Abstract
Objectives
Chronic widespread musculoskeletal pain (CWP) is a characteristic symptom of fibromyalgia, which has been shown to be associated with an altered gut microbiome. Microbiome studies to date have not examined the milder CWP phenotype specifically nor have they explored the role of raised BMI. The aim of this study was to investigate whether the microbiome is abnormal in CWP.
Methods
CWP was assessed using a standardized screening questionnaire in female volunteers from the TwinsUK cohort including 113 CWP cases and 1623 controls. The stool microbiome was characterized using 16S rRNA amplicon sequencing and amplicon sequence variants, and associations with CWP examined using linear mixed-effects models adjusting for BMI, age, diet, family relatedness and technical factors.
Results
Alpha diversity was significantly lower in CWP cases than controls (Mann–Whitney test, P-values 2.3e-04 and 1.2e-02, for Shannon and Simpson indices respectively). The species Coprococcus comes was significantly depleted in CWP cases (Padj = 3.04e-03). A genome-wide association study (GWAS) performed for C. comes in TwinsUK followed by meta-analysis with three Dutch cohorts (total n = 3521) resulted in nine suggestive regions, with the most convincing on chromosome 4 near the TRAM1L1 gene (rs76957229, P = 7.4e-8). A Mendelian randomization study based on the results of the GWAS did not support a causal role for C. comes on the development of CWP.
Conclusions
We have demonstrated reduced diversity in the microbiome in CWP, indicating an involvement of the gut microbiota in CWP; prospectively the microbiome may offer therapeutic opportunities for this condition.
Keywords: body mass index, chronic widespread pain, gut microbiome, healthy eating index
Rheumatology key messages
Chronic widespread pain (CWP) is characterized by decreased alpha diversity of the gut microbiome.
Coprococcus comes is the most significantly reduced in CWP.
No evidence for causal relationship between the gut microbiome and CWP is seen.
Introduction
Chronic widespread musculoskeletal pain (CWP) is a common disorder, affecting some 5–15% of the general population, and presents a sizeable economic burden in terms of disability, work absence and healthcare costs [1]. Risk factors for CWP include increasing age, female sex, increasing BMI and lower socioeconomic status [2, 3]. CWP has a complex aetiology, and forms part of the fibromyalgia syndrome that includes fatigue and sleep disturbance in addition to CWP. CWP also co-occurs with other functional somatic syndromes such as chronic fatigue syndrome (CFS), irritable bowel syndrome, and mood disorders such as depression and anxiety [4].
Previous work has shown that genetic and environmental influences are important in the development of CWP [5]. Genetic factors that have shown associations with CWP in various studies have been summarized in several reviews [6, 7]. Overall results indicate that the nociceptive signalling system is rewired, and possibly disrupted by an excessive stress response [7]. We have shown that CWP shares common genetic determinants with low back pain [8] and with frailty [9]. A genome-wide study of DNA methylation also showed that epigenetic modification of neurological pathways is implicated in CWP [10] .
Overweight, obesity and raised BMI are the well-established risk factors for CWP [11, 12]. Longitudinal studies have provided evidence that BMI is raised prior to the onset of pain, rather than being a result of pain-induced immobility [13, 14]. We have shown that the influence of BMI on CWP risk is mediated through increased fat mass [15].
The human gut microbiome plays an important role in human health and disease. The composition of the human gut microbiome is affected by host genetics as well as diet, age, sex, weight, medication use and other environmental factors [16, 17], and the gut microbiome can in turn affect host metabolism [18]. Various studies have shown that a certain proportion of the gut microbiome is heritable [19, 20]; however, a recent study concluded that environmental factors have a greater effect than host genetics on the gut microbiome [21] .
Obesity has been associated with changes in the gut microbiome both in human and in mouse studies [22, 23], and there is considerable interest in modulating the gut microbiota for the prevention of obesity-related disease [24]. Short-chain fatty acids such as acetate, propionate and butyrate produced by the gut microbiome act as signalling molecules [25–27]. Acetate, in particular, influences insulin secretion and may promote obesity [28].
Thus, hypothetically, there is a link between gut microbiome, obesity and CWP. Accordingly, the aim of this study was to investigate the association of the gut microbiome with CWP in a large population sample, taking into account diet and BMI, and to address causality in the relationship using Mendelian randomization.
Methods
Study sample
Participants were individuals from the UK Adult Twin Registry (TwinsUK) [29]. The TwinsUK registry comprises volunteers from the general population recruited through national media campaigns. The cohort is predominantly female (83%), middle-aged, mainly of Northern European descent, and nearly equal in numbers of monozygotic and dizygotic same-sex twins. Participants have been characterized for a variety of clinical and behavioural traits including CWP. Twins from this registry have been shown to be similar to age-matched singletons for a range of health and lifestyle factors [30]. Twins have not been specifically recruited for the purpose of the current study; instead, participants were selected based on the availability of gut microbiome and phenotype of CWP (see below) data with the TwinsUK database. BMI was calculated from height and weight measurements taken during clinical visits. Zygosity was ascertained with the use of a questionnaire and confirmed by genotyping. All subjects provided written informed consent in accordance with the St Thomas' Hospital Research Ethics Committee, and were unaware of the precise hypotheses being tested. Ethical approval for microbiome studies within TwinsUK was provided by the NRES Committee London—Westminster (REC Reference No.: EC04/015).
CWP phenotype
A modified version of the London Fibromyalgia Epidemiology Study Screening Questionnaire (LFESSQ) [31] was used to screen for the presence of CWP. Volunteers from TwinsUK first completed a web-based or postal screening questionnaire. Those who answered yes to the question ‘In the past 3 months, have you had pain in your muscles, bones and joints lasting at least 1 week?’ were sent a more detailed paper questionnaire, with questions adapted from the LFESSQ. The questionnaire contained questions about musculoskeletal pain lasting >1 week in the right or left shoulders, arms or hands, in the right or left legs or feet, and in the neck, chest or back. Participants were also asked whether the pain had lasted >3 months. Each twin completed the questionnaires without reference to the co-twin. CWP cases were defined as those reporting pain on both sides of the body, above and below the waist, and in the axial skeleton, present for at least 3 months. Controls were individuals reporting pain but not meeting the criteria for CWP. Co-twins of the CWP cases were excluded from the controls. This was done to reduce the possible impact of shared genetic factors underlying both CWP and microbiome. We also carried out a discordant twin analysis, in which the healthy co-twins of CWP cases were compared. This analysis did not produce significant results likely because the sample size was too small, so the results are not reported here.
Dietary intake
The Healthy Eating Index (HEI) is a measure of diet quality developed by the United States Department of Agriculture (USDA) (https://www.fns.usda.gov/resource/healthy-eating-index-hei) to assess the extent to which food intake aligns with key dietary recommendations for Americans. The HEI uses a scoring system from 0 to 100, with a higher score indicating greater alignment with the dietary recommendations. Food intake for the TwinsUK cohort was determined from validated Food Frequency Questionnaires (FFQ) converted to HEI-2010 scores [32].
Faecal microbiome sample collection and 16S rRNA sequencing
The collection and processing of the faecal samples have been described previously [19]. Briefly, the participants collected faecal samples at home and the samples were either posted to King's College London on ice or refrigerated for up to 2 days prior to the twin pair annual clinical visit. Samples were stored at –80°C before being shipped on dry ice to Cornell University, and stored at –80°C until they were processed.
Genomic DNA was extracted from the faecal samples, and the V4 section of the 16S rRNA genes was amplified by PCR using the 515F and 806R primers [33]. The resulting amplicons were multiplexed using the Caporaso 12-base Golay barcodes [34] and sequenced as 250 bp paired-end reads using the Illumina MiSeq platform. Sequencing data for the microbiome samples used in this study are a subset of a larger study, and have been deposited in the European Nucleotide Archive (ENA) as part of previously published work [19, 35] under accession numbers ERP006339, ERP006342 and ERP015317.
Sequencing reads were demultiplexed and split per sample using QIIME 1.9.0 [36] to produce fastq sequencing data files. Amplicon sequence variants (ASVs) were obtained by denoising the raw sequencing data (fastq files) using the R/Bioconductor package DADA2 [37], which was applied separately per sequencing run (n = 35). The data were merged across sequencing runs, and chimeras removed, before finally assigning taxonomy of ASVs using the phyloseq R/Bioconductor package [38] and the SILVA rRNA gene database version 1.3.9 [39]. Final quality control steps were undertaken: sequences that were unassigned at the kingdom/phylum level or that were assigned to Eukaryota were removed. Samples with a sequencing depth of <10 000 were removed from further analysis.
Statistical analysis
Association analysis of CWP and the microbiome
Association analysis of the gut microbiome with CWP was carried out using linear mixed-effects models with the lme4 package (version 1.1.12) in R-3.3.0 (R Foundation for Statistical Computing, Vienna, Austria). ASV counts were transformed using the variance stabilization method in the R/Bioconductor package DESeq2 (version 1.22.1 in R-3.5.2) [40]. ASVs were filtered to remove those not present in at least 5% of the samples, leaving 859 ASVs. Samples were filtered to remove those with fewer than 10 000 counts. Biological covariates included age, HEI, BMI and family relatedness. Technical covariates included sample sequencing depth, sequencing run, technician who extracted the DNA, technician who loaded the PCR plate, sample collection method, and FFQ date. Family relatedness and all technical covariates except sample sequencing depth were modelled as random effects. The variance stabilized ASV counts were used as the response variable in a linear model accounting for biological and technical factors. The Benjamini–Hochberg false discovery rate was used to control for multiple testing.
Causal inference
Causal inference was assessed using a combination of Mendelian randomization (MR) and polygenic risk score (PRS). Genome-wide association studies (GWAS) were carried out for bacteria of interest (see Results) and CWP. The bacteria counts were subjected to variance stabilizing transformation followed by an adjustment for technical covariates via residuals. The residuals have been further normalized using the qqnorm function in R statistical software to achieve a normal distribution. GWAS both for the bacterium and CWP was done using GEMMA software [41] adjusting for age, sex, BMI and family structure (via genomic relatedness matrix). The following filters were applied: minor allele frequency 0.03, Hardy–Weinberg equilibrium P-value 1e-6, imputation quality 0.7, and genotype missingness rate 0.95. To increase the precision of the bacterium GWAS, its results have been meta-analyzed with the results of GWAS of metagenomic data carried out in three Dutch cohorts—LifeLines-DEEP (n = 916), 500FG (n = 410), and MIBS-CO (n = 93)—as described elsewhere [42]. Meta-analysis was carried out for n = 4 841 651 overlapping single nucleotide polymorphisms (SNPs) using GWAMA software [43]. Following the completion of the GWAS, conditional and joint (COJO) analysis [44] was carried out to identify LD-independent association signals over the range of P-values: 2.5e-7, 5e-7, 2.5e-6, 5e-6, 2.5e-5, 5e-5, 2.5e-4, 5e-4, 2.5e-3, 5e-3, 2.5e-2 and 5e-2. For each set of SNPs resulting from the COJO analysis, we calculated PRSs as a weighted sum of genotype counts using regression coefficients from the GWAS as the weights. Predictive potential of the PRSs was assessed by regressing them on the phenotype of interest (the bacterium or CWP). The PRS with the highest coefficient of determination (R2) was then used to predict the exposure phenotype followed by regression of the predicted exposure on the outcome phenotype, adjusting for age, sex and BMI. Power of the MR analysis was calculated using the online tool https://shiny.cnsgenomics.com/mRnd/ [45].
Results
The characteristics of the study population are presented in Table 1. Briefly, there were 1736 women in the TwinsUK dataset having pain questionnaire, dietary intake and gut microbiome data available. CWP cases were statistically significantly older, had higher BMI and lower HEI scores than controls.
Table 1.
Summary statistics of TwinsUK sample
| Covariates | Controls (n = 1623) | CWP (n = 113) | Student’s t-test P-value |
|---|---|---|---|
| Age, years | 61.4 (11.3) | 63.5 (9.3) | 0.028 |
| BMI, kg/m2 | 25.7 (4.7) | 27.9 (5.7) | 6.1e-05 |
| Healthy eating index | 60.8 (10.0) | 57.8 (9.6) | 1.6e-03 |
Mean (s.d.) values are shown. CWP: chronic widespread pain.
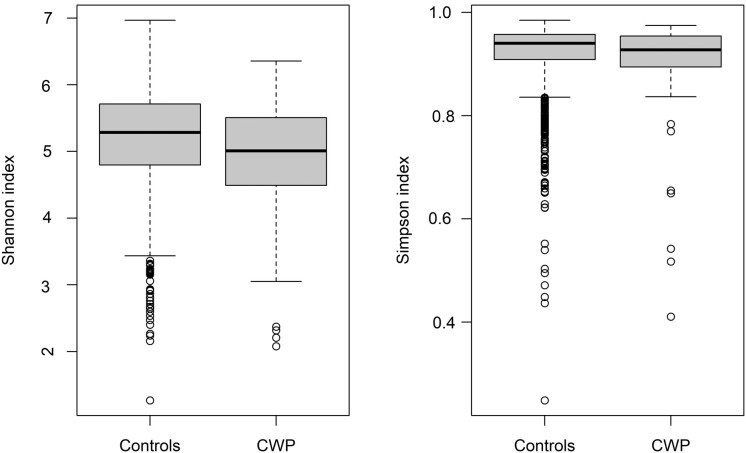
Alpha diversity was significantly lower in CWP cases than controls, as measured by Shannon and Simpson metrics (Fig. 1).
Fig. 1.
Alpha diversity values for Shannon and Simpson indices in CWP cases and controls
Alpha diversity was significantly lower in CWP cases (Mann–Whitney test, P-values 2.3e-04 and 1.2e-02, respectively). CWP: chronic widespread pain.
Using linear mixed-effects models, we performed the case–control analysis of association between CWP and the microbiome using 859 ASVs present in at least 5% of samples; adjustments were done for age, BMI, HEI, family structure and technical covariates. Sixty ASVs had nominal P-values <0.05 (Table 2), with one remaining statistically significant after correcting for multiple testing: β = –1.7227 (0.3703) (Padj = 3.04e-03).
Table 2.
Association between CWP and stool ASVs in TwinsUK
| Internal ASV ID | Effect (β) | s.e. | Nominal P-value | ASV taxonomy |
|---|---|---|---|---|
| ASVs decreased in CWP | ||||
| 85 | −1.7227 | 0.3703 | 3.54e-06 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Coprococcus_3; s_comes |
| 291 | −0.4056 | 0.1426 | 4.49e-03 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_; s_ |
| 367 | −0.2925 | 0.1051 | 5.44e-03 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Coprococcus_3; s_comes |
| 782 | −0.3320 | 0.1238 | 7.42e-03 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_GCA-900066575; s_ |
| 162 | −1.1984 | 0.4472 | 7.44e-03 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcaceae_UCG-003; s_ |
| 255 | −0.3490 | 0.1315 | 8.02e-03 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Roseburia; s_inulinivorans |
| 332 | −1.1333 | 0.4368 | 9.56e-03 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Lachnospiraceae_NK4A136_group; s_ |
| 118 | −1.2962 | 0.5061 | 1.05e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Faecalibacterium; s_ |
| 695 | −0.7161 | 0.2869 | 1.26e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcaceae_UCG-005; s_ |
| 631 | −0.3418 | 0.1385 | 1.37e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_; s_ |
| 658 | −0.6843 | 0.2836 | 1.59e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Howardella; s_ureilytica |
| 238 | −1.1532 | 0.4840 | 1.73e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Lachnospiraceae_UCG-010; s_ |
| 374 | −1.0080 | 0.4261 | 1.81e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Family_XIII; g_Family_XIII_AD3011_group; s_ |
| 29 | −0.8482 | 0.3597 | 1.85e-02 | k_Bacteria; p_Firmicutes; c_Erysipelotrichia; o_Erysipelotrichales; f_Erysipelotrichaceae; g_Erysipelotrichaceae_UCG-003; s_ |
| 42 | −1.2176 | 0.5202 | 1.94e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Lachnospiraceae_NK4A136_group; s_ |
| 482 | −0.6015 | 0.2577 | 1.97e-02 | k_Bacteria; p_Tenericutes; c_Mollicutes; o_Izimaplasmatales; f_; g_; s_ |
| 210 | −0.3135 | 0.1349 | 2.03e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Faecalibacterium; s_ |
| 166 | −0.7406 | 0.3201 | 2.08e-02 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Marinifilaceae; g_Odoribacter; s_splanchnicus |
| 403 | −0.7985 | 0.3509 | 2.30e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcaceae_NK4A214_group; s_ |
| 379 | −0.4238 | 0.1864 | 2.31e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_; s_ |
| 748 | −0.3009 | 0.1333 | 2.42e-05 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Family_XIII; g_Family_XIII_AD3011_group; s_ |
| 841 | −0.2326 | 0.1069 | 2.98e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_; s_ |
| 284 | −0.7409 | 0.3423 | 3.05e-02 | k_Bacteria; p_Proteobacteria; c_Alphaproteobacteria; o_Rhodospirillales; f_; g_; s_ |
| 609 | −0.0938 | 0.0439 | 3.27e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Dorea; s_formicigenerans |
| 343 | −0.9268 | 0.4344 | 3.30e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Lachnospiraceae_UCG-004; s_ |
| 169 | −0.9253 | 0.4341 | 3.32e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcus_1; s_ |
| 708 | −0.2969 | 0.1398 | 3.38e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminiclostridium_9; s_ |
| 587 | −0.7418 | 0.3511 | 3.48e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Oscillibacter; s_ |
| 310 | −0.1867 | 0.0886 | 3.52e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Blautia; s_obeum |
| 626 | −0.2869 | 0.1381 | 3.79e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Lachnospiraceae_NK4A136_group; s_ |
| 650 | −0.2989 | 0.1439 | 3.79e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Marvinbryantia; s_ |
| 429 | −0.3145 | 0.1518 | 3.85e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_; s_ |
| 464 | −0.6522 | 0.3149 | 3.85e-02 | k_Bacteria; p_Actinobacteria; c_Coriobacteriia; o_Coriobacteriales; f_Coriobacteriales_Incertae_Sedis; g_; s_ |
| 54 | −1.1981 | 0.5792 | 3.88e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Agathobacter; s_ |
| 409 | −0.8235 | 0.4020 | 4.07e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Lachnospiraceae_UCG-004; s_ |
| 474 | −0.3302 | 0.1621 | 4.18e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Faecalibacterium; s_ |
| 328 | −0.6931 | 0.3416 | 4.26e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcaceae_UCG-010; s_ |
| 143 | −0.9961 | 0.4934 | 4.37e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_; g_; s_ |
| 596 | −0.2854 | 0.1430 | 4.61e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcaceae_UCG-005; s_ |
| 110 | −0.9524 | 0.4781 | 4.65e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_; s_ |
| 594 | −0.5822 | 0.2928 | 4.69e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcaceae_UCG-010; s_ |
| 355 | −0.3037 | 0.1529 | 4.72e-02 | k_Bacteria; p_Proteobacteria; c_Gammaproteobateria; o_Betaproteobacteriales; f_Burkholderiaceae; g_Parasutterella; s_excrementihominis |
| 124 | −0.3410 | 0.1720 | 4.76e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcaceae_UCG-002; s_ |
| 123 | −0.2369 | 0.1208 | 5.00e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Subdoligranulum; s_ |
| ASVs increased in CWP | ||||
| 668 | 0.8838 | 0.2836 | 1.86e-03 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Shuttleworthia; s_ |
| 119 | 1.0217 | 0.3724 | 6.14e-03 | k_Bacteria; p_Firmicutes; c_Bacilli; o_Lactobacillales; f_Streptococcaceae; g_Streptococcus; s_ |
| 844 | 0.5121 | 0.1880 | 6.53e-03 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Sellimonas; s_ |
| 220 | 1.1548 | 0.4573 | 1.17e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminiclostridium_5; s_ |
| 746 | 0.5170 | 0.2056 | 1.20e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Eisenbergiella; s_massiliensis |
| 552 | 0.8018 | 0.3370 | 1.75e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminiclostridium_5; s_ |
| 338 | 0.3517 | 0.1516 | 2.05e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_; s_ |
| 20 | 1.2372 | 0.5413 | 2.24e-02 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Rikenellaceae; g_Alistipes; s_ |
| 461 | 0.6294 | 0.2791 | 2.43e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Lachnospira; s_ |
| 719 | 0.6488 | 0.2973 | 2.93e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_; g_; s_ |
| 441 | 0.7369 | 0.3461 | 3.34e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Ruminococcaceae; g_Ruminococcaceae_UCG-005; s_ |
| 173 | 0.7111 | 0.3375 | 3.52e-02 | k_Bacteria; p_Firmicutes; c_Negativicutes; o_Selenomonadales; f_Veillonellaceae; g_Dialister; s_ |
| 724 | 0.5483 | 0.2626 | 3.70e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Christensenellaceae; g_Christensenellaceae_R-7_group; s_ |
| 710 | 0.6325 | 0.3058 | 3.87e-02 | k_Bacteria; p_Actinobacteria; c_Actinobacteria; o_Actinomycetales; f_Actinomycetaceae; g_Actinomyces; s_odontolyticus |
| 589 | 0.5998 | 0.2908 | 3.93e-02 | k_Bacteria; p_Firmicutes; c_Clostridia; o_Clostridiales; f_Lachnospiraceae; g_Tyzzerella_3; s_ |
| 601 | 0.5768 | 0.2824 | 4.12e-02 | k_Bacteria; p_Bacteroidetes; c_Bacteroidia; o_Bacteroidales; f_Marinifilaceae; g_Butyricimonas; s_ |
Linear mixed-effects models were applied to test for association between ASVs and CWP in TwinsUK. Adjustment was done for age, BMI, HEI, family relatedness and technical covariates as detailed in the main text. ASV: amplicon sequence variant; CWP: chronic widespread pain.
The majority (44/60) of the ASVs with nominal P-values <0.05 were decreased in CWP cases compared with controls. The ASV that reached statistical significance after correction for multiple testing was assigned to Coprococcus comes, and it had a lower abundance in CWP cases than in controls.
The majority (38/44) of the ASVs nominally decreased in the CWP cases compared with the controls were assigned to Firmicutes of the order Clostridiales, with half assigned to the Lachnospiraceae family (19/38), and the rest mainly to the family Ruminococcaceae (16/38) (Table 2). Of the 60 ASVs with nominal P-values <0.05, the majority (48) were assigned at the genus level, but only 11 were assigned at the species level.
The majority (11/16) of the ASVs increased in the CWP cases compared with the controls with nominal P-values <0.05 were assigned to Firmicutes of the order Clostridiales, of which more than half were assigned to the Lachnospiraceae family (6/11) (Table 2).
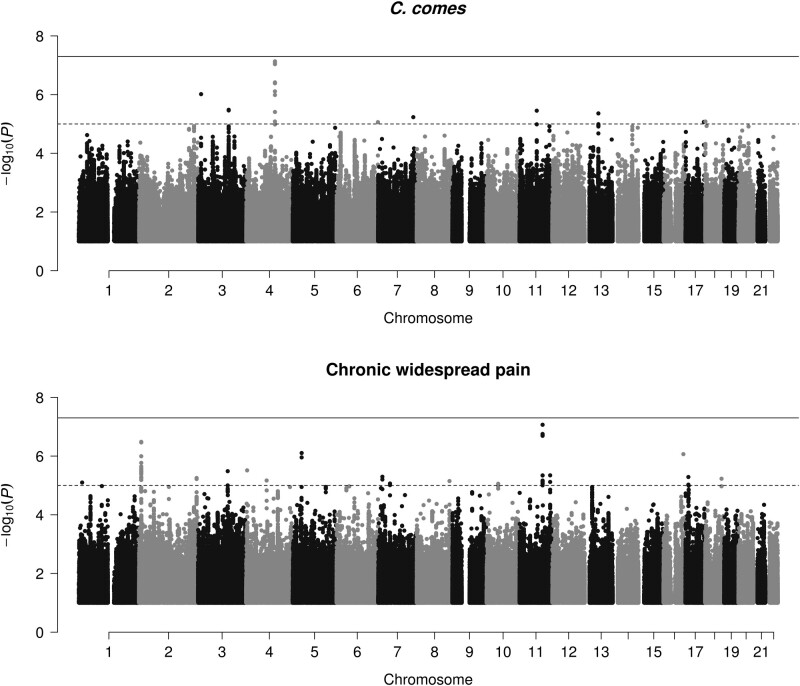
An association between C. comes and CWP raises a question of causality: in the absence of confounders, either CWP causes the decrease of the bacterium abundance or the bacterium depletion causes the increased risk of CWP. We addressed this using Mendelian randomization and polygenic risk score. Neither meta-GWAS for the bacterium nor GWAS for CWP produced genome-wide significant associations (Fig. 2). However, nine loci achieved a suggestive significance threshold of P < 1e-5 for C. comes, with the most convincing one on chromosome 4 near TRAM1L1 gene (Table 3; Fig. 2). Fifteen loci achieved suggestive significance for CWP, with the most convincing one on chromosome 11 near SRSF8 gene (Table 4; Fig. 2). The best PRS in terms of predictive capacity of the bacterium comprised 895 SNPs having P-value <5e-03 (R2 = 0.157, P = 6.06e-58). The levels of bacterium predicted using this PRS were not a statistically significant predictor for CWP (β = –0.293 (0.229); P = 0.201). The best PRS for CWP comprised 39 SNPs having P-value <2.5e-05 (R2 = 0.143, P = 9.49e-30). The case–control classes of CWP predicted using this PRS were not predictive of the levels of C. comes (β = –0.268 (0.181); P = 0.139). The results suggest that neither C. comes nor CWP is likely a causal factor for each other. However, power estimates showed that our study has 22% power to detect significant causal effect of C. comes on CWP, while the power to detect causal effect of CWP on C. comes was 33%.
Fig. 2.
Manhattan plots for genome-wide association study results of Coprococcus comes (A) and chronic widespread pain (CWP) (B)
Table 3.
Suggestive results of meta-GWAS for Coprococcus comes
| rs_number | Chr: position | Overlapped or nearest gene | Effect allele | Other allele | Effect size | s.e. | P-value | Q-statistic | P-value for Q-statistic | I 2 | Total sample | Direction of effects |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs11551661 | 3:11871215 | TAMM41 | C | T | −0.1119 | 0.0228 | 9.6e-07 | 1.14 | 0.768 | 0.000 | 3521 | −−−− |
| rs2735247 | 3:125477762 | GS1-388B5.8 / RP11-379B18.6 | G | A | −0.1123 | 0.0241 | 3.3e-06 | 6.89 | 0.076 | 0.564 | 3521 | −−−− |
| rs76957229 | 4:118043418 | TRAM1L1 / AC107399.1 | T | C | 0.1257 | 0.0233 | 7.4e-08 | 3.77 | 0.287 | 0.205 | 3521 | ++++ |
| rs4716409 | 6:170234376 | RP1-182D15.2 / RP11-302L19.1 | A | T | −0.0934 | 0.0210 | 8.7e-06 | 1.39 | 0.708 | 0.000 | 3521 | −−−− |
| rs59725556 | 7:145084002 | AC004911.2 / AC073055.2 | T | C | −0.1074 | 0.0237 | 5.9e-06 | 5.91 | 0.116 | 0.492 | 3521 | −−−− |
| rs72958391 | 11:71265701 | KRTAP5-9 / KRTAP5-10 | C | G | −0.1141 | 0.0246 | 3.5e-06 | 6.55 | 0.088 | 0.542 | 3521 | −−−− |
| rs4378518 | 13:56068607 | MIR5007 / HNF4GP1 | C | T | −0.0945 | 0.0206 | 4.4e-06 | 5.54 | 0.136 | 0.458 | 3521 | −−−− |
| rs56396930 | 17:75743839 | RP11-316M20.1 / FLJ45079 | T | G | 0.1079 | 0.0242 | 8.5e-06 | 1.92 | 0.589 | 0.000 | 3521 | ++++ |
| rs62082666 | 18:923474 | ADCYAP1 / RP11-672L10.1 | A | G | −0.0935 | 0.0210 | 8.3e-06 | 1.67 | 0.643 | 0.000 | 3521 | −−−− |
Genome-wide association study was carried out for C. comes in TwinsUK (n = 2118) using GEMMA software. Adjustments were made for age, sex, BMI and kinship via genetic relatedness matrix. Subsequent meta-analysis was performed including three Dutch cohorts described elsewhere [42]. Top SNPs from regions of suggestive associations are provided (P < 1e-5). Overlapped and nearest genes are identified using SNPnexus (https://www.snp-nexus.org/v4/). GWAS: genome-wide association study; SNP: single nucleotide polymorphism. s.e: standard error.
Table 4.
Suggestive results for GWAS for CWP
| SNPID | Chr: position | Overlapped or nearest gene | Effect allele | Other allele | Effect size | s.e. | P-value | EAF | Sample size |
|---|---|---|---|---|---|---|---|---|---|
| rs61777763 | 1:14477359 | RNU6-1265P / RP11-344F13.1 | T | C | 0.0935 | 0.0209 | 7.96e-06 | 0.062 | 3114 |
| rs13429284 | 2:237102161 | ASB18 | C | A | 0.0639 | 0.0141 | 5.57e-06 | 0.143 | 3114 |
| rs34057310 | 2:9423678 | ASAP2 | T | C | 0.0624 | 0.0122 | 3.27e-07 | 0.219 | 3114 |
| rs9821958 | 3:122009508 | CASR / HNRNPA1P23 | G | A | 0.0675 | 0.0145 | 3.27e-06 | 0.135 | 3114 |
| rs35020435 | 4:4076195 | AC116562.2 / RP11-489M13.1 | C | T | 0.0577 | 0.0123 | 3.07e-06 | 0.31 | 3114 |
| rs34698918 | 5:37421837 | WDR70 | A | G | 0.1084 | 0.0219 | 7.85e-07 | 0.061 | 3114 |
| rs55728567 | 7:17792341 | AC006482.1 / SNX13 | T | C | 0.0658 | 0.0144 | 5.05e-06 | 0.141 | 3114 |
| rs142024481 | 8:134637071 | RP11-629O1.2 / SNORA40 | G | A | 0.0948 | 0.0211 | 7.10e-06 | 0.061 | 3114 |
| rs11259826 | 10:47600049 | AHCYP1 / ANTXRLP1 | T | C | −0.1310 | 0.0294 | 8.76e-06 | 0.038 | 3114 |
| rs10893460 | 11:125867896 | CDON | A | G | −0.0512 | 0.0111 | 4.53e-06 | 0.269 | 3114 |
| rs78164056 | 11:94795234 | RP11-735A19.2 / SRSF8 | C | T | 0.0787 | 0.0147 | 8.56e-08 | 0.136 | 3114 |
| rs149091311 | 12:50885180 | LARP4 / DIP2B | T | C | 0.1805 | 0.0343 | 1.56e-07 | 0.036 | 3114 |
| rs56117891 | 16:81861570 | PLCG2 | T | C | 0.1296 | 0.0263 | 8.58e-07 | 0.039 | 3114 |
| rs4792257 | 17:12251758 | RP11-471L13.2 / LINC00670 | A | G | −0.0928 | 0.0203 | 5.17e-06 | 0.937 | 3114 |
| rs2849474 | 18:67569079 | CD226 | T | C | −0.0804 | 0.0177 | 5.90e-06 | 0.907 | 3114 |
GWAS for CWP was carried out in TwinsUK (n = 3273; 531 cases of CWP) using GEMMA software [41]. Adjustments were done for age, sex, BMI and kinship via genetic relatedness matrix. Top SNPs from regions of suggestive associations are provided (P < 1e-5). Overlapped and nearest genes are identified using SNPnexus (https://www.snp-nexus.org/v4/). CWP: chronic widespread pain; GWAS: genome-wide association study; SNP: single nucleotide polymorphism. s.e: standard error.
Discussion
The aim of this study was to examine the association between CWP and the gut microbiome while adjusting for the expected differences in BMI between cases and controls. Measures of species diversity were found to be significantly lower in CWP and we identified an ASV assigned to the species Coprococcus comes to be significantly decreased in CWP. Our Mendelian randomization study did not provide evidence for possible causal relationships between CWP and C. comes. However, this study was underpowered to show possible causal relationships (power achieved 22% and 33%).
C. comes is one of the most important butyrate-producing bacteria. Butyric acid produced by C. comes and other gut commensals exhibits remarkable anti-inflammatory effects in the gut. C. comes abundance was shown to decrease in inflammatory and immune-mediated disorders, such as Crohn’s disease [46] and type 1 diabetes [47]. Even though our MR analysis was not significant, it still does not fully rule out a causal relationship between C. comes and CWP, given the low statistical power. While CWP as a causal factor for reduced C. comes can be explained by an overall drop of microbial abundance, the alternative route whereby the bacterium influences disease risk is more difficult to fathom. One possible explanation rests on the fact that a high-fat diet causes a decrease of Coprococcus species abundance in the gut [48] and people with CWP are known to have on average a diet higher in fat [49]. This is consistent with our finding of a statistically significantly lower HEI and higher BMI in CWP cases compared with controls (Table 1). These data reinforce the hypothesis that a high-fat diet promotes the decrease of Coprococcus and other anti-inflammatory species that in turn results in a low-grade inflammation, supporting the development of CWP.
Higher diversity of the gut microbiome has generally been associated with good health [50, 51], and the accumulation of health deficits (frailty index) has been linked with lower alpha diversity of the gut microbiota [52]. There is an established relationship between CWP and frailty in our cohort and others [9, 53, 54], and studies suggest that CWP may precede the accumulation of ill health, and potentially contribute to it [55]. This makes understanding the role of the microbiome critical as it offers a potential therapeutic mediator in the prevention of both CWP and frailty. While lower diversity in itself may not represent a deficit of the gut microbiota in CWP, the significantly lower gut microbiota diversity demonstrated here is indicative of a diseased state. As the gut microbiota represents a therapeutic target—via interventions such as diet, pro-biotics, drugs to target the microbiota and faecal transfer—the results of our study are in accordance with the gut microbiome representing a future therapeutic target in CWP.
A study of the gut microbiome in myalgic encephalomyelitis (ME)/CFS [56] found that bacteria of the genus Coprococcus were among the taxa decreased in ME/CFS patients compared with controls. CWP is common in chronic fatigue and both conditions are considered to lie on a spectrum of non-specific pain and fatiguing illnesses [57]. Another study of the gut microbiome in ME/CFS found decreased microbial diversity, in particular a decrease in relative abundance and diversity of members of the Lachnospiraceae family, including Coprococcus species [58].
Two recent studies demonstrated variation of gut microbiome in individuals with fibromyalgia (FM), a condition characterized by CWP as a major symptom [59, 60]. The study by Minerbi et al. [59] compared stool microbiome in 77 FM patients and 79 controls and identified no difference in alpha-diversity between cases and controls and mildly reduced beta-diversity in patients. They also found differential abundance for 72 operational taxonomic units belonging to phyla Actinobacteria, Bacteroidetes, Firmicutes and Proteobacteria. The study by Clos-Garcia et al. [60] looked at 105 FM cases and 54 controls and identified reduced abundance of the Bifidobacterium and Eubacterium genera consistent with altered levels of neurotransmitters in the serum of the patients. Both studies provided evidence for potential of the gut microbiome as a biomarker of FM. However, the question of causality was not addressed.
Our study has limitations. For the case–control study, we looked at female twin volunteers only, so the results cannot be extrapolated to men. However, the prevalence of CWP is higher in women than in men so this middle-aged cohort is particularly relevant to the condition. The pain questionnaire used to diagnose CWP, the FFQ, and the faecal samples for gut microbiome analysis were obtained at different times, which in some cases may be several years apart. However, longitudinal studies have shown that the gut microbiota remain relatively stable in adults [61]. Also, longitudinal reporting of CWP in TwinsUK is reasonably consistent (Supplementary Table, available at Rheumatology online). Thus, we believe that even though the time difference between the stool sample acquisition and twins assessment might have played some role, our results still reflect true relationships between microbiome and CWP. Still, we cannot fully rule out the possibility of residual confounding due to longitudinal change of the diet, physical activity and other factors that may affect microbiota composition. Another potential limitation is the difference between cases and controls by age and BMI, so potentially the observed differences in microbiome can be attributable to these factors. However, the absolute differences in age is rather small (2 years, Table 1), so it is unlikely a significant factor in our study, also taking into account above-mentioned stability of microbiome with age. Concerning BMI, we cannot fully rule out its impact on the results; however, increased BMI is a characteristic feature of CWP, so it is inherently difficult to distinguish the impact of the two. As we statistically adjusted for BMI and age, the resultant differences in microbiome in CWP are likely attributable to CWP; however, collider bias cannot be excluded at this stage and this possibility needs to be explored further. Our analysis of the microbiome was limited by the use of 16S rRNA sequencing data, which did not enable assignment of genus or species to many of the ASVs. The use of shotgun metagenomics and metatranscriptomic data would provide more accurate species level assignment, as well as functional information about which genes and pathways are present and being expressed [62]. Finally, our Mendelian randomization study was underpowered (22% and 33%), so larger studies are warranted to look at possible causality between CWP and C. comes. Our estimates show that one needs a sample of at least 5500 individuals to achieve 80% to demonstrate causal impact of CWP on C. comes and 8700 to achieve 80% power to demonstrate causal impact of C. comes on the risk of CWP. While large GWAS for CWP using UK Biobank data are under way (Rahman et al. [unpublished results] Genome-wide association study identifies three loci associated with chronic widespread musculoskeletal pain; presented at the American Society of Human Genetics 69th Annual Meeting, October 15–19, 2019), GWAS of comparable size for C. comes are much more challenging.
In summary, our study establishes a suggestive connection between the gut microbiome and CWP. Lack of diversity of gut microbiome appears, in common with many conditions, and may potentially be addressed by dietary and other intervention. The role of Coprococcus merits further investigation and it may lie in a network of microbes which are indicative of poorer diet.
Funding statement: This work was supported by Arthritis Research UK grant number 20682 to F.M.K.W.
TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, Chronic Disease Research Foundation (CDRF), Zoe Global Ltd and the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London.
A.Z. is supported by ERC Starting Grant 715772, NWO-VIDI grant 016.178.056, the Netherlands Heart Foundation CVON grant 2018-27, and NWO Gravitation grant ExposomeNL 024.004.017.
Disclosure statement: The authors declare no conflict of interest.
Data availability statement
TwinsUK: Sequencing data for the microbiome samples used in this study have been deposited in the European Nucleotide Archive (ENA) under accession numbers ERP006339, ERP006342, and ERP015317. Dutch cohorts: Sequencing data for the microbiome samples of the The LifeLines-DEEP and MIBS metagenomics sequencing data are available at the European Genome phenome Archive (EGA); LifeLines-DEEP, EGAS00001001704; MIBS, EGAS00001001924. The 500FG data are available at the Sequence Read Archive (SRA): PRJNA319574.
Other data are available on request from the TwinsUK Resource Executive Committee (TREC) (https://twinsuk.ac.uk/resources-for-researchers/access-our-data/).
Supplementary data
Supplementary data are available at Rheumatology online.
Supplementary Material
References
- 1. Mansfield KE, Sim J, Jordan JL, Jordan KP.. A systematic review and meta-analysis of the prevalence of chronic widespread pain in the general population. Pain 2016;157:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Macfarlane GJ, Norrie G, Atherton K, Power C, Jones GT.. The influence of socioeconomic status on the reporting of regional and widespread musculoskeletal pain: results from the 1958 British Birth Cohort Study. Ann Rheum Dis 2009;68:1591–5. [DOI] [PubMed] [Google Scholar]
- 3. Macfarlane GJ, Pye SR, Finn JD. et al. ; EMAS Study Group. Investigating the determinants of international differences in the prevalence of chronic widespread pain: evidence from the European Male Ageing Study. Ann Rheum Dis 2009;68:690–5. [DOI] [PubMed] [Google Scholar]
- 4. Kato K, Sullivan PF, Evengard B, Pedersen NL.. Chronic widespread pain and its comorbidities: a population-based study. Arch Intern Med 2006;166:1649–54. [DOI] [PubMed] [Google Scholar]
- 5. Kato K, Sullivan PF, Evengard B, Pedersen NL.. Importance of genetic influences on chronic widespread pain. Arthritis Rheum 2006;54:1682–6. [DOI] [PubMed] [Google Scholar]
- 6. Ablin JN, Buskila D.. Update on the genetics of the fibromyalgia syndrome. Best Pract Res Clin Rheumatol 2015;29:20–8. [DOI] [PubMed] [Google Scholar]
- 7. Zorina-Lichtenwalter K, Meloto CB, Khoury S, Diatchenko L.. Genetic predictors of human chronic pain conditions. Neuroscience 2016;338:36–62. [DOI] [PubMed] [Google Scholar]
- 8. Malkin I, Williams FM, LaChance G. et al. Low back and common widespread pain share common genetic determinants. Ann Hum Genet 2014;78:357–66. [DOI] [PubMed] [Google Scholar]
- 9. Livshits G, Malkin I, Bowyer RCE. et al. Multi-OMICS analyses of frailty and chronic widespread musculoskeletal pain suggest involvement of shared neurological pathways. Pain 2018;159:2565–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Livshits G, Malkin I, Freidin MB. et al. Genome-wide methylation analysis of a large population sample shows neurological pathways involvement in chronic widespread musculoskeletal pain. Pain 2017;158:1053–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoo JJ, Cho NH, Lim SH, Kim HA.. Relationships between body mass index, fat mass, muscle mass, and musculoskeletal pain in community residents. Arthritis Rheumatol 2014;66:3511–20. [DOI] [PubMed] [Google Scholar]
- 12. Yunus MB, Arslan S, Aldag JC.. Relationship between body mass index and fibromyalgia features. Scand J Rheumatol 2002;31:27–31. [DOI] [PubMed] [Google Scholar]
- 13. McCarthy LH, Bigal ME, Katz M, Derby C, Lipton RB.. Chronic pain and obesity in elderly people: results from the Einstein aging study. J Am Geriatr Soc 2009;57:115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mork PJ, Vasseljen O, Nilsen TI.. Association between physical exercise, body mass index, and risk of fibromyalgia: longitudinal data from the Norwegian Nord-Trondelag Health Study. Arthritis Care Res (Hoboken) 2010;62:611–7. [DOI] [PubMed] [Google Scholar]
- 15. Livshits G, Macgregor AJ, Gieger C. et al. An omics investigation into chronic widespread musculoskeletal pain reveals epiandrosterone sulfate as a potential biomarker. Pain 2015;156:1845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kurilshikov A, Wijmenga C, Fu J, Zhernakova A.. Host genetics and gut microbiome: challenges and perspectives. Trends Immunol 2017;38:633–47. [DOI] [PubMed] [Google Scholar]
- 17. Luca F, Kupfer SS, Knights D, Khoruts A, Blekhman R.. Functional genomics of host-microbiome interactions in humans. Trends Genet 2018;34:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Janssen AW, Kersten S.. Potential mediators linking gut bacteria to metabolic health: a critical view. J Physiol 2017;595:477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goodrich JK, Waters JL, Poole AC. et al. Human genetics shape the gut microbiome. Cell 2014;159:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Turpin W, Espin-Garcia O, Xu W. et al. ; GEM Project Research Consortium. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet 2016;48:1413–7. [DOI] [PubMed] [Google Scholar]
- 21. Rothschild D, Weissbrod O, Barkan E. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210–5. [DOI] [PubMed] [Google Scholar]
- 22. Turnbaugh PJ, Hamady M, Yatsunenko T. et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ley RE, Backhed F, Turnbaugh P. et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005;102:11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brahe LK, Astrup A, Larsen LH.. Can we prevent obesity-related metabolic diseases by dietary modulation of the gut microbiota? Adv Nutr 2016;7:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perry RJ, Peng L, Barry NA. et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 2016;534:213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morrison DJ, Preston T.. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016;7:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trent CM, Blaser MJ.. Microbially produced acetate: a “missing link” in understanding obesity? Cell Metab 2016;24:9–10. [DOI] [PubMed] [Google Scholar]
- 28. Hernandez MAG, Canfora EE, Jocken JWE, Blaak EE.. The short-chain fatty acid acetate in body weight control and insulin sensitivity. Nutrients 2019;11:1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Verdi S, Abbasian G, Bowyer RCE. et al. Twins UK: the UK adult twin registry update. Twin Res Hum Genet 2019;22:523–9. [DOI] [PubMed] [Google Scholar]
- 30. Andrew T, Hart DJ, Snieder H. et al. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res 2001;4:464–77. [DOI] [PubMed] [Google Scholar]
- 31. White KP, Harth M, Speechley M, Ostbye T.. Testing an instrument to screen for fibromyalgia syndrome in general population studies: the London fibromyalgia epidemiology study screening questionnaire. J Rheumatol 1999;26:880–4. [PubMed] [Google Scholar]
- 32. Bowyer RCE, Jackson MA, Pallister T. et al. Use of dietary indices to control for diet in human gut microbiota studies. Microbiome 2018;6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caporaso JG, Lauber CL, Walters WA. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 2011;108: 4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Caporaso JG, Lauber CL, Walters WA. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Goodrich JK, Davenport ER, Beaumont M. et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 2016;19:731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caporaso JG, Kuczynski J, Stombaugh J. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Callahan BJ, McMurdie PJ, Rosen MJ. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McMurdie PJ, Holmes S.. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quast C, Pruesse E, Yilmaz P. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou X, Stephens M.. Genome-wide efficient mixed-model analysis for association studies. Nat Genet 2012;44:821–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bonder MJ, Kurilshikov A, Tigchelaar EF. et al. The effect of host genetics on the gut microbiome. Nat Genet 2016;48:1407–12. [DOI] [PubMed] [Google Scholar]
- 43. Magi R, Morris AP.. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J, Ferreira T, Morris AP, Medland SE. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brion MJ, Shakhbazov K, Visscher PM.. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 2013;42:1497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hall LJ, Walshaw J, Watson AJ.. Gut microbiome in new-onset Crohn's disease. Gastroenterology 2014;147:932–4. [DOI] [PubMed] [Google Scholar]
- 47. Pinto E, Anselmo M, Calha M. et al. The intestinal proteome of diabetic and control children is enriched with different microbial and host proteins. Microbiology (Reading) 2017;163:161–74. [DOI] [PubMed] [Google Scholar]
- 48. Claesson MJ, Jeffery IB, Conde S. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012;488:178–84. [DOI] [PubMed] [Google Scholar]
- 49. Vandenkerkhof EG, Macdonald HM, Jones GT, Power C, Macfarlane GJ.. Diet, lifestyle and chronic widespread pain: results from the 1958 British Birth Cohort Study. Pain Res Manag 2011;16:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gilbert JA, Blaser MJ, Caporaso JG. et al. Current understanding of the human microbiome. Nat Med 2018;24:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lloyd-Price J, Abu-Ali G, Huttenhower C.. The healthy human microbiome. Genome Med 2016;8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jackson MA, Jeffery IB, Beaumont M. et al. Signatures of early frailty in the gut microbiota. Genome Med 2016;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wade KF, Lee DM, McBeth J. et al. Chronic widespread pain is associated with worsening frailty in European men. Age Ageing 2016;45:268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Livshits G, Ni Lochlainn M, Malkin I. et al. Shared genetic influence on frailty and chronic widespread pain: a study from TwinsUK. Age Ageing 2018;47:119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jaremo P, Arman M, Gerdle B, Larsson B, Gottberg K.. Illness beliefs among patients with chronic widespread pain – associations with self-reported health status, anxiety and depressive symptoms and impact of pain. BMC Psychol 2017;5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nagy-Szakal D, Williams BL, Mishra N. et al. Fecal metagenomic profiles in subgroups of patients with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome 2017;5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McBeth J, Tomenson B, Chew-Graham CA. et al. Common and unique associated factors for medically unexplained chronic widespread pain and chronic fatigue. J Psychosom Res 2015;79:484–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Giloteaux L, Goodrich JK, Walters WA. et al. Reduced diversity and altered composition of the gut microbiome in individuals with myalgic encephalomyelitis/chronic fatigue syndrome. Microbiome 2016;4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Minerbi A, Gonzalez E, Brereton NJB. et al. Altered microbiome composition in individuals with fibromyalgia. Pain 2019;160:2589–602. [DOI] [PubMed] [Google Scholar]
- 60. Clos-Garcia M, Andres-Marin N, Fernandez-Eulate G. et al. Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine 2019;46:499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Flores GE, Caporaso JG, Henley JB. et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol 2014;15:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Franzosa EA, McIver LJ, Rahnavard G. et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018;15:962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TwinsUK: Sequencing data for the microbiome samples used in this study have been deposited in the European Nucleotide Archive (ENA) under accession numbers ERP006339, ERP006342, and ERP015317. Dutch cohorts: Sequencing data for the microbiome samples of the The LifeLines-DEEP and MIBS metagenomics sequencing data are available at the European Genome phenome Archive (EGA); LifeLines-DEEP, EGAS00001001704; MIBS, EGAS00001001924. The 500FG data are available at the Sequence Read Archive (SRA): PRJNA319574.
Other data are available on request from the TwinsUK Resource Executive Committee (TREC) (https://twinsuk.ac.uk/resources-for-researchers/access-our-data/).