Abstract
Cancer stem cells (CSCs) are a subpopulation of cancer that can self-renew and differentiate into large tumor masses. Evidence accumulated to date shows that CSCs affect tumor proliferation, recurrence, and resistance to chemotherapy. Recent studies have shown that, like stem cells, CSCs maintain cells with self-renewal capacity by means of asymmetric division and promote cell proliferation by means of symmetric division. This cell division is regulated by fate determinants, such as the NUMB protein, which recently has also been confirmed as a tumor suppressor. Loss of NUMB expression leads to uncontrolled proliferation and amplification of the CSC pool, which promotes the Notch signaling pathway and reduces the expression of the p53 protein. NUMB genes are alternatively spliced to produce six functionally distinct isoforms. An interesting recent discovery is that the protein NUMB isoform produced by alternative splicing of NUMB plays an important role in promoting carcinogenesis. In this review, we summarize the known functions of NUMB and NUMB isoforms related to the proliferation and generation of CSCs.
Keywords: Asymmetric cell division, Cancer stem cell (CSC), NOTCH signaling pathway, NUMB, NUMB isoforms
INTRODUCTION
Cancer is organized hierarchically and consists of a diverse population of functionally heterogeneous cells, with CSCs (also known as cancer stem-like cells) on top. CSCs can self-renew to maintain their proportions and their potential to differentiate into non-tumorigenic mass tumor cells (1). There are many studies suggesting that CSCs are responsible for cancer growth, recurrence, and resistance to chemo and radiation therapy (2, 3). CSC can be regulated by many intracellular and extracellular factors, which are drug targets for cancer treatment (3).
Many studies have reported that CSCs, like stem cells, maintain populations by means of self-renewal cell division. These cells can self-renew themselves through asymmetric division, in which one daughter cell has stem-cell properties and other cells undergo differentiation (4-6). In general, both great cellular heterogeneity and rapid cell proliferation are required for cancer development; so asymmetric and symme-tric division must coexist, especially for the survival of cancer cells in stress conditions, such as chemotherapy or metastasis (7). Therefore, efficient regulation of cell division (including both asymmetric and symmetric cell division) can be very important in effective cancer and CSCs therapy strategies.
In this review, we discuss the function of the NUMB protein, a cell-fate determinant and tumor suppressor, in the generation of cancer and CSCs. We also explore the role played by the NUMB isoforms produced by alternative splicing.
CSCs AND CELL DIVISION
Studies to date have shown that CSCs are a small population of self-renewing cells that are very likely to cause tumors and tumor recurrence (8, 9). Many studies have shown that conventional treatment methods, such as radiation and chemotherapy, promote CSC self-renewal and tumor recurrence (10), which suggests that CSC must be completely removed to eliminate the possibility of tumor recurrence (11).
Cell development occurs through cell proliferation, lineage determination, and final differentiation with early stem cells. Stem cells are capable of self-renew while producing differentiated cells. This mode of cell division plays an important role in the activity of stem cells (12). Individual stem cells produce two identical daughter cells (symmetric divisions, SD) or two unequal daughters, one that retains stem identity, the other that undergoes differentiation (asymmetric division, AD) (12). Therefore, in the process of stem-cell development, AD increases the diversity of cell types, and SD expands of cells.
For cancer development, AD and SD must co-exist, because both cellular heterogeneity and cellular proliferation are required. In addition, in stress conditions, such as tumor therapy and metastasis, cancer cells and CSC must co-exist with AD and SD for survival. Therefore, in order to design a more efficient cancer treatment strategy, many kinds of studies on the mechanism of cell-division regulation are required.
One characteristic of stem cells is that they undergo AD. This cell-fate determination can be accomplished extrinsically or intrinsically. The two daughter cells generated after AD are placed at different distances in the niche (extrinsically). AD is associated with the development of both invertebrates and vertebrates; most of what is known to date comes from genetically tractable invertebrate species, such as Drosophila melanogaster (5, 13-15) or Caenorhabditis elegans (15, 16). Asymmetric inheritance of endogenous cell-fate determinants such as proteins or RNA can lead to asymmetric cell-fate decisions (intrinsically) (17). In addition to macromolecules, organelles, such as centrosomes, intermediates, mitochondria, endoplasmic reticulum (ER) or lysosomes, have been described as being asymmetrically separated (18). However, the asymmetric inheritance of organelles is still unclear.
Prior to AD, the fate determinants differ in abundance at the apex or basal pole, where the mitotic spindle and centrosome are unevenly aligned. Proteins that promote self-renewal and stem are usually recruited into the spindles on the apical side facing the lumen of the body’s outer or inner cavity. In contrast, differentiation-promoting factors are recruited to the mitotic spindle placed basal toward the basement membrane (1).
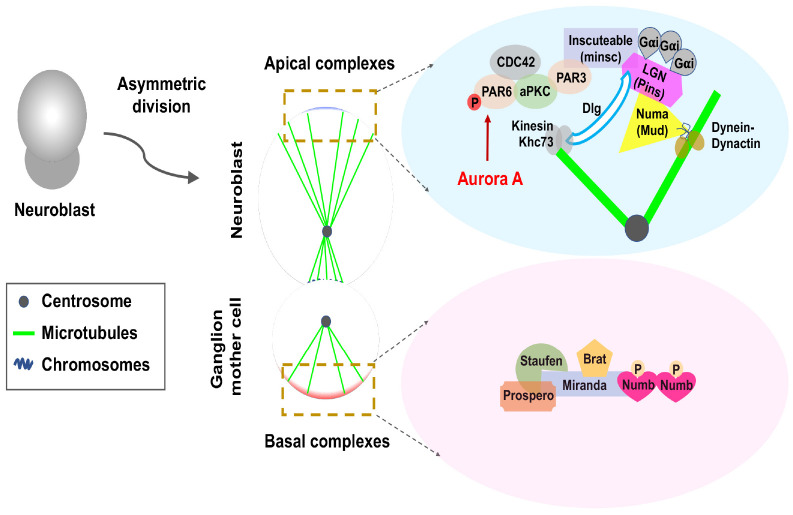
In Drosophila, neural stem cells divide asymmetrically to form neural stem cells and ganglion mother cells (Fig. 1). In this well-known model, we focus on NUMB among several proteins associated with AD.
Fig. 1.
Asymmetric division of Drosophila neuroblasts. Schematic description for a polarized mother cell during anaphase. Asymmetric division of Drosophila neuroblasts produces larger neuroblasts and smaller ganglion mother cells (GMCs) (left panel). The middle panel shows the asymmetric distribution of polarity proteins at the apical and basal cortex and the asymmetry of the spindle. The light-blue box highlights evolutionarily conserved apical complexes that are important for polarity establishment and spindle positioning in these two systems. In the apical cortex, the Cdc42/Par3/Par6/aPKC complex is linked with the Gαi/Pins/Mud complex by Inscuteable. Mud recruit’s dynein-dynactin activity to capture and pull astral microtubules, whereas Pin recruits’ kinesin Khc73 via Dlg to bind with astral microtubules. Phosphorylation of Par6 by Aurora-A plays an important role in regulating aPKC to induce fate-determining proteins, such as Brat, Prospero, Staufen, and NUMB, to the basement membrane and future GMCs. Miranda is an adapter protein for some basic proteins.
THE NUMB PROTEIN
NUMB was first identified in Drosophila as an important cell-fate determinant in which mutant embryos exhibit alterations in sensory-organ precursor lineage selection (19). The most important property of NUMB is its asymmetric cell distribution, which allows it to act as a fate determinant of cells. These properties are of importance in understanding the role of NUMB in physiological cell development and various diseases.
Drosophila Numb (dNumb) is a membrane-associated protein that separates asymmetrically from cell division (20) and consequently separates into one daughter cell, serving as an intrinsic determinant of cell fate (21). Numb, which is evolutionarily conserved from flies to mammals, is included in many cellular processes, and may serve as cell-fate determinants (19, 22). There are two mammalian homologs (encoded by two genes, mNUMB and mNUMB-like) (22, 23) and at least six major isoforms of mNUMB via alternative splicing (24-26).
Both mNUMB and mNUMB-like proteins are essential for AD. Although these proteins have functional overlaps, they function differently. The mNUMB-like protein is symmetrically distributed in cells and is expressed in neurons after mitosis of cortical plates rather than ventricular ancestors during mouse cortical neurogenesis. However, mNUMB is asymmetrically distributed to the apical membrane of dividing ventricular cells and separates into apical daughter cells that remain as a progenitor, which suggests that mNUMB plays a role in the maintenance of undifferentiated cells (23).
mNUMB encodes six alternatively spliced transcripts (25, 26). There are areas of functional overlap of these six isoforms, but physiological and pathological differences are distinct (24, 25, 27-29).
The NUMB protein consists of an amino-terminal PTB (phospho-tyrosine-binding domain), C-terminal PRR (proline-rich domain), and EH [Eps 15 homology region, including the DPF (Asp-Pro-Phe) and NPF (Asn-Pro-Phe) motif] domain (22, 25). The PTB domain usually mediates its association with the NPxY motif in transmembrane proteins (30, 31). Moreover, the NUMB protein interacts with intracellular adapters, such as alpha-adaptin and Eps15, which bind to the DPF and NPF motifs at the C-terminus of NUMB, respectively (32, 33).
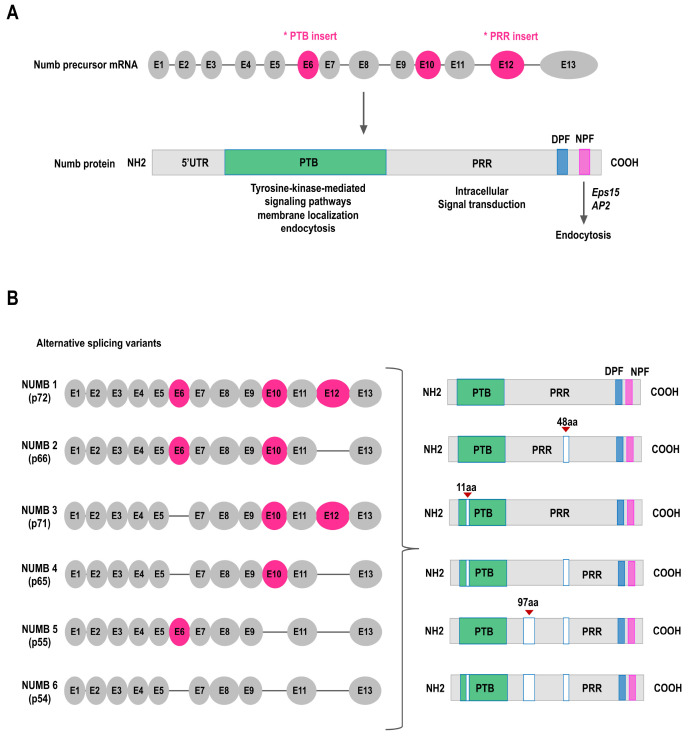
Mammalian NUMB genes are alternatively spliced to produce six functionally distinct isoforms (25, 26, 31, 34). Alternative splicing affects both PTB and PRR domains. The most abundantly expressed isoforms are clearly distinguished by the presence or absence of three coding exons (exon 6, E6; exon 10, E10; and exon 12, E12). The difference is that they contain two coding exons: E6, which corresponds to the 11 amino-acids region within the PTB domain, and E12, which corresponds to the 48 amino-acids region within the PRR domain. Alternative splicing of these exons produces the six isoform protein products: (1) NUMB 1 (p72), (2) NUMB 3 (p71, ΔE6), (3) NUMB 2 (p66, ΔE12), (4) NUMB 4 (p65, ΔE6 and ΔE12), (5) NUMB 5 (p55, ΔE10 and ΔE12), and (6) NUMB 6 (p54, ΔE6, ΔE10 and ΔE12) (Fig. 2).
Fig. 2.
NUMB structure and its alternative splicing variants. (A) NUMB PTB and PRR isoform – exons and protein domains. Up panel, the structure of NUMB precursor-mRNA, with exon 6, 10, and 12 depicted as an alternative cassette exon (top). Asterisks indicate exons that undergo alternative splicing during NUMB transcript maturation. Bottom panel, Schematic of full-length NUMB protein, which contains an amino-terminal PTB, a proline-rich region (PRR), two Eps 15 homology regions of aspartic acid–proline–phenylalanine (DPF) motifs and an asparagine–proline–phenylalanine (NPF) motif (bottom). The PTB domain facilitates tyrosine kinase signaling, endocytosis, and membrane localizing. The NPF domain can bind to the endocytic machinery components of Eps15. The PRR domains exist in certain isoforms containing Src homology binding sites involved in intracellular signal transduction. (B) As a result of three alternative splicing events, the NUMB locus can generate six different transcripts. All six isoforms of the human NUMB protein have a similar structure. NUMB isoforms 5/6 are only partial and cover the coding sequence.
THE ROLE OF NUMB ISOFORM PROTEINS: NUMB E12 OR E6 ALTERNATIVE SPLICING
The recently reported NUMB isoforms display different aspects of NUMB’s physiological and biochemical functions, along with a role that differentiates them from conventional NUMBs. However, the functions and mechanisms for NUMB isoforms are still not fully understood. We focus on analyzing the role of E12 and E6 alternative splicing events reported and describe the different roles of NUMB isoforms.
The NUMB isoform with E12 is 1/3 and the NUMB isoform without E12 is 2/4. The NUMB 1/3 isoforms are predominantly expressed in progenitor tissues, whereas the NUMB 2/4 isoforms are highly expressed in adult tissues (25, 34). In neural differentiation, a switch in NUMB isoform expression is mainly induced from NUMB 1/3 to NUMB 2/4 (34, 35). The different expressions of NUMB E12 isoforms also lead to functionally distinct differences. Overexpression of the NUMB isoforms including E12 in progenitor cells increase cell proliferation, whereas overexpression of the NUMB isoforms except for E12 promotes differentiation (24, 34, 36). The mechanisms underlying the different effects of NUMB isoforms on the balance between cell proliferation and differentiation are associated with Notch signaling (34). Especially, increased expression of NUMB 1/3 isoforms promotes Notch activation to proliferate cells, whereas increased expression of NUMB 2/4 isoforms decreases the Notch signal and promotes cell differentiation.
Alternative splicing is a tightly regulated process in which coding/non-coding sequences of pre-mRNA or protein are assembled and removed in various combinations to create new proteins with new functions (37). It has been reported that alternative splicings are poorly controlled in several cancer types (38-40). Ultimately, they make a huge contribution to many aspects of cancer, including cancer-causing modifications, cancer progression, and response / resistance to chemotherapy medications (41). It has been reported that there is an increased expression of the E12-containing NUMB isoform caused by alternative splicing of NUMB in breast, colon, and lung cancer (26, 27, 41-43).
The pre-mRNA splicing regulator has been described to affect NUMB E12 alternative splicing. Rbfox3 (pre-mRNA splicing regulator) promotes the skipping of NUMB E12, and RBM4 inhibits E12 inclusion by promoting the inclusion of E6 (44, 45).
Unlike E12, little is known about NUMB E6 alternative splicing. A NUMB isoform with E6 is 1/2 and a NUMB isoform without E6 is 3/4. In Alzheimer’s disease, a neurodegenerative disease, NUMB isoforms 3/4 affect the transport and processing of amyloids precursor proteins into cells to produce amyloid B peptide, the main component of neurogenic amyloid plaques (46, 47).
The difference between NUMB E6-containing and deficient isoforms is that they have different intracellular localizations. The NUMB PTB domain controls NUMB targeting to the cortical plasma membrane. NUMB isoforms 1/2 are located in the cell membrane, mainly by interaction with acidic membrane phospholipids (e.g., phosphoinositides, PIP). On the other hand, NUMB isoforms 3/4 are mainly distributed in the cytoplasm (48). However, the precise mechanisms governing the intracellular locating of NUMB isoforms are not yet clear.
It is known that the locating of the cortical plasma membrane of NUMB is effectively regulated by GPCR-activated phospholipid hydrolysis and aPKC-dependent phosphorylation (48, 49).
NUMB AND NUMB ISOFORMS IN CANCER/CSC: THE REGULATOR OF TUMOR PROGRESSION
Regulation of NUMB and NUMB isoforms in signaling pathways is key to their function as tumor suppressors in cancer development (50). The role of NUMB as a tumor suppressor is known in several types of tumors, including breast, lung, and liver cancer (50-52). Recent studies have shown that deletion of NUMB isoforms reduces cell growth and NUMB isoforms are regulators of EMT (Epithelial-mesenchymal transition) and ECM (extracellular matrix) protein networks in breast cancer cell (53). In addition, NUMB isoform 1 inhibits migration and invasion by preventing EMT in esophageal cancer. Furthermore, upregulation of NUMB isoform 1 expression suppresses growth of esophageal cancer cell and their cell cycle (53). The clinical association of NUMB in cancer incidence suggests that loss of the NUMB protein is found in about 30% of breast and lung tumors and is associated with poor prognosis and more aggressive tumors (51, 54). However, NUMB deficiency in tumors is accompanied by various mechanisms and promotes proteasome degradation (55). NUMB phosphorylation by aPKC (atypical protein kinase C) promotes the degradation of the p53 protein. As a result, reduced p53 protein expression and activity results in a pro-tumorigenic phenotype. In addition, aPKC-mediated NUMB phosphorylation plays an important role in promoting tumor formation and promoting proliferation and self-renewal of CSCs (55).
The role of the NUMB protein in inhibiting Notch signaling and simultaneously stabilizing the p53 signaling pathway is also implicated in the regulation of stem-cell homeostasis. In stem-cell mitosis, the asymmetric division of NUMB induces functional asymmetry of the NUMB-p53 and NUMB-Notch signaling pathways, which confer distinct developmental and proliferative fates on the two daughter cells (56). Collectively, the NUMB protein, which positively regulates the activity of p53, induces different p53 activities in the two daughter cells as a result of the expression of the differentiated NUMB protein, thereby regulating the mode of division of stem cells (56). At the stem-cell level of the mammary gland, the loss of expression of the NUMB protein and activation of p53 lead to a symmetric mode and acquires an unlimited self-renewal potential that can lead to the expansion of the stem-cell population. In contrast, loss of expression of the NUMB protein at the progenitor level activates the EMT program and acquires stem-cell properties. These effects lead to abnormal tissue formation and the appearance of CSCs. NUMB-deficient CSCs have more self-renewal potential than do normal breast stem cells or NUMB-proficient CSCs (56).
However, several reports have suggested mechanisms by which NUMB alternative splicing is regulated in response to oncogenic signaling pathways and contributes to the activation of downstream pathways that promote tumorigenesis. It has been reported that NUMB isoforms 2 and 4 are associated with the activation of the Notch pathway, and NUMB isoforms 2 and 4 will play an important role downstream of the oncogenic signaling pathway (57). NUMB isoforms 2 and 4 are highly expressed in non-small cell lung cancer, which activates the Notch target gene and promotes cell proliferation, thereby affecting cell signaling that contributes to tumor formation (42, 58).
CORRELATION BETWEEN NUMB AND NOTCH SIGNALING PATHWAYS
The Notch signaling pathway is a highly conserved developmental network involved in cell-fate determination, stem-cell homeostasis, and regulation of proliferation/differentiation balance during development (59). Trans-interactions between receptors and ligands present in opposite cells indicate activation events of the Notch signaling pathway (59, 60).
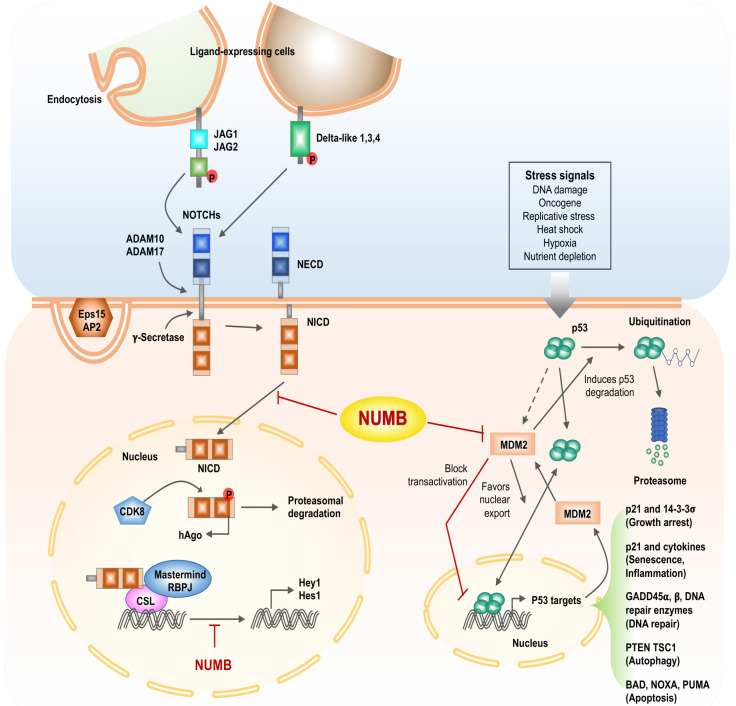
In Drosophila, the Numb protein plays an inhibitory role in the Notch signaling pathway. Many studies have shown that loss of expression of the Numb protein increases Notch function (61-63). During AD of stem cells, Numb proteins divide asymmetrically during mitosis. Daughter cells that have lost the Numb protein remain responsive to Notch and are involved in Notch activation, whereas daughter cells that have received the Numb protein do not respond to the Notch signal. Numb antagonizes Notch by inhibiting recycling to the plasma membrane via AP1 and regulating intracellular trafficking and stability of the Notch ligand Delta-like 4 (Dll4) to lysosomes for degradation (64). Translocated into the cell nucleus, NICD binds to the DNA-binding transcription factor RBPJ/Mastermind transcription-activating protein (65). This complex binds to the DNA in the nucleus and activate transcription. However, when NUMB and Notch are simultaneously expressed, the binding of Delta and Notch inhibits the translocation of RBPJ to the nucleus (66). In addition, NUMB binds directly to the PEST domain of NICD and inhibits Notch signaling, thereby inhibiting NICD from accessing the nucleus. At the same time, NUMB directly inhibits Notch by inducing polyubiquitination and degradation of Notch protein (67). A gamma-secretase inhibitor (DAPT) blocks Notch signal pathway by suppressing Notch receptor dissociation (68). As a result of studying the SHG-44 human glioblastoma cell line, DAPT treatment significantly reduced the expression of Hes1 and Notch1 genes and decreased the number of cancer cells. DAPT inhibits the dissociation of Notch receptors to regulate NUMB and Notch expression, thereby inhibiting Hes 1, a sub-target of Notch 1, to reduce cell proliferation and differentiation (69). Recent studies have shown that NUMB inhibits NICD (Notch intracellular domain) ubiquitination to regulate NICD protein expression (70). Activated NICD has a short activation of approximately four hours in cells. NUMB inhibits NICD activated during the Notch pathway, making it an effective strategy for developing treatments (67). In this way, Numb limits the transmission of Notch signals in signaling cells. In this complex scenario, the endocytic role of Numbs is essential for the removal or recycling of molecules from specific plasma membrane domains to execute their polarization function. In this regard, it has been suggested that abnormal activation of Notch is associated with carcinogenic signals associated with a variety of human cancers, including colon, pancreas, lung, breast, and glioblastoma (Fig. 3) (54, 67, 71-73).
Fig. 3.
Scheme of description for inhibition of the NOTCH signal pathway and regulation of p53 activity by NUMB. Left panel, NUMB interacts with AP2 and Eps15, interfering with the endocytosis of Notch ligands and receptors. The binding of ligands (Delta-like, JAGs) to Notch transmembrane receptors triggers a series of proteolytic cleavages by ADAM proteins (ADAM 10, ADAM 17) and a γ-secretase complex that leads to the release of the Notch intracellular domain (NICD), which is translocated into the nucleus, interacts with the RBPJ (recombining binding protein suppressor of hairless), CSL (CBF1, suppressor of hairless, Lag-1) transcription factor, and the transcriptional activator protein Mastermind to regulate target genes such as Hes1 and Hey1. NUMB can interfere with this transcriptional machinery and inhibit Notch-dependent gene expression. Phosphorylation of NICD by CDK8 leads to hAgo-dependent proteosomal degradation of NICD. NUMB inhibits translocation of NCID into the nucleus. Right panel, p53 activity is driven by several stress-related signals. p53-dependent transcription of MDM2 increases p53 degradation. MDM2 suppresses p53 activity by blocking p53 transcriptional activity and favoring p53 nuclear export. Transcription of p53 target genes promotes p53 tumor-suppressor properties.
In conclusion, tight regulation of the NUMB-Notch axis is essential to prevent potential tumor-inducing events that could result from deregulation. The main mechanisms of Notch-NUMB are described here, but the overall contextual scenario has not yet been fully understood.
CORRELATION BETWEEN NUMB AND P53 SIGNALING PATHWAYS
The p53 acts as an intracellular stress sensor that is activated in response to various stimuli such as DNA damage, oncogene activation, hypoxia, oxidative stress, and malnutrition (74). These stimuli are usually caused by tumor cells and play an important role in triggering the tumor suppressor function of p53 in vivo. Also, depending on the type of stimulation, p53 activation induces a variety of phenomena, such as temporary cell-cycle arrest, aging, DNA repair, autophagy, or other cellular consequences, such as cell death (75).
Numerous MDM2-p53 pathways have been studied to date and are representative cancer therapeutic targets. Suppression of the MDM2 or MDM2-p53 interaction refers to an indirect approach to reestablish p53 function and is therefore considered a viable therapeutic strategy to delay tumor progression (Fig. 3) (76).
NUMB stabilizes p53 protein levels by forming a trimer complex with p53 and MDM2 and interacting directly with the two proteins to inhibit MDM2-mediated ubiquitination of p53. As a result, NUMB-deficient tumors have reduced p53 expression and activity, respond to genotoxic treatment, and are less able to efficiently repair DNA damage (51, 56).
NUMB AND EMT
NUMB affects early EMT events by regulating cell-cell adhesion, cell polarity, and epithelial cell migration. Cell polarity is determined by the asymmetric composition of several cellular components. The polar complex protein (PAR) participated in coordinating this events and also serves to establish tight junctions, which suture adjacent epithelial cells, functionally separating the apical-basal lateral surface and restricting liquid flow between the intercellular spaces. Moreover, NUMB participated in the regulation of tight-junction kinetics by affecting and interacting with the localization of PAR3 (50, 77, 78). In summary, NUMB suppress the Notch signaling pathway by inhibiting EMT. The Notch pathway stimulates the EMT process by increasing the expression of Snail and Slug, the subsequences that prevent transcription of E-cadherin (79, 80). Downregulation of NUMB can induce an EMT phenotype, which can causally increase tumor invasion. Consequently, these data suggest the role of NUMB in driving all stages of the EMT process from loss of cell polarity and cell-cell contact (dynamic re-modulation of tight junctions) for cell migration.
CONCLUSION
NUMB was first discovered as a determinant of cell fate in Drosophila and has been reported as a tumor suppressor in various human cancers to date. NUMB protein functions play a pivotal role not only in controlling physiological processes, but also in disease when deregulated. NUMB’s ability to control stem/progenitor cell development has several effects on tumor formation. Mammalian NUMBs are structurally more complex than the Drosophila homologues in that they have more coding genes and a greater number of alternately spliced isoforms. Thus, it is showed that the NUMB protein has asymmetric cell division control and cell fate determination, cell migration, ubiquitination, and multiple signaling pathways (i.e., Notch, p53, EMT complex functions). AD is an efficient path to create heterogeneity while maintaining the self-renewal characteristic of CSC. However, to date, we still lack studies on the activity of CSCs and the role of ADs in different stages of human cancer development. Therefore, a complete description of all mechanisms involved in NUMB relief in cancer will be important in identifying new therapeutic targets and strategies for the treatment of NUMB-deficient tumors.
ACKNOWLEDGEMENTS
This paper was supported by Konkuk University Researcher Fund in 2019. This study was supported by grants from the National Research Foundation (NRF) funded by the Korean government (grant no. 2019M3A9H1030682 and NRF-2015R1A5A1009701). This paper was supported by Konkuk University Researcher Fund in 2019.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
References
- 1.Yoo YD, Kwon YT. Molecular mechanisms controlling asymmetric and symmetric self-renewal of cancer stem cells. J Anal Sci Technol. 2015;6:28. doi: 10.1186/s40543-015-0071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budillon A, Curley S, Fusco R, Mancini R. Identification and targeting of stem cell-activated pathways in cancer therapy. Stem Cells Int. 2019;2019:8549020. doi: 10.1155/2019/8549020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang L, Shi P, Zhao G, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clevers H. Stem cells, asymmetric division and cancer. Nat Genet. 2005;37:1027–1028. doi: 10.1038/ng1005-1027. [DOI] [PubMed] [Google Scholar]
- 5.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majumdar S, Liu ST. Cell division symmetry control and cancer stem cells. AIMS Mol Sci. 2020;7:82–98. doi: 10.3934/molsci.2020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badve S, Nakshatri H. Breast-cancer stem cells-beyond semantics. Lancet Oncol. 2012;13:e43–48. doi: 10.1016/S1470-2045(11)70191-7. [DOI] [PubMed] [Google Scholar]
- 9.Najafi M, Mortezaee K, Ahadi R. Cancer stem cell (a)symmetry & plasticity: tumorigenesis and therapy relevance. Life Sci. 2019;231:116520. doi: 10.1016/j.lfs.2019.05.076. [DOI] [PubMed] [Google Scholar]
- 10.Brooks MD, Burness ML, Wicha MS. Therapeutic implications of cellular heterogeneity and plasticity in breast cancer. Cell Stem Cell. 2015;17:260–271. doi: 10.1016/j.stem.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–238. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 13.Losick VP, Morris LX, Fox DT, Spradling A. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell. 2011;21:159–171. doi: 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santoro A, Vlachou T, Carminati M, Pelicci PG, Mapelli M. Molecular mechanisms of asymmetric divisions in mammary stem cells. EMBO Rep. 2016;17:1700–1720. doi: 10.15252/embr.201643021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkei ZG, Yamashita YM. Emerging mechan-isms of asymmetric stem cell division. J Cell Biol. 2018;217:3785–3795. doi: 10.1083/jcb.201807037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoblich JA. Asymmetric cell division during animal development. Nat Rev Mol Cell Biol. 2001;2:11–20. doi: 10.1038/35048085. [DOI] [PubMed] [Google Scholar]
- 17.Gehring WJ. Precis of Edwin G. Conklin's JEZ article, "Mosaic Development in Ascidian Eggs". J Exp Zool A Comp Exp Biol. 2004;301:461–463. doi: 10.1002/jez.a.87. [DOI] [PubMed] [Google Scholar]
- 18.Sunchu B, Cabernard C. Principles and mechanisms of asymmetric cell division. Development. 2020;147:dev167650. doi: 10.1242/dev.167650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN. Numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 20.Cayouette M, Raff M. Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat Neurosci. 2002;5:1265–1269. doi: 10.1038/nn1202-1265. [DOI] [PubMed] [Google Scholar]
- 21.Dho SE, Jacob S, Wolting CD, French MB, Rohrschneider LR, McGlade CJ. The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J Biol Chem. 1998;273:9179–9187. doi: 10.1074/jbc.273.15.9179. [DOI] [PubMed] [Google Scholar]
- 22.Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Exp Cell Res. 2010;316:900–906. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/S0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 24.Bani-Yaghoub M, Kubu CJ, Cowling R, et al. A switch in numb isoforms is a critical step in cortical development. Dev Dyn. 2007;236:696–705. doi: 10.1002/dvdy.21072. [DOI] [PubMed] [Google Scholar]
- 25.Dho SE, French MB, Woods SA, McGlade CJ. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem. 1999;274:33097–33104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 26.Karaczyn A, Bani-Yaghoub M, Tremblay R, et al. Two novel human NUMB isoforms provide a potential link between development and cancer. Neural Dev. 2010;5:31. doi: 10.1186/1749-8104-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karaczyn AA, Adams TL, Cheng RY, Matluk NN, Verdi JM. Human NUMB6 induces epithelial-mesenchymal transition and enhances breast cancer cells migration and invasion. J Cell Biochem. 2017;118:237–251. doi: 10.1002/jcb.25628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Wang D, Wang H, et al. A splicing factor switch controls hematopoietic lineage specification of pluripotent stem cells. EMBO Rep. 2021;22:e50535. doi: 10.15252/embr.202050535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei R, Liu X, Voss C, et al. NUMB regulates the endocytosis and activity of the anaplastic lymphoma kinase in an isoform-specific manner. J Mol Cell Biol. 2019;11:994–1005. doi: 10.1093/jmcb/mjz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kavanaugh WM, Turck CW, Williams LT. PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science. 1995;268:1177–1179. doi: 10.1126/science.7539155. [DOI] [PubMed] [Google Scholar]
- 31.Verdi JM, Schmandt R, Bashirullah A, et al. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr Biol. 1996;6:1134–1145. doi: 10.1016/S0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- 32.Salcini AE, Confalonieri S, Doria M, et al. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santolini E, Puri C, Salcini AE, et al. Numb is an endocytic protein. J Cell Biol. 2000;151:1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verdi JM, Bashirullah A, Goldhawk DE, et al. Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc Natl Acad Sci U S A. 1999;96:10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Revil T, Gaffney D, Dias C, Majewski J, Jerome- Majewska LA. Alternative splicing is frequent during early embryonic development in mouse. BMC Genomics. 2010;11:399. doi: 10.1186/1471-2164-11-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dooley CM, James J, Jane McGlade C, Ahmad I. Involvement of numb in vertebrate retinal development: evidence for multiple roles of numb in neural differentiation and maturation. J Neurobiol. 2003;54:313–325. doi: 10.1002/neu.10176. [DOI] [PubMed] [Google Scholar]
- 37.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Climente-Gonzalez H, Porta-Pardo E, Godzik A, Eyras E. The functional impact of alternative splicing in cancer. Cell Rep. 2017;20:2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Kahles A, Lehmann KV, Toussaint NC, et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell. 2018;34:211–224. doi: 10.1016/j.ccell.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, et al. The role of alternative splicing in cancer: from oncogenesis to drug resistance. Drug Resist Updat. 2020;53:100728. doi: 10.1016/j.drup.2020.100728. [DOI] [PubMed] [Google Scholar]
- 41.Lu Y, Xu W, Ji J, et al. Alternative splicing of the cell fate determinant Numb in hepatocellular carcinoma. Hepatology. 2015;62:1122–1131. doi: 10.1002/hep.27923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misquitta-Ali CM, Cheng E, O'Hanlon D, et al. Global profiling and molecular characterization of alternative splicing events misregulated in lung cancer. Mol Cell Biol. 2011;31:138–150. doi: 10.1128/MCB.00709-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao YJ, Han HZ, Liang Y, Shi CZ, Zhu QC, Yang J. Alternative splicing of VEGFA, APP and NUMB genes in colorectal cancer. World J Gastroenterol. 2015;21:6550–6560. doi: 10.3748/wjg.v21.i21.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim KK, Nam J, Mukouyama YS, Kawamoto S. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol. 2013;200:443–458. doi: 10.1083/jcb.201206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarn WY, Kuo HC, Yu HI, et al. RBM4 promotes neuronal differentiation and neurite outgrowth by modulating Numb isoform expression. Mol Biol Cell. 2016;27:1676–1683. doi: 10.1091/mbc.E15-11-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kyriazis GA, Wei Z, Vandermey M, et al. Numb endocytic adapter proteins regulate the transport and processing of the amyloid precursor protein in an isoform-dependent manner: implications for Alzheimer disease pathogenesis. J Biol Chem. 2008;283:25492–25502. doi: 10.1074/jbc.M802072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ntelios D, Berninger B, Tzimagiorgis G. Numb and Alzheimer's disease: the current picture. Front Neurosci. 2012;6:145. doi: 10.3389/fnins.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dho SE, Trejo J, Siderovski DP, McGlade CJ. Dynamic regulation of mammalian numb by G protein-coupled receptors and protein kinase C activation: structural determinants of numb association with the cortical membrane. Mol Biol Cell. 2006;17:4142–4155. doi: 10.1091/mbc.e06-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato K, Watanabe T, Wang S, et al. Numb controls E-cadherin endocytosis through p120 catenin with aPKC. Mol Biol Cell. 2011;22:3103–3119. doi: 10.1091/mbc.e11-03-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pece S, Confalonieri S, Romano PR, Di Fiore PP. NUMB-ing down cancer by more than just a NOTCH. Biochim Biophys Acta. 2011;1815:26–43. doi: 10.1016/j.bbcan.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 51.Colaluca IN, Tosoni D, Nuciforo P, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 52.Westhoff B, Colaluca IN, D'Ario G, et al. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong J, Liu Z, Zhu H, et al. The tumor suppressive role of NUMB isoform 1 in esophageal squamous cell carcinoma. Oncotarget. 2014;5:5602–5614. doi: 10.18632/oncotarget.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pece S, Serresi M, Santolini E, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siddique HR, Feldman DE, Chen CL, Punj V, Tokumitsu H, Machida K. NUMB phosphorylation destabilizes p53 and promotes self-renewal of tumor-initiating cells by a NANOG-dependent mechanism in liver cancer. Hepatology. 2015;62:1466–1479. doi: 10.1002/hep.27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tosoni D, Zecchini S, Coazzoli M, et al. The Numb/p53 circuitry couples replicative self-renewal and tumor suppression in mammary epithelial cells. J Cell Biol. 2015;211:845–862. doi: 10.1083/jcb.201505037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajendran D, Zhang Y, Berry DM, McGlade CJ. Regulation of Numb isoform expression by activated ERK signaling. Oncogene. 2016;35:5202–5213. doi: 10.1038/onc.2016.69. [DOI] [PubMed] [Google Scholar]
- 58.Langer W, Sohler F, Leder G, et al. Exon array analysis using re-defined probe sets results in reliable identification of alternatively spliced genes in non-small cell lung cancer. BMC Genomics. 2010;11:676. doi: 10.1186/1471-2164-11-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 60.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/S0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 62.Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–138. doi: 10.1016/j.cub.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 63.Wai P, Truong B, Bhat KM. Cell division genes promote asymmetric interaction between Numb and Notch in the Drosophila CNS. Development. 1999;126:2759–2770. doi: 10.1242/dev.126.12.2759. [DOI] [PubMed] [Google Scholar]
- 64.Shao X, Ding Z, Zhao M, et al. Mammalian Numb protein antagonizes Notch by controlling postendocytic trafficking of the Notch ligand Delta-like 4. J Biol Chem. 2017;292:20628–20643. doi: 10.1074/jbc.M117.800946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brou C. Intracellular trafficking of Notch receptors and ligands. Exp Cell Res. 2009;315:1549–1555. doi: 10.1016/j.yexcr.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 66.Frise E, Knoblich JA, Younger-Shepherd S, Jan LY, Jan YN. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci U S A. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flores AN, McDermott N, Meunier A, Marignol L. NUMB inhibition of NOTCH signalling as a therapeutic target in prostate cancer. Nat Rev Urol. 2014;11:499–507. doi: 10.1038/nrurol.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crawford TQ, Roelink H. The notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev Dyn. 2007;236:886–892. doi: 10.1002/dvdy.21083. [DOI] [PubMed] [Google Scholar]
- 69.Liu X, Xu QR, Xie WF, Wang MD. DAPT suppresses the proliferation of human glioma cell line SHG-44. Asian Pac J Trop Med. 2014;7:552–556. doi: 10.1016/S1995-7645(14)60092-4. [DOI] [PubMed] [Google Scholar]
- 70.Luo Z, Mu L, Zheng Y, et al. NUMB enhances Notch signaling by repressing ubiquitination of NOTCH1 intracellular domain. J Mol Cell Biol. 2020;12:345–358. doi: 10.1093/jmcb/mjz088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 72.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 73.Nowell CS, Radtke F. Notch as a tumour suppressor. Nat Rev Cancer. 2017;17:145–159. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 74.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nag S, Qin J, ivenugopal KS, Sr, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biomed Res. 2013;27:254–271. doi: 10.7555/JBR.27.20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Z, Sandiford S, Wu C, Li SS. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J. 2009;28:2360–2373. doi: 10.1038/emboj.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Steed E, Balda MS, Matter K. Dynamics and functions of tight junctions. Trends Cell Biol. 2010;20:142–149. doi: 10.1016/j.tcb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 79.Wang Z, Li Y, Kong D, Sarkar FH. The role of Notch signaling pathway in epithelial-mesenchymal transition (EMT) during development and tumor aggressiveness. Curr Drug Targets. 2010;11:745–751. doi: 10.2174/138945010791170860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qi S, Zhao X, Li M, et al. Aberrant expression of Notch1/numb/snail signaling, an epithelial mesenchymal transition related pathway, in adenomyosis. Reprod Biol Endocrinol. 2015;13:96. doi: 10.1186/s12958-015-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]