
Keywords: bioinformatic analysis, Ingenuity Pathway Analysis, mechanistic network, microRNA, peripheral nerve injury, peripheral nerve regeneration, RNA sequencing, sciatic nerve crush
Abstract
Studies have shown that microRNAs (miRNAs) mediate posttranscriptional regulation of target genes and participate in various physiological and pathological processes, including peripheral nerve injury. However, it is hard to select key miRNAs with essential biological functions among a large number of differentially expressed miRNAs. Previously, we collected injured sciatic nerve stumps at multiple time points after nerve crush injury, examined gene changes at different stages (acute, sub-acute, and post-acute), and obtained mRNA expression profiles. Here, we jointly analyzed mRNAs and miRNAs, and investigated upstream miRNAs of differentially expressed mRNAs using Ingenuity Pathway Analysis bioinformatic software. A total of 31, 42, 30, and 23 upstream miRNAs were identified at 1, 4, 7, and 14 days after rat sciatic nerve injury, respectively. Temporal expression patterns and biological involvement of commonly involved upstream miRNAs (miR-21, let-7, miR-223, miR-10b, miR-132, miR-15b, miR-127, miR-29a, miR-29b, and miR-9) were then determined at multiple time points. Expression levels of miR-21, miR-132, miR-29a, and miR-29b were robustly increased after sciatic nerve injury. Biological processes involving these miRNAs include multicellular organismal response to stress, positive regulation of the epidermal growth factor receptor signaling pathway, negative regulation of epithelial cell differentiation, and regulation of myocardial tissue growth. Moreover, we constructed mechanistic networks of let-7, miR-21, and miR-223, the most significantly involved upstream miRNAs. Our findings reveal that multiple upstream miRNAs (i.e., let-7, miR-21, and miR-223) were associated with gene expression changes in rat sciatic nerve stumps after nerve injury, and these miRNAs play an important role in peripheral nerve regeneration. This study was approved by the Experimental Animal Ethics Committee of Jiangsu Province of China (approval No. 20190303-18) on March 3, 2019.
Chinese Library Classification No. R452; R363; R364
Introduction
MicroRNAs (miRNAs) are evolutionarily conserved, small, non-coding RNAs that function in gene regulation (Fromm et al., 2015). In mammals, miRNAs generally pair with sequences complementary to the 3?-untranslated region of their target mRNAs, whereby they negatively regulate mRNA expression via degradation and/or translational inhibition (Bartel et al., 2004, 2009). miRNAs are critical for normal physiological functioning and play critical roles in the development, progression, diagnosis, treatment, and prognosis of various diseases (Jiang et al., 2009; Martinez and Peplow, 2019; Rojo Arias and Busskamp, 2019). Recent studies have shown that miRNAs can function as sponges in the competitive endogenous RNA network, bridge other non-coding RNAs with mRNAs, regulate target gene expression, and modulate a variety of biological activities (Salmena et al., 2011; Thomson et al., 2016).
Peripheral nerve injury (PNI) is a common situation that may happen through laceration, contusion, traction, gunshot wound, or other causes. Much effort has been dedicated to deciphering the molecular basis of PNI to advance the development of treatments (Yuan et al., 2020). Observed gene expression patterns have helped define the post-injury period after peripheral nerve crush injury to include an acute phase of cellular growth and proliferation-associated, sub-acute phase of cellular function and maintenance, and post-acute stage involving tissue formation (Yi et al., 2015). In addition to investigation of mRNA changes, rat sciatic nerve stumps have also been exposed, injured, and subjected to miRNA sequencing. Numerous miRNAs, including novel miRNAs, are differentially expressed after rat sciatic nerve injury (Li et al., 2011; Yu et al., 2011). Detailed investigations showed that many differentially expressed miRNAs after PNI, such as let-7, miR-30c, and miR-340, play important roles in modulating cellular behaviors and regulating nerve regeneration (Li et al., 2015, 2017; Yi et al., 2017).
Although miRNA sequencing provides an overview of miRNA profiles, it is difficult to select key molecules among a large number of differentially expressed miRNAs. Considering that miRNAs execute their biological functions by regulating target genes instead of directly investigating differentially expressed miRNAs, the current study focused on differentially expressed mRNA-associated miRNAs and screened for upstream miRNAs of differentially expressed mRNAs using an advanced bioinformatic software named Ingenuity Pathway Analysis (IPA).
Materials and Methods
RNA sequencing
RNA sequencing was previously performed using RNAs isolated from rat sciatic nerve stumps (Yi et al., 2015). Briefly, 75 adult (2-month-old) male, specific pathogen-free Sprague-Dawley rats (Rattus, weighting 180–220 g) were intraperitoneally injected with a mixture of narcotics containing 85 mg/kg trichloroacetaldehyde monohydrate (Energy-Chemical, Shanghai, China), 42 mg/kg magnesium sulfate (Honeywell, Phoenix, AZ, USA), and 17 mg/kg sodium pentobarbital (Sigma, St. Louis, MO, USA). The left sciatic nerves of anesthetized rats were exposed and crushed with small hemostatic forceps three times for 10 seconds each. Sciatic nerve segments of 5 mm in length at the crush site were collected at 1, 4, 7, and 14 days after crush injury. Rats in the Day 0 group were subjected to sham surgery without crush of the left sciatic nerve. Total mRNA was extracted from rat sciatic nerve stumps and fragmented into short pieces to synthesize cDNA. RNA sequencing was performed using a HiSeq™ 2000 platform (Illumina, San Diego, CA, USA). Sequencing data were conserved in NCBI database (https://www.ncbi.nlm.nih.gov) under accession number PRJNA394957 (SRP113121) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA394957) (Zhao et al., 2019). This study was approved by the Experimental Animal Ethics Committee of Jiangsu Province of China (approval No. 20190303-18) on March 3, 2019.
Bioinformatic analysis
Expression of mapped genes was determined using the reads per kilobase transcriptome per million mapped reads (RPKM) method (Mortazavi et al., 2008). Gene expression levels at 1, 4, 7, and 14 days after crush injury were compared with expression levels at 0 day to screen for differentially expressed genes with a fold change > 2 or < –2 with a false discovery rate ≤ 0.001 (Yi et al., 2015).
Differentially expressed genes were uploaded to IPA software (Ingenuity Systems Inc., Redwood City, CA, USA) for core analysis to determine upstream miRNAs. Discovered upstream miRNAs were subjected to Venn diagram analysis (Venny 2.1.0 online tool; http://bioinfogp.cnb.csic.es/tools/venny/index.html). Expression levels of upstream miRNAs after PNI identified in a previously published miRNA profiling of a rat sciatic nerve transection injury model (Yu et al., 2011) were displayed in a heatmap. Interactions between miRNAs, their target genes, and involved Gene Ontology (GO) terms were identified using Cytoscape software (Shannon et al., 2003). Mechanistic networks between miRNAs and mRNAs were built using the built-in Ingenuity Pathways Knowledge Base (IPKB) in IPA software (Ingenuity Systems Inc.).
Results
Identification of upstream miRNAs after PNI
To investigate miRNAs associated with differentially expressed mRNAs after PNI, previously obtained sequencing data of rat sciatic nerve stumps at 0, 1, 4, 7, and 14 days after crush injury (Yi et al., 2015) were subjected to IPA core analysis. Bioinformatic analyses using IPA upstream regulator function identified 31 upstream miRNAs for the cellular growth and proliferation-associated acute phase (1 day post nerve injury), 72 upstream miRNAs for the cellular function and maintenance-associated sub-acute phase (42 upstream miRNAs at 4 days post nerve injury and 30 upstream miRNAs at 7 days post nerve injury), and 23 upstream miRNAs at the tissue formation-associated post-acute stage (14 days post nerve injury). Upstream miRNAs were ranked by their P-values. Upstream miRNAs with P-values less than 0.05 are listed in Additional Table 1. To explore temporal characteristics, upstream miRNAs identified during the time course were displayed as a Venn diagram (Figure 1A). Ten miRNAs, including miR-21, miR-223, miR-10, miR-155, miR-132, miR-15, miR-127, miR-29, miR-135, and miR-9, were enriched in differentially expressed mRNAs at all time points. In addition, many other upstream miRNAs were involved at multiple time points. Seven miRNAs were enriched at 1, 4, and 7 days after PNI, including let-7, miR-208, miR-25, miR-34, miR-192, miR-17, and miR-19. Four miRNAs were enriched at 1, 4, and 14 days after PNI, including miR-146, miR-23, miR-27, and miR-218. Five miRNAs were enriched at 4, 7, and 14 days after PNI, including miR-8, miR-185, miR-133, miR-130, and miR-145.
Additional Table 1.
Upstream miRNAs at 1, 4, 7, and 14 days afterperipheral nerve injury
| 1d | |||
|---|---|---|---|
| Upstream | Predicted | Activation z | p-value of ove Target Molecules in Dataset |
| let-7 | Inhibited | -7.703 | 4.85E-19 ACVR1C,ALDH1B1,APC2,AR,ARID3A,AURKA,AURKB,BCAT1,BDNF,BOP1 |
| miR-21 | Inhibited | -6.82 | 6.10E-17 AIF1,AOAH,ARF6,ARNTL,ASPM,ATAD2,CACYBP,CCL17,CCL19,CCL2 |
| miR-223 | Activated | 3.06 | 2.13E-10 ABCA1,ABCA13,ABCB1,ALCAM,AMPD3,APOBEC1,ATM,BST1,C15orf48,C5AR1 |
| miR-10 | -1.461 | 2.30E-05 ACVR1C,APC2,AR,ARG1,BAX,CCL2,CD40,CD44,CD80,CD86 | |
| miR-155 | Inhibited | -3.731 | 3.58E-04 AICDA,ANXA2,APAF1,CCL2,CCND1,CD209,CD68,CD69,CDKN1A,CEBPB |
| miR-365 | Activated | 2 | 6.55E-04 AR,CKM,MYF6,MYOD1,MYOG |
| miR-208 | -0.25 | 1.17E-03 IL18BP,MYH7,MYH7B,MYL1,MYL3,MYL9,Nppb,TNNC1,TNNI2 | |
| miR-373 | Inhibited | -2.16 | 1.60E-03 ANGPTL4,CD44,CDH1,CDK2,CSDC2,IL6,RECK,SERPINE1 |
| miR-25 | Inhibited | -2.196 | 1.69E-03 BAX,BCL2L11,CDKN1A,E2F1,E2F3,ESR2,IL6,ITGA5,MAP2K4,MDM2 |
| miR-132 | 1.439 | 1.69E-03 BDNF,CCL2,Ccl2,CCNA2,FOXO3,FOXP2,GRIA1,GRIN2A,IRAK4,PCNA | |
| miR-15 | 1.592 | 2.20E-03 APP,AR,BACE1,BIRC5,CCL2,CCND1,CCNE1,CD69,CD82,CHEK1 | |
| miR-127 | -0.007 | 3.32E-03 ARG1,CLEC10A,IL10,IL1B,IL6,MRC1,NOS2,TNF | |
| miR-34 | Inhibited | -2.317 | 3.74E-03 AR,BIRC3,BIRC5,CCND1,CCNE1,CCNE2,CDC25A,CDK4,CPLX2,CTNND2 |
| miR-29 | 0.864 | 4.14E-03 ADAMTS9,AOX1,AR,ARPC3,BACE1,CARD9,CDH1,CLDN1,COL1A1,COL3A1 | |
| miR-146 | Inhibited | -3.027 | 4.75E-03 CCL2,CD40,CXCL10,ERBB4,FADD,FAF1,FOXP3,IL10,IL1B,IL6 |
| miR-181 | 0.305 | 8.31E-03 AICDA,APP,AR,BCL2L11,CD163,CD69,DUSP6,EZH2,GATA6,GRIA2 | |
| miR-320 | -0.499 | 8.99E-03 AQP1,AQP4,BIRC5,HSPB6,TFRC | |
| miR-154 | -0.749 | 1.06E-02 BAX,BCL2L11,BIRC5,CCND1,CDKN1B,HMOX1,PMP22,STAT3 | |
| miR-290 | 1.311 | 1.06E-02 CCNA1,CDK2,LIN28A,MYC,NANOG,POU5F1,PRKAA1,RECK | |
| miR-23 | -1.491 | 1.15E-02 CDH1,CKS1B,E2F1,GATM,HOXD10,PDE8B,PRDX3,PTK2B,RUNX2,SNCA | |
| miR-27 | -1.887 | 1.16E-02 ABCA1,ABCB1,BAG2,BIRC5,BMP2,CAD,E2F1,ESR1,IL10,IL1B | |
| miR-541 | 1.23E-02 AR,PTPRN,PTPRN2 | ||
| miR-148 | -1.258 | 1.73E-02 ABCA1,ALCAM,AQP4,BCL2L11,DNMT1,MMP15,PIK3IP1,PKM,RUNX3 | |
| miR-135 | -1.326 | 2.16E-02 ALOX5AP,AR,CASP1,Ccl7,CDH1,HIF1A,IL11,IL1B,IL1R1,MMP13 | |
| miR-33 | 1.043 | 2.42E-02 ABCA1,ABCG1,ACACA,ADIPOQ,AGT,FABP4,LPL,PPARGC1A,PRKAA1,SLC2A4 | |
| miR-9 | -1.352 | 2.52E-02 AR,BACE1,CXCR4,FOXG1,JAK2,JAK3,NFKB1,PMP22,RUNX1 | |
| miR-486 | -0.156 | 2.52E-02 ADARB1,AFF3,EMP1,FOXO1,PIK3AP1,TOB1,TWF1,UBASH3B,WT1 | |
| miR-218 | 1.506 | 3.55E-02 CXCR4,DKK2,IBSP,SFRP2,TOB1 | |
| miR-192 | -1.941 | 3.70E-02 ALCAM,CDC7,CDKN1A,CDKN1B,MAD2L1,MDM2,NOD2,TYMS | |
| miR-17 | Inhibited | -2.962 | 3.97E-02 ABCA1,APP,AR,ATM,BCL2L11,BIRC3,CASP7,CCND1,CDKN1A,DUSP2 |
| miR-19 | Inhibited | -2.349 | 4.77E-02 ABCA1,BCL2L11,CDKN1A,E2F1,E2F3,ESR1,PRKAA1,RAB13,TP63 |
| 4d | |||
| Upstream | Predicted | Activation z | p-value of ove Target Molecules in Dataset |
| miR-21 | Inhibited | -6.189 | 1.01E-20 ACTA2,AIF1,ALOX15,AOAH,ARF6,ARNTL,ASPM,ATAD2,CASP4,CCL17 |
| let-7 | Inhibited | -7.15 | 2.23E-17 ACTA2,ACVR1C,ALDH1B1,APC2,AR,ARID3A,AURKA,AURKB,BCAT1,BDNF |
| miR-223 | Activated | 2.585 | 1.35E-11 ABCA1,ABCB1,ACTA2,ALCAM,AMPD3,APOBEC1,ATM,BST1,C15orf48,C5AR1 |
| miR-10 | -0.817 | 3.57E-06 ACVR1C,APC2,AR,ARG1,BAX,BMF,CCL2,CD40,CD44,CD80 | |
| miR-29 | 0.833 | 6.81E-05 ADAMTS9,AOX1,AR,ARPC3,BACE1,CARD9,CD276,CDH1,CLDN1,COL1A1 | |
| miR-8 | -1.239 | 4.96E-04 ATF3,BAX,BCL2L15,BST1,CCNE2,CDH1,CDKN1A,CLDN7,DLX5,DUSP26 | |
| miR-15 | 1.539 | 1.30E-03 AR,BACE1,BIRC5,CCL2,CCL5,CCND1,CCNE1,CD69,CD82,CHEK1 | |
| miR-192 | -1.737 | 2.20E-03 ACTA2,ALCAM,CDC7,CDKN1A,CDKN1B,FN1,MAD2L1,MDM2,NOD2,TYMS | |
| miR-185 | Activated | 2.388 | 2.25E-03 AR,FDFT1,HMGCR,LDLR,NTRK2,SREBF2 |
| miR-24 | 0.103 | 2.58E-03 BCL2L11,CDK4,CDKN1A,CDKN1B,CDKN2B,CDX2,FN1,FURIN,IL12B,IL6 | |
| miR-155 | Inhibited | -3.581 | 2.94E-03 ANXA2,APAF1,CCL2,CCL5,CCND1,CD209,CD40LG,CD68,CD69,CDKN1A |
| miR-25 | Inhibited | -2.037 | 4.04E-03 BAX,BCL2L11,CDKN1A,E2F1,E2F3,ESR2,GADD45A,IL6,MAP2K4,MDM2 |
| miR-218 | 0.721 | 5.48E-03 CXCR4,DKK2,IBSP,SOST,SPP1,TOB1 | |
| miR-373 | -1.916 | 7.40E-03 ANGPTL4,CD44,CDH1,CDK2,IL6,RECK,SERPINE1 | |
| miR-27 | -1.669 | 7.88E-03 ABCA1,ABCB1,BIRC5,E2F1,ESR1,IL10,IL1B,IL6,MAP2K4,MET | |
| miR-126 | Inhibited | -2.351 | 8.24E-03 CDH1,GATA3,KIT,MMP7,MYOCD,PLAC1,PLK2,POU2AF1,PTPRC,SOX2 |
| miR-154 | -0.064 | 8.51E-03 BAX,BCL2L11,BIRC5,CCND1,CDKN1B,HMOX1,KIT,PMP22 | |
| miR-23 | -0.598 | 8.67E-03 CDH1,CKS1B,E2F1,FBXO32,GATM,PDE8B,PRDX3,PTK2B,RUNX2,SNCA | |
| miR-132 | 0.742 | 1.21E-02 BDNF,CCL2,Ccl2,CCNA2,FOXP2,GRIA1,IRAK4,NR4A2,PCNA,PEA15 | |
| miR-34 | -1.983 | 1.24E-02 AR,BIRC3,BIRC5,CCND1,CCNE1,CCNE2,CDC25A,CDK4,CNTN2,CPLX2 | |
| miR-127 | -0.244 | 1.30E-02 ARG1,CLEC10A,IL10,IL1B,IL6,MRC1,TNF | |
| miR-19 | -1.233 | 1.35E-02 ABCA1,BCL2L11,CDKN1A,E2F1,E2F3,ESR1,FZD4,MSMO1,PSAP,RAB13 | |
| miR-133 | -0.839 | 1.90E-02 CCNB1,CNN1,DNMT1,FN1,HCN2,HCN4,HLA-G,KCNH2,KLF15,MET | |
| miR-146 | Inhibited | -2.277 | 1.90E-02 CCL2,CD40,CXCL10,ERBB4,FADD,IL10,IL1B,IL6,IRAK2,KIT |
| miR-9 | -0.548 | 2.02E-02 AR,BACE1,CDKN2A,CXCR4,JAK2,JAK3,PMP22,PRDM1,RUNX1 | |
| miR-130 | Inhibited | -2.596 | 2.10E-02 ABCB1,CEBPE,GTF2H1,IRF8,MAFB,PPARA,TGFBR1 |
| miR-208 | -1.18 | 2.10E-02 IL18BP,MYH7,MYH7B,MYL1,MYL9,Nppb,TNNI2 | |
| miR-140 | -1.948 | 2.51E-02 CDH1,DNPEP,SOX2,VIM | |
| miR-194 | 3.08E-02 ACTA2,E2F1,MDM2,SLC7A5,THBS1 | ||
| miR-135 | -1.051 | 4.16E-02 AR,CASP1,Ccl7,CDH1,HIF1A,IL11,IL1B,IL1R1,RUNX2,SLC6A4 | |
| miR-221 | 1.261 | 4.26E-02 BMF,CDKN1B,Cdkn1c,KIT,MMP9,PIK3R1,TFAP2A,TIMP3 | |
| miR-148 | -0.998 | 4.26E-02 ABCA1,ALCAM,BCL2L11,DNMT1,LDLR,MMP15,PKM,RUNX3 | |
| miR-320 | 0.054 | 4.83E-02 AQP1,BIRC5,HSPB6,TFRC | |
| miR-17 | Inhibited | -2.466 | 4.92E-02 ABCA1,AR,ATM,BCL2L11,BIRC2,BIRC3,CASP7,CCND1,CDKN1A,DUSP2 |
| miR-338 | 4.97E-02 HIF1A,RUNX2 | ||
| miR-95 | 4.97E-02 AR,BRCA1 | ||
| miR-379 | 4.97E-02 HIF1A,PTPRN | ||
| miR-485 | 4.97E-02 GRIA2,SV2A | ||
| miR-585 | 4.97E-02 CASP3,PARP1 | ||
| 7 d | |||
| Upstream | Predicted | Activation z | p-value of ove Target Molecules in Dataset |
| miR-21 | Inhibited | -4.126 | 3.06E-15 ACTA2,AIF1,AOAH,ARNTL,ASPM,CASP4,CCL17,CCL19,CCL2,CCL3L3 |
| let-7 | Inhibited | -6.472 | 2.70E-10 ACTA2,ACVR1C,ALDH1B1,AURKA,AURKB,BDNF,BRCA1,BRCA2,BUB1,CASP3 |
| miR-223 | Activated | 3.998 | 6.16E-10 ABCA1,ABCB1,ACTA2,ALCAM,APOBEC1,ATM,BST1,C15orf48,C5AR1,CA4 |
| miR-155 | Inhibited | -3.04 | 1.17E-04 AICDA,BCL6,CCL2,CCL5,CCND1,CD209,CD68,CD69,CDKN1A,CLDN1 |
| miR-29 | 1.083 | 4.42E-04 AOX1,BDKRB2,CARD9,CDH1,CLDN1,COL1A1,DKK1,ELN,FKBP5,FLT3 | |
| miR-8 | Inhibited | -2.24 | 7.27E-04 ATF3,BCL2L15,BST1,CDC25B,CDH1,CDKN1A,E2F3,EPCAM,ERRFI1,ESM1 |
| miR-15 | 1.699 | 7.53E-04 BIRC5,CCL2,Ccl2,CCL5,CCND1,CCNE1,CD69,FGF1,IL1B,IL6 | |
| miR-10 | 0.18 | 1.98E-03 ACVR1C,ARG1,BMF,CCL2,CD40,CD44,CD80,CD86,CDH1,CXCL11 | |
| miR-127 | 1.313 | 2.46E-03 ARG1,BCL6,IL10,IL1B,IL6,NOS2,TNF | |
| miR-132 | 1.555 | 3.77E-03 BDNF,CCL2,Ccl2,CCNA2,GRIA1,GRIN2A,GRIN2B,PCNA,PEA15,STAT4 | |
| miR-208 | -0.295 | 4.21E-03 IL18BP,MYH7,MYL1,MYL9,Nppb,TNNC1,TNNI2 | |
| miR-204 | 0 | 6.76E-03 BDNF,CXCL2,IGF2R,IL1B,IL6,RUNX2,SPDEF | |
| miR-214 | -0.92 | 8.09E-03 ALCAM,ATM,BIRC5,CADM1,G6PC,PCK1,TFAP2A,TFAP2C | |
| miR-133 | -0.359 | 8.27E-03 CCNB1,CNN1,FN1,HCN2,HCN4,HLA-G,KCNH2,KCNQ1,KLF15,MET | |
| miR-194 | 9.12E-03 ACTA2,MDM2,SLC7A5,SOCS2,THBS1 | ||
| miR-17 | Inhibited | -2.754 | 9.26E-03 ABCA1,ATM,BCL2L11,BIRC3,CASP7,CCND1,CDKN1A,DUSP2,ESR1,FN1 |
| miR-9 | -0.851 | 1.15E-02 BCL6,CDKN2A,CXCR4,JAK2,JAK3,PMP22,PRDM1,RUNX1 | |
| miR-25 | Inhibited | -2.081 | 1.21E-02 BCL2L11,CDKN1A,E2F3,ESR2,GADD45A,IL6,MDM2,MYLIP,TGFBR1,TNF |
| miR-130 | Inhibited | -2.433 | 1.95E-02 ABCB1,GTF2H1,IRF8,MAFB,PPARA,TGFBR1 |
| miR-192 | -1.003 | 2.11E-02 ACTA2,ALCAM,CDKN1A,FN1,MDM2,NOD2,TYMS | |
| miR-19 | -1.372 | 2.14E-02 ABCA1,BCL2L11,CDKN1A,E2F3,ESR1,MSMO1,MYLIP,RAB13 | |
| miR-185 | 0.762 | 3.11E-02 EPHB2,FDFT1,HMGCR,NTRK2 | |
| miR-34 | -1.604 | 3.35E-02 BIRC3,BIRC5,CCND1,CCNE1,CDK4,CNTN2,CPLX2,CTNND2,E2F3,GRM7 | |
| miR-145 | Inhibited | -2.685 | 3.50E-02 ABCA1,ACTA2,ADAM17,CACNA1C,CCNA2,CD28,DMP1,E2F3,IL10,IL6 |
| miR-190 | 3.59E-02 CDH1,CLDN1,VIM | ||
| miR-506 | 3.59E-02 GFRA1,SPDEF,TNF | ||
| miR-28 | 3.59E-02 BTLA,HAVCR2,PDCD1 | ||
| miR-486 | -0.87 | 3.79E-02 ADARB1,FOXO1,GABRE,PIK3AP1,SLC4A8,UBASH3B,WT1 | |
| miR-135 | -1.311 | 4.23E-02 CASP1,Ccl7,CDH1,IL11,IL1B,IL1R1,MMP13,RUNX2,SLC6A4 | |
| 14 d | |||
| Upstream | Predicted | Activation z | p-value of ove Target Molecules in Dataset |
| miR-223 | Activated | 3.527 | 1.84E-14 ABCA1,ABCA13,ABCB1,ACTA2,ALCAM,APOBEC1,ATM,BST1,C15orf48,C5AR1 |
| miR-21 | Inhibited | -3.886 | 1.55E-12 ACTA2,AIF1,ALOX15,AOAH,ARNTL,CCL17,CCL19,CCL2,CCL3L3,CD180 |
| miR-214 | -0.92 | 1.46E-03 ALCAM,ATM,BIRC5,CADM1,G6PC,PCK1,TFAP2A,TFAP2C | |
| miR-146 | Inhibited | -2.342 | 2.12E-03 CCL2,CD40,CXCL10,ERBB4,IL10,IL1B,MMP2,NLGN1,RUNX1,SOCS1 |
| miR-135 | -0.5 | 2.31E-03 CASP1,Ccl7,CDH1,EDN1,IL11,IL1B,IL1R1,RUNX2,SLC6A4,SPP1 | |
| miR-126 | -1.71 | 3.07E-03 CDH1,MMP7,MYOCD,PLAC1,PLK2,POU2AF1,PTPRC,SOX2,SPI1,TAGLN2 | |
| miR-10 | -0.956 | 3.17E-03 ACVR1C,ARG1,BMF,CCL2,CD40,CD44,CD80,CD86,CDH1,CXCL11 | |
| miR-155 | Inhibited | -2.941 | 4.54E-03 CCL2,CCL5,CD209,CD68,CD69,CXCL10,DYNC1I1,EDN1,HK2,HMOX1 |
| miR-8 | -1.196 | 5.18E-03 ATF3,BBC3,BST1,CDC25B,CDH1,CLDN7,EPCAM,ERRFI1,FTH1,HMOX1 | |
| miR-29 | 0.634 | 8.36E-03 AOX1,CARD9,CDH1,DKK1,ELN,FKBP5,FLT3,IL12B,MMP2,OLR1 | |
| miR-9 | -1.192 | 9.50E-03 CDKN2A,CXCR4,JAK2,JAK3,PMP22,PRDM1,RUNX1 | |
| miR-185 | 0.762 | 1.22E-02 EPHB2,FDFT1,HMGCR,NTRK2 | |
| miR-23 | 0.574 | 1.67E-02 CDH1,GATM,PDE8B,PTK2B,RUNX2,SNCA,SPP1,TRIM63 | |
| miR-28 | 1.72E-02 BTLA,HAVCR2,PDCD1 | ||
| miR-15 | Activated | 2.266 | 1.80E-02 BIRC5,CCL2,Ccl2,CCL5,CD69,FGF1,IL1B,ISG15,MAPK3,MMP9 |
| miR-127 | 0.541 | 1.81E-02 ARG1,IL10,IL1B,NOS2,TNF | |
| miR218 | -1.188 | 1.98E-02 CXCR4,SFRP2,SOST,SPP1 | |
| miR-132 | 0.8 | 2.09E-02 BDNF,CCL2,Ccl2,GRIA1,GRIN2B,PEA15,STAT4,TNF | |
| miR-130 | Inhibited | -2.19 | 2.53E-02 ABCB1,CEBPE,IRF8,MAFB,TGFBR1 |
| miR-27 | -1.405 | 3.31E-02 ABCA1,ABCB1,BIRC5,EDN1,IL10,IL1B,MET,MYH7,MYOG,RUNX1 | |
| miR-133 | 1.117 | 4.18E-02 CNN1,HAND2,HCN2,HLA-G,KCNH2,KLF15,MET,SLC2A4,TAGLN,TRPS1 | |
Figure 1.
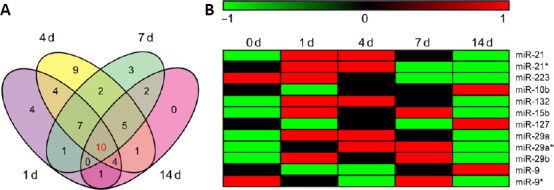
Differentially expressed mRNA-associated upstream miRNAs after peripheral nerve injury
(A) Venn diagram of upstream miRNAs in sciatic nerve stumps at 1, 4, 7, and 14 days after peripheral nerve injury. (B) Heatmap of ten overlapping upstream miRNAs at 1, 4, 7, and 14 days after peripheral nerve injury. Red indicates upregulation, while green indicates downregulation.
Expression patterns of overlapping upstream miRNAs were revealed at all time points based on previously obtained miRNA sequencing data of rats with sciatic nerve transection injury (Yu et al., 2011). According to miRNA sequencing data reported by Yu et al. (2011) 225 miRNAs were differentially expressed after PNI. Among the ten overlapping upstream miRNAs, only miR-155 and miR-135 were not found to be differentially expressed, while the other eight displayed altered expression levels after nerve injury. Two family members of miR-29 (miR-29a and miR-29b) were both differentially expressed after nerve injury. In addition, different isoforms of miR-21, miR-29a, and miR-9 were differentially expressed. miRNAs labeled with an asterisk indicate isoforms with relatively low expression levels. Temporal expression patterns of these miRNAs are displayed as a heatmap (Figure 1B). miR-21, miR-21*, miR-132, miR-15b, miR-29a, miR-29a*, and miR-29b were upregulated after nerve injury. miR-223 was downregulated after nerve injury; whereas miR-10b, miR-127, miR-9, and miR-9* were first downregulated and then upregulated.
Biological implications of differentially expressed upstream miRNAs
To evaluate underlying biological functions, differentially expressed upstream miRNAs were further analyzed by GluoGO and GluePedia tools using Cytoscape software. miRNA databases for human beings (Homo sapiens) and mice (Mus musculus) are much more comprehensive than those of rats (Rattus norveqicus). However, because miRNA sequences are extremely conserved among species, we used an miRNA database of mice (mmu-miRs) as a reference to explore differentially expressed upstream miRNAs. Among the eight differentially expressed miRNAs commonly involved in differentially expressed genes at 1, 4, 7, and 14 days after PNI, five (miR-21, miR-223, miR-132, miR-127, and miR-9) were identified by both GluoGO and GluePedia.
Target genes of these miRNAs were determined using the built-in miTarBase database in Cytoscape software and connected by importing networks. miR-21 targeted Mmp9 (matrix metallopeptidase 9), Smad7 (SMAD family member 7), Yy1 (YY1 transcription factor), Spry1/2/3 (Sprouty RTK signaling antagonist 1/2/3), Gt(ROSA)26Sor (ROSA26), Fasl (Fas Ligand), Pten (phosphatase and tensin homolog), and Tgfbi (transforming growth factor beta induced). miR-132 also targeted Mmp9, as well as Btg2 (BTG anti-proliferation factor 2), Nr4a2 (nuclear receptor subfamily 4 group a member 2), Ptbp2 (polypyrimidine tract binding protein 2), Ache (acetylcholinesterase), Arhgap32 (Rho GTPase activating protein 32), Ep300 (E1A binding protein p300), and Kdm5a (lysine demethylase 5a). miR-223 targeted Stat3 (signal transducer and activator of transcription 3), Mef2c (myocyte enhancer factor 2c), Gria2 (glutamate ionotropic receptor AMPA type subunit 2), Gzmb (granzyme b), Grin2b (glutamate ionotropic receptor NMDA type subunit 2b), and Fbxw7 (F-Box and WD repeat domain containing 7). miR-9 targeted Zfp521 (zinc finger protein 521), Camkk2 (calcium/calmodulin dependent protein kinase kinase 2), Stmn1 (stathmin 1), Runx1 (RUNX family transcription factor 1), Foxg1 (forkhead box g1), and Hes1 (Hes family BHLH transcription factor 1). miR-127 targeted Fcgr1 (Fc fragment of IgG receptor 1) (Figure 2).
Figure 2.

miRNA-mRNA-GO term network of miR-21, miR-132, miR-223, and miR-9
Dotted lines indicate associations of miRNAs and potential target genes, while solid lines indicate associations of genes and biological functions. Various biological functions are indicated by different colors, while node sizes reflect the enrichment of biological functions. Ache: Acetylcholinesterase; Arhgap32: Rho GTPase activating protein 32; Btg2: BTG anti-proliferation factor 2; Camkk2: calcium/calmodulin dependent protein kinase kinase 2; Ep300: E1A binding protein p300; Fasl: Fas Ligand; Fbxw7: F-Box And WD repeat domain containing 7; Foxg1: forkhead box g1; Gria2: glutamate ionotropic receptor AMPA type subunit 2; Grin2b: glutamate ionotropic receptor NMDA type subunit 2b; Gt (ROSA) 26Sor: ROSA26; Gzmb: granzyme b; Hes1: Hes family BHLH transcription factor 1; Kdm5a: lysine demethylase 5a; Mef2c: myocyte enhancer factor 2c; Mmp9: matrix metallopeptidase 9; Nr4a2: nuclear receptor subfamily 4 group a member 2; Ptbp2: polypyrimidine tract binding protein 2; Pten: phosphatase and tensin homolog; Runx1: RUNX family transcription factor 1; Smad7: SMAD family member 7; Spry1/2/3: Sprouty RTK signaling antagonist 1/2/3; Stat3: signal transducer and activator of transcription 3; Stmn1: stathmin 1; Tgfbi: transforming growth factor beta induced; Yy1: YY1 transcription factor; Zfp521: zinc finger protein 521.
Targets genes and their associated GO biological processes were then connected using bioinformatic tools. miR-127 and its target Fcgr1 did not interact with other genes and, therefore, were not included in the miRNA-mRNA-GO network. To identify the most significantly involved biological functions, GO biological processes with a P-value < 0.001 were selected and listed. Such GO biological processes were mainly categorized as multicellular organismal response to stress, positive regulation of epidermal growth factor receptor signaling pathway, negative regulation of epithelial cell differentiation, and regulation of cardiac muscle tissue growth (Figure 2).
Mechanistic networks of essential upstream miRNAs after PNI
Next, the most significantly involved upstream miRNAs after PNI were investigated in detail. The top five upstream miRNAs at each time point after PNI, as well as their P-values and targeted molecules, are displayed in Table 1. Let-7, miR-21, and miR-223 were commonly enriched at all time points.
Table 1.
Top 5 upstream miRNAs at 1, 4, 7, and 14 days after PNI
| Time after PNI (d) | miRNA | P-value | Target molecules in dataset |
|---|---|---|---|
| 1 | let-7 | 4.85E-19 | ACVR1C, ALDH1B1, APC2, AR, ARID3A, AURKA, AURKB, BCAT1, BDNF, BOP1 |
| miR-21 | 6.10E-17 | AIF1, AOAH, ARF6, ARNTL, ASPM, ATAD2, CACYBP, CCL17, CCL19, CCL2 | |
| miR-223 | 2.13E-10 | ABCA1, ABCA13, ABCB1, ALCAM, AMPD3, APOBEC1, ATM, BST1, C15orf48, C5AR1 | |
| miR-10 | 2.30E-05 | ACVR1C, APC2, AR, ARG1, BAX, CCL2, CD40, CD44, CD80, CD86 | |
| miR-155 | 3.58E-04 | AICDA, ANXA2, APAF1, CCL2, CCND1, CD209, CD68, CD69, CDKN1A, CEBPB | |
| 4 | miR-21 | 1.01E-20 | ACTA2, AIF1, ALOX15, AOAH, ARF6, ARNTL, ASPM, ATAD2, CASP4, CCL17 |
| let-7 | 2.23E-17 | ACTA2, ACVR1C, ALDH1B1, APC2, AR, ARID3A, AURKA, AURKB, BCAT1, BDNF | |
| miR-223 | 1.35E-11 | ABCA1, ABCB1, ACTA2, ALCAM, AMPD3, APOBEC1, ATM, BST1, C15orf48, C5AR1 | |
| miR-10 | 3.57E-06 | ACVR1C, APC2, AR, ARG1, BAX, BMF, CCL2, CD40, CD44, CD80 | |
| miR-29 | 6.81E-05 | ADAMTS9, AOX1, AR, ARPC3, BACE1, CARD9, CD276, CDH1, CLDN1, COL1A1 | |
| 7 | miR-21 | 3.06E-15 | ACTA2, AIF1, AOAH, ARNTL, ASPM, CASP4, CCL17, CCL19, CCL2, CCL3L3 |
| let-7 | 2.70E-10 | ACTA2, ACVR1C, ALDH1B1, AURKA, AURKB, BDNF, BRCA1, BRCA2, BUB1, CASP3 | |
| miR-223 | 6.16E-10 | ABCA1, ABCB1, ACTA2, ALCAM, APOBEC1, ATM, BST1, C15ORF48, C5AR1, CA4 | |
| miR-155 | 1.17E-04 | AICDA, BCL6, CCL2, CCL5, CCND1, CD209, CD68, CD69, CDKN1A, CLDN1 | |
| miR-29 | 4.42E-04 | AOX1, BDKRB2, CARD9, CDH1, CLDN1, COL1A1, DKK1, ELN, FKBP5, FLT3 | |
| 14 | miR-223 | 1.84E-14 | ABCA1, ABCA13, ABCB1, ACTA2, ALCAM, APOBEC1, ATM, BST1, C15ORF48, C5AR1 |
| miR-21 | 1.55E-12 | ACTA2, AIF1, ALOX15, AOAH, ARNTL, CCL17, CCL19, CCL2, CCL3L3, CD180 | |
| miR-214 | 1.46E-03 | ALCAM, ATM, BIRC5, CADM1, G6PC, PCK1, TFAP2A, TFAP2C | |
| miR-146 | 2.12E-03 | CCL2, CD40, CXCL10, ERBB4, IL10, IL1B, MMP2, NLGN1, RUNX1, SOCS1 | |
| miR-135 | 2.31E-03 | CASP1, CCL7, CDH1, EDN1, IL11, IL1B, IL1R1, RUNX2, SLC6A4, SPP1 |
Ache: Acetylcholinesterase; ACTA2: actin alpha 2, smooth muscle; AIF1: allograft inflammatory factor 1; Akt: AKT serine/threonine kinase 1; ALOX15: arachidonate 15-lipoxygenase; AOAH: acyloxyacyl hydrolase; AP-1: activator protein-1; ARF6: ADP ribosylation factor 6; Arhgap32: Rho GTPase activating protein 32; ARNTL: aryl hydrocarbon receptor nuclear translocator like; ASPM: abnormal spindle microtubule assembly; ATAD2: ATPase family AAA domain containing 2; Btg2: BTG anti-proliferation factor 2; Camkk2 : calcium/calmodulin dependent protein kinase kinase 2; CASP4: caspase 4; CCL17: C-C motif chemokine ligand 17; CCL3L3: C-C motif chemokine ligand 3 like 3; Ep300: E1A binding protein p300; Erk1/2: extracellular regulated protein kinases; Fasl: Fas Ligand; Fbxw7: F-Box And WD repeat domain containing 7; Fcgr1: Fc fragment of IgG receptor 1; Foxg1: forkhead box g1; GO: Gene ontology; Gria2: glutamate ionotropic receptor AMPA type subunit 2; Grin2b: glutamate ionotropic receptor NMDA type subunit 2b; Gzmb: granzyme b; Hes1: Hes family BHLH transcription factor 1; IFNG : interferon gamma; IL: interleukin; IPA: Ingenuity Pathway Analysis; IPKB: Ingenuity Pathways Knowledge Base; Jnk: JUN N-terminal kinase; Kdm5a: lysine demethylase 5a; MAPK3: mitogen-activated protein kinase 3; Mef2c: myocyte enhancer factor 2c; miRNAs: MicroRNAs; Mmp9: matrix metallopeptidase 9; NOS3: nitric oxide synthase 3; Nr4a2: nuclear receptor subfamily 4 group a member 2; PNI: peripheral nerve injury; Ptbp2: polypyrimidine tract binding protein 2; Pten: phosphatase and tensin homolog; ROSA26: Gt(ROSA)26Sor; RPKM: Reads per kilobase transcriptome per million mapped reads; Runx : RUNX family transcription factor 1; SD: Sprague-Dawley; Smad: SMAD family member; Spry1/2/3: Sprouty RTK signaling antagonist 1/2/3; Stat3: signal transducer and activator of transcription 3; Stmn1: stathmin 1; Tgfbi: transforming growth factor beta induced; Yy1: YY1 transcription factor; Zfp521: zinc finger protein 521.
Let-7 was most significantly enriched during the acute phase after nerve injury (1 day after PNI). Let-7 was predicted to decrease the activities of Stat3, Mapk3 (mitogen-activated protein kinase 3), Erk1/2 (extracellular regulated protein kinases), Smad3 (SMAD family member 3), and Akt (AKT serine/threonine kinase 1). Many downstream cascades of Mapk3, Stat3, Erk1/2, Smad3, and Akt, such as NF-κB (nuclear factor κB) and AP-1 (activator protein-1) would be affected by let-7 (Figure 3).
Figure 3.

Mechanistic network of let-7-associated genes at 1 day after PNI
For genes, blue indicates predicted inhibition and orange indicates predicted activation. Bold-type letters below genes indicate expression level and P-value according to sequencing data. For network shapes, inverted trapezoids indicate miRNA, ovals indicate transcription regulators, dual rings indicate complexes/groups, and inverted triangles indicates kinases. For relationships, orange lines indicate activation, gray lines indicate that an effect was not predicted, solid lines indicate a direct relationship, and dotted lines indicate an indirect relationship. For relationship labels, A indicates activation, P indicates phosphorylation/dephosphorylation, I indicates inhibition, LO indicates localization, RB indicates regulation of bindings, L indicates molecular cleavage (includes degradation for chemicals), and numbers in brackets indicate numbers of relationships identified by IPKB. Akt: AKT serine/threonine kinase 1; Ap1: activator protein-1; CEBPB: CCAAT enhancer binding protein beta; Creb: cAMP responsive element binding protein; Erk1/2: extracellular regulated protein kinases; FOS: Fos proto-oncogene, AP-1 transcription factor subunit; HIF1A: hypoxia inducible factor 1 subunit alpha; Jun: Jun proto-oncogene, AP-1 transcription factor subunit; MAPK3: mitogen-activated protein kinase 3; NFKB1: nuclear factor kappa b subunit 1; PNI: peripheral nerve injury; RELA: RELA proto-oncogene, NF-κB subunit; SMAD3/4/7: SMAD family member 3/4/7; STAT3: signal transducer and activator of transcription 3.
miR-21 was most significantly enriched at both 4 and 7 days after PNI, indicating that miR-21 was critical for the sub-acute phase after nerve injury. IPA analyses showed that miR-21 targeted ACTA2 (actin alpha 2, smooth muscle), AIF1 (allograft inflammatory factor 1), ALOX15 (arachidonate 15-lipoxygenase), AOAH (acyloxyacyl hydrolase), ARF6 (ADP ribosylation factor 6), ARNTL (aryl hydrocarbon receptor nuclear translocator like), ASPM (abnormal spindle microtubule assembly), ATAD2 (ATPase family AAA domain containing 2), CASP4 (caspase 4), and CCL17 (C-C motif chemokine ligand 17) at 4 days post nerve injury. At 7 days post nerve injury, ALOX15, ARF6, and ATAD2 were not predicted to be target molecules in the dataset; whereas, CCL19 (C-C motif chemokine ligand 19), CCL2 (C-C motif chemokine ligand 2), and CCL3L3 (C-C motif chemokine ligand 3 like 3) were included in the dataset. Constructed mechanistic networks of miR-21 at 4 and 7 days after PNI were also slightly different. miR-21 was predicted to affect IFNG (interferon gamma), IL4 (interleukin 4), IL6 (interleukin 6), and their downstream molecules at both time points (Figure 4). In addition to activation of these essential molecules, miR-21 might also inhibit NOS3 (nitric oxide synthase 3) (Figure 4A) at 4 days after PNI.
Figure 4.

Mechanistic network of miR-21-associated genes at (A) 4 days and (B) 7 days after PNI
For genes, blue indicates predicted inhibition and orange indicates predicted activation. Bold-type letters below genes indicate expression level and P-value according to sequencing data. For network shapes, inverted trapezoids indicate miRNA, ovals indicate transcription regulators, dual rings indicate complexes/groups, squares indicate cytokines, and rhombuses indicate enzymes. For relationships, blue lines indicate inhibition, orange lines indicate activation, gray lines indicate that an effect was not predicted, solid lines indicate a direct relationship, and dotted lines indicate an indirect relationship. For relationship labels, A indicates activation, P indicates phosphorylation/dephosphorylation, I indicates inhibition, LO indicates localization, RB indicates regulation of bindings, L indicates molecular cleavage (includes degradation for chemicals), M indicates biochemical modification, TR indicates translocation, and numbers in brackets indicate numbers of relationships identified by IPKB. CEBPB: CCAAT enhancer binding protein beta; IFNG: interferon gamma; IL4/6: interleukin 4/6; IRF1: interferon regulatory factor 1; NOS3: nitric oxide synthase 3; PNI: peripheral nerve injury; RELA: RELA proto-oncogene, NF-κB subunit; STAT1/3/6: signal transducer and activator of transcription 1/3/6.
The most significantly enriched miRNA at the post-acute stage (14 days after PNI) was miR-223. The mechanistic network of miR-223 was also created (Figure 5). The built-in Ingenuity ExpertAssist Finding in IPA software indicated that downregulation of miR-223 was involved in activation of Stat3 (Chen et al., 2012). Therefore, the Stat3 signaling pathway might be involved in miR-223-mediated biological functions after PNI.
Figure 5.

Mechanistic network of miR-223-associated genes at 14 days after PNI
For network shapes, inverted trapezoids indicate miRNA, ovals indicate transcription regulators, and dual rings indicate complexes/groups. For relationships, orange lines indicate activation, gray lines indicate that an effect was not predicted, solid lines indicate a direct relationship, and dotted lines indicate an indirect relationship. For relationship labels, A indicates activation, LO indicates localization, RB indicates regulation of bindings, L indicates molecular cleavage (includes degradation for chemicals), and numbers in brackets indicate numbers of relationships identified by IPKB. Genes in orange indicate predicted activation. Bold-type letters below genes indicate expression level and P-value according to sequencing data. NFκB1: Nuclear factor kappa b subunit 1; PNI: peripheral nerve injury; SMAD3: SMAD family member 3; STAT3: signal transducer and activator of transcription 3.
Discussion
Emerging high-throughput analyses, such as sequencing and microarray, are rapidly accelerating our understanding of cellular and molecular mechanisms of diverse biological processes, including PNI and repair (Yi et al., 2015, 2017; Yu et al., 2016; Cheng et al., 2020). Relative abundances and expression changes of miRNAs were also revealed, considering the important regulatory effects of miRNAs. For instance, in our laboratory, Yu et al. (2011) discovered 225 differentially expressed known miRNAs in rat sciatic nerve stumps after nerve transection. These identified differentially expressed miRNAs might participate in regulation of gene expression and cellular function. However, selecting key miRNAs among the large number of differentially expressed miRNAs was a major challenge. Simply selecting miRNAs with highest abundances or most robust expression changes might conceal useful information.
To reveal key miRNAs during peripheral nerve repair and regeneration, the current study applied IPA bioinformatics to discover upstream miRNAs of differentially expressed mRNAs at 1, 4, 7, and 14 days after PNI. IPA analyses recognized 53 upstream miRNAs after nerve injury, with some upstream miRNAs recognized at multiple time points. Among these miRNAs, miR-21, miR-223, miR-10, miR-155, miR-132, miR-15, miR-127, miR-29, miR-135, and miR-9 were found to be upstream miRNAs at all tested time points. Overlapping these common upstream miRNAs with differentially expressed known miRNAs at the same time points after nerve transection showed that miR-21, miR-223, miR-10b, miR-132, miR-15b, miR-127, miR-29a/b, and miR-9 were differentially expressed. These differentially expressed miRNAs might be critical for peripheral nerve repair and considered as potential biomarkers of neuroregeneration.
Biological enrichment of identified differentially expressed upstream miRNAs showed that these miRNAs and their associated target genes might regulate tissue development (multicellular organismal response to stress, and cardiac muscle tissue growth and development) and cellular status (negative regulation of epithelial cell differentiation, regulation of epithelial to mesenchymal transition, and negative regulation of epithelial-to-mesenchymal transition). Following PNI, Schwann cells undergo a dedifferentiation process that benefits their subsequent proliferation and migration, as well as regeneration of injured nerves (Painter et al., 2014; Stierli et al., 2019). Our bioinformatic analyses suggest that Mmp9, Spry1/2, and their upstream miR-21, as well as Hes1 and its upstream miR-9, might be essential for Schwann cell phenotype modulation and peripheral nerve regeneration. Epithelial-to-mesenchymal transition of Schwann cells is essential to the regeneration process (Arthur et al., 2017; Clements et al., 2017; Jessen et al., 2019). Our bioinformatic analyses suggest that miR-21 regulated Spry1, Spry2, Smad7, and Pten might contribute to nerve regeneration. In addition, many canonical signaling pathways were identified, including positive regulation of ERBB signaling pathway and negative regulation of neurotrophin TRK receptor signaling pathway. These results imply significant involvement of growth factors, such as neurotrophins, after PNI.
In addition to generating a miRNA-mRNA-GO biological process network, the most significantly enriched upstream miRNAs at different stages of peripheral nerve regeneration were identified and investigated in detail. Let-7 was found to be the most associated upstream miRNA at the acute post-injury phase, and was predicted to be inhibited. Previous functional studies in our laboratory showed that downregulation of let-7 after nerve injury leads to elevated expression of its target gene NGF (nerve growth factor), stimulation of Schwann cell proliferation and migration, and axon elongation (Li et al., 2015). In addition, decreased expression of let-7 was shown to increase the proliferation, migration, and tubulogenesis of endothelial cells (Ji et al., 2019). These findings suggest that altered expression of let-7 would affect the phenotypes of multiple cell types and, therefore, would contribute to initiation of peripheral nerve regeneration at the acute post-injury stage. Cell proliferation was also involved in the sub-acute phase after nerve injury. For example, a recent study indicated that miR-21, the most significantly involved upstream miRNA during the sub-acute phase, encouraged Schwann cell proliferation and axon regeneration (Ning et al., 2020). Our bioinformatic analyses suggest that miR-223 was the most significantly enriched upstream miRNA during the post-acute stage of PNI. Therefore, although the involvement of miR-223 in peripheral nerve regeneration has yet to be experimentally examined, miR-223 may also play important roles in peripheral nerve remodeling and tissue formation. In future studies, expression of the identified miRNAs could be modulated to determine their biological effects for potential applications in clinical therapies. Moreover, it is of great importance to specifically investigate the biological functions of miRNAs on various cell types (e.g., Schwann cells, macrophages, fibroblasts, endothelial cells, etc.) because multiple types of cells exist in injured peripheral nerve stumps.
Here, we analyzed enriched miRNAs based on previous sequencing data obtained from rats at 0, 1, 4, 7, and 14 days after PNI. It is worth noting that different parts of the sciatic nerve (proximal stump, injury site, and distal stump) underwent different pathological processes after nerve injury. Complete axonal regeneration is generally achieved 6 weeks after sciatic nerve crush injury in rodents, a much longer time period than our 14-day observation period. Thus, essential factors for different parts of the sciatic nerve at different time points might be dissimilar and should be further investigated. Another limitation of our current study is that we only used male adult rats; non-pregnant age-matched female rats should have also been taken into consideration to achieve a more comprehensive view. In addition, instead of comparing with Day 0 control rats, the uninjured site of each rat might be used as its own internal control to avoid biological differences between different cohorts.
In conclusion, in the current study, we identified essential upstream miRNAs and biological activities after PNI by screening upstream regulators and building gene-gene interactions using IPA software. Our results contribute to the understanding of essential regulatory roles of miRNAs in peripheral nerve regeneration. Moreover, our applied bioinformatic analysis methods provide a strategy for the recognition of essential genes.
Additional files:
Additional file 1: Open peer review reports 1 (92.2KB, pdf) and 2 (97.2KB, pdf) .
Additional Table 1: Upstream miRNAs at 1, 4, 7, and 14 days after peripheral nerve injury.
Footnotes
Conflicts of interest: The authors declare that there are no conflicts of interest associated with this manuscript.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 31971276 (to JHH) and the Natural Science Foundation of Jiangsu Higher Education Institutions of China (Major Program), No. 19KJA320005 (to JHH). The funding sources had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: This study was approved by the Experimental Animal Ethics Committee of Jiangsu Province of China (approval No. 20190303-18) on March 3, 2019. The experimental procedure followed the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996).
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewers: G.M. Smith, Temple University School of Medicine, USA; Eroboghene E. Ubogu, University of Alabama at Birmingham, UK.
P-Reviewers: Smith GM, Ubogu EE; C-Editor: Zhao M; S-Editor: Wang J; L-Editors: Deusen AV, Song LP; T-Editor: Jia Y
Funding: This study was supported by the National Natural Science Foundation of China, No. 31971276 (to JHH) and the Natural Science Foundation of Jiangsu Higher Education Institutions of China (Major Program), No. 19KJA320005 (to JHH).
References
- 1.Arthur-Farraj PJ, Morgan CC, Adamowicz M, Gomez-Sanchez JA, Fazal SV, Beucher A, Razzaghi B, Mirsky R, Jessen KR, Aitman TJ. Changes in the coding and non-coding transcriptome and DNA methylome that define the schwann cell repair phenotype after nerve injury. Cell Rep. 2017;20:2719–2734. doi: 10.1016/j.celrep.2017.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs:genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs:target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Q, Wang H, Liu Y, Song Y, Lai L, Han Q, Cao X, Wang Q. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1beta production in macrophages by targeting STAT3. PLoS One. 2012;7:e42971. doi: 10.1371/journal.pone.0042971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Z, Shen Y, Qian T, Yi S, He J. Protein phosphorylation profiling of peripheral nerve regeneration after autologous nerve grafting. Mol Cell Biochem. 2020;472:35–44. doi: 10.1007/s11010-020-03781-z. [DOI] [PubMed] [Google Scholar]
- 6.Clements MP, Byrne E, Camarillo Guerrero LF, Cattin AL, Zakka L, Ashraf A, Burden JJ, Khadayate S, Lloyd AC, Marguerat S, Parrinello S. The wound microenvironment reprograms schwann cells to invasive mesenchymal-like cells to drive peripheral nerve regeneration. Neuron. 2017;96:98–114, 117. doi: 10.1016/j.neuron.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ. A uniform system for the annotation of vertebrate microrna genes and the evolution of the human microRNAome. Annu Rev Genet. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jessen KR, Arthur-Farraj P. Repair Schwann cell update: Adaptive reprogramming, EMT, and stemness in regenerating nerves. Glia. 2019;67:421–437. doi: 10.1002/glia.23532. [DOI] [PubMed] [Google Scholar]
- 9.Ji X, Hua H, Shen Y, Bu S, Yi S. Let-7d modulates the proliferation, migration, tubulogenesis of endothelial cells. Mol Cell Biochem. 2019;462:75–83. doi: 10.1007/s11010-019-03611-x. [DOI] [PubMed] [Google Scholar]
- 10.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2 disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Josse C, Bouznad N, Geurts P, Irrthum A, Huynh-Thu VA, Servais L, Hego A, Delvenne P, Bours V, Oury C. Identification of a microRNA landscape targeting the PI3K/Akt signaling pathway in inflammation-induced colorectal carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G229–243. doi: 10.1152/ajpgi.00484.2012. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Wang X, Gu Y, Chen C, Wang Y, Liu J, Hu W, Yu B, Wang Y, Ding F, Liu Y, Gu X. Let-7 microRNAs regenerate peripheral nerve regeneration by targeting nerve growth factor. Mol Ther. 2015;23:423–433. doi: 10.1038/mt.2014.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Yu B, Wang Y, Yao D, Zhang Z, Gu X. Identification and functional annotation of novel microRNAs in the proximal sciatic nerve after sciatic nerve transection. Sci China Life Sci. 2011;54:806–812. doi: 10.1007/s11427-011-4213-7. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Zhang R, Yuan Y, Yi S, Chen Q, Gong L, Liu J, Ding F, Cao Z, Gu X. MiR-340 regulates fibrinolysis and axon regrowth following sciatic nerve injury. Mol Neurobiol. 2017;54:4379–4389. doi: 10.1007/s12035-016-9965-4. [DOI] [PubMed] [Google Scholar]
- 15.Martinez B, Peplow PV. MicroRNAs as biomarkers of diabetic retinopathy and disease progression. Neural Regen Res. 2019;14:1858–1869. doi: 10.4103/1673-5374.259602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 17.Ning XJ, Lu XH, Luo JC, Chen C, Gao Q, Li ZY, Wang H. Molecular mechanism of microRNA-21 promoting Schwann cell proliferation and axon regeneration during injured nerve repair. RNA Biol. 2020;17:1508–1519. doi: 10.1080/15476286.2020.1777767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Painter MW, Brosius Lutz A, Cheng YC, Latremoliere A, Duong K, Miller CM, Posada S, Cobos EJ, Zhang AX, Wagers AJ, Havton LA, Barres B, Omura T, Woolf CJ. Diminished Schwann cell repair responses underlie age-associated impaired axonal regeneration. Neuron. 2014;83:331–343. doi: 10.1016/j.neuron.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rojo Arias JE, Busskamp V. Challenges in microRNAs’ targetome prediction and validation. Neural Regen Res. 2019;14:1672–1677. doi: 10.4103/1673-5374.257514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi L, Fisslthaler B, Zippel N, Fromel T, Hu J, Elgheznawy A, Heide H, Popp R, Fleming I. MicroRNA-223 antagonizes angiogenesis by targeting beta1 integrin and preventing growth factor signaling in endothelial cells. Circ Res. 2013;113:1320–1330. doi: 10.1161/CIRCRESAHA.113.301824. [DOI] [PubMed] [Google Scholar]
- 23.Stierli S, Imperatore V, Lloyd AC. Schwann cell plasticity-roles in tissue homeostasis, regeneration, and disease. Glia. 2019;67:2203–2215. doi: 10.1002/glia.23643. [DOI] [PubMed] [Google Scholar]
- 24.Thomson DW, Dinger ME. Endogenous microRNA sponges:evidence and controversy. Nat Rev Genet. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Miao Y, Ni J, Wang Y, Qian T, Yu J, Liu Q, Wang P, Yi S. Peripheral nerve injury induces dynamic changes of tight junction components. Front Physiol. 2018;9:1519. doi: 10.3389/fphys.2018.01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi S, Tang X, Yu J, Liu J, Ding F, Gu X. Microarray and qPCR analyses of wallerian degeneration in rat sciatic nerves. Front Cell Neurosci. 2017;11:22. doi: 10.3389/fncel.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi S, Wang QH, Zhao LL, Qin J, Wang YX, Yu B, Zhou SL. miR-30c promotes Schwann cell remyelination following peripheral nerve injury. Neural Regen Res. 2017;12:1708–1715. doi: 10.4103/1673-5374.217351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi S, Zhang H, Gong L, Wu J, Zha G, Zhou S, Gu X, Yu B. Deep sequencing and bioinformatic analysis of lesioned sciatic nerves after crush injury. PLoS One. 2015;10:e0143491. doi: 10.1371/journal.pone.0143491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan YM, Wang Y, Cheng CC, Zhao MY, Pei F. Efficacy of exosomes in peripheral nerve injury. Zhongguo Zuzhi Gongcheng Yanjiu. 2020;24:5079–5084. [Google Scholar]
- 30.Yu B, Zhou S, Wang Y, Ding G, Ding F, Gu X. Profile of microRNAs following rat sciatic nerve injury by deep sequencing:implication for mechanisms of nerve regeneration. PLoS One. 2011;6:e24612. doi: 10.1371/journal.pone.0024612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J, Gu X, Yi S. Ingenuity pathway analysis of gene expression profiles in distal nerve stump following nerve injury: insights into wallerian degeneration. Front Cell Neurosci. 2016;10:274. doi: 10.3389/fncel.2016.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, Yi S. Transcriptional landscape of alternative splicing during peripheral nerve injury. J Cell Physiol. 2019;234:6876–6885. doi: 10.1002/jcp.27446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


