Abstract
The hierarchical composition and interactions of the labile thylakoid protein complexes can be assessed by sequential 2D-native gel-electrophoresis system. Mild non-ionic detergent digitonin is used to solubilize labile protein super-and megacomplexes, which are then separated with first-dimension blue native polyacrylamide gel electrophoresis (1D-BN-PAGE). The digitonin derived protein complexes are further solubilized with stronger detergent, β-DM, and subsequently separated on an orthogonal 2D-BN-PAGE to release smaller protein subcomplexes from the higher-order supercomplexes. Here we describe a detailed method for 2D-BN-PAGE analysis of thylakoid protein complexes from Arabidopsis thaliana.
Keywords: Native gel electrophoresis, Thylakoid membrane, Thylakoid protein complexes, 2D-BN-PAGE, Light harvesting complex, Photosystem, Photosynthesis
Background
Photosynthetic light reactions take place in the thylakoid membrane, which in higher plants is composed of appressed grana thylakoids and non-appressed stroma thylakoids. The light reactions are catalyzed by multi-subunit protein complexes photosystem (PS) I and II, cytochrome b6f and ATPase. PSII together with its light harvesting antenna complex (LHCII) is most abundant in grana-thylakoids and therefore spatially segregated from stroma thylakoid -located PSI-LHCI complexes (Andersson and Anderson, 1980). The interphase between the grana and stroma thylakoid is enriched in both photosystems (Albertsson, 2001; Suorsa et al., 2015 ). Mediated by light dependent reversible phosphorylation of LHCII and PSII proteins, the photosystems together with LHCII assemble into larger super- and megacomplexes. PSII core dimer together with two strongly and two moderately bound LHCII-trimers form large C2S2M2 supercomplexes, PSI together with loosely bound LHCII form PSI-LHCII supercomplexes, and finally, PSII and PSI together with L-LHCII form large PSII-LHCII-PSI megacomplexes ( Caffarri et al., 2009 ; Pesaresi et al., 2009 ; Rantala et al., 2017 ).
Non-ionic detergents are generally used for isolation of native protein complexes from biological membranes. Mild detergent digitonin maintains weak interactions between protein complexes, but due to its bulky structure, it selectively solubilizes only the non-appressed regions of the thylakoids. However, when supplemented with low ionic strength salt, aminocaproic acid (ACA), digitonin gets access to the partition gap between two grana appressions and solubilizes the entire thylakoid membrane allowing the analysis of the overall organization of labile thylakoid protein complexes ( Rantala et al., 2017 ). Solubilized protein complexes are supplemented with anionic Coomassie G-250 (CBB) dye that binds to the hydrophobic domains of protein complexes providing them with negative charge, and allows their electrophoretic separation according to molecular mass. Since the negative surface charges repel each other, CBB also prevents random protein complex aggregation.
Blue native PAGE (BN-PAGE) enables membrane protein complex separation in their native and functionally active form (Schägger and von Jagow, 1991). Coupling the 1D-BN-PAGE with a second (2D)-BN-PAGE allows the analysis of the subcomplex composition of the digitonin derived large protein super- and megacomplexes: The 1D-BN-gel lane containing the separated protein complexes is treated with slightly stronger detergent, n-dodecyl-β-D-maltoside (β-DM), which more effectively interferes with the interactions between protein complexes, particularly destroying the interaction of LHCII with the two photosystems. The lane is then subjected to the 2D-BN-PAGE for the separation of the dissociated subcomplexes. The composition of the subcomplexes can be further analyzed by electroblotting the 2D-gel and detecting specific proteins with antibodies or by cutting the lanes and subjecting the subcomplexes to denaturing 3D-SDS-PAGE.
This protocol describes optimized thylakoid protein complex isolation method and the analysis of the subcomplex composition of large protein super- and megacomplexes of Arabidopsis thaliana by 2D-BN-BN-PAGE. The method can be used for the analysis of the organization of the photosynthetic protein complexes and for the analysis of subcomplex composition of higher-order protein super- and megacomplexes.
Materials and Reagents
-
Consumables
Eppendorf microcentrifuge tubes 1.5 ml (Eppendorf, catalog number: 0030121694)
Falcon, Conical Centrifuge Tubes 15 ml (Corning, catalog number: 352096)
Culture tubes 5 ml (Lab Depot, catalog number: TLDT8301)
Finntip Pipette Tips (Finntip Flex 10, 250, 1,000 and 5 ml)
Whatman chromatographic paper (GE Healthcare, catalog number: 1001-931)
PVDF membrane (Merck, catalog number: IPVH304F0)
-
Plant material
Isolated thylakoids from 5-week old Arabidopsis thaliana (isolate thylakoids in the presence of 10 mM NaF in all buffers as described in Järvi et al., 2011 )
-
Reagents
6-aminocaproic acid (Sigma-Aldrich, catalog number: A2504)
Bis-Tris (Sigma-Aldrich, catalog number: B4429)
Glycerol (Avantor Performance Materials, J.T. Baker, catalog number: 7044)
Pefabloc SC (4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride) (Roche Diagnostics, catalog number: 11585916001)
Sodium Fluoride (NaF) (Avantor Performance Materials, J.T. Baker, catalog number: 3688)
EDTA disodium salt (Avantor Performance Materials, J.T. Baker, catalog number: 1073)
Digitonin (Merck, Calbiochem, catalog number: 300410)
n-dodecyl-β-D-maltoside (Sigma-Aldrich, catalog number: D4641)
Serva Coomassie Blue G (SERVA Electrophoresis, catalog number: 35050)
Sucrose (Sigma-Aldrich, catalog number: S0389)
Tricine (Sigma-Aldrich, catalog number: T0377)
TEMED (Tetramethylethylenediamine) (Bio-Rad Laboratories, catalog number: 1610801)
Liquid nitrogen
Methanol (VWR)
Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: A7030)
Acrylamide (AA) (Sigma-Aldrich, catalog number: A9099)
(N,N'-Methylene)-Bis-Acrylamide (Bis-AA) (Merck, Omnipur, catalog number: 2610)
APS (Ammonium persulfate) (Bio-Rad Laboratories, catalog number: 1610700)
Tris (Sigma-Aldrich, catalog number: T1503)
Glycine (Fisher Scientific, catalog number: 10070150)
SDS (VWR, catalog number: 442444H)
-
Antibodies
Lhcb1 (Agrisera, catalog number: AS01 004)
Phospho (P)-Lhcb1 (Agrisera, catalog number: AS13 2704)
Lhcb2 (Agrisera, catalog number: AS01 003)
Phospho (P)-Lhcb2 (Agrisera, catalog number: AS13 2705)
-
Stock solutions
Acrylamide solution A: 48% (w/v), 1.5% (w/v) Bis-acrylamide in MQ-water, store at 4 °C
Acrylamide solution B: 20% (w/v), 5% (w/v) Bis-acrylamide in MQ-water, store at 4 °C
APS (Ammonium persulfate): 5% (w/v) solution in MQ-water, store at 4 °C
Digitonin stock solution: 10% (w/v) in MQ-water, store at -20 °C
β-dodecyl maltoside: 10% (w/v) in MQ-water, store at -20 °C
Glycerol: 75% (w/v) solution in MQ-water, store at 4 °C
Pefabloc: 10 mg/ml (w/v) in MQ-water, store at -20 °C
Sucrose: 75% (w/v) in MQ-water, store at -20 °C
-
Buffers
3x Gel Buffer, store at 4 °C (see Recipes)
25BTH20G resuspension buffer, prepare fresh (see Recipes)
ACA buffer (see Recipes)
Detergent buffer A (4% Digitonin), prepare fresh (see Recipes)
Detergent buffer B (1% β-DM), prepare fresh (see Recipes)
CBB buffer, -20 °C (see Recipes)
Anode buffer for BN, store at 4 °C (see Recipes)
Blue cathode buffer for BN, store at 4 °C (see Recipes)
Clear cathode buffer for BN, store at 4 °C (see Recipes)
Transfer buffer, store at 4 °C (see Recipes)
BN-PA: 3.5-12.5% separation gel, 3% stacking gel (see Recipes)
Equipment
Dual gel caster with 10 x 8 cm plates (Hoefer, catalog number: SE215)
Hoefer gradient maker SG5 (or any gradient maker containing two 5 ml chambers)
0.75 mm T-spacers (Hoefer, catalog number: SE2119T-2-.75)
1 mm T-spacers (Hoefer, catalog number: SE2119T-2-1.0)
Sample gel comb, 0.75 mm, 10 wells (Hoefer, catalog number: SE211A-10-.75)
2D comb (flat) with a reference well, 1.0 mm thick
Mighty Small SE250 vertical electrophoresis system (Hoefer, catalog number: SE250)
Ismatec IPC-pump
Power supply, PowerPac HV (Bio-Rad Laboratories, catalog number: 164-5056)
Centrifuge (Eppendorf, model: 5424R)
FinnpipetteTM F2 Variable Volume Single-Channel Pipettes
Cold room (4 °C)
Freezer (-20 °C)
Photo scanner (e.g., Perfection V300 Photo, Epson, model: V300)
Rocker-Shaker (Biosan, model: MR-12, catalog number: BS-010130-AAI)
Semi-dry blotting system (Hoefer, catalog number: TE77X)
Procedure
-
Casting the BN-PA gel (linear gradient 3.5%-12.5% separation gel; 3% stacking gel)
Assemble gel caster (e.g., Hoefer) with 10 x 8 cm plates (Figure 1A) according to manufacturer’s instructions. Any vertical electrophoresis system with any compatible casting unit can be used.
Prepare the light (3.5% AA) and heavy (12.5% AA) buffers from the acrylamide stock solution A (48% acrylamide, 1.5% Bis-acrylamide) (see Recipes). Keep the solutions on ice to prevent untimely polymerization. Cast the gel between a glass plate and a notched aluminum oxide plate (10 x 8 cm) with a gradient maker (Figure 1B). Use 0.75 mm T-spacer for the 1D-BN-PAGE and 1 mm T-spacer for 2D-BN-PAGE. Allow the gradient gel to polymerize for 1-2 h at RT.
-
Cast the stacking gel (3% acrylamide) using the acrylamide stock solution B (20% acrylamide, 5% Bis-acrylamide). Use an appropriate sample gel comb (1D-BN-PA gel: standard comb with 10 wells, 2D-BN-PA gel: 2D-comb). Allow the stacking gel to polymerize for 30-60 min. Remove the sample gel comb under MQ-water. Store the gel at 4 °C.
PAUSE POINT: The gel can be stored at 4 °C at least for few days. The gel must not dry.
-
Solubilization of thylakoid membranes (Perform the solubilization under dim light)
Resuspend thylakoids equivalent to 3-7 µg chlorophyll in ACA buffer to a chlorophyll concentration of 1 mg/ml.
Add an equal volume of detergent buffer A (4% digitonin/ACA) on the sample and solubilize at RT with continuous gentle mixing for 10 min, e.g., in a shaker. The final chlorophyll concentration is 0.5 mg/ml.
Pellet the insolubilized material by centrifugation at 18,000 × g, at 4 °C, for 20 min, remove the supernatant to a new Eppendorf tube.
Add 1/10 of CBB-buffer to the sample and carefully mix with a pipet. Avoid making air bubbles.
-
Separation of the protein complexes with 1D-BN-PAGE
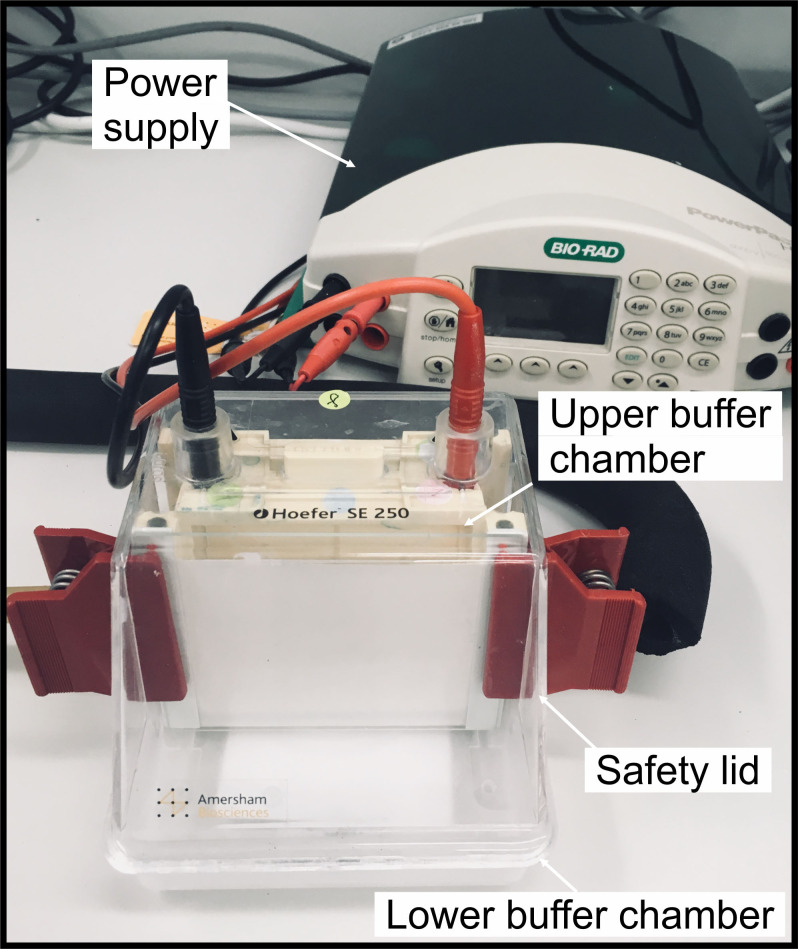
Assemble the gel to the vertical electrophoresis system and pour ~70 ml of blue cathode buffer to the gel tank (upper chamber), and ~100 ml of anode buffer to the lower chamber (Figure 2).
Load thylakoids equivalent to 2-5 µg of chlorophyll to the sample well.
-
Start the electrophoretic run by gradually increasing the voltage as follows:
75 V 30 min
100 V 30 min
125 V 30 min
150 V 60 min
175 V 30 min
Perform the electrophoresis at 4 °C
As the blue running front has moved about one third (at ~100-125 V) of the desired running distance, remove the blue cathode buffer from the upper chamber and replace it with a clear cathode buffer.
Stop the electrophoresis, when the protein complexes (B1-B9) (see Figure 6A) are separated and the gel is clear from the blue cathode.
-
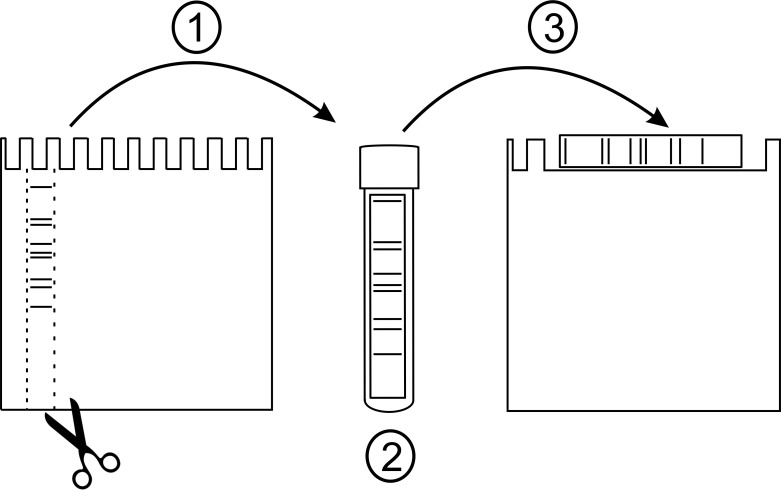
Scan the gel with a photo scanner (keep the gel between the glass and alumina plates while scanning), remove the glass plate and carefully cut out the lane containing the separated protein complexes using a spacer (see sketch in Figure 3). Do not use scalpel or any other sharp object for cutting to avoid scratching the alumina plate.
PAUSE POINT: It is possible to store the BN-gel strip at -80 °C, but do not shock-freeze the strip with liquid nitrogen.
Notes:
The amount of chlorophyll loaded on the gel depends on the subsequent analysis: for immunoblotting, load 2 µg (or less) of chlorophyll and for the 3D-SDS-PAGE, load 5 µg or more chlorophyll.
It is possible to also excise individual bands (complexes B1-B9) from the BN-PA-gel, re-solubilize the bands in Eppendorf-tubes with 1% β-DM and subject them to 2D-BN-PAGE.
-
2D-BN-PAGE
Place the gel strip in a culture tube (5 ml) and add 2 ml of detergent buffer B (1% β-DM).
Solubilize the strip for 40 min at 4 °C in gentle rocking to allow even solubilization of the strip (e.g., 20 rpm).
Place the strip on the second BN-PA gel (see sketch in Figure 3) and assemble the gel to the electrophoresis tank.
Perform the electrophoresis as described for the 1D-BN-PAGE.
Scan the gel after the electrophoresis whit a photo scanner (see sketch of the 2D-BN-PAGE in Figure 4, the flowchart demonstrating the follow-up paths in Figure 5 and scanned image of the representative gel in Figure 6B).
-
Western blotting of 2D-native gels
Place the 2D-BN-PA-gel to transfer buffer and incubate for 30 min.
Activate the PVDF membrane with 100% methanol and place the membrane to transfer buffer. Soak six Whatman filter papers in the transfer buffer.
Assemble three papers on the electrode (anode). Place the membrane on top of the papers and the gel on top of the membrane. Finally, place the three papers on top of the gel and mount the cathode on top. Set the current for 1 mA/cm2 gel area, and transfer for at least 1 h.
Destain the membrane with 100% methanol.
Block the membrane with 5% BSA for 1 h.
-
Perform immunostaining with specific antibodies (see representative blots from Figure 6C).
Lhcb1, Lhcb2, Lhcb3
P-Lhcb1 and P-Lhcb2
-
3D-SDS-PAGE
Cut the lanes containing the subcomplexes (corresponding to complexes B1-B9).
Protein solubilization and separation on SDS-PAGE are as described for the 2D-SDS-PAGE analysis in Järvi et al., 2011 .
After electrophoresis, visualize the protein with Sypro Ruby Stain or with silver staining according to Blum et al., 1987 (see representative 3D gels from Figure 6D, the nine gels correspond to complexes B1-B9).
Figure 1. Equipment for casting gradient gels.
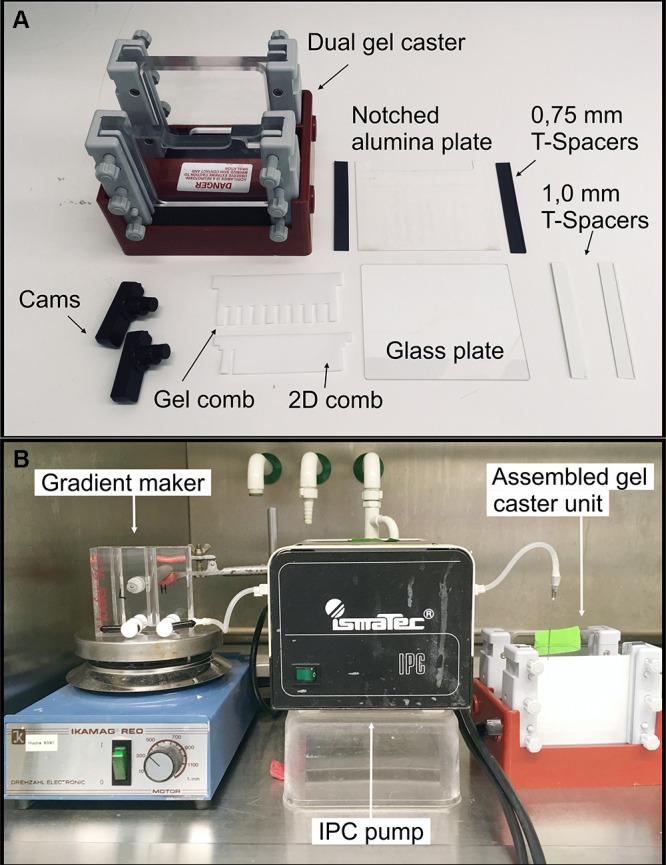
A. Hoefer dual gel caster with all the required equipment; B. Gradient maker, IPC-pump and gel assembled and ready for gel casting.
Figure 2. Example of the gradient gel assembly.
The gel is assembled in the SE 250 Mini-Vertical electrophoresis unit.
Figure 6. The representative results from the analysis of the subcomplex and subunit composition of thylakoid protein complexes.
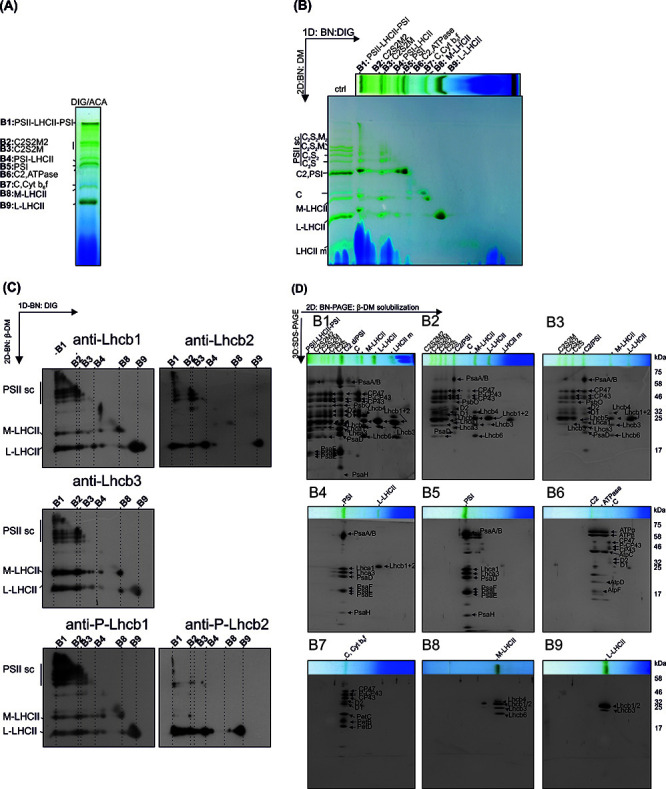
A. Protein complexes (B1-B9) of digitonin solubilized thylakoids after 1D-BN-PAGE separation. B. 2D-BN-PAGE separation of the subcomplexes obtained by re-solubilizing the 1D-strip with β-DM. Ctrl represents thylakoids directly solubilized with 1% DM. C. The Lhcb-protein localization in protein subcomplexes was analyzed by electroblotting the 2D-BN-gels and immunodecorating the blots with Lhcb1, Lhcb2, Lhcb3 protein specific antibodies and with Phospho-Lhcb1-2 protein specific antibodies. D. The protein composition of the subcomplexes (derived from complexes B1-B9) was analyzed by cutting the lanes from 2D-BN-gel and subjecting the gel strips to 3D-BN-PAGE. The proteins are visualized with Sypro Ruby stain. This figure has been originally published in Rantala et al. (2017) .
Figure 3. Preparation of 1D-BN-strip for 2D-BN-PAGE.
(1) After finishing 1D-BN-PAGE, remove the glass plate and excise the lane containing the protein complexes (B1-B9) using e.g., the T-spacer. (2) Carefully place the gel strip to a 5 ml culture tube containing 2 ml of detergent buffer B (1% β-DM). Solubilize in a shaker for 40 min. (3) Place the gel strip on the second BN-PA gel (the large well) and assemble the gel to the electrophoresis tank. Perform the 2D-electrophoresis as described for 1D-BN-PAGE.
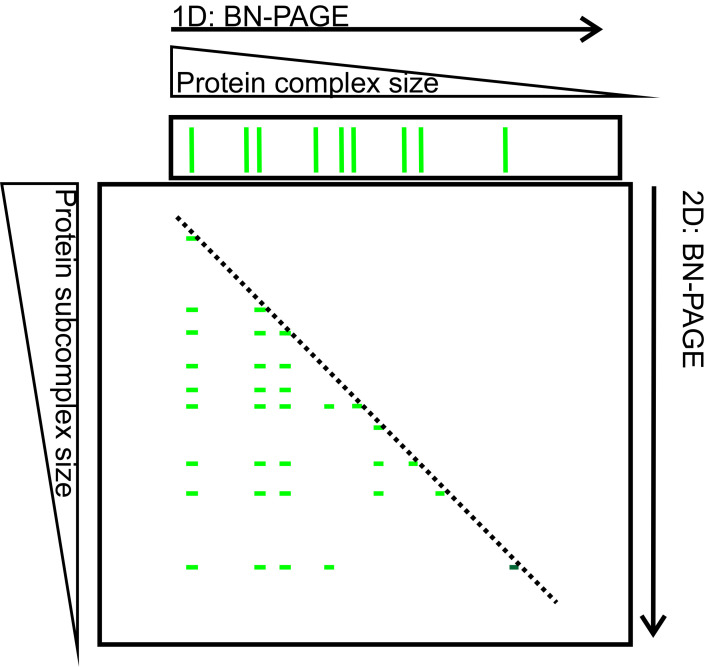
Figure 4. 2D-BN-PAGE separation of thylakoid protein complexes.
Protein complexes B1-B9 (green bands on the upper horizontal gel slice) were first separated according to their mass and shape on 1D-BN-PAGE. After separation the gel slice was subjected to 2D-BN-PAGE during which the complexes (B1-B9) are fractionated into subcomplexes (narrow green bands on the 2D-BN-gel). The complexes on the diagonal (dashed line) represent complexes that have preserved their mass, whereas the complexes below the diagonal are subcomplexes (of B1-B9) that have been disconnected during second solubilization and 2D-BN-PAGE.
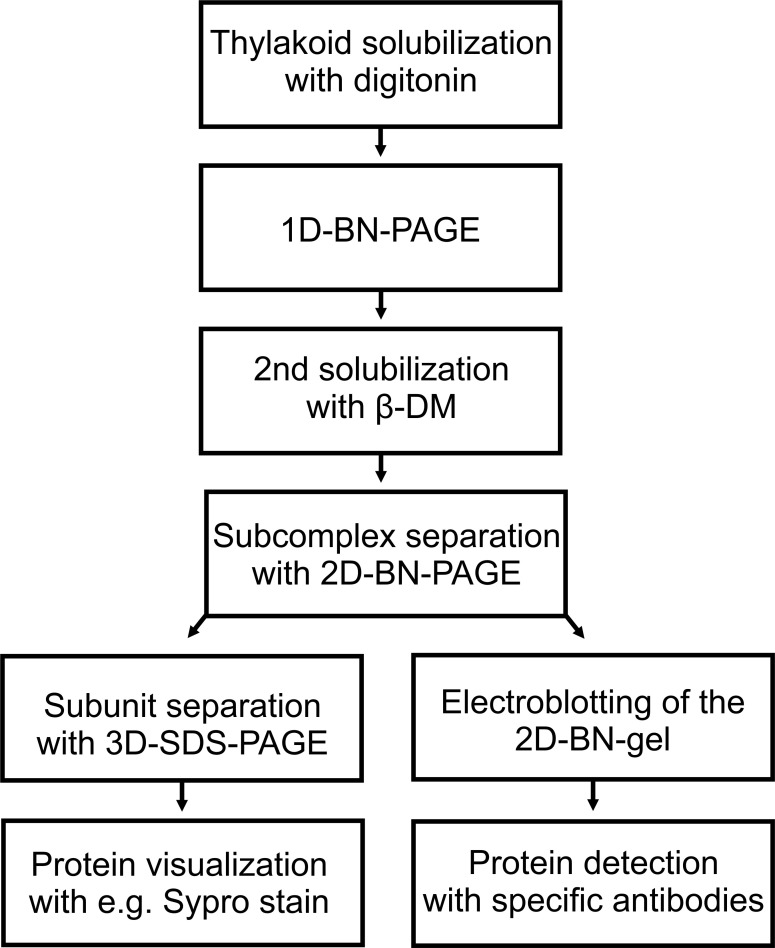
Figure 5. Flowchart of the experimental procedure.
Data analysis
For all the experiments (2D-BN-PAGE, 3D-SDS-PAGE, Western blotting) we use three biological replicates.
No image processing was performed. The identification of the protein complexes (on native gels) and individual proteins (on 3D-SDS gels) is based on mass spectrometry analysis according to ( Aro et al., 2005 ; Suorsa et al., 2015 ).
The representative data (Figure 6) is from the original research article ( Rantala et al., 2017 ).
Notes
The protocol is highly reproducible in our hands.
Digitonin must be purified according to the manufacturer’s instructions. We had no problems with digitonin, but the detergent stock may precipitate easily when melted.
The thylakoid isolation (protocol in Järvi et al., 2011 ) must be done from fresh leaves to obtain high-quality thylakoid protein complexes.
Freezing and thawing the thylakoid sample several times will affect the stability of the protein complexes.
Recipes
Note: All the solutions are prepared in MQ water.
-
3x Gel buffer
1.5 M 6-Aminocaproic acid
150 mM Bis-Tris
-
25BTH20G
25 mM Bis-Tris/HCl (pH 7.0)
20% (w/v) glycerol
0.25 mg/ml Pefabloc (add freshly)
10 mM NaF (add freshly)
-
ACA buffer
25 mM Bis-Tris/HCl (pH 7.0)
375 mM Aminocaproic acid
1 mM EDTA
0.25 mg/ml Pefabloc (add freshly from the stock solution)
10 mM NaF (add freshly)
-
Detergent buffer A
4 % digitonin (w/v)
25 mM Bis-Tris/HCl (pH 7.0)
375 mM Aminocaproic acid
1 mM EDTA
0.25 mg/ml Pefabloc (add freshly from the stock solution)
10 mM NaF (add freshly)
-
Detergent buffer B
1% β-dodecyl maltoside (w/v)
25 mM Bis-Tris/HCl (pH 7.0)
20% (w/v) glycerol and
0.25 mg/ml Pefabloc (add freshly from the stock solution)
10 mM NaF (add freshly)
-
CBB buffer
100 mM Bis-Tris/HCl (pH 7.0)
0.5 M ACA
30% (w/v) sucrose
50 mg/ml Serva Blue G
-
Anode buffer
50 mM Bis-Tris/HCl (pH 7.0)
-
Cathode buffer
50 mM Tricine
15 mM Bis-Tris/HCl (pH 7.0)
0.01% Serva Blue G
-
Transfer buffer
39 mM glysine
48 mM Tris
0.0375% SDS
20% methanol
-
BN-PA: 3.5-12.5% separation gel, 3% stacking gel
Light Heavy Stack 48% AA, 1.5% bis-AA 148 µl 530 µl - 20% AA, 5% bis-AA - - 180 µl 3x gel buffer 700 µl 700 µl 500 µl 75% glycerol 140 µl 560 µl - H2O 1,092 µl 290 µl 800 µl 5% APS 15 µl 11 µl 30 µl TEMED 3 µl 2 µl 3 µl Pipet to the chamber 1,830 µl 1,880 µl Notes:
10% aqueous APS must be made up fresh or stored frozen.
The recipe is suitable for 0.75 mm gel, multiply the recipe by 1.5 for 1 mm gel.
Prepare the solutions on ice to prevent untimely polymerization.
Acknowledgments
This research was financially supported by the Academy of Finland (project numbers 307335 and 303757), EU-funded Innovative Training Network (ITN) Solar Energy into Biomass (SE2B) Marie Skłodowska-Curie grant agreement (675006) and DPMLS. The protocol was adapted from publication ( Rantala et al., 2017 ). We declare no conflicting or competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Albertsson P.(2001). A quantitative model of the domain structure of the photosynthetic membrane. Trends Plant Sci 6: 349-358. [DOI] [PubMed] [Google Scholar]
- 2. Andersson B. and Anderson J. M.(1980). Lateral heterogeneity in the distribution of chlorophyll-protein complexes of the thylakoid membranes of spinach chloroplasts. Biochim Biophys Acta 593: 427-440. [DOI] [PubMed] [Google Scholar]
- 3. Aro E. M., Suorsa M., Rokka A., Allahverdiyeva Y., Paakkarinen V., Saleem A., Battchikova N. and Rintamäki E.(2005). Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J Exp Bot 56(411): 347-356. [DOI] [PubMed] [Google Scholar]
- 4. Blum H., Beier H. and Gross H. J.(1987). Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8: 93-99. [Google Scholar]
- 5. Caffarri S., Kouřil R., Kereïche S., Boekema E. J. and Croce R.(2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J 28(19): 3052-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Järvi S., Suorsa M., Paakkarinen V. and Aro E. M.(2011). Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem J 439: 207-214. [DOI] [PubMed] [Google Scholar]
- 7. Pesaresi P., Hertle A., Pribil M., Kleine T., Wagner R., Strissel H., Ihnatowicz A., Bonardi V., Scharfenberg M., Schneider A., Pfannschmidt T. and Leister D.(2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation . Plant Cell 21(8): 2402-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rantala M., Tikkanen M. and Aro E. M.(2017). Proteomic characterization of hierarchical megacomplex formation in Arabidopsis thylakoid membrane . Plant J 92: 951-962. [DOI] [PubMed] [Google Scholar]
- 9. Schägger H. and von Jagow G.(1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199: 223-231. [DOI] [PubMed] [Google Scholar]
- 10. Suorsa M., Rantala M., Mamedov F., Lespinasse M., Trotta A., Grieco M., Vuorio E., Tikkanen M., Jarvi S. and Aro E. M.(2015). Light acclimation involves dynamic re-organization of the pigment-protein megacomplexes in non-appressed thylakoid domains. Plant J 84: 360-373. [DOI] [PubMed] [Google Scholar]