Abstract
In this study, 22 Lactobacillus plantarum strains isolated from Siahmazgi traditional cheese were evaluated using different tests including resistance to low pH (1.50 and 2.50) and bile salt (0.50 and 1.00%), growth kinetic at low pH values and survival under simulated gastric and intestinal fluids. All the strains retained their viability at pH 2.50. However, the survival of all of the isolates was decreased at pH 1.50. Ten out of 22 strains which were able to tolerate low pH were selected for further investigations. All the selected isolates were able to grow at low pH. Strain F2 showed the highest specific growth rate. Five out of 10 isolates showed a significant decrease in bacterial count varied from 2.00 to 7.00 log CFU mL-1 during 3 hr exposure to 0.50% bile salt, while five isolates represented resistance to 0.50% bile during 3 hr. A significant reduction was observed in survival of all of the isolate at 1.00% bile salt concentration. Furthermore, viability of the selected isolates was lowered during 1 hr incubation under gastric conditions, while it remained unchanged within next 2 hr. Although, no significant changes were seen in bacterial count of the selected isolates during 1 hr of exposure to simulated intestinal condition, the survival of the isolates was relatively reduced after 3 hr. In conclusion, five out of 22 examined L. plantarum isolates showed appropriate resistance properties, therefore, could be good candidates for further examinations including functional and safety evaluation supporting their use as probiotics.
Key Words: Bile salt, Gastrointestinal condition, Lactobacillus plantarum, Probiotic, Siahmazgi cheese
Introduction
Probiotic food products are considered as functional foods which can improve consumers’ health. According to the World Health Organization (WHO) and the Food and Agriculture Organization (FAO), probiotics are live micro-organisms which when administered in adequate amounts, confer health benefits to their host.1 Several lactic acid bacteria (LAB) strains are known to be beneficial for their host and have been chosen for use as probiotics. They are human gastrointestinal (GI) micro-flora and are generally recognized safe (GRAS).2 Commonly used probiotic bacteria include several species of Lactobacillus, Bifidobacterium, Streptococcus, Lactococcus and some species of Enterococcus.3 Among them, Lactobacilli are one of the main genus of LAB known for probiotic characteristics. They are found in a wide range of food products. Lactobacillus plantarum is a common inhabitant of the human GI tract and some strains are used as ingredients of probiotic foods.1 In order to meet the growing demand of the market, especially in developed countries to produce the functional food products with active probiotic cultures, assessment of new probiotic strains is of great importance. To obtain an ideal probiotic culture, it is necessary to screen and characterize numerous strains since all lactic acid bacteria do not possess the health benefits for the host.4 In this way, there is a great interest in indigenous and traditional fermented foods from non-industrialized countries, especially dairy products, as a reservoir to search for new Lactobacillus strains with novel functional properties.5 Since dairy products are the most common carrier foods for delivery of probiotic bacteria, lactobacillus strains originally isolated from dairy products are probably the most suitable candidates for inclusion into several types of food as probiotics because they are well adapted to the conditions, therefore, they may be more competitive than probiotic strains from other sources.4 For the beneficial impact on the health of the host, probiotic cultures must be ingested in sufficient amount (106 to 107 CFU g-1). For being probiotic, the microorganism should overcome the physical and chemical barriers in the gastrointestinal tract, especially the acid and bile stress to reach in sufficient amount to the gut to confer their health benefits to the host.6 The low pH of stomach is the first defense barrier which helps the host to resist against pathogenic micro-organisms.7 The human stomach secretes about 2.50 liters of gastric juice per day and its pH is variable between 1.50 (while fasting) to 3.50 (post feeding) and food ingestion can take up to 3 hr.8 Bile salts are toxic to living cells and resistance to bile salts is considered as one of the essential features of lactic acid-bacteria needed to survive in the small intestine.9 Furthermore, the ability to tolerate in the presence of pancreatic enzymes has been proposed as a criterion for the selection of probiotic cultures.10 Siahmazgi cheese is a traditional cheese produced from raw sheep and goat milk in Northern provinces of Iran. This cheese ripens in bags made of sheep skin called Khik. It is a popular traditional product among northern people due to its delicious taste and belief that it improves health. In this study, we aimed to screen L. plantarum strains isolated from Siahmazgi cheese through their resistance to low pH, bile salt, and gastric and intestinal simulated conditions.
Materials and Methods
L. plantarum isolates. A total number of 22 L. plantarum strains used in this study were previously isolated from Siahmazgi traditional cheese and molecularly identified by the Department of Food Hygiene and Quality Control, Faculty of Veterinary Medicine, University of Tehran.11 The isolates were grown in de Man Rogosa Sharpe (MRS) broth (Merck, Darmstadt, Germany) at 37.00 ˚C for 24 hr.
Screening of resistance of L. plantarum isolates to acidic pH. All 22 L. plantarum isolates were examined for their resistance against low pH. Ten microliters of over-night culture (18 hr) of each isolate was added to 240 µL MRS broth, adjusted to pH 1.50 or 2.50 using hydrochloric acid 0.10 N (Merck), in a micro-well plate and were incubated at 37.00 ˚C for 3 hr. MRS broth with pH 5.70 was used as control. Bacterial count was carried out at hr 0 and 3, through surface plate count method. Briefly, 100 µL aliquots from each well was removed and ten-fold serial dilutions were prepared in 0.10% peptone water and inoculated on MRS agar (Merck) and incubated at 37.00 ˚C for 24 to 48 hr under aerobic conditions.12 The viability of the strains was calculated through colony counting.
Growth kinetics of the isolates at different pHs. The ability of strains to grow in acidic conditions in MRS broth was evaluated at four sublethal pH including 3.50, 4.00, 4.50, and 5.70. Inoculation dose was adjusted to 107 CFU mL-1 and growth of the isolates monitored by reading the optical density (OD) values at 600 nm during 72 hr incubation at 37ý.00 ˚C. The growth curve was obtained through plotting the OD values in logarithmic scale versus incubation time. The maximum specific growth rate (µmax) was calculated from the slop of the regression line during exponential phase multiplied by 2.303. The lag time was determined according to the distance from the extrapolated regression line to the y axis. The generation time was calculated according the following equation:13
Generation time (hr) = 0.693/µ max
Resistance of the selected isolates to bile salt. To assess the resistance of the selected acid tolerant isolates to bile salt, a modified method of Gao et al. was used.14 Briefly, overnight cultures (18 hr) of 10 selected strains which showed more resistance to acidic pH, were harvested by centrifugation at 5,000 g for 15 min at 4ý.00 ˚C and washed twice with phosphate-buffered saline (PBS) buffer (pH 7.20). Bacterial pellets were re-suspended in peptone water 0.10% and inoculated in 0, 0.50 or 1.00% (w/v) bile salt (Sigma, St. Louis, USA) solutions in PBS at final concentrations of 107 CFU mL-1. Spread plate method was carried out as mentioned above to determine bacterial counts at 0, 60, 120 and 180 min after exposure to bile salt.
Viability in simulated gastric and intestinal conditions. The L. plantarum isolates showing more resistance to bile salt were selected to evaluate their survival under gastric and intestinal condition. Briefly, the 18-hour cultures of bacterial cells were harvested by centrifugation at 5,000 g for 15 min at 4.00 ˚C and were washed two times with PBS. Bacterial pellets were re-suspended in peptone water 0.10% and inoculated in PBS pH 2.50 containing 3.00 mg mL-1 pepsin (Sigma) for gastric simulation or in PBS pH 8.00 containing 1.00 mg mL-1 pancreatin (Sigma) and 0.30% bile for simulated intestinal condition. Bacterial count was carried out at 0, 60, and 180 min after incubation at 37.00 ˚C, using surface plate count method as mentioned above.15
Statistical Analysis. Each experiment was carried out in triplicate and data were expressed as means ± SE. One-way ANOVA was used to compare different mean groups using SPSS (version 16.0; IBM Corp., Armonk, USAý). Tukey test used to assess significant differences among the groups. A p < 0.05 was considered as significant.
Results
In this study, the ability of 22 L. plantarum strains, isolated from Siahmazgi traditional cheese, to withstand acidic conditions at pH of 1.50 and 2.50 during 3 hr was screened. The results of the cell viability of the isolates at selected pHs are shown in Table 1. Different trends were observed in the survival of the isolates in acidic pH. At the pH of 1.50, the L. plantarum A11 strain showed the greatest decrease of viability which was lowered by 7.66 log CFU mL-1, while A33 strain remained unchanged.
Table 1.
Effect of low pH on the cell viability of Lactobacillus plantarum strains isolated from Siahmazgi cheese during 3 hr incubation. Data are presented as mean ± standard error of three independent experiments
| Isolates | Bacterial viability (log CFU mL -1 ) | ||
|---|---|---|---|
| pH 5.70 | pH 2.50 | pH 1.50 | |
| A2 | 7.71 ± 0.04j | 6.80 ± 0.03c | 2.53 ± 0.09c |
| A3 | 7.13 ± 0.09d | 7.06 ± 0.00fg | 0.00 ± 0.00a |
| A4 | 7.47 ± 0.02h | 7.50 ± 0.22l | 0.00 ± 0.00a |
| A7 | 7.42 ± 0.01h | 7.37 ± 0.07k | 3.45 ± 0.08g |
| A11 | 7.66 ± 0.25i | 7.23 ± 0.09ij | 0.00 ± 0.00a |
| A15 | 7.08 ± 0.12d | 6.92 ± 0.01d | 2.15 ± 0.21b |
| A16 | 7.22 ± 0.01f | 7.06 ± 0.04efg | 4.26 ± 0.37i |
| A17 | 7.18 ± 0.02ef | 7.11 ± 0.08gh | 0.00 ± 0.00a |
| A18 | 7.20 ± 0.09ef | 7.18 ± 0.02hi | 0.00 ± 0.00a |
| A19 | 7.22 ± 0.04f | 7.00 ± 0.00ef | 0.00 ± 0.00a |
| A24 | 7.14 ± 0.03de | 7.04 ± 0.07efg | 5.66 ± 0.06n |
| A27 | 7.63 ± 0.10i | 7.23 ± 0.09ij | 0.00 ± 0.00a |
| A31 | 7.45 ± 0.11h | 7.26 ± 0.05j | 2.72 ± 0.17d |
| A33 | 7.08 ± 0.06d | 7.15 ± 0.19h | 7.15 ± 0.19o |
| E4 | 7.34 ± 0.03g | 7.55 ± 0.11lm | 0.00 ± 0.00a |
| F1 | 7.66 ± 0.03ij | 7.62 ± 0.02mn | 0.00 ± 0.00a |
| F2 | 7.18 ± 0.06ef | 6.99 ± 0.06de | 5.58 ± 0.01m |
| F7 | 6.68 ± 0.12b | 6.21 ± 0.06b | 4.58 ± 0.06j |
| F16 | 7.75 ± 0.21k | 7.69 ± 0.08no | 4.73 ± 0.08k |
| F19 | 7.62 ± 0.01i | 7.72 ± 0.04o | 5.33 ± 0.01l |
| F21 | 6.99 ± 0.06c | 7.57 ± 0.05lm | 3.67 ± 0.03h |
| F22 | 7.76 ± 0.04k | 7.74 ± 0.00o | 3.31 ± 0.34f |
Values shown with different letters in each column are statistically different (p < 0.05).
However, all the strains retained their viability at pH 2.50. Among all the strains A33, A24, F2 and F19 revealed the highest resistance to acid with 0.70, 1.48, 1.60 and 2.29 log CFU mL-1 reduction, respectively. Ten of the 22 tested strains showing high survival at low pH were selected to evaluate their growth kinetic in acidic conditions. The results of the growth kinetic of the isolates in acidic conditions are shown in Table 2. Strains F21 and F22 showed the lowest lag times at pH 5.70 which was 4 hr. The lag times of all the studied isolates were increased by decreasing the pH of the growth medium. Strain F2 exhibited the highest specific growth rate of 0.68 at pH 5.70 followed by F19 and F21.
Table 2.
Growth kinetic of selected Lactobacillus plantarum strains isolated from Siahmazgi cheese at different pH during 72 hr
| Isolates | Lag time (hr) | µ max (hr-1) | Generation time (hr) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH 5.70 | pH 4.50 | pH 4.00 | pH 3.50 | pH 5.70 | pH 4.50 | pH 4.00 | pH 3.50 | pH 5.70 | pH 4.50 | pH 4.00 | pH 3.50 | |||
| F2 | 6.00 | 12.00 | 16.00 | >72.00 | 0.68 | 0.24 | 0.14 | ND | 1.00 | 2.90 | 4.89 | ND | ||
| F7 | 6.00 | 16.00 | 24.00 | >72.00 | 0.36 | 0.32 | 0.11 | ND | 1.94 | 2.19 | 6.37 | ND | ||
| F16 | 6.00 | 9.00 | 12.00 | 16.00 | 0.37 | 0.36 | 0.26 | 0.11 | 1.85 | 1.95 | 2.67 | 6.52 | ||
| F19 | 6.00 | 9.00 | 12.00 | 24.00 | 0.51 | 0.40 | 0.26 | 0.17 | 1.35 | 1.73 | 2.63 | 4.15 | ||
| F21 | 4.00 | 9.00 | 12.00 | 16.00 | 0.37 | 0.30 | 0.26 | 0.11 | 1.87 | 2.32 | 2.66 | 6.50 | ||
| F22 | 4.00 | 9.00 | 12.00 | 24.00 | 0.51 | 0.36 | 0.29 | 0.12 | 1.35 | 1.91 | 2.42 | 5.64 | ||
| A23 | 16.00 | 36.00 | 36.00 | >72.00 | 0.26 | 0.15 | 0.10 | ND | 2.62 | 4.49 | 7.17 | ND | ||
| A7 | 9.00 | 12.00 | 36.00 | >72.00 | 0.27 | 0.16 | 0.06 | ND | 2.53 | 4.35 | 10.68 | ND | ||
| A24 | 12.00 | 24.00 | 48.00 | >72.00 | 0.38 | 0.09 | 0.08 | ND | 1.81 | 7.40 | 8.85 | ND | ||
| A16 | 12.00 | 16.00 | 24.00 | >72.00 | 0.31 | 0.14 | 0.05 | ND | 2.23 | 5.11 | 13.82 | ND | ||
µmax: Maximum specific growth rate, ND: Not determined.
The maximum specific growth rates was decreased as a response to pH reduction. Furthermore, only four strains including F16, F19, F21 and F22 retained their growth at pH as low as 3.50. Ten L. plantarum isolates showing higher resistance to low pH were selected to evaluate their tolerance to bile salts. The results of the isolates resistance to different concentrations of bile salts are shown in Table 3. Five out of 10 isolates showed a significant decrease in bacterial count varied from 2.00 to 7.00 log CFU mL-1 during 3 hr incubation at 0.50% bile salt. The remaining five strains were relatively resistant to 0.50% bile, since they experienced lower than 0.50 log reduction in bacterial population.
Table 3.
Effect of bile on the cell viability of Lactobacillus plantarum strains isolated from Siahmazgi cheese during 3 hr incubation. Data are presented as mean ± standard error of three independent experiments
| Isolates | Bacterial viability (log CFU mL -1 ) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.50% bile | 1.00% bile | ||||||||
| 0 hr | 1 hr | 2 hr | 3 hr | 0 hr | 1 hr | 2 hr | 3 hr | ||
| F2 | 6.36 ± 0.10a | 6.21 ± 0.10a | 6.54 ± 0.01a | 6.21 ± 0.03a | 6.36 ± 0.10a | 1.00 ± 1.00b | 1.00 ± 1.00b | 1.00 ± 1.00b | |
| F7 | 6.68 ± 0.03a | 6.53 ± 0.12ab | 6.57 ± 0.04ab | 6.23 ± 0.10b | 6.78 ± 0.03a | 4.13 ± 0.01b | 4.20 ± 0.03b | 4.20 ± 0.03b | |
| F16 | 6.61 ± 0.02a | 6.54 ± 0.04a | 6.65 ± 0.03a | 6.35 ± 0.12a | 6.71 ± 0.02a | 3.09 ± 0.05b | 2.82 ± 0.22b | 2.81 ± 0.035b | |
| F19 | 6.50 ± 0.00a | 6.75 ± 0.13a | 6.56 ± 0.02a | 6.62 ± 0.10a | 6.50 ± 0.00a | 1.00 ± 1.00 b | 1.00 ± 1.00b | 1.00 ± 1.00b | |
| F21 | 7.09 ± 0.02a | 6.95 ± 0.03a | 6.73 ± 0.10ab | 6.73 ± 0.02b | 7.09 ± 0.02a | 4.69 ± 0.04b | 4.69 ± 0.04b | 4.73 ± 0.04b | |
| F22 | 6.59 ± 0.02a | 6.37 ± 0.01a | 5.73 ± 0.02b | 4.30 ± 0.02c | 6.59 ± 0.02a | 1.35 ± 1.34b | ND | ND | |
| A7 | 6.67 ±0.12a | 3.32 ± 0.02b | 2.79 ± 0.10c | 2.45 ± 0.06d | 6.67 ± 0.12 | ND | ND | ND | |
| A33 | 6.92 ± 0.07a | 5.12 ± 0.04b | 4.23 ± 0.02c | 2.21 ± 0.80d | 6.92 ± 0.07a | 1.64 ± 1.12b | ND | ND | |
| A24 | 7.20 ± 0.10a | 4.32 ± 0.03b | 4.10 ± 0.05b | 2.83 ± 0.05c | 7.20 ± 0.10 | ND | ND | ND | |
| A16 | 6.92 ± 0.04a | 5.75 ± 0.02b | 5.22 ± 0.03c | NDd | 6.92 ± 0.04 | ND | ND | ND | |
ND: No bacterial growth was observed by plate count method.
Different letters for each strain show statistically significant difference among various hours (p < 0.05).
However, the viability of the isolates was severely reduced when the concentration of bile was increased to 1.00%. Strains F7, F16 and F21 showed better resistance than the others with F21 strain and the most resistant one showed 2.40 log reduction in bacterial number. Furthermore, no statistically significant differences were observed in viability of each strain between 1, 2 and 3 hr of incubation in the presence of 1.00% bile salt (p > 0.05). Five pre-screened L. plantarum strains presented high viability against low pH and bile were selected to evaluate their survival under simulated gastrointestinal conditions.
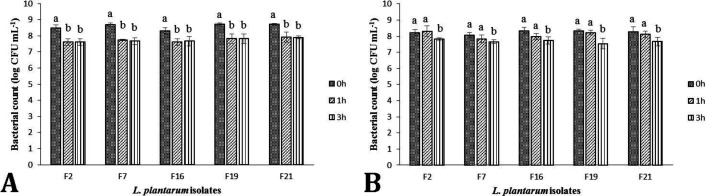
The results of gastrointestinal fluid simulation assay are shown in Fig. 1. All the strains showed suitable survival with maximum 1.00 log reduction during 3 hr incubation in simulated gastric condition (Fig. 1A). The L. plantarum F21 and F7 strains were the most resistant and the most sensitive strains with 0.83 and 1.02 log reduction, respectively. Furthermore, no statistically significant difference was observed in viability of each strain at hr 3 compared to that at hr 1 of incubation (p < 0.05). No significant changes were observed in bacterial viability of the selected isolate after 1 hr exposure to intestinal condition compared to the controls (Fig. 1B). All selected L. plantarum strains experienced a statistically significant decrease in bacterial count at hr 3 of incubation in intestinal fluid compared to the control groups (p < 0.05).
Fig. 1.
Cell viability of Lactobacillus plantarum strains isolated from Siahmazgi cheese during 3 hr incubation in simulated gastric (A) and intestinal (B) fluid. Different letters for each strain show statistically significant difference among various hours (p < 0.05).
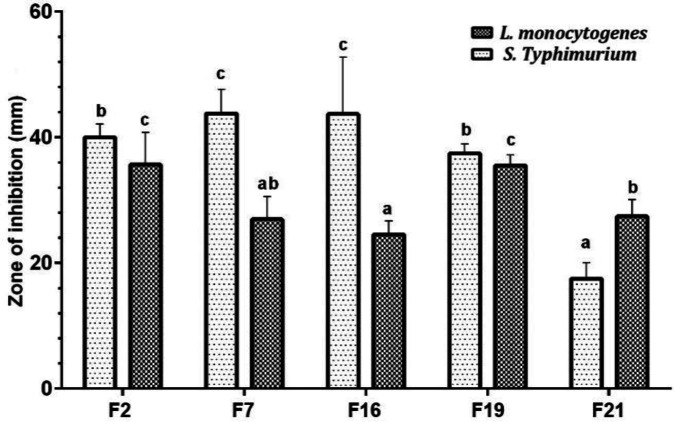
However, all the isolates effectively maintained their survival at intestinal conditions, since the highest reduction of viability seen in this experiment was as low as 0.78 log CFU mL-1 which was recorded for F19 isolate. The results of the isolates antibacterial effectiveness of the selected L. plantarum isolates are shown in Figure 2.
Fig. 2.
Inhibitory effect of the selected L. plantarum strains isolated from Siahmazgi cheese against L. monocytogenes and S. Typhimurium by agar spot assay. Values shown with different letters are statistically different (p < 0.05).
All selected L. plantarum strains represented an inhibitory capability against L. monocytogenes and S. Typhimurium. F16 and F7 strains generally demonstrated greater antimicrobial effectiveness against S. Typhimurium and F2 and F19 strains were more effective against L. monocytogenes.
Discussion
Microorganisms used as probiotics must bear some characteristics in order to reach the intestine alive and show their valuable properties and health benefits. The most important characteristics are their ability to overcome adverse gastrointestinal conditions, especially the acid and bile stress and the subsequent colonization in the digestive tract. Therefore, at the first, we evaluated the tolerance of the 22 L. plantarum strains, isolated from Siahmazgi cheese to pH as low as 1.50 and 2.50. The results of the present study showed that 20 out of 22 L. plantarum isolates experienced no reduction at pH 2.50 while only 10 isolates had viability over 3.00 log at pH 1.50 during 3 hr. It is reported that the pH value of 2.50 is very selective for the selection of potentially probiotic strains and even though it is not the most common pH value of thehuman stomach it assures the isolation of the very acid-tolerant strains.16 Furthermore, it is important to consider also the buffering capacity of the ingested food. Actually, ingested bacteria are rarely subjected to pH below 3.17 The viability of the present L. plantarum isolates are comparable to findings of Angmo et al., who reported that the number of viable LAB including L. plantarum isolated from fermented foods were significantly affected at pH 2.00 while no significant reduction was observed at pH 3.00.9 The L. plantarum isolates studied in the present study showed higher tolerance at low pH compared to the L. plantarum strains isolated from traditional Chinese cheese which showed survival rate of 31.00 - 80.00% at pH 2.50.18 Plessas et al. showed that 10 L. plantarum strains isolated from Caciotta and Sardo cheese were able to survive under pH 3.00 and 4.00 after 2 hr while considerable reduction of viability of the isolates was seen at pH 2.00.16 The results of this study were in agreement with those obtained from previous studies, where Lactobacillus strains isolated from fermented food were able to retain their viability when exposed to pH values of 2.00 to 4.00.9,19,20 The results of this study showed that the L. plantarum isolates were able to grow at low pH values. Oguntoyinbo et al. tested 2 L. plantarum strains isolated from fermented cereal foods for their growth ability in acidic conditions.21 The results showed that the strains were able to grow in low pH. Furthermore, Wu et al. found that L. casei Zhang strain showed proper growth at the pH of 4.30 during 60 hr.22 Bile salts are toxic to living cells and resistance to bile salts is considered as one of the essential features of lactic acid-bacteria needed to survive in the small intestine.9 The results of this study showed that 5 isolates had a relatively high resistance to 0.50% bile salt while were sensitive to 1.00% bile. The strains of the present study are more resistant compared to LAB strains of the study of Huang et al. which showed 2.27 to 5.42 log decrease during 3 hr at the presence of 0.30% bile salt.23 The results of this study were consistent with the study of Plessas et al. which showed that the viability of Lactobacillus strains isolate from Feta-type cheese was conserved at 0.50% bile salt.16 Furthermore, Owusu-Kwarteng et al. demonstrated that viability of Lactobacillus strains isolated from fermented millet dough was decreased significantly after 4 hr incubation in 1.00% of bile salt.24 It has been reported that bile salts lead to the leakage of cellular content and death of the cell by solving lipid membrane.25 Moreover, there is a significant positive correlation between resistance to bile and relative amounts of membrane fatty acid C16:0. It has been stated that, Lactobacilli derived from dairy products have relatively higher amount of fatty acid C16:0 in their membrane.14 The results of the simulated gastric passage exhibited good survival of all selected isolates. The results were in accordance with Archer and Halami who reported that Lactobacillus strains showed good survival under simulated gastric juice. 8 A proper probiotic must be able to endure the intestinal condition to reach the colon in sufficient numbers and operate meanwhile successfully. Bile salt concentration of human intestine is 0.03 to 0.30%, and 0.30% is the average concentration in the small intestine.26 The similar tolerance of LAB isolated from dairy products to GIT passage has been documented by Fang et al. who found that 13 different LAB strains isolated from traditional Chinese cheese displayed good survival in simulated intestinal condition during 3, 6, and 12 hr incubation.18 Also, Archer and Halami8 reported great survivability of 12 Lactobacillus strains isolated from dairy products in simulated intestinal condition during 3 and 6 hr incubations.
In conclusion, five out of 22 selected L. plantarum strains represented desirable properties according to bile and acid tolerance, growth kinetic at low pH, survival under simulated gastro intestinal fluids and antibacterial activity. Lactobacillus plantarum F2 and F7 isolates showed better resistance to undesirable conditions compared to other isolates. These isolates could be suitable candidates to be studied as potential probiotics. However, further experiments are still needed to evaluate their functional properties and safety, as well.
Acknowledgments
This work was supported by the Faculty of Veterinary Medicine, University of Tehran, Tehran, Iran, and also Iran National Science Foundation-Vice- Presidency for Science and Technology project (grant no. 91003935).
Conflict of interest
There is no conflict of interest to declare.
References
- 1.Bove P, Gallone A, Russo P, et al. Probiotic features of Lactobacillus plantarum mutant strains. Appl Microbiol Biotechnol. 2012;96(2):431–441. doi: 10.1007/s00253-012-4031-2. [DOI] [PubMed] [Google Scholar]
- 2.Fernández MF, Boris S, Barbés C. Safety evaluation of Lactobacillus delbrueckii subsp lactis UO 004 a pro-biotic bacterium. Res Microbiol. 2005;156(2):154–160. doi: 10.1016/j.resmic.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Argyri AA, Zoumpopoulou G, Karatzas KAG, et al. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013;33(2):282–291. doi: 10.1016/j.fm.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Turchi B, Mancini S, Fratini F, et al. Preliminary evaluation of probiotic potential of Lactobacillusplantarum strains isolated from Italian food products. World J Microbiol Biotechnol. 2013;29(10):1913–1922. doi: 10.1007/s11274-013-1356-7. [DOI] [PubMed] [Google Scholar]
- 5.Ayeni FA, Sánchez B, Adeniyi BA, et al. Evaluation of the functional potential of Weissella and Lactobacillus isolates obtained from Nigerian traditional fermented foods and cow's intestine. Int J Food Microbiol. 2011;147(2):97–104. doi: 10.1016/j.ijfoodmicro.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz-Moyano S, Martín A, Benito MJ, et al. Screening of lactic acid bacteria and Bifidobacteria for potential probiotic use in Iberian dry fermented sausages. Meat Sci. 2008;80(3):715–721. doi: 10.1016/j.meatsci.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Arokiyaraj S, Islam VIH, Bharanidharan R, et al. Antibacterial, anti-inflammatory and probiotic potential of Enterococcus hirae isolated from the rumen of Bos primigenius. World J Microbiol Biotechnol. 2014;30(7):2111–2118. doi: 10.1007/s11274-014-1625-0. [DOI] [PubMed] [Google Scholar]
- 8.Archer AC, Halami PM. Probiotic attributes of Lactobacillus fermentum isolated from human feces and dairy products. Appl Microbiol Biotechnol. 2015;99(19):8113–8123. doi: 10.1007/s00253-015-6679-x. [DOI] [PubMed] [Google Scholar]
- 9.Angmo K, Kumari A, Savitri , et al. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT- Food Sci Technol. 2016;66:428–435. [Google Scholar]
- 10.Verdenelli MC, Ghelfi F, Silvi S, et al. Probiotic properties of Lactobacillus rhamnosus and Lactobacillus paracasei isolated from human faeces. Eur J Nut. 2009;48(6):355–363. doi: 10.1007/s00394-009-0021-2. [DOI] [PubMed] [Google Scholar]
- 11.Partovi R, Gandomi H, Akhondzadeh Basti A, et al. Microbiological and chemical properties of Siahmazgi cheese, an Iranian artisanal cheese: isolation and identification of dominant lactic acid bacteria. J Food Process Preserv. 2014;39(6):871–880. [Google Scholar]
- 12.Lähteinen T, Malinen E, Koort JMK, et al. Probiotic properties of Lactobacillus isolates originating from porcine intestine and feces. Anaerobe. 2010;16(3):293–300. doi: 10.1016/j.anaerobe.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 13.McDonald LC, Shieh DH, Fleming HP, et al. Evaluation of malolactic-deficient strains of Lactobacillus plantarum for use in cucumber fermentations. Food Microbiol. 1993;10(6):489–499. [Google Scholar]
- 14.Gao Y, Li D, Liu S, et al. Probiotic potential of L sake C2 isolated from traditional Chinese fermented cabbage. Eur Food Res Technol. 2012;234:45–51. [Google Scholar]
- 15.Zhang Y, Zhang L, Du M, et al. Antimicrobial activity against Shigella sonnei and probiotic properties of wild lactobacilli from fermented food. Microbiol Res. 2011;167(1):27–31. doi: 10.1016/j.micres.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Plessas S, Nouska C, Karapetsas A, et al. Isolation, characterization and evaluation of the probiotic potential of a novel Lactobacillus strain isolated from Feta-type cheese. Food Chem. 2017;226:102–108. doi: 10.1016/j.foodchem.2017.01.052. [DOI] [PubMed] [Google Scholar]
- 17.Turková K, Mavrič A, Narat M, et al. Evaluation of Lactobacillus strains for selected probiotic properties. Folia Microbiol (Praha) 2015;58(4):261–267. doi: 10.1007/s12223-012-0208-4. [DOI] [PubMed] [Google Scholar]
- 18.Fang Z, Hongfei Z, Junyu Z, et al. Evaluation of probiotic properties of Lactobacillus strains isolated from traditional Chinese cheese. Ann Microbiol. 2015;65:1419–1426. [Google Scholar]
- 19.Arasu MV, Al-Dhabi NA, Rejiniemon TS, et al. Identification and characterization of Lactobacillus brevis P68 with antifungal, antioxidant and probiotic functional properties. Indian J Microbiol. 2015;55:19–28. [Google Scholar]
- 20.Park YU, Kim MD, Jung DH, et al. Probiotic properties of lactic acid bacteria isolated from Korean rice wine Makgeolli. Food Sci Biotechnol. 2015;24:1761–1766. [Google Scholar]
- 21.Oguntoyinbo FA, Narbad A. Multifunctional properties of Lactobacillus plantarum strains isolated from fermented cereal foods. J Funct Foods. 2015;17:621–631. [Google Scholar]
- 22.Wu C, Zhang J, Wang M, et al. Lactobacillus casei combats acid stress by maintaining cell membrane functionality. J Ind Microbiol Biotechnol. 2012;39:1031–1039. doi: 10.1007/s10295-012-1104-2. [DOI] [PubMed] [Google Scholar]
- 23.Huang HY, Hsieh HY, King VAE, et al. To pre-challenge lactic acid bacteria with simulated gastrointestinal conditions is a suitable approach to studying potential probiotic properties. J Microbiol Methods. 2014;107:138–146. doi: 10.1016/j.mimet.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Owusu-Kwarteng J, Tano-Debrah K, Akabanda F, et al. Technological properties and probiotic potential of Lactobacillus fermentum strains isolated from West African fermented millet dough. BMC Microbiol. 2015;15:261 . doi: 10.1186/s12866-015-0602-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das P, Khowala S, Biswas S. In vitro probiotic characterization of Lactobacillus casei isolated from marine samples. LWT- Food Sci Technol. 2016;73:383–390. [Google Scholar]
- 26.Lee YK, Salminen S. The coming of age of probiotics. Trends Food Sci Technol. 1995;6(7):241–245. [Google Scholar]