Abstract
Mucins are high molecular-weight epithelial glycoproteins and are implicated in many physiological processes, including epithelial cell protection, signaling transduction, and tissue homeostasis. Abnormality of mucus expression and structure contributes to biological properties related to human cancer progression. Tumor growth sites induce inhospitable conditions. Many kinds of research suggest that mucins provide a microenvironment to avoid hypoxia, acidic, and other biological conditions that promote cancer progression. Given that the mucus layer captures growth factors or cytokines, we propose that mucin helps to ameliorate inhospitable conditions in tumor-growing sites. Additionally, the composition and structure of mucins enable them to mimic the surface of normal epithelial cells, allowing tumor cells to escape from immune surveillance. Indeed, human cancers such as mucinous carcinoma, show a higher incidence of invasion to adjacent organs and lymph node metastasis than do non-mucinous carcinoma. In this mini-review, we discuss how mucin provides a tumor-friendly environment and contributes to increased cancer malignancy in mucinous carcinoma.
Keywords: Anti-cancer therapy, Mucin, Mucinous carcinoma, Preclinical mouse model, Tumorigenesis
INTRODUCTION
The mucosal surfaces of the body protect against various external environments. The intestinal tract is the guardian of the innate host defense because of the secretory factors of intestinal goblet cells (1). Colonization by intestinal bacteria is limited to an outer mucus layer and interacts with mucin glycoproteins, whereas an inner mucus layer is entirely devoid of bacteria (2). Thus, the defection of mucus layers increases bacterial interaction with the surface epithelium. Additionally, abnormality of mucins and mucin structure has occurred in mucinous colorectal carcinoma (MCC) (3). Since tumor growth sites induce inhospitable conditions for them to survive, mucins are suggested as an oncogenic microenvironment that avoids hypoxia, acidic, and other biological hurdles. The composition and structure of mucins enable them to mimic the surface of tumor cells like the surface of normal epithelial cells (4). Additionally, the mucus layer captures growth factors or cytokines, contributing to cell growth of the tumor. Alternatively, these properties interfere with the interaction bet-ween the immune system and tumor cells. Indeed, a high concentration of soluble mucins downregulates the motility and activation status of leukocytes (5). It was also reported that cell surface mucin contributes to cell proliferation and differentiation (6).
MCC shows a higher incidence of invasion to adjacent organs and lymph node metastasis than does non-mucinous colorectal carcinoma (7). Also, MCC is characterized by a large amount of extracellular mucin, and mucin pools contain malignant epithelium (4, 7). However, the function of mucin especially in MCC pathology is not completely understood. Thus, the unveiling of mucin’s role and molecular mechanisms for MCC tumorigenesis and understanding MCC mouse models is required for MCC study. In this mini-review, we briefly discuss major MCC-related mucins and their roles in MCC development. Further, we introduce the currently known MCC therapeutic drugs and mouse models proposed for MCC study.
GEL-FORMING AND TRNASMEMEBRANE MUCINS
The major constituent of mucus layers is mucins which are high-molecular-weight epithelial O-glycosylated glycoprotein (8) and are implicated in pathogenesis in cancer, especially mucinous adenocarcinomas. Currently, 21 mucin genes are known in humans. The mucins are classified into two groups based on their structure and functions: (i) secreted gel-forming mucins and (ii) transmembrane mucins. Gel-forming mucins including MUC2, MUC5AC, MUC5B, MUC6, and MUC19, cover epithelial cells in various organs (Table 1). Gel-forming mucins are secreted oligomeric mucin and might be responsible for the properties of mucus. Transmembrane mucins such as MUC1, MUC3, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17, MUC21, and MUC22, exhibit monomeric structural characteristics and mainly located on the cell surface, and might play a role in separating cells from the external environment (4). Mucins contain oligosaccharides, having excellent structural diversity (9), but their function is still unclear. However, polypeptide-attached oligosaccharide expands the volume of polypeptides through structural modification. Thus, mucins fill the space and raises its gel-forming character. Subsequently, it may lead to various physiological effects by increasing the chance of binding with a growth factor and cytokine for the survival of tumor cells (Fig. 1).
Table 1.
Mucin expression in human organs
| Organs | Gel-forming mucins | Transmembrane mucins | |
|---|---|---|---|
| Esophagus | MUC5B | MUC1, MUC4, MUC20 | |
| Stomach | MUC5AC, MUC6 | MUC1, MUC3, MUC13, MUC20 | |
| Liver | MUC2, MUC5AC, MUC5B, MUC6 | MUC1, MUC3 | |
| Pancreas | MUC5AC, MUC5B, MUC6 | MUC1, MUC11, MUC12, MUC20 | |
| Lung | MUC2, MUC5AC, MUC5B | MUC1, MUC3, MUC4, MUC11, MUC13, MUC20 | |
| Reproductive tract | Male | MUC1 | |
| Female | MUC5AC, MUC5B, MUC6 | MUC1, MUC4, MUC12 | |
| Intestine | Duodenum | MUC2, MUC6 | MUC1, MUC3, MUC17, MUC20 |
| Small intestine | MUC2 | MUC1, MUC3, MUC17, MUC20 | |
| Colorectum | MUC2 | MUC1, MUC3, MUC4, MUC11, MUC12, MUC13, MUC17, MUC20 | |
Fig. 1.
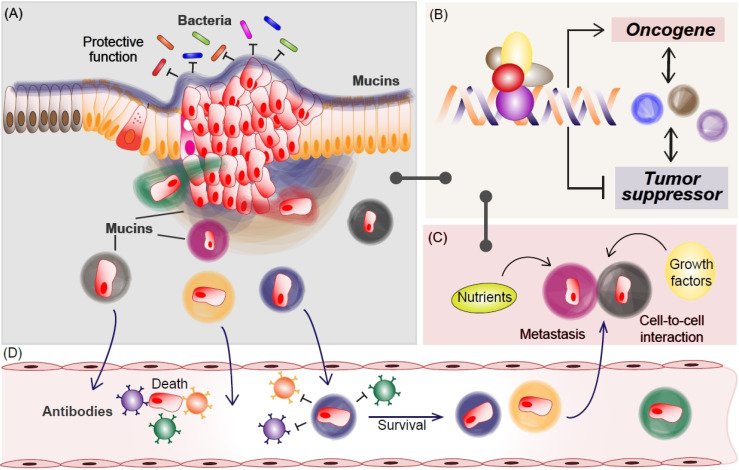
The role of mucins in MCC progression. In normal conditions, mucins protect epithelial cells from external environments such as bacteria (A). In MCC, various mucins transcriptionally increase oncogenic factors while inhibiting tumor suppressor (B), which might be mediated by mucin-captured nutrients or growth factors (C). Additionally, mucins protect MCC from immune surveillance via mimic epithelial surface during metastasis (A and D). For the journey to secondary tumor formation, many cancer cells are killed by the immune system in the circulating system (D). Metastasized mucin-cloaked cancer cells take advantage of including transcription, nutrients, and growth factors (C), for secondary MCC development, which would be shared via cell-to-cell interaction.
Gel-forming mucins in cancer
MUCIN2 (MUC2): Secreted protein MUC2 is a major com-ponent of the mucus layer in the small and large intestines. Decreased MUC2 is highly involved in the early stage of colorectal cancer (CRC) development, which is caused by methylation of the MUC2 promoter (10). Given that the Muc2-deficient mouse developments colitis (11), methylation of the MUC2 promoter might contribute to CRC initiation. However, the results of MUC2 gene promoter methylation analysis of MCC and non-MCC by narrowing the scope in the entire CRC analysis shows demethylation in the MUC2 promoter of MCC (10). MCC (10-15% of human CRC) is metastatic and therapeutically resistant (12) and shows accumulated genetic mutations including K-Ras, TOP-1, MAPK, and BRAF (12-18). However, MCC shows a low frequency of TP53 mutation and protein expression. Also, TP53 and p21 positively control MUC2 via transcriptional activation (19). These studies imply that MCC may show a genetic background difference from CRC for amplifying MUC2, at least for TP53 mutation or expression. Indeed, the Adenomatous polyposis coli (APC) mutation, with the highest frequency in CRC, is relatively low in MCC. In summary, CRC (non-MCC) downregulates MUC2 to foster an oncogenic environment by inflammation or colitis, whereas MCC might try to employ its unique genetic background for generating the mucinous environment including MUC2 secretion.
MUCIN5AC (MUC5AC): In normal physiological conditions, MUC5AC is barely secreted in intestinal mucus (20). But, similar to MUC2, MUC5AC is expressed at a high level in MCC and by microsatellite instability (MSI)-high tumors (21, 22). Cancer patient tissues show MUC5AC-positive tumor cells (35-100%), which depend on the tumor type (Adenocarcinoma: 147/420 [35%], Adenocarcinoma 1-49% mucinous component: 119/167 [71%], Mucinous > 50%: 46/49 [94%], Signet-ring cell carcinoma: 8/8 [100%]) (23). Signet-ring cells produce aberrant mucin and demonstrate high levels of MSI. The mechanism of MSI generation is involved in the dysfunction of the DNA mismatch repair protein. In normal tissue, DNA mismatch repair proteins correct errors during DNA replication. However, impaired DNA mismatch repair proteins in tumor cells trigger the possibility of MSI generation, subsequently resulting in chromosomal instability (CIN). Hypomethylation of the MUC5AC promoter is a predictor of MSI (24). Furthermore, elevated MUC5AC is associated with mismatch repair-deficiency (25) and downregulation of TP53 and its target gene p21 (26), which are strongly related to the maintenance of chromosomal stability. Since β-catenin has been reported to induce impaired DNA damage repair via LIG4 (27), MUC5AC-induced β-catenin (26) might be involved in MSI via radioresistance. Although Wnt/β-catenin signaling contributes to radioresistance, MSI does not happen under Wnt/β-catenin signaling hyperactivated conditions that do not show an increase in mucins (28-30). Moreover, MUC5AC shows a negative correlation with MLH1, a protein for DNA damage repair, in MCC (22). Thus, it is likely that MUC5AC is a key cooperator for MSI, but veiled a detailed molecular mechanism. Recently, it was reported that MUC5AC increases tumor heterogeneity (31), which may result from MSI. Increased tumor heterogeneity provides an advantage for escape from immune surveillance.
MUCIN5B (MUC5B): Consistent with MUC2 and MUC5AC, hypermethylation of the MUC5B promoter is a major regulatory mechanism for silencing it (32). Intriguingly, intronic regions regulate MUC5B (33-35). A 256bp segment of the first intron of the MUC5B contains eight tandem repeat GA boxes located in the GAGGG box, which interact with transcription factors such as Sp1, GATA-1, and AP-2 (35). These transcription factors suggest a role of MUC5B in cell differentiation during development. Moreover, the 37th intron of MUC5B interacts with Sp1 and NF1-MUC5B via the GC-rich region (36). These involvements of the intronic region in the regulation of the MUC gene are not found in other MUCs except to MUC5B. Long non-coding RNA also upregulates MUC5B and promotes cancer mobility and metastasis (37). The abnormal expression of the MUB5B on the biological behavior led to aggressive tumorigenesis including chemotherapeutic resistance and impairment of antitumor immune response (38). Although several studies reported that MUC5B knockdown reduces chemoresistance and decreases in Wnt/β-catenin signaling-induced cell proliferation and migration, the detailed molecular mechanism of how MUC5B contributes to MCC malignancy is still unclear. However, given that MUC5B-induced Wnt/β-catenin might contribute to cell migration, invasion, and chemoresistance (27, 39, 40), it is plausible that the interaction of Wnt/β-catenin signaling with MUC5AC and MUC5B may orchestrate the metastatic environment in MCC progression.
MUCIN6 (MUC6): MUC5AC and MUC6 are the major gastric mucins. Whereas MUC5AC is located on the epithelial cell surface, MUC6 is expressed in glandular structures. Staining in a subset of CRC displays exclusively cytoplasmic normal tissue (23). Thus, the expression of tumor cells might result from de novo synthesis by oncogenic signaling. The regulatory mechanism of MUC6 expression is related to the status of promoter methylation, BRAF-V600E mutation, a marker of MCC, as well as MLH1 methylation. Additionally, p53 overexpression shows an inverse association with MUC6 (23). These studies demonstrate a similar regulatory mechanism of gel-forming mucin. Investigation of transcriptional regulatory machinery reported that the Notch signaling pathway increases mRNA levels of the MUC6 and MUC5AC via Hath1, a crucial transcription factor for the Notch signaling pathway, in cancer cell lines (41). MUC6 is frequently associated with the nuclear β-catenin in gastric cancer and younger patients (less than or equal to 40 years old) are more likely to secrete MUC6 (42, 43). Inter-estingly, similar to MUC5, positive regulation of MUC6 is observed at the early stage of tumor development, but is diminished at late tumorigenesis, suggesting that MUC6 inhibits tumor cell migration (44). Indeed, MUC6 deficient patients showed short progression-free survival and cancer-specific survival, especially in stage II and III CRC patients (45), implying the involvement of MUC6 in inhibiting tumorigenesis. Conversely, a high level of MUC5AC increased progression-free survival in stage II and III CRC patients.
Together, gel-forming mucins are clustered in chromosome 11p15.5 and share similar regulatory machinery. However, their expression pattern leads to difficulty in targeting an MCC-specific therapeutic strategy. Additionally, they orchestrate diverse physiological events in MCC by communicating either with other signaling pathways or among mucins. Therefore, to resolve these scientific and translational medicine issues, the preclinical animal model of MCC mimicking is likely required.
Transmembrane mucins in cancer
MUCIN1 (MUC1): MUC1 is a single-pass transmembrane protein with a glycosylated extracellular domain. As a metabolic master regulator (46), MUC1 is mainly expressed in the epithelial cells of the stomach, intestine, and lungs. MUC1 competes with E-cadherin in the cytosol to bind β-catenin, by receptor tyrosine kinases (RTKs)-mediated MUC1 phosphorylation (47). Also, Protein Kinase C-d (PKC-d) promotes interaction between MUC1 and β-catenin (48, 49). As a negative regulator of β-catenin, GSK3β mediates the phosphorylation of β-catenin for proteasomal β-catenin degradation. However, since MUC1 blocks GSK3β-mediated β-catenin phosphorylation (50), the MUC1-β-catenin complex stably activates Wnt/β-catenin signaling. Besides, MUC1 binds to ErbB1 and increases p-ERK1/2 through the MAPK pathway (51). Anti-cancer immunity is frequently suppressed by oncogenic signals. MUC1 causes immunosuppression in CRC (52) and induces T-cell tolerance in vivo (53). Additionally, apoptosis activating pathway defects such as masking of death receptor (54) and attenuated cytochrome c releasing and caspase-3 activation (55) are led by MUC1. Also, MUC1 increases the survival rate response to cytotoxic or oxidative stress by activating phosphoinositol-3 kinase (PI3K) and the Akt pathway (55-57). These studies show that MUC1 might be a multifaced oncoprotein in MCC.
Epigenetic regulation of MUC1 shows the CpG island in the MUC1 promoter and methylation of histone H3K9, recovered by acetylation of histone H3K9 (47, 58). Thus, cancer cells increase MUC1 by demethylation of promoter and histone H3K9 and acetylation of H3K9. Further, the MUC1 promoter contains transcription factor binding sites for Sp1, AP1-4, NF-1, NF-κB, PPAR, and estrogen receptor (59). It was reported that proinflammatory cytokines such as TNF-α and IFN-γ, elevate MUC1 independently from NF-κB and STAT1α (60).
MUC1 is a single polypeptide chain that harbors three domains an extracellular domain, a transmembrane domain, and a Sea urchin sperm protein enterokinase and agrin (SEA) domain (47). MUC1 is autoproteolytically cleaved after translation at the GSVVV motif in the SEA domain, located within an extracellular domain, because of conformational stress (61). Cleaved MUC1 in GSVVV motif generates two peptide fragments, N-terminal subunit (MUC1-N) and C-terminal subunit (MUC1-C) (61). MUC1 exhibits seven exons that encode MUC1-N (exons 1-4) and MUC1-C (exons 4-7) (47). Alternative splicing of MUC1 produces about 80 isoforms of MUC1 with exon skipping and intron retention (62), the most common isoforms being MUC1-A, MUC1-B, MUC1-C, MUC1-D, MUC1-X, MUC1-Y, and MUC1-ZD (62). Especially, MUC1-C is associated with oncogenic effects through interaction with various transcription factors such as STAT3, NF-κB, p53, and β-catenin (63-66). Glycosylated MUC1-C provides a bridge between ErbB1 and epidermal growth factor receptor (EGFR), which stabilizes and improves the EGFR-associated signaling pathway for cell proliferation (51). Intriguingly, MUC1-C itself translocates into the nucleus and controls various physiological effects through β-catenin, FOXO3a, p53, and ER-α (64, 67-69). In summary, MUC1 plays a crucial role via various isoforms in constructing ‘a great wall’ to avoid anti-cancer therapeutic strategies.
MUCIN4 (MUC4): Like other transmembrane mucins, MUC4 also plays a role in protecting the epithelial surface (70). CRC patients show low expression of MUC4 (99/132 [75%]) (71), which might be mediated by Wnt/β-catenin signaling. Nuclear β-catenin promotes HES1, an antagonist of HATH1 (72). Given the increase of MUC4 by HATH1, CRC might display a low level of MUC4 by Wnt signaling. MUC4 controls cell proliferation, differentiation, apoptosis, and tumor progression via three EGF-like domains, which play as an intramembrane ligand to activate ErbB2 (73). The interaction between MUC4 and ErbB2 activates the downstream pathway of EGF signaling such as PI3K-Akt pathway, which related to proliferation and apoptosis in tumorigenesis. MCC might employ MUC4 for tumor progression instead of Wnt/β-catenin signaling activation.
MUCIN16 (MUC16): In normal tissue, MUC16, previously known as CA125, expresses in the epithelial lining of several organs (74, 75). It functions as a barrier against the external environment and supports the maintenance of the mucus layer (76). Given high expression in various cancer, MUC16 is exten-sively employed as a biomarker (77). It is one of the frequently mutated genes, resulting in increased tumor growth and malignancy (78-80). CA125 is a tandem repeated peptide (60+ repeats of 156 amino acids) of the MUC16, which promotes cancer cell proliferation and resistance to immune surveillance (81). MUC16 inhibits the function of Natural Killer (NK) cells via direct binding to the Siglec-9 receptor of NK cells, resulting in evasion of the innate immune response (82, 83). Also, the interaction may inhibit intimate interactions between NK and cancer cells (84). The interaction between MUC16 and mesothelin, a protein located in the mesothelial lining of the peritoneal cavity, triggers cancer metastasis (85) by facilitating attachment of cancer cells to the mesothelial lining (86, 87).
Because of MUC16-induced JAK-STAT, knockdown of MUC16 displays decreased in vitro and in vivo proliferation of cancer cells (88, 89). Furthermore, knockdown studies of MUC16 demonstrated that MUC16 affects caspase-dependent/-independent apoptosis, colony-forming ability, adhesion, migration, invasive ability, epithelial-mesenchymal transition (EMT), and chemoresistance of cancer cells (90-92), which suggest an increased cancer stage by MUC16 overexpression. Especially, the MUC16 C-terminal fragment (CT) leads to upregulation of the stem cell-related genes via interaction with JAK2. It may lead to the dedifferentiation of cancer cells to obtain malignant properties. Also, MUC16 CT disrupts deregulation of β-catenin and E-cadherin at the junctional complex, leading to EMT in cancer cells (93). Thus, MUC16 is a strong target for the development of anti-cancer therapy.
Together, transmembrane mucins such as MUC1 and MUC16 protect internal cells from the external environment. Moreover, it has the ability to play a crucial role in signaling pathway transduction while having various isoforms. Despite these indispensable roles in normal tissues, mucins are highly attractive functions for cancer cells and would be an essential factor for malignant tumors. Therefore, various anti-cancer therapies targeting transmembrane mucins have been attempted in various ways (Fig. 1).
CLINICAL TRIALS AND A PRECLINICAL MOUSE MODEL FOR MCC THERAPY
MCC is mainly characterized by the high expression of mucin, which is categorized into over 20 subtypes. However, their functions especially in MCC pathology are not completely understood. Additionally, MCC shows malignant features including highly accumulated DNA damage, resistance to cancer therapy, invasive characteristics, and poor prognosis (3, 4, 12). Nonetheless, the roles of mucins in MCC remain ambiguous, mainly because of the numerous subtypes of mucins, highly complex protein expression, and absence of proper mouse models. Recent studies suggested that MCC exhibits high mucin expression, MSI (3, 40), frequent mutation of KRAS, BRAF, MAPK, and TOP-1, hyperactivation of the PIK3CA signaling pathway (13, 14, 18, 40), and increased inflammation. Thus, it is imperative to unveil the molecular mechanism for MCC tumorigenesis and to establish MCC mouse models. In this section, we discuss past/current clinical trials for MCC treatment. Moreover, we introduce mucin-related mouse models and a novel mouse model for MCC research.
Therapeutic targeting of mucin-related oncogenic effects
EGFR receptor inhibitors: As pointed out above, mucins mainly upregulate the EGFR receptor to activate the MAPK pathway (Table 2). Thus, Cetuximab (NIH clinical trial numbers [clinicaltrials.gov]: NCT01198535, NCT00835679, and NCT00100841), Gefitinib (Iressa; NCT00052585), Erlotinib (Tarceva, NCT0006 0411), and Panitumumab (NCT01285102) were tested to downregulate MCC growth. Cetuximab is an EGFR specific monoclonal antibody, that competitively inhibits the binding of epidermal growth factor (EGF) and other ligands that are secreted by tumor cells (94, 95). Gefitinib is the first selective inhibitor of the EGFR tyrosine kinase, which it inhibits by binding to the adenosine triphosphate (ATP)-binding site of the enzyme. Gefitinib is also referred to as Her1 or ErbB-1. Erlotinib inhibits the intracellular phosphorylation of tyrosine kinase associated with the EGFR, but the mechanism of action has not been fully characterized (96). Panitumumab (ABX-EGF) is a recombinant human IgG2 monoclonal antibody, binding to EGFR. Panitumumab competes with the EGF ligand to bind EGFR and shows decreased VEGF production. Although EGRF inhibitors show anti-cancer effects, they exhibit several side effects such as acneform rash, vomiting, diarrhea, skin change, and loss of appetite (97, 98).
Table 2.
Current and past clinical trials for MCC-related therapy
| Drug | Mechanism of action | Phase | Indentifier |
|---|---|---|---|
| MK2206 | Akt inhibitor | Phase2 | NCT01802320 |
| Alisertib | Aurora A kinase inhibitor | Phase1 | NCT01923337 |
| Oxaliplatin | DNA synthesis inhibitor | Phase1 Phase2 Phase3 |
NCT00005036 NCT00060411 NCT00217737 NCT01643499 NCT01652196 |
| 6,8-Bis(benzylthio)octanoic acid | E1α PDH modulator | Phase1 | NCT02232152 |
| Cetuximab | EGFR inhibitor | Early phase1 Phase1 Phase2 |
NCT00100841 NCT00835679 NCT01198535 |
| Dasatinib | EGFR inhibitor | Early phase1 | NCT00835679 |
| Erlotinib | EGFR inhibitor | Phase1 | NCT00060411 |
| Gefitinib | EGFR inhibitor | Phase2 | NCT00052585 |
| Panitumumab | EGFR inhibitor | Phase2 | NCT01285102 |
| Gamma-seretase inhibitor RO4929097 | Gamma-seretase inhibitor | Phase1 | NCT01198535 |
| Recombinant inferferon gamma | Macrophage activation factor | Phase1 Phase2 |
NCT00002796 |
| Fluorouracil | Thymidylate synthase blocking | Phase1 Phase2 Phase3 |
NCT00002796 NCT00005036 NCT00052585 NCT00060411 NCT00217737 NCT01285102 NCT01643499 NCT01652196 NCT02232152 NCT02235324 |
| Irinotecan | Topoisomerase inhibitor | Phase1 Phase2 Phase3 |
NCT00005036 NCT00052585 NCT01285102 NCT01643499 NCT01923337 NCT04088786 |
| Aflibercept | VEGF inhibitor | Phase2 |
NCT01652196 NCT02235324 |
| Bevacizumab | VEGF inhibitor | Phase1 Phase2 Phase3 |
NCT00060411 NCT00100841 NCT00217737 |
Akt inhibitor: Mucins activate PI3K/Akt signaling to survive under various cytotoxicity conditions (55-57, 99). MK2206 (NCT01802320) is an orally available inhibitor of pan Akt (protein kinase B) that inhibits the activity of Akt in a non-ATP competitive manner, resulting in the inhibition of the PI3K/Akt signaling pathway and cell proliferation. Akt contains a Pleckstrin Homology (PH) domain, which binds with high affinity to phosphoinositides including PIP3 and PIP2. Although the mode of action of MK2206 is not clear, it may interfere with binding sites for Akt substrates (100). MK2206 also shows side effects such as skin rash and gastrointestinal upset (101).
Angiogenesis inhibitors: Vascular endothelial growth factor (VEGF) plays a crucial role in angiogenesis, lymphangio-genesis, and tumor growth. Thus, VEGF inhibitors are suggested as a potential therapeutic target for anti-cancer therapies. Bevaci-zumab (Avastin; NCT00217737, NCT00060411, and NCT00100841) and Aflibercept (Ziv-aflibercept; NCT01652196 and NCT0223 5324) are employed to test the effect of VEGF inhibitors on MCC progression. Bevacizumab gained FDA approval as the first antiangiogenic agent, which is a humanized monoclonal IgG antibody and neutralizes VEGF-A (102, 103). As a recombinant protein, Aflibercept acts as a decoy receptor for VEGF-A and placental growth factor (PIGF), resulting in suppression of VEGFR-1 and VEGFR-2. However, VEGF inhibitors increase the risk of stroke, and myocardial infarction with various side effects including diarrhea, neutropenia, and thrombocytopenia.
Topoisomerase I inhibitor: MCC shows a frequent amplifying mutation in TOP-1, which might increase genetic instability such as MSI. Irinotecan (Onyvyde; NCT00005036, NCT0005 2585, NCT01285102, NCT01643499, NCT01923337, and NCT04088786) is an antineoplastic enzyme inhibitor, which is derived from camptothecin which inhibits the action of TOP-1 (104). Irinotecan prevents re-ligation of the DNA strand by binding to the TOP-1-DNA complex, which results in lethal double-strand DNA breakage. Irinotecan exhibits toxicity such as nausea, vomiting, abdominal cramping, diarrhea, and infection. Other anti-cancer drugs: Fluorouracil, an inhibitor of DNA synthesis, is a classical anti-cancer drug. Fluorouracil blocks the formation of thymidylate from uracil, leading to the inhibition of DNA and RNA synthesis. RO4929097 is an orally available gamma-secretase (GS) inhibitor, which directly binds to GS and blocks activation of the Notch receptor. These drugs also lead to severe side effects including trouble sleeping, irritability, temporary hair loss, and abnormal taste.
Preclinical mouse model for MCC research
Despite the poor prognosis and metastatic characteristics of MCC (7), the genetic mechanism of MCC development is unknown. Several genetically engineered mouse models (GEMMs) of Muc1, Muc2, Muc5ac, Muc5b, Muc6, and Muc16 have been employed for MCC study (Table 3). However, those GEMMs are highly involved in the maintenance of tissue homeostasis and inflammation without mutations of genes frequently found in the human MCC (KRAS, BRAF, MAPK, and TOP-1) (12-16). It is noteworthy that despite the hyperactivation of the PIK3CA signaling pathway in human MCC, genetic mutations in KRAS, BRAF, MAPK, and TOP-1 do not lead to MCC development in mouse models (105-107). For example, the KRASG12D mouse displays tumorigenesis such as non-small cell lung cancer and pancreatic cancer without MCC development (106).
Table 3.
Mucin-related GEMMs for MCC study
| Gene | Allele symbol | Allele attributes | Reported phenotypes | Reference |
|---|---|---|---|---|
| Muc1 | Muc1<em1Smoc> | Null/knockout | No abnormal phenotype observed | Shanghai Model Organisms Center |
| Muc1<tm1(cre/ERT2)Lcm> | Inducible recombinase | No abnormal phenotype observed | Kopinke and Murtaugh, 2010 BMC Dev Biol | |
| Muc1<tm1(KOMP)Vlcg> | Null/knockout, reporter | No abnormal phenotype observed | Velocigene MGI Direct Data Submission | |
| Muc1<tm1.1(cre/ERT2)Lcm> | Inducible recombinase | No abnormal phenotype observed | Kopinke and Murtaugh, 2010 BMC Dev Biol | |
| Muc1<tm1a(EUCOMM)Wtsi> | Conditional ready, null/knockout, reporter | No abnormal phenotype observed | Skarnes et al., 2011 Nature | |
| Muc1<tm1e(EUCOMM)Wtsi> | Null/knockout, reporter | No abnormal phenotype observed | Skarnes et al., 2011 Nature | |
| Muc1<tm1Gend> | Null/knockout | Digestive/alimentary, homeostasis, liver/biliary, neoplasm | Spicer et al., 1995 J Biol Chem | |
| Muc1<tm1Smoc> | Conditional ready | No abnormal phenotype observed | Shanghai Model Organisms Center | |
| Muc2 | Muc2<eey> | Chemically induced (ENU) | Cellular, digestive/alimentary, endocrine/exocrine, hematopoietic, immune, mortality/aging | Heazlewood et al., 2008 PLoS Med |
| Muc2<keny> | Chemically induced (ENU) | Digestive/alimentary, immune | The Australian Phenomics Facility at The Australian National University | |
| Muc2<M1Btlr> | Chemically induced (ENU) | Digestive/alimentary, immune | Brandl K et al., MGI Direct Data Submission | |
| Muc2<m2Btlr> | Chemically induced (ENU) | Digestive/alimentary, immune | Brandl K et al., MGI Direct Data Submission | |
| Muc2<m3Btlr> | Chemically induced (ENU), no specific | Digestive/alimentary, immune | McAlpine W et al., MGI Direct Data Submission | |
| Muc2<tm1a(KOMP)Wtsi> | Conditional ready, null/knockout, reporter | No abnormal phenotype observed | Skarnes et al., 2011 Nature | |
| Muc2<tm1Avel> | Null/knockout | Velcich et al., 2002 Science | ||
| Muc2<tm1e(KOMP)Wtsi> | Null/knockout, reporter | No abnormal phenotype observed | Skarnes et al., 2011 Nature | |
| Muc2<wnn> | Chemically induced (ENU) | Cardiovascular, cellular, digestive/ alimentary, endocrine/exocrine, growth/size/body, hematopoietic, homeostasis, immune, mortality/aging | Robinson et al., 2017 Am J Physiol Gastrointest Liver Physiol | |
| Muc5ac | Muc5ac<em1Smoc> | Null/knockout | No abnormal phenotype observed | Shanghai Model Organisms Center |
| Muc5ac<tm1.1Evns> | Null/knockout | Digestive/alimentary, homeostasis, immune, vision/eye | Morgan et al., 2021 Nat Commun | |
| Muc5ac<tm2a(EUCOMM)Hmgu> | Conditional ready, null/knockout, reporter | No abnormal phenotype observed | Helmholtz Zentrum Muenchen GmbH | |
| Muc5ac<tm2b(EUCOMM)Hmgu> | Null/knockout, reporter | No abnormal phenotype observed | International Knockout Mouse Consortium | |
| Muc5ac<tm2e(EUCOMM)Hmgu> | Null/knockout, reporter | No abnormal phenotype observed | Helmholtz Zentrum Muenchen GmbH | |
| Muc5b | Muc5b<em1(IMPC)Mbp> | Null/knockout | No abnormal phenotype observed | International Mouse Phenotyping Consortium (IMPC) Database Release |
| Muc5b<Gt(EUCE0173a08)Hmgu> | Gene trapped | No abnormal phenotype observed | Mouse Genome Informatics (MGI) and National Center for Biotechnology Information (NCBI) | |
| Muc5b<tm1(NCOM)Mfgc> | Null/knockout, reporter | No abnormal phenotype observed | Mammalian Functional Genomics Centre | |
| Muc5b<tm1.2Evns> | Null/knockout | Cellular, growth/size/body, hearing/vestibular/ear, hematopoietic, homeostasis, immune, mortality/aging, respiratory | Roy et al., 2014 Nature | |
| Muc6 | Muc6<tm1(Hbegf)Koo> | Inserted expressed sequence, reporter | No abnormal phenotype observed | Han et al., 2019 Cell Stem Cell |
| Muc16 | Muc15<tm1Lex> | Null/knockout | No abnormal phenotype observed | Tang et al., 2010 Nat Biotechnol |
| Muc16<em1Smoc> | Null/knockout | No abnormal phenotype observed | Shanghai Model Organisms Center | |
| Muc16<m1Mhda> | Null/knockout | No abnormal phenotype observed | Sabrautzki S et al., MGI Direct Data Submission | |
| Muc16<tm1Bhr> | Null/knockout | Reproductive | Shirai et al., 2014 Invest Ophthalmol Vis Sci | |
| Muc16<tm1Bhr> | Null/knockout | No abnormal phenotype observed | Tang et al., 2010 Nat Biotechnol |
Recently, it was shown that Cancer-related Regulator of Actin Dynamic (CRAD) is highly mutated in small cell lung cancer, CRC, and melanoma (40, 108). Additionally, the Crad KO mouse initiates tumorigenesis in the pancreas and lung with MCC development (40). Thus, it is highly likely that inactivation of the CRAD gene might be associated with MCC tumorigenesis. The oncogenic mutations and signaling amplification such as KRAS, BRAF, MAPK, and TOP-1, might result from concomitant MCC progression in the MCC-specific tumor suppressor-deficient condition. Therefore, the genetic and molecular basis of MCC development and MCC-specific tumor suppressor needs to be resolved. Currently, anti-MCC therapies, including IR and chemotherapy, are not successful, because of the high resistance of MCC. However, the anti-cancer therapy resistance mechanism of MCC is not understood.
A major reason that these issues have not been successfully addressed was the lack of a preclinical MCC animal model. Mucins interact with each other and utilize another signaling pathway to develop MCC, but the GEMM of each mucin does not mimic the environment of MCC patients. However, Crad KO displays overexpression of several mucins at the same time, and MCC markers such as MSI and Top-I are upregulated (40). Therefore, Crad KO is proposed as a novel preclinical mouse model for MCC study.
DISCUSSION
Metastasis is accompanied by multiple events and requires ideal timing. Further, tumor suppression mechanisms including the immune system, tightly function in the organism to kill cancer cells, thus it is difficult to acquire a wealth of growth factors and nutrients for growth. Hence even malignant tumor cells would often fail to metastasize. The selected malignant tumor cells are more likely to succeed in metastasis. MCC may be the selected cells. Given that MCC maintains enough mucins that could be called a ‘stealth cloak’, MCC can take a stealth strategy and metastasize while being protected by a cloak (Fig. 1). Mucins interact and support MCC efficiently acquire factors necessary for growth and metastasis. Furthermore, mucin provides sanctuary to escape from the surveillance of the immune system. These demonstrate that MCC thoroughly exploits the superior abilities of mucins. For example, mucins are tightly controlled by a regulatory mechanism such as promoter methylation and transcription factor (10, 11). The expression patterns of MUC2 and MUC5 are similar, but the function appears to be independent, which could be utilized by MCC. Transcription factors such as Sp1, commonly regulate the expression of MUC2 and MUC5, but MUC5-induced β-catenin inhibits the expression of MUC2 (43). In the early stage of MCC tumorigenesis, the expression of MUC2 and MUC5 indicates an incompatible pattern. Reduced MUC2 might lead to the inflammatory response that is necessary for MCC development or promote oncogenic mucins. Subsequently upregulated MUC5 might add suppression force for MUC2 via β-catenin. During MCC progression, MUC5 is downregulated as MSI completion, resulting in MUC2 upregulation to escape from immune surveillance through MUC2’s protective function. Additionally, following the loss of cell polarity during MCC tumorigenesis, mucins are expressed all over the cell surface and become available to interplay with several growth factor receptors to modulate their downstream signaling.
CRAD stabilizes the cadherin-catenin-actin filament (CCA) complex (40), which means control of cell adhesion and Wnt/β-catenin signaling by CRAD. We have already discussed the role of mucin in cell adhesion and Wnt/β-catenin signaling. The destabilized CCA complex disrupts epithelial cell polarity, which would trigger an inflammatory response and cell proliferation. It is plausible that abnormal polarity, inflammation, and Wnt/β-catenin signaling might foster an oncogenic environment for MCC via elevated mucins. It still unclear how inactivated CRAD increases mucins, but it is clear that a GEMM in which several mucins are simultaneously overexpressed would helpful for future MCC research and the development of anti-MCC therapeutic strategies.
ACKNOWLEDGEMENTS
This work was supported by the CHUNG-ANG UNIVERSITY Grant in 2020 and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1F1A1075419) to Y-.S. Jung.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
References
- 1.Gum JR, Jr, Hicks JW, Gillespie AM, et al. Goblet cell-specific expression mediated by the MUC2 mucin gene promoter in the intestine of transgenic mice. Am J Physiol. 1999;276:G666–676. doi: 10.1152/ajpgi.1999.276.3.G666. [DOI] [PubMed] [Google Scholar]
- 2.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wahrenbrock M, Borsig L, Le D, Varki N, Varki A. Selectin-mucin interactions as a probable molecular explanation for the association of Trousseau syndrome with mucinous adenocarcinomas. J Clin Invest. 2003;112:853–862. doi: 10.1172/JCI200318882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smagghe BJ, Stewart AK, Carter MG, et al. MUC1* ligand, NM23-H1, is a novel growth factor that maintains human stem cells in a more naive state. PLoS One. 2013;8:e58601. doi: 10.1371/journal.pone.0058601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mekenkamp LJ, Heesterbeek KJ, Koopman M, et al. Mucinous adenocarcinomas: poor prognosis in metastatic colorectal cancer. Eur J Cancer. 2012;48:501–509. doi: 10.1016/j.ejca.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Tran DT, Ten Hagen KG. Mucin-type O-glycosylation during development. J Biol Chem. 2013;288:6921–6929. doi: 10.1074/jbc.R112.418558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okudaira K, Kakar S, Cun L, et al. MUC2 gene promoter methylation in mucinous and non-mucinous colorectal cancer tissues. Int J Oncol. 2010;36:765–775. doi: 10.3892/ijo_00000552. [DOI] [PubMed] [Google Scholar]
- 11.Niv Y. Mucin gene expression in the intestine of ulcerative colitis patients: a systematic review and meta-analysis. Eur J Gastroenterol Hepatol. 2016;28:1241–1245. doi: 10.1097/MEG.0000000000000707. [DOI] [PubMed] [Google Scholar]
- 12.Hugen N, Brown G, Glynne-Jones R, de Wilt JH, Nagtegaal ID. Advances in the care of patients with mucinous colorectal cancer. Nat Rev Clin Oncol. 2016;13:361–369. doi: 10.1038/nrclinonc.2015.140. [DOI] [PubMed] [Google Scholar]
- 13.Day FL, Jorissen RN, Lipton L, et al. PIK3CA and PTEN gene and exon mutation-specific clinicopathologic and molecular associations in colorectal cancer. Clin Cancer Res. 2013;19:3285–3296. doi: 10.1158/1078-0432.CCR-12-3614. [DOI] [PubMed] [Google Scholar]
- 14.Garrido-Laguna I, Hong DS, Janku F, et al. KRASness and PIK3CAness in patients with advanced colorectal cancer: outcome after treatment with early-phase trials with targeted pathway inhibitors. PLoS One. 2012;7:e38033. doi: 10.1371/journal.pone.0038033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunal A, Hui P, Kilic S, et al. KRAS mutations are associated with specific morphologic features in colon cancer. J Clin Gastroenterol. 2013;47:509–514. doi: 10.1097/MCG.0b013e3182703030. [DOI] [PubMed] [Google Scholar]
- 16.Negri FV, Azzoni C, Bottarelli L, et al. Thymidylate synthase, topoisomerase-1 and microsatellite instability: relationship with outcome in mucinous colorectal cancer treated with fluorouracil. Anticancer Res. 2013;33:4611–4617. [PubMed] [Google Scholar]
- 17.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pai RK, Jayachandran P, Koong AC, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 2012;36:744–752. doi: 10.1097/PAS.0b013e31824430d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ookawa K, Kudo T, Aizawa S, Saito H, Tsuchida S. Transcriptional activation of the MUC2 gene by p53. J Biol Chem. 2002;277:48270–48275. doi: 10.1074/jbc.M207986200. [DOI] [PubMed] [Google Scholar]
- 20.Pereira MB, Dias AJ, Reis CA, Schmitt FC. Immunohistochemical study of the expression of MUC5AC and MUC6 in breast carcinomas and adjacent breast tissues. J Clin Pathol. 2001;54:210–213. doi: 10.1136/jcp.54.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biemer-Huttmann AE, Walsh MD, McGuckin MA, et al. Mucin core protein expression in colorectal cancers with high levels of microsatellite instability indicates a novel pathway of morphogenesis. Clin Cancer Res. 2000;6:1909–1916. [PubMed] [Google Scholar]
- 22.Losi L, Scarselli A, Benatti P, et al. Relationship between MUC5AC and altered expression of MLH1 protein in mucinous and non-mucinous colorectal carcinomas. Pathol Res Pract. 2004;200:371–377. doi: 10.1016/j.prp.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Walsh MD, Clendenning M, Williamson E, et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol. 2013;26:1642–1656. doi: 10.1038/modpathol.2013.101. [DOI] [PubMed] [Google Scholar]
- 24.Renaud F, Vincent A, Mariette C, et al. MUC5AC hypomethylation is a predictor of microsatellite instability independently of clinical factors associated with colorectal cancer. Int J Cancer. 2015;136:2811–2821. doi: 10.1002/ijc.29342. [DOI] [PubMed] [Google Scholar]
- 25.Rico SD, Hoflmayer D, Buscheck F, et al. Elevated MUC5AC expression is associated with mismatch repair deficiency and proximal tumor location but not with cancer progression in colon cancer. Med Mol Morphol. 2020;54:156–165. doi: 10.1007/s00795-020-00274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pothuraju R, Rachagani S, Krishn SR, et al. Mole-cular implications of MUC5AC-CD44 axis in colorectal cancer progression and chemoresistance. Mol Cancer. 2020;19:37. doi: 10.1186/s12943-020-01156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jun S, Jung YS, Suh HN, et al. LIG4 mediates Wnt signalling-induced radioresistance. Nat Commun. 2016;7:10994. doi: 10.1038/ncomms10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung YS, Jun S, Kim MJ, et al. TMEM9 promotes intestinal tumorigenesis through vacuolar-ATPase-activated Wnt/beta-catenin signalling. Nat Cell Biol. 2018;20:1421–1433. doi: 10.1038/s41556-018-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung YS, Stratton SA, Lee SH, et al. TMEM9-v-ATPase activates Wnt/beta-catenin signaling via APC lysosomal degradation for liver regeneration and tumo-rigenesis. Hepatology. 2021;73:776–794. doi: 10.1002/hep.31305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Jung YS, Jun S, et al. PAF-Wnt signaling-induced cell plasticity is required for maintenance of breast cancer cell stemness. Nat Commun. 2016;7:10633. doi: 10.1038/ncomms10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Y, Zhou L, Zhao D, et al. MUC5AC enhances tumor heterogeneity in lung adenocarcinoma with mucin production and is associated with poor prognosis. Jpn J Clin Oncol. 2020;50:701–711. doi: 10.1093/jjco/hyaa016. [DOI] [PubMed] [Google Scholar]
- 32.Vincent A, Perrais M, Desseyn JL, Aubert JP, Pigny P, Van Seuningen I. Epigenetic regulation (DNA methylation, histone modifications) of the 11p15 mucin genes (MUC2, MUC5AC, MUC5B, MUC6) in epithelial cancer cells. Oncogene. 2007;26:6566–6576. doi: 10.1038/sj.onc.1210479. [DOI] [PubMed] [Google Scholar]
- 33.Pigny P, Van Seuningen I, Desseyn JL, et al. Identification of a 42-kDa nuclear factor (NF1-MUC5B) from HT-29 MTX cells that binds to the 3' region of human mucin gene MUC5B. Biochem Biophys Res Commun. 1996;220:186–191. doi: 10.1006/bbrc.1996.0378. [DOI] [PubMed] [Google Scholar]
- 34.Desseyn JL, Aubert JP, Van Seuningen I, Porchet N, Laine A. Genomic organization of the 3' region of the human mucin gene MUC5B. J Biol Chem. 1997;272:16873–16883. doi: 10.1074/jbc.272.27.16873. [DOI] [PubMed] [Google Scholar]
- 35.Van Seuningen I, Perrais M, Pigny P, Porchet N, Aubert JP. Sequence of the 5'-flanking region and promoter activity of the human mucin gene MUC5B in different phenotypes of colon cancer cells. Biochem J. 2000;348(Pt 3):675–686. doi: 10.1042/bj3480675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Seuningen I, Pigny P, Perrais M, Porchet N, Aubert JP. Transcriptional regulation of the 11p15 mucin genes. Towards new biological tools in human therapy, in inflammatory diseases and cancer? Front Biosci. 2001;6:D1216–1234. doi: 10.2741/A675. [DOI] [PubMed] [Google Scholar]
- 37.Yuan S, Liu Q, Hu Z, et al. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis. 2018;9:450. doi: 10.1038/s41419-018-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia EP, Tiscornia I, Libisch G, et al. MUC5B silencing reduces chemo-resistance of MCF-7 breast tumor cells and impairs maturation of dendritic cells. Int J Oncol. 2016;48:2113–2123. doi: 10.3892/ijo.2016.3434. [DOI] [PubMed] [Google Scholar]
- 39.Lahdaoui F, Messager M, Vincent A, et al. Depletion of MUC5B mucin in gastrointestinal cancer cells alters their tumorigenic properties: implication of the Wnt/beta-catenin pathway. Biochem J. 2017;474:3733–3746. doi: 10.1042/BCJ20170348. [DOI] [PubMed] [Google Scholar]
- 40.Jung YS, Wang W, Jun S, et al. Deregulation of CRAD-controlled cytoskeleton initiates mucinous colorectal cancer via beta-catenin. Nat Cell Biol. 2018;20:1303–1314. doi: 10.1038/s41556-018-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekine A, Akiyama Y, Yanagihara K, Yuasa Y. Hath1 up-regulates gastric mucin gene expression in gastric cells. Biochem Biophys Res Commun. 2006;344:1166–1171. doi: 10.1016/j.bbrc.2006.03.238. [DOI] [PubMed] [Google Scholar]
- 42.Silva EM, Begnami MD, Fregnani JH, et al. Cadherin-catenin adhesion system and mucin expression: a comparison between young and older patients with gastric carcinoma. Gastric Cancer. 2008;11:149–159. doi: 10.1007/s10120-008-0468-5. [DOI] [PubMed] [Google Scholar]
- 43.Pai P, Rachagani S, Dhawan P, Batra SK. Mucins and Wnt/beta-catenin signaling in gastrointestinal cancers: an unholy nexus. Carcinogenesis. 2016;37:223–232. doi: 10.1093/carcin/bgw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leir SH, Harris A. MUC6 mucin expression inhibits tumor cell invasion. Exp Cell Res. 2011;317:2408–2419. doi: 10.1016/j.yexcr.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Betge J, Schneider NI, Harbaum L, et al. MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: expression profiles and clinical significance. Virchows Arch. 2016;469:255–265. doi: 10.1007/s00428-016-1970-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mehla K, Singh PK. MUC1: a novel metabolic master regulator. Biochim Biophys Acta. 2014;1845:126–135. doi: 10.1016/j.bbcan.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Ren J, Yu W, et al. The epidermal growth factor receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and beta-catenin. J Biol Chem. 2001;276:35239–35242. doi: 10.1074/jbc.C100359200. [DOI] [PubMed] [Google Scholar]
- 49.Schroeder JA, Thompson MC, Gardner MM, Gendler SJ. Transgenic MUC1 interacts with epidermal growth factor receptor and correlates with mitogen-activated protein kinase activation in the mouse mammary gland. J Biol Chem. 2001;276:13057–13064. doi: 10.1074/jbc.M011248200. [DOI] [PubMed] [Google Scholar]
- 50.Schroeder JA, Adriance MC, Thompson MC, Camenisch TD, Gendler SJ. MUC1 alters beta-catenin-dependent tumor formation and promotes cellular invasion. Oncogene. 2003;22:1324–1332. doi: 10.1038/sj.onc.1206291. [DOI] [PubMed] [Google Scholar]
- 51.Pochampalli MR, el Bejjani RM, Schroeder JA. MUC1 is a novel regulator of ErbB1 receptor trafficking. Oncogene. 2007;26:1693–1701. doi: 10.1038/sj.onc.1209976. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Dong X, Bai L, Shang X, Zeng Y. MUC1-induced immunosuppression in colon cancer can be reversed by blocking the PD1/PDL1 signaling pathway. Oncol Lett. 2020;20:317. doi: 10.3892/ol.2020.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rowse GJ, Tempero RM, VanLith ML, Hollingsworth MA, Gendler SJ. Tolerance and immunity to MUC1 in a human MUC1 transgenic murine model. Cancer Res. 1998;58:315–321. [PubMed] [Google Scholar]
- 54.Agata N, Ahmad R, Kawano T, Raina D, Kharbanda S, Kufe D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008;68:6136–6144. doi: 10.1158/0008-5472.CAN-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ren J, Agata N, Chen D, et al. Human MUC1 carcinoma-associated protein confers resistance to genotoxic anticancer agents. Cancer Cell. 2004;5:163–175. doi: 10.1016/S1535-6108(04)00020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279:20607–20612. doi: 10.1074/jbc.M310538200. [DOI] [PubMed] [Google Scholar]
- 57.Yin L, Wu Z, Avigan D, et al. MUC1-C onco-protein suppresses reactive oxygen species-induced terminal differentiation of acute myelogenous leukemia cells. Blood. 2011;117:4863–4870. doi: 10.1182/blood-2010-10-296632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada N, Nishida Y, Tsutsumida H, et al. MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res. 2008;68:2708–2716. doi: 10.1158/0008-5472.CAN-07-6844. [DOI] [PubMed] [Google Scholar]
- 59.Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia. 2001;6:339–353. doi: 10.1023/A:1011379725811. [DOI] [PubMed] [Google Scholar]
- 60.Lagow EL, Carson DD. Synergistic stimulation of MUC1 expression in normal breast epithelia and breast cancer cells by interferon-gamma and tumor necrosis factor-alpha. J Cell Biochem. 2002;86:759–772. doi: 10.1002/jcb.10261. [DOI] [PubMed] [Google Scholar]
- 61.Levitin F, Stern O, Weiss M, et al. The MUC1 SEA module is a self-cleaving domain. J Biol Chem. 2005;280:33374–33386. doi: 10.1074/jbc.M506047200. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Vlad A, Milcarek C, Finn OJ. Human mucin MUC1 RNA undergoes different types of alternative splicing resulting in multiple isoforms. Cancer Immunol Immunother. 2013;62:423–435. doi: 10.1007/s00262-012-1325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Bharti A, Chen D, Gong J, Kufe D. Interaction of glycogen synthase kinase 3beta with the DF3/MUC1 carcinoma-associated antigen and beta-catenin. Mol Cell Biol. 1998;18:7216–7224. doi: 10.1128/MCB.18.12.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7:167–178. doi: 10.1016/j.ccr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 65.Ahmad R, Raina D, Joshi MD, et al. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-kappaB p65 transcription factor. Cancer Res. 2009;69:7013–7021. doi: 10.1158/0008-5472.CAN-09-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahmad R, Rajabi H, Kosugi M, et al. MUC1-C oncoprotein promotes STAT3 activation in an auto-inductive regulatory loop. Sci Signal. 2011;4:ra9. doi: 10.1126/scisignal.2001426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang L, Ren J, Chen D, Li Y, Kharbanda S, Kufe D. MUC1 cytoplasmic domain coactivates Wnt target gene transcription and confers transformation. Cancer Biol Ther. 2003;2:702–706. doi: 10.4161/cbt.2.6.610. [DOI] [PubMed] [Google Scholar]
- 68.Wen Y, Caffrey TC, Wheelock MJ, Johnson KR, Hollingsworth MA. Nuclear association of the cytoplasmic tail of MUC1 and beta-catenin. J Biol Chem. 2003;278:38029–38039. doi: 10.1074/jbc.M304333200. [DOI] [PubMed] [Google Scholar]
- 69.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 70.Pelaseyed T, Hansson GC. Membrane mucins of the intestine at a glance. J Cell Sci. 2020;133 doi: 10.1242/jcs.240929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shanmugam C, Jhala NC, Katkoori VR, et al. Prognostic value of mucin 4 expression in colorectal adenocarcinomas. Cancer. 2010;116:3577–3586. doi: 10.1002/cncr.25095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peignon G, Durand A, Cacheux W, et al. Complex interplay between beta-catenin signalling and Notch effectors in intestinal tumorigenesis. Gut. 2011;60:166–176. doi: 10.1136/gut.2009.204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carraway KL, 3rd, Rossi EA, Komatsu M, et al. An intramembrane modulator of the ErbB2 receptor tyrosine kinase that potentiates neuregulin signaling. J Biol Chem. 1999;274:5263–5266. doi: 10.1074/jbc.274.9.5263. [DOI] [PubMed] [Google Scholar]
- 74.Govindarajan B, Gipson IK. Membrane-tethered mucins have multiple functions on the ocular surface. Exp Eye Res. 2010;90:655–663. doi: 10.1016/j.exer.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haridas D, Ponnusamy MP, Chugh S, Lakshmanan I, Seshacharyulu P, Batra SK. MUC16: molecular analysis and its functional implications in benign and malignant conditions. FASEB J. 2014;28:4183–4199. doi: 10.1096/fj.14-257352. [DOI] [PubMed] [Google Scholar]
- 76.Gipson IK, Spurr-Michaud S, Tisdale A, Menon BB. Comparison of the transmembrane mucins MUC1 and MUC16 in epithelial barrier function. PLoS One. 2014;9:e100393. doi: 10.1371/journal.pone.0100393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aithal A, Rauth S, Kshirsagar P, et al. MUC16 as a novel target for cancer therapy. Expert Opin Ther Targets. 2018;22:675–686. doi: 10.1080/14728222.2018.1498845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim N, Hong Y, Kwon D, Yoon S. Somatic mutaome profile in human cancer tissues. Genomics Inform. 2013;11:239–244. doi: 10.5808/GI.2013.11.4.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Das S, Rachagani S, Torres-Gonzalez MP, et al. Carboxyl-terminal domain of MUC16 imparts tumorigenic and metastatic functions through nuclear translocation of JAK2 to pancreatic cancer cells. Oncotarget. 2015;6:5772–5787. doi: 10.18632/oncotarget.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giannakouros P, Matte I, Rancourt C, Piche A. Transformation of NIH3T3 mouse fibroblast cells by MUC16 mucin (CA125) is driven by its cytoplasmic tail. Int J Oncol. 2015;46:91–98. doi: 10.3892/ijo.2014.2707. [DOI] [PubMed] [Google Scholar]
- 81.Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi: 10.1146/annurev.physiol.70.113006.100659. [DOI] [PubMed] [Google Scholar]
- 82.Belisle JA, Gubbels JA, Raphael CA, et al. Peritoneal natural killer cells from epithelial ovarian cancer patients show an altered phenotype and bind to the tumour marker MUC16 (CA125) Immunology. 2007;122:418–429. doi: 10.1111/j.1365-2567.2007.02660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Belisle JA, Horibata S, Jennifer GA, et al. Identification of Siglec-9 as the receptor for MUC16 on human NK cells, B cells, and monocytes. Mol Cancer. 2010;9:118. doi: 10.1186/1476-4598-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Felder M, Kapur A, Gonzalez-Bosquet J, et al. MUC16 (CA125): tumor biomarker to cancer therapy, a work in progress. Mol Cancer. 2014;13:129. doi: 10.1186/1476-4598-13-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rump A, Morikawa Y, Tanaka M, et al. Binding of ovarian cancer antigen CA125/MUC16 to mesothelin mediates cell adhesion. J Biol Chem. 2004;279:9190–9198. doi: 10.1074/jbc.M312372200. [DOI] [PubMed] [Google Scholar]
- 86.Gubbels JA, Belisle J, Onda M, et al. Mesothelin-MUC16 binding is a high affinity, N-glycan dependent interaction that facilitates peritoneal metastasis of ovarian tumors. Mol Cancer. 2006;5:50. doi: 10.1186/1476-4598-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen SH, Hung WC, Wang P, Paul C, Konstantopoulos K. Mesothelin binding to CA125/MUC16 promotes pancreatic cancer cell motility and invasion via MMP-7 activation. Sci Rep. 2013;3:1870. doi: 10.1038/srep01870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Komatsu M, Arango ME, Carraway KL. Synthesis and secretion of Muc4/sialomucin complex: implication of intracellular proteolysis. Biochem J. 2002;368:41–48. doi: 10.1042/bj20020862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Albrecht H, Carraway KL., 3rd MUC1 and MUC4: switching the emphasis from large to small. Cancer Biother Radiopharm. 2011;26:261–271. doi: 10.1089/cbr.2011.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boivin M, Lane D, Piche A, Rancourt C. CA125 (MUC16) tumor antigen selectively modulates the sensitivity of ovarian cancer cells to genotoxic drug-induced apoptosis. Gynecol Oncol. 2009;115:407–413. doi: 10.1016/j.ygyno.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 91.Comamala M, Pinard M, Theriault C, et al. Downregulation of cell surface CA125/MUC16 induces epithelial-to-mesenchymal transition and restores EGFR signalling in NIH:OVCAR3 ovarian carcinoma cells. Br J Cancer. 2011;104:989–999. doi: 10.1038/bjc.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reinartz S, Failer S, Schuell T, Wagner U. CA125 (MUC16) gene silencing suppresses growth properties of ovarian and breast cancer cells. Eur J Cancer. 2012;48:1558–1569. doi: 10.1016/j.ejca.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 93.Theriault C, Pinard M, Comamala M, et al. MUC16 (CA125) regulates epithelial ovarian cancer cell growth, tumorigenesis and metastasis. Gynecol Oncol. 2011;121:434–443. doi: 10.1016/j.ygyno.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 94.Snyder LC, Astsaturov I, Weiner LM. Overview of monoclonal antibodies and small molecules targeting the epidermal growth factor receptor pathway in colorectal cancer. Clin Colorectal Cancer. 2005;5 Suppl 2:S71–80. doi: 10.3816/CCC.2005.s.010. [DOI] [PubMed] [Google Scholar]
- 95.Vincenzi B, Zoccoli A, Pantano F, Venditti O, Galluzzo S. Cetuximab: from bench to bedside. Curr Cancer Drug Targets. 2010;10:80–95. doi: 10.2174/156800910790980241. [DOI] [PubMed] [Google Scholar]
- 96.Ling YH, Li T, Yuan Z, Haigentz M, Jr, Weber TK, Perez-Soler R. Erlotinib, an effective epidermal growth factor receptor tyrosine kinase inhibitor, induces p27KIP1 up-regulation and nuclear translocation in association with cell growth inhibition and G1/S phase arrest in human non-small-cell lung cancer cell lines. Mol Pharmacol. 2007;72:248–258. doi: 10.1124/mol.107.034827. [DOI] [PubMed] [Google Scholar]
- 97.Yano S, Kondo K, Yamaguchi M, et al. Distribution and function of EGFR in human tissue and the effect of EGFR tyrosine kinase inhibition. Anticancer Res. 2003;23:3639–3650. [PubMed] [Google Scholar]
- 98.Sipples R. Common side effects of anti-EGFR therapy: acneform rash. Semin Oncol Nurs. 2006;22:28–34. doi: 10.1016/j.soncn.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 99.Yin L, Li Y, Ren J, Kuwahara H, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J Biol Chem. 2003;278:35458–35464. doi: 10.1074/jbc.M301987200. [DOI] [PubMed] [Google Scholar]
- 100.Cherrin C, Haskell K, Howell B, et al. An allosteric Akt inhibitor effectively blocks Akt signaling and tumor growth with only transient effects on glucose and insulin levels in vivo. Cancer Biol Ther. 2010;9:493–503. doi: 10.4161/cbt.9.7.11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yap TA, Yan L, Patnaik A, et al. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. J Clin Oncol. 2011;29:4688–4695. doi: 10.1200/JCO.2011.35.5263. [DOI] [PubMed] [Google Scholar]
- 102.Kazazi-Hyseni F, Beijnen JH, Schellens JH. Bevacizumab. Oncologist. 2010;15:819–825. doi: 10.1634/theoncologist.2009-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stacker SA, Achen MG. The VEGF signaling pathway in cancer: the road ahead. Chin J Cancer. 2013;32:297–302. doi: 10.5732/cjc.012.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wood JP, Smith AJ, Bowman KJ, Thomas AL, Jones GD. Comet assay measures of DNA damage as biomarkers of irinotecan response in colorectal cancer in vitro and in vivo. Cancer Med. 2015;4:1309–1321. doi: 10.1002/cam4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu Y, Her C. Inhibition of topoisomerase (DNA) I (TOP1): DNA damage repair and anticancer therapy. Biomolecules. 2015;5:1652–1670. doi: 10.3390/biom5031652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]