Abstract
The budding of membranes and curvature generation is common to many forms of trafficking in cells. Clathrin-mediated endocytosis, as a prototypical example of trafficking, has been studied in great detail using a variety of experimental systems and methods. Recently, advances in experimental methods have led to great strides in insights on the molecular mechanisms and the spatio-temporal dynamics of the protein machinery associated with membrane curvature generation. These advances have been ably supported by computational models, which have given us insights into the underlying mechanical principles of clathrin-mediated endocytosis. On the other hand, targeted experimental perturbation of membranes has lagged behind that of proteins in cells. In this area, modeling is especially critical to interpret experimental measurements in a mechanistic context. Here, we discuss the contributions made by these models to our understanding of endocytosis and identify opportunities to strengthen the connections between models and experiments.
Keywords: clathrin-mediated endocytosis, bending modulus, Helfrich, budding, snapthrough instability, membrane tension, multiscale modeling
Graphical Abstract
Model development for the different subprocesses of clathrin-mediated endocytosis (CME) can span different length scales. Like the pieces of a puzzle, these different modeling approaches at different scales contribute to our understanding of the mechanisms underlying CME.
Introduction
In the 1970s, two separate developments in two disparate fields took place: cell biologists discovered receptor-mediated endocytosis [1, 2]; meanwhile other researchers, investigating the mechanics of lipid bilayers (not biological membranes), generated a model that expresses the energy of a lipid bilayer as a function of its local curvatures [3, 4, 5]. While these two developments stem from what appear to be independent fields and methods, molecular and cell biological approaches to elucidate the molecular mechanisms of clathrin-mediated endocytosis (CME) and the non-trivial mathematics of differential geometry and thin shell theory for lipid mechanics models [6, 7], they have converged over the years to result in experimentally-informed models that have given us insight into the mechanics and energetics of CME [8, 9, 10, 11, 12, 13, 14]. Here, we summarize those models from the context of their predictive capabilities and discuss the value of modeling CME. We have organized these models, with a focus on bud formation, based on specific biophysical themes including identifying the role of membrane tension in CME, the spatial organization of endocytic proteins, and accounting for the heterogeneous composition of the plasma membrane (Figure 1A). Throughout our discussion we consider the following question: what is the value of the model?
Figure 1:
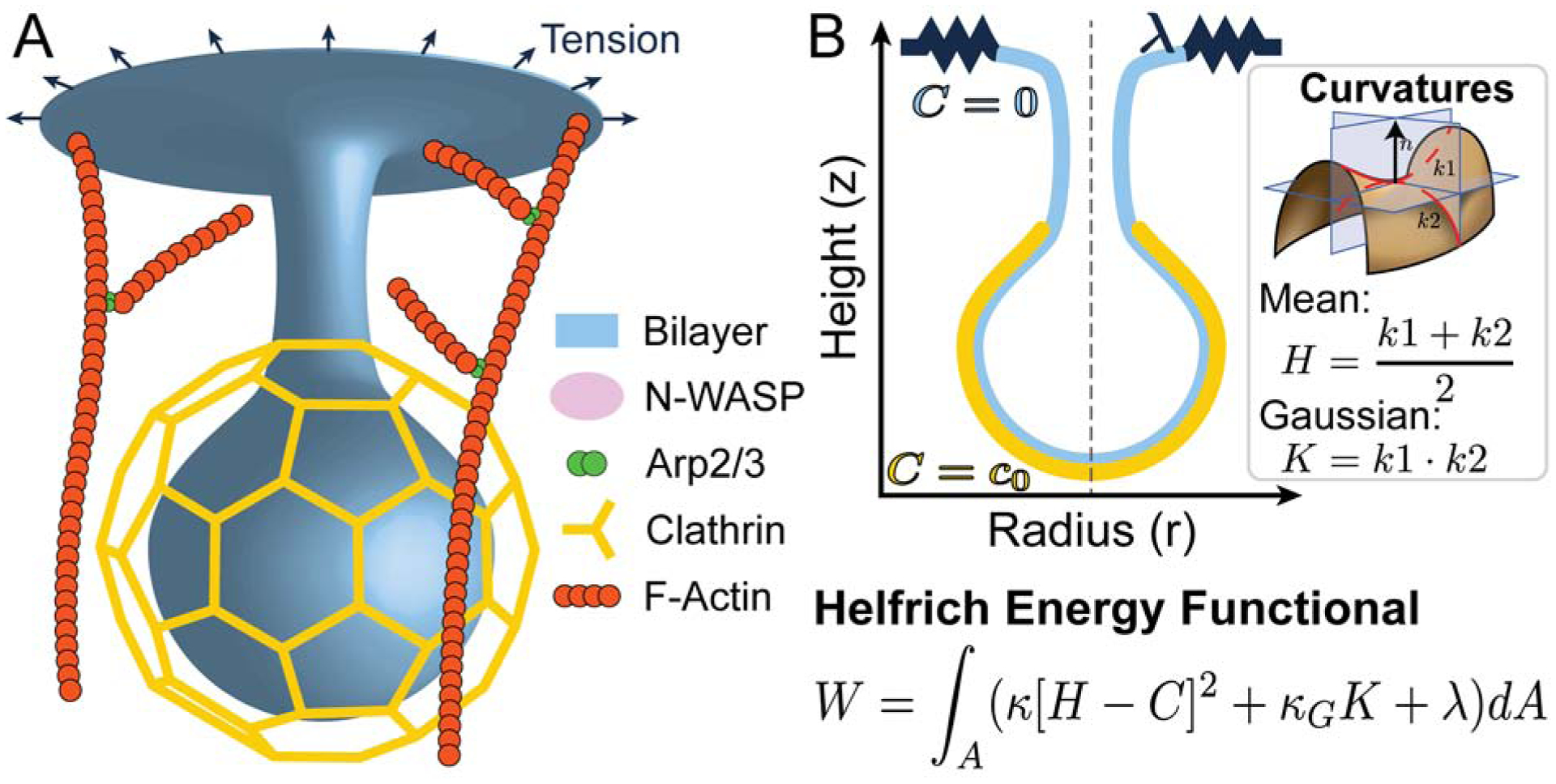
Schematic of endocytosis. A) 3D cartoon of an endocytic bud with protein machinery of interest. The localization of many of these molecules is being elucidated by advances in experimental imaging techniques. B) Equivalent 2D axisymmetric representation of the endocytic bud in A. In the Helfrich model, the contributions of the clathrin scaffold to membrane bending can be represented by a localized non-zero spontaneous curvature c0. Here W is the total energy of the membrane, k is the bending modulus, H is the mean curvature of the membrane, which is the average of the two principal curvatures, C is the spontaneous curvature, kG is the Gaussian modulus, K is the Gaussian curvature, which is the product of the two principal curvatures, λ is the membrane tension, and A is the total membrane surface area [15].
Membrane curvature generation in clathrin-mediated endocytosis - a mathematical description
As noted in a comprehensive review by Seifert [15], the mechanical models for membrane curvature generation were primarily biologically inspired physics (focused on lipid bilayer mechanics) as opposed to biophysically inspired (focused on cell membrane mechanics). Although such models make many simplifications, they lay the theoretical and computational foundations for biophysically inspired models. We first summarize the Helfrich model in the context of membrane budding in CME because it is the foundation for a vast majority of the current models of membrane curvature generation.
The principle behind the Helfrich model is as follows: while lipids can flow in the plane of the membrane, the bilayer is elastic in bending. Compared to the observed curvatures of membrane buds, the thickness of the bilayer (~ 5 nm) is very small and so the membrane can be thought of as a surface with negligible thickness. In this case, the bending energy can be written as a function of the curvatures of the surface [16, 17]. This bending energy density was proposed by Helfrich [4], Canham [3], and Evans [5] and is shown in (Figure 1B). To obtain the shape of the membrane for a given load, the bending energy must be minimized, often under the assumption is that the membrane is under mechanical equilibrium. This approach is commonly used to model buds in CME [8, 18, 12, 9, 11]; indeed one of the energy minimizing configurations that can be obtained from this model is the spherical vesicle associated with a bud in CME. We illustrate how this model has played an important role in addressing some critical phenomena in CME.
The role of membrane tension in CME
In general, the plasma membrane of animal cells is under tension as a result of in-plane stresses in the bilayer and connections between the membrane and the underlying actomyosin cortex [19, 20, 21, 22•]. Recent studies have shown that the membrane tension is heterogeneous in cells [23] and can be measured using a fluorescent sensor [22•]. Furthermore, membrane tension varies across cell types and plays an important role locally and globally [21, 23]. In CME, in particular, membrane tension plays a critical role in the progression of bud formation. Experiments have demonstrated that membrane tension opposes deformations to the membrane by curvature-generating proteins [24]. Elevated tension in combination with actin inhibitors causes clathrin-coated pits (CCPs) to exhibit longer lifetimes and stall at an open, U-shaped intermediate conformation [25, 26•, 27]. Similar observations have been made in a reconstituted system [28]. However, these observations did not explain the mechanical principles relating membrane tension to bud formation.
Using the Helfrich model described above, the role of membrane tension in regulating budding was investigated in recent studies by us and others [12, 10, 29•]. The curvature induced by the coat proteins was modeled using the spontaneous curvature term (Figure 1B). Such model representation allowed us to simulate a clathrin coat by tuning the area of the coat, spontaneous curvature, and the bending modulus of the coated region with respect to the uncoated membrane. Most of these values have been measured experimentally either in vitro or in cells [12, 18, 21, 20]. This model predicted that high tension is energetically unfavorable for bud formation, while low tension is favorable. More importantly, as the membrane tension was tuned, the model also predicted that there is a “jump” from the U-shaped bud to an Ω-shaped bud at intermediate, physiologically relevant [30], membrane tensions (Figure 2).
Figure 2:
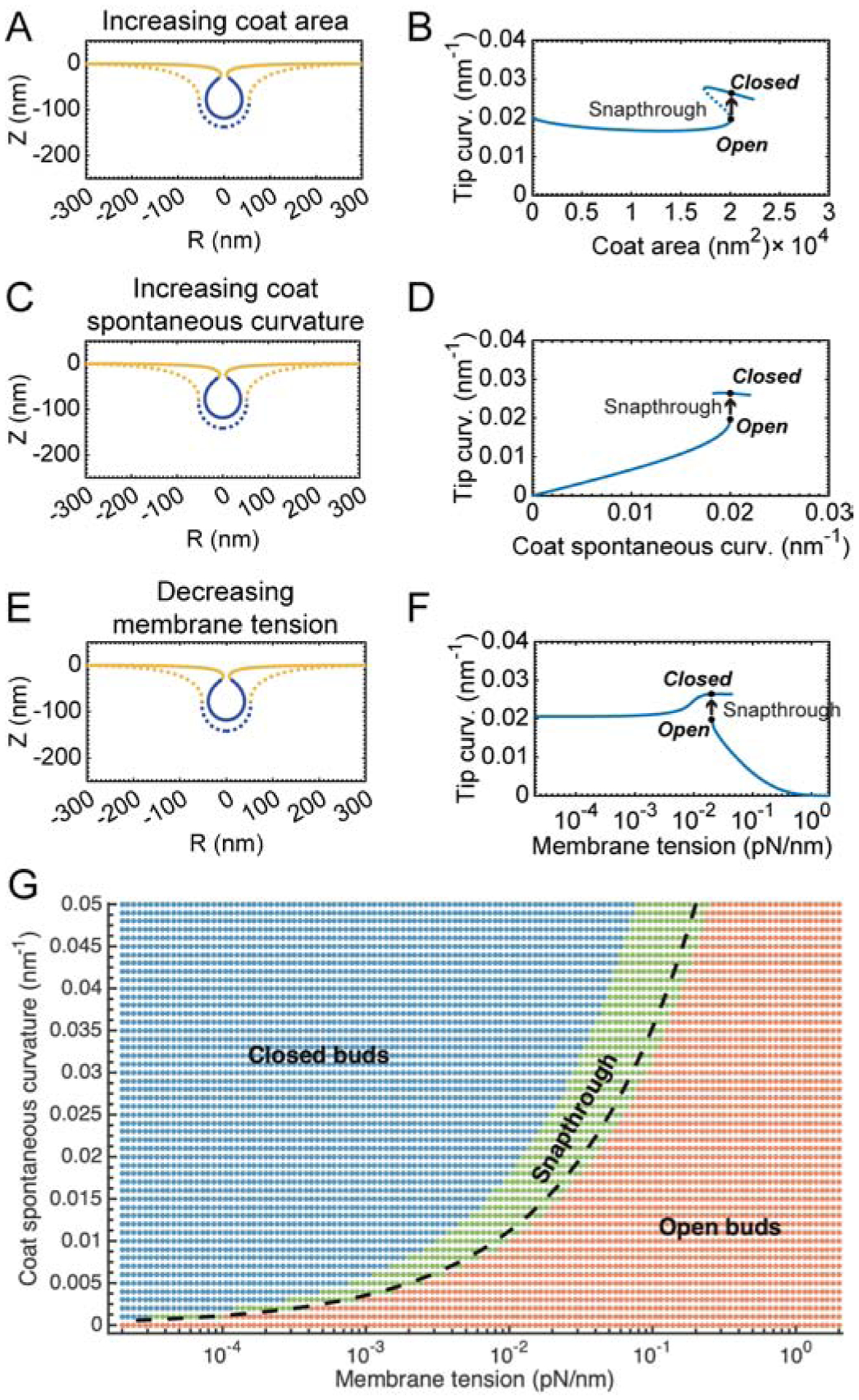
Transition from open to closed buds in CME is mediated by a snapthrough instability. This instability is observed as a function of coat area (A, B), coat curvature (C, D), and membrane tension (E,F). Membrane profiles of the bud morphology before (dashed line, Acoat = 20,065 nm2) and after (solid line, Acoat = 20,105 nm2) addition of a small amount of area to the coat, C0 = 0.02 nm−1 are shown in (A, B). Bud morphologies before (dashed line) and after (solid line) a snapthrough instability with increasing spontaneous curvature, Acoat = 20, 106 nm2, C0 = 0.02nm−1 are shown in (C, D). Bud morphology before (dashed line) and after (solid line) a snapthrough instability with decreasing membrane tension, Acoat = 20, 106 nm2, C0 = 0.02nm−1, λ0 = 0.02 pN/nm is shown in (E, F). Phase diagram showing the range of membrane tension and spontaneous curvature variations and the regimes in which the membrane shape corresponds to open and closed buds (G). The green region indicates transitions with a “jump”. Adapted from Hassinger et al. [12] with permission pending.
This “jump” can be understood as follows. There are two stable branches of solutions of the equilibrium membrane shape equations. The lower branch consists of open, U-shaped buds while the upper branch consists of closed, Ω-shaped buds. The dashed portion of the curve indicates “unstable” solutions that are not accessible by simply increasing and decreasing the area of the coat. The marked positions on the curve denote the membrane profiles shown in (Fig. 2A). The transition between these two shapes is a snapthrough instability, in which the bud “snaps” closed upon a small addition to area of the coat. In other words, a small addition to the coat area, coat curvature, or tension does thermodynamic work on the system to enable access to a closed bud configuration. Thus, a continuum description of membrane budding synthesizes multiple factors that can deform the membrane and helps us interpret the transition from the open to closed bud shape from an energetic standpoint.
The spatial organization of endocytic proteins
The above models were useful in predicting the effect of membrane tension; however, their predictive power was limited to regimes where the molecular organization of the protein composition on the membrane could be simplified in terms of a spontaneous curvature. A critical step in CME is the assembly of a multicomponent protein coat that clusters cargo and bends the membrane into a budded morphology [31, 32, 33••, 34••]. The initiation of an endocytic patch is thought to be random and likely involves fluctuation-driven molecular binding and unbinding until a critical concentration is achieved [35]. Clathrin assembles into a lattice-like cage on the membrane with the assistance of adaptor proteins which directly bind lipids and membrane receptors [36, 37, 38, 29•]. This assembly is generally thought to act as a scaffold which restricts the fluctuations of the membrane curvature, similar to a Brownian ratchet, while the adaptor proteins such as AP2 and epsin may impose membrane curvature [39, 38, 40]. Recent work suggests that other components of the coat can also contribute to membrane bending through scaffolding by F-BAR domains, amphipathic helix insertion into the bilayer, and adaptor protein crowding [41, 42, 37,43]. The BAR domain proteins associated with CME and the details of their recruitment have been identified but how they synergize with coat components and their unique role relative to isotropic coat proteins (coat proteins that induce the same curvature in both principal directions) is yet to be investigated in detail or understood. From a structural standpoint, proteins containing BAR domains can induce two different curvatures along the membrane, allowing them to form tubes or neck-like (catenoidal) structures [44].
We consider two critical questions based on the above information: how can this level of detail be incorporated in a model, and what can be learned from such modeling efforts? From a modeling perspective, the simplest way to include the contributions of the many proteins discussed above has been to combine the contributions into a single measure of the curvature generating capability of the coat, or spontaneous curvature (Table 1), with an effective strength that depends on the local protein composition, density, and area coverage [4, 45]. Even with this simplification, when the localization of this spontaneous curvature on the membrane surface is considered, we and others have shown that this local protein-induced spontaneous curvature alters the membrane tension locally [46, 45, 24]. Spontaneous curvature only accounts for the isotropic curvature on the membrane surface such that the curvature induced in both principal directions is the same [4,45], such that the protein coats result in spherical buds or vesicles [4]. The curvatures induced by BAR-domain proteins, on the other hand, can be captured by using anisotropic spontaneous curvature [47, 48]. As summarized in Table 1, the radius of curvature induced by these proteins and their molecule numbers gives us a critical set of parameters to constrain more detailed models. Going beyond the spontaneous curvature term, the energetics of membrane-protein interactions can be directly considered in a more quantitative model for curvature generation in CME [49, 50, 51, 52, 53], where the contributions to the total energy of the system can be formulated to include the energy of the membrane-protein interactions and the energy of bending the membrane. While the exact form of the interaction energy with these proteins remains to be experimentally verified, based on thermodynamic arguments, a quadratic dependence of the energy on the local protein density has been proposed [50, 54, 51, 55, 56, 57, 52]. Using these thermodynamic arguments, the protein density on the membrane (number of molecules per unit area), the curvature generation capability (both isotropic and anisotropic), binding and unbinding kinetics, and diffusion of proteins on the membrane can be modeled. Over the years, we have extended this formulation to include heterogeneity in the membrane due to proteins [58, 50], flow of lipids along the plane of the membrane [46], membrane-protein interactions [59, 60], and diffusion of proteins in the plane of the membrane [52]. These studies have highlighted how adding layers of complexity to the models can give rise to emergent properties due to the interaction of multiple aspects in CME. Therefore, these heterogeneous, dynamic features are important not only because they are one way to capture the compositional complexity of biological membranes in CME but also because heterogeneity implies that membrane tension is a local and dynamic variable [23, 58, 12, 50, 45].
Table 1:
Preferred curvature and molecule numbers of select endocytic proteins.
| Protein | Protein Class | Type of Curvature | Radius of Curvature (nm) | No. of Molecules |
|---|---|---|---|---|
| Clathrin | Coat scaffold | Isotropic | >15[61] | 120 (40 triskelia)[61] |
| FCHo | F-BAR | Anisotropic | 9–40[62] | 20 (10 dimers)[63, 64••] |
| Epsin | Coat protein | Both | 19[65] | 20[63, 64••, 66] |
| Endophilin | N-BAR | Anisotropic | 5[24] | 10–20[67] |
| Amphiphysin | N-BAR | Anisotropic | 3[68] | 10–20[69] |
| SNX9 | BAR | Anisotropic | 5–10[70] | 22–40[64••] |
| Dynamin | GTPase | Anisotropic | 10 (unconstricted) 3.5 (constricted) 1.9 (superconstricted)[73] |
20–60[71, 72] |
Estimation of material properties to account for compositional heterogeneity of the membrane
Another important parameter in membrane curvature generation in CME is the bending modulus. This bending modulus, or rigidity, of the plasma membrane is a material property of the lipid bilayer describing its resistance to bending, and is function of the molecular composition [74]. Estimates of the membrane bending modulus for specific lipid compositions range from ~10–50 kBT [75, 76]. Actin and microtubules, for reference, have bending rigidities on the order of ~2500 and ~350 000 kBT respectively (given measured rigidities of 7.3 × 10−26 Nm2 and 2.1 × 10−23 Nm2 divided by approximate filament widths of 7 nm and 25 nm for actin and microtubules respectively [77]). Although cellular membranes have a heterogeneous composition that can vary dynamically with location, cell type, and cell stage. Particularly for CME, PIP2 is important for the progression of endocytosis since it interacts with binding sites in AP2 and other proteins involved in the coat assembly [78••, 79•]. The dynamics of phosphoinositide conversion specify the site of endocytosis [78••] and have been modeled using line tension as a model parameter [80]. Lipid composition variations can thus alter the membrane bending and Gaussian moduli along the membrane.
How can models incorporate the variation in membrane physical properties? If experimental measurements exist, then the modulus can be input as a heterogeneous parameter for the simulations [12, 18]. In the absence of such measurements, one method to estimate the membrane bending modulus is to use molecular dynamics simulations [75]. Recent advances in computational power and complexity of lipid force fields now allow for the simulation of realistic bilayers with complex lipid compositions and asymmetry at atomistic and/or coarse grained detail [81]. While MD simulations can be computationally intensive, Coarse Grained Molecular Dynamics (CGMD) models such as the MARTINI force field among others, coupled with advanced simulation and analysis schemes [82, 75, 83, 76], provide a good compromise between chemical detail and computational tractability to estimate properties such as area per lipid and bending moduli [81]. Given the diversity and heterogeneity of lipids in cell membranes, modeling the individual properties of the myriad compositions will be a challenge. Scaling laws for lipid mixtures such as those proposed by Kelley et al. [84•] which related the area per lipid to the bending modulus, area compression modulus, and the viscosity of the bilayer, can help create improved approaches for extracting membrane material properties from MD simulations while helping to interpolate the material properties between known lipid compositions in large length scale models of membrane bending.
Image-based modeling for CME
Finally, we discuss an emerging avenue for image-based modeling in CME. The explosive advances in microscopy have now given us high resolution and 3D images of an endocytic site. Can such data be used to inform quantitative models of CME? Conceptually, the shape of the membrane in an endocytic site can be considered as a reporter of applied forces [59]. This concept has been used to calculate the axial forces in membrane tethers drawn from a vesicle [85] and to estimate the magnitude of the Gaussian modulus [86]. 3D reconstructions of CME [87, 88, 33••] sites can be processed using image analysis and meshing software such as GAMer 2 [89] and the curvatures of the membrane along the endocytic pit can be calculated. From these curvatures, the traction forces acting along different portions of the membrane can be calculated [59] assuming that the Helfrich model is valid. Such calculations can be coupled to the molecular mechanism either from CGMD simulations [90, 91] or experimental analysis. We note that while it is difficult to assign a reaction coordinate or a temporal scheme from still images, the progression of endocytosis can be inferred from the changes to the membrane geometry [88]. One of the advantages of using electron micrographs of membrane structures in cells is that we can now bridge the gap between membrane mechanics, curvature studies, and realistic geometries in CME.
Summary
We are the cusp of a very exciting time in cell biology - quantitative experimental biology methods can be ably supported by quantitative models, especially in CME given its state of maturity. Success in such an endeavor requires a community-wide acknowledgment of the expanding role played by models and modelers in enhancing our understanding of cellular processes.
As a community, we also need to invest in two fronts to realize the promise of model-experiment collaboration in quantitative biology. The first investment requires a wider recognition that while models are not “real”, they are necessary to understand the physical mechanisms governing these cellular processes. A predictive model generally does not include all the known molecular details; after all, the goal is not to build the entire cell in a computer. With reasonable assumptions, a well-constrained model can generate experimentally testable hypotheses and, critically, eliminate hypotheses that are not physically plausible. These two features are what make for “good” and useful models. In order to achieve such a value proposition, we need to have more conversations about the bounds for what models can predict and what experiments can measure. Establishing metrics that can be compared between models and experiments, and between models of different scales will strengthen the connection and confidence in the iterative loop between models and experiments.
The second investment needed from the community, in order to realize the opportunities identified here, is to acknowledge the jargon barrier that arises from the differences in technical language used by cell biologists and theoreticians, which can be sometimes compounded by field-specific cultural differences. It is the corresponding author’s own experience in transitioning between engineering, biology, and back to engineering that while such communication barriers do exist, with good faith effort and interdisciplinary training, such barriers can be lowered. Ultimately, such efforts will bring fresh ideas and insights and strengthen our understanding of CME and membrane trafficking.
Highlights.
Membrane curvature generation is a readout of the underlying biomechanical events
Computational models help identify the physical principles behind membrane budding
Tight connection between experiments and models will aid study of design principles
New experiments should consider how measurements can be related to model parameters
Acknowledgements
This work is supported by NIH R01-GM132106 to P.R. C.T.L. also acknowledges support from a Hartwell Foundation Postdoctoral Fellowship. M.A. is funded by NIH grant K99-GM132551. The authors would like to thank their many collaborators over the years for the exchange of interdisciplinary ideas in this field.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors declare no conflicts of interest.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- [1].Goldstein JL, Anderson RG, and Brown MS: Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature 1979, 279:679–685. [DOI] [PubMed] [Google Scholar]
- [2].Schmid SL, Sorkin A, and Zerial M: Endocytosis: Past, present, and future. Cold Spring Harb. Perspect. Biol 2014, 6:a022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Canham PB: The minimum energy of bending as a possible explanation of the biconcave shape of the human red blood cell. J. Theor Biol 1970, 26:61–81. [DOI] [PubMed] [Google Scholar]
- [4].Helfrich W: Elastic properties of lipid bilayers: theory and possible experiments. Zeitschrift für Naturforschung C 1973, 28:693–703. [DOI] [PubMed] [Google Scholar]
- [5].Evans E and Skalak R Mechanics and Thermodynamics of Biomembranes. In: Mechanics and Thermodynamics of Biomembranes. Boca Raton: CRC Press, 1980. [Google Scholar]
- [6].Kreyszig E Introduction to differential geometry and Riemannian geometry. Mathematical Expositions. Toronto, ON, Canada: University of Toronto Press, 1969. [Google Scholar]
- [7].Ciarlet PG: An introduction to differential geometry with applications to elasticity. Journal of Elasticity 2005, 78:1–215. [Google Scholar]
- [8].Liu J, Kaksonen M, Drubin DG, and Oster G: Endocytic vesicle scission by lipid phase boundary forces. Proceedings of the National Academy of Sciences 2006, 103:10277–10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Liu J, Sun Y, Drubin DG, and Oster GF: The mechnochemistry of endocytosis. PLoS Biology 2009, 7:e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Walani N, Torres J, and Agrawal A: Endocytic proteins drive vesicle growth via instability in high membrane tension environment. Proceedings of the National Academy of Sciences 2015, 112:E1423–E1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dmitrieff S and Nédélec F: Membrane Mechanics of Endocytosis in Cells with Turgor. PLoS Comput. Biol 2015, 11:e1004538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hassinger JE, Oster G, Drubin DG, and Rangamani P: Design principles for robust vesiculation in clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A 2017, 114:E1118–E1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Carlsson AE and Bayly PV: Force Generation by Endocytic Actin Patches in Budding Yeast. Biophysical Journal 2012, 106:1596–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Akamatsu M, Vasan R, Serwas D, Ferrin MA, Rangamani P, and Drubin DG: Principles of Self-Organization and Load Adaptation by the Actin Cytoskeleton during Clathrin-Mediated Endocytosis. eLife 2020, 9:e49840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Seifert U: Configurations of fluid membranes and vesicles. Advances in physics 1997, 46:13–137. [Google Scholar]
- [16].Steigmann D: Fluid films with curvature elasticity. Archive for Rational Mechanics and Analysis 1999, 150:127–152. [Google Scholar]
- [17].Deserno M: Fluid lipid membranes: from differential geometry to curvature stresses. Chem. Phys. Lipids 2015, 185:11–45. [DOI] [PubMed] [Google Scholar]
- [18].Walani N, Torres J, and Agrawal A: Endocytic proteins drive vesicle growth via instability in high membrane tension environment. Proceedings of the National Academy of Sciences of the United States of America 2015, 112:E1423–E1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hochmuth F, Shao JY, Dai J, and Sheetz MP: Deformation and flow of membrane into tethers extracted from neuronal growth cones. Biophysical journal 1996, 70:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Diz-Muñoz A, Fletcher D, and Weiner OD: Use the force: membrane tension as an organizer of cell shape and motility. Trends in Cell Biology 2013, 23:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sitarska E and Diz-Muñoz A: Pay attention to membrane tension: Mechanobiology of the cell surface. Current opinion in cell biology 2020, 66:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22•].Colom A, Derivery E, Soleimanpour S, Tomba C, Molin MD, Sakai N, González-Gaitán M, Matile S, and Roux A: A Fluorescent Membrane Tension Probe. Nature Chem 2018, 10:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper presents a fluorescent probe for membrane tension measurement in cells.
- [23].Shi Z, Graber ZT, Baumgart T, Stone HA, and Cohen AE: Cell Membranes Resist Flow. Cell 2018, 175:1769. –1779.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shi Z and Baumgart T: Membrane tension and peripheral protein density mediate membrane shape transitions. Nature communications 2015, 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Boulant S, Kural C, Zeeh JC, Ubelmann F, and Kirchhausen T: Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nature Cell Biology 2011, 13:1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26•].Ferguson JP, Huber SD, Willy NM, Aygün E, Goker S, Atabey T, and Kural C: Mechanoreg-ulation of clathrin-mediated endocytosis. J. Cell Sci 2017, 130:3631–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elegant experimental study that used different methods to manipulate membrane tension and showed that endocytosis depends on local membrane tension.
- [27].Yarar D, Waterman-Storer CM, and Schmid SL: A dynamic actin cytoskeleton functions at multiple stages of clathrin-mediated endocytosis. Mol. Biol. Cell 2005, 16:964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Saleem M, Morlot S, Hohendahl A, Manzi J, Lenz M, and Roux A: A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nature Communications 2015, 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29•].Bucher D, Frey F, Sochacki KA, Kummer S, Bergeest JP, Godinez WJ, Kräusslich HG, Rohr K, Taraska JW, Schwarz US, et al. : Clathrin-adaptor ratio and membrane tension regulate the flat-to-curved transition of the clathrin coat during endocytosis. Nat. Commun 2018, 9:1109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper uses a combination of correlative electron and light microscopy and mathematical models of growth laws to identify the flat-to-curved transition of clathrin coats.
- [30].Sens P and Plastino J: Membrane tension and cytoskeleton organization in cell motility. Journal of Physics: Condensed Matter 2015, 27:273103. [DOI] [PubMed] [Google Scholar]
- [31].Kaksonen M, Sun Y, and Drubin DG: A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 2003, 115:475–487. [DOI] [PubMed] [Google Scholar]
- [32].Kaksonen M, Toret CP, and Drubin DG: A Modular Design for the Clathrin- and Actin-Mediated Endocytosis Machinery. Cell 2005, 123:305–320. [DOI] [PubMed] [Google Scholar]
- [33••].Mund M, van der Beek JA, Deschamps J, Dmitrieff S, Hoess P, Monster JL, Picco A, Nédélec F, Kaksonen M, and Ries J: Systematic Nanoscale Analysis of Endocytosis Links Efficient Vesicle Formation to Patterned Actin Nucleation. Cell 2018, 174:884–896.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper uses high-throughput super resolution imaging to identify the spatial locations of 23 proteins and finds that they organize themselves into distinct radial zones. They used this experimental data to inform a computational model of actin dynamics at endocytic sites to calculate the dynamics of internalization.
- [34••].Skruzny M, Pohl E, Gnoth S, Malengo G, and Sourjik V: The protein architecture of the endocytic coat analyzed by FRET microscopy. Mol. Syst. Biol 2020, 16:e9009. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses FRET microscopy to identify the organization of endocytic proteins in the radial and normal direction to the membrane.
- [35].Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, and Kirchhausen T: Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 2004, 118:591–605. [DOI] [PubMed] [Google Scholar]
- [36].McMahon HT and Boucrot E: Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nature reviews. Molecular Cell Biology 2011, 12:517–533. [DOI] [PubMed] [Google Scholar]
- [37].Kirchhausen T, Owen D, and Harrison SC: Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harbor perspectives in biology 2014, 6:a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sochacki KA, Dickey AM, Strub MP, and Taraska JW: Endocytic proteins are partitioned at the edge of the clathrin lattice in mammalian cells. Nat. Cell Biol 2017, 19:352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dannhauser P and Ungewickell E: Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nature Cell Biology 2012, 14:634–9. [DOI] [PubMed] [Google Scholar]
- [40].Frey F, Bucher D, Sochacki KA, Taraska JW, Boulant S, and Schwarz US: Eden growth models for flat clathrin lattices with vacancies. New J. Phys 2020, 22:073043. [Google Scholar]
- [41].Ford M, Mills I, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, and McMahon HT: Curvature of clathrin-coated pits driven by epsin. Nature 2002, 419:361–366. [DOI] [PubMed] [Google Scholar]
- [42].Stachowiak J, Schmid E, Ryan C, Ann HS, Sasaki D, Sherman M, Geissler P, Fletcher D, and Hayden C: Membrane bending by protein-protein crowding. Nature Cell Biology 2012, 14:944–949. [DOI] [PubMed] [Google Scholar]
- [43].Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM, and Stachowiak JC: Intrinsically disordered proteins drive membrane curvature. Nat Communications 2015, 6: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Simunovic M, Voth GA, Callan-Jones A, and Bassereau P: When Physics Takes Over: BAR Proteins and Membrane Curvature. Trends Cell Biol. 2015, 25:780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lipowsky R: Spontaneous tubulation of membranes and vesicles reveals membrane tension generated by spontaneous curvature. Faraday discussions 2013, 161:305–331. [DOI] [PubMed] [Google Scholar]
- [46].Rangamani P, Agrawal A, Mandadapu KK, Oster G, and Steigmann DJ: Interaction between surface shape and intra-surface viscous flow on lipid membranes. Biomechanics and modeling in mechanobiology 2013, 12:833–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Iglič A, Babnik B, Gimsa U, and Kralj-Iglič V: On the role of membrane anisotropy in the beading transition of undulated tubular membrane structures. J. Phys. A Math. Gen 2005, 38:8527. [Google Scholar]
- [48].Kabaso D, Bobrovska N, Góźdź W, Gov N, Kralj-Iglič V, Veranič P, and Iglič A: On the role of membrane anisotropy and BAR proteins in the stability of tubular membrane structures. J. Biomech 2012, 45:231–238. [DOI] [PubMed] [Google Scholar]
- [49].Agrawal A and Steigmann DJ: A model for surface diffusion of trans-membrane proteins on lipid bilayers. Z. Angew. Math. Phys. (ZAMP) 2011, 62:549–563. [Google Scholar]
- [50].Chabanon M and Rangamani P: Gaussian curvature directs the distribution of spontaneous curvature on bilayer membrane necks. Soft Matter 2018, : [DOI] [PubMed] [Google Scholar]
- [51].Gov NS: Guided by curvature: shaping cells by coupling curved membrane proteins and cytoskeletal forces. Philos. Trans. R. Soc. Lond. B Biol. Sci 2018, 373: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mahapatra A, Saintillan D, and Rangamani P: Transport Phenomena in Fluid Films with Curvature Elasticity. J. Fluid Mech 2020, 905: [Google Scholar]
- [53].Molina RR, Liese S, Alimohamadi H, Rangamani P, and Carlson A: Diffuso-Kinetic Membrane Budding Dynamics. Soft Matter 2020, : [DOI] [PubMed] [Google Scholar]
- [54].Steigmann DJ. Mechanics and Physics of Lipid Bilayers. In: The Role of Mechanics in the Study of Lipid Bilayers. Ed. by Steigmann DJ. Cham: Springer International Publishing, 2018, 1–61. [Google Scholar]
- [55].Landau LD and Lifshitz EM. Course of Theoretical Physics. en. Elsevier, 2013. [Google Scholar]
- [56].Veksler A and Gov NS: Phase transitions of the coupled membrane-cytoskeleton modify cellular shape. Biophys. J 2007, 93:3798–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Argudo D, Bethel NP, Marcoline FV, Wolgemuth CW, and Grabe M: New Continuum Approaches for Determining Protein-Induced Membrane Deformations. Biophys.J 2017, 112:2159–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rangamani P, Mandadapu K, and Oster G: Protein-induced membrane curvature alters local membrane tension. Biophysical Journal 2014, 107:751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Alimohamadi H, Vasan R, Hassinger JE, Stachowiak J, and Rangamani P: The role of traction in membrane curvature generation. Mol. Biol. Cell 2018, 29:2024–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Alimohamadi H, Ovryn B, and Rangamani P Modeling membrane nanotube morphology: the role of heterogeneity in composition and material properties. en. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cheng Y, Boll W, Kirchhausen T, Harrison SC, and Walz T: Cryo-electron tomography of clathrin-coated vesicles: structural implications for coat assembly. Journal of molecular biology 2007, 365:892–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, and McMahon HT: FCHo proteins are nucleators of clathrin-mediated endocytosis. Science 2010, 328:1281–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Borner GH, Antrobus R, Hirst J, Bhumbra GS, Kozik P, Jackson LP, Sahlender DA, and Robinson MS: Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J Cell Biol 2012, 197:141–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64••].Schöneberg J, Lehmann M, Ullrich A, Posor Y, Lo WT, Lichtner G, Schmoranzer J, Haucke V, and Noé F: Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nature communications 2017, 8:15873. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using a combination of super-resolution imaging and computational modeling, this study shows how PIP2 synthesis and the PX-BAR domain protein SNX9 are coupled and lead to membrane constriction.
- [65].Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, and McMahon HT: Curvature of clathrin-coated pits driven by epsin. Nature 2002, 419:361. [DOI] [PubMed] [Google Scholar]
- [66].Lai CL, Jao CC, Lyman E, Gallop JL, Peter BJ, McMahon HT, Langen R, and Voth GA: Membrane binding and self-association of the epsin N-terminal homology domain. J. Mol. Biol 2012, 423:800–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bertot L, Grassart A, Lagache T, Nardi G, Basquin C, Olivo-Marin JC, and Sauvonnet N: Quantitative and statistical study of the dynamics of clathrin-dependent and-independent endocytosis reveal a differential role of EndophilinA2. Cell Reports 2018, 22:1574–1588. [DOI] [PubMed] [Google Scholar]
- [68].Sorre B, Callan-Jones A, Manzi J, Goud B, Prost J, Bassereau P, and Roux A: Nature of curvature coupling of amphiphysin with membranes depends on its bound density. Proceedings of the National Academy of Sciences 2012, 109:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Meinecke M, Boucrot E, Camdere G, Hon WC, Mittal R, and McMahon HT: Co-operative recruitment of dynamin and BIN/amphiphysin/Rvs (BAR) domain-containing proteins leads to GTP-dependent membrane scission. Journal of Biological Chemistry 2013, 288:6651–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wang Q, Kaan HYK, Hooda RN, Goh SL, and Sondermann H: Structure and plasticity of Endophilin and Sorting Nexin 9. Structure 2008, 16:1574–1587. [DOI] [PubMed] [Google Scholar]
- [71].Grassart A, Cheng AT, Hong SH, Zhang F, Zenzer N, Feng Y, Briner DM, Davis GD, Malkov D, and Drubin DG: Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J. Cell. Biol 2014, 205:721–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Cocucci E, Gaudin R, and Kirchhausen T: Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Molecular biology of the cell 2014, 25:3595–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sundborger AC, Fang S, Heymann JA, Ray P, Chappie JS, and Hinshaw JE: A dynamin mutant defines a superconstricted prefission state. Cell reports 2014, 8:734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Dimova R: Recent developments in the field of bending rigidity measurements on membranes. Advances in colloid and interface science 2014, 208:225–234. [DOI] [PubMed] [Google Scholar]
- [75].Venable RM, Brown FLH, and Pastor RW: Mechanical Properties of Lipid Bilayers from Molecular Dynamics Simulation. Chemistry and Physics of Lipids 2015, 192:60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Eid J, Razmazma H, Jraij A, Ebrahimi A, and Monticelli L: On Calculating the Bending Modulus of Lipid Bilayer Membranes from Buckling Simulations. J. Phys. Chem. B 2020, 124:6299–6311. [DOI] [PubMed] [Google Scholar]
- [77].Gittes F, Mickey B, Nettleton J, and Howard J: Flexural Rigidity of Microtubules and Actin Filaments Measured from Thermal Fluctuations in Shape. J Cell Biol 1993, 120:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78••].He K, Marsland R, Upadhyayula S, Song E, Dang S, Capraro BR, Wang W, Skillern W, Gaudin R, Ma M, et al. : Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature 2017, 1298:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors developed ‘coincidence detector’ sensors and report the phosphoinositide composition of the lipid bilayer in an endocytic site. They further track the composition as a function of endocytic progression and show that distinct stages such as vesicle closure are related to specific phosphoinositide conversion.
- [79•].Garcia-Alai MM, Heidemann J, Skruzny M, Gieras A, Mertens HDT, Svergun DI, Kaksonen M, Uetrecht C, and Meijers R: Epsin and Sla2 form assemblies through phospholipid interfaces. Nat. Commun 2018, 9:328. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses mass spectrometry to show that ENTH and ANTH domain containing proteins form clusters by sharing PIP2 molecules and that this assembly could be relevant for endocytosis.
- [80].Liu J, Kaksonen M, Drubin DG, and Oster G: Endocytic vesicle scission by lipid phase boundary forces. Proceedings of the National Academy of Sciences of the United States of America 2006, 103:10277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Marrink SJ, Corradi V, Souza PC, Ingólfsson HI, Tieleman DP, and Sansom MS: Computational Modeling of Realistic Cell Membranes. Chem. Rev 2019, 119:6184–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hu M, Briguglio JJ, and Deserno M: Determining the Gaussian curvature modulus of lipid membranes in simulations. Biophys. J 2012, 102:1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Doktorova M, Harries D, and Khelashvili G: Determination of Bending Rigidity and Tilt Modulus of Lipid Membranes from Real-Space Fluctuation Analysis of Molecular Dynamics Simulations. Phys. Chem. Chem. Phys 2017, 19:16806–16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84•].Kelley EG, Butler PD, Ashkar R, Bradbury R, and Nagao M: Scaling Relationships for the Elastic Moduli and Viscosity of Mixed Lipid Membranes. PNAS 2020, 117:23365–23373. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors measure the elastic and viscous properties of lipid membranes as a function of their composition and identify scaling laws that relate the area per lipid and these material properties.
- [85].Lee HJ, Peterson EL, Phillips R, Klug WS, and Wiggins PA: Membrane shape as a reporter for applied forces. Proceedings of the National Academy of Sciences 2008, 105:19253–19257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Baumgart T, Das S, Webb W, and Jenkins J: Membrane elasticity in giant vesicles with fluid phase coexistence. Biophysical journal 2005, 89:1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Avinoam O, Schorb M, Beese CJ, Briggs JA, and Kaksonen M: Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science 2015, 348:1369–1372. [DOI] [PubMed] [Google Scholar]
- [88].Kukulski W, Schorb M, Kaksonen M, and Briggs JA: Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell 2012, 150:508–520. [DOI] [PubMed] [Google Scholar]
- [89].Lee CT, Laughlin JG, Beaumelle NAdL, Amaro RE, McCammon JA, Ramamoorthi R, Holst M, and Rangamani P: 3D Mesh Processing Using GAMer 2 to Enable Reaction-Diffusion Simulations in Realistic Cellular Geometries. PLOS Computational Biology 2020, 16:e1007756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Goetz R and Lipowsky R: Computer simulations of bilayer membranes: Self-assembly and interfacial tension. J. Chem. Phys 1998, 108:7397–7409. [Google Scholar]
- [91].Vanegas JM, Torres-Sánchez A, and Arroyo M: Importance of Force Decomposition for Local Stress Calculations in Biomembrane Molecular Simulations. J. Chem. Theory Comput 2014, 10:691–702. [DOI] [PubMed] [Google Scholar]


