Abstract
Monitoring the genetic signal of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) through RNA titers in wastewater has emerged as a promising strategy for tracking community-scale prevalence of coronavirus disease 2019 (COVID-19). Although many studies of SARS-CoV-2 in wastewater have been conducted around the world, a uniform procedure for concentrating the virus in wastewater is lacking. The goal of this study was to comprehensively evaluate how different methods for concentrating the suspended solids in wastewater affect the associated SARS-CoV-2 RNA signal and the time required for processing samples for wastewater-based epidemiology efforts. We additionally consider the effects of sampling location in the wastewater treatment train (i.e., following preliminary or primary treatment), pasteurization, and RNA extraction method. Comparison of the liquid phase to suspended solids obtained via centrifugation or vacuum filtration suggests that the RNA signal of SARS-CoV-2 preferentially occurs in the solids. Therefore, we assert that the recovery of SARS-CoV-2 from wastewater should focus on suspended solids. Our data indicate that the measured SARS-CoV-2 signal is higher among samples taken from the primary clarifier effluent, as opposed to those taken after preliminary treatment. Additionally, we provide evidence that sample pasteurization at 60 °C for 90 min reduces the SARS-CoV-2 signal by approximately 50-55%. Finally, the results indicate that a magnetic bead approach to RNA extraction leads to a higher SARS-CoV-2 signal than does a silica membrane approach.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Wastewater-based epidemiology (WBE), Virus concentration
Graphical abstract
1. Introduction
The continuing coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has catalyzed the development of methods to monitor the prevalence of the disease. Many approaches utilize data obtained from individual clinical samples, but relying solely on such data provides an incomplete picture of COVID-19 prevalence in communities. Because individual COVID-19 testing is often dependent on voluntary participation and resource availability, cases of COVID-19, including asymptomatic and pre-symptomatic cases, likely are underreported (Wu et al., 2020b). Evidence has shown that SARS-CoV-2 can be detected in the feces of infected individuals, independent of the presence or absence of symptoms (Jiang et al., 2020; Xu et al., 2020). Additionally, recent studies have suggested that detectable concentrations of SARS-CoV-2 RNA in nasopharyngeal swabs can lag behind the onset of symptoms and the appearance of detectable concentrations of SARS-CoV-2 RNA in fecal samples (Lo et al., 2020). Because SARS-CoV-2 is shed fecally, researchers have heralded the use of wastewater-based epidemiology (WBE) during the COVID-19 pandemic (Venugopal et al., 2020), and many groups track the SARS-CoV-2 signal in wastewater as an indicator of COVID-19 in communities (Baldovin et al., 2021; Agrawal et al., 2021; Cao and Francis, 2021; Bivins et al., 2020a; Peccia et al., 2020). A uniform methodology for collecting and processing wastewater samples for SARS-CoV-2 RNA quantification is lacking among existing WBE studies; however, Pecson et al. (2021) notes that multiple methods can yield reproducible SARS-CoV-2 concentrations and that the chosen method should be used consistently at a given wastewater treatment plant.
WBE has been used previously for the successful monitoring of human pathogens including poliovirus, hepatitis A, and norovirus (Hellmér et al., 2014; Xagoraraki and O’Brien, 2019). Typical protocols for concentrating poliovirus from wastewater include an initial step to remove the suspended solids via centrifugation (which are retained for downstream analysis) while the supernatant is further processed (Lago et al., 2003). The supernatant is subject to a filtration or precipitation step to concentrate viral particles from the liquid phase prior to recombination with the suspended solids (Lago et al., 2003; Nakamura et al., 2015). Concentration of hepatitis A and norovirus from the liquid phase has been accomplished by using additives such as beef extract solution to elute the virus captured on a filter (Morace et al., 2002) or milk powder to precipitate the virus (Hellmér et al., 2014). Such existing viral protocols can be used to guide the development of effective methods for SARS-CoV-2 recovery from wastewater.
Current SARS-CoV-2 WBE studies vary widely in the method used to concentrate SARS-CoV-2 from wastewater. Quantifiable concentrations of SARS-CoV-2 RNA from wastewater have been obtained by polyethylene glycol (PEG) precipitation (Kocamemi et al., 2020; Hata et al., 2021), aluminum hydroxide precipitation (Randazzo et al., 2020), centrifugal ultrafiltration (Jafferali et al., 2021; Baldovin et al., 2021), centrifugation (Westhaus et al., 2021), and ethanol, salt, and silica extraction (Whitney et al., 2021). Experiments to evaluate and compare viral signal recovery via different concentration methods have been conducted using surrogate enveloped viruses such as murine hepatitis virus (Ahmed et al., 2020b), as well as porcine epidemic diarrhea virus and mengovirus (Randazzo et al., 2020). Several studies have specifically evaluated the SARS-CoV-2 signal obtained via different concentration methods (LaTurner et al., 2021; Forés et al., 2021; Barril et al., 2021). However, none of these studies examined centrifugation as a stand-alone method to concentrate the SARS-CoV-2 signal in wastewater samples; rather, for some of the methods tested, a centrifugation step was utilized to remove suspended solids from samples prior to the capture of the SARS-CoV-2 RNA signal from the liquid phase. Thus, the aforementioned studies cannot be used to select a method specifically for the recovery of SARS-CoV-2 RNA from the suspended solids of wastewater.
Current SARS-CoV-2 WBE studies also vary with respect to the sampling location within the wastewater treatment train, the use of sample pasteurization, and the RNA extraction procedure. Several studies have been conducted using untreated wastewater (Ahmed et al., 2020a; Hata et al., 2021), but others focus on primary effluent (Balboa et al., 2021), secondary effluent (Randazzo et al., 2020), or primary sludge (Peccia et al., 2020). Randazzo et al. (2020) utilized unpasteurized samples while Wu et al. (2020a) and Weidhaas et al. (2021) utilized samples that had been pasteurized at 60 °C for 90 min and 65 °C for 60 min, respectively. Bivins et al. (2020b) assessed the reduction of infectious SARS-CoV-2 resulting from sample pasteurization; however, that study used a different pasteurization temperature and aliquot size as compared to those used by Wu et al. (2020a) and Weidhaas et al. (2021), and that study did not assess the effect of pasteurization on the SARS-CoV-2 RNA signal. Finally, RNA extraction methods have varied among SARS-CoV-2 WBE studies. For example, some studies have utilized automated magnetic processes (Kocamemi et al., 2020) while others have utilized a silica membrane approach (Ahmed et al., 2020a). To date, there have been few studies (Pérez-Cataluña et al., 2021) assessing the effect of RNA extraction method on SARS-CoV-2 RNA signal.
The current study provides a direct comparison among wastewater sample collection points and processing methods. In particular, the study focuses on the (1) viral concentration step, with additional consideration of the (2) sample collection point within the wastewater treatment train, (3) pasteurization of the sample, and (4) RNA extraction method. These results are intended to select a streamlined workflow that produces a reproducible SARS-CoV-2 signal from wastewater samples.
2. Materials and methods
2.1. Wastewater collection and characterization
One-liter, flow-weighted, 24-h composite samples were collected from two Austin, Texas, wastewater treatment plants. Six samples were taken between November and December 2020 from the approximately 75 million gallons per day (MGD) South Austin Regional (SAR) wastewater treatment plant, and 14 samples were taken between May 2020 and January 2021 from the approximately 60 MGD Walnut Creek (WAL) wastewater treatment plant. Using data provided by Austin Water, the influent compositions of the SAR and WAL wastewater treatment plants from 2020 were compared (minimum of 58 samples for each analyte). An independent sample t-test was conducted on alkalinity, 5-day biochemical oxygen demand (BOD5), and ammonia (NH3), and a non-parametric Mann-Whitney U test was conducted on chemical oxygen demand (COD) and total suspended solids (TSS). All statistical tests were performed in RStudio (Boston, Massachusetts).
Plant influent (INF) samples were collected following the bar screen, and primary clarifier effluent (PCE) samples were collected following the primary clarifier. Samples were transported on ice to the laboratory, mixed, and divided into 50-mL aliquots.
We did not utilize recovery of an exogenous viral spike for normalization of SARS-CoV-2 in our samples. Feng et al. (2021) found that the recovery of exogenous spikes of the bovine coronavirus did not correlate with the recovery of SARS-CoV-2. Furthermore, Forés et al. (2021) - who found that the recovery of spiked murine hepatitis virus and SARS-CoV-2 did not vary in the same manner between two concentration methods - suggests that testing real environmental samples for native SARS-CoV-2 is a better way to assess various concentration methods.
2.2. Sample pasteurization
All 50-mL aliquots, except those used as non-pasteurized controls, were pasteurized in a water bath at 60 °C for 90 min before concentration and RNA extraction. A comparison of unpasteurized and pasteurized samples was conducted on triplicate 50-mL aliquots of six PCE samples from the SAR wastewater treatment plant. These aliquots were concentrated via centrifugation, and the pellet was extracted using ThermoFisher's MagMax Microbiome Ultra Nucleic Acid Isolation kit (Waltham, Massachusetts). A similar analysis was conducted using one PCE and one INF sample from the WAL wastewater treatment plant. These aliquots were concentrated via centrifugation, and the pellet was extracted using QIAGEN's RNeasy PowerMicrobiome kit (Hilden, Germany).
2.3. Sample concentration and homogenization
2.3.1. Concentration procedure
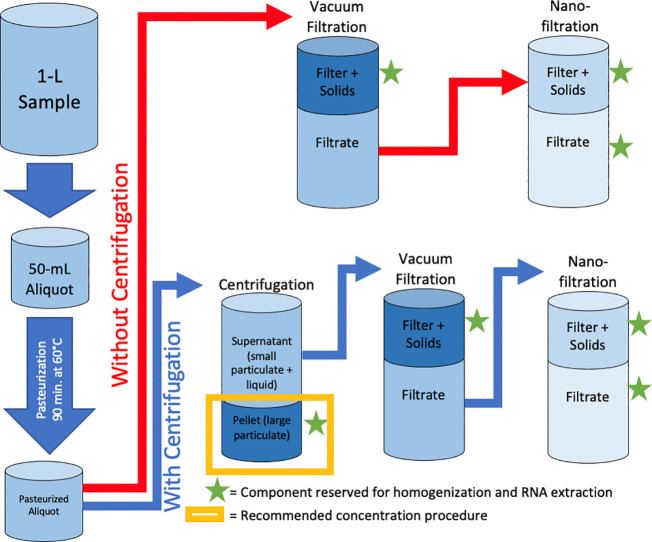
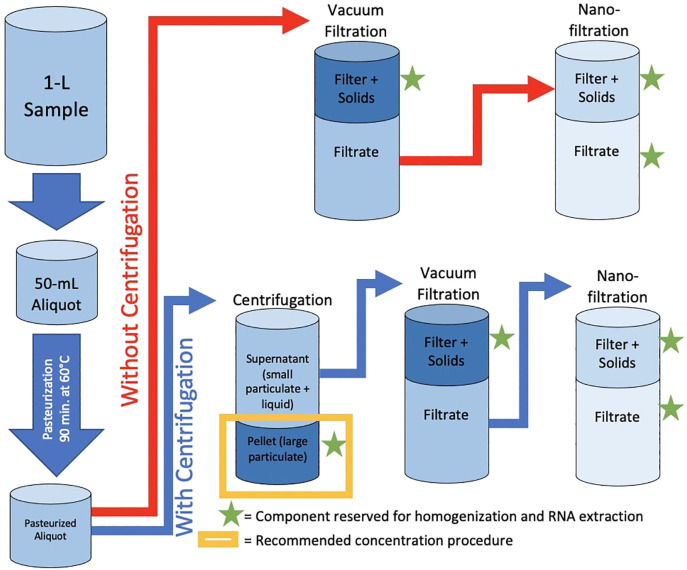
Two wastewater concentration pathways (summarized in Fig. 1 ) were modified from methods previously described in the literature (Ahmed et al., 2020a). The modified methods are described as follows, where sub-samples were reserved for SARS-CoV-2 analysis at each step of the method (noted by green stars in Fig. 1):
Fig. 1.
Schematic summarizing the two tested SARS-CoV-2 concentration pathways: one with centrifugation and one without centrifugation, both followed by vacuum filtration and nanofiltration.
For concentration without centrifugation, triplicate 50-mL aliquots of wastewater were subjected to vacuum filtration through 0.45-μm pore-size, 47-mm diameter, electronegative membranes (HAWP04700; Merck Millipore Ltd., Sydney, Australia). The membrane and captured solids were combined in a lysis tube and reserved for homogenization and RNA extraction. This concentration method is hereafter referred to as “vacuum filtration without centrifugation”. Then, the filtrate was subjected to high-pressure (45 psi) filtration through a 1-nm pore-size, 43-mm diameter, hydrophobic nanofilter (FilmTec NF270; DuPont, Wilmington, Delaware) using an Amicon stirred cell (UFSC05001; Merck KGaA, Darmstadt, Germany). The nanofilter and up to 1.5-mL of retentate were combined in a lysis tube and reserved for homogenization and RNA extraction. This component is hereafter referred to as “nanofilter”. A 1-mL aliquot of the nanofilter filtrate was reserved for homogenization and RNA extraction. This component is hereafter referred to as “nanofilter filtrate”.
For concentration with centrifugation, triplicate 50-mL aliquots of wastewater were subjected to centrifugation at 4 °C, 4,500 xg for 5 min to pellet large, suspended solids from the sample. Following centrifugation, the majority of the supernatant was decanted and reserved. The remaining supernatant (4-7 mL) was used to resuspend the pellet via gentle inversion of the tube. The resuspended pellet was transferred to a 15-mL centrifuge tube and centrifuged at 4 °C, 4,500 xg for 5 min. Following this centrifugation, all supernatant was decanted and combined with the previously reserved supernatant. Then, the centrifuged supernatant (a total of approximately 49 mL) was vacuum-filtered through the above-described electronegative membrane. The membrane and captured solids were combined in a lysis tube and reserved for homogenization and RNA extraction. This concentration method is hereafter referred to as “vacuum filtration with centrifugation”. Similar to the concentration without centrifugation procedure, high-pressure nanofiltration was conducted on the filtrate from vacuum filtration; this produced “nanofilter” and “nanofilter filtrate” components that were reserved for homogenization and RNA extraction. The aforementioned pellet was resuspended in the lysis buffer included in each of the extraction kits listed below (containing guanidine thiocyanate) and reserved for homogenization and RNA extraction. This concentration method is hereafter referred to as “centrifugation”.
For both concentration pathways (with and without centrifugation), all reserved components were stored in 2-mL lysis tubes at -80 °C until homogenization and lysis.
2.3.2. Homogenization and lysis
Three lysing matrices were used, depending on the sample concentration method and the downstream RNA extraction method. The lysing matrix consisting of 0.56- to 0.7-mm garnet flakes (MP Biomedicals, Santa Ana, California) generally was used for samples concentrated via filtration to encourage membrane break up. The garnet lysing matrix provided the sharp edges necessary for tearing the membrane so that solids trapped on the membrane could be accessed. The lysing matrix consisting of 0.1-mm glass beads provided in the RNeasy PowerMicrobiome kit was used for solids concentrated via centrifugation prior to extraction via the RNeasy kit. The lysing matrix consisting of zirconia beads provided in the MagMax Ultra Nucleic Acid Isolation kit was used for solids concentrated via centrifugation prior to extraction via the MagMax kit.
Prior to homogenization and lysis, frozen concentrates were thawed at room temperature (~20 °C) for approximately 30 min. Then, if not done previously for pellet resuspension, an appropriate amount of lysis buffer (as directed by the RNA extraction kit) was added to each tube. An aliquot (6.5 μL) of β-mercaptoethanol was added to concentrates that were to be processed with the RNeasy PowerMicrobiome kit, per the manufacturer's instructions. All lysis tubes were subjected to 4-6 alternating rounds of homogenization using MP Biomedicals’ FastPrep-24 (4.0 m/s for 20 s) and centrifugation (13,000 xg for 15-20 s).
2.4. RNA extraction
Following the final round of homogenization, lysis tubes were centrifuged at 13,000 xg for 1 min. The supernatant was decanted and immediately subjected to RNA extraction using QIAGEN's RNeasy PowerMicrobiome kit (a silica membrane approach) or ThermoFisher's MagMax Microbiome Ultra Nucleic Acid Isolation kit (a magnetic bead approach), following the respective manufacturer's instructions. Extracts were not treated with DNase. Extraction using the MagMax kit was automated using ThermoFisher's KingFisher Flex system. RNA extracts were stored at -20 °C prior to analysis via reverse transcription, quantitative, real-time polymerase chain reaction (RT-qPCR).
2.5. RT-qPCR
The concentration of SARS-CoV-2 in each RNA extract was measured in triplicate on a ViiA7 Real-Time PCR System (ThermoFisher). RT-qPCR targeted the nucleocapsid gene of SARS-CoV-2 using the CDC nCOV_N1 primer/probe set (Integrated DNA Technologies [IDT], Coralville, Iowa). Standard curves were developed using the plasmid 2019-nCoV_N_Positive Control (IDT) at concentrations of 20,000, 2,000, 200, 20, and 2 N gene copies/μL. RT-qPCR reactions (20 μL) contained 5 μL of RNA extract, 5 μL of TaqMan™ Fast Virus 1-Step Master Mix (ThermoFisher), 1.49 μL of the CDC nCOV_N1 primer/probe set mix (IDT), and 8.51 μL of PCR-grade water. The concentration of each primer in the reaction was 500 nM, and the concentration of probe in the reaction was 125 nM. RT-qPCR was run under the following conditions, per the CDC's recommendations: 50 °C for 5 min, 95 °C for 20 s, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. The limit of detection (LOD) was determined by using six serial dilutions of the plasmid 2019-nCoV_N_Positive Control SARS-CoV-2 standard (from 100,000 to 1 copy per reaction). After narrowing the initial range, 20 replicates of standards with 10, 5, 2.5, and 1 copy per reaction were analyzed. Following the procedure of Klymus et al. (2019), the LOD for the N1 assay was calculated as 10.6 N gene copies per reaction when samples are analyzed in triplicate. For our final, selected workflow (50-mL wastewater sample and RNA extraction into 200 μL of elution buffer), this translates to an LOD of 8,480 N gene copies/L of wastewater.
Similar to the method of Pecson et al. (2021), only RT-qPCR reactions that produced results above the LOD were included in downstream analysis; if one or two of the triplicate RT-qPCR reactions for a wastewater aliquot produced results below the LOD, those were removed from the analysis. Care was taken to ensure the quality of the results: all RT-qPCR reactions were performed in triplicate, and positive controls (2019-nCoV_N_Positive Control, IDT) and negative controls (PCR-grade water) were used in every RT-qPCR plate. All negative controls indicated an absence of SARS-CoV-2 RNA in the materials and reagents used.
2.6. Statistical analysis
Triplicate RT-qPCR measurements were used to calculate the average SARS-CoV-2 concentration in each RNA extract in units of N gene copies/μL extract. The volume of wastewater concentrated, the volume of eluent used during the RNA extraction, and the extract's SARS-CoV-2 concentration were used to calculate the concentration of SARS-CoV-2 in units of N gene copies/L wastewater.
Each variation of the sample processing method was performed on triplicate aliquots of the same wastewater sample. For comparison across sample processing methods, the values of SARS-CoV-2 concentrations across triplicate aliquots were averaged, and the standard deviation was calculated. The coefficient of variation (CV) was calculated by dividing the standard deviation by the average SARS-CoV-2 concentration across the triplicate aliquots. When applicable, a Wilcoxon signed-rank test was performed in MatLab (MathWorks, Natick, Massachusetts) at a 95% confidence level to determine if two sample processing methods yielded significantly different SARS-CoV-2 concentrations.
2.7. Selected workflow
After examining the effects of the sampling location within the wastewater treatment train, pasteurization, concentration protocol, and RNA extraction method on the SARS-CoV-2 RNA signal, a workflow was selected to increase both the magnitude and reproducibility of the SARS-CoV-2 concentration measured in wastewater samples while decreasing the time required for processing samples. The selected workflow includes 1) sampling of PCE, 2) pasteurization of triplicate 50-mL aliquots at 60 °C for 90 min, 3) concentration via centrifugation, and 4) automated RNA extraction using ThermoFisher's MagMax kit and KingFisher Flex system.
The selected workflow is summarized as follows: triplicate 50-mL aliquots of PCE were pasteurized at 60 °C for 90 min and centrifuged at 4 °C, 4,500 xg for 5 min. A volume of 43-46 mL of supernatant was decanted and discarded, while the remaining supernatant (4-7 mL) was reserved and used for resuspension of pelleted solids via gentle inversion. The supernatant and resuspended solids were transferred to a 15-mL centrifuge tube and centrifuged at 4 °C, 4,500 xg for 5 min. The supernatant was decanted, and the pelleted solids were resuspended in lysis buffer provided in the MagMax kit. This suspension was transferred to 2-mL zirconia bead lysis tubes included in the MagMax kit. Lysis tubes were subjected to four alternating rounds of homogenization using MP Biomedicals’ FastPrep-24 (4.0 m/s for 20 s) and centrifugation (13,000 xg for 15-20 s). Samples were then subjected to automated RNA extraction using ThermoFisher's MagMax kit and KingFisher Flex system, following the manufacturer's instructions.
3. Results and discussion
3.1. Wastewater characterization
As shown in Table 1 , the mean values between the influent to the SAR and WAL wastewater treatment plants are significantly different for alkalinity, NH3-N, COD, and TSS. However, the mean values between the two influents are not significantly different for BOD5.
Table 1.
Influent composition for the South Austin Regional (SAR) and Walnut Creek (WAL) wastewater treatment plants in 2020. The average (standard deviation) are shown. Significant differences between the mean values of an analyte at the two wastewater treatment plants are indicated with a * by the p-value.
| TSS (mg/L) | NH3-N (mg/L) | Alkalinity (mg/L as CaCO3) | COD (mg/L) | BOD5 (mg/L) | |
|---|---|---|---|---|---|
| SAR | 260 (76.2) | 34.3 (5.28) | 283 (19.5) | 501 (118) | 248 (45.2) |
| WAL | 220 (83.3) | 46.5 (5.25) | 252 (19.9) | 385(78.2) | 235 (62.8) |
| p-value | <4.113e-6* | <2.2e-16* | <2.2e-16* | <2.2e-16* | p = 0.137 |
3.2. Sample concentration and homogenization effects
A preliminary experiment was conducted using one PCE and one INF sample collected from the WAL wastewater treatment plant on the same date. The SARS-CoV-2 concentrations measured from the reserved components (Fig. 1) of the PCE and INF samples are shown in SI Figs. 1-3. This experiment indicated unreliable or no recovery of SARS-CoV-2 signal from the nanofilter of PCE and INF samples (SI Figs. 1 and 2) when samples were concentrated with or without centrifugation. Additionally, no SARS-CoV-2 signal was detected in the nanofilter filtrate of PCE and INF samples concentrated with or without centrifugation, as indicated by triplicate non-detect RT-qPCR results for all aliquots processed in this way. The preferential occurrence of SARS-CoV-2 RNA in association with the solid components of wastewater samples (Li et al., 2021), as opposed to the liquid phase, emphasized the need to maximize the capture of SARS-CoV-2 from the suspended solids. Thus, our subsequent analysis focused on determining if concentration via centrifugation or vacuum filtration without centrifugation would be a more suitable and time-efficient method for our final workflow.
A comparison of the total SARS-CoV-2 signal recovered from all reserved components with and without centrifugation indicated that signal loss occurs when concentrating via centrifugation (SI Fig. 3). Because the sharp edges of the garnet lysing matrix were not expected to be necessary when a filter membrane was not in the lysis tube, a trial was conducted using the glass bead lysing matrix on samples concentrated via centrifugation (i.e., producing a pellet). Additionally, evidence indicates that enveloped viruses might be sensitive to shear force (Kim and Lim, 2017), so it was hypothesized that the garnet lysing matrix might be unnecessarily harsh on samples concentrated via centrifugation. The results (SI Fig. 4) indicate that more gentle lysing matrices (i.e., glass beads rather than garnet) should be used when homogenizing samples concentrated via centrifugation for SARS-CoV-2 detection.
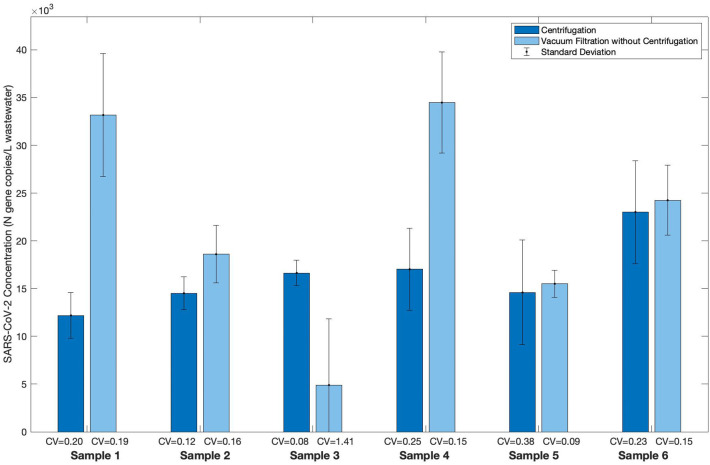
A further comparison of concentration via centrifugation and vacuum filtration without centrifugation was conducted using three PCE samples taken from the WAL wastewater treatment plant (SI Fig. 5) and six PCE samples taken from the SAR wastewater treatment plant (Fig. 2 ). The garnet lysing matrix was used for concentrates obtained via vacuum filtration without centrifugation. The lysing matrix included in the extraction kit was used for concentrates obtained via centrifugation; specifically, RNA extraction of WAL wastewater concentrates was performed using the RNeasy PowerMicrobiome kit with the glass bead lysing matrix, while RNA extraction of the SAR wastewater concentrates was performed using the MagMax Ultra Nucleic Acid Isolation kit with the zirconia bead lysing matrix. A Wilcoxon signed-rank test of the SARS-CoV-2 concentrations from the SAR samples indicates that there was no significant difference between the SARS-CoV-2 concentration from a sample concentrated via centrifugation to that concentrated via vacuum filtration without centrifugation (p = 0.219). Additionally, the median CVs for the two concentration techniques (Fig. 2) are similar: 0.22 (centrifugation) and 0.16 (vacuum filtration without centrifugation); these CVs compare favorably to those from other viral concentration protocols in the literature, indicating method reproducibility: 0.21 for SARS-CoV-2 concentrated with magnesium chloride addition and vacuum filtration (Feng et al., 2021), 0.15 and 0.39 for bovine coronavirus with direct extraction and concentrated with PEG, respectively (LaTurner et al., 2021), and 0.58-0.68 for MS2 and murine hepatitis virus concentrated by ultrafiltration methods (Forés et al., 2021). Furthermore, the SARS-CoV-2 concentrations from the WAL samples indicate that concentration via centrifugation, as compared to vacuum filtration without centrifugation, yielded fewer non-detect RT-qPCR results during a period of relatively low COVID-19 prevalence (Austin Public Health, 2020).
Fig. 2.
Concentrating SARS-CoV-2 from wastewater samples via centrifugation or vacuum filtration without centrifugation. WWTP: South Austin Regional (SAR); Sampling location: Primary Clarifier Effluent (PCE); Sample details: six composite samples were retrieved on six different dates between November and December 2020; Pasteurization: yes; Concentration Method: centrifugation vs. vacuum filtration without centrifugation; RNA Extraction Method: MagMax with zirconia lysis beads for samples concentrated via centrifugation or garnet lysis beads for samples concentrated via vacuum filtration without centrifugation. Samples concentrated via centrifugation were processed using the final selected workflow. Each column represents the average of triplicate aliquots processed for a sample, where triplicate RT-qPCR reactions were run for each aliquot; error bars represent the standard deviation among the aliquots; the coefficient of variation (CV) normalizes the standard deviation to the average SARS-CoV-2 concentration.
For routine monitoring, the selected workflow for concentrating SARS-CoV-2 from wastewater should consider the time required for sample processing. Concentration via centrifugation is preferable to concentration via vacuum filtration because it requires less processing time. For example, processing ten wastewater samples in triplicate would take approximately four hours for concentration via vacuum filtration without centrifugation (assuming an ability to run three filter units in parallel) but only 30 min for concentration via centrifugation.
3.3. Sample collection point effects
The effect of sample collection point in the wastewater treatment train on the SARS-CoV-2 signal was assessed using two sets of paired PCE and INF samples from both the WAL and SAR wastewater treatment plants. PCE and INF samples taken on the same day at the same plant were concentrated from triplicate 50-mL aliquots via centrifugation. An examination of the SAR data (Fig. 3 ) indicates that PCE samples yielded higher average concentrations of SARS-CoV-2 than did INF samples. Additionally, two of the aliquots from both SAR wastewater INF samples yielded non-detect RT-qPCR results. An examination of the WAL data (SI Fig. 6) indicates that INF samples yielded higher average concentrations of SARS-CoV-2 than did PCE samples; however, the CV of INF samples were higher than those of PCE samples, indicating better reproducibility with PCE samples. Based on the SAR and WAL data, PCE was selected for the final workflow to decrease the likelihood of non-detects in RT-qPCR and to improve reproducibility.
Fig. 3.
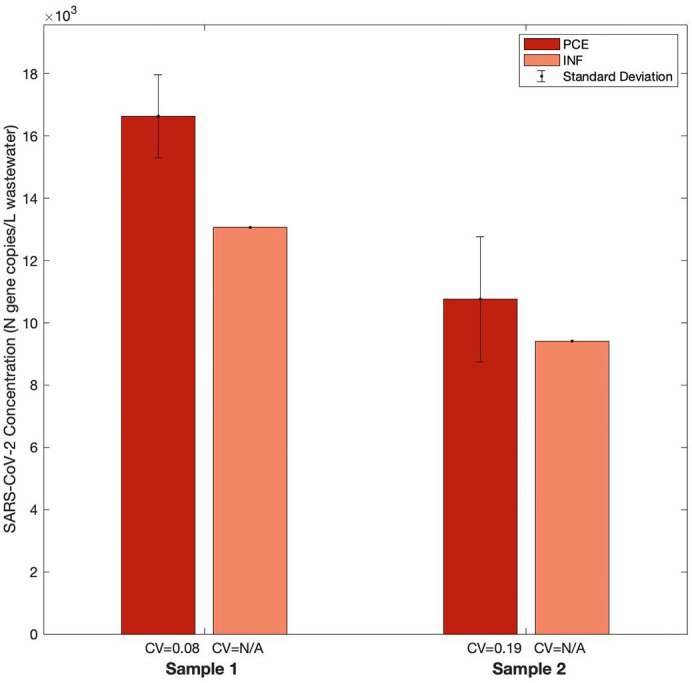
Concentrating SARS-CoV-2 from wastewater samples taken following primary treatment (PCE) or preliminary treatment (INF). WWTP: South Austin Regional (SAR); Sampling location: Primary Clarifier Effluent (PCE) and Influent (INF); Sample details: two pairs of PCE and INF composite samples were retrieved on two different dates between November and December 2020; Pasteurization: yes; Concentration Method: centrifugation; RNA Extraction Method: MagMax with zirconia lysis beads. PCE samples were processed using the final selected workflow. Each column represents the average of triplicate aliquots processed for a sample, where triplicate RT-qPCR reactions were run for each aliquot; error bars represent the standard deviation among the aliquots; the coefficient of variation (CV) normalizes the standard deviation to the average SARS-CoV-2 concentration. A CV value noted as “N/A” indicates that two of the triplicate aliquots processed yielded triplicate non-detect RT-qPCR results.
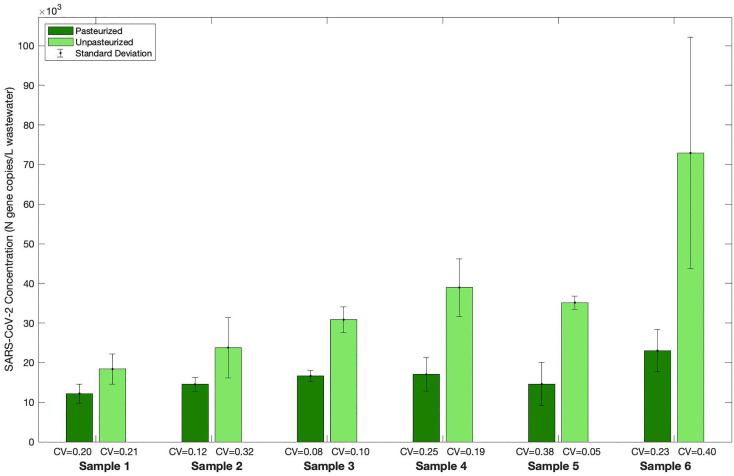
3.4. Sample pasteurization effects
The use of pasteurization in WBE studies for SARS-CoV-2 varies (Pecson et al., 2021), and its use depends on the safety requirements of the organization or the comfort-level of the principal investigator and/or laboratory personnel. Thus, the effect of sample pasteurization was assessed using one PCE and one INF sample taken from the WAL wastewater treatment plant (SI Fig. 7) and six PCE samples taken from the SAR wastewater treatment plant (Fig. 4 ). A Wilcoxon signed-rank test of the SARS-CoV-2 concentrations from the SAR samples indicates that pasteurization (90 min at 60 °C) yielded a significantly lower SARS-CoV-2 signal (p = 0.0312), with an average decrease of approximately 50% as compared to no pasteurization. The CV values of pasteurized and unpasteurized SAR samples were similar (0.08-0.38 and 0.05-0.40, respectively). A similar decrease in average SARS-CoV-2 concentrations (55%) was observed in pasteurized as compared to unpasteurized WAL samples. Additionally, pasteurized WAL samples had lower CV values than did unpasteurized samples. Because pasteurization of wastewater reduces the potential risk to laboratory personnel from a variety of potentially infectious agents, a SARS-CoV-2 signal loss of 50-55% might be acceptable to a given lab. However, Pecson et al. (2021) determined that pasteurization of 60 min at 60 °C did not result in a statistically significant loss of SARS-CoV-2 signal. Thus, for those labs who want or are required to pasteurize wastewater samples prior to processing for SARS-CoV-2 RNA, 60 min at 60 °C might be considered.
Fig. 4.
Concentrating SARS-CoV-2 from wastewater samples with and without pasteurization at 60 °C for 90 min. WWTP: South Austin Regional (SAR); Sampling location: Primary Clarifier Effluent (PCE); Sample details: six composite samples were retrieved on six different dates between November and December 2020; Pasteurization: some samples were pasteurized (see legend); Concentration Method: centrifugation; RNA Extraction Method: MagMax with zirconia lysis beads. Pasteurized samples were processed using the final selected workflow. Each column represents the average of triplicate aliquots processed for a sample, where triplicate RT-qPCR reactions were run for each aliquot; error bars represent the standard deviation among the aliquots; the coefficient of variation (CV) normalizes the standard deviation to the average SARS-CoV-2 concentration.
3.5. RNA extraction
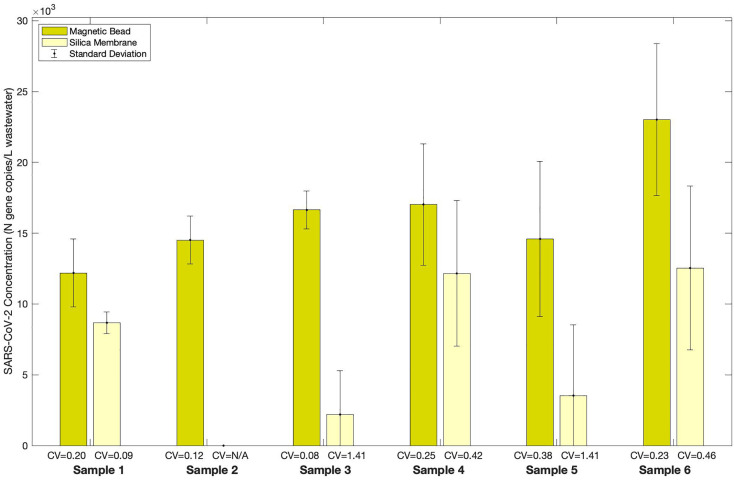
We compared the SARS-CoV-2 signal after RNA extraction utilizing a silica membrane approach and a micro-spherical magnetic bead approach using six PCE samples from the SAR wastewater treatment plant (Fig. 5 ). As compared to Pérez-Cataluña et al. (2021), who found no statistically significant difference in SARS-CoV-2 concentrations when extracted with a manual, column-based RNA extraction approach versus an automated, magnetic bead approach, a Wilcoxon signed-rank test of the SARS-CoV-2 concentrations from our SAR samples indicates that the magnetic bead approach yielded significantly higher SARS-CoV-2 concentrations than did the silica membrane approach (p = 0.0312). Additionally, samples extracted with the magnetic bead approach had a lower median CV (0.22) as compared to the silica membrane approach (0.42). Because the magnetic bead approach to RNA extraction can be automated, adopting this approach can decrease the personnel time required for routine SARS-CoV-2 WBE monitoring.
Fig. 5.
Extracting SARS-CoV-2 from wastewater using a magnetic bead or silica membrane approach. WWTP: South Austin Regional (SAR); Sampling location: Primary Clarifier Effluent (PCE); Sample details: six composite samples were retrieved on six different dates between November and December 2020; Pasteurization: yes; Concentration Method: centrifugation; RNA Extraction Method: MagMax with zirconia lysis beads (magnetic bead approach) and RNeasy with glass lysis beads (silica membrane approach). Samples that were extracted using a magnetic bead approach were processed using the final selected workflow. Each column represents the average of triplicate aliquots processed for a sample, where triplicate RT-qPCR reactions were run for each aliquot; error bars represent the standard deviation among the aliquots; the coefficient of variation (CV) normalizes the standard deviation to the average SARS-CoV-2 concentration. A CV value noted as “N/A” indicates that all of the triplicate aliquots processed yielded triplicate non-detect RT-qPCR results.
4. Conclusion
This study utilized wastewater samples from two wastewater treatment plants to directly compare SARS-CoV-2 RNA concentration as a function of wastewater sample collection point and subsequent processing methods. The preferential occurrence of SARS-CoV-2 viral particles in the suspended solids of wastewater emphasizes the need for a concentration method focused on the suspended solids. SARS-CoV-2 WBE workflows that include a solids removal step (e.g., filtration or centrifugation) prior to processing the liquid phase for SARS-CoV-2 detection are unlikely to achieve complete capture of the SARS-CoV-2 signal. We recommend that the suspended solids of wastewater samples be captured via centrifugation and analyzed for SARS-CoV-2 signal. The data produced in this study indicate that concentration via centrifugation, when paired with an appropriate homogenization method, yields a comparable SARS-CoV-2 signal as compared to the other concentration methods tested. While workflows that include concentration via vacuum filtration are common, we argue that a concentration via centrifugation approach is more time efficient and yields similar SARS-CoV-2 signal recovery.
The results of this study indicate that PCE samples should be utilized to improve the reproducibility of the SARS-CoV-2 signal, particularly at low concentrations. Additionally, given the reduced time required for extraction and the increased SARS-CoV-2 signal, a magnetic bead approach to RNA extraction is preferable to a silica membrane approach. Finally, although pasteurization of wastewater samples mitigates risk of infectious disease to laboratory personnel, this study provides evidence that pasteurization (60 °C for 90 min) leads to an average 50-55% decrease in SARS-CoV-2 RNA signal.
CRediT authorship contribution statement
Emma J. Palmer: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Juan P. Maestre: Conceptualization, Methodology, Investigation, Writing – review & editing. David Jarma: Methodology, Investigation. Alisa Lu: Investigation, Writing – review & editing. Elisabeth Willmann: Investigation, Writing – review & editing. Kerry A. Kinney: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition. Mary Jo Kirisits: Conceptualization, Methodology, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
From Austin Water, we thank Lisa Boatman, Gary Gilmer, Trinity O'Neal, Robert Moss, and the lab personnel of the SAR and WAL wastewater treatment plants for providing data and samples. From the University of Texas at Austin, we thank Jim Walker, Eric Kong, and Suzanne Pierce for picking up wastewater samples, Chris Singh for his guidance in ensuring the safety of laboratory personnel during protocol development, and Paola Passalacqua for advice on statistical methods. We thank our WBE colleagues who shared protocols and other information; in particular, we thank Aaron Bivins from the University of Notre Dame for his advice on protocol development.
Funding sources
This work was supported by the University of Texas at Austin through the Fluor Centennial Teaching Fellowship in Engineering #1, the Office of the Vice President for Research, Whole Communities-Whole Health, and Planet Texas 2050 and the National Science Foundation through the Graduate Research Fellowship Program.
Editor: Jay Gan
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.149405.
Appendix A. Supplementary data
Supplementary figures
References
- Agrawal S., Orschler L., Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in Southern Germany. Sci. Rep. 2021;11(1):1. doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J., Choi P., Kitajima M., Simpson S., Li J., Tscharke B., Verhagen R., Smith W., Zaugg J., Dierens L., Hugenholtz P., Thomas K., Mueller J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B., Mueller J., Simpson S., Smith W., Symonds E., Thomas K., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin Public Health COVID-19 Dashboards. COVID-19 Dashboards|AustinTexas.gov. 2020. https://www.austintexas.gov/page/covid-19-dashboards
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F., Regueiro B., Lema J. The fate of SARS-CoV-2 in WWTPs points out the sludge line as a suitable spot for monitoring. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldovin T., Amoruso I., Fonzo M., Buja A., Baldo V., Cocchio S., Bertoncello C. SARS-CoV-2 RNA detection and persistence in wastewater samples: an experimental network for COVID-19 environmental surveillance in Padua, Veneto Region (NE Italy) Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barril P.A., Pianciola L.A., Mazzeo M., Ousset M.J., Jaureguiberry M.V., Alessandrello M., Sánchez G., Oteiza J.M. Evaluation of viral concentration methods for SARS-CoV-2 recovery from wastewaters. Sci. Total Environ. 2021;756 doi: 10.1016/j.scitotenv.2020.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Gurol Z.C., Chakraborty S., Costa F., Curcio S., de los Reyes F.L., Vela J.D., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Francis R. On forecasting the community-level COVID-19 cases from the concentration of SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS ES&T Water. 2021 doi: 10.1021/acsestwater.1c00160. [DOI] [Google Scholar]
- Forés E., Bofill-Mas S., Itarte M., Martínez-Puchol A., Hundesa A., Calvo M., Borrego C.M., Corominas L.L., Girones R., Rusiñol M. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration for wastewater. Sci. Total Environ. 2021;768 doi: 10.1016/j.scitotenv.2020.144786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Hara-Yamamura H., Meuchi Y., Imai S., Honda R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis a virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Luo M., Zou Z., Wang X., Chen C., Qiu J. Asymptomatic SARS-CoV-2 infected case with viral detection positive in stool but negative in nasopharyngeal samples lasts for 42 days. J. Med. Virol. 2020;92(10):1807. doi: 10.1002/jmv.25941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-H., Lim K.-I. Stability of retroviral vectors against ultracentrifugation is determined by the viral internal core and envelope proteins used for pseudotyping. Molecules and Cells. 2017;40(5):339. doi: 10.14348/molcells.2017.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymus K.E., Merkes C.M., Allison M.J., Goldberg C.S., Helbing C.C., Hunter M.E., Jackson C.A., Lance R.F., Mangan A.M., Monroe E.M., Piaggio A.J., Stokdyk J.P., Wilson C.C., Richter C.A. Reporting the limits of detection and quantification for environmental DNA assays. eDNA. 2019;2:271. [Google Scholar]
- Kocamemi B.A., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. medRxiv. 2020 doi: 10.1101/2020.05.12.20099358. [DOI] [Google Scholar]
- Lago P.M., Gary H.E., Pérez L.S., Cáceres V., Olivera J.B., Puentes R.P., Corredor M.B., Jímenez P., Pallansch M.A., Cruz R.G. Poliovirus detection in wastewater and stools following an immunization campaign in Havana, Cuba. Int. J. Epidemiol. 2003;32:772. doi: 10.1093/ije/dyg185. [DOI] [PubMed] [Google Scholar]
- LaTurner Z.W., Zong D.M., Kalvapalle P., Reyes-Gamas K., Terwilliger A., Crosby T., Ali P., Avadhanula V., Santos H.H., Weesner K., Hopkins L., Piedra P.A., Maresso A.W., Stadler L.B. Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Di D.Y.W., Saingam P., Jeon M.K., Yan T. Fine-scale temporal dynamics of SARS-CoV-2 RNA abundance in wastewater during a COVID-19 lockdown. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I., Lio C., Cheong H., Lei C., Cheong T., Zhong X., Tian Y., Sin N. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int. J. Biol. Sci. 2020;16:1698. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morace G., Aulicino F., Costanzo L., Donadio F., Rapicetta M. Microbial quality of wastewater: detection of hepatitis a virus by reverse transcriptase-polymerase chain reaction. J. Appl. Microbiol. 2002;92(5):828. doi: 10.1046/j.1365-2672.2002.01587.x. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Hamasaki M., Yoshitomi H., Ishibashi T., Yoshiyama C., Maeda E., Sera N., Yoshida H. Environmental surveillance of poliovirus in sewage water around the introduction period for inactivated polio vaccine in Japan. Appl. Environ. Microbiol. 2015;81:1859. doi: 10.1128/AEM.03575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: finding from an interlaboratory methods evaluation in the U.S. Environ. Sci.: Water Res. Technol. 2021;7:504. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Cuevas-Ferrando E., Randazzo W., Falcó I., Allende A., Sánchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;758 doi: 10.1016/j.scitotenv.2020.143870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal A., Ganesan H., Raja S.S.S., Govindasamy V., Arunachalam M., Narayanasamy A., Sivaprakash P., Rahman P.K.S.M., Gopalakrishnan A.V., Siama Z., Vellingiri B. Novel wastewater surveillance strategy for early detection of coronavirus disease 2019 hotspots. Curr. Opin. Environ. Sci. Health. 2020;17:8. doi: 10.1016/j.coesh.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J.V., LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany - suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney O.N., Kennedy L.C., Fan V.B., Hinkle A., Kantor R., Greenwald H., Crits-Christoph A., Al-Shayeb B., Chaplin M., Maurer A.C., Tjian R., Nelson K.L. Sewage, salt, silica, and SARS-CoV-2 (4S): an economical kit-free method for direct capture of SARS-CoV-2 RNA from wastewater. Environ. Sci. Technol. 2021;55(8):4880. doi: 10.1021/acs.est.0c08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A., Erickson T., Chai P., Thompson J., Alm E. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4):614. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Mertens A., Crider Y., Nguyen A., Pokpongkiat N., Djajadi S., Seth A., Hsiang M., Colford J., Reingold A., Arnold B., Hubbard A., Benjamin-Chung J. Substantial underestimation of SARS-CoV-2 infection in the United States. Nat. Commun. 2020;11:4507. doi: 10.1038/s41467-020-18272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xagoraraki I., O’Brien E. Women in Water Quality. Vol. 75. 2019. Wastewater-based epidemiology for early detection of viral outbreaks. [Google Scholar]
- Xu Y., Li X., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020;26:502. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures