Abstract
Background
Astragalus Radix (AR)-Panax notoginseng (PN), a classical herb pair, has shown significant effects in treating diabetic nephropathy (DN). However, the intrinsic mechanism of AR-PN treating DN is still unclear. This study aims to illustrate the mechanism and molecular targets of AR-PN treating DN based on network pharmacology combined with bioinformatics.
Materials and Methods
The Traditional Chinese Medicine Systems Pharmacology database was used to screen bioactive ingredients of AR-PN. Subsequently, putative targets of bioactive ingredients were predicted utilizing the DrugBank database and converted into genes on UniProtKB database. DN-related targets were retrieved via analyzing published microarray data (GSE30528) from the Gene Expression Omnibus database. Protein-protein interaction networks of AR-PN putative targets and DN-related targets were established to identify candidate targets using Cytoscape 3.8.0. GO and KEGG enrichment analyses of candidate targets were reflected using a plugin ClueGO of Cytoscape. Molecular docking was performed using AutoDock Vina software, and the results were visualized by Pymol software. The diagnostic capacity of hub genes was verified by receiver operating characteristic (ROC) curves.
Results
Twenty-two bioactive ingredients and 189 putative targets of AR-PN were obtained. Eight hundred and fifty differently expressed genes related to DN were screened. The PPI network showed that 115 candidate targets of AR-PN against DN were identified. GO and KEGG analyses revealed that candidate targets of AR-PN against DN were mainly involved in the apoptosis, oxidative stress, cell cycle, and inflammation response, regulating the PI3K-Akt signaling pathway, cell cycle, and MAPK signaling pathway. Moreover, MAPK1, AKT1, GSK3B, CDKN1A, TP53, RELA, MYC, GRB2, JUN, and EGFR were considered as the core potential therapeutic targets. Molecular docking demonstrated that these core targets had a great binding affinity with quercetin, kaempferol, isorhamnetin, and formononetin components. ROC curve analysis showed that AKT1, TP53, RELA, JUN, CDKN1A, and EGFR are effective in discriminating DN from controls.
Conclusions
AR-PN against DN may exert its renoprotective effects via various bioactive chemicals and the related pharmacological pathways, involving multiple molecular targets, which may be a promising herb pair treating DN. Nevertheless, these results should be further validated by experimental evidence.
1. Introduction
Diabetes mellitus (DM) is a common metabolic disorder characterized by chronic hyperglycemia due to defective insulin secretion and/or utilization, which has been considered an enormous global health concern [1]. According to the International Diabetes Federation data, 352 million people between the ages of 20 and 65 years are suffering from DM worldwide, which will occur in up to 486 million by 2045 [2]. Diabetic nephropathy (DN), a major microvascular complication of DM, is a leading cause of end-stage renal disease [3]. Research studies indicate that 20%–40% of patients suffering from DM develop DN and patients with DN have a higher risk of death with 3 to 12 times than those with DM alone [4, 5]. Advances have been made in the treatment of DN in recent years with the introduction of lifestyle changes, including exercise and diet, and therapies such as angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, sodium-glucose cotransporter 2 inhibitors, and glucagon-like peptide-1 receptor [3, 6]. Unfortunately, these drugs were reported to have adverse effects, such as diabetic ketoacidosis, gastrointestinal, and hepatotoxicity [7, 8], and there are still no pharmacotherapies available to cure or reverse disease progression. Highlighting the requirement for novel therapeutic with better efficacy and safe treating DN is still necessary.
Chinese herbal medicine with various bioactive chemicals and the related pharmacological pathways, involving multiple cellular and molecular targets, and fewer side effects has been frequently and extensively used for thousands of years in China [9], which may be to provide beneficial and effective therapy for DN. According to the theory of traditional Chinese medicine (TCM), the basic pathology of DN is qi deficiency and blood stasis [10]. Hence, tonifying qi and activating blood circulation are the basic therapeutic principles for DN. Astragalus Radix (AR, Huang qi in Chinese) was initially recorded in Shennong Bencao Jing, which has been extensively known to exert the tonifying Qi effect. Panax notoginseng (PN, San qi in Chinese) was found in Bencao Gangmu with multiple beneficial effects, especially activating blood circulation and decreasing swelling to relieving pain. The herb pair is considered the most basic form of TCM therapy containing two special Chinese herbal medicines, which plays a vital role in connecting mutual enhancement, assistance, and restraint [11]. AR-PN, a classic highly valued and widely used herb pair, is well known to possess functions with tonifying qi and activating blood circulation. Modern pharmacological research shows that AR treatment ameliorated the severity of DN by inhibiting inflammation-related gene IL-1β and IL-18 expression and fibrosis indexes in DN rats [12]. Likewise, it has been confirmed that AR possesses antioxidative stress effects [13]. Previous research demonstrated that PN can significantly reduce albuminuria, proteinuria, and serum creatinine and improve the metabolism of serum lipids in patients with DN [14]. In addition, both in vivo and in vitro research studies confirmed that the combination of AR and PN could strengthen the renoprotection effects by inhibiting inflammation, alleviating oxidative stress, and upregulating autophagy [15]. Taken together, the AR-PN herb pair playing a renoprotection role in DN is mainly aimed at anti-inflammation and antioxidative stress. However, the underlying pharmacological mechanism of the AR-PN herb pair in the treatment of DN remains to be illustrated. A novel approach to clarify the pharmacological mechanisms of the efficacy of the AR-PN herb pair on DN is necessary.
Network pharmacology was first proposed by Hopkins in 2007, which provides a convenient and systematic method to reveal the complex mechanisms of drug treating disease through identifying core targets based on the theory of systems biology and network pharmacology [16]. Moreover, network pharmacology is also contributed to integrate and extract possible signaling pathways that drugs prevent and treat diseases to improve the therapeutic effect of drugs, and the pattern is “drug-target-disease.” It has been applied to explore the mechanisms of TCM treating some diseases, including diabetes mellitus [17] and ischemic stroke [18], showing that applying network pharmacology to elucidate the mechanisms of bioactive components and the potential of TCM is an emerging trend. Bioinformatics is the application of tools of computation and analysis to the capture and interpretation of biological data, which is an interdisciplinary field of computer science, mathematics, physics, and biology [19]. Currently, bioinformatics has been increasingly recommended in the study of biological fields, particularly in genomics and proteomics. miRNAs involve the fine-tuning of mRNA abundance via binding to the 3′ untranslated region of a target mRNA and results in translational repression. Increasing evidence suggests that the downregulation or upregulation of miRNAs is closely associated with podocyte injury, inflammation, accumulation of the extracellular matrix, fibrosis, and so on in DN, concerning their potential as biomarkers and miRNA modulation as a therapeutic option for DN [20, 21]. Besides, some microarray data analysis studies related to DN have been performed and numerous DEGs have been identified, which may be potential biomarkers and target candidates in DN [22]. Except for DN, bioinformatics strategies are also involved in identifying the core genes of other diseases, such as diabesity [23], familial hypercholesterolemia [24], lung squamous cell carcinoma [25], and prostate cancer [26], indicating that bioinformatics plays vital roles in predicting potential biomarkers and providing therapeutic options. Collectively, network pharmacology integrated with bioinformatics is a novel strategy to study ingredient identification, target prediction, and therapeutic options.
Consequently, we integrated network pharmacology with Gene Expression Omnibus (GEO) database to further identify the comprehensive pharmacological mechanisms, screen for potential molecular targets, and explore underlying pathways of AR-PN herb pair on treating DN, which may provide a guideline in the further research and development. The flowchart of this study is shown in Figure 1.
Figure 1.
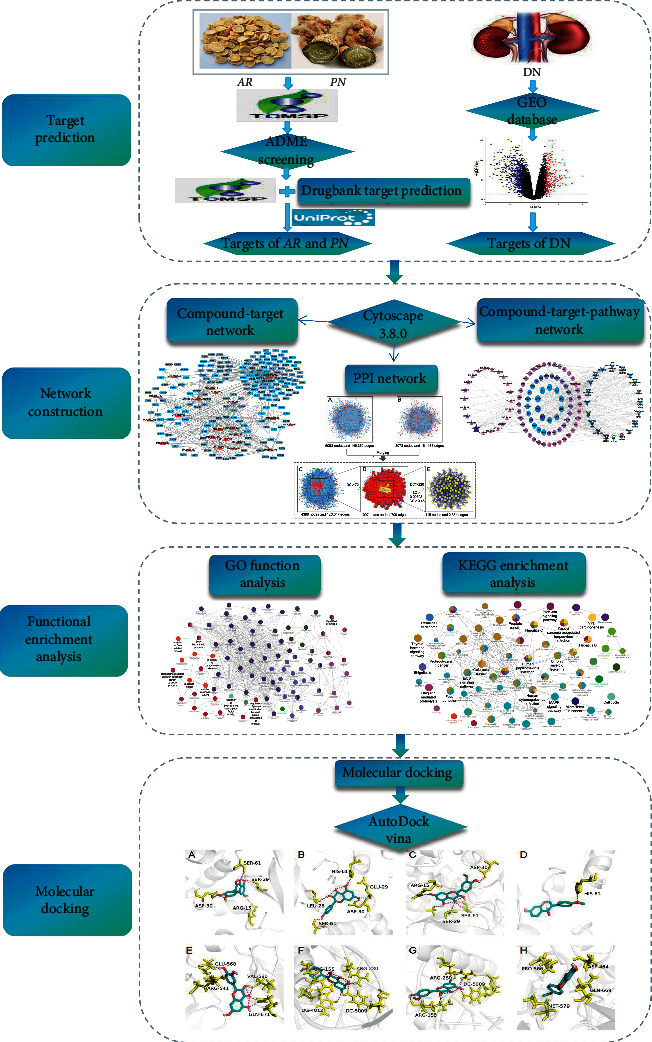
The flowchart of network pharmacology study. AR: Astragalus Radix; PN: Panax notoginseng; DN: diabetic nephropathy.
2. Material and Methods
2.1. Screening of Active Ingredients of AR-PN Herb Pair
All active ingredients of the AR-PN herb pair were collected from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database (http://tcmspw.com/tcmsp.php), which is a unique systematic pharmacology platform to display the relationships between components, targets, and diseases [27]. The absorption, distribution, metabolism, and excretion (ADME) model [28], which was an important contributor to predict the pharmacokinetic properties of chemical ingredients, was used to screen the compounds of the AR-PN herb pair. Two vital parameters, i.e., oral bioavailability (OB) and druglikeness (DL), among all ADME-related properties were used to identify chemical ingredients of the AR-PN herb pair. The OB refers to the relative amount of a drug absorbed into the systemic circulation after extravascular administration [29]. DL is used to estimate the similitude physiochemical properties or/and structural features between an ingredient and certified drug candidates [30], which is calculated by the following formula:
| (1) |
where “X” represents the active ingredients of the AR-PN herb pair and “Y” stands for the average DL index of all molecules that are collected from the DrugBank database (https://www.drugbank.ca) [31]. In our present study, bioactive compounds were collected according to the threshold values of OB ≥ 30% and DL ≥ 0.18.
2.2. Prediction of the AR-PN Herb Pair-Related Targets
Identification of putative targets of the AR-PN herb pair bioactive ingredients was performed with DrugBank. Subsequently, all proteins of each bioactive ingredient of the AR-PN herb pair were converted into genes in the UniProt database (http://www.uniprot.org/) [32] with species as “Homo sapiens” limited. Ultimately, all putative targets of the AR-PN herb pair were obtained after removing duplicated targets. Moreover, we used Cytoscape 3.8.0 software (http://www.cytoscape.org/) to establish and visualize the component-target network of the AR-PN herb pair.
2.3. Differentially Expressed Gene Search, Identification, and Analysis of DN
Expression profiling data and Raw CEL data from the GSE30528 dataset [33] were acquired from the GEO database according to Affymetrix GPL570 platform and Affymetrix Human Genome U133 Plus 2.0 Array, which included nine samples from DN glomeruli tissues and 13 samples from controlled normal glomeruli tissues. Then, probe IDs were applied to identify the corresponding genes. Subsequently, differentially expressed genes (DEGs) between DN glomeruli tissues and controlled normal glomeruli tissues were screened by the limma package of R software. DEGs with P < 0.05 and |log2 fold change (FC)| > 1 were selected as genes of DN and were visualized using a volcano plot.
2.4. Construction of Protein-Protein Interaction Network
Protein-protein interaction (PPI) is the core of almost all biological processes, which present the interaction of proteins and may provide potential targets for therapeutic intervention [34]. The PPI networks of the AR-PN herb pair putative targets as well as DN-related DEGs were constructed and visualized using the BisoGenet [35], a plugin of Cytoscape 3.8.0, which were established based on the currently available PPI databases, including Biological General Repository for Interaction Datasets (BioGRID), Database of Interacting Proteins (DIP), Biomolecular Interaction Network Database (BIND), IntAct Molecular Interaction Database (IntAct), Human Protein Reference Database (HPRD), and Molecular INTeraction Database (MINT). Subsequently, a merged network was performed according to the intersection data of the two networks.
2.5. Network Topological Characteristics Analysis
The three crucial topological properties, including betweenness centrality (BC), closeness centrality (CC), and degree centrality (DC), were used to calculate and analyze topological properties of every node in the merged interaction network using the CytoNCA [36], a plugin of Cytoscape 3.8.0. The node is more important, and the received quantitative value is higher. First, the degree of centrality was calculated and the network was extracted according to more than twice the median degree of DC. Then, more than the median values of DC, BC, and CC as the threshold values were used to further extract hub nodes in the network. Ultimately, a core subnetwork was obtained for the ensuing analysis.
2.6. GO and KEGG Pathway Enrichment Analysis
GO, a freely available public resource, provides a standardized language to systematically describe the functional properties of gene products from all species, which consists of three ontologies, namely, biological process (BP), cellular component (CC), and molecular functional (MF) [37]. BP describes the functioning of biological objectives to which they contribute, CC describes the place of a gene product within the cell and their extracellular environments, and MF represents the biochemical activities, such as binding and catalysis, of gene products [38]. KEGG is a highly complex network structure, which has been an exceptionally comprehensive site for describing metabolic pathways, gene signaling networks, and practical application of genomic information [39]. The biological interpretations of the hub genes in the core network were reflected by GO and KEGG pathway enrichment analysis using ClueGO, a plugin of Cytoscape 3.8.0, which is utilized to visualize nonrepetitive biological terms as functionally grouped networks. It is worth noting that the P value less than or equal to 0.01 was set as the threshold in both the GO or KEGG functional categories. Furthermore, the component-target-pathway network was established and visualized using Cytoscape 3.8.0 software.
2.7. Molecular Docking
Molecular docking, a method of analyzing the binding site and affinity between the conformation of molecules and macromolecular targets, has been considered a crucial technique for structure-based drug discovery [40]. The core DN-related genes targeted by the AR-PN herb pair bioactive components was identified according to the component-target-pathway network, which were further validated by molecular docking with the experimentally verified AR-PN herb pair bioactive components possessing renoprotective effects on DN, including quercetin [41], kaempferol [42], formononetin [43], and isorhamnetin [44]. First, the mol2 formats of these bioactive components were obtained from the TCMSP database and PubChem database, which were converted to the format of pdb and pdbqt via Pymol software and AutoDock Tools 1.5.6, respectively. Consequently, the 3-dimensional structures of potential receptors saved in pdb format were downloaded from the PDB database (https://www.wwpdb.org/). The removal of the ligand and water in the protein was performed via AutoDock Tools 1.5.6., which was saved in pdbqt format. Subsequently, molecular docking was performed to evaluate the combination mode and affinity between the ligand-receptor and molecules by AutoDock Vina, which was run with default settings. The smaller the lowest energy value is, the more stable and the stronger affinity between ligand and receptor binding. Ultimately, we selected the top two receptor proteins with the lowest energy value and the ligand for visualization and construction by Pymol software.
2.8. Receiver Operating Characteristic (ROC) Curves for Hub Genes
To evaluate the diagnostic accuracy of each hub gene for DN, ROC curves were plotted and area under the curve (AUC) values were calculated, which were performed with “pROC” package of R software 4.0.3. P < 0.05 was considered to be significant.
3. Results
3.1. Screening of Bioactive Components and Putative Targets of the AR-PN Herb Pair
A total of 27 bioactive components, including 19 components in AR, seven components in PN, and one component in both AR and PN, were identified which met the criterion of OB ≥ 30% and DL ≥ 0.18 as the threshold from the TCMSP database. The information of bioactive components of AR-PN herb pair is shown in Table 1. Five bioactive ingredients without any corresponding target, whose mol IDs were MOL000374, MOL000398, MOL000438, MOL000439, and MOL007475, were excluded. A total of 461 target proteins and 252 target proteins of AR and PN active components were collected, respectively. After deleting duplicate targets, 189 putative targets of AR-PN herb pair were identified. The compound-target network of AR-PN herb pair consisted of 211 nodes and 583 edges, which is shown in Figure 2.
Table 1.
Information of bioactive components of AR and PN.
| Herb | Mol ID | Molecule name | OB (%) | DL |
|---|---|---|---|---|
| AR | MOL000211 | Mairin | 55.38 | 0.78 |
| AR | MOL000239 | Jaranol | 50.83 | 0.29 |
| AR | MOL000296 | Hederagenin | 36.91 | 0.75 |
| AR | MOL000033 | (3S,8S,9S,10R,13R,14S,17R)-10,13-Dimethyl-17-[(2R,5S)-5-propan-2-yloctan-2-yl]-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol | 36.23 | 0.78 |
| AR | MOL000354 | Isorhamnetin | 49.6 | 0.31 |
| AR | MOL000371 | 3,9-di-O-Methylnissolin | 53.74 | 0.48 |
| AR | MOL000374 | 5'-Hydroxyiso-muronulatol-2',5'-di-O-glucoside | 41.72 | 0.69 |
| AR | MOL000379 | 9,10-Dimethoxypterocarpan-3-O-beta-D-glucoside | 36.74 | 0.92 |
| AR | MOL000380 | (6aR,11aR)-9,10-Dimethoxy-6a,11a-dihydro-6H-benzofurano[3,2-c]chromen-3-ol | 64.26 | 0.42 |
| AR | MOL000387 | Bifendate | 31.1 | 0.67 |
| AR | MOL000392 | Formononetin | 69.67 | 0.21 |
| AR | MOL000398 | Isoflavanone | 109.99 | 0.3 |
| AR | MOL000417 | Calycosin | 47.75 | 0.24 |
| AR | MOL000422 | Kaempferol | 41.88 | 0.24 |
| AR | MOL000433 | FA | 68.96 | 0.71 |
| AR | MOL000438 | (3R)-3-(2-Hydroxy-3,4-dimethoxyphenyl)chroman-7-ol | 67.67 | 0.26 |
| AR | MOL000439 | Isomucronulatol-7,2'-di-O-glucosiole | 49.28 | 0.62 |
| AR | MOL000442 | 1,7-Dihydroxy-3,9-dimethoxy pterocarpene | 39.05 | 0.48 |
| PN | MOL001792 | DFV | 32.76 | 0.18 |
| PN | MOL001494 | Mandenol | 42 | 0.19 |
| PN | MOL007475 | Ginsenoside F2 | 36.43 | 0.25 |
| PN | MOL002879 | Diop | 43.59 | 0.39 |
| PN | MOL005344 | Ginsenoside Rh2 | 36.32 | 0.56 |
| PN | MOL000358 | Beta-sitosterol | 36.91 | 0.75 |
| PN | MOL000449 | Stigmasterol | 43.83 | 0.76 |
| AR and PN | MOL000098 | Quercetin | 46.43 | 0.28 |
AR: Astragalus Radix; PN: Panax notoginseng; OB: oral bioavailability; DL: druglikeness.
Figure 2.
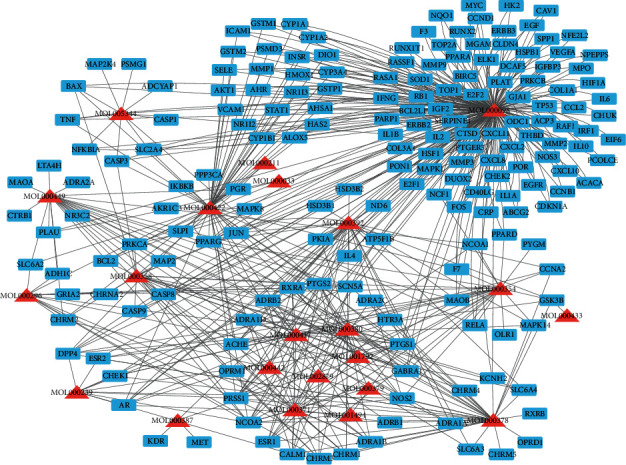
Compound-target network of AR-PN herb pair. Blue squares represent targets, and red triangles represent ingredients of AR-PN herb pair.
3.2. Identification of DN-Related DEGs
Ultimately, 850 DN-related DEGs were identified between DN glomeruli tissues and controlled normal glomeruli tissues with |log2 FC| > 1 and P < 0.05 by analyzing the sequencing data downloaded from the GEO database. A volcano plot of the distribution of DEGs is exhibited in Figure 3, which includes 253 upregulated genes dotted in red, 597 downregulated genes dotted in blue, and other genes without significant differences dotted in black.
Figure 3.
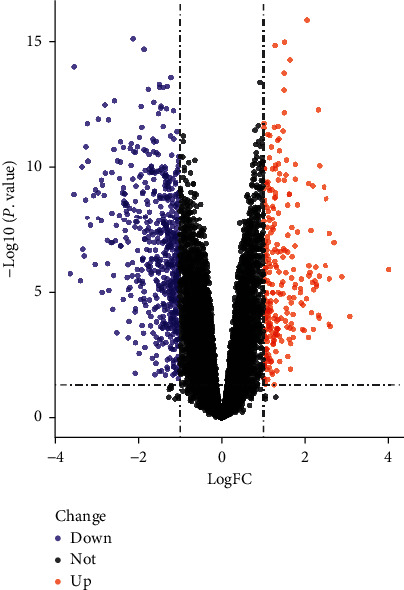
A volcano plot of differentially expressed genes of DN. The red dots represent significantly upregulated genes, and the blue dots represent significantly downregulated genes.
3.3. PPI Network Construction, Merging, and Topology Analysis
To illustrate the interactions and underlying mechanisms between AR-PN herb pair putative targets and DN-related DEGs, we visually constructed PPI networks of AR-PN herb pair putative targets and DN-related DEGs, respectively. The PPI network of AR-PN herb pair putative targets contained 6063 nodes and 149,352 edges (Figure 4(a)), while the PPI network of DN-related DEGs contained 8073 nodes and 181,168 edges (Figure 4(b)). In the PPI network graph, nodes represent interacting proteins, and edges represent interactions. A new PPI network was generated after merging these two PPI networks to identify the candidate targets for the AR-PN herb pair in the treatment of DN, which comprised 4390 nodes and 123,047 edges (Figure 4(c)). Subsequently, we analyzed the topological properties of the merged PPI network based on BC, CC, and DC key parameters and extracted targets above twofold median values of DC as well as more than the median values of BC and CC. Therefore, nodes with DC that were above twofold median values (DC > 72) of all nodes were the first extracted, which included 997 nodes and 44,709 edges (Figure 4(d)). Whereafter, nodes with DC > 226, BC > 0.000528, and CC > 0.45 of these 997 nodes were the second identified core targets containing 115 nodes and 2,334 edges (Figure 4(e)). The information of 115 candidate targets sorted in descending order based on the value of the degree is presented in Table 2.
Figure 4.
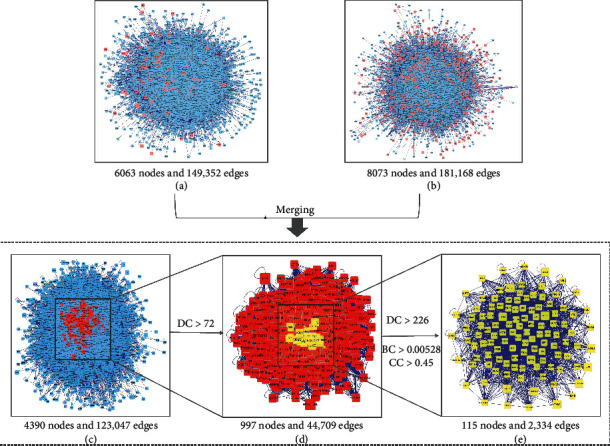
Protein-protein interaction (PPI) network construction and topology analysis. (a) The PPI network of AR-PN herb pair putative targets. (b) The PPI network of DN-related DEGs. (c) The interactive PPI network of AR-PN herb pair putative targets and DN-related DEGs. (d) The PPI network of significant proteins screened from (c). (e) The PPI network of candidate targets of AR-PN herb pair against DN screened from (d). BC: betweenness centrality; CC: closeness centrality; DC: degree centrality; DEGs: differentially expressed genes.
Table 2.
The information of 115 candidate targets.
| No. | Genes | Protein name | Betweenness centrality | Closeness centrality | Degree |
|---|---|---|---|---|---|
| 1 | NTRK1 | Neurotrophic receptor tyrosine kinase 1 | 0.05711884 | 0.56600852 | 1199 |
| 2 | TP53 | Tumor protein p53 | 0.02958689 | 0.5339014 | 768 |
| 3 | APP | Amyloid-beta precursor protein | 0.04325808 | 0.53585828 | 746 |
| 4 | CUL3 | Cullin 3 | 0.01621362 | 0.52767084 | 744 |
| 5 | EGFR | Epidermal growth factor receptor | 0.03631785 | 0.53435673 | 738 |
| 6 | MCM2 | Minichromosome maintenance complex component 2 | 0.01374128 | 0.52022299 | 639 |
| 7 | ESR1 | Estrogen receptor 1 | 0.01715955 | 0.52376403 | 634 |
| 8 | XPO1 | Exportin 1 | 0.02022198 | 0.52090261 | 631 |
| 9 | FN1 | Fibronectin 1 | 0.01700008 | 0.51837844 | 612 |
| 10 | UBC | Ubiquitin C | 0.01612385 | 0.52282751 | 583 |
| 11 | CDK2 | Cyclin-dependent kinase 2 | 0.01028813 | 0.50905292 | 554 |
| 12 | COPS5 | COP9 signalosome subunit 5 | 0.00808418 | 0.51083159 | 517 |
| 13 | CUL7 | Cullin 7 | 0.0048695 | 0.50431183 | 515 |
| 14 | HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | 0.0130468 | 0.5220807 | 510 |
| 15 | GRB2 | Growth factor receptor bound protein 2 | 0.01540288 | 0.51268264 | 485 |
| 16 | RNF2 | Ring finger protein 2 | 0.00618846 | 0.4979564 | 484 |
| 17 | MYC | V-myc avian myelocytomatosis viral oncogene homolog | 0.01293694 | 0.50705202 | 482 |
| 18 | SIRT7 | Sirtuin 7 | 0.00526073 | 0.49220065 | 470 |
| 19 | CUL1 | Cullin 1 | 0.00576927 | 0.50257821 | 469 |
| 20 | YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | 0.00881384 | 0.51503053 | 466 |
| 21 | OBSL1 | Obscurin-like 1 | 0.00370032 | 0.49442002 | 454 |
| 22 | CAND1 | Cullin-associated and neddylation dissociated 1 | 0.00443969 | 0.49280899 | 450 |
| 23 | NPM1 | Nucleophosmin | 0.00555129 | 0.51059371 | 447 |
| 24 | ITGA4 | Integrin subunit alpha 4 | 0.00477184 | 0.49671574 | 444 |
| 25 | VCP | Valosin-containing protein | 0.00713559 | 0.51005931 | 413 |
| 26 | CCDC8 | Coiled-coil domain containing 8 | 0.00480319 | 0.49453151 | 412 |
| 28 | EP300 | E1A binding protein p300 | 0.00792101 | 0.50367478 | 410 |
| 27 | HSP90AB1 | Heat shock protein 90 alpha family class B member 1 | 0.00696006 | 0.51089109 | 410 |
| 29 | VCAM1 | Vascular cell adhesion molecule 1 | 0.0048008 | 0.4901654 | 404 |
| 30 | CDC5L | Cell division cycle 5 like | 0.00445741 | 0.4891268 | 399 |
| 31 | FBXO6 | F-box protein 6 | 0.00660866 | 0.4884731 | 393 |
| 32 | BRCA1 | BRCA1, DNA repair associated | 0.00689339 | 0.49897611 | 391 |
| 33 | TRAF6 | TNF receptor-associated factor 6 | 0.00575486 | 0.49914647 | 378 |
| 34 | HNRNPU | Heterogeneous nuclear ribonucleoprotein U | 0.00359109 | 0.49694086 | 371 |
| 35 | SNW1 | SNW domain containing 1 | 0.00370573 | 0.49164892 | 370 |
| 36 | HDAC1 | Histone deacetylase 1 | 0.00504406 | 0.48798398 | 362 |
| 37 | EED | Embryonic ectoderm development | 0.00288746 | 0.47673913 | 359 |
| 38 | HUWE1 | HECT, UBA, and WWE domain containing 1, E3 ubiquitin protein ligase | 0.00326087 | 0.49508974 | 358 |
| 39 | EWSR1 | EWS RNA binding protein 1 | 0.00617052 | 0.49931694 | 356 |
| 40 | HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | 0.00376562 | 0.49660326 | 352 |
| 41 | MDM2 | MDM2 proto-oncogene | 0.0054897 | 0.49744811 | 346 |
| 42 | UBE2I | Ubiquitin conjugating enzyme E2 I | 0.00515795 | 0.4938077 | 344 |
| 43 | YWHAQ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta | 0.00453158 | 0.49643463 | 338 |
| 47 | RPA1 | Replication protein A1 | 0.00447185 | 0.4904943 | 329 |
| 46 | VHL | Von Hippel-Lindau tumor suppressor | 0.0044112 | 0.49126344 | 329 |
| 44 | PARK2 | Parkin RBR E3 ubiquitin protein ligase | 0.00331119 | 0.48923592 | 329 |
| 45 | HSPA5 | Heat shock protein family A (Hsp70) member 5 | 0.002573 | 0.50292398 | 329 |
| 48 | HDAC5 | Histone deacetylase 5 | 0.00203062 | 0.48282695 | 327 |
| 49 | HIST1H3E | Histone cluster 1 H3 family member e | 0.00117386 | 0.4791348 | 320 |
| 50 | HIST1H3B | Histone cluster 1 H3 family member b | 0.00117386 | 0.4791348 | 320 |
| 51 | HIST1H3G | Histone cluster 1 H3 family member g | 0.00117386 | 0.4791348 | 320 |
| 52 | HIST1H3C | Histone cluster 1 H3 family member c | 0.00117386 | 0.4791348 | 320 |
| 53 | HIST1H3H | Histone cluster 1 H3 family member h | 0.00117386 | 0.4791348 | 320 |
| 54 | HIST1H3J | Histone cluster 1 H3 family member j | 0.00117386 | 0.4791348 | 320 |
| 55 | HIST1H3A | Histone cluster 1 H3 family member a | 0.00117386 | 0.4791348 | 320 |
| 56 | HIST1H3D | Histone cluster 1 H3 family member d | 0.00117386 | 0.4791348 | 320 |
| 57 | HIST1H3I | Histone cluster 1 H3 family member i | 0.00117386 | 0.4791348 | 320 |
| 58 | HIST1H3F | Histone cluster 1 H3 family member f | 0.00117386 | 0.4791348 | 320 |
| 59 | HSPB1 | Heat shock protein family B (small) member 1 | 0.00950276 | 0.49475465 | 319 |
| 60 | HSPA8 | Heat shock protein family A (Hsp70) member 8 | 0.00598969 | 0.50361695 | 312 |
| 61 | SRC | SRC proto-oncogene, non-receptor tyrosine kinase | 0.00389245 | 0.50017106 | 312 |
| 62 | RPA2 | Replication protein A2 | 0.00253078 | 0.48961822 | 310 |
| 63 | CUL2 | Cullin 2 | 0.00217148 | 0.48171334 | 309 |
| 64 | CTNNB1 | Catenin beta 1 | 0.00495783 | 0.5011426 | 304 |
| 65 | EEF1A1 | Eukaryotic translation elongation factor 1 alpha 1 | 0.00334497 | 0.49660326 | 304 |
| 66 | YWHAG | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma | 0.00349418 | 0.49570524 | 300 |
| 67 | AKT1 | AKT serine/threonine kinase 1 | 0.00665037 | 0.49682827 | 296 |
| 68 | AR | Androgen receptor | 0.00533699 | 0.49264293 | 294 |
| 70 | CREBBP | CREB binding protein | 0.00344521 | 0.47960634 | 292 |
| 69 | U2AF2 | U2 small nuclear RNA auxiliary factor 2 | 0.00188521 | 0.4802365 | 292 |
| 71 | IKBKG | Inhibitor of nuclear factor kappa B kinase subunit gamma | 0.00421812 | 0.49126344 | 291 |
| 72 | MAPK1 | Mitogen-activated protein kinase 1 | 0.00590832 | 0.493086 | 287 |
| 73 | YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon | 0.00319492 | 0.49253229 | 283 |
| 74 | PAN2 | PAN2 poly(A) specific ribonuclease subunit | 0.00154999 | 0.46525936 | 281 |
| 75 | SUZ12 | SUZ12 polycomb repressive complex 2 subunit | 0.00168493 | 0.46605037 | 279 |
| 76 | IKBKE | Inhibitor of nuclear factor kappa B kinase subunit epsilon | 0.00234006 | 0.47493232 | 276 |
| 77 | ARRB2 | Arrestin beta 2 | 0.00295499 | 0.48351891 | 274 |
| 78 | CUL5 | Ccullin 5 | 0.00161801 | 0.47421343 | 274 |
| 79 | FUS | FUS RNA binding protein | 0.00199535 | 0.48673843 | 271 |
| 80 | SMURF1 | SMAD specific E3 ubiquitin protein ligase 1 | 0.00239525 | 0.48288011 | 270 |
| 81 | RPA3 | Replication protein A3 | 0.00151664 | 0.47575659 | 268 |
| 82 | STAU1 | Staufen double-stranded RNA binding protein 1 | 0.00121324 | 0.47908247 | 261 |
| 83 | RELA | RELA proto-oncogene, NF-kB subunit | 0.00499948 | 0.48700866 | 260 |
| 84 | GSK3B | Glycogen synthase kinase 3 beta | 0.00641109 | 0.48641455 | 256 |
| 85 | SMAD3 | SMAD family member 3 | 0.00358228 | 0.48956357 | 255 |
| 86 | BMI1 | BMI1 proto-oncogene, polycomb ring finger | 0.00156804 | 0.46619898 | 254 |
| 87 | COMMD3-BMI1 | COMMD3-BMI1 readthrough | 0.00156804 | 0.46619898 | 254 |
| 88 | BAG3 | BCL2-associated athanogene 3 | 0.00385077 | 0.47819451 | 252 |
| 89 | YWHAB | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta | 0.00260668 | 0.48722506 | 250 |
| 90 | HSPA4 | Heat shock protein family A (Hsp70) member 4 | 0.00289896 | 0.49548125 | 248 |
| 91 | PARP1 | Poly(ADP-ribose) polymerase 1 | 0.00322874 | 0.48086833 | 247 |
| 92 | SMAD2 | SMAD family member 2 | 0.00305262 | 0.48002627 | 246 |
| 93 | RPS27A | Ribosomal protein S27a | 0.00243271 | 0.4884731 | 246 |
| 94 | FLNA | Filamin A | 0.00324118 | 0.4884731 | 245 |
| 95 | MYH9 | Myosin heavy chain 9 | 0.00289354 | 0.48673843 | 244 |
| 96 | CDKN1A | Cyclin-dependent kinase inhibitor 1A | 0.00408751 | 0.48123766 | 241 |
| 98 | ACTB | Actin beta | 0.00348475 | 0.49325236 | 238 |
| 97 | TARDBP | TAR DNA binding protein | 9.56E-04 | 0.47436729 | 238 |
| 99 | HDAC2 | Histone deacetylase 2 | 0.00315062 | 0.47055037 | 236 |
| 100 | ABL1 | ABL proto-oncogene 1, nonreceptor tyrosine kinase | 0.00149438 | 0.49076871 | 236 |
| 101 | CUL4B | Cullin 4B | 0.00369003 | 0.47788189 | 234 |
| 102 | FYN | FYN proto-oncogene, Src family tyrosine kinase | 0.00105834 | 0.46619898 | 234 |
| 103 | EZH2 | Enhancer of zeste 2 polycomb repressive complex 2 subunit | 0.00157335 | 0.46884019 | 233 |
| 104 | CRK | CRK proto-oncogene, adaptor protein | 0.00664489 | 0.48218997 | 232 |
| 105 | PRKCA | Protein kinase C alpha | 0.00314723 | 0.47971125 | 232 |
| 107 | ARRB1 | Arrestin beta 1 | 0.00432736 | 0.48018393 | 231 |
| 106 | JUN | Jun proto-oncogene, AP-1 transcription factor subunit | 0.00304336 | 0.47762169 | 231 |
| 108 | TUBB | Tubulin beta class I | 0.00214432 | 0.49464306 | 231 |
| 109 | HNRNPK | Heterogeneous nuclear ribonucleoprotein K | 0.00139431 | 0.48787542 | 230 |
| 110 | RPL10 | Ribosomal protein L10 | 0.0011806 | 0.46314678 | 229 |
| 112 | CLTC | Clathrin heavy chain | 0.00232751 | 0.48652246 | 228 |
| 111 | FBXW11 | F-box and WD repeat domain containing 11 | 0.00221635 | 0.47298609 | 228 |
| 113 | HLA-B | Major histocompatibility complex, class I, B | 0.00338256 | 0.45549901 | 227 |
| 114 | PRKDC | Protein kinase, DNA-activated, catalytic polypeptide | 0.00185628 | 0.48950893 | 227 |
| 115 | BTRC | Beta-transducin repeat-containing E3 ubiquitin protein ligase | 0.00175312 | 0.47793397 | 227 |
3.4. GO and KEGG Pathway Enrichment Analysis
To further evaluate better the molecular mechanism of the AR-PN herb pair on DN, GO and KEGG pathway enrichment analyses associated with 115 candidate targets were performed using the Cytoscape plugin ClueGO, which included GO biological process and the signaling pathway. The GO biological processes were mainly involved in regulating DNA-binding transcription factor activity, regulating protein modification by small protein conjugation or removal, rhythmic process, proteasome-mediated ubiquitin-dependent protein catabolic process, and cellular response to heat, and so on (Figure 5(a)). According to the KEGG pathway analysis, the signaling pathways were mainly focused on the PI3K-Akt signaling pathway, viral carcinogenesis, cell cycle, microRNAs in cancer, and hepatitis B, and so on (Figure 5(b)).
Figure 5.
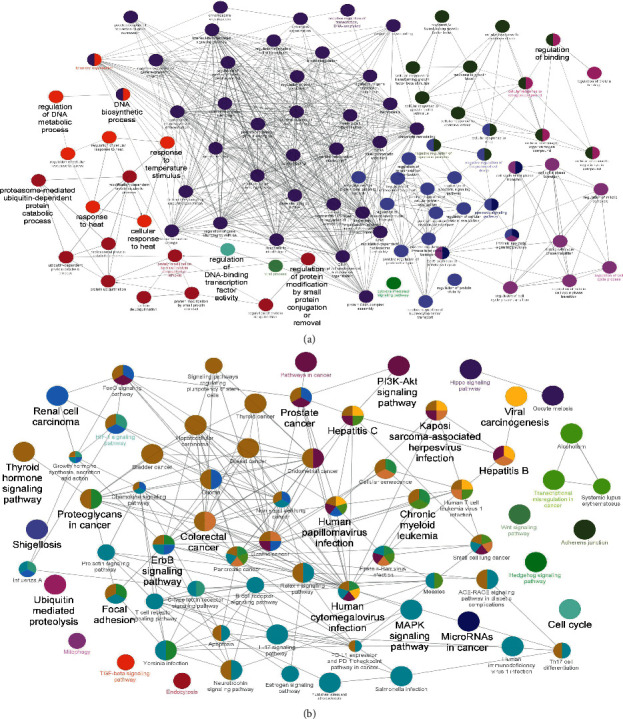
Enrichment analysis of candidate targets based on ClueGO. (a) Possible enriched targets of AR-PN herb pair in typical biological processes. (b) Possible enriched targets of AR-PN herb pair in signaling pathways.
3.5. Construction of the Component-Target-Pathway Network
To further explore the interaction between components, candidate targets, and the top 20 signaling pathways, the network map of component-target-pathway was constructed by Cytoscape 3.8.0, which included 113 nodes and 395 edges (Figure 6). The larger and darker the color of nodes was shown, the higher the degree value was represented, the thicker the edges were displayed, and the greater closeness and betweenness were found. Hence, we speculated that quercetin (MOL000098), kaempferol (MOL000422), isorhamnetin (MOL000354), and formononetin (MOL000392) regulate the PI3K-Akt signaling pathway, viral carcinogenesis, microRNAs in cancer, cell cycle, and MAPK signaling pathway, and so on, via core target genes MAPK1 (PDB ID: 4iz5), AKT1 (PDB ID: 1unq), GSK3B (PDB ID: 4afj), CDKN1A (PDB ID: 2zvw), TP53 (PDB ID: 2k8f), RELA (PDB ID: 1nfi), MYC (PDB ID: 5g1x), GRB2 (PDB ID: 1gri), JUN (PDB ID: 1s9k), and EGFR (PDB ID: 5wb7).
Figure 6.
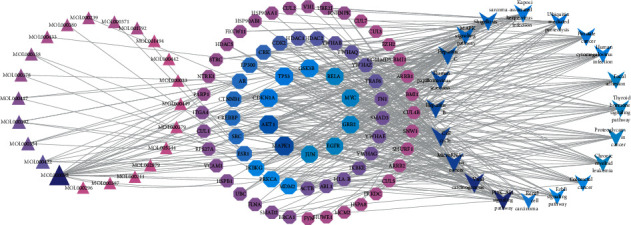
Component-target-pathway network of AR-PN herb pair against DN. The larger size and darker the color of nodes were shown, the higher the degree value was represented. Rhombus represents targets, triangles represent ingredients of AR-PN herb pair, and arrowhead represents signal pathways.
3.6. Molecular Docking Analysis
According to the network of component-target-pathway, the core targets (MAPK1, AKT1, GSK3B, CDKN1A, TP53, RELA, MYC, GRB2, JUN, and EGFR) were selected for molecular docking with quercetin, kaempferol, isorhamnetin, and formononetin bioactive components verified experimentally via AutoDock Vina 1.1.2 software. As shown in Table 3, all the core targets mentioned above had a great binding affinity with quercetin, kaempferol, isorhamnetin, and formononetin. Furthermore, the MAPK1 and JUN target proteins were molecularly docked with quercetin, kaempferol, isorhamnetin, and formononetin active components with the lowest energy value that was less than –8.0 kcal·mol−1. Therefore, we selected the MAPK1 and JUN receptor proteins with the lowest energy value and the ligands for visualization and construction by Pymol software (Figure 7).
Table 3.
Docking scores of targets with components (kcal·mol−1).
| Target name | PDBID | Components | |||
|---|---|---|---|---|---|
| Quercetin | Kaempferol | Isorhamnetin | Formononetin | ||
| MAPK1 | 4iz5 | −8.4 | −8.4 | −8.4 | −8 |
| AKT1 | 1unq | −6.4 | −6 | −6.3 | −6.6 |
| GSK3B | 4afj | −8.3 | −8.4 | −8.4 | −7.6 |
| CDKN1A | 2zvw | −7.5 | −7.5 | −7.5 | −7.4 |
| TP53 | 2k8f | −6.9 | −6.4 | −6.7 | −7.4 |
| RELA | 1nfi | −7.7 | −8.5 | −8.6 | −8.3 |
| MYC | 5g1x | −7.9 | −7.9 | −7.5 | −7.5 |
| GRB2 | 1gri | −7.5 | −7.6 | −7.7 | −7.6 |
| JUN | 1s9k | −8.6 | −9.7 | −9.7 | −8.3 |
| EGFR | 5wb7 | −7.7 | −7.9 | −8 | −8 |
Figure 7.
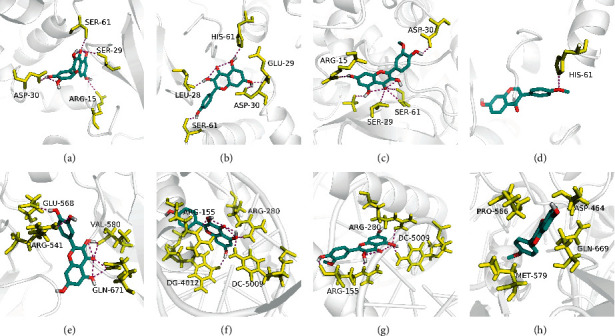
Molecular models of the binding of quercetin, kaempferol, isorhamnetin, and formononetin with MAPK1 (a–d) and JUN (e–h), respectively.
3.7. Verification of Hub Genes by ROC Curve Analysis
ROC curves were applied to evaluate the diagnostic value of the hub genes for DN. This analysis showed that AKT1 (AUC = 0.93162; P < 0.001), TP53 (AUC = 0.97436; P < 0.001), RELA (AUC = 0.92308; P < 0.001), JUN (AUC = 0.86325; P < 0.001), CDKN1A (AUC = 0.79487; P = 0.033), and EGFR (AUC = 0.82906; P = 0.012) are effective in discriminating between DN patients and controls in the GSE30528 dataset. However, MAPK1 (AUC = 0.71795; P = 0.051), GSK3B (AUC = 0.65812; P = 0.345), MYC (AUC = 0.70940; P = 0.102), and GRB2 (AUC = 0.51282; P = 0.948) were proved to be no diagnostic capability for DN (Figure 8).
Figure 8.
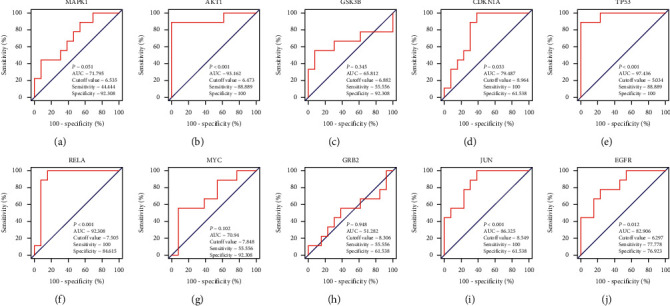
Receiver operating characteristic curves of hub genes, including MAPK1, AKT1, GSK3B, CDKN1A, TP53, RELA, MYC, GRB2, JUN, and EGFR in the GSE30528 dataset.
4. Discussion
Microarrays are a powerful and effective high-throughput technology and provide information on gene profiling data to researchers to explore biomarker, molecular pathways, target selectivity, and development of prognostic, and so on involved in complex disorders [45]. They have been widely applied to investigate the key molecule biomarkers and pathways of various diseases, including ovarian cancer [46], multiple myeloma [47], systemic lupus erythematosus (SLE) [48], and idiopathic pulmonary fibrosis [49]. DN is the most common microvascular complication of DM, which has been considered the major cause of ESRD worldwide [3]. Nevertheless, effective treatments remain scarce. Previous studies have been demonstrated that AR and PN play a critical role in treating DN [12–14, 50], suggesting AR-PN may be a potential herb pair for the treatment of DN. For all we know, this is the first study to perform a comprehensive analysis of network pharmacology combined with gene expression profiling to further explore the underlying pharmacological mechanisms and therapeutic targets of the AR-PN herb pair on DN. Our findings demonstrated that a total of 22 bioactive compounds and 115 core targets for the AR-PN herb pair in the treatment of DN were identified. According to the KEGG pathway enrichment, the AR-PN herb pair played a therapeutic role in DN through regulating the PI3K-Akt signaling pathway, cell cycle, and MAPK signaling pathway, and so on. Furthermore, MAPK1, AKT1, GSK3B, CDKN1A, TP53, RELA, MYC, GRB2, JUN, and EGFR were regarded as core targets of the AR-PN herb pair against DN according to the component-target-pathway network. What is more, molecular docking analysis validated that these core target genes showed strong binding interactions and affinity with quercetin, kaempferol, isorhamnetin, and formononetin bioactive compounds of the AR-PN herb pair.
As shown in the component-target network (Figure 2), 22 compounds in the AR-PN herb pair with possible efficacy against DN were identified, many of which shared targets and had a synergistic effect, indicating that the AR-PN herb pair exerts effects in various bioactive components and multiple cellulars and molecular targets way. Quercetin, a natural flavonoid, is well known to possess multiple pharmacological functions, including but not limited to antioxidant, anti-inflammatory, anticancer, antidiabetic, neuroprotective, and antiobesity [51, 52]. The use of quercetin contributes to a meaningful improvement in disease symptoms, renal hypertrophy index, renal histopathology, oxidative stress, and blood glucose in a rat model of DN [41]. Furthermore, quercetin treatment is beneficial to antioxidant capacity and vasoprotective effects against STZ-induced ER stress of diabetic rats via reducing the lipid peroxidation and increasing the expression of vascular endothelial growth factor (VEGF) and its receptor, VEGFR2, and nitric oxide (NO) [53]. Accumulating evidence also demonstrated that the nephroprotective effects of quercetin are related to inhibiting renal tubular epithelial-mesenchymal transition and renal fibrosis and reducing the production of cytokines in DN, including IL-6, TNF-α, and IL-1β [54–56]. What is more, quercetin inhibited glomerular mesangial cell proliferation and epithelial-mesenchymal transition in DN via reactivation of the Hippo pathway and inhibition of the TGF-β/PI3K/Akt pathway, respectively [57, 58]. Kaempferol, a natural polyflavonol, also exhibits nephroprotective effects in addition to its antioxidant and anti-inflammatory properties [59]. Studies demonstrated that kaempferol plays vital nephroprotective roles associating with inhibiting oxidative stress, proinflammatory cytokines (TNF-α and IL-1β), and fibrosis in DN via inhibiting hyperglycemia-induced activation RhoA/Rho-kinase [42]. Moreover, kaempferol exerts antioxidant potential in DN rats by the upregulation of the Nrf-2/HO-1 axis [60]. Formononetin possesses multiple biological activities, such as antioxidant, anti-inflammatory, and regulating immune responses [61, 62]. Formononetin improved oxidative stress and prevented the progression of renal fibrosis in DN by activating the Nrf2/ARE signaling pathway [43]. Experimental studies have shown that isorhamnetin has renoprotective effects on DN through improving fasting blood glucose, increasing autophagosomes in renal tissues, and suppressing miRNA regulation of autophagy genes [44]. Besides, isorhamnetin inhibits oxidative stress, inflammation, and apoptosis in STZ-induced diabetic rats [63]. Considering this, quercetin, kaempferol, formononetin, and isorhamnetin are the potential components of the AR-PN herb pair treating DN, associated with their various bioactive actions, such as antioxidant, anti-inflammation, and antiapoptosis.
Subsequently, 115 nodes of the AR-PN herb pair acting on DN were identified based on the merger PPI network, which were considered as core targets that the AR-PN herb pair plays therapeutic roles in DN. To further illustrate the mechanism of AR-PN herb pair on DN, we conducted GO and KEGG pathway enrichment analyses of 115 core targets. As shown in Figure 5(a), the BP terms of AR-PN herb pair on DN were mainly related to DNA synthesis, transcription, protein catabolism, cell cycle, apoptosis, oxidative stress, and inflammation regulation. Interestingly, apoptosis, oxidative stress, and inflammation are deemed as the important pathogenesis of DN [3]. KEGG enrichment analysis showed that the AR-PN herb pair may alleviate the development of DN through the action of core targets in multiple signaling pathways (Figure 5(b)), including PI3K-Akt signaling pathway, cell cycle, and MAPK signaling pathway. Among them, the enriched targets of the PI3K-Akt signaling pathway are markedly higher than others. Phosphoinositide-3-kinase (PI3K) can promote the phosphorylation of protein kinase B (PKB, Akt), which involves diversified cell activities, e.g., cell proliferation, protein synthesis, glycolysis, autophagy, and apoptosis [64]. Previous studies demonstrated that restoring podocyte autophagy, attenuating podocyte apoptosis, and inhibiting MC proliferation of DN were closely associated with inhibiting the PI3K/AKT signaling pathway [65–67]. Furthermore, notoginsenoside R1, which is extracted from PN, increased the phosphorylation of both PI3K and Akt and ameliorated podocyte injury in DN rats by activating the PI3K/Akt signaling pathway [68]. Cell cycle refers to order progression from one cell cycle phase to the next, involving a family of cyclins and their associated kinases, and mediates cell proliferation, which will be ceased if its checkpoint secures, such as Rad3-related protein kinase and checkpoint kinase 1, causing severe problems [69]. Moreover, the cell cycle is regulated by several cellular pathways, e.g., PI3K/Akt signaling pathway that is famous for regulating cell proliferation [64]. MAPK pathway acts on inhibiting cell apoptosis and promoting proliferation and anti-inflammatory effects in DN rats, which is associated with MAPK1 [70]. Viral carcinogenesis accelerates DNA damage and virus integration, which are closely related to the progression of human cancers [71], as well as microRNAs in cancer [72], suggesting AR-PN herb pair also exerts anticancer effects. Thus, our identified core targets are highly related to the signaling pathways associated with cell proliferation, apoptosis, and cell cycle.
Furthermore, the network of component-target-pathway showed that MAPK1, AKT1, GSK3B, CDKN1A, TP53, RELA, MYC, GRB2, JUN, and EGFR were core targets in the network due to possessing the higher degree (Figure 6), suggesting that these core targets were identified as core genes that were strongly associated with the pathogenesis of AR-PN herb against DN. MAPK1, also known as an extracellular signal-regulated protein kinase, is a member of the MAPKs family that is contributed to enhancing cellular proliferation and participating in cell metabolism, proliferation, apoptosis, differentiation, and survival in physiological and pathological processes [73]. It has been demonstrated that downregulated expression of MAPK1 plays a protective role in lipopolysaccharide-induced podocyte damage in DN [74]. Many studies have observed that the expression of MAPK1 was increased in various cancers, such as breast cancer [75], hepatocellular carcinoma [76], and gastric cancer [77], which are involved in inhibiting cell proliferation, cell cycle, and cell apoptosis. Furthermore, they found that the upregulation of MAPK1 was negatively regulated by miR-20a [75], miR-193b [76], and miR-378 [77], respectively. This suggests that MAPK1 vital gene plays essential roles in the development and progression of DN, which could participate in cell metabolism, cell proliferation, and apoptosis of podocyte, mesangial cell, etc. AKT1 is an important factor to regulate inflammation and apoptosis in kidney disease [78]. The phosphorylation of AKT1 can directly promote mesangial cell (MC) proliferation, basement membrane thickening, renal tubular epithelial cell transdifferentiation, and podocyte injury that are well known to play a critical role in the pathogenic mechanisms of DN [67, 79]. Evidence revealed that GSK3B is phosphorylated by AKT at serine 9, resulting in facilitating the expression of inflammatory cytokines [80]. Moreover, the inhibition of GSK3B induces human cell proliferation [81]. Kumar et al. found that AKT1 was one of the essential genes in the development and progression of SLE, with regard to increasing autoimmune responses via different signaling pathways [48]. TP53, a tumor suppressor, mediates glucose and lipotoxicity and causes a stop of the cell cycle, apoptosis, DNA repair, and metabolic changes, which is inhibited by microRNA-770-5p through regulating podocyte apoptosis [82]. This indicates that AKT1, GSK3B, TP53, and RELA might be associated with cell proliferation, cell cycle, and apoptosis of DN. The activation of JUN N-terminal kinase is associated with inflammatory factor secretion during DN [83]. Furthermore, JUN is a highly unstable protein due to polyubiquitination by GSK3 at Ser-243, and its level is inversely associated with GSK3 activity in mammalian cells that have entered the cell cycle [84], showing that JUN interacts with GSK3 via polyubiquitination. Nevertheless, the roles of JUN in DN require further research. Epidermal growth factor receptor (EGFR) belongs to the family of receptor tyrosine kinase ErbB receptors, and the activation of which can result in phosphorylation on specific tyrosine residues, such as ErbB2, ErbB3, or ErbB4, and further regulate cell proliferation, differentiation, and apoptosis [85]. The activation of EGFR leads to podocyte injury and loss in DN, while EGFR deletion in podocytes ameliorates glomerular injury and attenuates the progression of DN [86]. Previous studies also demonstrated that EGFR inhibition plays a nephroprotective role in DN via decreasing reactive oxygen species (ROS), inhibiting endoplasmic reticulum(ER) stress, alleviating podocyte injury, and increasing autophagy [87, 88]. RELA is one of the transcription factor genes in the NF-κB signaling pathway that contributes to human disease processes, notably inflammatory diseases and cancer [89]. Site-specific phosphorylation has been proven to be a prerequisite for additional modifications regulating RELA activity, especially acetylation and ubiquitination [90]. A previous study showed that the phosphorylation of RELA at S536 may be related to regulating NF-kB activity, which was further corroborated in a mouse model that knocked down RELA S534A, a nonphosphorylatable mutant version of the human RELA S536 homolog in murine [91]. Besides, RELA also participates in the process of cell proliferation [92]. However, the role of RELA in DN remains unclear and requires further research.
To explore the direct or indirect interactions between core target genes, an interrelation analysis for them was performed. As shown in Figure 9, the core target genes interacted with each other via multiple signaling pathways mainly associated with the PI3K-Akt signaling pathway, MAPK signaling pathway, cell cycle, apoptosis, microRNAs in cancer, and viral carcinogenesis. The core genes MAPK1, AKT1, TP53, RELA, MYC, GRB2, and EGFR are involved directly or indirectly in the PI3K-Akt signaling pathway regulating cell proliferation and MAPK signaling pathway resulting in apoptosis [93–96]. The cell cycle interacts with cell proliferation, differentiation, DNA damage, and cell apoptosis through TP53, GSK3B, CDKN1A, and MYC [97, 98]. Collectively, these findings indicate that the core genes could be critical factors for AR-PN herb pair against the development of DN, which require to be further experimentally validated to prove this hypothesis.
Figure 9.
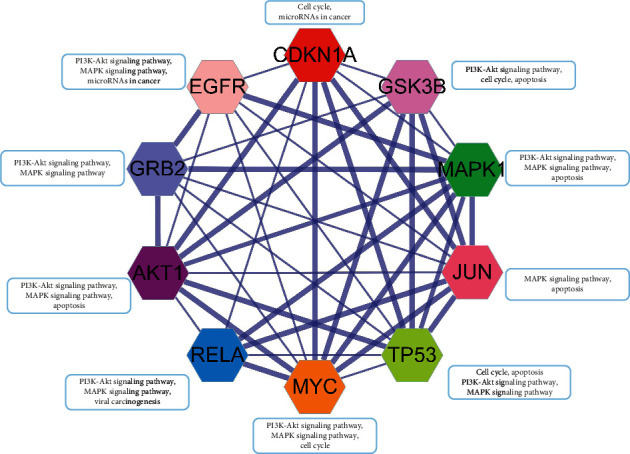
Interrelation analysis of the core genes MAPK1, AKT1, GSK3B, CDKN1A, TP53, RELA, MYC, GRB2, JUN, and EGFR related to AR-PN herb pair against DN. Genes are directly or indirectly involved in different pathways. The gene network was performed on the STRING and visualized in Cytoscape 3.8.0.
Molecular docking was performed to validate the binding interactions and affinity between active components (quercetin, kaempferol, isorhamnetin, and formononetin) and the core targets (MAPK1, AKT1, GSK3B, CDKN1A, TP53, RELA, MYC, GRB2, JUN, and EGFR). It has been demonstrated that a docking score between bioactive components and targets structures less than −7, −5, and −4.25 kcal·mol−1 represents a strong binding affinity, a good binding affinity, and binding affinity, respectively [99], which means the smaller the docking score, the more stable and stronger the affinity between components and targets (ligand and receptor) binding. As shown in Table 3, the binding energies of all docking are less than −5 kcal·mol−1, indicating that these target genes have great binding interactions and affinity with quercetin, kaempferol, formononetin, and isorhamnetin bioactive components. Moreover, the energy value of MAPK1 and JUN docked with quercetin, kaempferol, isorhamnetin, and formononetin was less than −8.0 kcal·mol−1 among the core target proteins. MAPK1 had the least binding affinity with quercetin, kaempferol, and formononetin at −8.4 kcal/mol. JUN docked with kaempferol and isorhamnetin had the lowest energy value at 9.7 kcal/mol. The docking results provide an efficient method to estimate the binding modes of AR-PN herb pair with DN-related core target proteins and provide us with sound directions for further validated.
Subsequently, ROC curves were applied to evaluate the diagnostic value of the core genes for DN. TP53, AKT1, RELA, and JUN with an AUC of 0.97436, 0.93162, 0.92308, and 0.86325, respectively, indicate that these hub genes exhibit high diagnostic values to distinguish DN patients and controls. Of course, their expression and related function also need to be further validated by mutation analysis, immune infiltration analysis, and RT-PCR.
Our study still has some limitations. Firstly, we only explored the potential functional mechanism of AR-PN herb pair on DN and have not identified the pharmacologically mutual interferences in bioactive components of AR-PN herb pair. Secondly, the DEGs of DN are acquired by analyzing published microarray data from the GEO database. Nevertheless, the sample size of DN-related DEGs is small, which needs to be enhanced in the future. Furthermore, the application of network pharmacology and bioinformatics in this study is just initial screening of the mechanisms and molecular targets of AR-PN herb pair against DN, and the results of this study have not been validated through experimental data. Hence, further studies, especially pharmacological experiments and clinical validations, are still required to be carried out to explain the complex bioactive chemical interactions. Moreover, we should provide larger groups and experimental evidence to validate these results with the interdisciplinary research works integrating computer science, network science, mathematics, and pharmacology in the future.
5. Conclusion
In summary, we utilized network pharmacology combined with bioinformatics to explore the mechanisms and molecular targets of AR-PN herb pair against DN in the present study. Our findings revealed that AR-PN herb pair exerts pharmacological effects on DN with various bioactive chemicals and the related pharmacological pathways, involving molecular targets and multiple cellulars, including antioxidant, anti-inflammatory, and antiapoptosis, and regulating the cell cycle. The AR-PN herb pair and its components may be promising drugs in the treatment of DN in the future.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (no. 81860838), the Postdoctoral Science Foundation of China (no. 2017M612987), and the Studio of Shi Wei for the Heritage of Famous Traditional Chinese Medicine Practitioners (no. [2017] 2).
Data Availability
The data used to support the findings of this study are included within the article.
Disclosure
Jie Zhao and Chao Mo are joint first authors of this article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Jie Zhao and Chao Mo contributed equally to this work. All authors have contributed to this study and approved its submission.
References
- 1.Cole J. B., Florez J. C. Genetics of diabetes mellitus and diabetes complications. Nature Reviews Nephrology. 2020;16(7):377–390. doi: 10.1038/s41581-020-0278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Federation I. D. International Diabetes Federation Diabetes Atlas. Brussels, Belgium: International Diabetes Federation; 2019. https://www.idf.org/ [Google Scholar]
- 3.Selby N. M., Taal M. W. An updated overview of diabetic nephropathy: Diagnosis, prognosis, treatment goals and latest guidelines. Diabetes, Obesity and Metabolism. 2020;22(1):3–15. doi: 10.1111/dom.14007. [DOI] [PubMed] [Google Scholar]
- 4.Chen C., Wang C., Hu C., et al. Normoalbuminuric diabetic kidney disease. Frontiers of Medicine. 2017;11(3):310–318. doi: 10.1007/s11684-017-0542-7. [DOI] [PubMed] [Google Scholar]
- 5.Alicic R. Z., Johnson E. J., Tuttle K. R. SGLT2 inhibition for the prevention and treatment of diabetic kidney disease: a review. American Journal of Kidney Diseases. 2018;72(2):267–277. doi: 10.1053/j.ajkd.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 6.Dong L., Li J., Lian Y., et al. Long-term intensive lifestyle intervention promotes improvement of stage III diabetic nephropathy. Medical Science Monitor. 2019;25:3061–3068. doi: 10.12659/msm.913512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., He K., Ge J., Li C., Jing Z. Efficacy and safety of the glucagon-like peptide-1 receptor agonist oral semaglutide in patients with type 2 diabetes mellitus: a systematic review and meta-analysis. Diabetes Research and Clinical Practice. 2021;172(9) doi: 10.1016/j.diabres.2021.108656.108656 [DOI] [PubMed] [Google Scholar]
- 8.Fadini G. P., Bonora B. M., Avogaro A. SGLT2 inhibitors and diabetic ketoacidosis: data from the FDA Adverse Event Reporting System. Diabetologia. 2017;60(8):1385–1389. doi: 10.1007/s00125-017-4301-8. [DOI] [PubMed] [Google Scholar]
- 9.Xu H.-Y., Zhang Y.-Q., Liu Z.-M., et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Research. 2019;47(D1):D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mou X., Zhou D.-y., Liu W.-h., et al. Study on the relationship between Chinese medicine constitutive susceptibility and diversity of syndrome in diabetic nephropathy. Chinese Journal of Integrative Medicine. 2013;19(9):656–662. doi: 10.1007/s11655-013-1411-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang S., Hu Y., Tan W., et al. Compatibility art of traditional Chinese medicine: from the perspective of herb pairs. Journal of Ethnopharmacology. 2012;143(2):412–423. doi: 10.1016/j.jep.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Tao C., Xuan C., Jiang J, Cao W. Transcriptomic analysis reveals the protection of astragaloside IV against diabetic nephropathy by modulating inflammation. Oxidative Medicine and Cellular Longevity. 2020;2020(5) doi: 10.1155/2020/9542165.9542165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang E., Wang L., Ding R., et al. Astragaloside IV acts through multi-scale mechanisms to effectively reduce diabetic nephropathy. Pharmacological Research. 2020;157(7) doi: 10.1016/j.phrs.2020.104831.104831 [DOI] [PubMed] [Google Scholar]
- 14.Tang X., Huang M., Jiang J., et al. Panax notoginseng preparations as adjuvant therapy for diabetic kidney disease: a systematic review and meta-analysis. Pharmaceutical Biology. 2020;1(58):138–145. doi: 10.1080/13880209.2020.1711782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen D., Tan R.-Z., Zhao C.-Y., et al. Astragalus mongholicus bunge and Panax notoginseng (burkill) F. H. Chen formula for renal injury in diabetic nephropathy-in vivo and in vitro evidence for autophagy regulation. Frontiers in Pharmacology. 2020;11(6):p. 732. doi: 10.3389/fphar.2020.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.AL H. Network pharmacology. Nature Biotechnology. 2007;10(25):1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 17.Yin B., Bi Y. M., Fan G. J., Xia Y. Q. Molecular mechanism of the effect of huanglian jiedu decoction on type 2 diabetes mellitus based on network pharmacology and molecular docking. Journal of Diabetes Research. 2020;2020 doi: 10.1155/2020/5273914.5273914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casas A. I., Hassan A. A., Larsen S. J., et al. From single drug targets to synergistic network pharmacology in ischemic stroke. Proceedings of the National Academy of Sciences. 2019;116(14):7129–7136. doi: 10.1073/pnas.1820799116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayat A. Science, medicine, and the future: Bioinformatics. BMJ. 2002;324(7344):1018–1022. doi: 10.1136/bmj.324.7344.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarahmadi A., Shahrokhi S. Z., Mostafavi-Pour Z., Azarpira N. MicroRNAs in diabetic nephropathy: from molecular mechanisms to new therapeutic targets of treatment. Biochemical Pharmacology. 2020;2020 doi: 10.1016/j.bcp.2020.114301.114301 [DOI] [PubMed] [Google Scholar]
- 21.Ishii H., Kaneko S., Yanai K., et al. MicroRNAs in podocyte injury in diabetic nephropathy. Frontiers in Genetics. 2020;11:p. 993. doi: 10.3389/fgene.2020.00993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z., Liu J., Wang W., Zhao Y., Yang D., Geng X. Investigation of hub genes involved in diabetic nephropathy using biological informatics methods. Annals of Translational Medicine. 2020;8(17):p. 1087. doi: 10.21037/atm-20-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar U., Rajan B., Kumar T., et al. Involvement of essential signaling cascades and analysis of gene networks in diabesity. Genes (Basel) 2020;11(11) doi: 10.3390/genes11111256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Udhaya Kumar S., Thirumal Kumar D., Bithia R., et al. Analysis of differentially expressed genes and molecular pathways in familial hypercholesterolemia involved in atherosclerosis: a systematic and bioinformatics approach. Frontiers in Genetics. 2020;11:p. 734. doi: 10.3389/fgene.2020.00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu D., Zhang B., Yang L., et al. Development of an immune-related risk signature for predicting prognosis in lung squamous cell carcinoma. Frontiers in Genetics. 2020;11:p. 978. doi: 10.3389/fgene.2020.00978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan J., Jiang S., Jiang Y., et al. Data mining and expression analysis of differential lncRNA ADAMTS9-AS1 in prostate cancer. Frontiers in Genetics. 2019;10:p. 1377. doi: 10.3389/fgene.2019.01377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ru J., Li P., Wang J., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(1):p. 13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barton H. A., Pastoor T. P., Baetcke K., et al. The acquisition and application of absorption, distribution, metabolism, and excretion (ADME) data in agricultural chemical safety assessments. Critical Reviews in Toxicology. 2006;36(1):9–35. doi: 10.1080/10408440500534362. [DOI] [PubMed] [Google Scholar]
- 29.Wang J., Hou T. Advances in computationally modeling human oral bioavailability. Advanced Drug Delivery Reviews. 2015;86(4):11–16. doi: 10.1016/j.addr.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian S., Wang J., Li Y., et al. The application of in silico drug-likeness predictions in pharmaceutical research. Advanced Drug Delivery Reviews. 2015;86(3):2–10. doi: 10.1016/j.addr.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Law V., Knox C., Djoumbou Y., et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Research. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woroniecka K. I., Park A. S., Mohtat D., et al. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60(9):2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syafrizayanti, Betzen C., Hoheisel J. D., et al. Methods for analyzing and quantifying protein-protein interaction. Expert Review of Proteomics. 2014;11(1):107–120. doi: 10.1586/14789450.2014.875857. [DOI] [PubMed] [Google Scholar]
- 35.Martin A., Ochagavia M. E., Rabasa L. C., et al. BisoGenet: a new tool for gene network building, visualization and analysis. BMC Bioinformatics. 2010;11:p. 91. doi: 10.1186/1471-2105-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Y., Li M., Wang J., et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Ashburner M., Ball C. A., Blake J. A., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L. L., Dai B., Sun Y. H., et al. The activation of IL-17 signaling pathway promotes pyroptosis in pneumonia-induced sepsis. Annals of Translational Medicine. 2020;8(11):p. 674. doi: 10.21037/atm-19-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wixon J., Kell D. The Kyoto encyclopedia of genes and genomes--KEGG. Yeast. 2000;17(1):48–55. doi: 10.1155/2000/981362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres P. H. M., Sodero A. C. R., Jofily P., et al. Key topics in molecular docking for drug design. International Journal of Molecular Sciences. 2019;20(18):p. 4574. doi: 10.3390/ijms20184574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang L., Li K., Zhang Y., et al. Quercetin liposomes ameliorate streptozotocin-induced diabetic nephropathy in diabetic rats. Scientific Reports. 2020;10(1):p. 2440. doi: 10.1038/s41598-020-59411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma D., Kumar Tekade R., Kalia K. Kaempferol in ameliorating diabetes-induced fibrosis and renal damage: an in vitro and in vivo study in diabetic nephropathy mice model. Phytomedicine. 2020;76 doi: 10.1016/j.phymed.2020.153235.153235 [DOI] [PubMed] [Google Scholar]
- 43.Zhuang K., Jiang X., Liu R., et al. Formononetin activates the Nrf2/ARE signaling pathway via Sirt1 to improve diabetic renal fibrosis. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.616378.616378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matboli M., Ibrahim D., Hasanin A. H., et al. Epigenetic modulation of autophagy genes linked to diabetic nephropathy by administration of isorhamnetin in type 2 diabetes mellitus rats. Epigenomics. 2021;13(3):187–202. doi: 10.2217/epi-2020-0353. [DOI] [PubMed] [Google Scholar]
- 45.Butte A. The use and analysis of microarray data. Nature Reviews Drug Discovery. 2002;1(12):951–960. doi: 10.1038/nrd961. [DOI] [PubMed] [Google Scholar]
- 46.Kumar S. U., Kumar D. T., Siva R., et al. Integrative bioinformatics approaches to map potential novel genes and pathways involved in ovarian cancer. Frontiers in Bioengineering and Biotechnology. 2019;7:p. 391. doi: 10.3389/fbioe.2019.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan H., Zheng G., Qu J., et al. Identification of key candidate genes and pathways in multiple myeloma by integrated bioinformatics analysis. Journal of Cellular Physiology. 2019;234(12):23785–23797. doi: 10.1002/jcp.28947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Udhaya Kumar S., Thirumal Kumar D., Siva R., et al. Dysregulation of signaling pathways due to differentially expressed genes from the B-cell transcriptomes of systemic lupus erythematosus patients - a bioinformatics approach. Frontiers in Bioengineering and Biotechnology. 2020;8:p. 276. doi: 10.3389/fbioe.2020.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mishra S., Shah M. I., Udhaya Kumar S., et al. Network analysis of transcriptomics data for the prediction and prioritization of membrane-associated biomarkers for idiopathic pulmonary fibrosis (IPF) by bioinformatics approach. Advances in Protein Chemistry and Structural Biology. 2021;123:241–273. doi: 10.1016/bs.apcsb.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Xie L., Zhai R., Chen T., et al. Panax notoginseng ameliorates podocyte EMT by targeting the wnt/beta-catenin signaling pathway in STZ-induced diabetic rats. Drug Design, Development and Therapy. 2020;14(1):527–538. doi: 10.2147/dddt.s235491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ebrahimpour S., Zakeri M., Esmaeili A. Crosstalk between obesity, diabetes, and Alzheimer’s disease: introducing quercetin as an effective triple herbal medicine. Ageing Research Reviews. 2020;62 doi: 10.1016/j.arr.2020.101095.101095 [DOI] [PubMed] [Google Scholar]
- 52.de Oliveira M. R., Nabavi S. M., Braidy N., et al. Quercetin and the mitochondria: a mechanistic view. Biotechnology Advances. 2016;34(5):532–549. doi: 10.1016/j.biotechadv.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 53.Suganya N., Dornadula S., Chatterjee S., et al. Quercetin improves endothelial function in diabetic rats through inhibition of endoplasmic reticulum stress-mediated oxidative stress. European Journal of Pharmacology. 2018;819:80–88. doi: 10.1016/j.ejphar.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 54.Maciel R. M., Costa M. M., Martins D. B., et al. Antioxidant and anti-inflammatory effects of quercetin in functional and morphological alterations in streptozotocin-induced diabetic rats. Research in Veterinary Science. 2013;95(2):389–397. doi: 10.1016/j.rvsc.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 55.Gomes I. B., Porto M. L., Santos M. C., et al. Renoprotective, anti-oxidative and anti-apoptotic effects of oral low-dose quercetin in the C57BL/6J model of diabetic nephropathy. Lipids in Health and Disease. 2014;13:p. 184. doi: 10.1186/1476-511x-13-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Q., Ji X. J., Zhou Y. X., et al. Quercetin inhibits the mTORC1/p70S6K signaling-mediated renal tubular epithelial-mesenchymal transition and renal fibrosis in diabetic nephropathy. Pharmacological Research. 2015;99:237–247. doi: 10.1016/j.phrs.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Lei D., Cheng C. L., Xuan Q., et al. Quercetin inhibited mesangial cell proliferation of early diabetic nephropathy through the Hippo pathway. Pharmacological Research. 2019;146 doi: 10.1016/j.phrs.2019.104320.104320 [DOI] [PubMed] [Google Scholar]
- 58.Du L., Hao M., Li C., et al. Quercetin inhibited epithelial mesenchymal transition in diabetic rats, high-glucose-cultured lens, and SRA01/04 cells through transforming growth factor-β2/phosphoinositide 3-kinase/Akt pathway. Molecular and Cellular Endocrinology. 2017;452:44–56. doi: 10.1016/j.mce.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 59.Yao H., Sun J., Wei J., et al. Kaempferol protects blood vessels from damage induced by oxidative stress and inflammation in association with the Nrf2/HO-1 signaling pathway. Frontiers in Pharmacology. 2020;11:p. 1118. doi: 10.3389/fphar.2020.01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alshehri A. S. Kaempferol attenuates diabetic nephropathy in streptozotocin-induced diabetic rats by a hypoglycaemic effect and concomitant activation of the Nrf-2/Ho-1/antioxidants axis. Archives of Physiology and Biochemistry. 2021;127:1–14. doi: 10.1080/13813455.2021.1890129. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Z., Zhou X., Dong Y., et al. Formononetin ameliorates high glucose-induced endothelial dysfunction by inhibiting the JAK/STAT signaling pathway. Molecular Medicine Reports. 2019;20:2893–2901. doi: 10.3892/mmr.2019.10512. [DOI] [PubMed] [Google Scholar]
- 62.Aladaileh S. H., Hussein O. E., Abukhalil M. H., et al. Formononetin upregulates Nrf2/HO-1 signaling and prevents oxidative stress, inflammation, and kidney injury in methotrexate-induced rats. Antioxidants (Basel) 2019;8(10) doi: 10.3390/antiox8100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jamali-Raeufy N., Baluchnejadmojarad T., Roghani M., et al. Isorhamnetin exerts neuroprotective effects in STZ-induced diabetic rats via attenuation of oxidative stress, inflammation and apoptosis. Journal of Chemical Neuroanatomy. 2019;102 doi: 10.1016/j.jchemneu.2019.101709.101709 [DOI] [PubMed] [Google Scholar]
- 64.Chen J., Wang F., Xu H., et al. Long non-coding RNA SNHG1 regulates the Wnt/β-catenin and PI3K/AKT/mTOR signaling pathways via EZH2 to affect the proliferation, apoptosis, and autophagy of prostate cancer cell. Frontiers in Oncology. 2020;10 doi: 10.3389/fonc.2020.552907.552907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang F., Qu Q., Zhao C., et al. Paecilomyces cicadae-fermented Radix astragali activates podocyte autophagy by attenuating PI3K/AKT/mTOR pathways to protect against diabetic nephropathy in mice. Biomed Pharmacother. 2020;129 doi: 10.1016/j.biopha.2020.110479.110479 [DOI] [PubMed] [Google Scholar]
- 66.Huang W. J., Fu Q., Xiao Y. H., et al. Effect of qufengtongluo decoction on PI3K/Akt signaling pathway in the kidney of type 2 diabetes mellitus rat (GK rat) with diabetic nephropathy. Evidence-Based Complementary and Alternative Medicine. 2018;2018 doi: 10.1155/2018/8421979.8421979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qian X., He L., Hao M., et al. YAP mediates the interaction between the Hippo and PI3K/Akt pathways in mesangial cell proliferation in diabetic nephropathy. Acta Diabetologica. 2021;58(1):47–62. doi: 10.1007/s00592-020-01582-w. [DOI] [PubMed] [Google Scholar]
- 68.Huang G., Lv J., Li T., et al. Notoginsenoside R1 ameliorates podocyte injury in rats with diabetic nephropathy by activating the PI3K/Akt signaling pathway. International Journal of Molecular Medicine. 2016;38(4):1179–1189. doi: 10.3892/ijmm.2016.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Panagopoulos A., Altmeyer M. The hammer and the dance of cell cycle control. Trends in Biochemical Sciences. 2020;46(4) doi: 10.1016/j.tibs.2020.11.002. [DOI] [PubMed] [Google Scholar]
- 70.Peng L., Li J., Xu Y., et al. The protective effect of beraprost sodium on diabetic nephropathy by inhibiting inflammation and p38 MAPK signaling pathway in high-fat diet/streptozotocin-induced diabetic rats. International Journal of Endocrinology. 2016;2016 doi: 10.1155/2016/1690474.1690474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elgui de Oliveira D., Müller-Coan B. G., Pagano J. S. Viral carcinogenesis beyond malignant transformation: EBV in the progression of human cancers. Trends in Microbiology. 2016;24(8):649–664. doi: 10.1016/j.tim.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yi M., Xu L., Jiao Y., et al. The role of cancer-derived microRNAs in cancer immune escape. Journal of Hematology & Oncology. 2020;13(1):p. 25. doi: 10.1186/s13045-020-00848-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X., Jiang S., Huang S. Maduramicin-activated protein phosphatase 2A results in extracellular signal-regulated kinase 1/2 inhibition, leading to cytotoxicity in myocardial H9c2 cells. Toxicology Letters. 2018;284:96–102. doi: 10.1016/j.toxlet.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao D., Liu Z., Zhang H. The protective effect of the TUG1/miR-197/MAPK1 axis on lipopolysaccharide-induced podo-cyte injury. Molecular Medicine Reports. 2019;5(20):49–56. doi: 10.3892/mmr.2019.10216. [DOI] [PubMed] [Google Scholar]
- 75.Si W., Shen J., Du C., et al. A miR-20a/MAPK1/c-Myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death & Differentiation. 2018;25(2):406–420. doi: 10.1038/cdd.2017.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ye Y., Guo J., Xiao P., et al. Macrophages-induced long noncoding RNA H19 up-regulation triggers and activates the miR-193b/MAPK1 axis and promotes cell aggressiveness in hepatocellular carcinoma. Cancer Letters. 2020;469:310–322. doi: 10.1016/j.canlet.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 77.Diao L., Wang S., Sun Z. Long noncoding RNA GAPLINC promotes gastric cancer cell proliferation by acting as a molecular sponge of miR-378 to modulate MAPK1 expression. OncoTargets and Therapy. 2018;11:2797–2804. doi: 10.2147/ott.s165147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lou Z., Li Q., Wang C., et al. The effects of microRNA-126 reduced inflammation and apoptosis of diabetic nephropathy through PI3K/AKT signalling pathway by VEGF. Archives of Physiology and Biochemistry. 2020;126:1–10. doi: 10.1080/13813455.2020.1767146. [DOI] [PubMed] [Google Scholar]
- 79.Ribback S., Cigliano A., Kroeger N., et al. PI3K/AKT/mTOR pathway plays a major pathogenetic role in glycogen accumulation and tumor development in renal distal tubules of rats and men. Oncotarget. 2015;6(15):13036–13048. doi: 10.18632/oncotarget.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ge D., Dauchy R. T., Liu S., et al. Insulin and IGF1 enhance IL-17-induced chemokine expression through a GSK3B-dependent mechanism: a new target for melatonin’s anti-inflammatory action. Journal of Pineal Research. 2013;55(4):377–387. doi: 10.1111/jpi.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen W., Taylor B., Jin Q., et al. Inhibition of DYRK1A and GSK3B induces human β-cell proliferation. Nature Communications. 2015;6:p. 8372. doi: 10.1038/ncomms9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang S. Z., Qiu X. J., Dong S. S., et al. MicroRNA-770-5p is involved in the development of diabetic nephropathy through regulating podocyte apoptosis by targeting TP53 regulated inhibitor of apoptosis 1. European Review for Medical and Pharmacological Sciences. 2019;3(23):1248–1256. doi: 10.26355/eurrev_201902_17018. [DOI] [PubMed] [Google Scholar]
- 83.Qiao Y., Tian X., Men L., et al. “Spleen tyrosine kinase promotes NLR family pyrin domain containing 3 inflammasome-mediated IL-1β secretion via c-Jun N-terminal kinase activation and cell apoptosis during diabetic nephropathy”. Molecular Medicine Reports. 2018;2(18):1995–2008. doi: 10.3892/mmr.2018.9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei W., Jin J., Schlisio S., et al. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8(1):25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 85.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110(6):669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 86.Chen J., Chen J. K., Harris R. C. EGF receptor deletion in podocytes attenuates diabetic nephropathy. Journal of the American Society of Nephrology. 2015;26(5):1115–1125. doi: 10.1681/asn.2014020192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang M. Z., Wang Y., Paueksakon P., et al. Epidermal growth factor receptor inhibition slows progression of diabetic nephropathy in association with a decrease in endoplasmic reticulum stress and an increase in autophagy. Diabetes. 2014;63(6):2063–2072. doi: 10.2337/db13-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Y., Pan Y., Cao S., et al. Podocyte EGFR inhibits autophagy through upregulation of rubicon in type 2 diabetic nephropathy. Diabetes. 2021;70(2):562–576. doi: 10.2337/db20-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kabacaoglu D., Ruess D. A., Ai J., et al. NF-κB/Rel transcription factors in pancreatic cancer: focusing on RelA, c-rel, and RelB. Cancers (Basel) 2019;11(7) doi: 10.3390/cancers11070937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lu X., Yarbrough W. G. Negative regulation of RelA phosphorylation: Emerging players and their roles in cancer. Cytokine Growth Factor Reviews. 2015;26(1):7–13. doi: 10.1016/j.cytogfr.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 91.Pradère J. P., Hernandez C., Koppe C., et al. Negative regulation of NF-κB p65 activity by serine 536 phosphorylation. Science Signaling. 2016;9(442):p. ra85. doi: 10.1126/scisignal.aab2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao J., Tian M., Zhang S., et al. Deamidation shunts RelA from mediating inflammation to aerobic glycolysis. Cell Metabolism. 2020;31(5):937–955 e937. doi: 10.1016/j.cmet.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wei L., Yi Z., Guo K., et al. Long noncoding RNA BCAR4 promotes glioma cell proliferation via EGFR/PI3K/AKT signaling pathway. Journal of Cellular Physiology. 2019;234(12):23608–23617. doi: 10.1002/jcp.28929. [DOI] [PubMed] [Google Scholar]
- 94.Huang C., Zhang Y., Qi H., Xu X., Yang L., Wang J. Myc is involved in genistein protecting against LPS-induced myocarditis in vitro through mediating MAPK/JNK signaling pathway. Bioscience Reports. 2020;40(6) doi: 10.1042/bsr20194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stramucci L., Pranteda A., Bossi G. Insights of crosstalk between p53 protein and the MKK3/MKK6/p38 MAPK signaling pathway in cancer. Cancers (Basel) 2018;10(5) doi: 10.3390/cancers10050131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kouhara H., Hadari Y. R., Spivak-Kroizman T., et al. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89(5):693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 97.Wang Z., Yang B., Zhang M., et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33(4):706–720.e709. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee I. H., Kawai Y., Fergusson M. M., et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336(6078):225–228. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hsin K. Y., Ghosh S., Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0083922.e83922 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.


