Abstract
As the sister clade of seed plants, ferns are significant materials for plant phylogeny research. However, the genomic DNA extraction protocol for fern samples like modified CTAB method still lacks robustness. Here, we found that the amount and condition of the pinnae samples are critical for gDNA extraction in fern, Adiantum capillus-veneris L. In 500 μl CTAB solution, the recommended amount of pinnae is about 10-20 mg (2-3 pieces). The condition of the pinnae must be instantly-picked from a plant cultivated in a suitable environment. With these factors under control, it is highly reproducible to get the high-quality gDNA with low degradation rate.
Keywords: Fern, DNA extraction, Adiantum capillus-veneris L. , CTAB method, Fern pinnae
Background
The CTAB method has been applied to gDNA extraction from Adiantum capillus-veneris L. ( Han et al., 2012 ; Li et al., 2017 ). However, the protocol still shows instability and high degradation rate in our experiment. The pinnae of ferns accumulate large quantities of secondary metabolites, such as polysaccharides and polyphenols, which is adverse to DNA extraction ( Ponnusamy et al., 2015 ).
To increase the robustness of gDNA extraction, we optimized the protocol basically from two aspects: 1) Reduce the amount of material and use only 2-3 pinnae (about 10-20 mg). 2) The pinnae must be freshly picked from a plant cultured in suitable cultivation environment (temperature: 25 °C, humidity: 65%, 16-h white light/8-h dark treatment) and instantly used for DNA extraction. Compared with the classical method, we can get gDNA of higher quality and lower degradation rate from ferns with this optimized protocol.
Materials and Reagents
1.5 ml Eppendorf tubes (Eppendorf®)
Steel beads (Diameter: 3.175 mm)
Nutrient soil
Vermiculite
Pipette tips (Corning, Axygen®, catalog number: T-1000-B; T-200-Y; T-300)
-
Freshly-picked pinnae of Adiantum capillus-veneris
Note: The plant is cultured in the cultivation room of PKU for several years and generations, which makes it domesticated.
Liquid nitrogen
β-mercapto-ethanol (BME) (Thermo Fisher Scientific, Gibco®, catalog number: 21985023)
PVP-40 (AMRESCO, catalog number: 0507-500G)
Phenol:Chloroform:Isoamyl alcohol (25:24:1) (Coolaber, catalog number: SL2040-100ML) (purity: AR)
Chloroform, purity: AR
Isopropanol, purity: AR
75% ethanol, purity: AR
Ultrapure water
CTAB (AMRESCO, catalog number: 0833-1KG)
Sodium chloride, purity: GR
EDTA (AMRESCO, catalog number: 0322)
Tris (AMRESCO, catalog number: 0497-5KG)
RNase I
HCl, purity: AR
M3 (Marker III) (TIANGEN Biotech, catalog number: MD103)
2% CTAB (see Recipes)
10% RNaseI (TaKaRa, catalog number: 2158-1) (see Recipes)
Equipment
Pipettes (Eppendorf®)
Stainless steel spoon
Mortar (diameter: 100 mm) and pestle (size: 117 mm)
Tissue grinder (Retsch®, TissueLyser II)
Water bath (65 °C)
High-speed centrifuges (Refrigeration is not necessary)
Refrigerator (-20 °C)
Oven (37 °C)
Magnetic stirrer
Electrophoresis equipment
Procedure
-
Sample Preparation
Here, we describe two methods for sample preparation: One is liquid nitrogen method, and the other is steel beads method. Both of them require the pinnae from plants growing under a proper condition. Moreover, the amount of sample is also a critical factor. The steel beads method is suitable for high-throughput gDNA extraction.
Notes:
The condition of pinnae is critical for gDNA yield rate. Pinnae from a plant growing under suitable environment are required for this method (temperature: 25 °C, humidity: 65 %, 16-h white light and 8-h dark treatment; The soil condition is not a critical factor in this protocol. For reference, we usually mix nutrient soil with vermiculite (about 10:1) as the culture medium.
Avoid using too much sample for gDNA isolation. The amount of sample is critical for the performance. For 500 μl CTAB solution, 10-20 mg pinnae is the suggested mass. Figure 1 provides a reference size of pinnae in different phases.
Liquid nitrogen method
Cool stainless steel spoon, mortar and pestle in liquid nitrogen.
-
Harvest 2-3 freshly picked pinnae (total wet mass: 10-20 mg) and grind in liquid nitrogen to fine powder using the thoroughly cooled mortar and pestle. And then transfer the powder into a tube.
Note: All equipment should be fully cooled in liquid nitrogen during this procedure. Be careful not to let the liquid nitrogen dry while grinding, and the grinded tissue must remain frozen prior to extraction with CTAB solution. Accidental thawing may result in DNA degradation.
-
Add 500 μl 2% CTAB solution (see Recipes) to the tube. Mix by flicking the bottom of the tube until the sample is thoroughly suspended.
Note: Make sure adding BME and PVP to the 2% CTAB solution and the final concentration is 2% BME and 0.2% PVP.
Steel beads method
-
Add 2-3 steel beads in a new clean tube. Add 500 μl 2% CTAB solution (see Recipes) to the tube.
Note: Make sure adding BME and PVP to the 2% CTAB solution and the final concentration is 2% BME and 0.2% PVP.
Harvest 2-3 freshly picked pinnae (total mass: 10-20 mg) and transfer into the tube. Grind the material with the tissue grinder at room temperature (25.0 hz, 5 min).
Briefly centrifuge for 3-5 sec to avoid the samples sticking to the lid of the tube. Let stand in a 65 °C water bath for 10 min to soften the tissue and make it easier to break. Cool them at room temperature for about 2 min.
Grind the material again with the tissue grinder (25.0 hz, 10 min). See if the pinnae are fully grinded. If so, then continue with the following steps. If not, repeat Steps 3 and 4. The sample before and after grinding is shown in Figure 2.
-
DNA extraction
-
Briefly centrifuge the CTAB treated samples for 3-5 sec. Incubate in a 65 °C water bath for 1 h. Mix by inverting the tube for 3-4 times during incubation in the water bath. Take out the samples and cool at room temperature for 1 min.
Note: The samples can be cooled on ice or at RT. You can do the cooling step while preparing the reagents and materials and there is no need to get ice for this step.
Add 500 μl phenol:chloroform:isoamyl alcohol (25:24:1) to the tube. Shake vigorously to mix thoroughly. Centrifuge the samples at 12,000 × g for 15 min.
Transfer the supernatant to a new clean tube with pipettes. Be careful not to transfer the white protein in the middle layer.
Add to the aqueous phase an equal volume of chloroform. Mix by shaking and inverting. Centrifuge the sample at 12,000 × g for 15 min.
Transfer the supernatant to a new clean tube with pipettes. Be careful not to transfer the white protein in the middle layer.
Add to the aqueous phase an equal volume of isopropyl alcohol. Mix by gently inverting the tube and let it stand at -20 °C for at least 1 h. Centrifuge the sample at 12,000 × g for 15 min.
-
Decant the supernatant and be careful not to lose the pellet.
Note: Pellet may be difficult to see.
Add 700 μl 75% ethanol to the tube.
Centrifuge the sample at 12,000 × g for 5 min. Decant the supernatant carefully. Briefly centrifuge for 3-5 sec to collect the residual liquid and remove it with a pipette. Open the lid of the tube, lay it down horizontally and let stand overnight to air-dry the pellet.
-
Add 30 μl ultrapure water or TE to the tube and let it stand for 10 min to dissolve the gDNA sample.
Note: Dissolve the sample in ultrapure water for subsequent DNA sequencing. For long long-term storage, dissolve the sample in 30 μl TE.
Add 1 μl 10% RNaseI to the tube and incubate at 37 °C for 1 h.
Store gDNA sample at -20 °C.
-
Figure 1. Different phases of Ac pinnae to show available samples for gDNA extraction.
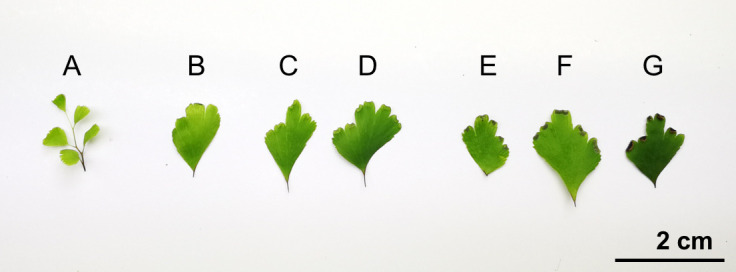
A-B. Juvenile pinnae; C-D. Pinnae with green and premature sporangia; E-F. Pinnae with brown and mature sporangia; G. Pinna with dehiscent sporangia.
Figure 2. The material before the grinding process (Before) and the fully grinded material (After).

Data analysis
The result of gel electrophoresis is shown in Figure 3. When the amount of pinnae is excessive, all the gDNA samples show high degradation rate (Figure 3A). When we decreased the amount of material, the degradation rate is dramatically reduced (Figure 3B). However, if we draw the material from a fern cultured in an unsuitable environment, the result still shows erratic degradation rate. And picking the material from a plant in a suitable environment can solve this problem.
Figure 3. Gel electrophoresis of gDNA samples extracted from material with different amount or growing condition.
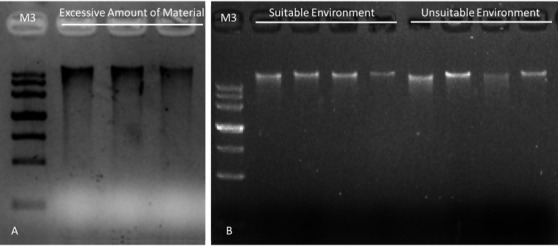
A. gDNA samples extracted from excessive amount of material (about 60 mg each). B. gDNA samples extracted from moderate amount of material (10-20 mg each). Suitable cultivation environment: temperature: 25 °C, humidity: 65%, 16-h white light and 8-h dark treatment. Unsuitable cultivation environment: temperature: 20 °C, humidity: 21% (natural sunlight treatment: about 12-h light and 12-h dark) time: 24 h.
Recipes
-
2% CTAB solution (400 ml)
CTAB 8 g
Sodium chloride 8 g
0.5 M EDTA 16 ml
1.0 M Tris-HCl (pH 7.5) 40 ml
PVP-40 0.8 g
ddH2O to 400 ml
Mix with magnetic stirrer overnight
Add 8 ml BME immediately before use
-
10% RNase I (10 μl)
RNase I 1 μl
Ultrapure water 9 μl
Acknowledgments
This protocol was optimized from procedures published in Han et al., (2012). We thank the Institute of Vegetables and Flowers, CAAS for providing a greenhouse for Adiantum cultivation. The authors declare that there are no conflicts of interest or competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Han J. D., Qin M. and Wang X.(2012). Comparison of different DNA extraction protocols in Adiantum capillus-veneris L . J Henan Agric Univ 02: 70–75..(in Chinese) [Google Scholar]
- 2. Li X., Han J. D., Fang Y. H., Bai S. N. and Rao G. Y.(2017). Expression analyses of embryogenesis-associated genes during somatic embryogenesis of Adiantum capillus-veneris L. In vitro: New insights into the evolution of reproductive organs in land plants . Front Plant Sci 8: 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ponnusamy Y., Chear N. J., Ramanathan S. and Lai C. S.(2015). Polyphenols rich fraction of Dicranopteris linearis promotes fibroblast cell migration and proliferation in vitro . J Ethnopharmacol 168: 305-314. [DOI] [PubMed] [Google Scholar]


