Abstract
Background:
The microbiota interacts with the brain through the gut–brain axis, and a distinct dysbiosis may lead to major depressive episodes. Bacteria can pass through the gut barrier and be found in the blood. Using a multiomic approach, we investigated whether a distinct blood microbiome and metabolome was associated with major depressive episodes, and how it was modulated by treatment.
Methods:
In this case–control multiomic study, we analyzed the blood microbiome composition, inferred bacterial functions and metabolomic profile of 56 patients experiencing a current major depressive episode and 56 matched healthy controls, before and after treatment, using 16S rDNA sequencing and liquid chromatography coupled to tandem mass spectrometry.
Results:
The baseline blood microbiome in patients with a major depressive episode was distinct from that of healthy controls (patients with a major depressive episode had a higher proportion of Janthinobacterium and lower levels of Neisseria) and changed after antidepressant treatment. Predicted microbiome functions confirmed by metabolomic profiling showed that patients who were experiencing a major depressive episode had alterations in the cyanoamino acid pathway at baseline. High baseline levels of Firmicutes and low proportions of Bosea and Tetrasphaera were associated with response to antidepressant treatment. Based on inferred baseline metagenomic profiles, bacterial pathways that were significantly associated with treatment response were related to xenobiotics, amino acids, and lipid and carbohydrate metabolism, including tryptophan and drug metabolism. Metabolomic analyses showed that plasma tryptophan levels are independently associated with response to antidepressant treatment.
Limitations:
Our study has some limitations, including a lack of information on blood microbiome origin and the lack of a validation cohort to confirm our results.
Conclusion:
Patients with depression have a distinct blood microbiome and metabolomic signature that changes after treatment. Dysbiosis could be a new therapeutic target and prognostic tool for the treatment of patients who are experiencing a major depressive episode.
Introduction
Major depressive disorder (MDD) is the second most frequent cause of disability worldwide,1 and it is among the diseases with the greatest effect on public health.2 It can be driven by neuroendocrine,3 neuroimmune,4 metabolic5 or neurotransmitter (particularly serotonin) dysregulation.6 The main treatment for major depressive episodes is antidepressant drugs, but these show limited efficacy: half of patients experiencing a major depressive episode do not achieve a response, and two-thirds do not achieve remission after 3 months of treatment.7 Moreover, the mechanisms of action of these drugs are poorly understood.
Recent evidence suggests that the intestinal microbiota interacts with the brain through the so-called microbiota–gut–brain axis, and that it may be a factor associated with MDD.8,9 Altered composition (dysbiosis) of the intestinal microbiota increases intestinal permeability, the translocation of bacterial products and inflammatory responses as a result of immune activation in several diseases, including MDD.10–14 Several studies have reported changes in the intestinal microbiota15–17 in patients with depression, and intestinal permeability in other psychiatric disorders.18–20 Communication between the microbiota and the brain may involve neural, endocrine and immune pathways, including translocated bacterial products.8 Water-avoidance stress in rats increases intestinal permeability,21 and chronic mild stress leads to the translocation of bacterial products and neuroinflammatory responses by immune activation.13 This mechanism, known as “leaky gut,” may also occur in MDD14 and is involved in the inflammatory process of major depressive episodes. Indeed, mice deficient in segmented filamentous bacteria (bacteria that produce serum amyloid proteins, which increase the production of T helper 17 cells) are resilient to the induction of depression-like behaviour.22 Moreover, humanization of germ-free mice with microbiota derived from patients with MDD resulted in depression-like behaviours in recipient mice. These mice also showed changes in microbial genes and host metabolites involved in carbohydrate and amino acid metabolism, suggesting that alterations in the intestinal microbiota play a causal role in the development of depression-like behaviours.17
New DNA-sequencing technologies have also led to the discovery of an altered blood microbial DNA profile or blood microbiome23,24 that has been associated with several diseases.25,26 The blood microbiome mostly reflects the translocation of bacteria to the bloodstream from different sites (gut, oral cavity, skin), especially in diseases related to increased intestinal permeability such as MDD. However, despite the potential involvement of the intestinal microbiota in major depressive episodes, no data are available on the presence or composition of the circulating microbiome in patients who experience a major depressive episode.
In this exploratory multiomic study, we first investigated whether a distinct blood microbiome with a distinct inferred functional metagenome and metabolomic profile was associated with major depressive episodes in a cohort of well-characterized patients with MDD compared to matched healthy controls. We then studied changes in the circulating microbiome, its inferred functional metagenome, and the metabolomic profile after antidepressant treatment, and we explored whether distinct signatures might be associated with treatment response.
Methods
Study population
This omics-based, prospective, case–control, exploratory study was nested in the METADAP cohort27 with no a priori expectations. The METADAP cohort was a 6-month prospective, multicentre treatment study in a real-world setting that included patients with a current unipolar major depressive episode in the context of MDD (DSM-IV-TR criteria). Participants were enrolled between November 2009 and March 2013 from 6 university psychiatry departments in France, based on the following inclusion criteria: consecutive in- or outpatients; age 18 to 65 years; research-confirmed diagnosis of a current major depressive episode in the context of MDD (DSM-IV-TR criteria), based on the Mini International Neuropsychiatric Interview, with a minimum depression score of 18 on the 17-item Hamilton Depression Rating Scale (HDRS)28; and requiring first-time antidepressant treatment or a change in treatment. Depression was assessed by clinical psychologists, independently from the clinicians who treated the patients. The Mini International Neuropsychiatric Interview provided the main diagnosis. The 17-item HDRS and the Clinical Global Impressions Scale29 were administered at baseline and 1, 3 and 6 months after the initiation of the current antidepressant treatment. Patients were not included in this cohort if they had psychotic symptoms, bipolar disorder, a psychotic disorder, an eating disorder, current substance abuse or dependence (DSM-IV-TR criteria), pregnancy, organic brain syndromes or severe unstable medical conditions (including liver disease), or if they were being treated with antipsychotics or mood stabilizers before inclusion and/or for 4 months or more during the preceding year.
Patients from the cohort included in the present analysis were antidepressant-free for at least 1 year at baseline; initiated treatment with venlafaxine (n = 25), citalopram (n = 19) or escitalopram (n = 12); and had a blood sample at baseline and after 3 months of treatment. All patients were evaluated for major depression at the start of antidepressant treatment (M0) and 3 months later (M3). Further details are provided in Appendix 1, available at jpn.ca/200159-a1.
Response to treatment was defined by a decrease of greater than 50% in HDRS score after 3 months of treatment. Remitters were those with an HDRS score of less than 7 after 3 months of treatment.
Healthy controls were matched to the patients with a major depressive episode based on age, sex and body mass index (BMI), and were selected from the VARIETE study, a population-based, cross-sectional study designed to recruit a reference population in 10 university hospitals in France between January 2011 and February 2012.30 For these patients, only baseline samples were available.
All patients and controls provided written informed consent. The METADAP (ClinicalTrials.gov NCT00526383) and VARIETE (ClinicalTrials.gov Identifier: NCT01831648) studies were approved by the Ethics Committee of Paris-Boulogne and Ile de France VII, respectively, and conformed to international ethical standards.
Blood microbiota and metabolomics
Blood samples were obtained from patients with a major depressive episode in the METADAP cohort and healthy controls in the VARIETE study using the same procedure. The microbiome composition was characterized quantitatively (16S quantitative polymerase chain reaction) and qualitatively (16S targeted sequencing; Vaiomer). Bacterial DNA from plasma samples was isolated and amplified in a strictly controlled environment at Vaiomer using a stringent contamination-aware approach as described previously.31–33
Details of the blood microbiota and the metabolomics and statistical analysis are available in Appendix 1.
Results
Patient characteristics
In total, we analyzed findings for 112 participants (56 patients with a major depressive episode and 56 matched healthy controls). The mean (± standard deviation) age of patients with a major depressive episode was 41.9 ± 11.6 years, and 68% were female. In the patient group, 55% had a recurrent major depressive episode; the mean (± standard deviation) HDRS score at baseline was 23.6 ± 3.8; and 57% were responders and 43% remitters after 3 months of antidepressant treatment. Response and remission were not associated with patient characteristics (Table 1).
Table 1.
Patient characteristics
| Characteristic | Healthy controls (n = 56) | Patients with an MDE; mean ± SD or n (%) | |||
|---|---|---|---|---|---|
|
| |||||
| Before treatment (M0) (n = 56) | After treatment (M3) (n = 56) | Responders (M3) (n = 32) | Nonresponders (M3) (n = 24) | ||
| Age, yr | 41.9 ± 12.7 | 41.9 ± 11.6 | — | 40.7 ± 11.99 | 43.7 ± 10.99 |
| Male | 17 (30.4) | 18 (32.1) | 11 (34.4) | 7 (29.1) | |
| BMI, kg/m2 | — | 24 ± 5.3 | 25.2 ± 5.3 | 24.5 ± 4.65 | 24.4 ± 6.24 |
| HDRS score | — | 23.6 ± 3.8 | 12.3 ± 8.93§ | 23.0 ± 3.93 | 24.3 ± 3.67 |
| History of an MDE | — | 31 (55.4) | — | 19 (59.3) | 12 (50.0) |
| No. of previous MDEs | — | 1.65 ± 1.02 | — | 1.5 ± 0.9 | 1.8 ± 1.2 |
| History of antidepressant use | — | 25 (44.6) | — | 13 (40.6) | 12 (50.0) |
| Current antidepressant use (SSRI/SNRI)* | 31 (55.4)/25 (44.6) | — | 20 (62.5)/12 (37.5) | 11 (45.8)/13 (54.2) | |
| Tobacco use | 5 (8.9) | 16 (28.6) | — | 8 (25.0) | 8 (33.3) |
| Diabetes | — | 3 (5.4) | 1 (3.1) | 2 (8.3) | |
| Responders† | — | — | 32 (57.1) | — | — |
| Remitters‡ | — | — | 24 (42.8) | — | — |
BMI = body mass index; HDRS = Hamilton Depression Rating Scale (17 items); MDE = major depressive episode; SD = standard deviation; SSRI = selective serotonin reuptake inhibitor, SNRI = serotonin noradrenergic reuptake inhibitor.
Current antidepressant use refers to the treatment patients received after study inclusion and sample collection.
Response to treatment was defined by a > 50% decrease in score on the HDRS after 3 months of treatment.
Remitters were those who had an HDRS score < 7.
p < 0.05.
Patients with major depressive episodes had a distinct blood microbiome and metabolomic signature
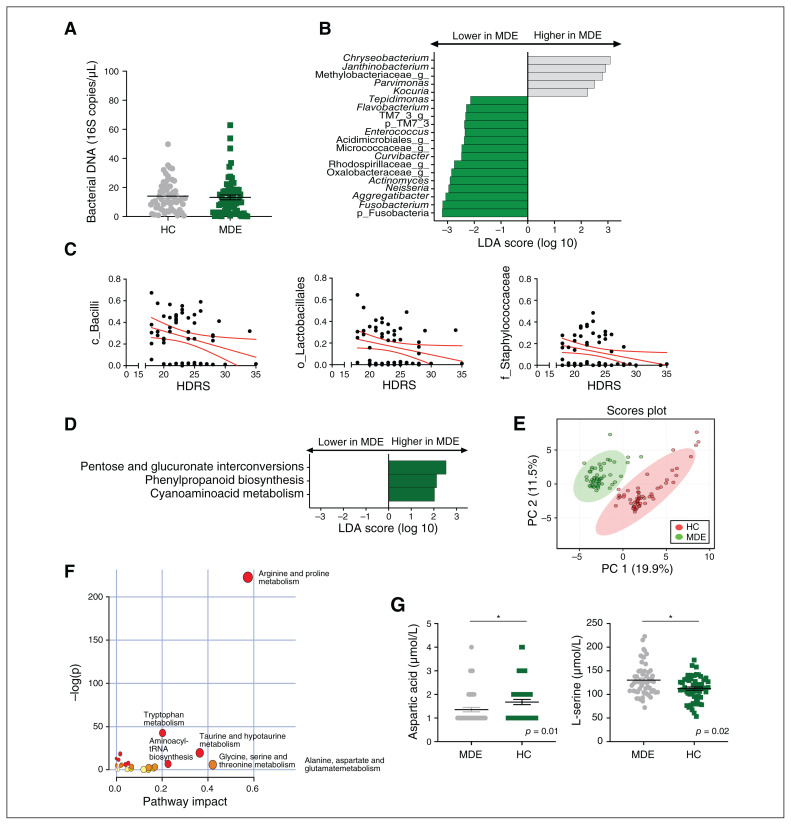
We found no difference in the absolute quantity of blood bacterial 16S DNA in patients with a major depressive episode compared to healthy controls (Figure 1A).
Fig. 1.
Blood microbiota and metabolomic profiles of patients with a major depressive episode (green; n = 56) and matched healthy controls (grey; n = 56). (A) Bacterial 16S rDNA assessed by quantitative polymerase chain reaction in healthy controls and patients with a major depressive episode (p = 0.3, unpaired Mann–Whitney test). (B) LDA effect size for the taxa enriched in patients with a major depressive episode and healthy controls. (C) Correlations between HDRS score and baseline relative abundance of Bacilli, Lactobacillales and Staphylococcaceae (Spearman correlation, p < 0.05 after FDR correction for multiple comparisons). (D) LDA effect size for the predicted metagenome metabolic pathways (KEGG modules) for patients with a major depressive episode versus healthy controls. Only taxa with an LDA score greater than 2 and p < 0.05 (determined by Wilcoxon signed rank test) are shown. (E) PCA score plots of the metabolomic profiles of healthy controls and patients with a major depressive episode. (F) Summary of the pathway analysis using MetaboAnalyst 2.0, which shows all matched pathways according to the p values from the pathway enrichment analysis and pathway impact values from the pathway topology analysis. (G) Absolute plasma concentrations of the metabolites aspartic acid and L-serine; *p < 0.05. FDR = false discovery rate; HC = healthy control; HDRS = Hamilton Depression Rating Scale; KEGG = Kyoto Encyclopedia of Genes and Genomes; LDA = linear discriminant analysis; MDE = major depressive episode; PC = principal component; PCA = principal component analysis.
The interindividual diversity (presence or absence of operational taxonomic units) assessed by 16S targeted sequencing was significantly different between patients and healthy controls (unweighted UniFrac, p = 0.01), suggesting a difference in the composition of the blood microbiome (Appendix 1, Figure S1A). We found no difference in the homogeneity of the blood microbiome between the 2 groups (PERMDISP, p = 0.5), or in interindividual diversity, reflecting the structure of the blood microbiome (weighted UniFrac; Appendix 1, Figure S1B), or intraindividual α diversity (Appendix 1, Figure S1C).
The blood microbiome of patients showed a lower relative abundance of bacterial DNA from the phyla Fusobacteria (p = 0.03) and Saccharibacteria (previously known as Candidate division TM7; p = 0.02) and 13 taxa at the genus level, including Actinomyces, Flavobacterium, Enterococcus, Neisseria, Tepidimonas, Aggregatibacter, Curvibacter, Fusobacterium (Figure 1B; Appendix 1, Table S1) and several unidentified taxa. The blood microbiome of patients was also significantly enriched for 5 taxa at the genus level, including Kocuria, Chryseobacterium, Parvimonas and Janthinobacterium (Appendix 1, Table S1). Scores on the HDRS, reflecting the severity of depression at baseline, were negatively correlated with the baseline relative abundance of Bacilli (at the class level, p = 0.02), Lactobacillales (at the order level, p = 0.04) and Staphylococcaceae (at the family level, p = 0.02, Figure 1C).
With respect to the predicted metagenomic functions of the blood microbiota, patients showed relative enrichment of pathways involved in pentose and glucoronate interconversions (p = 0.005), phenylpropanoid biosynthesis (p = 0.002) and cyanoamino acid metabolism (p = 0.02) relative to healthy controls (Figure 1D).
We confirmed these predicted functions by studying the metabolomic profile of patients using liquid chromatography coupled to tandem mass spectrometry, and patients with a major depressive episode showed a different metabolomic profile (Figure 1E; Appendix 1, Figure S2). Seventeen pathways were significantly different between patients and healthy controls (Figure 1F; Appendix 1, Table S2). In the cyanoamino acid metabolism pathway (also modified in the circulating microbiome), aspartic acid levels were lower in patients than in healthy controls (p = 0.01), but L-serine levels were higher (Figure 1G, p = 0.02). In addition, patients showed significantly lower kynurenine levels (tryptophan metabolism, p < 0.001) than healthy controls (Appendix 1, Figure S2).
Changes in the blood microbiome and metabolome after antidepressant treatment depended on treatment response
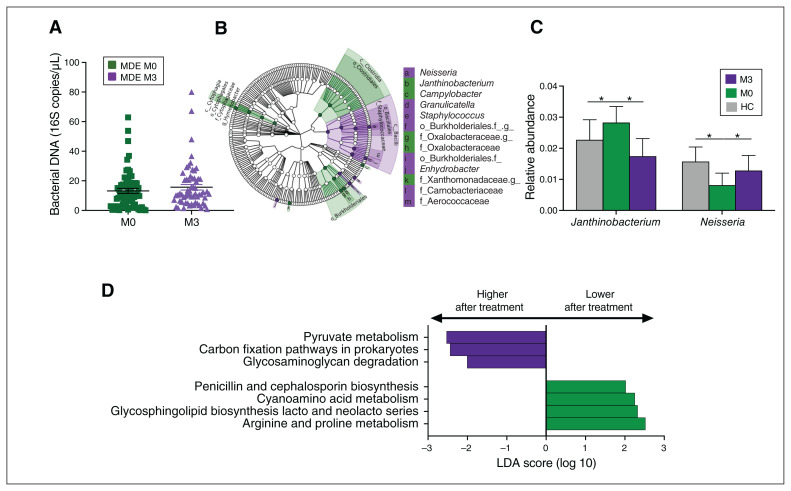
The amount of bacterial 16S DNA between groups remained unchanged after 3 months of antidepressant treatment (Figure 2A). However, the composition of the blood microbiome changed significantly (unweighted UniFrac, p = 0.01). Indeed, the relative abundance of Bacilli increased (p = 0.016) and that of Clostridia (p = 0.049) and Cytophagia (p = 0.004) decreased at the class level (Figure 2B). Moreover, at the genus level, the relative abundance of 5 taxa increased (including Neisseria, Enhydrobacter, Granulicatella and Staphylococcus), whereas that of 5 other taxa decreased (including Janthinobacterium, Campylobacter and Hymenobacter; Figure 2B; Appendix 1, Table S3). Thus, the initial lower relative abundance of Neisseria and higher abundance of Janthinobacterium was modified in patients after treatment (Figure 2C).
Fig. 2.
Blood microbiota profile of patients with a major depressive episode before (n = 56) and after (n = 56) treatment. (A) Bacterial 16S rDNA assessed by quantitative polymerase chain reaction in patients with a major depressive episode before and after treatment (p = 0.2, paired Mann–Whitney test). (B) Cladogram showing the taxa with the largest differences in abundance in LDA effect size analysis between M0 (green) and M3 (purple). The size of the circle in the cladogram plot is proportional to the bacterial abundance. From inside to outside, the circles represent phylum, class, order, family and genus. Only taxa with an LDA score greater than 2 and p < 0.05 (determined by Wilcoxon signed rank test) are shown. (C) Comparison of the relative abundance at the genus level of taxa that were modified in healthy controls (grey) and patients with a major depressive episode before (M0, green) and after (M3, purple) treatment. (D) LDA effect size analysis for the predicted metagenome metabolic pathways (KEGG modules) in patients with a major depressive episode before (M0, green) and after (M3, purple) treatment; only pathways with an LDA score greater than 2 and p < 0.05 (determined by Wilcoxon signed rank test) are shown. HC = healthy control; KEGG = Kyoto Encyclopedia of Genes and Genomes; LDA = linear discriminant analysis; M0 = before treatment; M3 = after treatment; MDE = major depressive episode.
Several metabolic pathways of the blood microbiome involved in glycosaminoglycan (p = 0.047), pyruvate (p = 0.034) and carbon (p = 0.018) metabolism, were enriched after treatment. Pathways involved in the metabolism of cyanoamino acid (p = 0.02) and arginine and proline (p = 0.047) were decreased (Figure 2D; Appendix 1, Table S4).
We found no overall difference in the global metabolomic profile or metabolic pathways of patients with a major depressive episode before and after treatment. However, based on the predicted pathways, metabolomics studies confirmed the occurrence of significant and interesting changes in the levels of several metabolites after treatment (Appendix 1, Figure S3), including an increase in L-tyrosine (p = 0.04), a decrease in L-isoleucine (p = 0.03, from the cyanoamino acid pathway) and an increase in N-acetylornithine (p < 0.001, arginine and proline pathway, Figure 2D). Moreover, kynurenine levels, which were lower in patients than in healthy controls, increased after antidepressant treatment (p = 0.045, Appendix 1, Figure S3).
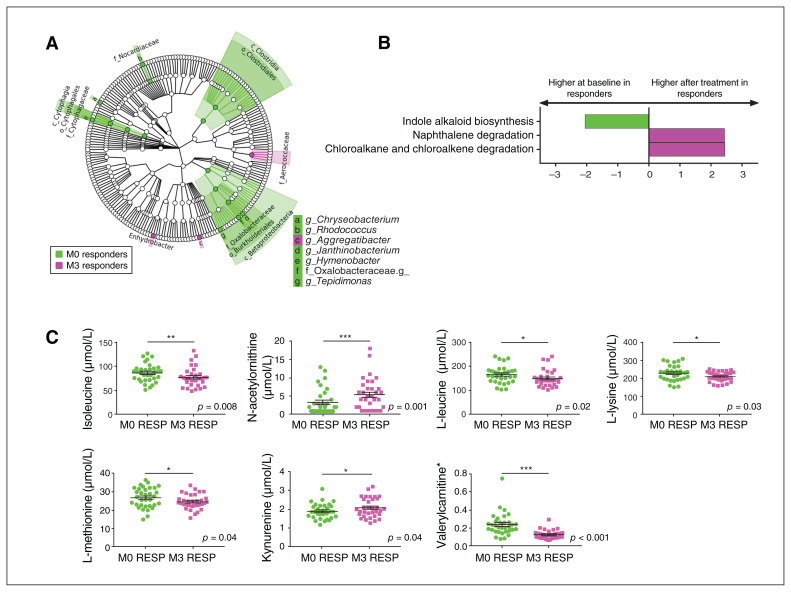
Interestingly, antidepressant treatment modified the blood microbiome in both responders and nonresponders. At baseline, responders showed higher levels of Clostridia (p = 0.03), Cytophagia (p = 0.01) and Betaproteobacteria (p = 0.04, class level), 6 taxa at the genus level and a lower relative abundance of 2 taxa than post-treatment (Figure 3A; Appendix 1, Table S5). Notably, the relative abundance of Janthinobacterium decreased after treatment (p = 0.004).
Fig. 3.
Blood microbiota profile in patients with a major depressive episode who responded to treatment (n = 32), before and after treatment. (A) Cladogram showing the taxa with the largest differences in abundance of the microbiota before (green) and after (pink) treatment in patients who responded to treatment. From inside to outside, the circles represent phylum, class, order, family and genus. Only taxa with an LDA score greater than 2 and p < 0.05 (determined by Wilcoxon signed rank test) are shown. (B) LDA effect size analysis for the predicted metagenome metabolic pathways (KEGG modules) in the baseline microbiota (green) compared to the microbiota after treatment (pink) in patients who responded to treatment. Only pathways with an LDA score greater than 2 and p < 0.05 (determined by Wilcoxon signed rank test) are shown. (C) Absolute plasma concentrations of the significant single metabolites that were different between patients with a major depressive episode who responded to treatment, before and after treatment; *p < 0.05, ** p < 0.01, ***p < 0.001. HC = healthy control; KEGG = Kyoto Encyclopedia of Genes and Genomes; LDA = linear discriminant analysis; M0 = before treatment; M3 = after treatment; RESP = responders.
In terms of predicted metagenomic functions, future responders showed higher levels of pathways involved in indole alkaloid biosynthesis at baseline (p = 0.02) and higher levels of pathways involved in the metabolism of naphthalene (p = 0.04) and chloroalkane and chloroalkene (p = 0.04) after treatment (Figure 3B).
Concerning the metabolic profile of responders before and after treatment, we found changes in the levels of several metabolites involved in amino acid metabolism, including increases in kynurenine (p = 0.04) and N-acetylornithine (p < 0.001) after treatment and decreases in isoleucine (p = 0.008), L-leucine (p = 0.02), L-lysine (p = 0.03), L-methionine (p = 0.04) and valerylcarnitine (p < 0.001) after treatment (Figure 3C).
Nonresponders showed a different composition of the blood microbiome. At baseline, they showed a higher abundance of Proteobacteria (phylum level) and Alphaproteobacteria (class level). After treatment, they showed a higher abundance of Firmicutes (phylum level); Bacilli and Rubrobacteria (class level); and Rubrobacter, Staphylococcus, Granulicatella and Neisseria (genus level; Appendix 1, Figure S4A, Table S6). Nonresponders also showed a different profile of predicted microbial metagenomic functions (Appendix 1, Figure 4B, Table S7). However, we found no differences in the metabolomic profile in nonresponders before and after treatment.
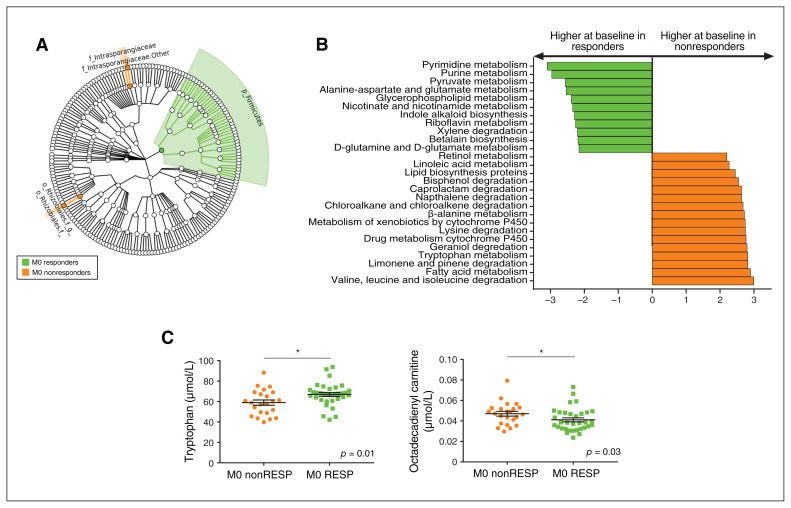
Fig. 4.
Baseline blood microbiota profile associated with treatment response in patients with a major depressive episode (responders, n = 32; nonresponders, n = 24). (A) Cladogram showing the taxa with the largest differences in abundance of the baseline microbiota according to response to treatment: responders (green) and nonresponders (orange). From inside to outside, the circles represent phylum, class, order, family and genus. Only taxa with an LDA score greater than 2 and p < 0.05 (determined by Wilcoxon signed rank test) are shown. (B) LDA effect size analysis for the predicted metagenome metabolic pathways (KEGG modules) in the baseline blood microbiota according to response to treatment for responders (M0 responders, green) and nonresponders (M0 nonresponders, orange). Only pathways with an LDA score greater than 2 and p < 0.05 (determined by Wilcoxon signed rank test) are shown. (C) Absolute plasma concentrations of the metabolites tryptophan and octadecadienyl-carnitine; *p < 0.05. KEGG = Kyoto Encyclopedia of Genes and Genomes; LDA = linear discriminant analysis; M0 = before treatment; M3 = after treatment; nonRESP = nonresponders; RESP = responders.
Pretreatment blood microbiome and baseline plasma tryptophan levels were independently associated with response to antidepressant treatment
Three taxa were associated with response to treatment: the Firmicutes phylum (higher in responders at baseline; p = 0.047) and 2 unidentified taxa at the genus level (lower in responders at baseline), 1 belonging to the Actinobacteria phylum (identified as the Tetrasphaera genus using BLAST [https://blast.ncbi.nlm.nih.gov/Blast.cgi]; p = 0.035) and the other to the Proteobacteria phylum (identified as the Bosea genus using BLAST; p = 0.012; Figure 4A).
Several predicted metagenomic functions were associated with response to treatment. Responders showed lower levels of 20 metabolic pathways and higher levels of 2 pathways (Figure 4B; Appendix 1, Table S8). Pretreatment pathways that were lower in responders were involved in the metabolism of amino acids (tryptophan, β-alanine, lysine, valine, leucine and isoleucine), lipids (fatty acids, lipid protein and linoleic acid), xenobiotics (bisphenol, caprolactam, naphthalene, chloroalkane and chloroalkene, drug metabolism [cytochrome P450] and metabolism of xenobiotics by cytochrome P450), terpenoids and polyketides (geraniol, limonene and pinene degradation), and cofactors and vitamins (retinol).
Nonresponders showed lower levels of predicted pathways involved in the metabolism of amino acids (alanine aspartate, glutamate, D-glutamine, D-glutamate and β-alanine), xenobiotics (xylene), cofactors and vitamins (riboflavin, nicotinate and nicotinamide), secondary metabolites (indole alkaloid), lipids (glycerophospholipid), carbohydrates (pyruvate) and nucleotides (purine and pyrimidine; Figure 4B; Appendix 1, Table S8).
We found a similar difference in the microbiota (Appendix 1, Table S9) and predicted metagenomic function profiles (Appendix 1, Table S10) between remitters and non-remitters (Appendix 1, Figure S5).
We performed metabolomic analyses to confirm the modifications of the predicted pathways. We found no global difference between responders and nonresponders in the metabolomic profile at baseline. However, distinct modifications consistent with the changes observed in the metagenomic functions of the circulating microbiome were associated with response to treatment: an increase in tryptophan plasma levels and the modulation of lipid metabolism, and a decrease in octadecadienyl-carnitine levels in responders (Figure 4C). We observed similar changes in the metabolomic profile at baseline between remitters and non-remitters (Appendix 1, Figure S5C).
After adjusting for sex, age, BMI, diabetes, smoking status, history of major depressive episodes, HDRS at baseline and the 3 taxa (Firmicutes, Tetrasphaera, Bosea) and 2 metabolites (tryptophan, octadecadienyl-carnitine) that were different at baseline between responders and nonresponders, we found that levels of Bosea and tryptophan were independently associated with response to treatment (Appendix 1, Table S11).
Discussion
There is growing evidence for a role of microbiota in depression. In the present study, using a unique cohort, we showed that unmedicated patients experiencing a major depressive episode had a distinct blood microbiome. Moreover, we identified some of the bacteria and metabolites that were associated with response to antidepressant treatment.
One of our major findings was a lower proportion of bacteria from the Saccharibacteria and Fusobacteria phyla in patients. This was consistent with the findings from an animal model of depression, in which rats that were transplanted with feces from patients with depression showed decreased levels of Saccharibacteria and Actinobacteria in their feces.16 Similar changes in circulating microbiota profiles were described in patients with obesity and alcoholic hepatitis.25,26 The changes observed in our study were not related to these conditions, because there were no differences in BMI between the groups, and patients with liver disease (including alcoholic liver disease) were excluded.
We also observed a lower relative abundance of Fusobacterium in patients experiencing a major depressive episode. A higher overall proportion of Fusobacteria has been reported in the intestinal microbiota of patients with a major depressive episode. However, in that study, patients who responded to treatment showed less Fusobacteria in their feces than healthy controls.15 Moreover, mice transplanted with the intestinal microbiota from patients experiencing a major depressive episode developed depression-like behaviour and showed a lower proportion of Fusobacterium than mice that received the intestinal microbiota of healthy controls,17 in accordance with our results.
Patients experiencing a major depressive episode also showed higher levels of Janthinobacterium than healthy controls, but these levels decreased after antidepressant treatment. Elevated relative levels of these taxa were also reported in the circulating microbiome of patients with alcohol dependence.26 We also found that the abundance of Neisseria (a pathobiont) was lower in patients and increased after treatment. Most Neisseria species are benign commensal members of the oral and nasopharyngeal microbiota. Their presence and abundance have been correlated with the onset and progression of several inflammatory diseases34 and have been shown to be in the circulating microbiome of patients with alcoholic hepatitis.26 Such lower levels in patients with inflammation may also be related to the presence of a major depressive episode as a comorbidity of other disorders. Indeed, Neisseria shows neuroactive potential, because it can produce tryptophan, the precursor of serotonin and other neuroactive molecules.9
The circulating microbiome could also be related to the severity of depression. Major depressive episodes were negatively correlated with the abundance of several bacterial taxa, including Lactobacillales, an order of bacteria that includes Lactobacillus and Lactococcus — 2 probiotic strains. A meta-analysis that studied the effect of probiotic-based interventions on depression found that probiotics (including Lactobacillus and Lactococcus) significantly decreased scores on a depression scale.35
We found a distinct signature of the blood microbiota associated with clinical response to antidepressant treatment: responders showed a higher relative pretreatment abundance of Firmicutes compared to nonresponders. Similar findings have been reported in the intestinal microbiota,15 and reinforce our findings. Conversely, the abundance of Bosea was independently associated with nonresponse to antidepressant treatment. An elevated relative abundance of circulating Bosea has been reported for patients with obesity and fibrosis.25 The difference we observed was not related to obesity, because the patients in our study had a median BMI of 24 kg/m2 and there were no significant differences in BMI between groups. However, Bosea may be associated with MDD and metabolic syndromes.
Our results in terms of predicted functional metagenomics must be viewed with caution, because we were unable to determine whether circulating bacterial DNA belonged to viable or dormant bacteria, or if they were merely bacterial DNA fragments. Therefore, metabolites present in the blood may have come from other body sites, the gut being one of the main potential sources. Nevertheless, the changes we found in metabolic pathways based on untargeted metabolomic profiling were consistent with those predicted for the circulating microbiome. Carbohydrate metabolism was higher in the predicted circulating microbiota of depressed patients. Interestingly, symptoms related to elevated carbohydrate metabolism, such as asthenia, lower energy and decreased activity, are among the clinical symptoms of major depression. Furthermore, humanized mice that developed depression-like features after a transfer of intestinal microbiota from patients with MDD also showed an increase in the carbohydrate pathway in the intestinal microbiota, serum and hippocampus, similar to our results.17 The increase in carbohydrate metabolism pathways suggests lower available energy in depressed mice.
We also observed elevated cyanoamino acid metabolism in the microbiome of patients experiencing a major depressive episode, with elevated levels of L-serine (a metabolite involved in cyanoamino acid metabolism) in the metabolomic analyses. Interestingly, serine is a co-agonist of the N-methyl-d-aspartate receptor, which is involved in the pathophysiology of major depression. Our results were consistent with those of other studies, which also found similar profiles of higher levels of serine36 and lower levels of kynurenine37 in patients with major depression. Furthermore, the differences in levels of kynurenine, isoleucine, L-leucine and N-acetyl-ornithine observed between patients with a major depressive episode and healthy controls were modified after treatment.
We also observed elevated levels of the tryptophan metabolism predicted pathway in the circulating microbiome of nonresponders. We confirmed this result by showing a lower level of plasma tryptophan at baseline in nonresponders and nonremitters than in responders and remitters. Therefore, the changes in the microbiota could have been associated with impaired serotonin metabolism in these patients, because increased metabolism of tryptophan by bacteria would decrease the availability of tryptophan for serotonin synthesis. Indeed, tryptophan is the sole precursor of peripherally and centrally produced serotonin. In animal models of depression, germ-free mice show higher levels of plasma tryptophan and lower anxiety than conventional mice. However, plasma tryptophan levels decreased after colonization with the intestinal microbiota of normal healthy mice, and the mice showed more anxiety-like behaviour.38 Moreover, mice fed a tryptophan-deficient diet show exacerbated inflammation of the central nervous system.39
Limitations
Our study had several limitations. Although the presence of a blood “living” microbiome is a subject of debate, the presence of bacteria in the blood of healthy people has been confirmed by various techniques, including imaging techniques such as transmission electron microscopy and dark-field microscopy.40 However, studying the blood microbiome is a technical challenge, with potential artifacts and contaminants.32 Circulating bacterial DNA can reflect living bacteria, bacterial content (resulting from immune degradation) or contamination. Dormant bacteria can return to a state of replication depending on experimental conditions. Several arguments reinforce the relevance of a blood microbiome under healthy conditions and disease (reviewed in Potgieter and colleagues23 and Castillo and colleagues40). The presence of a specific blood microbiome, at least as bacterial DNA, has already been confirmed in both health and disease.25,26,31,33 The technique we used was carefully designed to study blood bacterial DNA using a stringent contamination-aware approach described previously.31–33 We cannot exclude the possibility that part of the blood microbiome measured signal was affected by environmental and technical contaminants, but because of the specific protocol and high number of samples, possible contaminants should not have substantially influenced the comparisons between groups.32 Moreover, 16S sequencing is compositional, especially when comparing relative abundance between data sets with significant differences in the total microbial load.41 In our study there was no difference in total microbial load between groups, but some technical biases could have persisted. The origin of the blood microbiome has not yet been fully elucidated but is thought to be derived mostly from the mother preceding birth, or from the translocation of microorganisms derived from other sources after birth and during the normal human life cycle (gut, oral cavity, skin).40 Therefore, our findings could reflect changes in microbiota from different sources. Our cohort did not include data on the microbiome from sites other than the blood. It has also been reported that contamination by skin bacteria is a major challenge when using small volumes of blood (20 μL) taken by skin puncture.32 Because we collected a volume of 30 mL, contamination by the skin microbiome was unlikely or at least negligible (incidentally, we did not observe a substantial amount of bacterial taxa known as commensals of the skin). Another limitation was the absence of information about previous antibiotic use and diet. Nevertheless, it has been shown in patients with alcoholic hepatitis that the use of antibiotics had no effect on the principal findings in the circulating microbiome.26 Although we did not have comprehensive data on dietary habits, patients in the 2 groups had similar BMIs and none was following a vegetarian or vegan diet. In addition, 55% of the patients in our cohort had a previous major depressive episode, and 45% had already been exposed to antidepressant treatment. Because both major depressive episodes and antidepressant treatment can modify the microbiota, their long-term effects on the intestinal microbiota and blood microbiota are unknown. Moreover, although they are quite similar, future studies will have to use the current DSM-5 criteria for major depressive episodes rather than the criteria from DSM-IV-TR. Finally, our study lacked a validation cohort to confirm our results.
Conclusion
Our study showed that major depression is associated with a distinct blood microbiome signature and a shift in its predicted functions. The predicted functions were confirmed by the distinct metabolomic profiles of these patients. Finally, we identified a distinct circulating microbiome signature, distinct predicted bacterial metabolic pathways and a distinct measured metabolic profile associated with the response to antidepressant treatment.
Although there is growing evidence of the role of intestinal microbiota in major depressive episodes and mood disorders, there are few data on the bacteria or bacterial fragments that can pass through mucosal barriers and reach the bloodstream. Our study brings new insights into the microbiota landscape in major depressive episodes and provides a basis for further studies to better understand the physiological and pathophysiological functions of the microbiota in depression. They also pave the way for new therapeutic approaches in the treatment of major depression, suggesting that targeting the microbiome itself — using pre-, pro- or symbiotics, or their metabolic pathways — may help to improve the treatment of patients experiencing a major depressive episode.
Acknowledgments
This study was funded by the Programme Hospitalier de Recherche Clinique National of the French Ministry of Health (AOM06022 for the METADAP study and AOM09122 for the VARIETE study) and sponsored by the Assistance Publique-Hôpitaux de Paris (NCT01831648). The biobank METADAP is stored in CRB Paris South (BRIF: BB-0033-00089).
Footnotes
Competing interests: D. David serves as a consultant for and receives compensation from Lundbeck. G. Perlemuter reports grants and personal fees from Servier (IRIS); personal fees and non-financial support from Gilead; personal fees from Bicodex, Pileje, Elsevier Masson, Flammarion and Solar; and non-financial support from Abbvie, all outside the submitted work. No other competing interests declared.
Contributors: D. Ciocan, A.-M. Cassard, L. Becquemont, C. Verstuyft, S. Trabado, G. Perlemuter and E. Corruble designed the study. D. Ciocan, C. Verstuyft, C. Voican, P. Chanson and E. Corruble acquired the data, which D. Ciocan, A.-M. Cassard, K. El Asmar, R. Colle, D. David, B. Feve, G. Perlemuter and E. Corruble analyzed. D. Ciocan, L. Becquemont, D. David and E. Corruble wrote the article, which A.-M. Cassard, C. Verstuyft, C. Voican, K. El Asmar, R. Colle, S. Trabado, B. Feve, P. Chanson, G. Perlemuter and E. Corruble reviewed. All authors approved the final version to be published and can certify that no other individuals not listed as authors have made substantial contributions to the paper.
References
- 1.Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015;386:743–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vos T, Flaxman AD, Naghavi M, et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2163–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stetler C, Miller GE. Depression and hypothalamic-pituitary-adrenal activation: a quantitative summary of four decades of research. Psychosom Med 2011;73:114–26. [DOI] [PubMed] [Google Scholar]
- 4.Wohleb ES, Franklin T, Iwata M, et al. Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci 2016;17:497–511. [DOI] [PubMed] [Google Scholar]
- 5.Jokela M, Hamer M, Singh-Manoux A, et al. Association of metabolically healthy obesity with depressive symptoms: pooled analysis of eight studies. Mol Psychiatry 2014;19:910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blokhin IO, Khorkova O, Saveanu RV, et al. Molecular mechanisms of psychiatric diseases. Neurobiol Dis 2020;146:105136. [DOI] [PubMed] [Google Scholar]
- 7.Trivedi MH, Rush AJ, Wisniewski SR, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry 2006; 163:28–40. [DOI] [PubMed] [Google Scholar]
- 8.Dinan TG, Cryan JF. Gut–brain axis in 2016: Brain–gut–microbiota axis—mood, metabolism and behaviour. Nat Rev Gastroenterol Hepatol 2017;14:69–70. [DOI] [PubMed] [Google Scholar]
- 9.Valles-Colomer M, Falony G, Darzi Y, et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol 2019;4:623–32. [DOI] [PubMed] [Google Scholar]
- 10.Cassard A-M, Ciocan D. Microbiota, a key player in alcoholic liver disease. Clin Mol Hepatol 2018;24:100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciocan D, Voican CS, Wrzosek L, et al. Bile acid homeostasis and intestinal dysbiosis in alcoholic hepatitis. Aliment Pharmacol Ther 2018; 48:961–74. [DOI] [PubMed] [Google Scholar]
- 12.Ciocan D, Rebours V, Voican CS, et al. Characterization of intestinal microbiota in alcoholic patients with and without alcoholic hepatitis or chronic alcoholic pancreatitis. Sci Rep 2018;8:4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martín-Hernández D, Caso JR, Bris ÁG, et al. Bacterial translocation affects intracellular neuroinflammatory pathways in a depression-like model in rats. Neuropharmacology 2016;103:122–33. [DOI] [PubMed] [Google Scholar]
- 14.Maes M, Kubera M, Leunis J-C, et al. Increased IgA and IgM responses against gut commensals in chronic depression: further evidence for increased bacterial translocation or leaky gut. J Affect Disord 2012;141:55–62. [DOI] [PubMed] [Google Scholar]
- 15.Jiang H, Ling Z, Zhang Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun 2015;48:186–94. [DOI] [PubMed] [Google Scholar]
- 16.Kelly JR, Borre Y, O’ Brien C, et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res 2016;82:109–18. [DOI] [PubMed] [Google Scholar]
- 17.Zheng P, Zeng B, Zhou C, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry 2016;21:786–96. [DOI] [PubMed] [Google Scholar]
- 18.Kılıç F, Isık Ü, Demirdas A, et al. Serum zonulin and claudin-5 levels in patients with bipolar disorder. J Affect Disord 2020;266:37–42. [DOI] [PubMed] [Google Scholar]
- 19.Isık Ü, Aydogan Avsar P, Aktepe E, et al. Serum zonulin and claudin-5 levels in children with obsessive-compulsive disorder. Nord J Psychiatry 2020;74:346–51. [DOI] [PubMed] [Google Scholar]
- 20.Aydogan Avsar P, Isık Ü, Aktepe E, et al. Serum zonulin and claudin-5 levels in children with attention-deficit/hyperactivity disorder. Int J Psychiatry Clin Pract 2021;25 49–55. [DOI] [PubMed] [Google Scholar]
- 21.Santos J, Yang PC, Söderholm JD, et al. Role of mast cells in chronic stress induced colonic epithelial barrier dysfunction in the rat. Gut 2001;48:630–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina-Rodriguez EM, Madorma D, O’Connor G, et al. Identification of a signaling mechanism by which the microbiome regulates Th17 cell-mediated depressive-like behaviors in mice. Am J Psychiatry 2020;177:974–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potgieter M, Bester J, Kell DB, et al. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev 2015; 39:567–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perlemuter G. Circulating bugs in blood in alcoholic liver disease! Hepatology 2018;67:1207–9. [DOI] [PubMed] [Google Scholar]
- 25.Lelouvier B, Servant F, Païssé S, et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: a pilot analysis. Hepatology 2016;64:2015–27. [DOI] [PubMed] [Google Scholar]
- 26.Puri P, Liangpunsakul S, Christensen JE, et al. The circulating microbiome signature and inferred functional metagenomics in alcoholic hepatitis. Hepatology 2018;67:1284–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corruble E, El Asmar K, Trabado S, et al. Treating major depressive episodes with antidepressants can induce or worsen metabolic syndrome: results of the METADAP cohort. World Psychiatry 2015; 14:366–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry 1960;23:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guy W. Clinical global impression. ECDEU assessment manual for psychopharmacology, revised. Rockville (MD): National Institute of Mental Health; 1976. [Google Scholar]
- 30.Souberbielle J-C, Massart C, Brailly-Tabard S, et al. Prevalence and determinants of vitamin D deficiency in healthy French adults: the VARIETE study. Endocrine 2016;53:543–50. [DOI] [PubMed] [Google Scholar]
- 31.Païssé S, Valle C, Servant F, et al. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfus 2016;56:1138–47. [DOI] [PubMed] [Google Scholar]
- 32.Schierwagen R, Alvarez-Silva C, Servant F, et al. Trust is good, control is better: technical considerations in blood microbiome analysis. Gut 2019;69:1362–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anhê FF, Jensen BAH, Varin TV, et al. Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat Metab 2020;2:233–42. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, Tang CM, Exley RM. Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology 2015;161:1297–312. [DOI] [PubMed] [Google Scholar]
- 35.Huang R, Wang K, Hu J. Effect of probiotics on depression: a systematic review and meta-analysis of randomized controlled trials. Nutrients 2016;8:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashimoto K, Yoshida T, Ishikawa M, et al. Increased serum levels of serine enantiomers in patients with depression. Acta Neuropsychiatr 2016;28:173–8. [DOI] [PubMed] [Google Scholar]
- 37.Pan J-X, Xia J-J, Deng F-L, et al. Diagnosis of major depressive disorder based on changes in multiple plasma neurotransmitters: a targeted metabolomics study. Transl Psychiatry 2018;8:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke G, Grenham S, Scully P, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry 2013; 18: 666–73. [DOI] [PubMed] [Google Scholar]
- 39.Rothhammer V, Mascanfroni ID, Bunse L, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 2016;22:586–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Castillo DJ, Rifkin RF, Cowan DA, et al. The healthy human blood microbiome: fact or fiction? Front Cell Infect Microbiol 2019; 9:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morton JT, Marotz C, Washburne A, et al. Establishing microbial composition measurement standards with reference frames. Nat Commun 2019;10:2719. [DOI] [PMC free article] [PubMed] [Google Scholar]