin (0.175 mM), or salt (50 mM NaCl). Arabidopsis plants were exposed to catechin and H2O2 for two and one and a half days, respectively. Tobacco plants were exposed to catechin, H2O2, and NaCl for four, two, and six days, respectively. Fresh weight was recorded for each plant. Each plate corresponded to 12 control and 12 treated plants, used for direct comparison. The same procedure was followed to perform catechin treatments on the wild‐type and the double gapc1 gapc2 k.o. plants. Plants were exposed to catechin for three days and pretreated with Tyr‐Asp. Leaf series were performed as described in Gonzalez et al (2010). An additional stress experiment was performed using solid MS medium supplemented with 3% sucrose. Tobacco seedlings were grown on a nylon mesh for 2 weeks. Subsequently, seedlings (by lifting the mesh) were transferred for 24 h to new plates with or without (mock) Tyr‐Asp, and afterward to plates supplemented with different combination of catechin (0.175 mM), salt (50 mM), and Tyr‐Asp.
Feeding experiments with 14C‐labeled glucose
Feeding experiments were performed according to Obata et al (2017). The assay was performed in the absence of photosynthetically active radiation. For more details, see Appendix Information.
Glyceraldehyde‐3‐phosphate dehydrogenase enzymatic assays
Native protein extract
Proteins were extracted from 20 mg of Arabidopsis leaf material by addition of approximately 10 mg (w/v) polyvinylpolypyrrolidone and 1 ml ice‐cold extraction buffer, followed by vigorous shaking. Extraction buffer was composed of 50 mM HEPES/KOH, pH 7.5, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM benzamidine, 1 mM ɛ‐aminocaproic acid, 0.25% (w/v) BSA, 10 µM leupeptin, 0.5 mM DTT, 0.1% (v/v) Triton X‐100, 20% (v/v) glycerol, and 1 mM phenylmethylsulfonyl fluoride. The lysate was centrifuged for 10 min at 14,000 g and 4°C (Gibon et al, 2004).
Activity measurements
The enzymatic activity of GAPC, GAPN, and GAPA/B was determined as described by Rius et al (2006), with minor modifications. GAPC was assayed in 50 mM Tricine‐KOH pH 8.5, 4 mM NAD+, 1.2 mM fructose 1,6‐bisphosphate (FBP), 10 mM sodium arsenate, and 1 U/ml aldolase from rabbit muscle (Sigma). GAPN was assayed in 50 mM Tricine‐KOH pH 8.5, 0.4 mM NADP+, 1.2 mM FBP, and 1 U/ml aldolase from rabbit muscle (Sigma). GAPA/B was assayed in 50 mM Tricine‐KOH pH 8.5, 0.4 mM NADP+, 1.2 mM FBP, 10 mM sodium arsenate, and 1 U/ml aldolase from rabbit muscle (Sigma). All measurements were performed in a final volume of 0.25 ml at 30°C under control (mock) conditions or with the addition of 100 µM Tyr‐Asp. Assays were performed with 4 biological replicates from 3‐week‐old Arabidopsis leaves, obtained either from wild‐type plants or the gapc1 gapc2 double mutant. One unit (U) is defined as the amount of enzyme that catalyzes the formation of one μmol NAD(P)H in one min under the specified assay conditions.
Phosphoenolpyruvate carboxykinase enzymatic assay
Recombinant protein assay
AthPEPCK1 was produced in Escherichia coli, and its activity was measured in the decarboxylation direction of the reaction, as previously described (Rojas et al, 2019). Kinetic parameters were obtained by fitting data of enzyme activity versus substrate concentration using a modified Hill equation (Rius et al, 2006), with the software Origin Pro 8 (OriginLab Corporation).
Native protein extract
Considering that PEPCK from cucumber cotyledons is subject to proteolysis (Walker & Leegood, 1995), we optimized the extraction protocol to maintain Arabidopsis PEPCK as stable as possible during the assays (Appendix Fig S11). Plant material was homogenized in a precooled mortar with liquid nitrogen, and protein extraction was performed with an extraction buffer consisting of 100 mM Bicine/NaOH pH 9.0, 10% (v/v) glycerol, 0.1% (v/v) Triton X‐100, 5 mM β‐mercaptoethanol, 1 mM EDTA, 1 mM EGTA, 2 mM phenylmethylsulfonyl fluoride, and 1× Set III protease cocktail (Merck). After addition of extraction buffer, samples were vortexed, incubated on ice for 10 min, and then centrifuged at 21,000 g for 10 min to remove tissue debris. In all cases, protein was quantified with the Bradford reagent (Bradford, 1976), using bovine serum albumin as standard.
Crude extract enzymatic assay
PEPCK activity in Arabidopsis leaves is considerably low (˜ 0.05 µmol min−1 g FW−1) (Malone et al, 2007). Therefore, we adapted a fluorometric method initially developed to measure protein kinase activity to assay PEPCK activity in crude extracts (Rojas et al, 2018). Reactions were linear in the 0–15 min range with 1.0–4.5 µg of crude extract (Appendix Fig S11). Activity was assayed in the carboxylation direction in 100 mM HEPES/NaOH pH 7.0, 4 mM β‐mercaptoethanol, 0.2 mM NADH, 4 mM MgCl2, 1 mM MnCl2, 100 mM KHCO3, 0.5 mM ADP, 10 mM PEP, and 1 U malate dehydrogenase (Sigma). Data were adjusted for PEPC activity by performing a blank without ADP, as previously described (Martín et al, 2007). Reactions were performed in a final volume of 20 µl in black, 96‐well microplates (Thermo Scientific) at room temperature, and were initiated with a proper amount of the crude extract diluted in a buffer consisting of 25 mM HEPES/NaOH pH 7.0, 10% (v/v) glycerol, and 1 mM β‐mercaptoethanol. Reactions were stopped with 20 µl of 0.3 M HCl, and the following procedures to form and detect the NAD+‐alkali derivatives were done as previously described (Rojas et al, 2018).
NAD(H) and NADP(H) measurements in the feeding experiment
NAD(H) and NADP(H) measurements were performed as described by Gibon and Larher (1997). For more details, see Appendix Information.
Preparation of a native cellular lysate
Native lysate was prepared as described before (Veyel et al, 2018). For more details, see Appendix Information.
Affinity purification
Affinity purification experiments were performed and analyzed as described before (Veyel et al, 2018). For more details, see Appendix Information.
Thermal proteome profiling (TPP)
Native Arabidopsis lysate was filtered using a 10‐kD Amicon®Ultra centrifugal filter (Merck, Darmstadt, Germany) to remove free (protein unbound) small molecules. Protein concentration was measured using the Bradford assay, and the lysate was diluted to reach protein concentration of 5 mg/ml. The obtained lysate was incubated with (treatment) or without (control) 10 μM Tyr‐Asp for 30 min at room temperature with mixing. Further steps were adapted from (Franken et al, 2015). Briefly, a 3‐min temperature treatment was applied to treated (Tyr‐Asp) and non‐treated (control) samples using a thermocycler (40.4; 45.8; 51.1; 56.2; 60.1; 65.6; 70.9; 76.0; 79.9; RT, room temperature). Denatured proteins were pelleted by centrifugation at 20,817 g, and the remaining soluble proteins were precipitated with precooled acetone (1:4). Proteins were dried in the vacuum concentrator and then stored at −20°C for further proteomic analysis. Data are from three independent replicas. For more details, see Appendix Information.
MTBE protein and metabolite extraction
Proteins and metabolites were extracted using a methyl‐tert‐butyl ether (MTBE)/methanol/water solvent system as described before (Giavalisco et al, 2011). For more details, see Appendix Information.
Metabolomics
The dried aqueous phase was measured using ultra‐performance liquid chromatography coupled to an Exactive mass spectrometer (Thermo Fisher Scientific) in positive and negative ionization modes, as described in Giavalisco et al (2011). For more details, see Appendix Information.
Proteomics
Samples for proteomics were prepared using Trypsin/Lys‐C Mix (Promega) digestion and peptide desalting as described in Kosmacz et al (2019) using C18 Empore® extraction disk (3 M, Maplewood, MN) STAGE tips (Rappsilber et al, 2003). Measurements were executed on a Q Exactive HF coupled to an Easy nLC1000 HPLC (Thermo Scientific) or on a Q Exactive HF coupled to nanoACQUITY UPLC System (Waters). Raw files were submitted to MaxQuant software for protein identification and quantification (Cox & Mann, 2008). For more details, see Appendix Information.
Estimation of Tyr‐Asp content
The content of Tyr‐Asp in Arabidopsis seedlings was estimated by spiking different amounts of non‐labeled Tyr‐Asp (from 100 nM to 1 mM) into metabolic extract prepared via MTBE method (see Appendix Information) from 15N labeled Arabidopsis leaf material (Giavalisco et al, 2011). Peak intensities corresponding to spiked Tyr‐Asp were used to prepare a calibration curve. The obtained calibration curve had a linear trend from 100 nM to 100 µM and was used to calculate the amount of Tyr‐Asp in the Arabidopsis seedling material. The intracellular concentration of Tyr‐Asp was estimated using volume data measured for a mesophyll cell located at the center of a young leaf: 621.15 and 23.42 µl g−1 FW−1 for the whole cell and the cytosol, respectively (Koffler et al, 2013).
Analysis of the protein and metabolite data from the feeding experiment
GeneData‐derived raw metabolite intensities were normalized to the median intensity of all mass features detected in a given chromatogram. MaxQuant‐derived LFQ intensities were used for further analysis. Both metabolite and protein data were subjected to log2 transformation prior two‐way analysis of variance (ANOVA) implemented in MeV software (Howe et al, 2011) using treatment (treated versus untreated) and time (15, 30 min, 1, 6, 24 h) as variables. Obtained P‐values were subjected to FDR correction to select metabolites and proteins significantly affected by the treatment.
Docking analysis
The crystal structures of GAPC1 with bound NAD+ of Arabidopsis thaliana (PDB‐ID of reduced variant: 4z0h, oxidized variant: 6qun, and glutathionylated variant: 6quq) (Berman et al, 2000; Zaffagnini et al, 2016; Zaffagnini et al, 2019) were taken as the target for molecular docking of Tyr‐Asp, taken as a 3D‐model downloaded in.mol‐format from http://molview.org/ and considered with appropriately set charges (smeared negative charge on carboxyl group of tyrosine residue). The respective protein model was loaded as a receptor into the docking software package LeadIT FlexX (version 2.3.2) (BioSolveIT GmbH, Sankt Augustin, Germany, 2017, www.biosolveit.de/LeadIT). All amino acid residues within a 20 Å radius around NAD+ were selected to constitute the candidate binding site. Sulfate ions with reported positions in the crystal structure, also those reported to be near the active site, have been ignored; i.e., they have been removed, as they are likely associated with the crystallization solution (3–3.5 M ammonium sulfate) and/or have uncertain positions (occupancy = 0.5) (Zaffagnini et al, 2016). Tyr‐Asp was docked to the receptor, and all docked conformations were extracted. The docking process was repeated with the binding pocket detection and definition performed by LeadIT (automatic pocket detection). The approximate binding affinities of all conformations were evaluated in SeeSAR (version 10.0) (BioSolveIT GmbH, Sankt Augustin, Germany, 2018, www.biosolveit.de/SeeSAR) using the protein model as the receptor. The best conformations with the highest affinities were selected to be displayed on the surface of the protein model using PyMol (version 2.1.1) (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC, Open source license). Cp12‐2 was taken from PDB‐ID: 3qv1 and was overlaid.
Author contributions
AS conceived the project and designed experimental strategy with input from JCM; JCM performed AP, oxidative stress, Tyr‐Asp feeding experiments, and prepared metabolomic and proteomic samples; BER and CMF performed GAPC enzymatic assays and analyzed the data; JSP‐A provided technical assistance; AS analyzed metabolomic and proteomic data from the feeding experiment, and JCM built the protein/metabolite overview (MapMan) and Tyr‐Asp interaction network (Cytoscape); MG ran proteomic samples and analyzed the data; JCM and MC performed thermal proteome profiling experiments, and DC analyzed the data; SA and JCM performed flux experiments under supervision of ARF; YZ and JCM performed NADP(H) and NAD(H) measurements; RV and ML provided valuable discussion regarding enzymatic assays and dipeptides; ZN provided scientific discussion; BER and MDH performed PEPCK experiments under supervision of AAI and CMF; TM performed docking analysis under supervision of DW and initial input from RZ; AS and JCM wrote the manuscript with input from ARF, AAI, and CMF. Open Access funding enabled and organized by ProjektDEAL.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Appendix
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Dataset EV8
Acknowledgements
We would like to thank Prof. Lothar Willmitzer for support and discussion, Änne Michaelis and Gudrun Wolter for excellent technical assistance, Prof. Roc Ros from Valencia University, and Prof. Sam Wang from Donald Danforth Center for providing the GFP‐GAPCP1 and GFP‐GAPCP2, and YFP‐GAPC1 and gapc1 gapc2 seeds, respectively.
The EMBO Journal (2021) 40: e106800.
Data availability
Proteomics data can be found in PRIDE (Perez‐Riverol et al, 2019) with accession number: PXD019332. http://www.ebi.ac.uk/pride/archive/projects/PXD019332.
References
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M et al (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32: D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avin‐Wittenberg T, Bajdzienko K, Wittenberg G, Alseekh S, Tohge T, Bock R, Giavalisco P, Fernie AR (2015) Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27: 306–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedhomme M, Adamo M, Marchand CH, Couturier J, Rouhier N, Lemaire SD, Zaffagnini M, Trost P (2012) Glutathionylation of cytosolic glyceraldehyde‐3‐phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro . Biochem J 445: 337–347 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL (2015) Biochemistry and molecular biology of plants. Chichester, West Sussex; Hoboken, NJ: Wiley Blackwell; [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Dick TP, Ralser M (2015) Metabolic remodeling in times of stress: who shoots faster than his shadow? Mol Cell 59: 519–521 [DOI] [PubMed] [Google Scholar]
- Diether M, Nikolaev Y, Allain FH, Sauer U (2019) Systematic mapping of protein‐metabolite interactions in central metabolism of Escherichia coli . Mol Syst Biol 15: e9008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doppler M, Kluger B, Bueschl C, Steiner B, Buerstmayr H, Lemmens M, Krska R, Adam G, Schuhmacher R (2019) Stable isotope‐assisted plant metabolomics: investigation of phenylalanine‐related metabolic response in wheat upon treatment with the fusarium virulence factor deoxynivalenol. Front Plant Sci 10: 1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond PJ, Astley HM, Parsley K, Aubry S, Williams BP, Menard GN, Craddock CP, Nunes‐Nesi A, Fernie AR, Hibberd JM (2015) Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nat Commun 6: 6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini G, Fermani S, Ripamonti A, Sabatino P, Sparla F, Pupillo P, Trost P (2003) Dual coenzyme specificity of photosynthetic glyceraldehyde‐3‐phosphate dehydrogenase interpreted by the crystal structure of A4 isoform complexed with NAD. Biochemistry 42: 4631–4639 [DOI] [PubMed] [Google Scholar]
- Fermani S, Trivelli X, Sparla F, Thumiger A, Calvaresi M, Marri L, Falini G, Zerbetto F, Trost P (2012) Conformational selection and folding‐upon‐binding of intrinsically disordered protein CP12 regulate photosynthetic enzymes assembly. J Biol Chem 287: 21372–21383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Roscher A, Ratcliffe RG, Kruger NJ (2001) Fructose 2,6‐bisphosphate activates pyrophosphate: fructose‐6‐phosphate 1‐phosphotransferase and increases triose phosphate to hexose phosphate cycling in heterotrophic cells. Planta 212: 250–263 [DOI] [PubMed] [Google Scholar]
- Franken H, Mathieson T, Childs D, Sweetman GMA, Werner T, Tögel I, Doce C, Gade S, Bantscheff M, Drewes G et al (2015) Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat Protoc 10: 1567–1593 [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Kohl K, Willmitzer L (2011) Elemental formula annotation of polar and lipophilic metabolites using (13) C, (15) N and (34) S isotope labelling, in combination with high‐resolution mass spectrometry. Plant J 68: 364–376 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JH, Palacios N, Cross J, Selbig J, Stitt M (2004) A Robot‐based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Larher F (1997) Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal Biochem 251: 153–157 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, De Bodt S, Sulpice R, Jikumaru Y, Chae E, Dhondt S, Van Daele T, De Milde L, Weigel D, Kamiya Y et al (2010) Increased leaf size: different means to an end. Plant Physiol 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, Wang X (2012) Cytosolic glyceraldehyde‐3‐phosphate dehydrogenases interact with phospholipase Ddelta to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24: 2200–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Ma F, Wei F, Fanella B, Allen DK, Wang X (2014) Cytosolic phosphorylating glyceraldehyde‐3‐phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell 26: 3023–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habenicht A, Hellman U, Cerff R (1994) Non‐phosphorylating GAPDH of higher plants is a member of the aldehyde dehydrogenase superfamily with no sequence homology to phosphorylating GAPDH. J Mol Biol 237: 165–171 [DOI] [PubMed] [Google Scholar]
- Han S, Wang Y, Zheng X, Jia Q, Zhao J, Bai F, Hong Y, Liu Y (2015) Cytoplastic glyceraldehyde‐3‐phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27: 1316–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry E, Fung N, Liu J, Drakakaki G, Coaker G (2015) Beyond glycolysis: GAPDHs are multi‐functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet 11: e1005199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt T, Knuesting J, Berndt C, Morgan B, Scheibe R (2015) Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol Chem 396: 523–537 [DOI] [PubMed] [Google Scholar]
- Howe EA, Sinha R, Schlauch D, Quackenbush J (2011) RNA‐Seq analysis in MeV. Bioinformatics 27: 3209–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias AA (1990) On the metabolism of triose‐phosphates in photosynthetic cells ‐ their involvement on the traffic of Atp and Nadph. Biochem Educ 18: 2–5 [Google Scholar]
- Jung DW, Kim WH, Williams DR (2014) Chemical genetics and its application to moonlighting in glycolytic enzymes. Biochem Soc Trans 42: 1756–1761 [DOI] [PubMed] [Google Scholar]
- Kanegawa N, Suzuki C, Ohinata K (2010) Dipeptide Tyr‐Leu (YL) exhibits anxiolytic‐like activity after oral administration via activating serotonin 5‐HT1A, dopamine D1 and GABAA receptors in mice. FEBS Lett 584: 599–604 [DOI] [PubMed] [Google Scholar]
- Kaushik S, Bais HP, Biedrzycki ML, Venkatachalam L (2010) Catechin is a phytototoxin and a pro‐oxidant secreted from the roots of Centaurea stoebe. Plant Signal Behav 5: 1088–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly GJ, Gibbs M (1973) A mechanism for the indirect transfer of photosynthetically reduced nicotinamide adenine dinucleotide phosphate from chloroplasts to the cytoplasm. Plant Physiol 52: 674–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Guo L, Wang X (2013) Phosphatidic acid binds to cytosolic glyceraldehyde‐3‐phosphate dehydrogenase and promotes its cleavage in Arabidopsis. J Biol Chem 288: 11834–11844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffler BE, Bloem E, Zellnig G, Zechmann B (2013) High resolution imaging of subcellular glutathione concentrations by quantitative immunoelectron microscopy in different leaf areas of Arabidopsis. Micron 45: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova NY, Thor K, Gubler A, Meier S, Dietrich D, Weichert A, Suter Grotemeyer M, Tegeder M, Rentsch D (2008) AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol 148: 856–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmacz M, Gorka M, Schmidt S, Luzarowski M, Moreno JC, Szlachetko J, Leniak E, Sokolowska EM, Sofroni K, Schnittger A et al (2019) Protein and metabolite composition of Arabidopsis stress granules. New Phytol 222: 1420–1433 [DOI] [PubMed] [Google Scholar]
- Luzarowski M, Vicente R, Kiselev A, Wagner M, Schlossarek D, Erban A, de Souza LP, Childs D, Wojciechowska I, Luzarowska U et al (2021) Global mapping of protein‐metabolite interactions in Saccharomyces cerevisiae reveals that Ser‐Leu dipeptide regulates phosphoglycerate kinase activity. Commun Biol 4: 181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone S, Chen ZH, Bahrami AR, Walker RP, Gray JE, Leegood RC (2007) Phosphoenolpyruvate carboxykinase in Arabidopsis: changes in gene expression, protein and activity during vegetative and reproductive development. Plant Cell Physiol 48: 441–450 [DOI] [PubMed] [Google Scholar]
- Marri L, Trost P, Trivelli X, Gonnelli L, Pupillo P, Sparla F (2008) Spontaneous assembly of photosynthetic supramolecular complexes as mediated by the intrinsically unstructured protein CP12. J Biol Chem 283: 1831–1838 [DOI] [PubMed] [Google Scholar]
- Martín M, Plaxton WC, Podestá FE (2007) Activity and concentration of non‐proteolyzed phosphoenolpyruvate carboxykinase in the endosperm of germinating castor oil seeds: effects of anoxia on its activity. Physiol Plantarum 130: 484–494 [Google Scholar]
- Michels S, Scagliarini S, Della Seta F, Carles C, Riva M, Trost P, Branlant G (1994) Arguments against a close relationship between non‐phosphorylating and phosphorylating glyceraldehyde‐3‐phosphate dehydrogenases. FEBS Lett 339: 97–100 [DOI] [PubMed] [Google Scholar]
- Mizushige T, Kanegawa N, Yamada A, Ota A, Kanamoto R, Ohinata K (2013) Aromatic amino acid‐leucine dipeptides exhibit anxiolytic‐like activity in young mice. Neurosci Lett 543: 126–129 [DOI] [PubMed] [Google Scholar]
- Moreno JC, Martinez‐Jaime S, Kosmacz M, Sokolowska EM, Schulz P, Fischer A, Luzarowska U, Havaux M, Skirycz A (2021) A multi‐OMICs approach sheds light on the higher yield phenotype and enhanced abiotic stress tolerance in tobacco lines expressing the carrot lycopene β‐cyclase1 gene. Front Plant Sci 12: 624365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz‐Bertomeu J, Cascales‐Minana B, Mulet JM, Baroja‐Fernandez E, Pozueta‐Romero J, Kuhn JM, Segura J, Ros R (2009) Plastidial glyceraldehyde‐3‐phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol 151: 541–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Noctor G, Reichheld JP, Foyer CH (2018) ROS‐related redox regulation and signaling in plants. Semin Cell Dev Biol 80: 3–12 [DOI] [PubMed] [Google Scholar]
- Nunes‐Nesi A, Carrari F, Lytovchenko A, Smith AMO, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Rosado‐Souza L, Fernie AR (2017) Coupling radiotracer experiments with chemical fractionation for the estimation of respiratory fluxes, In Plant respiration and internal oxygen, Jagadis Gupta K (ed) pp 17–30. New York: Humana Press; [DOI] [PubMed] [Google Scholar]
- Perez‐Riverol Y, Csordas A, Bai J, Bernal‐Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M et al (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47: D442–D450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Brinkmann H, Cerff R (2003) Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J Mol Evol 57: 16–26 [DOI] [PubMed] [Google Scholar]
- Piattoni CV, Guerrero SA, Iglesias AA (2013) A differential redox regulation of the pathways metabolizing glyceraldehyde‐3‐phosphate tunes the production of reducing power in the cytosol of plant cells. Int J Mol Sci 14: 8073–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47: 185–214 [DOI] [PubMed] [Google Scholar]
- Plaxton WC, Podesta FE (2006) The functional organization and control of plant respiration. Crit Rev Plant Sci 25: 159–198 [Google Scholar]
- Raineri J, Hartman MD, Chan RL, Iglesias AA, Ribichich KF (2016) A sunflower WRKY transcription factor stimulates the mobilization of seed‐stored reserves during germination and post‐germination growth. Plant Cell Rep 35: 1875–1890 [DOI] [PubMed] [Google Scholar]
- Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, Klipp E, Jakobs C, Breitenbach M, Lehrach H et al (2007) Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol 6: 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralser M, Wamelink MM, Latkolik S, Jansen EE, Lehrach H, Jakobs C (2009) Metabolic reconfiguration precedes transcriptional regulation in the antioxidant response. Nat Biotechnol 27: 604–605 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M (2003) Stop and go extraction tips for matrix‐assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez‐Casati DF (2006) Characterization of an Arabidopsis thaliana mutant lacking a cytosolic non‐phosphorylating glyceraldehyde‐3‐phosphate dehydrogenase. Plant Mol Biol 61: 945–957 [DOI] [PubMed] [Google Scholar]
- Rius SP, Casati P, Iglesias AA, Gomez‐Casati DF (2008) Characterization of Arabidopsis lines deficient in GAPC‐1, a cytosolic NAD‐dependent glyceraldehyde‐3‐phosphate dehydrogenase. Plant Physiol 148: 1655–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas BE, Hartman MD, Figueroa CM, Leaden L, Podesta FE, Iglesias AA (2019) Biochemical characterization of phosphoenolpyruvate carboxykinases from Arabidopsis thaliana. Biochem J 476: 2939–2952 [DOI] [PubMed] [Google Scholar]
- Rojas BE, Santin F, Ulloa RM, Iglesias AA, Figueroa CM (2018) A fluorometric method for the assay of protein kinase activity. Anal Biochem 557: 120–122 [DOI] [PubMed] [Google Scholar]
- Rylott EL, Gilday AD, Graham IA (2003) The gluconeogenic enzyme phosphoenolpyruvate carboxykinase in Arabidopsis is essential for seedling establishment. Plant Physiol 131: 1834–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini SK, Ostermeir K, Ramnarayan VR, Schuster H, Zacharias M, Springer S (2013) Dipeptides promote folding and peptide binding of MHC class I molecules. Proc Natl Acad Sci USA 110: 15383–15388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski MM, Reinhard FBM, Franken H, Werner T, Savitski MF, Eberhard D, Molina DM, Jafari R, Dovega RB, Klaeger S et al (2014) Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346: 1255784 [DOI] [PubMed] [Google Scholar]
- Scarpeci TE, Zanor MI, Carrillo N, Mueller‐Roeber B, Valle EM (2008) Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol Biol 66: 361–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R (2019) Maintaining homeostasis by controlled alternatives for energy distribution in plant cells under changing conditions of supply and demand. Photosynth Res 139: 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Knuesting J, Birkholz O, Heinisch JJ, Scheibe R (2018) Cytosolic GAPDH as a redox‐dependent regulator of energy metabolism. BMC Plant Biol 18: 184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinski J, Scheibe R (2019) Malate valves: old shuttles with new perspectives. Plant Biol 21(Suppl 1): 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Sonnewald U (1995) Regulation of metabolism in transgenic plants. Annu Rev Plant Physiol Plant Mol Biol 46: 341–368 [Google Scholar]
- Strehmel N, Hoehenwarter W, Monchgesang S, Majovsky P, Kruger S, Scheel D, Lee J (2017) Stress‐related mitogen‐activated protein kinases stimulate the accumulation of small molecules and proteins in Arabidopsis thaliana root exudates. Front Plant Sci 8: 1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P et al (2017) The STRING database in 2017: quality‐controlled protein‐protein association networks, made broadly accessible. Nucleic Acids Res 45: D362–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Shiomi H, Ueda H, Amano H (1979) Morphine‐like analgesia by a new dipeptide, L‐Tyrosyl‐L‐Arginine (Kyotorphin) and its analog. Eur J Pharmacol 55: 109–111 [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user‐driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thirumalaikumar VP, Wagner M, Balazadeh S, Skirycz A (2020) Autophagy is responsible for the accumulation of proteogenic dipeptides in response to heat stress in Arabidopsis thaliana . FEBS J 288: 281–292. [DOI] [PubMed] [Google Scholar]
- Veyel D, Kierszniowska S, Kosmacz M, Sokolowska EM, Michaelis A, Luzarowski M, Szlachetko J, Willmitzer L, Skirycz A (2017) System‐wide detection of protein‐small molecule complexes suggests extensive metabolite regulation in plants. Sci Rep 7: 42387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyel D, Sokolowska EM, Moreno JC, Kierszniowska S, Cichon J, Wojciechowska I, Luzarowski M, Kosmacz M, Szlachetko J, Gorka M et al (2018) PROMIS, global analysis of PROtein‐metabolite interactions using size separation in Arabidopsis thaliana . J Biol Chem 293: 12440–12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RP, Leegood RC (1995) Purification, and phosphorylation in vivo and in vitro, of phosphoenolpyruvate carboxykinase from cucumber cotyledons. FEBS Lett 362: 70–74 [DOI] [PubMed] [Google Scholar]
- Wolosiuk RA, Buchanan BB (1978) Activation of chloroplast NADP‐linked glyceraldehyde‐3‐phosphate dehydrogenase by the ferredoxin/thioredoxin system. Plant Physiol 61: 669–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Loscalzo J (2019) Metabolic responses to reductive stress. Antioxid Redox Signal 32: 1330–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W (2008) NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal 10: 179–206 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Fermani S, Costa A, Lemaire SD, Trost P (2013) Plant cytoplasmic GAPDH: redox post‐translational modifications and moonlighting properties. Front Plant Sci 4: 450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Fermani S, Calvaresi M, Orru R, Iommarini L, Sparla F, Falini G, Bottoni A, Trost P (2016) Tuning cysteine reactivity and sulfenic acid stability by protein microenvironment in glyceraldehyde‐3‐phosphate dehydrogenases of Arabidopsis thaliana . Antioxid Redox Signal 24: 502–517 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Marchand CH, Malferrari M, Murail S, Bonacchi S, Genovese D, Montalti M, Venturoli G, Falini G, Baaden M et al (2019) Glutathionylation primes soluble glyceraldehyde‐3‐phosphate dehydrogenase for late collapse into insoluble aggregates. Proc Natl Acad Sci USA 116: 26057–26065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhao Y, Wang X, Lin R, Zhang Y, Ma H, Guo Y, Xu L, Zhao B (2016) The novel dipeptide Tyr‐Ala (TA) significantly enhances the lifespan and healthspan of Caenorhabditis elegans. Food Funct 7: 1975–1984 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File
Appendix
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Dataset EV8
Data Availability Statement
Proteomics data can be found in PRIDE (Perez‐Riverol et al, 2019) with accession number: PXD019332. http://www.ebi.ac.uk/pride/archive/projects/PXD019332.


