Abstract
This protocol can be applied to analyze the direct interaction between a soluble protein and a target ligand molecule using Isothermal Titration Calorimetry (ITC, Malvern). ITC allows the biophysical characterization of binding between label-free, non-immobilized and in-solution biomolecules by providing the stoichiometry of the interaction, the equilibrium binding constants and the thermodynamic parameters. ITC monitors heat changes (released and/or absorbed) caused by macromolecular interactions with no restrictions of buffer and molecular weight of the macromolecules.
Keywords: ITC, Calorimetry, Macromolecular interaction, Binding affinity, KAT1 channels, 14-3-3 proteins
Background
Macromolecular interactions are critical cellular events as they form the basis for signal transduction pathways. Thus, macromolecular interactions are an essential field of research, as they allow a deeper understanding of the molecular mechanisms which underlie both physiological and pathophysiological processes, and the rational design of drugs able to modulate disease-causing macromolecular binding events.
In this context, Isothermal Titration Calorimetry (ITC) is a powerful technique for the characterization of macromolecular interactions. ITC determines the heat change that occurs upon the binding of two molecules. Heat can be absorbed (endothermic reaction) or released (exothermic reaction). ITC monitors such heat changes by determining the differential power, provided by heaters of the instrument to both the reference and the sample cells, needed for counteracting any temperature difference between the two cells during the binding reaction such that no difference in temperature arises between the reference and sample cells (Figure 1).
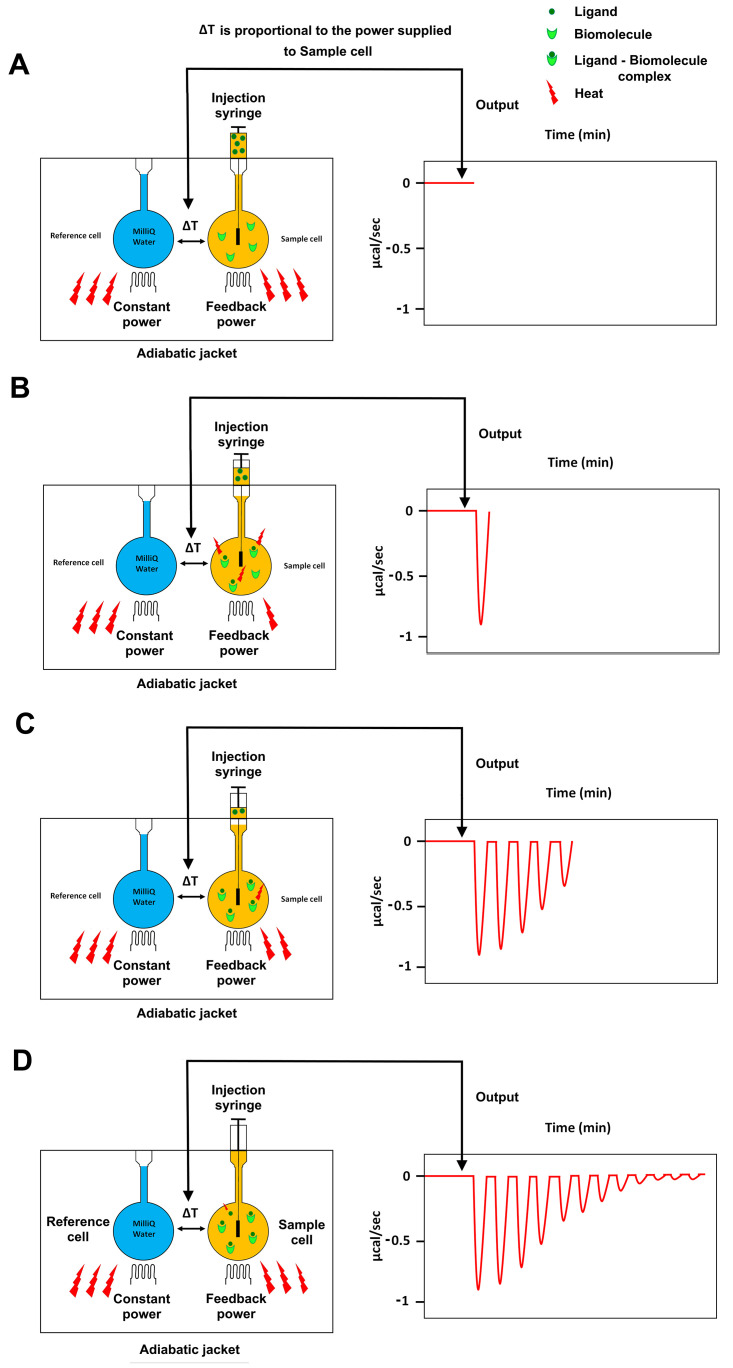
Figure 1. Principle of Isothermal Titration Calorimetry (ITC).
A. Cartoon representation of an isothermal titration calorimetry instrument composed of: a reference cell filled with MilliQ water; a sample cell containing a biomolecule; and an automated injection syringe containing the other binding molecule (ligand) used to titrate the ligand into the sample cell. The sample and reference cells are surrounded by an adiabatic jacket. The system is able to detect temperature differences between the reference and sample cells and to maintain an absence of temperature difference between them (ΔT = 0) by supplying power to both the reference and the sample cell via two heaters. The output of the instrument is the power (μcal/sec) required to maintain ΔT = 0 between the reference and sample cells. B-D. The temperature difference between the reference and the sample cell, induced by the ligand–biomolecule binding, is converted into the power needed to bring the two cells back to the same temperature during the binding reaction. As the titration proceeds, the biomolecule in the sample cell becomes saturated with the ligand, so that less interactions occur and consequently the heat change decreases (C) until the biomolecule is fully saturated and the instrument detects only heat change due to the dilution of the ligand (D).
ITC provides important information about the nature of the macromolecular interaction: the binding stoichiometry (N); the thermodynamic parameters of the binding reaction (enthalpy, H, entropy, ∆S, and Gibbs free energy, ∆G); the strength of the interaction (the equilibrium association constant KA, from which the more commonly used equilibrium dissociation constant KD can be derived).
Among the methods used to characterize macromolecular interactions, ITC has two major advantages: i) the biomolecules are free to move in solution and are not labelled, which insures a direct characterization of the binding event, unbiased by labelling and/or by limitation on molecule motions due to their immobilization on a surface; ii) ITC is the only method that allows a detailed characterization of the binding event by providing not only the binding affinity, but also other critical information, i.e., the binding stoichiometry and the thermodynamic parameters. This information can help significantly in the understanding of the molecular mechanism of the binding reaction, even when no structural data are yet available. Furthermore, they can be used as complementary data to validate structural results.
Recently, I presented crystallographic and functional data showing that the K+ inward rectifier KAT1 (K+ Arabidopsis thaliana 1) channel is regulated by the direct binding of 14-3-3 proteins ( Saponaro et al., 2017 ). In particular, I identified a 14-3-3 mode III binding site at the very C-terminus of KAT1 and co-crystallized it with tobacco 14-3-3 proteins (14-3-3c) to describe the protein complex in atomic detail. The structural results were complemented/supported by measuring, through ITC, the interaction between a synthetic KAT1 C-terminal phosphopeptide (CPP) and 14-3-3c. ITC was employed to quantify the stoichiometry, the equilibrium binding affinity and the thermodynamic parameters of the 14-3-3c-CPP binding reaction.
The aim of this protocol is to provide a detailed description of the setting procedure of an ITC experiment, highlighting the crucial steps and related concerns, and providing, at the same time, a well-established strategy to overcome such problems. Moreover, the present protocol describes the analysis of an ITC measurement of the single binding event in a 14-3-3c/CPP interaction.
Materials and Reagents
Pipette tips (20 μl, 200 μl and 1,000 μl)*
Glass syringe 2.5 ml, 18GA, 8.5IN, PT3 (Malvern Panalytical, catalog number: SYN161714)
Disposable borosilicate glass tubes, 0.7 ml, 6 x 50 mm (O.D. x Height) (DWK Life Sciences, Kimble Chase, catalog number: 73500-650)
0.22 μm vacuum filter*
-
Purified Nicotiana tabacum 14-3-3c protein recombinantly expressed in E. coli. Protein stock concentration is 100 μM
Note: The protein was expressed and purified as described in Saponaro et al. (2017).
Synthetic phosphopeptide* (corresponding to the last 5 residues of Arabidopsis thaliana KAT1 channel (hereafter CPP) dissolved in MilliQ Water. Peptide stock concentration is 15 mM) (Peptide was purchased from CASLO ApS as a lyophilized powder, purity ≥ 98%)
NaCl*, stock concentration 5 M (Sigma-Aldrich, catalog number: 746398)
HEPES*, stock concentration 1 M, pH 7.4 (Sigma-Aldrich, catalog number: RDD002)
NaOH*, stock concentration 10 M (Sigma-Aldrich, catalog number: S8045)
MgCl2*, stock concentration 2 M (Sigma-Aldrich, catalog number: M8266)
β-mercaptoethanol*, stock concentration 100 mM (Sigma-Aldrich, catalog number: M6250)
MilliQ Water*
Sample buffer (see Recipes)
*Note: These items can be purchased from any suitable vendor.
Equipment
Pipettors (P10, P20, P200, P1000)*
Microcalorimeter (Malvern Panalytical, model: MicroCal VP-ITC)
Vacuum pump (provided together with MicroCal VP-ITC, Malvern Panalytical, model: ThermoVac, catalog number: 29013182)
Filling syringe (provided together with MicroCal VP-ITC, Malvern Panalytical)
Plastic tubes, 3 ml (provided together with MicroCal VP-ITC, Malvern Panalytical)
Benchtop refrigerated centrifuge with rotor FA-45-48-1 (Eppendorf, model: 5427 R, catalog number: 5409000210)
Stir bars 7 mm for mixing the solutions within the vacuum pump (ThermoVac) (Malvern Panalytical, catalog number: BAR150020-005)
UV spectrometer (Eppendorf, BioSpectrometer® basic supplied with Eppendorf μCuvette® G1.0, that is suitable for volumes of 1.5-10 μl, catalog number: 6135000904)
pH meter*
*Note: These items can be purchased from any suitable vendor.
Software
Origin software (version7, MicroCal, Malvern Instruments Ltd, RRID: SCR_014212)
Procedure
Measurements were carried out at 25 °C using a VP-ITC Microcalorimeter (Malvern Instruments Ltd). The volume of both the reference and the sample cell was 1.4 ml.
-
The solutions containing the titrant (CPP) and the titrated (14-3-3c) molecules have to be identical to avoid heat changes due to the dilution of the titrant into the sample cell.
The following table describes the appropriate dilutions required to prepare the solutions containing the titrant (CPP) and the titrated (14-3-3c) molecules at 500 μM and 50 μM concentration respectively:
Titrant (CPP, 500 μM) Titrated molecule (14-3-3c, 50 μM) --- 1,000 μl 14-3-3c (stock 100 μM) 16.7 μl (stock 15 mM in MilliQ water)1/30 of the total volume66.7 μl MilliQ water1/30 of the total volume483.3 μl buffer (see Recipes) 933.3 μl buffer (see Recipes) Total volume: 500 μl Total volume: 2,000 μl As highlighted in the table, since CPP stock is in MilliQ water, in preparing 14-3-3c dilution it is necessary to use the same proportion of MilliQ water used in the preparation of the CPP dilution to have identical solutions for both titrant (CPP) and titrated molecule (14-3-3c).
Prior to loading MilliQ water and sample solutions into the machine, load them into the 3 ml plastic tubes (see equipment) together with stir bars 7 mm and degas them for 5 min by using the Vacuum pump (ThermoVac). This instrument is indeed designed to allocate the 3 ml plastic tubes and favorite the degassing by combining the vacuum with a gently stirring.
Centrifuge both protein and titrant at 16,000 × g, for 5 min, at 4 °C.
-
Filling procedure of reference/sample cell (Video 1):
Insert the 2.5 ml glass syringe until you gently touch the bottom of the cell with the tip of the syringe.
Inject slowly and constantly the solution, removing, at the same time, the needle from the cell. In this way, the cell is filled from the bottom to the top, and the operation avoids the formation of bubbles.
Position the tip of the syringe on the ledge present just below the visible portion of the cell port and draw the liquid to remove the excess of liquid from the cell.
Fill the reference cell with MilliQ water.
-
Fill the sample cell with the protein (in this case 14-3-3c, 50 μM).
Note: In order to obtain a reliable measurement, both reference and sample cells must be precisely filled. This operation can be performed by loading a larger volume of solution (2 ml recommended) compared to the maximum capacity of the cells. Often, the low yield of purified protein limits this operation. Nevertheless, use at least 1.6 ml to be sure to appropriately load the sample. The proper loading of the solutions is a critical step in order to avoid the formation of bubbles, which severely impair the measurements.
The volume of the injection syringe is 300 μl. Thus, to appropriately load the titrant, prepare at least 500 μl of the titrant sample.
-
Load the titrant (in this case CPP, 500 μM) into the injection syringe with the following procedure (Video 2):
Connect the plastic tubing of a filling syringe to the filling port of the injection syringe.
Place the titrant sample in a glass filling tube, and insert it into the pipette stand of the machine.
Slowly withdraw the plunger of the filling syringe to draw up the solution containing the titrant.
Purge the bottom "close fill port" as soon as the titrant solution begins to exit the filling port.
Perform a refill step in order to remove any air bubbles from the injection syringe.
-
Titrate CPP using injection volumes of 10 μl. The spacing between consecutive injections is 150 sec. A total of 29 injections are performed.
Note: The volume of the first injection is 2 μl, 1/5 of the settled injection volume. This is needed to equilibrate the interface between the tip of the injection syringe and the surrounding solution of the sample cell. The heat change caused by this first injection has to be discarded during data analysis. Because of this first 2 μl injection, a total of 292 μl of titrant out of the 300 μl loaded into the syringe will be used and 8 μl will be left into the syringe after the experiment.
-
It is worth noting that the concentration of the titrant and the titrated molecules, as well as the injection volumes and the spacing time between consecutive injections, is appropriate for the molecules used in this protocol. These parameters must be optimized for every sample.
Note: In the initial phase of setting parameters, a 10-fold difference in the concentration between the titrant and the titrated molecule should be maintained. This usually ensures a complete saturation of the binding reaction for those interactions having a binding affinity in the nM to μM range. If there is evidence that the binding affinity of the system under study is in the hundred μM to mM range, however, the fold difference of the concentration between the titrant and the titrated molecule should be increased.
Moreover, the spacing should be set at 180 sec, which is usually a long enough time to ensure the complete recovery of the heat baseline upon titrant injection. In case your measurement requires a different time to return to the baseline, adapt the spacing time to your system. Data extrapolated from measurements performed with different spacing times can be combined, as long as the baseline is completely recovered and the concentration of titrant/titrated molecules and the injection volumes are the same.
Video 1. Filling procedure of reference/sample cell.
The video shows the steps necessary for the proper loading of the solutions into the reference/sample cells (see Steps 5a-5c). For simplicity, the video describes only the filling of the sample cell.
Video 2. Filling procedure of the injection syringe.
The video shows the steps necessary for the proper loading of the titrant into the injection syringe (see Steps 9a-9e).
Data analysis
Calorimetric data were analysed with Origin software (version7, MicroCal, RRID:SCR_014212) and equations were described for the single-site binding model ( Wiseman et al., 1989 ). It is worth noting that the procedure generally follows the “Instrument Handbook” of MicroCal VP-ITC system (Malvern Instruments Ltd).
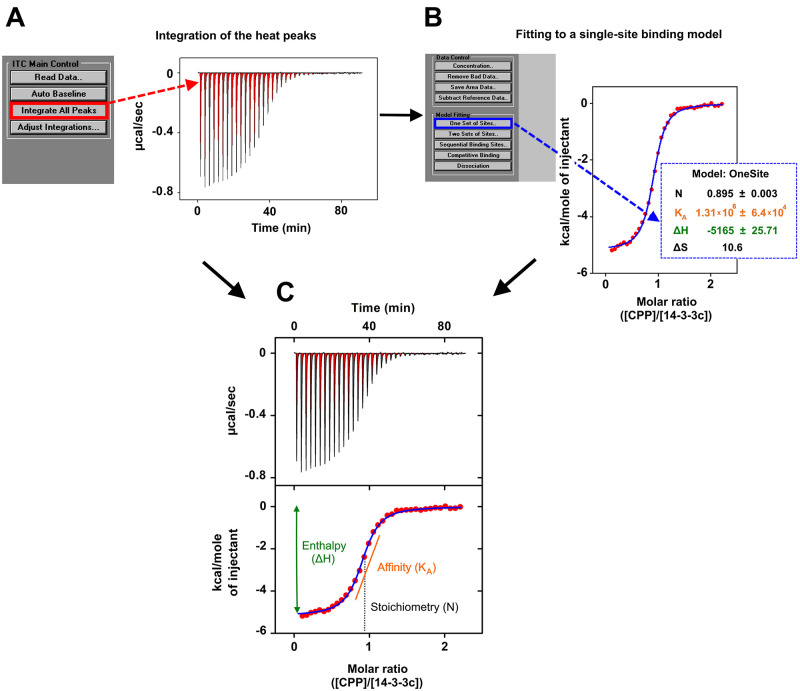
Figure 2 summarizes the processing of the ITC raw data and their subsequent analysis by using a single-binding model needed to obtain the binding curve for the CPP–14-3-3c interaction. Panel A shows negative heat changes (μcal/sec) during successive injections of 10 μl of CPP (500 μM) into the sample cell containing 14-3-3c (50 μM). This behavior, according to the basic ITC principles described in the Background section and illustrated in Figure 1, indicates that the CPP–14-3-3c interaction releases heat, so that the binding reaction is exothermic.
Figure 2. ITC measurement of CPP–14-3-3c interaction.
A. Raw ITC data of CPP (500 μM) binding to 14-3-3c (50 μM). The area of the negative heat changes (μcal/sec) during successive injections of 10 μl of CPP into the sample cell containing 14-3-3c was calculated by selecting the option “integrate all peaks” from the ITC main control list in order to measure the amount of heat released by each CPP injection of the titration. The area of the peaks is colored in red. B. The quantified heat changes (red filled circles), normalized per mole of CPP injected and plotted against the molar ratio of CPP : 14-3-3c, were fitted to a single-site binding model by selecting the option “One set of sites” in the model fitting list implemented in the software. This generates the fitting curve (blue line) that interpolates the heat changes (red filled circles), and the related values of stoichiometry (N, black labelled), association equilibrium constant (KA, orange labelled), enthalpy (∆H, green labelled), and entropy (∆S black labelled) of the binding reaction. All these values are shown in the blue dotted box. C. Final ITC figure composed by an upper panel (corresponding to panel A) and lower panel (corresponding to panel B). Lower panel shows also the schematic representation of the calculations performed by the software to obtain: ∆H as the difference between the initial H value and the plateau H value (green line with double arrowheads); KA, which is the slope value of the orange line that intercepts the exponential phase of the isothermal curve (blue line); N, which is to the molar ratio (CPP:14-3-3c) at the centre of the isothermal curve (indicated by the black dotted line).
The software automatically integrates the peaks. This operation is graphically represented in Figure 2, Panel A, by the peaks colored in red. A critical aspect in the data analysis is the correct assessment of the integration details: the baseline and the integration range. The software automatically sets the integration details and integrates the peaks. However, there are cases where the signal to noise ratio is low and the automatic calculation of the integration details is not accurate enough. In these cases, both the baseline and the integration range need to be manually adjusted, following the “Instrument Handbook” instructions of MicroCal VP-ITC.
Moreover, the software automatically normalizes the integrated peaks to the CPP concentration, and plots them against the molar ratio of CPP (titrant) : 14-3-3c (titrated molecule) (red filled circles in Figure 2, Panel B). In Panel B, the solid blue line interpolating the red circles represents a nonlinear least-squares fit to a single-site binding model ( Wiseman et al., 1989 ). The software calculates the stoichiometry (N), the association equilibrium constant (KA), the enthalpy (∆H), and the entropy (∆S) of the binding reaction. In Figure 2, Panel B, these values are shown within the blue dotted box. N, KA, and ∆H are directly derived from the isothermal curve, and for this reason they yield a fitting error. On the other hand, ∆S, which is indirectly calculated, does not yield a fitting error.
The Gibbs free energy (∆G) is not calculated by the software, but it can be calculated by the operator (see below).
Figure 2C, which is composed by panels A and B, corresponds to the final ITC figure most commonly used for publication.
For a better comprehension of the operations performed by the software, panel C includes a graphic representation of how the software calculates, starting from the isothermal curve, ∆H, N, and KA.
The binding enthalpy (∆H), which is due to hydrogen bonds and van der Waals interactions, is calculated as the difference between the starting H value and the plateau H value (see green line in Figure 2C). As stated above, the ∆H value is normalized per mole of the titrant.
The stoichiometry (N) corresponds to the molar ratio at the center of the isothermal curve (see the black dotted line in Figure 2C).
Note: An accurate assessment of the concentration of the biomolecules is very important, because the stoichiometry parameter is extremely sensitive to their concentration.
The association equilibrium constant (KA) corresponds to the slope of the line that intercepts the exponential phase of the isothermal curve (see blue line in Figure 2C). It is possible to manually calculate the dissociation equilibrium constant (KD), which is the equilibrium constant more commonly used to describe the affinity of biomolecules, by using the following equation:
KD = 1/KA
The binding entropy (∆S), which is due to the desolvation of biomolecules and eventually to conformational changes induced by the complex formation, is indirectly calculated using the following equation:
The Gibbs free energy (∆G) is indirectly calculated using the following equation:
KA
Notes
As already stated in the "Data analysis" section, the concentration of the biomolecules is extremely important for the calculation of the stoichiometry of the binding relation. Therefore, the first time that the concentration of the biomolecules under study is assessed, the recommendation is to use at least two different methods in order to be sure of the accuracy of their concentration. For example, in this specific case, the first time I quantified the concentration of the 14-3-3c stock, I used two different methods: a Bradford-based method, and a 280 nm absorption-based method using a molar extinction coefficient value (ε) of 14-3-3c, which I theoretically calculated using the ProtParam tool (https://web.expasy.org/protparam/).
The ITC measurements should be repeated at least three times to have enough data for statistics.
A control measurement should be performed by titrating the titrant against the buffer, in order to evaluate the heat change caused by the titrant injection per se. If the titrant injection causes a significant heat change, this measurement should be used to subtract the heat change originating from the dilution of the titrant into the buffer from the titrant - titrated molecule binding relation.
Recipes
-
Sample buffer
100 mM NaCl
30 mM HEPES (pH 7.4)
10 mM MgCl2
2 mM β-mercaptoethanol
Notes:
Do not use Tris-HCl and DTT because they can severely affect the measurements by producing intense heat changes. If it is necessary to maintain a reductive state, the recommendation is to use β-mercaptoethanol.
The above buffer works with the protein and the molecule used in this specific protocol. Buffer must be optimized for other samples.
Acknowledgments
This protocol was adapted from Saponaro et al. (2017), and generally follows the “Instrument Handbook” of the MicroCal VP-ITC system (Malvern Instruments Ltd). This work was supported by Lincean Academy (Italian Science Academy), Giuseppe Levi foundation.
Conflicts of interest or competing interests: none.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Saponaro A., Porro A., Chaves-Sanjuan A., Nardini M., Rauh O., Thiel G. and Moroni A.(2017). Fusicoccin activates KAT1 channels by stabilizing their interaction with 14-3-3 proteins. Plant Cell 29(10): 2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wiseman T., Williston S., Brandts J. F. and Lin L. N.(1989). Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem 179(1): 131-137. [DOI] [PubMed] [Google Scholar]