Abstract
Inflammatory immune cells play direct pathological roles in cases of acute kidney injury (AKI) and chronic kidney disease (CKD). However, the identification and characterization of distinct populations of leukocytes in human kidney biopsies have been confounded by the limitations of immunohistochemical (IHC)-based techniques used to detect them. This methodology is not amenable to the combinations of multiple markers necessary to unequivocally define discrete immune cell populations. We have developed a multi-parameter, flow cytometric-based approach that addresses the need for panels of cell-specific markers in the identification of immune cell populations, allowing both the accurate detection and quantitation of leukocyte subpopulations from a single, clinical kidney biopsy specimen. In this approach, fresh human kidney tissue is dissociated into a single cell suspension followed by antibody-labeling and flow cytometric-based acquisition and analysis. This novel technique provides a major step forward in identifying and enumerating immune cell subpopulations in human kidney disease and is a powerful platform to complement traditional histopathological examinations of clinical kidney biopsies.
Keywords: Leukocytes, Kidney, Flow cytometry, Kidney biopsy, Immunology
Background
Diseases of the kidney are rising in incidence and prevalence, placing a significant burden on the healthcare community worldwide and significantly affecting patients’ quality of life (World Kidney Day: Chronic Kidney Disease, 2015). Kidney damage can arise from a wide range of insults including infections, ischemia, toxins, hypertension, genetic and metabolic disorders (Imig and Ryan, 2013). The immediate response to insult functionally is acute kidney injury (AKI), a clinical syndrome characterised physiologically by a rapid (hours to days) decrease in kidney function and histomorphologically by infiltration of inflammatory immune cells into the renal tubulointerstitium, the tissue compartment adjoining the tubules of the kidney ( Basile et al., 2012 ). The subsequent biological responses aim to limit ongoing injury and repair damaged tissue. If unresolved, this inflammation may lead to chronic kidney disease (CKD). CKD is defined as a loss in kidney function that persists for at least 3 months in duration and is characterized histomorphologically, regardless of its cause, by tubulointerstitial fibrosis and chronic inflammation ( Kawakami et al., 2014 ). The presence of inflammatory immune cells in human AKI and CKD is suggestive of functional roles in disease progression. However, the identification and enumeration of these CD45+ leukocyte populations in human kidney tissue remain poorly defined.
Until now, the identification of immune cells in human kidney disease has primarily been made using immunohistochemistry (IHC). Numerous IHC-based studies of kidney biopsies report strong associations between the loss of renal function/tubulointerstitial injury and the prominent accumulation of inflammatory immune cell populations of distinct leukocyte lineages, including: T cells (CD3+ cells [ Hoffmann et al., 2006 ]), T helper cells (CD4+ cells [ Liu et al., 2012 ; Pei et al., 2014 ]), cytotoxic T cells (CD8+ cells [ Pei et al., 2014 ]), B cells (CD19+ or CD20+ cells [ Heller et al., 2007 ]), natural killer (NK) cells (CD16+ or CD56+ cells [ Furuichi et al., 2001 ; Hidalgo et al., 2010 ; Shin et al., 2015 ]), monocytes/macrophages (CD14+ cells [ Zhou et al., 2010 ]), dendritic cells (DC) (HLA-DR+ cells [ Hart et al., 1981 ; Markovic-Lipkovski, 1990] or DC-SIGN+ cells [ Segerer et al., 2008 ; Pei et al., 2014 ]) and granulocytes (neutrophils, basophils, eosinophils; CD15+ or CD16+ cells [ Weidner et al., 2004 ; Segerer et al., 2006 ]).
However, IHC staining for single antigens is not sufficiently specific to directly identify these immune cell populations, given the broader expression of many of these markers on several leukocyte and non-hematopoietic cell types. For example, single staining for DC-SIGN to identify DC will be non-specific given its co-expression on monocytes/macrophages in kidney tissue ( Figel et al., 2011 ). Similarly, using CD16 or CD56 to identify NK cells will be inadequate, with both markers also expressed on T cells (Markey and MacDonald, 1989; Trinchieri, 1989). Furthermore, IHC-based methods are unable to effectively distinguish expression levels of these antigens, required, for instance, to distinguish subsets of functionally specialized NK cells – CD56bright CD16-/low NK cells and CD56dim CD16+ NK cells. Clearly, a progression to multi-parameter staining methodologies is required to more accurately detect and quantify immune cell populations in human kidney disease.
We have developed a novel protocol for obtaining single cell suspensions from fresh kidney biopsies to allow the identification and enumeration of immune cells subsets via multi-color flow cytometry ( Kassianos et al., 2013 ; Law et al., 2017 and 2018; Muczynski et al., 2018 ). Biopsy tissue (excess to diagnostic purposes) is obtained with informed patient consent, followed by enzymatic digestion into single cells and finally, antibody labeling for acquisition by flow cytometry. This process enables precision in detecting discrete immune cell populations based on the absence/presence and intensity of multiple markers. This approach also allows differentiation of populations based on cell size (forward scatter; FSC), granularity (side scatter; SSC), cell viability (Near-IR) and absolute immune cell counts (fluorescent counting bead standard). This single platform approach is proving versatile in answering basic research questions in kidney disease and is readily translatable to human diagnostics of the future, where immune cell populations may become prognostic markers for the progressive loss of kidney function.
Materials and Reagents
Samco Extra Fine-tip polyethylene transfer pipettes (thermo fisher scientific, Fisher ScientificTM, Molecular BioProducts, catalog number: 231)
1.5 ml microcentrifuge tubes (Eppendorf, catalog number: 0030125150)
5 ml polystyrene round-bottom tubes (flow cytometry tubes) (Corning, Falcon®, catalog number: 352008)
Flow-Count Fluorospheres (Beckman Coulter, catalog number: 7547053)
Aluminum foil
Hank’s balanced salt solution (HBSS + Calcium + Magnesium) (thermo fisher scientific, GibcoTM, catalog number: 14025076)
Hank’s balanced salt solution (HBSS, no calcium, no magnesium) (thermo fisher scientific, GibcoTM, catalog number: 14175095)
Collagenase P (Roche Molecular Systems, catalog number: 11213857001) (2 mg/ml stock)
DNase I (Roche Molecular Systems, catalog number: 11284932001) (10 mg/ml stock)
Trypsin-EDTA (0.05%), phenol red (Thermo Fisher Scientific, GibcoTM, catalog number: 25300054)
Sodium Azide (Sigma-Aldrich, catalog number: S2002)
Fetal Bovine Serum (FBS) (thermo fisher scientific, GibcoTM, catalog number: 1009141)
RPMI 1640 Medium (Thermo Fisher scientific, GibcoTM, catalog number: 11875119)
Bovine Serum Albumin (Sigma-Aldrich, catalog number: A7906-500G)
Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: 158127)
Phosphate buffered saline (PBS)
LIVE/DEADTM Fixable Near-IR Dead Cell Stain Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: L10119)
Human TruStain FcXTM (BioLegend, catalog number: 422302)
Brilliant Violet 510TM anti-human CD45 Antibody clone HI30 (e.g., BioLegend, catalog number: 304036)
Phycoerythrin (PE) Mouse Anti-Human CD19 Antibody clone HIB19 (e.g., BD, catalog number: 555413)
PE-CF594 Mouse Anti-Human CD16 clone 3G8 (e.g., BD, catalog number: 562320)
Alexa Fluor® 700 anti-human CD14 Antibody clone M5E2 (e.g., BioLegend, catalog number: 301822)
Brilliant Violet 605TM anti-human CD4 Antibody clone OKT4 (e.g., BioLegend, catalog number: 317437)
Brilliant Violet 650TM anti-human CD3 Antibody clone OKT3 (e.g., BioLegend, catalog number: 317323)
FITC anti-human CD8a Antibody clone RPA-T8 (e.g., BioLegend, catalog number: 301006)
PerCP/Cy5.5 anti-human CD56 (NCAM) Antibody clone HCD56 (e.g., BioLegend, catalog number: 318322)
Brilliant Violet 785TM anti-human HLA-DR Antibody clone L243 (e.g., BioLegend, catalog number: 307642)
Digestion solution I (see Recipes)
Digestion solution II (see Recipes)
FACS buffer (see Recipes)
1% PFA (see Recipes)
Equipment
Vortex
Benchtop centrifuge
Timer
FACS flow cytometer (BD, model: LSRFortessaTM, 4 laser)
Conventional cell culture incubator (37 °C/5% CO2)
Software
FlowJo version 10.1.0 (FlowJo LLC, https://www.flowjo.com)
BD FACSDivaTM software
Procedure
-
Biopsy preparation
Obtain 16-gauge human kidney biopsy core with informed donor consent (see Note 1).
Measure the length of the kidney biopsy core prior to processing. Note the measurement; it is used to determine absolute numbers of immune cell populations (as outlined in the Notes section).
Place the portion of the kidney biopsy core (approx. 1-5 mm) in 0.25 ml Digestion Solution I (see Recipes) in a 1.5 ml microfuge tube.
Incubate at 37 °C for a total of 15 min. At 7 min, and at the end of the 15 min, gently pipette up and down several times with a Samco pipette to mechanically dissociate the cells. The diameter of the Samco pipettes listed above facilitates this dissociation.
Centrifuge at low speed (600 × g) for 5 min at 4 °C to pellet cells.
Harvest supernatant for analysis of soluble products from the biopsy. Immediately aliquot and store at -80 °C until required (see Note 2).
Add 0.5 ml Digestion Solution II (see Recipes) and incubate at 37 °C for 10 min. At the beginning and end of this period, gently pipette up and down several times with a Samco pipette to mechanically dissociate the cells.
Add 0.5 ml cell culture media containing calcium (e.g., RPMI 1640 medium) to neutralize the trypsin (see Note 3). At this point, the biopsy should be completely digested with minimal cell clumps present. However, if sufficient tissue is used, a white fibrous cord may remain which does not contain cells. This adheres to the Samco pipette tip and is easily removed.
Centrifuge the resulting cells at 600 × g for 5 min at 4 °C. Remove supernatant.
-
Antibody labeling of cells
Dilute LIVE/DEADTM Fixable Near-IR Dead Cell Stain 1:4 in PBS.
Resuspend cells in 0.5 ml PBS and add 0.5 μl diluted LIVE/DEADTM Fixable Near-IR Dead Cell Stain (final conc. 1:4,000; see Note 4).
Stain 15 min on ice.
Wash cells by adding 0.5 ml FACS buffer (see Recipes), centrifuge at 600 × g for 5 min at 4 °C and remove supernatant.
Resuspend cell pellet in 100 μl FACS buffer.
Add 5 μl of Human TruStain FcXTM (see Note 5) to 100 μl cells in FACS buffer (5 min at RT) prior to adding antibodies of interest.
-
Prepare antibody master mix for surface staining (see panel below; Notes 6-8).
Antigen Fluorophore Volume CD45 BV510 2.5 μl CD14 AF700 0.5 μl CD16 PE-CF594 1 μl CD19 PE 2 μl CD3 BV650 2.5 μl CD8 FITC 1.25 μl CD4 BV605 2 μl CD56 PerCPCy5.5 5 μl HLA-DR BV785 2.5 μl -
Add antibody master mix to cells, vortex for 5 sec and incubate for 30 min on ice.
Note: This panel can be modified depending upon cells of interest and can also include activation markers, chemokine receptors or other molecules of interest.
Wash cells by adding 0.5 ml FACS buffer, centrifuge at 600 × g for 5 min at 4 °C and remove supernatant.
Fix cells in 1% PFA to a final volume of 270 μl and transfer into polystyrene FACS tube (see Note 11). Cover with aluminum foil and keep on ice/refrigerate until acquisition. Cells should be analyzed on the flow cytometer within four days. It is possible to run cells without fixation if the acquisition is performed within two hours.
Immediately prior to analysis, add 30 μl Flow-Count fluorospheres (see Note 12).
Analyse by flow cytometry.
-
Flow cytometry
Set up compensation on a flow cytometer.
Acquire entire sample.
Data analysis
-
Identify and analyze cells of interest. Samples are analyzed with FlowJo.
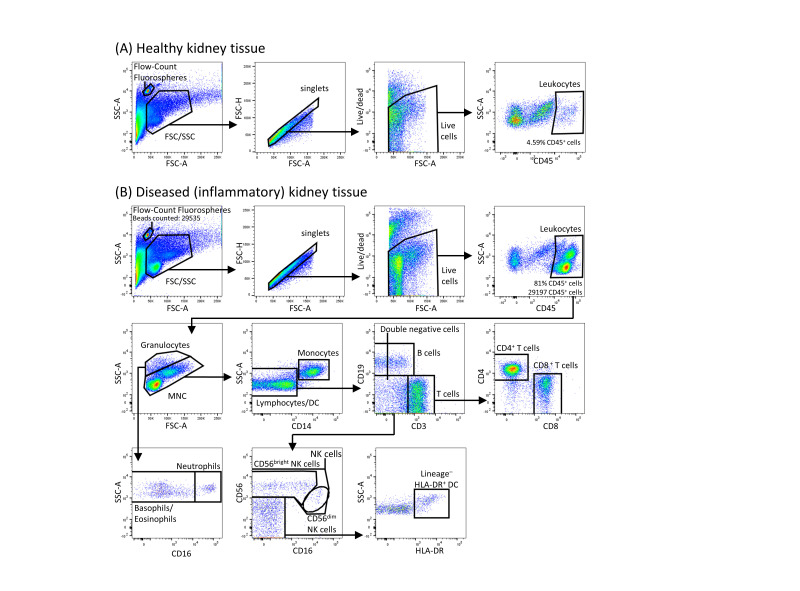
Within the single, live, CD45+ immune cells, gates are first set on polymorphonuclear (increased SSC) and mononuclear cells.
The polymorphonuclear cells are further gated into neutrophils (CD16+) or basophils/eosinophils (CD16-).
The mononuclear cell population is gated into monocytes and lymphocytes. The monocytes are separated from the lymphocytes based on their increased side scatter and expression of CD14.
The lymphocyte population is further gated into B cells (CD19+) and T cells (CD3+) cells. Cells that are both negative for CD19 and CD3 (double negative) are further gated into NK cell subsets and DC by expression of CD56 (NK cells) or HLA-DR (DC; see Figure 1).
Calculate cell numbers using Flow-Count fluorospheres (see Note 14).
Figure 1. Gating strategy used to identify immune cell subsets in human kidney tissue by flow cytometry.
Flow-Count fluorospheres are identified and gated as the unique 10 μm bead population on the forward scatter (FSC)/side scatter (SSC) plot. Single, live, CD45+ mononuclear cells (MNC) and granulocytes are gated on an FSC/SSC plot. Total lymphocytes are distinguished from granulocytes and monocytes based on low side scatter and absent CD14 expression. Total lymphocytes are further separated into T cells or B cells by their expression of CD3 and CD19 respectively. NK subpopulations, CD56bright and CD56dim NK cells, are identified based on CD56 intensity and CD16 expression. DC are gated as lineage (CD3, CD14, CD19, CD56)- HLA-DR+ cells. A. Healthy kidney biopsy tissue; B. Diseased (inflammatory) kidney biopsy tissue. The gating strategy used to identify immune cell subsets in diseased kidney tissue is identical between inflammatory AKI and CKD biopsies.
Notes
Acquisition of human tissue requires approval from an appropriate ethics committee and written informed consent from the donor. When working with human tissue appropriate biosafety practices (PC2 facilities) must be followed. Biopsies should be transported from the procedure room to laboratory in RPMI media containing 10% FBS on ice. Immune cells should be isolated within 12 h of the biopsy being taken.
Harvested supernatants collected after the first tissue dissociation step may be used to assess cytokines and chemokines within the interstitium. We have assessed inflammatory proteins in the supernatant by cytometric bead array and ELISA.
As trypsin is inhibited by divalent cations (calcium and magnesium), when preparing the digestion solution II, the trypsin is diluted in HBSS without calcium or magnesium. The trypsin is then neutralized by the addition of cell culture media containing calcium.
The LIVE/DEADTM Fixable Near-IR Dead Cell Stain Kit is used to determine the viability of cells. The Near-IR fluorescent reactive dye works by binding to free amines on the cell surface and in the interior of cells with compromised membranes. Viable cells exhibit reduced fluorescent intensity as the dye can only bind to cell surface amines. The difference in fluorescent intensity allows viable cells to be easily discriminated from dead cells.
Human TruStain FcXTM is utilized to block antibodies non-specifically binding to Fc receptors expressed on some leukocytes (such as monocytes, granulocytes, B cells and dendritic cells). Human TruStain FcXTM works by blocking FcR-mediated Ig Fc binding without interfering with antibody-mediated specific staining of human cells.
It is important to recognize that some antigens may be cleaved during the process of tissue dissociation. Therefore, we recommend performing a digestion test for the antigens being assessed in your panel. This is performed by staining peripheral blood mononuclear cells (PBMC) that have undergone the dissociation process with the selected antibody panel and comparing it to untreated PBMC. In some cases where an epitope is lost, a change of antibody clone directed to a different epitope of the same antigen may be useful.
Antibody concentrations are optimal for starting populations of ≤ 5 x 105 cells. Do not exceed this cell number as it reduces binding efficiencies. However, for optimal results we recommend titrating your antibodies and testing their compatibility with the other antibodies in your panel.
For antibody staining steps use pre-cooled solutions; this will prevent capping of antibodies on the cell surface and non-specific cell labeling.
PBMC are used to set up compensation on the flow cytometer. To prepare single-color compensation control tubes, PBMC is stained with individual antibodies listed in the panel (step 16) and incubated for 30 min on ice. PBMC are then washed with 0.5 ml FACS buffer, centrifuged at 600 × g for 5 min at 4 °C and resuspended in 300 μl 1% PFA. Each compensation control tube is acquired on a flow cytometer, with automatic compensation values for each fluorochrome combination accurately calculated using BD FACSDivaTM software. Compensation values will vary between flow cytometers due to differences between instruments, including laser intensity and detector sensitivity.
Unlabeled cells from biopsy tissue can be used as controls to eliminate autofluorescence from analyses.
For the calculation of absolute cell numbers, it is extremely important that the cells are resuspended after the final wash to a total volume of 270 μl. To achieve this, after aspirating the supernatant, we initially add 100 μl of 1% PFA to resuspend cells. We measure the resulting volume by adjusting the volume on the pipette until all the liquid is withdrawn from the tube in one action (this would normally be between 140-170 μl). We then add the remaining volume of 1% PFA to make an exact final volume of 270 μl.
It is important to follow the manufacturer’s recommendations when using the Flow-Count fluorospheres. The fluorospheres must be properly resuspended to ensure the aliquot reflects the specific concentration indicated on the product insert. This is important as the ratio of biological cells in the specimen to fluorospheres is used in determining the absolute cell count. To resuspend, the fluorospheres are vortexed for 10-12 sec; longer vortexing times are not recommended to avoid forming excessive bubbles. Add the fluorospheres just prior to analysis, making sure to accurately pipette the desired volume into the sample. The fluorospheres must also be thoroughly mixed with the test sample. It is recommended to do this by holding the FACS tube at 50% of the tube height and vortexing for 5 sec immediately before flow cytometric analysis.
Flow-Count fluorospheres can be detected in a fluorescence emission range of 525 nm to 700 nm when excited at 488 nm or alternatively by FSC versus SSC as presented in Figure 1. In this methodology, we gate closely around the discrete bead population, based on 10 μm size, identified in the FSC versus SSC analysis.
-
Absolute numbers of each gated immune cell population can be determined using the ratio of the biological cells in the specimen to the Flow-Count fluorospheres following the manufacturer’s recommendations. This can be normalized to the volume of starting tissue to allow a comparison of absolute numbers between multiple specimens. Briefly, the concentrations of target cell populations (in cells/μl) are calculated using the following formula:
The total number of target cells counted/total number of fluorospheres counted x Flow-Count fluorosphere concentration (noted on product insert) x dilution factor
The concentrations of each target cell population are then multiplied by the total sample volume to obtain absolute counts. The absolute cell counts can then be normalized to cell numbers per cm3 of kidney tissue, by dividing the absolute cell count by the volume of renal tissue (in cm3), which is calculated as follows:
πr2 x length of biopsy tissue (in cm) where, the radius (r) of a 16-gauge biopsy is 0.08 cm.
A representative calculation of the absolute number of CD45+ leukocytes from Figure 1B is presented.
Note: A 0.5 cm 16-gauge human kidney biopsy core was processed for flow cytometric analysis, with 29197 CD45+ leukocytes and 29535 fluorospheres (at a stock concentration of 968 fluorospheres/μl) counted.
The flow cytometry gating figures depicted are donor dependent and are highly variable.
-
Additional markers have been assessed in extended panels within our laboratory by utilizing antibodies conjugated with the following fluorochromes: Allophycocyanin (APC), Brilliant Violet 421TM, Brilliant Violet 711TM and PE-Cy7.
.
Recipes
-
Digestion Solution I (prepare and use immediately)
0.125 ml HBSS (+ Calcium + Magnesium)
0.125 ml Collagenase P (final conc. 1 mg/ml)
0.5 μl DNase I (final conc. 20 μg/ml)
-
Digestion Solution II (prepare and use immediately)
10 μl Trypsin-EDTA (0.05%), phenol red
490 μl HBSS (no calcium, no magnesium)
-
FACS buffer (prepare and store long-term at 4 °C)
1x PBS (1 L)
5 g BSA Powder (final conc. 0.5%)
200 mg Sodium Azide (final conc. 0.02%)
-
1% PFA
1x PBS (1 L)
10 g Paraformaldehyde
Prepare the PFA solution by heating to 60 °C while stirring and adding 1-2 drops of 1 M NaOH to help the paraformaldehyde to dissolve. Aliquot and store at -20 °C
Acknowledgments
The authors would like to thank the tissue donors and clinicians. The work was funded by Pathology Queensland, Royal Brisbane and Women’s Hospital (RBWH) Research Grant, Kidney Research Foundation and National Health and Medical Research Council (NHMRC) Project Grant GNT1099222.
Competing interests
All authors declare no conflict of interest or competing interests of any kind.
Ethics
Kidney tissue was obtained with informed patient consent following approval by the Royal Brisbane and Women’s Hospital Human Research Ethics Committee (2002/011 and 2006/072).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Basile D. P., Anderson M. D. and Sutton T. A.(2012). Pathophysiology of acute kidney injury. Compr Physiol 2(2): 1303-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Figel A. M., Brech D., Prinz P. U., Lettenmeyer U. K., Eckl J., Turqueti-Neves A., Mysliwietz J., Anz D., Rieth N., Muenchmeier N., Buchner A., Porubsky S., Siegert S. I., Segerer S., Nelson P. J. and Noessner E.(2011). Human renal cell carcinoma induces a dendritic cell subset that uses T-cell crosstalk for tumor-permissive milieu alterations. Am J Pathol 179(1): 436-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Furuichi K., Wada T., Iwata Y., Sakai N., Yoshimoto K., Shimizu M., Kobayashi K., Takasawa K., Kida H., Takeda S., Matsushima K. and Yokoyama H.(2001). Upregulation of fractalkine in human crescentic glomerulonephritis. Nephron 87(4): 314-320. [DOI] [PubMed] [Google Scholar]
- 4. Hart D. N., Fuggle S. V., Williams K. A., Fabre J. W., Ting A. and Morris P. J.(1981). Localization of HLA-ABC and DR antigens in human kidney. Transplantation 31(6): 428-433. [DOI] [PubMed] [Google Scholar]
- 5. Heller F., Lindenmeyer M. T., Cohen C. D., Brandt U., Draganovici D., Fischereder M., Kretzler M., Anders H. J., Sitter T., Mosberger I., Kerjaschki D., Regele H., Schlondorff D. and Segerer S.(2007). The contribution of B cells to renal interstitial inflammation. Am J Pathol 170(2): 457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hidalgo L. G., Sis B., Sellares J., Campbell P. M., Mengel M., Einecke G., Chang J. and Halloran P. F.(2010). NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection. Am J Transplant 10(8): 1812-1822. [DOI] [PubMed] [Google Scholar]
- 7. Hoffmann U., Segerer S., Rummele P., Kruger B., Pietrzyk M., Hofstadter F., Banas B. and Kramer B. K.(2006). Expression of the chemokine receptor CXCR3 in human renal allografts--a prospective study. Nephrol Dial Transplant 21(5): 1373-1381. [DOI] [PubMed] [Google Scholar]
- 8. Imig J. D. and Ryan M. J.(2013). Immune and inflammatory role in renal disease. Compr Physiol 3(2): 957-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kassianos A. J., Wang X., Sampangi S., Muczynski K., Healy H. and Wilkinson R.(2013). Increased tubulointerstitial recruitment of human CD141hi CLEC9A+ and CD1c+ myeloid dendritic cell subsets in renal fibrosis and chronic kidney disease . Am J Physiol Renal Physiol 305(10): F1391-1401. [DOI] [PubMed] [Google Scholar]
- 10. Kawakami T., Mimura I., Shoji K., Tanaka T. and Nangaku M.(2014). Hypoxia and fibrosis in chronic kidney disease: crossing at pericytes. Kidney Int Suppl(2011) 4(1): 107-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Law B. M. P., Wilkinson R., Wang X., Kildey K., Lindner M., Rist M. J., Beagley K., Healy H. and Kassianos A. J.(2017). Interferon-γ production by tubulointerstitial human CD56bright natural killer cells contributes to renal fibrosis and chronic kidney disease progression . Kidney Int 92(1): 79-88. [DOI] [PubMed] [Google Scholar]
- 12. Law B. M., Wilkinson R., Wang X., Kildey K., Lindner M., Beagley K., Healy H. and Kassianos A. J.(2018). Effector γδ T cells in human renal fibrosis and chronic kidney disease. Nephrol Dial Transplant. [DOI] [PubMed] [Google Scholar]
- 13. Liu L., Kou P., Zeng Q., Pei G., Li Y., Liang H., Xu G. and Chen S.(2012). CD4+ T Lymphocytes, especially Th2 cells, contribute to the progress of renal fibrosis . Am J Nephrol 36(4): 386-396. [DOI] [PubMed] [Google Scholar]
- 14. Markey A. C. and MacDonald D. M.(1989). HNK-1 antigen is not specific for natural killer cells. J Invest Dermatol 92(5): 774-775. [DOI] [PubMed] [Google Scholar]
- 15. Markovic-Lipkovski J., Muller C. A., Risler T., Bohle A. and Muller G. A.(1990). Association of glomerular and interstitial mononuclear leukocytes with different forms of glomerulonephritis. Nephrol Dial Transplant 5(1): 10-17. [DOI] [PubMed] [Google Scholar]
- 16. Muczynski K. A., Leca N., Anderson A. E., Kieran N. and Anderson S. K.(2018). Multicolor fow cytometry and cytokine analysis provides enhanced information on kidney transplant Biopsies. Kidney International Reports.(in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pei G., Zeng R., Han M., Liao P., Zhou X., Li Y., Zhang Y., Liu P., Zhang C., Liu X., Yao Y. and Xu G.(2014). Renal interstitial infiltration and tertiary lymphoid organ neogenesis in IgA nephropathy. Clin J Am Soc Nephrol 9(2): 255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Segerer S., Heller F., Lindenmeyer M. T., Schmid H., Cohen C. D., Draganovici D., Mandelbaum J., Nelson P. J., Grone H. J., Grone E. F., Figel A. M., Nossner E. and Schlondorff D.(2008). Compartment specific expression of dendritic cell markers in human glomerulonephritis. Kidney Int 74(1): 37-46. [DOI] [PubMed] [Google Scholar]
- 19. Segerer S., Henger A., Schmid H., Kretzler M., Draganovici D., Brandt U., Noessner E., Nelson P. J., Kerjaschki D., Schlondorff D. and Regele H.(2006). Expression of the chemokine receptor CXCR1 in human glomerular diseases. Kidney Int 69(10): 1765-1773. [DOI] [PubMed] [Google Scholar]
- 20. Shin S., Kim Y. H., Cho Y. M., Park Y., Han S., Choi B. H., Choi J. Y. and Han D. J.(2015). Interpreting CD56+ and CD163+ infiltrates in early versus late renal transplant biopsies . Am J Nephrol 41(4-5): 362-369. [DOI] [PubMed] [Google Scholar]
- 21. Trinchieri G.(1989). Biology of natural killer cells. Adv Immunol 47: 187-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weidner S., Carl M., Riess R. and Rupprecht H. D.(2004). Histologic analysis of renal leukocyte infiltration in antineutrophil cytoplasmic antibody-associated vasculitis: importance of monocyte and neutrophil infiltration in tissue damage. Arthritis Rheum 50(11): 3651-3657. [DOI] [PubMed] [Google Scholar]
- 23. Zhou J., Ouyang X., Cui X., Schoeb T. R., Smythies L. E., Johnson M. R., Guay-Woodford L. M., Chapman A. B. and Mrug M.(2010). Renal CD14 expression correlates with the progression of cystic kidney disease. Kidney Int 78(6): 550-560. [DOI] [PMC free article] [PubMed] [Google Scholar]