Abstract
This protocol was developed to functionalize styrene maleic acid (SMA) by direct fluorescent labeling in an easy way, accessible to biochemistry laboratories. This novel method is based on the coupling of carboxylic acids to primary amines using a carbodiimide, a reaction commonly used for protein chemistry. The procedure uses the hydrolyzed styrene-maleic acid copolymer and occurs entirely in aqueous solution with mild conditions compatible with many biomolecules.
Keywords: SMALPs, Membrane solubilization, Nanodiscs, Fluorescent labeling, Nanodiscs modification
Background
Characterization of membrane proteins in-vitro can be very challenging (Grisshammer and Tate, 1995). In addition to difficulties with over-expression and membrane isolation, membrane proteins need to be extracted from their native environment. The necessary solubilization step commonly requires the use of detergent to replace the native lipid environment, this can often lead to the loss of structure and/or activity of the membrane protein ( Duquesne et al., 2016 ). For this reason, several alternative options such as amphipols (Popot, 2010) have been developed to avoid some of the difficulties associated with detergents and maintain the solubility of membrane protein. A few years ago Styrene Maleic Acid (SMA) copolymer became more frequently used as an alternative to low molecular weight detergent strategies ( Dorr et al., 2014 ; Jamshad et al., 2015 ; Prabudiansyah et al., 2015 ). SMA has been shown to be able to spontaneously solubilize biological membranes and give disc-shape particles with an average size of 10 nm. These nanodiscs (called SMALPs) contain a mixture of protein embedded in lipids from membrane and SMA copolymer maintaining the particle in solution ( Knowles et al., 2009 ). SMALPs are compatible with most common biochemical approaches such as affinity purification or size exclusion chromatography. One important advantage of using SMA is the “almost native” environment they provide to membrane proteins, allowing preservation of key lipids often involved in maintaining membrane protein structure or function. Protein characterization sometimes requires labeled material, but fluorescent tags can also alter protein structure. Chemically modified SMA allows labeling of nanodiscs without protein modification and so could be of interest for various types of analysis. Previous studies have used labeled amphipols with different chemistries such as poly-histidine ( Giusti et al., 2015 ); biotin ( Charvolin et al., 2009 ); and DNA oligonucleotides tags (Le Bon et al., 2014 ) to immobilize membrane proteins. Fluorescent-labeled SMA can be employed in a similar way with lipid environment being preserved allowing for the study of membrane proteins in a native-like environment using fluorescence correlation spectroscopy or energy transfer measurements.
This protocol aims to chemically modify SMA in solution based on the reaction coupling of carboxylic acids to primary amines using a carbodiimide, a reaction commonly used for protein cross-linking (Carraway and Koshland, 1972). This experiment was previously done with a 2:1 styrene-maleic anhydride form as the starting point (Lindhoud et al, 2016). Our protocol maintains mild conditions compatible with many biomolecules throughout the procedure. This makes the chemistry easily accessible to biological laboratories, and will allow a wide range of molecules to be attached to the SMA.
Materials and Reagents
50 ml sterile Falcon tubes (SARSTEDT, catalog number: 62.657.254)
Disposable pipet tips 10-200 µl (Ultratip, Greiner Bio One International, catalog number: 739290)
Disposable pipet tips 100-1,000 µl (Ultratip, Greiner Bio One International, catalog number: 686290)
8,000 MW cut-off membrane (Spectra/Por membrane, Spectrum Laboratories)
1.5 ml sterile Eppendorf tubes (Eppendorf, catalog number: 0030125150)
0.5 ml sterile Eppendorf PCR tubes (Eppendorf, catalog number: 0030124537)
0.2 ml sterile Eppendorf PCR tubes (Eppendorf, catalog number: 0030124332)
MicroBioSpin chromatography columns (Bio-Rad Laboratories, catalog number: 7326221)
E. coli total lipid extract (Avanti Polar Lipids, catalog number: 100500P)
Ethanol 96% (VWR, catalog number: VWRC20823.362)
dH2O in lab wash bottle
NaCl (Sigma-Aldrich, catalog number: S3014-5KG)
Tris base (Roche Diagnostics, catalog number: 10708976001)
SMA 3:1 solution at 25% w/v (Polyscope Polymers, XIRAN®, catalog number: SL25010 S25)
MES dry powder (Sigma-Aldrich, catalog number: M2933)
Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) (Sigma-Aldrich, catalog number: E7750)
N-hydroxysulfosuccinimide (Sulfo-NHS) (Sigma-Aldrich, catalog number: 56485)
Cystamine dihydrochloride (Sigma-Aldrich, catalog number: 30050)
1,4-Dithiothreitol (DTT) (Sigma-Aldrich, Fluka BioChemika, catalog number: 43817)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418-250ML)
Atto488 maleimide (Atto-TEC, catalog number: AD 488-41)
Atto532 maleimide (Atto-TEC, catalog number: AD 532-41)
100 mM MES buffer pH 7.0 (see Recipes)
25 mM MES buffer pH 7.0 (see Recipes)
75 mM MES buffer pH 5.8, EtOH 25% (see Recipes)
Tris-HCl 20 mM pH 8.0 (see Recipes)
Tris-HCl 20 mM pH 8.0, NaCl 200 mM (see Recipes)
Equipment
P10, P20, P200 and P1000 Pipetman pipettes (Gilson, catalog number: F167300)
Centrifuge for 1.5 and 2.0 ml Eppendorf tubes (Eppendorf, model: 5424 R)
Magnetic stirrer hot plate
10 mm stir bar
pH meter (Fisher Scientific)
Thermomixer comfort (Eppendorf, model: ThermoMixer® comfort, catalog number: 5355 000.011)
Precision cell quartz cuvettes (Hellma, catalog number: 105.202-QS)
UV Spectrophotometer (Shimadzu)
Procedure
-
SMA modification
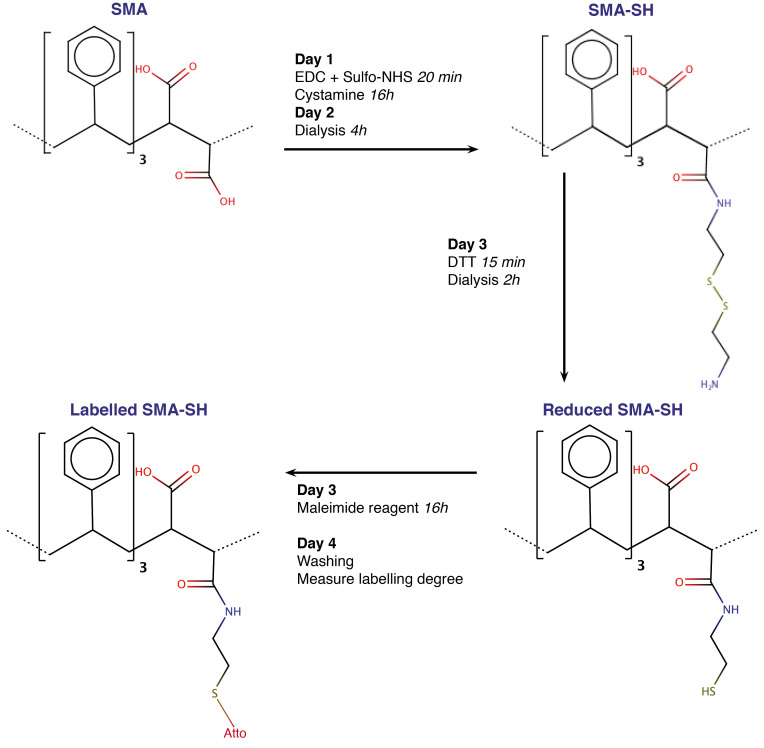
We developed this protocol to obtain thiol modified SMA (SMA-SH). The aim is to form an amide bond between carboxyl groups of the maleic acid and the amine groups of cystamine. The di-thiol bridge is then reduced using dithiothreitol (DTT) before labeling (Figure 1). We target a small proportion of the carboxyl groups (here 1%) to maintain the hydrophilic/hydrophobic balance and thus solubility and solubilizing ability. The reaction of carboxyl groups increases the hydrophobicity and thus a proportion of the maleic acid decreases the solubility and the efficiency of membrane solubilization. We have used the commercial XIRAN® resin with a 3:1 styrene to maleic acid ratio, however the procedure is compatible with other copolymer compositions.
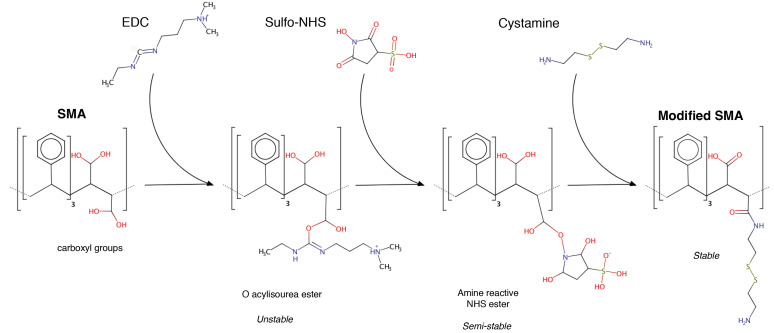
Each step of the chemical reaction can be followed in Figure 2. The reaction can be performed in plastic tube either in glass beaker, but to cover hermetically the reaction and prevent EtOH evaporation we choose 50 ml Falcon tube.
-
EDC activation and Sulfo-NHS stabilization
During this first step, EDC activates carboxyl groups forming an unstable reaction intermediate. This intermediate can be stabilized by addition of Sulfo-NHS. EDC catalyzed cross-linking is normally performed at low pH (~4.5) for improved efficiency, but due to the pH dependence of SMA 3:1 solubility we performed the reaction at pH 6.5 ( Scheidelaar et al., 2016 ). The solution has a tendency to change from a transparent liquid to a slightly milky gel if the pH decreases below 6.0, so it is important to pay attention to the clarity of the solution.
The main experimental concern with the chemical modification of SMA is avoiding either a too low pH or a too high local concentration of one of the reagents. That is why we dissolved powders in a small volume of MES buffer before their addition to the SMA-containing solution.
To modify 1% of carboxyl groups in 3:1 styrene maleic acid copolymer, we calculate that 1 g of copolymer corresponds to 4.47 mM of carboxylate groups, so 1% modification is equivalent to 45 μmol/g of polymer.
Dilute 4 ml of SMA 25% (w/v) stock solution in 6 ml of 100 mM MES buffer pH 7.0 with gentle stirrer agitation.
Dissolve both 45 μm of EDC and Sulfo-NHS at a 20% molar excess (54 μmol/g polymer) in 500 µl MES 75 mM pH 5.8 and 25% ethanol. Add them drop-wise in the solution and incubate for 20 min at room temperature under agitation. The solution should stay liquid and transparent.
-
Cystamine addition
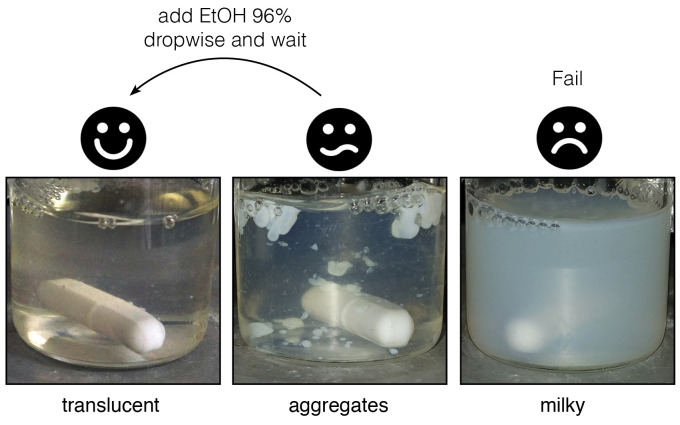
For reaction efficiency it is necessary to avoid the formation of white granular aggregates particularly during cystamine addition. To reduce precipitate formation and increase reaction efficiency it proved useful to increase the solvent hydrophobicity slightly, for this we added 25% ethanol if milky white aggregates appear.
Add a 2-fold excess (108 μmol/g polymer) of cystamine dihydrochloride dissolved in 500 µl 25 mM MES pH 7.0 in 25% EtOH to the SMA solution drop-wise. As this step is critical, it should be done very carefully. Wait 1 min between each drop to watch the solution behavior and add EtOH 96% drop-wise if the solution becomes less transparent or if aggregates appear, until it is clear (Figure 3).
When the solution is stabilized, i.e., it does not become milky or jellified 2 h after cystamine addition, incubate for 12 h at room temperature with stirring covered with a 50 ml Falcon cap to avoid EtOH evaporation.
-
Dialysis
The solution now contains SMA, SMA-SH but also chemical reagents and other low molecular weight products in MES buffer. Dialysis allows buffer exchange for a buffer suitable for next reactions and removal of low molecular weight products.
Dialyze the solution for at least 1 h at room temperature against 100 fold excess volume of Tris-HCl 20 mM pH 8.0 with an 8,000 MW cut-off membrane with gentle stirring.
Change the dialysis buffer after 1 h and dialyze for 3 additional hours.
Collect the SMA-SH solution and store it at 4 °C until use.
-
-
SMA-SH labeling
SMA-SH must be reduced prior to covalent labeling by maleimide. The key to covalent bond formation between maleimide and SMA-SH is the availability and accessibility of the reduced thiol groups. Therefore, it is important to reduce contact between the atmosphere and the solution to the minimum. Oxygen will oxidize thiol groups and thus block the reaction with the maleimide.
-
Add a five-fold excess of DTT (0.27 μmol/g polymer) to the appropriate amount of SMA-SH–depending of the quantity of labeled SMA-SH desired–and incubate for 15 min at room temperature.
Note: Use a small tube for doing this experiment to limit contact between atmosphere and the solution, e.g., 1.5, 0.5 or 0.2 ml Eppendorf plastic tubes respectively for more than 500 µl, more than 100 µl or below 100 µl of final volume.
Dialyse in an 8,000 MW cut-off membrane against 10 fold volume Tris-HCl pH 8.0, NaCl 200 mM for 2 h at room temperature.
Immediately after reduction and dialysis, add a 1.3 fold excess of maleimide (1 mg previously dissolved in 200 μl DMSO) to the SMA-SH solution and incubated for 24 h at 22 °C in Tris-HCl 20 mM pH 8.0 NaCl 200 mM and protected from light and air.
Remove any unreacted fluorophores using MicroBioSpin Chromatography Columns (Promega). Briefly, wash the column buffer by centrifugation for 1 min at 1,000 × g, add 500 µl of Tris-HCl 8.0, NaCl 200 mM, and centrifuge again for 2 min at 1,000 × g. Finally, add the labeling reaction and centrifuge for 4 min at 1,000 × g.
-
-
Labeled SMALPs formation
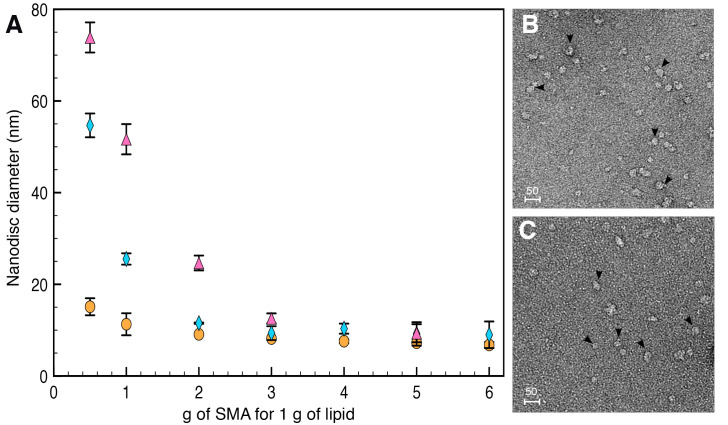
Due to carboxylate group replacement and maleimide group grafting, labeled SMA-SH solubilizes membranes more slowly than unmodified SMA. However, at the proportion typically used for nanodisc formation (i.e., 3 g SMA for 1 g of lipids), no differences in nanodisc properties were observed between SMA, SMA-SH or labeled SMA-SH (Figure 4).
Dry 500 μg of E. coli total lipid extract (Avanti Polar Lipids) under nitrogen flow and evaporate it for 15 more minutes under vacuum to eliminate any traces of solvent.
Disperse the dried lipid film in Tris-HCl 20 mM pH 8.0, NaCl 200 mM by strong vortexing.
Then add an appropriate amount of fluorescent SMA to the lipid solution to get the desired SMA to lipid ratio, and incubate for 1 h at room temperature with mixing. Typically 3 g of labeled SMA is mixed with 1 g of dispersed lipids (w/w).
Remove any unsolubilized material by ultracentrifugation for 1 h at 140,000 × g.
Collect the supernatant containing labeled nanodiscs and have fun!
Figure 1. Workflow of the SMA modification and labeling protocol.
Each step is described in detail in text. SMA-SH and labeled SMA-SH can be stored at 4 °C. From modification to labeling, the protocol takes 4 days and result in replacement of a carboxyl group for a thiol group with finally a covalently bonded maleimide dye.
Figure 2. General scheme of the EDC-mediate reaction.
Only the expected products are depicted. The reaction is performed in solution at pH 6.5.
Figure 3. What a “good” reaction looks like.
The translucent liquid can form drop shaped white aggregates when compounds are added. If these do not disappear after few minutes, add EtOH 96% dropwise and wait ~15 min. The reaction mixture can return to the translucent state, or sometimes retain aggregates forming a milky state.
Figure 4. Diameter and shape of nanodiscs formed by SMA or SMA-SH.
A. Nanodiscs diameter formed by SMA (orange circle), SMA-SH (blue diamond) or labeled SMA-SH (pink triangle) analyzed using DLS (Dynamic Light Scattering). At the 3 g SMA/1 g membrane ratio use, nanodiscs have a comparable diameter. B and C. The nanodiscs formed by SMA (B) or SMA-SH (C) also exhibit similar sizes observed by TEM. Scale bars = 50 nm.
Data analysis
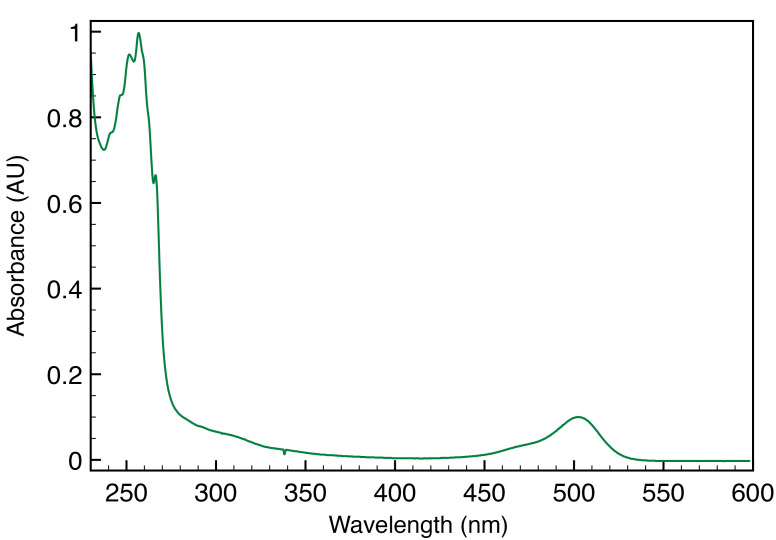
The degree of SMA labeling is calculated based on the decomposition of the absorption spectrum of the washed labeled SMA-SH reaction (Figure 5). The quantity of SMA is calculated from the absorption at 260 nm and fluorophore from absorption at its absorption maximum (500 for Atto488 and 550 nm for Atto532, respectively). An absorption spectrum of the labeled SMA-SH solution is recorded from 700 to 200 nm (with 1 nm increment at medium speed) in a quartz cuvette on a UV spectrometer (Shimadzu). It can be necessary to dilute labeled SMA-SH solution prior to measure absorbance to avoid intensity saturation. Spectra are extracted in .csv format. The degree of labeling is then calculated using the following formula:
Figure 5. Absorption spectra of SMA-SH labeled with Atto-488.
Absorption at 260 nm, corresponding to SMA-SH and 500 nm for Atto488 are used to calculate the degree of labeling as mentioned in the formula.
The extinction coefficient for SMA was ϵSMA of 6989M-1·cm-1 ( Grethen et al., 2017 ; Oluwole et al., 2017 ). The extinction carboxylate coefficients of Atto dyes were ϵAtto of 9 x 104 M-1.cm-1 and 1.5 x 105 M-1·cm-1 for Atto488 and Atto532 respectively, based on the Atto-TEC literature. Dye absorbance is measured at it maximum, 500 nm for Atto488 in our example.
Recipes
-
100 mM MES buffer pH 7.0 (100 ml)
Dissolve 1.952 g of MES dry powder in 90 ml of dH2O by stirring
Titrate to pH 7.0 with HCl
Adjust volume to 100 ml with dH2O
-
25 mM MES buffer pH 7.0 (100 ml)
Dissolve 0.488 g of MES free acid in 90 ml of dH2O by stirring
Titrate to pH 7.0 with HCl
Adjust volume to 100 ml with dH2O
-
75 mM MES buffer pH 5.8, EtOH 25% (100 ml)
Dissolve 1.463 g of MES dry powder in 90 ml of dH2O by stirring
Add 26 ml of EtOH 96%
Titrate to pH 7.0 with HCl
Adjust volume to 100 ml with dH2O
-
Tris-HCl 20 mM pH 8.0 (1 L)
Dissolve 2.423 g of Tris base with 700 ml of dH2O by stirring
Titrate to pH 8.0 with HCl
Adjust volume to 1 L with dH2O
-
Tris-HCl 20 mM pH 8.0, NaCl 200 mM (1 L)
Dissolve 2.423 g of Tris base and 11.688 g NaCl with 700 ml of dH2O by stirring
Titrate to pH 8.0 with HCl
Adjust volume to 1 L with dH2O
Acknowledgments
The authors thank V. Prima, JP Duneau and Y. Rhamani for valuable technical assistance and very helpful discussions. This work was supported by CNRS and Espoir contre la Mucoviscidose.
This protocol has been used in Schmidt and Sturgis, 2018.
Competing interests
The authors declare no conflicts of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Carraway K. L. and Koshland D. E., Jr (1972).[56] Carbodiimide modification of proteins. Methods Enzymol 25: 616-623. [DOI] [PubMed] [Google Scholar]
- 2. Charvolin D., Perez J. B., Rouviere F., Giusti F., Bazzacco P., Abdine A., Rappaport F., Martinez K. L. and Popot J. L.(2009). The use of amphipols as universal molecular adapters to immobilize membrane proteins onto solid supports. Proc Natl Acad Sci U S A 106(2): 405-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dorr J. M., Koorengevel M. C., Schafer M., Prokofyev A. V., Scheidelaar S., van der Cruijsen E. A., Dafforn T. R., Baldus M. and Killian J. A.(2014). Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs . Proc Natl Acad Sci U S A 111(52): 18607-18612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Duquesne K., Prima V. and Sturgis J. N.(2016). Membrane protein solubilization and composition of protein detergent complexes. Methods Mol Biol 1432: 243-260. [DOI] [PubMed] [Google Scholar]
- 5. Giusti F., Kessler P., Hansen R. W., Della Pia E. A., Le Bon C., Mourier G., Popot J. L., Martinez K. L. and Zoonens M.(2015). Synthesis of a polyhistidine-bearing amphipol and its use for immobilizing membrane proteins. Biomacromolecules 16(12): 3751-3761. [DOI] [PubMed] [Google Scholar]
- 6. Grethen A., Oluwole A. O., Danielczak B., Vargas C. and Keller S.(2017). Thermodynamics of nanodisc formation mediated by styrene/maleic acid(2:1) copolymer. Sci Rep 7(1): 11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grisshammer R. and Tate C. G.(1995). Overexpression of integral membrane proteins for structural studies. Q Rev Biophys 28(3): 315-422. [DOI] [PubMed] [Google Scholar]
- 8. Jamshad M., Grimard V., Idini I., Knowles T. J., Dowle M. R., Schofield N., Sridhar P., Lin Y., Finka R., Wheatley M., Thomas O. R. T., Palmer R. E., Overduin M., Govaerts C., Ruysschaert J. M., Edler K. J. and Dafforn T. R.(2015). Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Research 8(3):774-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Knowles T. J., Finka R., Smith C., Lin Y. P., Dafforn T. and Overduin M.(2009). Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc 131(22): 7484-7485. [DOI] [PubMed] [Google Scholar]
- 10. Le Bon C., Della Pia E. A., Giusti F., Lloret N., Zoonens M., Martinez K. L. and Popot J. L.(2014). Synthesis of an oligonucleotide-derivatized amphipol and its use to trap and immobilize membrane proteins. Nucleic Acids Res 42(10): e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lindhoud S., Carvalho V., Pronk J. W. and Aubin-Tam M. E.(2016). SMA-SH: Modified styrene-maleic acid copolymer for functionalization of lipid nanodiscs. Biomacromolecules 17(4): 1516-1522. [DOI] [PubMed] [Google Scholar]
- 12. Oluwole A. O., Danielczak B., Meister A., Babalola J. O., Vargas C. and Keller S.(2017). Solubilization of membrane proteins into functional lipid-bilayer nanodiscs using a diisobutylene/maleic acid copolymer. Angew Chem Int Ed Engl 56(7): 1919-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Popot J. L.(2010). Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem 79: 737-775. [DOI] [PubMed] [Google Scholar]
- 14. Prabudiansyah I., Kusters I., Caforio A. and Driessen A. J.(2015). Characterization of the annular lipid shell of the Sec translocon. Biochim Biophys Acta 1848(10Pt A): 2050-2056. [DOI] [PubMed] [Google Scholar]
- 15. Scheidelaar S., Koorengevel M. C., van Walree C. A., Dominguez J. J., Dorr J. M. and Killian J. A.(2016). Effect of polymer composition and pH on membrane solubilization by styrene-maleic acid copolymers. Biophys J 111(9): 1974-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt V. and Sturgis J. N.(2018). Modifying styrene-maleic acid co-polymer for studying lipid nanodiscs. Biochim Biophys Acta 1860(3): 777-783. [DOI] [PubMed] [Google Scholar]