Abstract
Background
Epidemiological studies of adult glioma have identified genetic syndromes and 25 heritable risk loci that modify individual risk for glioma, as well increased risk in association with exposure to ionizing radiation and decreased risk in association with allergies. In this analysis, we assess whether there is a shared genome-wide genetic architecture between glioma and atopic/autoimmune diseases.
Methods
Using summary statistics from a glioma genome-wide association studies (GWAS) meta-analysis, we identified significant enrichment for risk variants associated with gene expression changes in immune cell populations. We also estimated genetic correlations between glioma and autoimmune, atopic, and hematologic traits using linkage disequilibrium score regression (LDSC), which leverages genome-wide single-nucleotide polymorphism (SNP) associations and patterns of linkage disequilibrium.
Results
Nominally significant negative correlations were observed for glioblastoma (GB) and primary biliary cirrhosis (rg = −0.26, P = .0228), and for non-GB gliomas and celiac disease (rg = −0.32, P = .0109). Our analyses implicate dendritic cells (GB pHM = 0.0306 and non-GB pHM = 0.0186) in mediating both GB and non-GB genetic predisposition, with GB-specific associations identified in natural killer (NK) cells (pHM = 0.0201) and stem cells (pHM = 0.0265).
Conclusions
This analysis identifies putative new associations between glioma and autoimmune conditions with genomic architecture that is inversely correlated with that of glioma and that T cells, NK cells, and myeloid cells are involved in mediating glioma predisposition. This provides further evidence that increased activation of the acquired immune system may modify individual susceptibility to glioma.
Keywords: allergies, autoimmune disease, genetic architecture, glioma, heritability
Key Points.
There is an anticorrelation between the genetic architecture of glioma and autoimmune conditions.
There is a significant enrichment of the heritability of glioma in variants affecting immune cell-specific gene expression.
This work supports prior studies which have found acquired immune traits modify risk for glioma.
Importance of the Study.
This analysis identifies anticorrelation between the genetic architecture of glioma and autoimmune conditions, as well as significant enrichment of the heritability of these tumors in variants affecting immune cell-specific gene expression. The findings of this research support prior studies which find acquired immune traits modify risk for glioma, and may identify specific immune cells and pathways that are implicated in this association.
Glioma is the most commonly occurring primary malignant brain tumor in the United States, with an average of ~10 000 cases diagnosed per year.1 Though uncommon, these tumors cause significant morbidity and mortality. While ionizing radiation exposure has been shown to increase risk of glioma, most cases cannot be attributed to an underlying exposure and the vast majority of cases occur in people without significant family history of cancer.2,3 Allergies and other atopic conditions have been consistently shown to decrease risk of glioma in case-control studies, though Mendelian randomization studies assessing the causality of this association have not been significant.4–6 Reported history of any autoimmune condition has also been shown to decrease risk of these tumors.7,8 This suggests that increased immune activation protects against glioma development. Due to the rarity of many autoimmune traits, they have often been pooled in glioma association studies, making it difficult to determine the nature of the association with specific autoimmune conditions.
Complete blood count (CBC) values (eg, red and white blood cell counts and proportions, platelet count, and hemoglobin), cytokines, and other blood and immune cell factors are frequently dysregulated in cancer (including glioma) due both to tumor and treatment effects.9 Prior genome-wide association studies (GWAS) have identified multiple loci that modify these hematologic traits and indicate these loci have pleiotropic effects on immune phenotypes.10–12 While leukocyte traits associated with increased systemic inflammation have been shown to be prognostic in glioma and multiple CBC values have been previously associated with cancer prognosis, including in glioblastoma (GB), there has been no evaluation of how heritable variation in these traits contributes to glioma etiology.13,14 The majority of glioma case-control studies have collected blood samples after diagnosis, where blood composition may already be dysregulated due to systemic immunosuppression caused by the tumor or by steroidal treatments.
A prior GWAS meta-analysis in glioma identified 25 independent loci associated with glioma in European ancestry populations, most of which have relatively small effect sizes15 and none of which have been previously identified as being associated with autoimmune disease. These associations are estimated to account for 30% of the glioma heritability, leaving 70% of the variation in genetic risk in glioma still unaccounted for.16,17 A recent GWAS completed in IDH1/2-mutant adult diffuse glioma identified in D2HGDH, a region known to be associated with allergy.18 The power of GWAS methods to detect additional common variants of small effect sizes and resolve this “missing heritability” is limited by sample size and sample resources, particularly in rare diseases such as glioma. However, many risk loci identified by GWAS are pleiotropic and are implicated in risk for multiple complex diseases.19–21
Linkage disequilibrium score regression (LDSC) is a statistical methodology that uses genome-wide single-nucleotide polymorphism (SNP) association data and patterns of LD to estimate heritability and correlations between traits, while minimizing the effect of confounding and population stratification.22 It can also be extended to estimate the polygenic contribution of specific gene annotations (eg, cell-type-specific transcriptomic networks) to heritability in GWAS. In this analysis, we used LDSC to partition glioma heritability by cell type and to estimate the genetic correlation between glioma and atopic, autoimmune, and hematologic traits in a set of 12 455 European ancestry cases and 18 169 European ancestry controls from eight case-control studies.
Methods
Data Sources and Quality Control (QC)
Glioma data were generated from a meta-analysis conducted using eight prior glioma GWAS studies, for all glioma, GB, and non-GB glioma (see Supplementary Table 1 for an overview of study characteristics) and betas, standard errors, and P values were used to generate z scores for use in LDSC. These studies were previously combined as part of the meta-analysis presented in Melin et al.15 and do not include overlapping samples. We obtained summary statistics (including betas, standard errors, and P values) for 13 immune- and atopy-related traits from the prior case-control studies and the UK Biobank and CBC values from the UK Biobank (Supplementary Table 2). QC of phenotypes and genotypes is documented in prior publications and documentation.23–30 We filtered the data to include autosomal SNPs with minor allele frequency (MAF) of at least 1%. The SNPs for glioma and UK Biobank datasets were imputed and filtered to have imputation quality >0.9. Since glioma is heterogeneous with distinct pathways of gliomagenesis and genetic susceptibility, we performed all analyses in all glioma, as well as for GB and non-GB glioma separately.
Estimation of Heritability and Genetic Correlation Between Traits
Pairwise genetic correlation between these traits was generated using LDSC (https://github.com/bulik/ldsc) using 1000 Genomes Project European (EUR) samples as a reference for patterns of genome-wide LD.22 Briefly, LDSC is a method that regresses summary statistics from GWAS on the LD score, or the strength with which each individual variant tags other variants in the genome. This method allows for calculation of genetic correlation without bias due to population stratification or cryptic relatedness. We used LDSC v1.0.1 to estimate genetic correlation between traits (rg), the 95% confidence intervals (95% CI) and P values for each pairing, and heritability for each trait. LDSC intercept, estimated lambda (λ, an estimate of genetic inflation), maximum χ 2 statistic, and intercept of genetic covariance. The ratio of regression intercept to χ2 statistic, which estimates the proportion of inflation in χ 2 statistic that is not due to polygenic heritability. In the absence of inflation, this ratio should be equal to zero. Due to the complex and unusual genetic architecture of the human leukocyte antigen (HLA) complex region (chromosome 6 from 29 Mb to 33 Mb), the LDSC method excludes SNPs within this region from all analyses. SNPs were limited to those included in HapMap. After QC, 1 290 028 SNPs were used from glioma datasets. Total number of SNPs used in each pairwise analysis varied due to variation in array used by study and study-specific imputation quality scores. P values were corrected using the Benjamini-Hochberg procedure, and associations were considered significant at P < .05. Glioma heritability estimates we converted to the liability scale using an overall lifetime risk of malignant brain tumor of 0.58%, with lifetime risk of GB estimated to be 0.32% and non-GB estimated to be 0.26%.31 Autoimmune disease heritability estimated from case-control studies was also adjusted to liability scale using prior published prevalence estimates (Supplementary Table 2). Heritability for binary traits from the United Kingdom was adjusted to the liability scale using sample prevalence, which approximates the prevalence of these conditions in the population.
Partitioned heritability is an extension of the LDSC method described by Finucane et al. that uses genotyping and gene expression reference datasets to identify cell types which are significant enriched for variants contributing to the heritability of a trait.32,33 Cell-type-specific heritability was estimated for glioma and glioma subtypes using LDSC v1.0.1 using provided reference data from GTex and ImmGen,34,35 and further details of this data preparation are provided in prior publications.32 All available tissue types and immune cell types were used. Briefly, these reference datasets (previously prepared by Finucane and colleagues, available at http://data.broadinstitute.org/alkesgroup/LDSCORE/LDSC_SEG_ldscores/) were generated using a matrix of normalized gene expression values across genes, for which a t statistic was calculated for specific expression in the specific tissue or cell type. Briefly, Finucane et al. then ranked genes by these t statistics and the top 10% of genes with the highest t statistic to be the gene set for each tissue or cell type, which includes genes that are specifically expressed and genes that are weakly specifically expressed. LDSC is then used to evaluate contribution of the genome annotation to trait heritability. Specific tissue and immune cell samples were grouped into broader categories.32,33 Proportion of heritability due to tissue or cell type was defined as the proportion of SNP-heritability in the category divided by the proportion of total SNPs. P values within immune tissue types were combined using the harmonic mean P value (pHM)36 using R package harmonicmeanp.37 Associations were considered statistically significant at P < .05.
Results
Estimation of Glioma Heritability
We estimated that the overall liability-scaled heritability for glioma was 6.69% (95% CI: 6.67%-6.71%), while estimates for GB and non-GB were 5.62% (95% CI: 5.58%-5.67%) and 6.45% (95% CI: 6.40%-6.50%), respectively.
Cell-Type-Partitioned Heritability
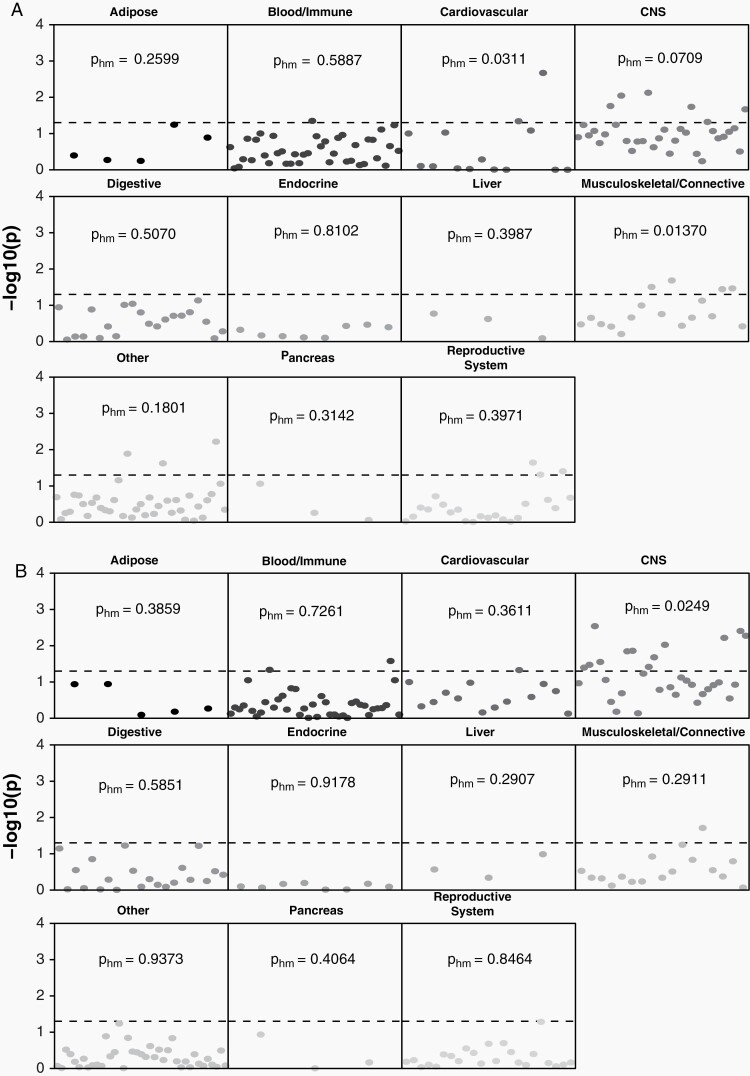
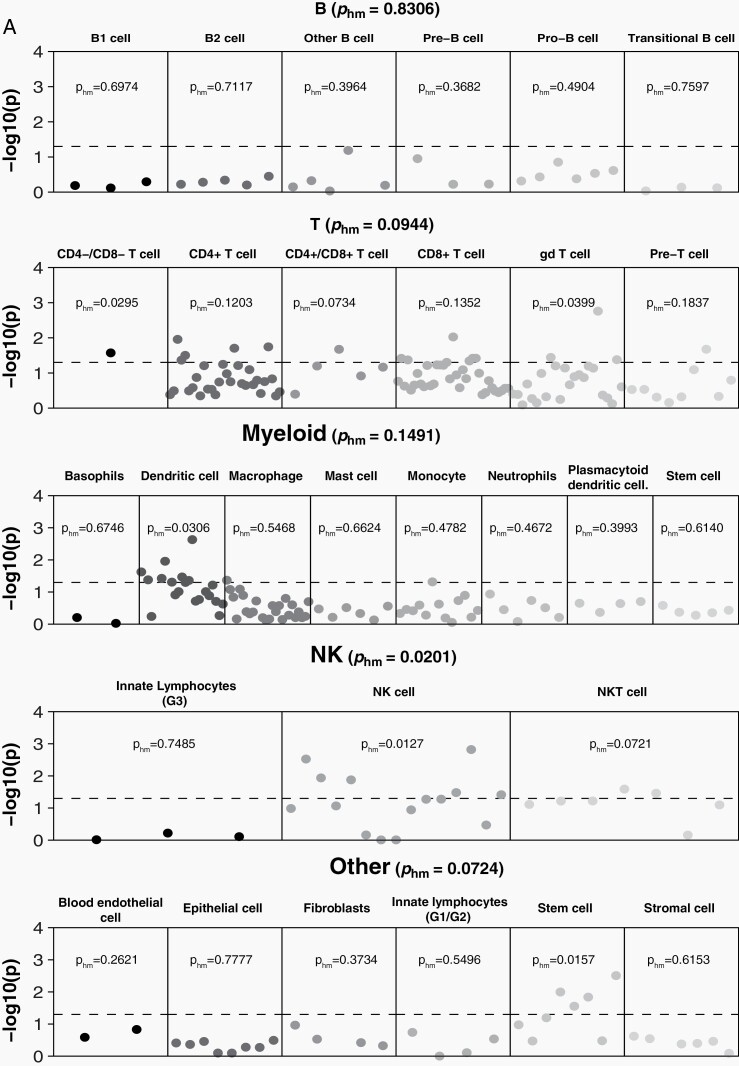
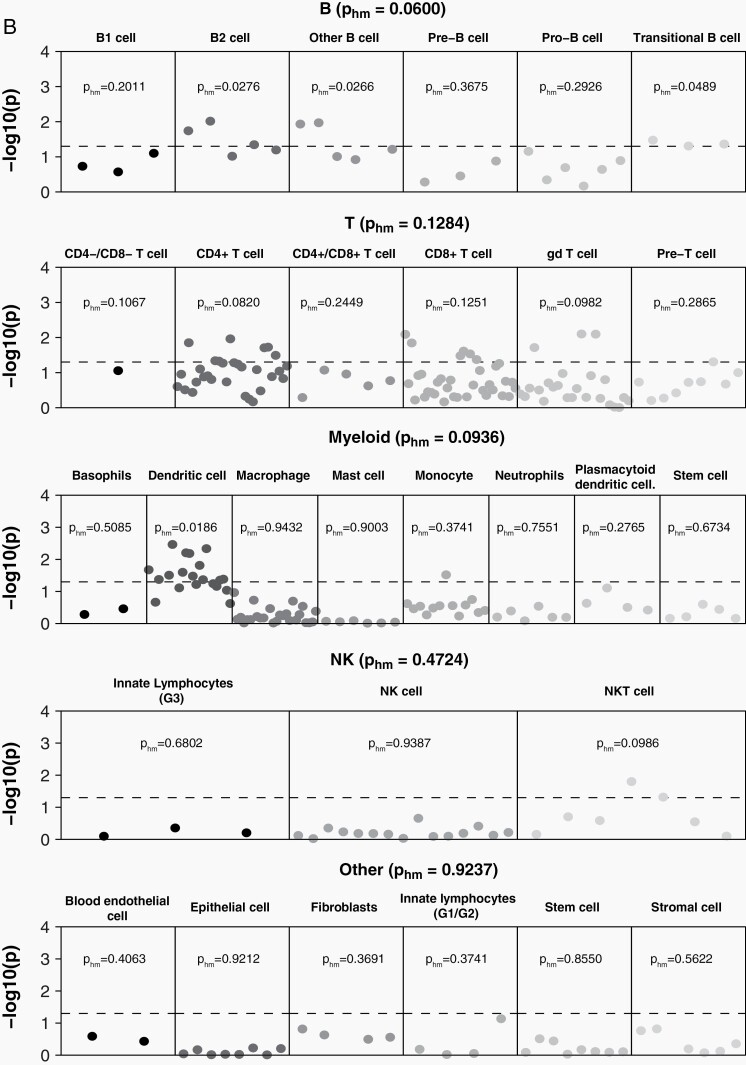
In order to identify tissue and cell types where function was likely to be affected by variants associated with risk for glioma, heritability partitioned by tissue and cell type was estimated using GTex and ImmGen reference data (Supplementary Tables 3–5). Across all tissue types, heritability for glioma was significantly enriched in CNS (pHM = 0.0221), (Figure 1, Supplementary Table 3). Within GTex brain regions only, there was significant enrichment for GB in cortex (pHM = 0.0061), Supplementary Table 4) that was not observed in non-GB (pHM = 0.2498). There was heritability enrichment in multiple immune cell types overall and for both GB and non-GB (Figure 2, Supplementary Table 5). There was a marginally significant association between non-GB only and B cells (pHM = 0.0600), with the strongest association in B2 cells (pHM = 0.0276) and transitional B cells (pHM = 0.0489). There was no significant enrichment with myeloid cells in GB (pHM = 0.1491) or non-GB (pHM = 0.0936), though there was specific enrichment in dendritic cells in both GB (pHM = 0.0306) and non-GB (pHM = 0.0186). Natural killer (NK) cells were significantly associated with GB (pHM = 0.0201). There was a marginally significant association with T cells for GB (pHM = 0.0944), with enrichment of association in γδ T cells in GB (pHM = 0.0399). Among cell types included in a broad “other” category, we also identified stem cell-specific enrichment in GB (pHM = 0.0265). See Supplementary Table 5 for all cell-specific coefficients and P values.
Fig. 1.
−log10(P) for individual-specific tissue sample and harmonic mean P within tissue categories for (A) GB and (B) non-GB based on GTex reference data (dotted line indicates P < .05). Abbreviation: GB: glioblastoma.
Fig. 2.
log10(P) by individual cell line sample and harmonic mean P within immune cell category for (A) GB and (B) non-GB based on ImmGen reference data (dotted line indicates P < .05). Abbreviation: GB: glioblastoma.
Genome-Wide Genetic Correlations With Autoimmune, Hematologic and Atopic Traits
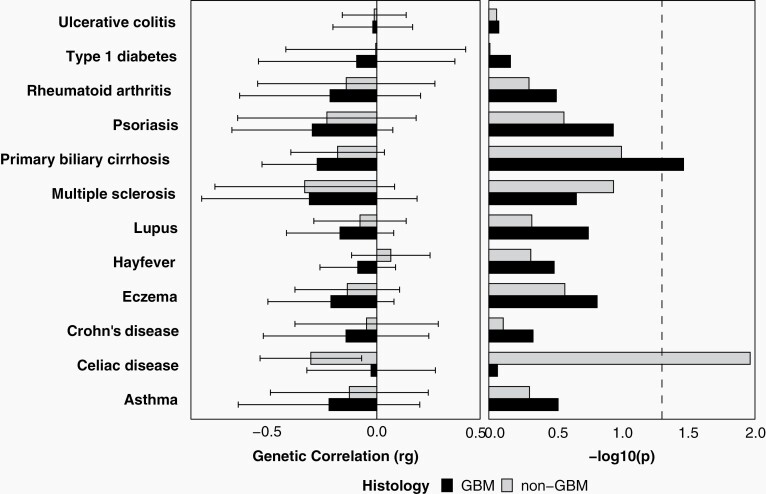
We evaluated genome-wide correlations between the genetic architecture of glioma and multiple atopic, allergic, and autoimmune conditions, as well as hematologic traits. In general, λ estimates were higher than intercepts, suggesting that signal was driven by polygenicity rather than population stratification. Ratios were elevated for many traits obtained from case-control studies, including celiac disease, lupus, multiple sclerosis, and ulcerative colitis, suggesting that population stratification may have a significant impact on observed results. After correcting for multiple testing, no associations were significant. A nominally significant negative genetic correlation was identified between glioma and primary biliary cirrhosis (PBC, rg = −0.26, P = .0228) (Figure 3, Supplementary Table 6). When correlations were estimated by broad glioma subtypes, the scale and direction of associations were generally consistent in both GB and non-GB, though most associations were not statistically significant. There was a negative correlation with celiac disease in non-GB only (rg = −0.32, P = .0109). No associations remained significant after false discovery rate (FDR) adjustment of P values.
Fig. 3.
Estimated heritability for autoimmune and atopic phenotypes, and genetic correlation between atopic and autoimmune phenotypes and glioma subgroups, dotted line indicates P < .05.
Due to the high variability in the HLA region, LDSC excludes this by default. There is a significant enrichment for germline variation associated with the autoimmune disease within this region. As a sensitivity analysis, genetic correlation was also estimated for these traits and glioma with a reference including the HLA region. Several phenotypes (particularly lupus and type 1 diabetes) showed significant increases in their SNP-based heritability estimates (Supplementary Table 2). The direction and scale of genetic correlation did not change substantially for traits other than lupus and type 1 diabetes (Supplementary Table 7).
There was a positive correlation between glioma and mean platelet volume (rg = −0.09, P = .0337) (Supplementary Table 4), mostly driven by the effect of non-GB (rg = −0.12, P = .0096). There was also a negative correlation observed between non-GB and lymphocyte percentage (rg = −0.11, P = .0312). We did not find any significant associations between GB and any hematologic traits. Although epidemiologic studies have repeatedly observed inverse associations between glioma and atopic conditions, glioma was not correlated with either eosinophil count (rg = 0.01, P = .9060) or eosinophil percentage (rg = 0.02, P = .6137), nor was it correlated with atopic traits including hay fever, asthma, and eczema (Supplementary Table 6). A weak inverse correlation was observed between GB and eczema in the anticipated direction (rg = −0.25, P = .0991). No associations remained significant after FDR adjustment of P values. In general, λ estimates were higher than intercepts, suggesting that signal was due to polygenicity as compared to solely population stratification. The intercept for LDSC has been observed to increase linearly with sample size,38 which may lead to artificial inflation of these statistics in these continuous traits with very large sample sizes.
Discussion
LDSC is a hypothesis-generating approach that we used to identify (1) cell types that mediate glioma predisposition and (2) traits with correlated or anticorrelated genomic architecture. This methodology enables researchers to identify traits that are correlated with their phenotype of interest (eg, glioma) which are otherwise not amenable to study in case-control designs due to the rarity of such traits (eg, the inverse association of PBC with glioma). When based on validly constructed GWAS estimates, LDSC can also minimize many sources of bias from case-control studies related to differential participation rates and recall biases.39 While this analysis did not identify shared genomic architecture underlying allergic/atopic traits and glioma risk, it did identify associations with specific autoimmune phenotypes that varied by GB status.
We identified significant heritability enrichment for glioma in multiple brain regions as well as immune cell types. This supports previous evidence that conditions affecting individual immune function may be associated with risk of glioma.7,8,40 Gliomagenesis also involves significant manipulation of the immune system, and is associated with local and global immunosuppression that “sequesters” T cells to the bone marrow and “reprograms” other immune cells within the tumor microenvironment.41 Increased prevalence of markers associated with dysregulated immune function are strongly associated with increased tumor growth and decreased glioma survival, suggesting that innate immune function influences risk and prognosis and leading to a current focus on immune-stimulating treatments for this disease.42–46 Together, these factors demonstrate the significant role of immune function in both glioma susceptibility and patient prognosis.
This analysis identified significant negative correlations between glioma and several autoimmune traits, with the strongest associations identified with PBC and celiac disease. The estimate of rg for multiple sclerosis was similar in scale to both PBC and celiac disease in GB and non-GB, respectively, but this association was not statistically significant. Most of these traits have not been investigated in association with glioma previously. While prior research has suggested a protective association between long-term diagnosis of (primary type 2 or adult-onset) diabetes and glioma,47 we did not find a correlation between the genetic architecture of type 1 diabetes and glioma. There is an increasing body of scientific literature documenting the biological connections between the gut and the brain. The permeable tissue of the digestive system is the most direct way for compounds to enter the bloodstream, and if small enough, these compounds can cross the blood-brain barrier. The gut microbiome has been shown to significantly affect neuropsychiatric disorders, as well as response to cancer immunotherapies.48,49 Celiac disease and PBC are organ-specific autoimmune diseases, in contrast to systematic autoimmune conditions such as rheumatoid arthritis and lupus. The autoimmune component of celiac disease arises when gliadin (a component of gluten) peptides cross the intestinal epithelial barrier and activate CD4+ T-lymphocytes, stimulating the expansion of B cells that secrete anti-gliadin and auto-antibodies.50 Gliadin also activates antigen-presenting cells (APCs or dendritic cells) and intestinal epithelial cells, leading to the expansion of CD8+ T-lymphocytes. Immune cell-type-specific heritability analyses found a significant enrichment for GB and non-GB heritability in dendritic cells. PBC is also a T-cell-mediated autoimmune disease that primarily affects biliary epithelial cells, and biopsy specimens from patients with this condition show significant invasion of CD4+ and CD8+ T cells.51 While it is not clear what role NK cells play in the pathogenesis of PBC, these cells have been found in increased quantity in blood and liver specimens of PBC patients. Our analysis found significant GB-specific enrichment for heritability in NK cells. NK cells are an essential component of tumor surveillance in the healthy innate immune system, as they do not need to be activated and will respond to cells based on the absence of appropriate MHC (major histocompatibility complex) receptors. While the primary damaging immune response in autoimmune diseases is usually B- and/or T-cell-mediated, both quantitative and qualitative variation in NK cells has been observed in multiple autoimmune conditions.52 NK cells are currently under investigation for potential role in cancer immunotherapy, as their ability to perform their function in the absence of a “danger” signal makes them less susceptible to the immunosuppressive tumor environment present in GB.53 GB-specific enrichment was also identified in stem cells (pHM = 0.0157). Stem-like behavior is observed in the subpopulation of glioma cells referred to as glioma stem cells, which exert significant control over the glioma immune microenvironment.54
LDSC identified few genetic correlations with the hematologic factors, with the exception of mean platelet volume and lymphocyte percentage. These values all have the potential to be markers of underlying immune function, and further research is necessary to determine their association with glioma risk. The lack of consistent association across related phenotypes and lack of statistical significance after FDR correction, also suggests that these may not be genuine associations. Estimates of heritability for glioma were notably lower than previously estimated using GCTA (genome-wide complex trait analysis).55 Heritability estimates for autoimmune diseases and allergy/atopy traits estimated by this analysis are generally also lower than what has previously been reported.56,57 Heritability estimates in LDSC are known to be biased downwards when data have been previously corrected for genomic control, as the glioma data used in this analysis have been based on estimated principal components. Heritability estimates may also be biased when cases and controls are not from the same source population, as is the case in several included glioma GWAS (Supplementary Table 1). Despite adjustment to the liability scale, heritability estimates from case-control studies may be inflated as compared to cohort studies such as the UK Biobank. As a result, the reported heritability estimates may be lower than “true” heritability for these traits. Using genome-wide complex trait analysis (GCTA) to estimate genome-wide heritability has been shown to result in inflated estimates, particularly when there are measurement errors in the underlying phenotype,58 which may explain our lower heritability estimates using LDSC when compared to previous estimates using GCTA. Molecular characterization of glioma has demonstrated that the presence or absence of IDH1/2 mutation differentiates two distinct lineages of glioma, which were pooled in glioma GWAS data.59 While the majority of GB (~95%) are IDH1/2 wild type and non-GB are IDH1/2 mutant (80%),59 these groups likely contain nontrivial phenotypic misclassification. Relatively small sample sizes within some phenotypes (particularly for cases with autoimmune traits in the UK Biobank) may also lead to some tests being underpowered. LDSC recommends a minimum sample size of 3000 for estimates of heritability, and 5000 cases for partitioning heritability. Glioma data meet this threshold, but do not substantially exceed it.
This analysis uses LDSC to identify significant enrichment for glioma heritability in immune cell types and novel inverse associations between several autoimmune traits and glioma risk. A strength of this methodology is that it does not require having collected prior information on medical history. This method has been shown to be robust under multiple different genetic architecture, as well as when the underlying summary statistics used may be biased.60 This method requires that a trait have a sufficient portion of risk that is attributable to genetic factors, and is not appropriate for traits that are entirely affected by environment. Prior glioma case-control studies are inconsistent in how these data were collected and which diseases were included on questionnaires, and this approach allows for use of all glioma GWAS regardless of questionnaire data. Both glioma and specific autoimmune diseases are uncommon, and as a result, very large sample sizes would be necessary to have adequate power to identify these associations with traditional case-control study designs. There are also several limitations to this approach. Summary statistics are used and therefore it is not able to conduct sensitivity analyses using specific groups of controls within each study, and the sharing of these controls between glioma subsets may bias results. The LDSC method excludes the HLA region, which is known to be enriched for risk variants in atopic and autoimmune disease as well as a punitive haplotype association with non-GB glioma,61 and may be a significant source of shared heritability between these traits, though our sensitivity analysis including the HLA region shows that the significance of inclusion varies by trait.
While this analysis does not identify a genetic basis for the previously identified protective effect of atopic traits on glioma risk, it does suggest that the genomic architecture of glioma predisposition may manifest in part via its activity in immune cells. This is consistent with prior research demonstrating the protective effect of allergy and atopic disease on glioma risk, as well as the systemic immune suppression that occurs in the context of glioma. Atopic traits had low heritability, suggesting that the protective effect due to these traits may be influenced mostly by environmental factors. Autoimmune conditions, on the other hand, did show association with glioma and are more strongly influenced by intrinsic (eg, genetic) factors than extrinsic (eg, environmental) exposures, much like glioma. Further studies are necessary in order to confirm these associations and to identify the mechanism through which increased immune activation may reduce the risk of glioma.
Supplementary Material
Acknowledgments
*The GLIOGENE Consortium is: Christopher I. Amos (Baylor College of Medicine, Houston, Texas, USA), Jill S. Barnholtz-Sloan (Case Western Reserve University School of Medicine, Cleveland, Ohio, USA), Jonine L. Bernstein (Memorial Sloan Kettering Cancer Center, New York, New York, USA), Melissa L. Bondy (Stanford University School of Medicine, Stanford, California, USA), Elizabeth B. Claus (Yale University, New Haven, Connecticut, USA), Richard S. Houlston (Institute of Cancer Research, London, UK), Dora Il’yasova (Georgia State University, Atlanta, Georgia, USA), Robert B. Jenkins (Mayo Clinic, Rochester, Minnesota, USA), Christoffer Johansen (University of Copenhagen, Denmark), Daniel Lachance (Mayo Clinic, Rochester, Minnesota, USA), Rose Lai (University of Southern California, Los Angeles, California, USA), Beatrice S. Melin (Umeå University, Umeå, Sweden), Ryan T. Merrell (NorthShore University HealthSystem, Chicago, Illinois, USA), Sara H. Olson (Memorial Sloan Kettering Cancer Center, New York, New York, USA), Siegal Sadetzki (Tel-Aviv University, Tel-Aviv, Israel), Joellen Schildkraut (Emory University, Atlanta, Georgia, USA), Sanjay Shete (MD Anderson Cancer Center, Houston, Texas, USA).
Contributor Information
GLIOGENE Consortium:
Christopher I Amos, Jill S Barnholtz-Sloan, Jonine L Bernstein, Melissa L Bondy, Elizabeth B Claus, Richard S Houlston, Dora Il’yasova, Robert B Jenkins, Christoffer Johansen, Daniel Lachance, Rose Lai, Beatrice S Melin, Ryan T Merrell, Sara H Olson, Siegal Sadetzki, Joellen Schildkraut, and Sanjay Shete
Conflict of interest statement. There are no conflicts of interest to report.
Funding
Q.T.O. is supported by a Research Training Grant from the Cancer Prevention and Research Institute of Texas (CPRIT; RP160097T). K.M.W. is supported by a Distinguished Scientist Award from the Sontag Foundation. C.I.A. is a Cancer Prevention Research Institute of Texas Research Scholar. Partial support for this work was provided by CPRIT RR170048. This work was supported by grants from the NIH, Bethesda, MD: R01CA139020 (to M.L.B.). Additional support was provided by the McNair Medical Institute and the Population Sciences Biorepository at Baylor College of Medicine (P30CA125123).
Author contributions. Q.T.O., J.E., J.B., Y.H., K.M.W., C.I.A., and M.L.B. designed the analysis. GLIOGENE Consortium provided the data. Q.T.O. and J.E. analyzed the data. Q.T.O. wrote the paper. Q.T.O., J.E., J.B., Y.H., B.M., R.S.H., M.M., K.M.W., C.I.A., and M.L.B. provided feedback and approved the final version of the paper.
References
- 1. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20(suppl_4):iv1–iv86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wrensch M, Lee M, Miike R, et al. Familial and personal medical history of cancer and nervous system conditions among adults with glioma and controls. Am J Epidemiol. 1997;145(7):581–593. [DOI] [PubMed] [Google Scholar]
- 4. Amirian ES, Zhou R, Wrensch MR, et al. Approaching a Scientific Consensus on the Association between Allergies and Glioma Risk: a report from the Glioma International Case-Control Study. Cancer Epidemiol Biomarkers Prev. 2016;25(2):282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao H, Cai W, Su S, Zhi D, Lu J, Liu S. Allergic conditions reduce the risk of glioma: a meta-analysis based on 128,936 subjects. Tumour Biol. 2014;35(4):3875–3880. [DOI] [PubMed] [Google Scholar]
- 6. Disney-Hogg L, Cornish AJ, Sud A, et al. Impact of atopy on risk of glioma: a Mendelian randomisation study. BMC Med. 2018;16(1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Safaeian M, Rajaraman P, Hartge P, et al. Joint effects between five identified risk variants, allergy, and autoimmune conditions on glioma risk. Cancer Causes Control. 2013;24(10):1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brenner AV, Linet MS, Fine HA, et al. History of allergies and autoimmune diseases and risk of brain tumors in adults. Int J Cancer. 2002;99(2):252–259. [DOI] [PubMed] [Google Scholar]
- 9. Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50(7):906–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahola-Olli AV, Würtz P, Havulinna AS, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet. 2017;100(1):40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roederer M, Quaye L, Mangino M, et al. The genetic architecture of the human immune system: a bioresource for autoimmunity and disease pathogenesis. Cell. 2015;161(2):387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang RF, Li M, Yang Y, Mao Q, Liu YH. Significance of pretreatment red blood cell distribution width in patients with newly diagnosed glioblastoma. Med Sci Monit. 2017;23:3217–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wiencke JK, Koestler DC, Salas LA, et al. Immunomethylomic approach to explore the blood neutrophil lymphocyte ratio (NLR) in glioma survival. Clin Epigenetics. 2017;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Melin BS, Barnholtz-Sloan JS, Wrensch MR, et al. ; GliomaScan Consortium . Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kinnersley B, Mitchell JS, Gousias K, et al. Quantifying the heritability of glioma using genome-wide complex trait analysis. Sci Rep. 2015;5:17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eckel-Passow JE, Drucker KL, Kollmeyer TM, et al. Adult diffuse glioma GWAS by molecular subtype identifies variants in D2HGDH and FAM20C. Neuro Oncol. 2020;22(11):1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu YH, Graff RE, Passarelli MN, et al. Identification of pleiotropic cancer susceptibility variants from genome-wide association studies reveals functional characteristics. Cancer Epidemiol Biomarkers Prev. 2018;27(1):75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bien SA, Peters U. Moving from one to many: insights from the growing list of pleiotropic cancer risk genes. Br J Cancer. 2019;120(12):1087–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chesmore K, Bartlett J, Williams SM. The ubiquity of pleiotropy in human disease. Hum Genet. 2018;137(1):39–44. [DOI] [PubMed] [Google Scholar]
- 22. Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cordell HJ, Han Y, Mells GF, et al. ; Canadian-US PBC Consortium; Italian PBC Genetics Study Group; UK-PBC Consortium . International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun. 2015;6:8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jostins L, Ripke S, Weersma RK, et al. ; International IBD Genetics Consortium (IIBDGC) . Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium, Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bentham J, Morris DL, Graham DSC, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47(12):1457–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dubois PC, Trynka G, Franke L, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42(4):295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abbott L, Anttila V, Aragam K, et al. Neale Lab - UK Biobank GWAS. 2018. http://www.nealelab.is/uk-biobank/. Accessed September 22, 2020.
- 29. Details and considerations of the UK Biobank GWAS. 2017. http://www.nealelab.is/blog/2017/9/11/details-and-considerations-of-the-uk-biobank-gwas. Accessed September 22, 2020.
- 30. Selecting primary UKB round 2 phenotypes for LDSR analysis. 2019. https://nealelab.github.io/UKBB_ldsc/select_topline.html. Accessed September 23, 2020.
- 31. Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88(3):294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Finucane HK, Bulik-Sullivan B, Gusev A, et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium . Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finucane HK, Reshef YA, Anttila V, et al. ; Brainstorm Consortium . Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50(4):621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heng TSP, Painter MW, Immunological Genome Project Consortium . The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. [DOI] [PubMed] [Google Scholar]
- 35. Gamazon ER, Segrè AV, van de Bunt M, et al. ; GTEx Consortium . Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet. 2018;50(7):956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wilson DJ. The harmonic mean p-value for combining dependent tests. Proc Natl Acad Sci USA. 2019;116(4):1195–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson DJ. harmonicmeanp: harmonic mean p-values and model averaging by mean maximum likelihood. R package version 3.0.2019. https://CRAN.R-project.org/package=harmonicmeanp. Accessed September 14, 2020.
- 38. Loh PR, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50(7):906–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johansen C, Schüz J, Andreasen AS, Dalton SO. Study designs may influence results: the problems with questionnaire-based case-control studies on the epidemiology of glioma. Br J Cancer. 2017;116(7):841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schwartzbaum J, Jonsson F, Ahlbom A, et al. Cohort studies of association between self-reported allergic conditions, immune-related diagnoses and glioma and meningioma risk. Int J Cancer. 2003;106(3):423–428. [DOI] [PubMed] [Google Scholar]
- 41. Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B. Immune microenvironment of gliomas. Lab Invest. 2017;97(5):498–518. [DOI] [PubMed] [Google Scholar]
- 42. Chongsathidkiet P, Jackson C, Koyama S, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med. 2018;24(9):1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15(7):422–442. [DOI] [PubMed] [Google Scholar]
- 44. Alban TJ, Alvarado AG, Sorensen MD, et al. Global immune fingerprinting in glioblastoma patient peripheral blood reveals immune-suppression signatures associated with prognosis. JCI Insight. 2018;3(21):e122264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li X, Wang B, Gu L, et al. Tim-3 expression predicts the abnormal innate immune status and poor prognosis of glioma patients. Clin Chim Acta. 2018;476:178–184. [DOI] [PubMed] [Google Scholar]
- 46. Feng E, Liang T, Wang X, et al. Correlation of alteration of HLA-F expression and clinical characterization in 593 brain glioma samples. J Neuroinflammation. 2019;16(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Seliger C, Ricci C, Meier CR, et al. Diabetes, use of antidiabetic drugs, and the risk of glioma. Neuro Oncol. 2016;18(3):340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gracie DJ, Guthrie EA, Hamlin PJ, Ford AC. Bi-directionality of brain-gut interactions in patients with inflammatory bowel disease. Gastroenterology. 2018;154(6):1635–1646.e3. [DOI] [PubMed] [Google Scholar]
- 49. Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. [DOI] [PubMed] [Google Scholar]
- 50. Parzanese I, Qehajaj D, Patrinicola F, et al. Celiac disease: from pathophysiology to treatment. World J Gastrointest Pathophysiol. 2017;8(2):27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lleo A, Invernizzi P, Mackay IR, Prince H, Zhong RQ, Gershwin ME. Etiopathogenesis of primary biliary cirrhosis. World J Gastroenterol. 2008;14(21):3328–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schleinitz N, Vély F, Harlé JR, Vivier E. Natural killer cells in human autoimmune diseases. Immunology. 2010;131(4):451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Golán I, Rodríguez de la Fuente L, Costoya JA. NK cell-based glioblastoma immunotherapy. Cancers. 2018;10(12):522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ma Q, Long W, Xing C, et al. Cancer stem cells and immunosuppressive microenvironment in Glioma. Front Immunol. 2018;9:2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kinnersley B, Mitchell JS, Gousias K, et al. Quantifying the heritability of glioma using genome-wide complex trait analysis. Sci Rep. 2015;5:17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Selmi C, Lu Q, Humble MC. Heritability versus the role of the environment in autoimmunity. J Autoimmun. 2012;39(4):249–252. [DOI] [PubMed] [Google Scholar]
- 57. Ferreira MAR, Mathur R, Vonk JM, et al. ; 23andMe Research Team; eQTLGen Consortium; BIOS Consortium . Genetic architectures of childhood- and adult-onset asthma are partly distinct. Am J Hum Genet. 2019;104(4):665–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krishna Kumar S, Feldman MW, Rehkopf DH, Tuljapurkar S. Limitations of GCTA as a solution to the missing heritability problem. Proc Natl Acad Sci USA. 2016;113(1):E61–E70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. The Cancer Genome Atlas Research Network, Brat DJ, Verhaak RGW, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee JJ, McGue M, Iacono WG, Chow CC. The accuracy of LD Score regression as an estimator of confounding and genetic correlations in genome-wide association studies. Genet Epidemiol. 2018;42(8):783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang C, de Smith AJ, Smirnov IV, et al. Non-additive and epistatic effects of HLA polymorphisms contributing to risk of adult glioma. J Neurooncol. 2017;135(2):237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.