Abstract
Accurate measurements of an organism’s fitness are crucial for measuring evolutionary change. Methods of fitness measurement are most accurate when incorporating an individual’s survival and fecundity, as well as accounting for any ecological interactions or environmental effects experienced by the organism. Here, we describe a protocol for measuring the relative mean fitness of Caenorhabditis elegans populations, or strains, through an assay that accounts for individual survival, fecundity, and intraspecific competitive ability in the presence of a bacterial parasite. In this competitive fitness assay nematodes from a focal population or strain are mixed with a GFP-marked tester strain in equal proportions, the mixture of nematodes are then exposed to a parasite, and the relative competitive fitness of the focal strain is determined by measuring the change in the ratio of focal nematodes to GFP-marked nematodes after one generation. Specifically, this protocol can be implemented to measure changes in nematode host fitness after experimental evolution by determining the relative competitive fitness of evolved versus ancestral nematode populations.
Keywords: Fitness assay, Experimental evolution, C. elegans, S. marcescens, Competition
Background
Accurate measurements of fitness and changes in fitness over time are critical for determining a population’s response to natural selection. Nonetheless, fitness is notoriously difficult to measure because it incorporates an individual’s survival, fecundity, reproductive timing, and must account for ecological and environmental effects on individuals. Although no protocol for measuring fitness is optimal under all possible conditions, measures of fitness that account for survival and fecundity, while holding ecological and environmental effects constant, are likely to provide reliable overall estimates of fitness for a given scenario. Here we describe a protocol for measuring the relative fitness differences between C. elegans populations or strains and for determining the change in relative fitness over evolutionary time in the presence of a bacterial parasite. We utilized the gram-negative bacterium, Serratia marcescens, as a virulent parasite when consumed by C. elegans. Especially, S. marcescens strain SM2170 is capable of killing C. elegans hosts within 24 to 48 h of ingestion ( Penley et al., 2017 ). This procedure makes use of Competitive Fitness Assays (CFAs) ( Lenski et al., 1991 ; Wiser and Lenski, 2015), utilizing intraspecific competition to compare the relative fitness between different nematode populations or strains ( Morran et al., 2009 ). Measurements of relative fitness, as determined via the CFA, incorporate survival and reproduction with intraspecific competition in a controlled environment to provide a comprehensive measure of fitness ( Penley et al., 2017 ).
Relative fitness in the presence of the bacterial parasite is determined by competing a focal strain with an isogenic GFP-labeled tester strain over the course of one generation and measuring the reproductive success of the focal strain against the tester strain ( Morran et al., 2009 ). Thus, this CFA accounts for survival against the parasite and host reproduction over the course of the nematode’s lifecycle. One single tester strain is used to measure the relative fitness of each focal nematode population or strain to facilitate comparisons of relative fitness between populations or strains. Importantly, the tester is marked with pharyngeal GFP to allow easy visualization of tester strain offspring versus focal population or strain offspring after one generation of competition. CFAs are initialized with a 50:50 mix of focal and tester strain individuals, and therefore, any deviation from 50:50 mix in the offspring indicates unequal competitive fitness between the focal and tester strains. An increase in the proportion of focal nematodes in the offspring indicates greater competitive fitness relative to the tester, while a decrease indicates reduced competitive fitness relative to the tester. The proportion of focal hosts in the offspring can be compared across multiple populations to measure the relative competitive fitness between focal strains or populations of interest. Importantly, competitive fitness measures are most effective when competing approximately equal numbers of individuals between two populations or strains with minor to moderate differences in competitive fitness. Uneven or variable starting ratios of strains or populations can confound measurements of relative fitness, while major differences in fitness between competing strains or populations are often difficult to accurately quantify (Wiser and Lenski, 2015).
This protocol is particularly useful for measuring evolutionary change after experimental evolution of C. elegans hosts in the presence of a bacterial parasite. First, the relative fitness of experimental host populations can be directly compared with the relative fitness of the ancestral population. Ancestral C. elegans populations can be stored at -80 °C during experimental evolution and then revived for CFAs to assess changes in the experimental population fitness over time (Gray and Cutter, 2014; Teotonio et al., 2017 ). Second, during experimental evolution, hosts may adapt to parasite exposure through altered life histories and/or increased levels of host defense. Therefore, measuring only survival in the presence of the parasite may not fully account for changes in host fitness. This CFA can account for changes in both life history and resistance that alter reproductive output in the presence of the parasite. Importantly, this procedure was originally developed to measure the change in C. elegans’ competitive fitness after multiple generations of evolution in presence of the bacterial parasite, Serratia marcescens ( Morran et al., 2009 ; Morran et al., 2014 ; Parrish et al., 2016 ; Penley et al., 2017 ). Nonetheless, this protocol can be adapted to measure the relative competitive fitness of any two or more C. elegans populations or strains in the presence of any relevant bacterial parasite. Further, it can be used to measure the change in relative competitive fitness over the course of experimental evolution for any C. elegans populations evolved in the presence of a bacterial parasite.
Materials and Reagents
1.5 ml micro-centrifuge tube (MIDSCI, catalog number: MID15C)
1,000 μl pipette tips (MIDSCI, catalog number: AVR4)
200 μl pipette tips (MIDSCI, catalog number: AVR1)
1000 μl wide bore pipette tips (Genesee Scientific, catalog number: 22-426)
200 μl wide bore pipette tips (Genesee Scientific, catalog number: 22-423)
Microscope slides (Fisher Scientific, catalog number: 12-550-19)
Semimicro spatula (Fisher Scientific, catalog number: 14-374)
Disposable inoculating loops, 10 μl (VWR, catalog number: 12000-810)
100 x 15 mm Petri dishes (Tritech Research, catalog number: T3301)
0.22 μm sterile syringe filter (Spectrum Chemical Manufacturing, catalog number: 882-66597)
Disposable plastic syringe (Thermo Fisher Scientific, catalog number: S7510-10)
Serratia marcescens strain SM2170, BSL2 (Sue Katz, Rogers State University)
Escherichia coli strain OP50, BSL1 (Caenorhabditis Genetics Center)
GFP-labeled C. elegans (strain JK2735) (Caenorhabditis Genetics Center)
Nematode Growth Media Lite powder (United States Biological, catalog number: N1005)
LB granules (Fisher Scientific, catalog number: BP9723-500)
Potassium Phosphate Monobasic (KH2PO4) (Fisher Scientific, catalog number: P288-100)
Sodium Chloride (NaCl) (Fisher Scientific, catalog number: S671-500)
Sodium Phosphate Dibasic Anhydrous (Na2HPO4) (Fisher Scientific, catalog number: S374-500)
Magnesium Sulfate Anhydrous (MgSO4) (Fisher Scientific, catalog number: M65-500)
Ampicillin Sodium Salt (Dot Scientific, catalog number: DSA40040-25)
Household bleach
LB Broth (see Recipes)
NGM Lite plates (see Recipes)
Escherichia coli (OP50)-seeded NGM Lite plates (see Recipes)
M9 Buffer (see Recipes)
1 M MgSO4 Solution (see Recipes)
Ampicillin 200 mg/ml (see Recipes)
Equipment
Hand tally counters (United Scientific Supplies, catalog number: HTCP01)
2 L flask (Corning, PYREX®, catalog number: 5320-2L)
P1000 μl pipetman (Eppendorf, model: Research® plus, catalog number: 3121000120)
P100 μl pipetman (Eppendorf, model: Research® plus, catalog number: 3121000074)
-80 °C freezer (Eppendorf, New BrunswickTM, model: Innova® U725)
Tabletop centrifuge for 1.5 ml micro-centrifuge tubes (Eppendorf, model: 5424)
20 °C controlled environment chamber (Percival Scientific, model: I36NLC8)
28 °C shaker incubator (Eppendorf, New BrunswickTM, model: Innova® 42R, catalog number: M1335-0010)
Stereomicroscope (Olympus, model: SZX16)
LED transmitted light illumination base (Olympus, model: SZX2-ILLT)
GFP filter for stereomicroscope (Olympus, model: SZX2-FGFP)
Stereomicroscope objective 7x-115x (Olympus, model: SDFPLAPO1XPF)
Fluorescence illumination lamp (Excelitas Technologies, model: X-Cite® 120Q)
X-Cite® Liquid Light Guide (Bulbtronics, Excelitas Technologies, model: 805-00038)
Autoclave (STERIS, model: SG-120)
Chemical fume hood (Kewaunee Scientific, model: H05)
Software
JMP Pro 12.0.1 (SAS Institute Inc., Cary, NC)
Procedure
Notes:
This protocol specifically describes measuring the change in a population’s competitive fitness after experimental evolution in the presence of Serratia marcescens (SM2170). However, it can be adapted to measure relative fitness between any two or more nematode populations of interest in the presence of any parasite that can grow and transmit under the conditions described. No specific experimental nematode strains or experimental evolution protocols are necessary to use this protocol simply as a means of measuring fitness. However, a clearly marked GFP strain is required.
Before experimental evolution, the generation 0 ancestor nematodes should be frozen at -80 °C. Similarly, at the end of experimental evolution, it will be necessary to freeze the experimentally evolved populations for use in these and any other subsequent assays. The freezing protocol can be found in Stiernagle (2006) (https://www.ncbi.nlm.nih.gov/books/NBK19649/).
This procedure spans over ~2 weeks. See Table 1 for an example of the procedure schedule.
Table 1. Example of Procedure Daily Schedule.
| Day | 1 | 8 | 10 | 11 | 12 | 15 |
|---|---|---|---|---|---|---|
| Procedure Step | A | B | C and D | D | E and F | G |
| Description | Thaw Experimental and Tester Populations | Prep for Synchronization |
Synchronize Nematode Populations and Inoculate Bacteria |
Prep Serratia Selection Plate |
Calculate Nematode Concentration and Compete Nematode Populations | Score SSP Assay |
-
Thawing experimental and tester populations
-
Remove one frozen tube for each of the following strains from the -80 °C freezer:
Generation 0 ancestor
Experimentally evolved population of interest
GFP-labeled tester strain (JK2735)
Note: If measuring relative competitive fitness, thaw 2 or more nematode populations of interest and the GFP-labeled tester strain.
Allow the tubes to thaw at room temperature.
After thawing, gently invert the tubes several times to mix the solution.
Pour the entire contents of the tube onto a labeled 100 mm E. coli OP50-seeded NGM Lite plate.
Allow the plates to absorb all the liquid content. If the liquid content is not fully absorbed, dry the plates briefly in the chemical fume hood with lids off.
Incubate these plates at 20 °C for 24 h.
Examine the plates to see that at least 50 nematodes survived thawing. If so, incubate the plates at 20 °C to allow the population to recover from thawing and expand (about 1 week). If not, thaw another freezer tube for the population of interest and repeat the thawing process.
-
-
Prepping for Synchronization
Once the generation 0 ancestor, experimentally evolved, and GFP-labeled tester populations have expanded to the point that hundreds of individuals are observed on each plate, you may proceed.
For each population, use a flame-sterilized metal spatula to cut approximately 2.5 cm2 of NGM Lite from the plate containing nematodes.
For each population, remove this piece of NGM Lite from the original plate and place it face down onto a new 100 mm OP50-seeded NGM Lite plate.
Incubate these new plates at 20 °C for 48 h.
After 48 h, view the plates from Step B4 under the microscope to check for eggs. If at least ~1,000 eggs are observed on the surface of the plate, proceed to Procedure C. If not, incubate the plates at 20 °C and check the next day. If the plate is exhausted of OP50, the nematodes will no longer produce eggs. In this case, repeat Steps B2-B5.
-
Synchronization
Note: The synchronization protocol is adapted from Stiernagle (2006). The purpose of synchronization is to kill all nematodes, leaving only the eggs that have been deposited on the plate to survive. This will age synchronize the population at the egg stage.
-
Working one plate at a time, add 2 ml of M9 buffer to the plate.
Note: Some of the M9 buffer will be absorbed into the plate. Therefore, adding 2 ml of M9 buffer to the plate will result in ~1 ml to be collected from the plate.
-
Tilt the plate side to side to wash the entire surface of the plate with M9 buffer.
Note: If the bacterial lawn on the plate appears thick and sticky, use a pipette tip to gently scrape the surface of the plate. This will help loosen the eggs.
Tilt the plate to allow all of the M9 buffer to pool in one area.
Use a pipette to remove the M9 buffer and move it into a 1.5 ml micro-centrifuge tube.
Repeat Steps C1-C4 so that each population is in its own micro-centrifuge tube.
Centrifuge tubes at 845 × g for 30 sec to form a pellet.
Remove the supernatant from each tube, using a new pipette tip between tubes.
-
Add 800 μl M9 buffer to each tube containing the pellets.
Note: Steps C9-C17 should be completed in 3 min or less to prevent the bleach from damaging the eggs.
-
Add 200 μl household bleach to each centrifuge tube containing the pellets and 800 μl M9 buffer.
Note: This will yield a 20% household bleach solution in each tube.
Close and invert the tubes several times to mix the bleach and M9 buffer, and to loosen the pellet.
Centrifuge the tubes at 845 × g for 30 sec.
Remove the supernatant from each tube, using a new pipette tip between tubes.
Wash each pellet with 1 ml M9 buffer. Close and invert the tubes several times to loosen the pellet.
Centrifuge the tubes at 845 × g for 30 sec.
Remove supernatant from each tube, using a new pipette tip between tubes.
-
Repeat Steps C13-C15.
Note: You will have washed the pellet twice.
Pipette the entire pellet from each tube onto its own new 100 mm OP50-seeded NGM Lite plate, being sure to change tips between tubes.
Incubate the plate at 20 °C for 48 h, or until the nematodes are at larval stage L4.
-
-
Preparation of Serratia Selection Plates (SSPs)
On the same day of synchronization (see Table 1–Day 10), inoculate OP50 and SM2170 (or relevant parasite) each into 5 ml LB. Incubate overnight at 28 °C, shaking at 160 rpm.
-
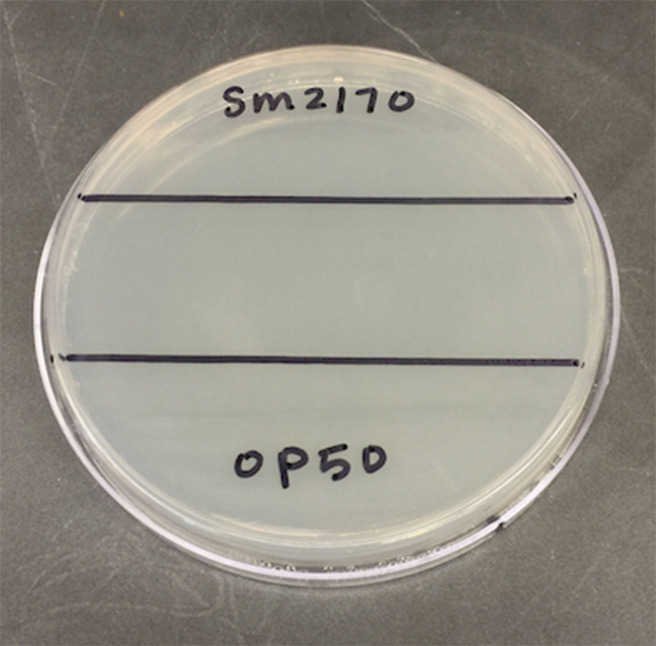
The next day, on the bottom of a 100 mm Petri dish filled with 25 ml NGM Lite, draw a ~2.5 cm stripe across the middle of the dish, dividing the dish into 3 sections (see Figure 1).
Notes:
It is important to use plates that are not damp. In order to dry the surface of the plates, they can be placed in a chemical fume hood with lids off for ~20 min. Be sure to remove excess condensation from the plate lid as well.
Make enough SSPs to give proper replication of the ancestor and each experimentally evolved population. We recommend at least 3 replicates for each.
Pipette 35 μl of OP50 (~ 2.9 x 109 CFU/ml) onto one of the far thirds of the NGM Lite and use a sterile inoculation loop to evenly spread the OP50 within the section. It is important to plate the OP50 first to prevent SM2170 from contaminating and being spread in this section of the plate. When pipetting the bacteria onto the SSP, discharge only to the first stop of the pipette to prevent splattering on the plate.
On the opposite far third of the NGM Lite, pipette 35 μl of SM2170 (~ 7.8 x 108 CFU/ml) and use a sterile inoculation loop to evenly spread the bacteria within this section. Again, be sure to discharge to only the first pipette stop. The middle third will be left blank.
Keep the plates level to prevent bacteria from spreading into other sections, incubate the SSPs overnight at 28 °C.
-
Calculating Nematode Concentration
-
48 h after bleach synchronizing the nematode populations, add 2 ml M9 buffer to the plates containing synchronized nematodes.
Notes:
Nematodes should be at L4 larval stage.
Some of the M9 buffer will absorb into the plate. Therefore, adding 2 ml of M9 buffer to the plate will result in approximately 1 ml to be collected from the plate.
Tilt the plate with M9 buffer side to side to wash the entire surface of the plate.
-
Keeping the plate tilted to pool the M9 buffer, and using a new wide bore pipette tip between populations, collect each population of nematodes into its own 1.5 ml micro-centrifuge tube.
Note: Wide bore tips are used when transferring live nematodes to avoid damaging the C. elegans and to increase the precision of measurements.
Centrifuge the tubes containing nematodes at 94 × g for 30 sec to form a pellet.
Wash the pellet 2 times with M9 buffer (as described in Steps C12-C16, except use 94 × g when centrifuging).
After washing the populations twice, resuspend the nematodes in 1 ml M9 buffer.
Invert the micro-centrifuge tube several times to mix.
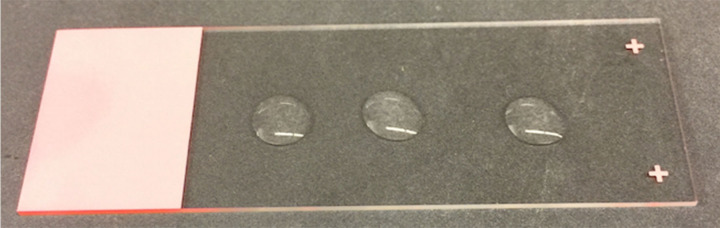
Working one population at a time, and using a wide bore pipette tip, take 3-20 μl samples, and place onto a microscope slide (see Figure 2).
-
Under the microscope, count the number of nematodes in each 20 μl sample using a hand tally counter.
Note: In order to count accurately, there should be approximately 30-50 nematodes in each sample. If the number of nematodes is outside this range, appropriately dilute or concentrate the centrifuge tubes containing the nematodes, and take 3 new 20 μl samples onto a microscope slide. This may need to be done more than once to get the correct approximate range.
Calculate the average of the 3 samples and divide by 20 to get the average concentration of nematodes per 1 μl.
-
Use the following equation to calculate the volume needed to give 100 nematodes:
Volume =
where x is the average concentration of nematodes per 1 μl, calculated in Step E10.
-
Repeat Steps E7-E11 for the generation 0 ancestor, each experimentally evolved C. elegans population and the GFP-labeled tester strain.
Note: The nematode population of interest will be competed against the GFP-labeled tester strain on the same plate. Therefore, if the calculated volumes of the focal strain and the GFP-tester strain total to more than ~150 μl, it will be necessary to concentrate the nematodes, repeat Steps E7-E11.
-
-
Competing either ancestral or experimental “focal” populations vs. a tester strain on the SSP
-
Prior to introducing nematodes to the SSP, add a stripe of ampicillin across the middle section of the plate. (20 μl of 200 mg/ml ampicillin) (see Figure 3)
Note: SM2170 is very motile. Ampicillin prevents SM2170 from spreading into the OP50 portion of the SSP.
To the parasite side of the SSP, add both the focal host population of interest and the GFP-labeled tester strain in the volumes calculated in Step E11. Replicate as needed for each experimental population and the ancestral population to generate multiple pseudo replicate CFAs for each population.
-
Pipette onto 3 OP50-seeded NGM Lite plates the volume calculated in Step E11 for the following:
The generation 0 ancestor
The experimentally evolved strain
The GFP-labeled tester strain
Note: These plates will be counted in Procedure G to determine the actual average number of hosts added to the SSP assay plates.
Allow the SSPs to remain level and dry at room temperature for approximately 20 min.
Incubate SSPs at 20 °C for 48-72 h, or enough time for the competed populations to reproduce and for the offspring to be at larval stage L1/L2.
-
-
Scoring the SSP assays
Note: It is necessary to use the GFP filter with the stereomicroscope and fluorescence illumination lamp in order to visualize the pharyngeal GFP marker.
-
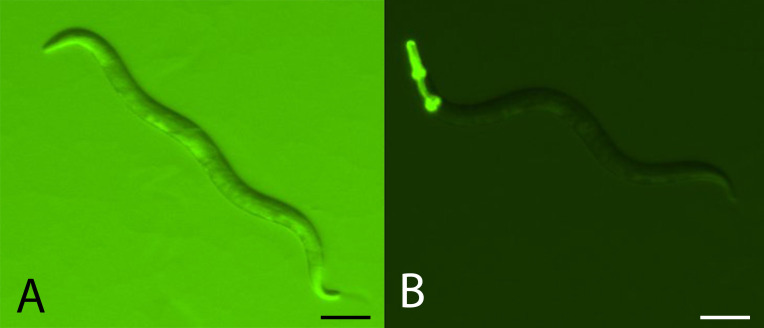
Within the OP50 third of the SSP, count the number of focal and GFP-tester offspring out of a sample of 200 total offspring in a cross-section of the SSP (see Figure 4).
Note: It is necessary to alternate between high and low backlighting during counting. High background light will allow the counter to visualize all the offspring, while low background light will allow the counter to differentiate between focal and GFP-labeled strains.
Count the number of adult hosts on each of the 3 OP50-seeded plates for each population from Step F3. Take the average of the 3 counts and use this number to calculate the initial ratio of focal to tester strain parents.
-
-
Calculate the mean relative fitness and the percent change in fitness after experimental evolution
-
The relative competitive fitness of a focal population or strain (experimental population or ancestral population) to the tester strain can be calculated as the relative change in the ratio of non-GFP to GFP expressing individuals over the course of the assay. Specifically, this is determined as:
WX =
where WX is the relative fitness of the focal strain, x is the proportion of focal strain offspring, and y is the proportion of focal strain parents initially plated on the SSP. See Supplementary Data for a sample data set from competitive fitness assays conducted on C. elegans host populations evolved in the presence of SM2170 (Evolution) and host populations evolved in the presence of heat-killed SM2170 (Control) for 30 generations. Relative fitness values greater than one indicate greater fitness for the focal strain relative to the tester strain. Whereas, relative fitness values of less than one indicate greater tester strain fitness relative to the focal strain. Finally, calculate the mean relative fitness for each focal strain by averaging the relative fitness values across each pseudo replicate.
-
After experimental evolution, the percent change in mean competitive fitness for an experimental population can be calculated as:
(E – A)/A
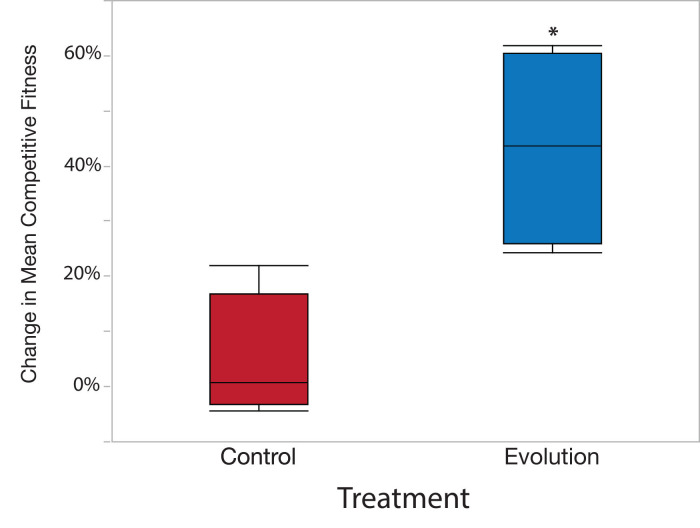
where, E is the mean relative fitness of the experimental population across all pseudo replicates and A is the relative fitness of the ancestral population across all pseudo replicates (Supplementary Data). Positive percent change values indicate that the experimental population increased in fitness during experimental evolution. Conversely, negative percent change values indicate a loss of fitness in the experimental population. Importantly, the percent change in mean fitness should be calculated separately for each independently evolved replicate population. We found a greater percent change in mean fitness in populations evolved under the Evolution treatment than those evolved under the Control treatment (Supplementary Data and Figure 5).
-
Figure 1. View of the bottom of a 100 mm Petri dish with ~2.5 cm strip drawn across the middle.
Figure 2. Microscope slide with 3-20 μl samples of nematodes in M9 buffer.
Figure 3. Serratia selection plate (SSP) design including ampicillin stripe.
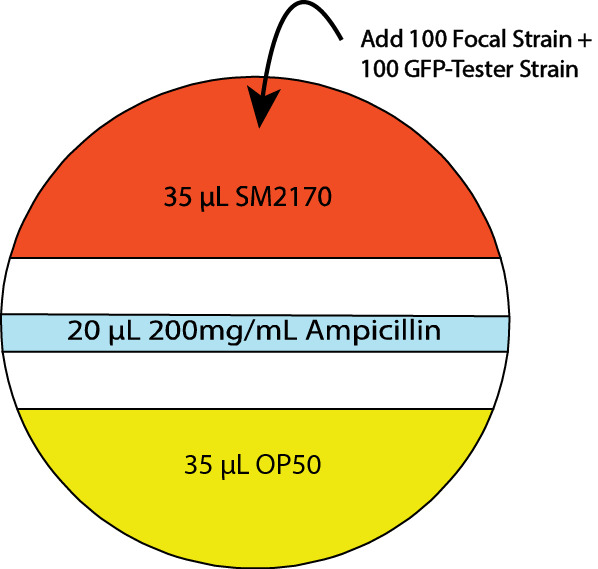
Figure 4. Comparing focal and GFP-tester strains.
A. Example of how both non-GFP and GFP expressing C. elegans individuals will appear under high backlight conditions. Scale bar = 0.1 mm. B. Example of GFP expressing C. elegans under low backlight conditions. Focal strains do not express GFP. Scale bar = 0.1 mm.
Figure 5. Sample data results using example data from Supplementary Data.
Data analysis
These analyses were previously described in Penley et al. (2017) with the exception that the mean proportion of experimental to tester strain (GFP) offspring was analyzed as opposed to the relative competitive fitness measure. All analyses were performed in JMP Pro 12.0.1.
The values of mean relative competitive fitness and the percent change in mean competitive fitness can be compared between particular strains, populations, or treatments using Analysis of Variance (ANOVA) and subsequent least squares mean linear contrast tests to test factors within different fixed effects (0.05 alpha value). However, the data must conform to the ANOVA assumptions of normality and homogeneity of variance. Shapiro-Wilk test (normality) and Levene’s test (homogeneity of variance) can be applied to the dataset to test the appropriateness of using ANOVA to analyze the data. If the data do not conform to the assumption of ANOVA despite transformation, then the data can be analyzed via a nonparametric Kruskal Wallis test. We tested the main effect of treatment (Evolution vs. Control) on values of the percent change in mean competitive fitness (Table 2), and found a significant treatment effect (Table 2 and Figure 5).
Table 2. ANOVA table.
Testing the main effect of treatment (Evolution vs. Control) on the mean percent change in competitive fitness.
| Source | Sum of Squares | df | Mean Square | F | p |
| Model | 0.297 | 1 | 0.297 | 12.17 | = 0.013 |
| Error | 0.147 | 7 | 0.024 | ||
| Total | 0.444 | 8 |
It is important to note that independent replicate populations within experimental evolution treatments serve as the true replicates for analysis. Thus, the mean of all pseudo replicates is used for statistical analysis as the value representing each independently evolved replicate population. Accordingly, replicate population serves as a random effect and is nested with the relevant treatment in the ANOVA model.
Notes
The GFP marker carried by the tester strain is dominant. This CFA works most effectively when assaying only C. elegans hermaphrodites from both the focal strain/population and the test strain. C. elegans males can outcross with hermaphrodites. Focal population males may outcross with tester strain hermaphrodites. If this occurs the offspring from these hybrid crosses will express GFP, resulting in underestimates of the focal strain relative fitness.
Recipes
-
LB Broth (200 ml)
Add 5 g LB granules to 200 ml dH2O and stir until dissolved
Aliquot 5 ml into each test tube and add a cap to each
Autoclave for 20 min and store at 4 °C for up to 3 months
-
NGM Lite plates (1 L)
Add 29 g NGM Lite powder to 2 L flask containing 1 L dH2O
Cover with aluminum foil and autoclave for 20 min
Once cooled enough to touch, pour ~25 ml NGM Lite into 100 mm Petri dishes
Store at 4 °C for up to 3 months
-
Escherichia coli (OP50)-seeded NGM Lite plates
Inoculate E. coli OP50 in LB, incubate at 28 °C overnight, shaking at 160 RPM
The next day add 200 μl OP50 culture to each NGM Lite plate
Spread the culture over the entire surface of the plate
Incubate at 28 °C overnight
Store at 4 °C for up to 1 month
-
M9 Buffer (1 L) (adapted from Stiernagle, 2006)
Add 3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, and 1 ml 1 M MgSO4 solution to a graduated cylinder
Add dH2O to 1 L and stir until dissolved
Aliquot into bottles and keep lids loose
Autoclave for 20 min
Store at room temp for up to 6 months
-
1 M MgSO4 Solution (100 ml)
Add 12.037 g MgSO4 to 100 ml dH2O
Stir until dissolved and store at room temp for up to 1 year
-
Ampicillin 200 mg/ml (10 ml)
Add 2 g ampicillin sodium salt into a 15 ml tube and fill dH2O up to 10 ml
Stir until dissolved and filter-sterilize (0.22 μm)
Store in 1 ml aliquots at -20 °C for up to 1 year
Acknowledgments
This protocol was adapted from previous work ( Morran et al., 2009 ; Morran et al., 2011 ; Morran et al., 2014 ; Parrish et al., 2016 ; Penley et al., 2017 ). We thank S. Scholz, M. Parmenter, J. Anderson, and P. Phillips for assistance in developing this protocol. Additionally, we thank R. Parrish II, O. Schmidt, M. Allen, and A. Khalid for help in refining the protocol. We also thank three anonymous reviewers for improving this manuscript. Funding for this work was provided by Emory University to LTM. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
Competing interests
The authors declare that they have no conflicts of interest or competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Supplementary Data.
References
- 1. Gray J. C. and Cutter A. D.(2014). Mainstreaming Caenorhabditis elegans in experimental evolution . Proc Biol Sci 281(1778): 20133055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lenski R. E., Rose M. R., Simpson S. C. and Tadler S. C.(1991). Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations . American Naturalist 138(6): 1315-1341. [Google Scholar]
- 3. Morran L. T., Parmenter M. D. and Phillips P. C.(2009). Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature 462(7271): 350-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Morran L. T., Parrish R. C. 2nd, Gelarden I. A., Allen M. B. and Lively C. M.(2014). Experimental coevolution: rapid local adaptation by parasites depends on host mating system. Am Nat 184 Suppl 1: S91-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Morran L. T., Schmidt O. G., Gelarden I. A., Parrish R. C, 2nd, and Lively C. M.(2011). Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science 333(6039): 216-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parrish R. C., 2nd, Penley M. J. and Morran L. T.(2016). The integral role of genetic variation in the evolution of outcrossing in the Caenorhabditis elegans-Serratia marcescens host-parasite system . PLoS One 11(4): e0154463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Penley M. J., Ha G. T. and Morran L. T.(2017). Evolution of Caenorhabditis elegans host defense under selection by the bacterial parasite Serratia marcescens . PLoS One 12(8): e0181913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stiernagle, T. 2006. Maintenance of C. elegans in The C. elegans research community, ed. WormBook: the online review of C. elegans biology. [DOI] [PMC free article] [PubMed]
- 9. Teotonio H., Estes S., Phillips P. C. and Baer C. F.(2017). Experimental evolution with Caenorhabditis nematodes . Genetics 206(2): 691-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wiser M. J. and Lenski R. E.(2015). A comparison of methods to measure fitness in Escherichia coli . PLoS One 10(5): e0126210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.