Abstract
Stable-isotope labeled metabolic analysis is an essential methodology to characterize metabolic regulation during biological processes. However, the method using stable-isotope-labeled tracer (e.g., 13C-glucose) in live animal is only beginning to be developed. Here, we contribute a qualitative metabolic labeling experiment protocol in Drosophila melanogaster using stable-isotope-labeled 13C-glucose tracer followed by liquid chromatography-mass spectrometry (LC-MS) analysis. Detailed experimental setup, data acquisition and analysis are provided to facilitate the application of in vivo metabolic labeling analysis that might be applied in a wide range of biological studies.
Keywords: Stable-isotope labeling, 13C-glucose tracer , Metabolic analysis, Qualitative analysis, Liquid chromatography-mass spectrometry, Drosophila melanogaster
Background
Metabolomics is a newly emergent omic-level study aiming to profile small molecule metabolites in a complex biological system. It has been applied in diverse research areas pertaining to human health and disease, such as biomarker discovery, disease pathogenesis, and assessment of drug toxicity. Measurement of metabolites is important to determine alterations in metabolic pathway in response to endogenous and exogenous changes. To accurately characterize metabolic pathway activity, isotope-labeled tracers (e.g., 13C and 15N) have been used ( Park et al., 2016 ; Jang et al., 2018 ). There are many such studies (both quantitatively and qualitatively) in cultured cells ( Buescher et al., 2015 ; Liu et al., 2018 ), however, stable-isotope based metabolic labeling experiment in live animal remain largely unexplored. In the current protocol, we describe a qualitative metabolic labeling analysis by using the labeled 13C-glucose as a tracer, and we have successfully applied this protocol to comparatively analyze the activity of glycolysis pathway in Drosophila melanogaster, during aging and between wild-type and mutant animals.
Materials and Reagents
-
Consumables
Pipette tips (Eppendorf, catalog number: 0030073428)
Ceramic beads (Aoran, catalog number: 150010C)
Eppendorf tube (2 ml) (Eppendorf, catalog number: 0030120094)
HPLC glass vial (Agilent Technologies, catalog number: 5182-0716)
Injection needle (Agilent Technologies, catalog number: G4226-87201)
Kimwipe filter paper (KCWW, Kimberly-Clark, catalog number: 34120)
-
Biological material
-
Drosophila melanogaster
The Drosophila strain used was 5905 (FlyBase ID: FBst0005905, w1118). Flies were cultured in standard media (Recipe 1) at 25 °C with 60% humidity in a 12 h light and 12 h dark cycle.
Prior to the test, flies were starved on 1% Agar media for 6 h before transferred to the vials containing a small piece of Kimwipe filter paper (KCWW, Kimberly-Clark, catalog number: 34120) pre-soaked in 1 ml of 10% U-13C6-glucose (U-13C6-glucose was added to phosphate buffer at a final concentration of 10%). Flies were treated for 3 days, and then transferred to new vials with fresh U-13C6-glucose for additional 2 days. Fly heads were dissected from anesthetized flies with CO2 for subsequent metabolic analysis. For each experiment, 8 biological repeats were conducted, with 20 heads for each repeat. One hundred and sixty male flies were used, with 20 flies per vial.
-
-
Chemicals
LC-MS chemicals:
Methanol (MeOH), LC-MS grade (Honeywell, catalog number: LC230-2.5HC). Store at the room temperature (20 °C-25 °C)
Acetonitrile (ACN), LC-MS grade (Merck, catalog number: 1.00029.2500). Store at the room temperature (20 °C-25 °C)
Water (H2O) (Honeywell, catalog number: LC365-2.5HC). Store at the room temperature (20 °C-25 °C)
Ammonium acetate, LC-MS grade (Sigma-Aldrich, catalog number: 73594-25G-F). Store at 4 °C
Ammonium hydroxide, LC-MS grade (Sigma-Aldrich, catalog number: 44273-100mL-F). Store at 4 °C
Liquid nitrogen
Labeled chemicals:
D-Glucose (U-13C6, 99%) (Cambridge Isotope Laboratories, catalog number: CLM-1396-PK). Store at the room temperature (20 °C-25 °C)
Drosophila standard media:
Sucrose
Maltose
Yeast
Agar
Maizena
Soybean flour
438 sodium benzoate
Methyl-p-hydroxybenzoate
Propionic acid
-
Mobile phase setup
Mobile phase A (see Recipes)
Mobile phase B (see Recipes)
Equipment
Pipettes
Homogenizer (BERTIN, model: Precellys® 24)
Incubator
Sonicator
Centrifuge
Vacuum concentrator (Labconco, German)
Merck SeQuant ZIC-pHILIC column [particle size, 5 μm; 100 mm (length) x 2.1 mm (i.d.)]
UHPLC system (Agilent Technologies, model: 1290 Infinity)
Quadruple time-of-flight mass spectrometer (Agilent Technologies, model: 6550 Series)
Software
Pathways to PCDL (version B.07.00, Agilent Technologies)
PCDL Manager (version B.07.00, Agilent Technologies)
Profinder (version B.08.00, Agilent Technologies)
MassHunter software (version B.07.00, Agilent Technologies)
Procedure
-
Metabolites extraction
Quickly freeze the animal tissues (head of Drosophila) in liquid nitrogen immediately after dissection.
Homogenize the tissue sample with 200 μl of H2O and 5 ceramic beads using the homogenizer.
Add 800 μl ACN:MeOH (1:1, v/v) to homogenized solution for subsequent metabolite extraction.
Incubate the samples for 1 h at -20 °C to precipitate proteins.
Proceed with 15 min centrifugation at 15,000 × g under 4 °C.
Transfer the resulting supernatant to a new Eppendorf tube (2 ml), then evaporate to dryness in a vacuum concentrator under 4 °C.
Reconstitute the dry extracts with 100 µl of ACN:H2O (1:1, v/v).
Sonicate the reconstitution solution for 10 min, and centrifuge for 15 min at 15,000 × g under 4 °C to remove insoluble debris.
Transfer the supernatant to an HPLC glass vial and store at -80 °C if the samples will be subjected to LC-MS analysis within 3 h. For extracted samples that require long time (over 12 h) stored prior to being analyzed, we suggest storing the samples after Step A6 and then proceeding with A8-A9 before LC-MS analysis.
-
LC-MS analysis
-
Liquid chromatography
Load worklist with method embedded using MassHunter software. Please note that LC-MS operation (both instrument and software) requires specialized training.
-
Run batch sequence with following LC parameters:
Wash injection needle one time with needle washing solvent MeOH:H2O (1:1, v/v).
Load sample and inject 2 μl of sample.
Run LC method using the LC gradient as described in Table 1.
-
Mass spectrometry
Set MS parameters as described below:
-
ESI source parameters:
Sheath gas temperature, 300 °C.
Dry gas temperature, 250 °C.
Sheath gas flow, 12 L/min.
Dry gas flow, 16 L/min.
Capillary voltage, 2,500 V (+) and -2,500 V (-), respectively. Please note that the same sample is analyzed twice for each ionization mode.
Nozzle voltage, 0 V.
Nebulizer pressure, 20 psi.
-
Time of Flight (TOF) parameters:
TOF scan range: m/z 60-1,200 Da.
MS1 acquisition frequency: 4 Hz.
-
-
Table 1. The gradient elution method for LC-MS analysis.
| Time (min) | Flow rate (ml/min) | Eluent A (vol. %) | Eluent B (vol. %) |
| 0.0 | 0.2 | 20.0 | 80.0 |
| 2.0 | 0.2 | 20.0 | 80.0 |
| 17.0 | 0.2 | 80.0 | 20.0 |
| 17.1 | 0.2 | 20.0 | 80.0 |
| 22.1 | 0.4 | 20.0 | 80.0 |
| 22.2 | 0.2 | 20.0 | 80.0 |
Data analysis
-
Extraction of isotopologues
-
Metabolite library construction
Use Pathways to PCDL software (version B.07.00, Agilent Technologies) and PCDL Manager software (version B.07.00, Agilent Technologies) to build a metabolite library for metabolites in both glycolysis and citric acid cycle. Specifically, each metabolite standard is analyzed under the same LC-MS condition as biological samples. The ion chromatograph of each metabolite is extracted to obtain the retention time information. Then, the retention time together with formula value is used to construct a metabolite library using PCDL manager. The input example is provided as below (Table 2):
-
Raw data loading
Load the acquired LC-MS raw data files (.d) into Profinder (version B.08.00, Agilent Technologies) for the extraction of metabolite isotopologues using the constructed metabolite library.
-
Feature extraction parameters:
Ion abundance criterion: peak core area 20% of peak height.
Mass tolerance: ± 15 ppm + 2.00 mDa.
Retention time tolerance: ± 0.20 min.
Anchor ion height threshold: 250 counts.
Sum of ion heights threshold: 1,000 counts.
Correlation coefficient threshold: 0.5.
-
-
Stable-isotope-labeled metabolic analysis
-
Peak integration result manual check
After isotopologues extraction in Profinder, peak integration result need to be reviewed and manually curated for subsequent accurate stable-isotope labeled metabolic analysis. Make sure the peak integration range is consistent across multiple samples. Figure 1A illustrates the extracted ion chromatography (EIC) of the key metabolite 13C3-lactate and the peak integration range.
-
Calculation of tracer incorporation
For each targeted metabolite, different isotope pattern will be obtained corresponding to the number of incorporated 13C atoms. For example, the isotopologues of lactate are m + 0, m + 1, m + 2, and m + 3 (Figure 1B).
-
Abundance of individual isotopologue is the integrated peak area.
Taken metabolite lactate as an example, Figure 1C shows the abundance level of one isotopologue of lactate M + 3 between two groups (wild type and PRC2 mutant). In the article by Ma et al., 2018 , Figure 6G is generated using this calculated data.
-
Total metabolite abundance
Total metabolite abundance is calculated using the following formula:
Mn is the labeling pattern of the isotopologue with all atoms (C or N) labeled.
Figure 1D shows the total abundance level of lactate between two groups (wild type and PRC2 mutant). In the article by Ma et al., 2018 , Figure S5E was generated using this calculated data.
-
Proportion of individual isotopologue
Proportion of individual isotopologue is calculated using the following formula:
Mi is the labeling pattern of individual isotopologue.
Mn is the labeling pattern of the isotopologue with all atoms (C or N) labeled.
Figure 1E shows the percentage of total pool level of one isotopologue of lactate M + 3 between two groups (wild type and PRC2 mutant). In the article by Ma et al., 2018 , Figure S5B and Figure S5D were generated using this calculated data.
-
Total tracer incorporation
Total tracer incorporation is calculated using the following formula:
Figure 1F shows the percentage of total tracer incorporation between two groups (wild type and PRC2 mutant).
Above results demonstrated that lactate, the end product of glycolysis pathway, significantly increased in PRC2 mutants.
-
-
Table 2. The metabolite library for metabolites in both glycolysis and citric acid cycle.
| Metabolite name | Formula | Retention time (min) |
| Glucose | C6H11NaO6 | 5.1 |
| F6P | C6H13O9P | 7.8 |
| G6P | C6H13O9P | 8.2 |
| GADP | C3H7O6P | 7.4 |
| PEP | C3H5O6P | 8.7 |
| Pyruvate | C3H4O3 | 7.3 |
| Lactate | C3H6O3 | 3.2 |
| Citrate | C6H8O7 | 8.9 |
| Malate | C4H6O5 | 7.8 |
| Fumarate | C4H4O4 | 7.8 |
| Cis-aconitate | C6H6O6 | 8.9 |
| α-KG | C5H6O5 | 7.5 |
| Succinate | C4H6O4 | 7.4 |
Figure 1. Stable-isotope-labeled metabolic analysis strategy.
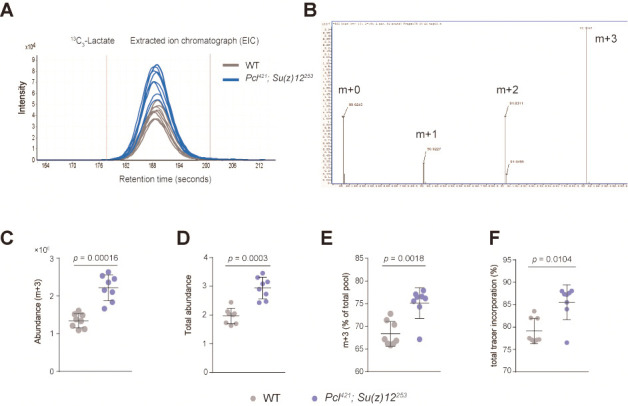
A. The extracted ion chromatography (EIC) of isotopologue 13C3-lactate (m + 3). (mean ± SD of 8 biological repeats with 10 flies for each measurement; Student’s t-test; n.s.: not significant). Test was from muscle tissues of 30 d old male flies. Genotypes: WT: 5905. Mut: Pclc421/+; Su(z)12c253/+. B. The labeling pattern of lactate demonstrated in mass spectrum (m + 0, m + 1, m + 2, and m + 3). C. The abundance level of lactate isotopologue m + 3 between two groups (mean ± SD of 8 biological repeats with 10 flies for each measurement; Wilcox test). Genotypes: WT: 5905. Mut: Pclc421/+; Su(z)12c253/+. D. The total abundance level of lactate between two groups (wild type and PRC2 mutant) (mean ± SD of 8 biological repeats with 10 flies for each measurement; Wilcox test). Genotypes: WT: 5905. Mut: Pclc421/+; Su(z)12c253/+. E. The percentage of total pool level of one isotopologue of lactate m + 3 between two groups (mean ± SD of 8 biological repeats with 10 flies for each measurement; Wilcox test). Genotypes: WT: 5905. Mut: Pclc421/+; Su(z)12c253/+. F. The percentage of total tracer incorporation between two groups (mean ± SD of 8 biological repeats with 10 flies for each measurement; Wilcox test). Genotypes: WT: 5905. Mut: Pclc421/+; Su(z)12c253/+.
Recipes
-
Standard Drosophila food
Sucrose 36 g/L
Maltose 38 g/L
Yeast 22.5 g/L
Agar 5.4 g/L
Maizena 60 g/L
Soybean flour 8.25 g/L
438 sodium benzoate 0.9 g/L
Methyl-p-hydroxybenzoate 0.225 g/L
Propionic acid 6.18 ml/L
ddH2O to make up 1 L
-
Mobile phase A
25 mM ammonium acetate
25 mM ammonium hydroxide
For the preparation of 1 L mobile phase A, firstly weigh 1.9271 g CH3COONH4. Dissolve the CH3COONH4 in 1 L H2O. Then add 3.5 ml NH4OH (25%) to generate the mobile phase A. Store the solution at 4 °C for up to 2 weeks
-
Mobile phase B
Acetonitrile
Store at the room temperature (20-25 °C)
Acknowledgments
We thank the financial support provided by the startup funding from Interdisciplinary Research Center on Biology and Chemistry (IRCBC), and Agilent Technologies Thought Leader Award. N.L. and Z.-J. Z. are also supported by Thousand Youth Talents Program. This protocol is also a part of our previous work by Ma et al., 2018 .
Competing interests
The authors declare no competing financial interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Buescher J. M., Antoniewicz M. R., Boros L. G., Burgess S. C., Brunengraber H., Clish C. B., DeBerardinis R. J., Feron O., Frezza C., Ghesquiere B., Gottlieb E., Hiller K., Jones R. G., Kamphorst J. J., Kibbey R. G., Kimmelman A. C., Locasale J. W., Lunt S. Y., Maddocks O. D., Malloy C., Metallo C. M., Meuillet E. J., Munger J., Noh K., Rabinowitz J. D., Ralser M., Sauer U., Stephanopoulos G., St-Pierre J., Tennant D. A., Wittmann C., Vander Heiden M. G., Vazquez A., Vousden K., Young J. D., Zamboni N. and Fendt S. M.(2015). A roadmap for interpreting 13C metabolite labeling patterns from cells . Curr Opin Biotechnol 34: 189-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jang C., Chen L. and Rabinowitz J. D.(2018). Metabolomics and isotope tracing. Cell 173(4): 822-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu L., Su X., Quinn W. J. 3rd Hui S., Krukenberg K., Frederick D. W., Redpath P., Zhan L., Chellappa K., White E., Migaud M., Mitchison T. J., Baur J. A. and Rabinowitz J. D.(2018). Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab 27(5): 1067-1080 e1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ma Z., Wang H., Cai Y., Wang H., Niu K., Wu X., Ma H., Yang Y., Tong W., Liu F., Liu Z., Zhang Y., Liu R., Zhu Z. J. and Liu N.(2018). Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila . Elife 7: e35368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park J. O., Rubin S. A., Xu Y. F., Amador-Noguez D., Fan J., Shlomi T. and Rabinowitz J. D.(2016). Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat Chem Biol 12(7): 482-489. [DOI] [PMC free article] [PubMed] [Google Scholar]


