Keywords: COVID-19, lumisterol, RNA-dependent RNA polymerase, SARS-CoV-2 main protease, vitamin D metabolites
Abstract
Vitamin D deficiency significantly correlates with the severity of SARS-CoV-2 infection. Molecular docking-based virtual screening studies predict that novel vitamin D and related lumisterol hydroxymetabolites are able to bind to the active sites of two SARS-CoV-2 transcription machinery enzymes with high affinity. These enzymes are the main protease (Mpro) and RNA-dependent RNA polymerase (RdRP), which play important roles in viral replication and establishing infection. Based on predicted binding affinities and specific interactions, we identified 10 vitamin D3 (D3) and lumisterol (L3) analogs as likely binding partners of SARS-CoV-2 Mpro and RdRP and, therefore, tested their ability to inhibit these enzymes. Activity measurements demonstrated that 25(OH)L3, 24(OH)L3, and 20(OH)7DHC are the most effective of the hydroxymetabolites tested at inhibiting the activity of SARS-CoV-2 Mpro causing 10%–19% inhibition. These same derivatives as well as other hydroxylumisterols and hydroxyvitamin D3 metabolites inhibited RdRP by 50%–60%. Thus, inhibition of these enzymes by vitamin D and lumisterol metabolites may provide a novel approach to hindering the SARS-CoV-2 infection.
NEW & NOTEWORTHY Active forms of vitamin D and lumisterol can inhibit SARS-CoV-2 replication machinery enzymes, which indicates that novel vitamin D and lumisterol metabolites are candidates for antiviral drug research.
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic has brought tremendous socioeconomic losses, causing great adversity with some intriguing and complex scientific questions to be answered. The variations in mortality and severity of patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in different regions of the world is a challenging conundrum that still remains unanswered. A plethora of reports has made a striking and intreguing corelation that vitamin D deficiency influences the risk of mortality in patients infected with SARS-CoV-2 (1). Vitamin D is a pleotropic hormone, which exerts genomic and nongenomic effects on expression of a large number of genes that modify many important biological functions (2). The genomic effects initially involve the binding of 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] to the vitamin D receptor (VDR). The ligand-based activation of this nuclear transcription factor can modulate the expression of genes positively or negatively. However, a variety of signaling pathways can be activated in a nongenomic manner by 1,25(OH)2D3 (2). Multiple mechanisms have been proposed by which vitamin D protects against infection and reduces mortality when present in sufficient physiological levels (3–5). Vitamin D modulates the innate and adaptive immune response (6), and hence vitamin D deficiency is a recognized risk factor for the cytokine storm and acute respiratory distress syndrome (ARDS; for review, see Refs. 7 and 8). Vitamin D also has been reported to affect the expression of angiotensin-converting enzyme 2 (ACE2) and transmembrane protease, serine 2 (TMPRSS2), membrane receptors responsible for the cellular entry of the virus (9). In addition, active forms of vitamin D can regulate the expression of the antiviral pathways and also viral load in the system, when used at high concentrations (5, 10). However, as of 2021, there has been only one report confirming a direct relationship between vitamin D and inhibition of the SARS-CoV-2 viral replication where it was shown that calcitriol [1,25(OH)2D3] reduces the viral load in the cytopathogenic effect (CPE) assay (11). Although the authors did not provide the mechanism behind this action, their study indicates that active forms of vitamin D3 (D3) may have the potential to inhibit or neutralize SARS-CoV-2 growth and reduce the severity of the infection.
The exposure of skin to ultraviolet radiation (UVB) leads to conversion of 7-dehydrocholesterol (7-DHC) to previtamin D3 (pre-D3), which then undergoes thermal isomerization to form D3 (6, 12). With a high UVB dose, pre-D3 undergoes photoisomerization to lumisterol (L3) and tachysterol (T3). T3, being the most photoreactive product, undergoes UVB-driven conversion to L3 via pre-D3. The resulting product, L3, is the major photoisomer observed in skin after prolonged UVB exposure (13). Traditionally, only D3 was regarded as the important biological regulator from these photochemical reactions, whereas L3 was considered to not affect calcium metabolism nor display any other significant biological activity (14). It was postulated that transformation of pre-D3 into metabolically inactive L3 explained why UVB-induced cutaneous production of pre-D3 does not lead to systemic D3 intoxication (12). It is now apparent that this assumption is erroneous since new CYP11A1-dependent and CYP27A1-dependent pathways of hydroxylation of L3 have been discovered with the products such as 20(OH)L3, 22(OH)L3, 24(OH)L3, and 25(OH)L3 displaying biological activity (14–17). CYP11A1-mediated pathways of both L3 and vitamin D metabolism have been shown to occur in vivo with their products having phenotypical/biological activities determined by their structures and cellular targets (14).
To evaluate whether these novel L3 and D3 hydroxyderivatives possess antiviral potential, we screened them for effects on viral replication and transcription machinery proteins that are the most promising drug targets against SARS-CoV-2. The proteins tested in the study were RNA-dependent RNA polymerase (RdRP or nsp12) and 3C-like protease (3CLpro or Mpro). SARS-CoV-2 contains a ∼30-kb RNA genome encoding two large overlapping polyprotein precursors (pp1a and pp1ab), four structural proteins (spike, envelope, membrane, and nucleocapsid), and several accessory proteins (18, 19). It is essential for SARS-CoV-2 replication that the two polyproteins (pp1a/pp1ab) be cleaved into individual nonstructural proteins. Mpro exclusively cleaves polypeptides after a glutamine (Gln) residue and no other human protease has the same cleavage specificity as Mpro (19, 20). This critical function of SARS-CoV-2 Mpro in replication and transcriptional regulation has made it a prime target for drug discovery purposes (18, 19). It shows a 96% similarity at the amino acid sequence level with SARS-CoV Mpro (20, 21). RdRP is another conserved protein of retroviruses and is also a proven target for the development of antiviral drugs (22). RdRP has two binding sites for ligands, one is the active site which binds nucleoside inhibitors (NIs) and the other is an alosteric site that binds non-nucleoside inhibitors (NNIs). Remdesivir mimics a nucleoside triphosphate substrate for the polymerase becoming covalently linked to the replicating RNA, which interferes with further synthesis of the RNA (23).
In this study, we used a systematic approach for structure-based drug discovery in the search for potential therapeutic candidates from a pool of vitamin D and lumiterol metabolites, targeting the SARS-CoV-2 replication machinery enzymes Mpro and RdRP. Structure-based virtual screening was used to predict analogs that could bind to these two proteins followed by enzyme inhibition studies to confirm these interactions.
MATERIALS AND METHODS
Materials
Methods of production of the compounds tested in this study and their structures are described in the supplemental material including Supplemental Table S1; see https://doi.org/10.6084/m9.figshare.14547747.v1.
Structure-Based Virtual Screening
In this study, a systematic in-silico approach was used, using a number of bioinformatics software, such as MGL AutoDock Tools (22), AutoDock Vina (23), and Discovery Studio Visualizer (24). The three-dimensional coordinates of SARS-CoV-2 Mpro and RdRP were retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB). The structure of SARS-CoV-2 Mpro in complex with a Michael acceptor inhibitor known as N3 provides a basis to design high-affinity inhibitors with improved pharmacological properties (17). The atomic coordinates of SARS-CoV-2 Mpro were taken from the PDB (ID: 6LU7) for molecular docking-based virtual screening. The structural coordinates of SARS-CoV-2 RdRP in apo form and in complex with remdesivir were taken from the PDB (ID: 7BV1 and 7BV2, respectively). The structure was prepared by removing cocrystallized inhibitors and adding hydrogens, and assigning appropriate atom types. AutoDock Vina was used to carry out molecular docking, where the search space was structurally blind for all the compounds with default docking parameters. High-affinity compounds were selected, and their docked conformations were generated for analyzing their possible interaction toward SARS-CoV-2 Mpro and RdRP. Only those compounds which specifically interact with the critical residues of the binding pockets of SARS-CoV-2 Mpro and RdRP were selected from the interaction analysis.
Enzymatic Assay for Mpro
The SARS-CoV-2 3CLPro (Mpro), MBP-tagged Assay Kit (BPS Biosciences, San Diego, CA) was used to measure 3CL Protease (Mpro) activity for screening and profiling applications. The 3CL inhibitor GC376 was included as a positive control and also used as a standard. The assay was performed according to the protocol provided by the manufacturer with the concentration of compounds tested being 10−7 M.
Enzymatic Assay for RdRP
The assay for RdRP activity was outsourced to BPS Biosciences (San Jose, California) using the following protocol. Dilutions of the compounds tested were prepared in assay buffer containing 5% ethanol, then 2 µL of this was added to the reaction mix to give a final volume of 10 µL, thus making the final ethanol concentration 1% in all reactions (controls without compounds similarly contained 1% ethanol). The RdRP reactions were performed in triplicates at 37°C for 60 min in a 10 µL mixture containing Assay buffer, RNA duplex, ATP substrate and enzyme, and the test compound. The reactions were carried out in wells of 384-well Optiplate (PerkinElmer). After enzymatic reactions, 10 µL of anti-Dig Acceptor beads (PerkinElmer, diluted 1:500 with 1× detection buffer) were added to the reaction mix. After brief shaking, the plate was incubated for 30 min. Finally, 10 µL of AlphaScreen Streptavidin-conjugated donor beads (Perkin, diluted 1:125 with 1× detection buffer) were added. After 30 min, the samples were measured using an AlphaScreen microplate reader (EnSpire Alpha 2390 Multilabel Reader, PerkinElmer). The values of % activity versus the concentrations of the compound tested were then plotted using nonlinear regression analysis of a Sigmoidal dose-response curve generated with the equation Y = B + (T−B)/1 + 10[(LogEC50 − X) × Hill Slope], where Y = percent activity, B = minimum percent activity, T = maximum percent activity, X = logarithm of compound, and Hill Slope = slope factor or Hill coefficient. The IC50 value was determined as the concentration causing half-maximal activity.
RESULTS AND DISCUSSION
Structure-based virtual screening of an in-house library of D3, L3, and 7-DHC analogs was performed to search for high-affinity binding partners of SARS-CoV-2 Mpro and RdRP that could be used in the design of potential therapeutics against COVID-19 (24). We first calculated the binding affinities of the available compounds in the library when bound to SARS-CoV-2 Mpro and RdRP, and then performed interaction analysis to identify better hits. Based on the specific interaction, we identified a set of 10 D3, L3, and 7-DHC derivatives out of 35 compounds tested that had substantial affinity and specific interactions with the active site pocket of SARS-CoV-2 Mpro and RdRP (Table 1 and Table 2). The predicted binding affinities of the 10 compounds compared well with those of recognized antivirals such as danoprevir, lopinavir, and ritonavir (Table 1 and Table 2).
Table 1.
Binding affinities to the SARS-CoV-2 Mpro in comparison with danoprevir, lopinavir, and ritonavir
| No. | Name of the Ligand | Binding Free Energy (kcal/mol) |
|---|---|---|
| 1 | 24(OH)L3 | −8.3 |
| 2 | 25(OH)L3 | −8.0 |
| 3 | 20S(OH)7DHC | −8.0 |
| 4 | 22R(OH)7DHC | −7.8 |
| 5 | 20S(OH)L3 | −7.8 |
| 6 | 25(OH)7DHC | −7.7 |
| 7 | 1,25(OH)2D3 | −7.6 |
| 8 | 20S(OH)D3 | −7.6 |
| 9 | 7-DHP | −7.5 |
| 10 | 25(OH)D3 | −7.3 |
| 11 | Danoprevir | −8.5 |
| 12 | Lopinavir | −8.3 |
| 13 | Ritonavir | −7.2 |
Table 2.
Binding affinities of to the SARS-CoV-2 RdRP in comparison with ritonavir, danoprevir, lopinavir, and remdesivir
| No. | Name of the Ligand | Binding Free Energy (kcal/mol) |
|---|---|---|
| 1 | 25(OH)D3 | −8.5 |
| 2 | 20S(OH)D3 | −8 |
| 3 | Ritonavir | −8 |
| 4 | Danoprevir | −7.9 |
| 5 | 20S(OH)L3 | −7.8 |
| 6 | 25(OH)L3 | −7.7 |
| 7 | 1,25(OH)2D3 | −7.7 |
| 8 | 20S(OH)7DHC | −7.6 |
| 9 | 22R(OH)L3 | −7.5 |
| 10 | 24(OH)L3 | −7.5 |
| 11 | 25(OH)7DHC | −7.5 |
| 12 | Lopinavir | −7.3 |
| 13 | Remdesivir | −7.3 |
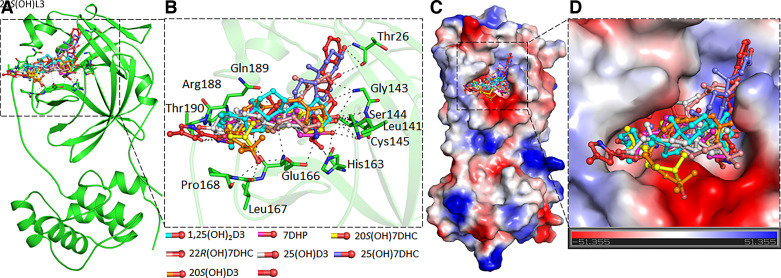
A detailed analysis of all the docked conformers of the top 10 hits was performed to find compounds binding specifically to the SARS-CoV-2 Mpro and RdRP substrate-binding pockets (Supplemental Figs. S1 and S2). We selected seven compounds that form interactions to a set of critical residues of SARS-CoV-2 Mpro to study in detail. The critical active site residue Cys145 of SARS-CoV-2 Mpro along with the bonding pocket residues Thr26, His41, Leu141, Asn142, Gly143, Ser144, Cys145, His163, Glu166, Leu167, Pro168, Arg188, Gln189, and Thr190 provide a significant number of interactions with each of the seven compounds (Fig. 1 and Supplemental Fig. S1). Importantly, all of these sterols and secosteroids mimic the same binding pose and show a similar pattern as N3, a cocrystallized inhibitor of SARS-CoV-2 Mpro. The binding pattern of the selected compounds indicates a virtuous complementarity with the Mpro binding pocket, which may hinder the substrate accessibility, thus inhibiting the enzymatic activity. Each of the seven compounds make significant interactions with critically important residues of the Mpro substrate-binding pocket (Fig. 1B and Supplemental Fig. S1) and blocks the substrate-binding pocket of COVID-19 Mpro (Fig. 1, C and D). The compounds docked to the Mpro binding pocket were analyzed further for their detailed interactions with the critical residues, including Cys145 (Supplemental Fig. S1) (18).
Figure 1.
The binding pattern of identified compounds with SARS-CoV-2 Mpro. A: structural representation of the protein in complex with selected sterols and secosteroids. B: selected compounds blocking the binding pocket and making significant interactions with the functionally important residues of SARS-CoV-2 Mpro. C: surface representation of conserved substrate-binding pocket of SARS-CoV-2 Mpro in complex with selected compounds. D: zoomed view of the substrate-binding pocket of SARS-CoV-2 Mpro in complex with selected compounds.
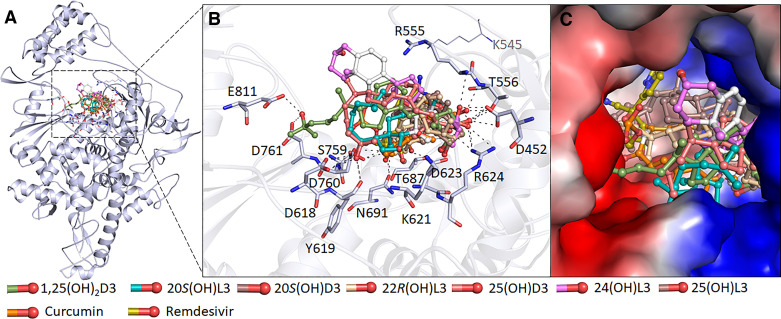
Similar to Mpro molecular docking, we selected top 10 hits based on interaction with the SARS-CoV-2 RdRP active site pocket, from which we identified seven compounds that form interactions with a set of critically essential residues of SARS-CoV-2 RdRP. The residues Lys545, Arg555, Asp623, Ser682, Thr687, Asn691, Ser759, Asp760, and Asp761 of SARS-CoV-2 in RdRP substrate binding pocket that interact with remdesivir, also offered a significant number of interactions with each of the seven compounds (Fig. 2 and Supplemental Fig. S2). Notably, these sterols and secosteroids have the same binding pattern as reported for the inhibitor remdesivir with SARS-CoV-2 RdRP (23). The binding prototype of the compounds indicates a virtuous complementarity to the SARS-CoV-2 RdRP binding pocket indicating that they have the capability to inhibit its enzymatic activity (Fig. 2). The compounds, including curcumin, interact with residues within the RdRP active site pocket (Fig. 2, A and B and Supplemental Fig. S2) that are critical for substrate binding (Fig. 2C).
Figure 2.
The binding pattern of identified sterols and secosteroids with SARS-CoV-2 RdRP. A: structural representation of the protein in complex with selected compounds. B: active site residues of the RdRP-binding pocket making significant interactions with each of the identified compounds. C surface view of the RdRP active site with the electrostatic potential from red (negative) to blue (positive) in complex with selected compounds. RdRP, RNA-dependent RNA polymerase.
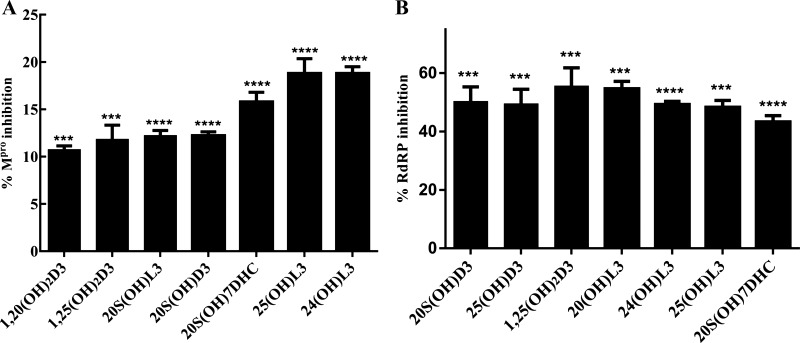
The binding data predict that the selected sterols and secosteroids are likely to function as inhibitors of Mpro and RdRP enzyme activity and potentially serve as therapeutics against COVID-19. Consistent with this prediction, we found that the compounds inhibited Mpro enzyme activity using the 3CL Protease, MBP-tagged (SARS-CoV-2) Assay kit (Fig. 3A). The results show that all seven sterols and secosteroids tested inhibited Mpro activity with 25(OH)L3, 24(OH)L3, and 20S(OH)7DHC being most effective, inhibiting it by 16%–19% at a concentration of 2 × 10−7 M (Fig. 3A).
Figure 3.
Enzyme inhibition by the selected sterols and secosteroids. A: the Mpro enzyme inhibition by the selected metabolites at concentration of 2 × 10−7 M. The inhibition percentages were calculated using the formula: % inhibition = 100 × [1 − (X − Minimum)/(Maximum − Minimum)]. Minimum = negative control without any enzyme (0% enzyme activity), Maximum = positive control with enzyme and substrate (100% enzyme activity). The test sets included enzymes, substrates, and the test compounds, excitation at a wavelength of 360 nm, and detection of emission at a wavelength of 460 nm was observed for change in enzyme activity. The statistical significance of differences was evaluated by one-way ANOVA, ***P < 0.001 and ****P < 0.0001 for all conditions relative to ethanol blank, n = 3. B: the RdRP enzyme activity inhibition by selected sterols and secosteroids. The inhibition percentages were calculated using the formula: % inhibition = 100 × [1 − (X − Minimum)/(Maximum − Minimum)]. Minimum = negative control without any enzyme (0% enzyme activity), Maximum = positive control with enzyme and substrate (100% enzyme activity). The statistical significance of differences was evaluated by one-way ANOVA, ***P < 0.001 and ****P < 0.0001 for all conditions relative to the ethanol blank, n = 3. RdRP, RNA-dependent RNA polymerase.
RdRP catalyzes the replication of RNA from an RNA template and it is known that SARS-CoV-2 RdRP only functions when all three subunits are present (nsp12, nsp7, and nsp8). Hence, establishing the assay to measure its activity was difficult so this was out sourced to BPS biosciences who had already developed an in-house assay to measure enzyme activity. Although the inhibition of RdRP activity did not precisely reflect the pattern of docking energies, all of the compounds tested exhibited inhibitory activity ranging from 40% to 60% at a concentration of 2 × 10−7 M (Fig. 3B). The IC50 was also calculated for each compound, which revealed that the most potent was 25(OH)L3 with an IC50 of 0.5 µM followed by 1,25(OH)2D3 and 20S(OH)L3, which had an IC50 of 1 µM (Supplemental Table S2).
Overall, these results provide strong support for the ability of D3, L3, and 7-DHC hydroxy-metabolites to reduce the viral load in infected cells or the blood stream. A deficiency of these hydroxyometabolites may play a vital role in enabling the transition of patients with SARS-CoV-2 from becoming asymptomatic to symptomatic. For Mpro, 25(OH)D3, the major form of vitamin D3 present in blood, significant inhibition was observed at a concentration of 100 nM, which compares with a plasma concentration in nondeficient people typically between 50 to 100 nM (15). For RdRP, the IC50 for 25(OH)D3 was 1.3 µM, approximately one order of magnitude above its plasma concentration. For the hydroxylumisterols tested, plasma concentrations are unknown except for 20(OH)L3 where a value of 25 nM has been reported (17), but based on the enzymology, 25(OH)L3, which had the lowest IC50 (0.5 µM) for inhibition of RdRP, is likely to be substantially higher (15).
A plethora of reports strongly suggests that vitamin D plays a vital role in protection against SARS-COV-2, which includes preventing infected patients from developing severe disease. Here, we report for the first time that a range of vitamin D3-related compounds, including 7-DHC and L3 hydroxyderivaties, display anti-SARS-CoV-2 activities and we provide a possible target on which they may act directly. Vaccines against SARS-CoV-2 are clearly a major advance in controlling COVID-19; however, new viral variants emphasize the need for alternative therapeutic approaches. This study presents novel vitamin D and L3 metabolites as candidates for antiviral drugs.
SUPPLEMENTAL DATA
Supplemental Tables S1 and S2 and Supplemental Figs. S1 and S2: https://doi.org/10.6084/m9.figshare.14547747.v1
GRANTS
This study was supported by the National Institutes of Health Grants 1R01AR073004-01A1 and R01AR071189-01A1 (to A.T.S.), Veteran Administration merit Grants 1I01BX004293-01A1 (to A.T.S.) and R21 AI149267-01A1 (to C.R. and A.T.S.), and the Indian Council of Medical Research Grant ISRM/12(22)/2020 (to T.M.).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.Q., T.M., R.M.S., M.I.H., C.R., and A.T.S. conceived and designed research; S.Q., T.M., and M.I.H. performed experiments; S.Q., T.M., R.M.S., M.I.H., R.C.T., C.R., and A.T.S. analyzed data; S.Q., T.M., R.M.S., M.I.H., R.C.T., C.R., and A.T.S. interpreted results of experiments; S.Q., T.M., and M.I.H. prepared figures; S.Q., T.M., M.I.H., C.R., and A.T.S. drafted manuscript; S.Q., R.M.S., R.C.T., C.R., and A.T.S. edited and revised manuscript; S.Q., T.M., R.M.S., M.I.H., R.C.T., C.R., and A.T.S. approved final version of manuscript.
ACKNOWLEDGMENTS
T.M. and M.I.H. acknowledge the Indian Council of Medical Research, Government of India, for financial assistance (ISRM/12(22)/2020). We also acknowledge the support of National Institutes of Health Grants 1R01AR073004-01A1, R01AR071189-01A1 and of a Veteran Administration merit grant (No. 1I01BX004293-01A1) to A.T.S. and R21 AI149267-01A1) to C.R. and A.T.S.
REFERENCES
- 1.Maghbooli Z, Sahraian MA, Ebrahimi M, Pazoki M, Kafan S, Tabriz HM, Hadadi A, Montazeri M, Nasiri M, Shirvani A, Holick MF. Vitamin D sufficiency, a serum 25-hydroxyvitamin D at least 30 ng/mL reduced risk for adverse clinical outcomes in patients with COVID-19 infection. PLoS One 15: e0239799, 2020. doi: 10.1371/journal.pone.0239799. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Haussler MR, Jurutka PW, Mizwicki M, Norman AW. Vitamin D receptor (VDR)-mediated actions of 1α, 25 (OH) 2vitamin D3: genomic and non-genomic mechanisms. Best Pract Res Clin Endocrinol Metab 25: 543–559, 2011. doi: 10.1016/j.beem.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Grant WB, Al Anouti F, Moukayed M. Targeted 25-hydroxyvitamin D concentration measurements and vitamin D3 supplementation can have important patient and public health benefits. Eur J Clin Nutr 74: 366–376, 2020. doi: 10.1038/s41430-020-0564-0. [DOI] [PubMed] [Google Scholar]
- 4.Shirvani A, Kalajian TA, Song A, Holick MF. Disassociation of vitamin D’s calcemic activity and non-calcemic genomic activity and individual responsiveness: a randomized controlled double-blind clinical trial. Sci Rep 9: 17685, 2019. doi: 10.1038/s41598-019-53864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teymoori-Rad M, Shokri F, Salimi V, Marashi SM. The interplay between vitamin D and viral infections. Rev Med Virol 29: e2032, 2019. doi: 10.1002/rmv.2032. [DOI] [PubMed] [Google Scholar]
- 6.Bikle DD. Vitamin D: newer concepts of its metabolism and function at the basic and clinical level. J Endocr Soc 4: bvz038, 2020. doi: 10.1210/jendso/bvz038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quesada-Gomez JM, Entrenas-Castillo M, Bouillon R. Vitamin D receptor stimulation to reduce acute respiratory distress syndrome (ARDS) in patients with coronavirus SARS-CoV-2 infections: revised Ms SBMB 2020_166. J Steroid Biochem Mol Biol 202: 105719, 2020. doi: 10.1016/j.jsbmb.2020.105719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slominski RM, Stefan J, Athar M, Holick MF, Jetten AM, Raman C, Slominski AT. COVID-19 and Vitamin D: a lesson from the skin. Exp Dermatol 29: 885–890, 2020. doi: 10.1111/exd.14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhodes JM, Subramanian S, Laird E, Griffin G, Kenny RA. Perspective: vitamin D deficiency and COVID‐19 severity–plausibly linked by latitude, ethnicity, impacts on cytokines, ACE2 and thrombosis. J Intern Med 289: 97–115, 2021. doi: 10.1111/joim.13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coussens AK, Naude CE, Goliath R, Chaplin G, Wilkinson RJ, Jablonski NG. High-dose vitamin D3 reduces deficiency caused by low UVB exposure and limits HIV-1 replication in urban Southern Africans. Proc Natl Acad Sci USA 112: 8052–8057, 2015. doi: 10.1073/pnas.1500909112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mok CK, Ng YL, Ahidjo BA, Lee RCH, Loe MWC, Liu J, Tan KS, Kaur P, Chng WJ, Wong JEL. Calcitriol, the active form of vitamin D, is a promising candidate for COVID-19 prophylaxis. bioRxiv, 2020. doi: 10.1101/2020.06.21.16239. [DOI]
- 12.Holick MF. Vitamin D: a millenium perspective. J Cell Biochem 88: 296–307, 2003. doi: 10.1002/jcb.10338. [DOI] [PubMed] [Google Scholar]
- 13.Zu S, Deng Y-Q, Zhou C, Li J, Li L, Chen Q, Li X-F, Zhao H, Gold S, He J, Li X, Zhang C, Yang H, Cheng G, Qin C-F. 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor. Cell Res 30: 1043–1045, 2020. doi: 10.1038/s41422-020-00398-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slominski AT, Chaiprasongsuk A, Janjetovic Z, Kim TK, Stefan J, Slominski RM, Hanumanthu VS, Raman C, Qayyum S, Song Y, Song Y, Panich U, Crossman DK, Athar M, Holick MF, Jetten AM, Zmijewski MA, Zmijewski J, Tuckey RC. Photoprotective properties of vitamin D and lumisterol hydroxyderivatives. Cell Biochem Biophys 78: 165–180, 2020. doi: 10.1007/s12013-020-00913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuckey RC, Cheng CYS, Slominski AT. The serum vitamin D metabolome: what we know and what is still to discover. J Steroid Biochem Mol Biol 186: 4–21, 2019. doi: 10.1016/j.jsbmb.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slominski AT, Li W, Kim TK, Semak I, Wang J, Zjawiony JK, Tuckey RC. Novel activities of CYP11A1 and their potential physiological significance. J Steroid Biochem Mol Biol 151: 25–37, 2015. doi: 10.1016/j.jsbmb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slominski AT, Kim TK, Hobrath JV, Janjetovic Z, Oak ASW, Postlethwaite A, Lin Z, Li W, Takeda Y, Jetten AM, Tuckey RC. Characterization of a new pathway that activates lumisterol in vivo to biologically active hydroxylumisterols. Sci Rep 7: 11434, 2017. doi: 10.1038/s41598-017-10202-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368: 409–412, 2020. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai W, Zhang B, Jiang X-M, Su H, Li J, Zhao Y, Xie X, Jin Z, Peng J, Liu F, Li C, Li Y, Bai F, Wang H, Cheng X, Cen X, Hu S, Yang X, Wang J, Liu X, Xiao G, Jiang H, Rao Z, Zhang L-K, Xu Y, Yang H, Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368: 1331–1335, 2020. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, Atif SM, Hariprasad G, Hasan GM, Hassan MI. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol Basis Dis 1866: 165878, 2020. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA 100: 13190–13195, 2003. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lung J, Lin YS, Yang YH, Chou YL, Shu LH, Cheng YC, Liu HT, Wu CY. The potential chemical structure of anti‐SARS‐CoV‐2 RNA‐dependent RNA polymerase. J Med Virol 92: 693–697, 2020. [Erratum in J Med Virol 92: 2248, 2020]. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin W, Mao C, Luan X, Shen D-D, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, Chang S, Xie Y-C, Tian G, Jiang H-W, Tao S-C, Shen J, Jiang Y, Jiang H, Xu Y, Zhang S, Zhang Y, Xu HE. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368: 1499–1504, 2020. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammad T, Shamsi A, Anwar S, Umair M, Hussain A, Rehman MT, AlAjmi MF, Islam A, Hassan MI. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: towards the development of effective COVID-19 therapy. Virus Res 288: 198102, 2020. doi: 10.1016/j.virusres.2020.198102. [DOI] [PMC free article] [PubMed] [Google Scholar]