Abstract
Therapeutic efficiency and toxicity are two of the three critical factors in molecular therapy and pharmaceutical drug development. Specific tumor targeting and rapid renal excretion contribute to improving efficiency and reducing toxicity. We recently found that RNA nanoparticles display rubber-like properties, enabling them to deliver therapeutics to cancer with high efficiency. Off-target RNA nanoparticles were rapidly cleared by renal excretion, resulting in nontoxicity. However, previous biodistribution studies relied mainly on fluorescent markers, which can cause interference from fluorophore quenching and auto-fluorescence. Thus, the quantification of biodistribution requires further scrutiny. In this study, radionuclide [3H] markers were used for quantitative pharmacokinetic (PK) studies to elucidate the favorable PK profile of RNA nanoparticles. Approximately 5% of [3H]-RNA nanoparticles accumulated in tumors, in contrast to the 0.7% tumor accumulation reported in the literature for other kinds of nanoparticles. The amount of [3H]-RNA nanoparticles accumulated in tumors was higher than that in the liver, heart, lung, spleen, and brain throughout the entire process after IV injection. [3H]-RNA nanoparticles rapidly reached the tumor vasculature within 30 min and remained in tumors for more than 2 days. Nontargeting [3H]-RNA nanoparticles were found in the urine 30 min after IV injection without degradation and processing, and more than 55% of the IV-injected radiolabeled RNA nanoparticles were cleared from the body within 12 h, while the other 45% includes the radiative counts that cannot be recovered due to whole-body distribution and blood dilution after intravenous injection. The high specificity of tumor targeting, fast renal excretion, and low organ accumulation illustrate the high therapeutic potential of RNA nanoparticles in cancer treatment as efficient cancer-targeting carriers with low toxicity and side effects.
Keywords: RNA nanoparticles, tritium labeling, quantitative biodistribution
Graphical Abstract
INTRODUCTION
RNA nanotechnology allows the construction of programmable nanostructures by exploiting intra- and intermolecular interactions, enabling the design of complex structures with well-defined dimensions, shapes, and sizes.1–7 The advancement of RNA nanotechnology in recent decades has demonstrated the uniqueness of RNA nanoparticles in biomedical applications.2,3,7–10 In particular, the three-way junction (3WJ) originating from the packaging RNA (pRNA) of the phi29 DNA packaging motor has been engineered to incorporate various functional modules, including imaging probes, therapeutic agents, and targeting ligands.9–12 The constructed RNA nanoparticles have been extensively explored for in vivo cancer imaging and therapeutic drug delivery.4,8,13–17
Targeted drug delivery can increase therapeutic efficacy and reduce toxicity. A favorable pharmacological profile is necessary for an ideal targeted delivery platform. However, after surveying publications in the past 10 years and averaging the widely disparate data, it was reported that only 0.7% of the administered nanoparticles were delivered to solid tumors.18 Concerns have been raised regarding the clinical translation potential of nanotechnology because of low efficacy, undesirable side effects, and toxicity.18 Recently, using optical tweezers and in vivo imaging technologies, we found that RNA nanoparticles possess rubber-like properties.19 RNA nanoparticles were stretchable and shrinkable by optical tweezers with multiple repeated extensions and relaxations, similar to rubber. These findings explain two advantages of RNA nanotechnology: (1) RNA nanoparticles have high tumor targeting efficiency since their rubber- or amoeba-like deformation ability enables them to squeeze out of the leaky vasculature to improve the enhanced permeability and retention (EPR) effect;20–22 (2) RNA nanoparticles remain nontoxic since they can be cleared from the body via renal excretion into the urine with little accumulation in the body.19,23 Evidence from in vivo fluorescence imaging studies has demonstrated the target specificity of nanoparticles in various tumor models, such as breast cancer, glioblastoma, gastric cancer, and prostate cancer.9,24–27 However, a quantitative biodistribution profile is still challenging to determine due to the limitations of fluorophore label stability, autofluorescence, and quenching effects.28,29 To date, few studies have been reported to exploit radionucleotides as a more sensitive and quantitative imaging tool for RNA nanoparticles, limiting our understanding of their in vivo performance. Therefore, it is helpful to develop radioactive marker-labeled RNA nanoprobes for more sensitive and accurate bioimaging and pharmacokinetic (PK) studies.
In this study, we investigated two RNA nanoparticles harboring different targeting ligands, small molecules, and nucleic acid aptamers by measuring the radioactivity from harvested organs as well as by fluorescence imaging in vivo. Folic acid (FA) is a high-affinity binding ligand of an endogenous folate receptor that is upregulated in many types of cancer cells.30–32 The CL4 RNA aptamer is a nucleic acid aptamer with high affinity and specificity to EGFR, which is overexpressed in triple-negative breast cancer mouse xenografts.33–36 3WJ nanoparticles harboring these two targeting ligands were labeled with [3H] to elucidate the quantitative biodistribution of RNA nanoparticles.
For the first time, we quantitatively determined the biodistribution profile of RNA nanoparticles. The quantitative biodistribution study revealed significantly high tumor uptake and tumor retention, providing insights into the promising anticancer efficacy. The study also proved RNA nanoparticles to have low toxicity and limited accumulation in healthy metabolic organs. Our results demonstrate that RNA nanoparticles with favorable biodistribution profiles are an ideal delivery platform for tumor-specific drug delivery and gene therapy.
MATERIALS AND METHODS
RNA Synthesis.
Solid-phase RNA oligomer synthesis was performed on a 1 μmol scale. An automated oligo synthesizer ASM-800E from Biosset was used to start the synthesis from a universal 1000 Å long-chain amino alkyl-controlled pore glass (LCAA-CPG) solid support. Coupling efficiency was monitored after removal of the dimethoxytrityl (DMT) protecting groups. Sequences of the RNA oligomers used are listed in a 5′ to 3′ orientation (uppercase denotes a 2′-OH base; lowercase denotes a 2′-F modified base).
3WJa: 5′- uuG ccA uGu GuA uGu GGG -3′
3WJb: 5′- ccc AcA uAc uuu Guu GAu ccu uuG Cga cuG Guu Acc cGG ucG-3′
3WJb-FA: 5′- ccc AcA uAc uuu Guu GAu cc (Folic Acid) -3′
3WJb-CL4: 5′- ccc AcA uAc uuu Guu GAu ccG ccu uAG uAA cGu Gcu uuG AuG ucG Auu cGA cGA GAG Gc -3′
3WJc: 5’- GGA ucA Auc AuG GcA A -3′
3WJc-AFdye 647: 5′- GGA ucA Auc AuG GcA A(C6-NH) (AFdye 647) -3′
Each strand was synthesized using 2′-fluorinated pyrimidine. Following synthesis, oligomers were cleaved from beads and deprotected in a 1:1 mixture (v/v) of ammonium hydroxide and methylamine (AMA) solution at room temperature for 2 h. The 2′-TBDMS protecting groups were removed by triethylamine trihydrofluoride (TEA·3HF) followed by desalting using a Glen Gel-Pak desalting column. The collected fraction was dried under speed vacuum. Then, the synthesized strands were analyzed by 8 M urea 16% denaturing polyacrylamide gel electrophoresis (PAGE). 3WJc-FA was provided from Exonano RNA Pharmaceutical Co., Ltd. (Columbus, OH).
Radiolabeling of Oligonucleotides by Hydrogen-Tritium Exchange.
Radiolabeling of oligonucleotides was performed using a published procedure.36 Briefly, 1 mg of HPLC-purified oligonucleotide was dissolved in 20 μL of reaction buffer (50 mM sodium phosphate, 0.1 mM EDTA, pH 7.8) and dried in speed vacuum. The oligonucleotide was then resuspended in 20 μL of tritiated water (Moravek, molar activity: 1 Ci/g) containing 0.83 μL of 2-mercaptoethanol (beta-ME) and incubated at 90 °C for 6 h. Following incubation, the sample was dried to remove unexchanged tritiated water. The sample was resuspended in 100 μL of water and incubated at room temperature for 1 h followed by two cycles of dry and resuspension in 100 μL of diethylpyrocarbonate (DEPC) H2O. The oligonucleotide was applied to the 0.2 mL desalting column (Glen Gel-Pak) for final purification. The oligonucleotide was washed and eluted with 200 μL of DEPC water and dried for further evaluation.
Fluorophore Conjugation on RNA.
NHS ester of AFDye 647 was purchased from Chematech and Click Chemistry Tools. Conjugation reactions were carried out by mixing a 1:10 molar ratio of amine-labeled 3WJc and NHS ester in 0.1 M sodium bicarbonate buffer (pH 8.5). The mixture was incubated at a dark room temperature for 16 h, allowing for conjugation reactions. The reaction mixtures were ethanol precipitated, washed twice with 75% ethanol to remove the unreacted fluorophore, and purified by HPLC.
Reverse-Phase HPLC Purification of the RNA-AFDye 647 Strand.
Fluorophore-conjugated RNA was purified by ion-pairing reverse phase HPLC. The gradient mobile phase was used to separate dye-modified RNA from unmodified strands. Buffer A was 0.1 M triethylamine acetate (TEAA) (Glen Research) in water, and buffer B was 0.1 M TEAA in 75% acetonitrile and 25% water. All purifications were performed on an Agilent PLRP-S column (Agilent cat. #: PL1512-5500) using an Agilent 1260 Infinity HPLC system. A flow rate of 1.0 mL/min was used throughout all HPLC methods, and absorbance was monitored at 260 nm (RNA) and 650 nm (Cy5). Fractions were collected and dried under vacuum and then resuspended in DEPC water. Urea (8 M) 16% denaturing PAGE was done to characterize the purified strands.
RNA Nanoparticle Assembly and Purification.
An equimolar ratio of composite strands (3WJa, 3WJb, and 3WJc) was mixed in TBE buffer (100 mM Tris-borate, 2 μM EDTA, pH 7.4). By denaturing the strands at 95 °C, the mixture solution was slowly cooled down to 4 °C at a rate of −2 °C/min. The assembled nanoparticles were desalted using a Sephadex G10 column to remove salts and small-molecule contaminants.
Dynamic Light Scattering Measurements of RNA Nanoparticles.
Apparent hydrodynamic sizes and zeta potentials of RNA nanoparticles were characterized using a Zetasizer nano-ZS (Malvern Instrument, Malvern, UK). The RNA nanoparticles were measured at 50 μmol/L in PBS buffer (137 mM NaCl, 2.7 mM KCl, 100 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) at 25 °C.
Temperature Gradient Gel Electrophoresis (TGGE) Characterization.
The RNA nanoparticles were run in a 10% (w/v) native PAGE in TMS buffer (50 mM Tris, 100 mM NaCl, and 0.2 mM MgCl2, pH 8.0) at 100 V for 10 min at room temperature. TGGE analysis was then done as previously reported.37 A gradient temperature (40–80 °C) was applied perpendicular to the electrical current, and the electrophoresis was run for 1 h. Next, the gel was stained by ethidium bromide and visualized using a Typhoon (GE Healthcare). Quantified values of bands for each nanoparticle from ImageJ were divided by the sum of the total values in corresponding lanes. Melting curves were plotted with quantified data points using GraphPad Prism 8. Tm values were defined as the temperature at which 50% of the RNA nanoparticles dissociated.
Tm Measurement by Quantitative RT-PCR.
The RNA nanoparticles were aliquoted to a 96-well plate at a 10 μM final concentration. SYBR Green II dye (Invitrogen) was added to each well as a reporter molecule for RNA nanoparticles formation and dissociation. Samples were heated to 95 °C for 5 min to denature the RNA nanoparticles followed by a slow cooling ramp to 20 °C at a rate of 0.11 °C per second as a reannealing process. Annealing curves of RNA nanoparticles were characterized using a Roche LightCycler 480 RT-PCR machine and were plotted with data points using GraphPad 8. Tm was determined by LightCycler 480 Software using the first derivative of the annealing profile.
Serum Stability Assay.
The RNA nanoparticles were incubated at a concentration of 1 μM in a cell culture medium containing 20% (v/v) fetal bovine serum (FBS) at 37 °C for different time points. The samples were collected and treated with 10% SDS for 10 min followed by examination using a 2% agarose gel run, stained with EB, and visualized using the Typhoon (GE Healthcare). Quantification analysis of the RNA band was performed using ImageJ software. The percentage of intact nanoparticles (intensity of the band at a time point/intensity of the band at 0 h for each time point) was calculated accordingly. A degradation curve was plotted with quantified data points using GraphPad Prism 8.
Radioactive Marker Stability Assay.
For each RNA sample, 100 μL of 20 μM RNA was dried and resuspended with 100 μL of PBS buffer and RPMI-1640 medium (Gibco) with 10% FBS. At varying time points, a 10 μL sample was collected and assayed by liquid scintillation counting (LSC). In a size exclusion chromatography assay, RNA nanoparticles and controls were applied to a 1 × 10 cm column containing Sephadex G10 (Sigma-Aldrich). Oligonucleotide was eluted with DEPC water at a flow rate of 0.5 mL/min; 50 μL of each fraction was collected in 96-well plates. The fluorescence was assayed using a plate reader. Next, each fraction was pipetted to a scintillation counting vial and assayed by LSC (Beckman Coulter LS6500).
Cell Culture.
MDA-MB-231 cells from human breast cancer cell lines were grown and cultured in a DMEM/F-12 medium (Invitrogen, Grand Island, NY) containing both 10% (v/v) FBS in a 37 °C incubator with a 5% CO2 and a humidified atmosphere. KB cells from human cancer cell lines were grown and cultured in an RPMI 1640 medium (Gibco) containing both 10% (v/v) FBS in a 37 °C incubator with a 5% CO2 and a humidified atmosphere.
Flow Cytometry Assay of pRNA-3WJ Nanoparticle Binding Affinity.
KB and MDA-MB-231 cells were cultured on 24-well plates at 37 °C overnight. AFDye 647-labeled 3WJ nanoparticles at concentrations from 50 to 800 nM were incubated with 2 × 105 cells KB and MDA-MB-231 cells, respectively, at 37 °C for 2 h. The cells were washed by PBS buffer twice and then trypsinized and resuspended in PBS buffer. Flow cytometry analysis was performed to compare the binding affinity and efficacy of the RNA nanoparticles to different cells. The data was analyzed by FlowJo 7.6.1 software.
Confocal Microscopy Imaging.
KB and MDA-MB-231 cells were cultured on glass cover slips at 37 °C overnight followed by treatment with AFDye 647-labeled RNA nanoparticles at a 100 nM concentration for 1 h at 37 °C. The glass slips were washed twice with PBS buffer followed by fixation using 4% formaldehyde. Triton X-100 (0.1%) (Sigma-Aldrich) in PBS buffer was applied to treat the slips for 5 min followed by cytoskeleton staining using Alexa Fluor 488 phalloidin (ThermoFisher Scientific) for 30 min at room temperature. The cells were then mounted with a ProLong Gold Antifade Reagent (Life Technologies Corp.) containing DAPI for cell nucleus staining. The confocal imaging was performed on an Olympus FV3000 confocal microscope (Olympus Corp.). Data were acquired using a Fluoview FV31S-SW.
Cellular Uptake of Tritium-Labeled RNA Nanoparticles.
KB cells and MDA-MB-231 cells were seeded on a 24-well plate at 70% confluence 1 day prior to the treatment. Two hundred microliters of 100 nM [3H]-labeled RNA nanoparticles was added to cells, and experiments were performed by incubating the plate at 37 °C for 8 h. Cells were washed 3 times with PBS buffer to remove the unbound RNA and trypsinized at serial time points. Cells were pelleted by centrifugation for 5 min at 2500g. The pellet was removed from the tube. Scintillation counting was performed.
In Vivo Tritium PK/Biodistribution Assay.
To compare the biodistribution profile, a fluorescence imaging study and a scintillation counting assay were performed. When tumor sizes reach 400 mm3, a total of 100 μL of 20 μM tritium and AFDye 647-labeled 3WJ-FA, 3WJ-CL4, and 3WJ nanoparticles were injected into mice bearing KB and MDA-MB-231 tumors by tail vein injection. PBS-injected mice were used as negative controls. Blood samples were collected via cardio puncture at different time points (5 min and 1, 2, 4, 8, 12, and 24 h). Pharmacokinetic parameters were calculated using noncompartmental analysis with the software package WinNonlin 8.1 (Phoenix).
The mice were then sacrificed by the inhalation of CO2 followed by cervical dislocation. Major organs including the brain, heart, lungs, liver, spleen, and kidneys together with tumors were collected and subjected to fluorescence imaging using an IVIS Spectrum station (Caliper Life Sciences) with excitation 640 nm and emission at 680 nm for the assessment of fluorescence biodistribution profiles. Organs and tumors were then dissolved using SOLVABLE (Perkin Elmer) following the official scintillation counting preparation method and subjected to scintillation counting for the assessment of radiation biodistribution profiles.
RESULTS
Construction of RNA Nanoparticles.
We investigated three RNA nanoparticles harboring different ligands, including small molecules and nucleic acid aptamers. Tritium-labeled RNA strands were synthesized via the tritiated water exchange method, achieving high labeling radioactivity (Figure 1). Instead of in vitro transcription labeling, which is costly and time-consuming in large-scale production and is limited in incorporating functional groups into RNA strands, the hydrogen-tritium exchange technology was used to efficiently achieve high radioactivity oligonucleotide labeling. Due to the 2′F modification of RNA nucleotides, the RNA strand could remain intact at 90 °C for 6 h, which facilitated the rapid exchange between tritiated water and the hydrogen on purine at the C8 position. After purification, no significant degradation was observed, and the recovery yield was higher than 90% (Figure 1b). We mixed the corresponding strands in TES buffer using an equal molar ratio of each strand and performed a stepwise assembly characterization by 12% native PAGE, demonstrating high assembly efficiency (Figure 1c). A gel mobility shift was observed from the monomer and dimer to the 3WJ trimer complex.
Figure 1.
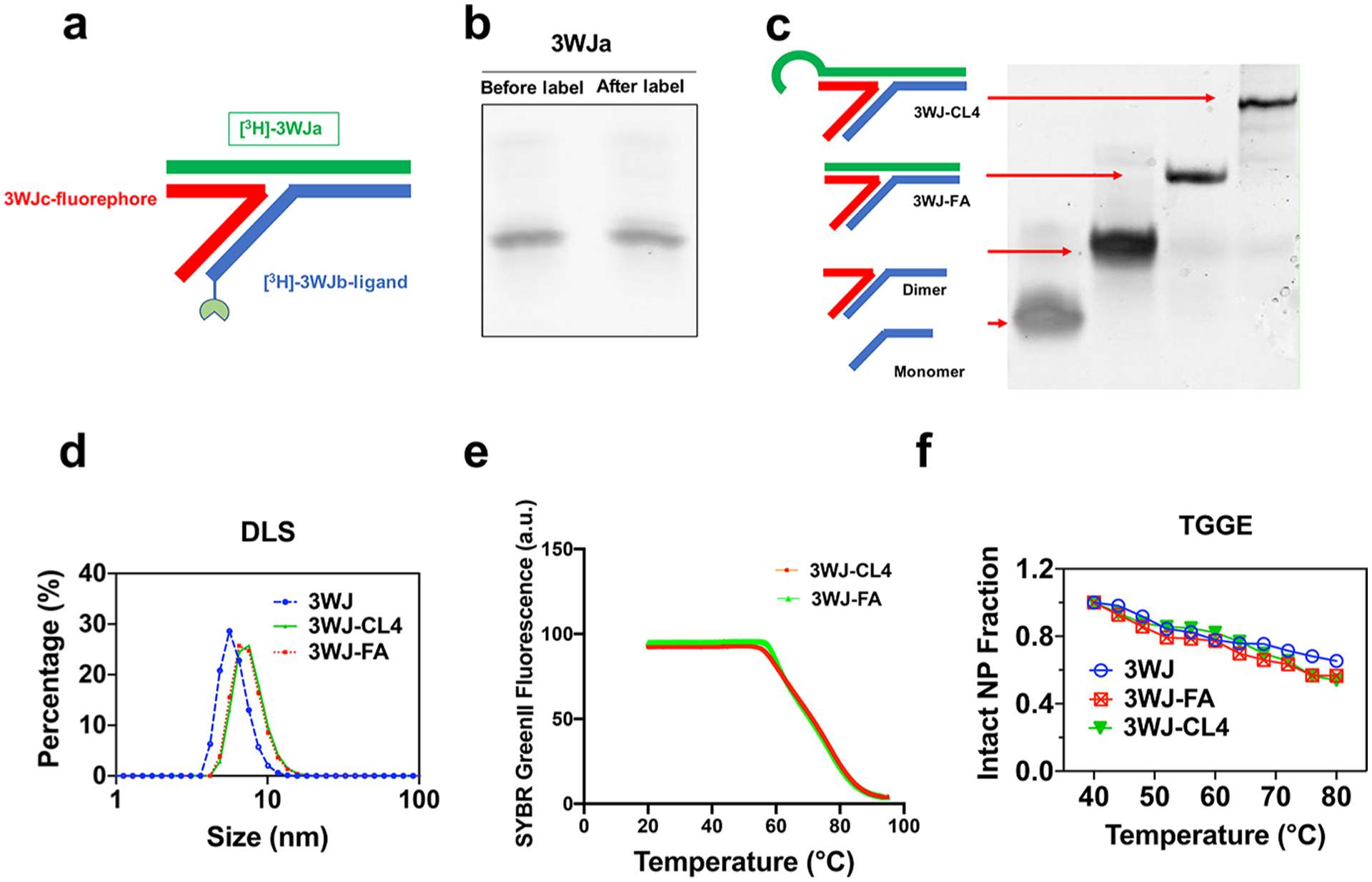
Design and characterization of the RNA nanoparticles. (a) 2D structure of the 3WJ RNA nanoparticles. (b) Comparison of the tritiated and unlabeled RNA strand shown by 16% denaturing gel electrophoresis. (c) Stepwise assembly of RNA nanoparticles shown by 12% native PAGE. (d) Size of the RNA nanoparticles shown by dynamic light scattering. (e) Melting temperature of 3WJ nanoparticles in annealing curves shown by real-time PCR (RT-PCR). (f) Melting temperature of 3WJ nanoparticles shown by TGGE.
Characterization of RNA Nanoparticles.
As the physicochemical properties of RNA nanoparticles play critical roles in governing the in vivo behavior, we performed further characterizations of these RNA nanoparticles in terms of size and thermodynamic stability. Dynamic light scattering measurements revealed that the sizes of 3WJ, 3WJ-FA, and 3WJ-CL4 were 5.8 ± 1.1, 6.0 ± 0.9, and 14.8 ± 2.6 nm, respectively (Figure 1d). To confirm that tritium labeling did not affect the physical properties and thermodynamic stability of the 3WJ nanoparticles, Tm values for each particle were measured at approximately 70 °C by quantitative RT-PCR (Figure 1e) and TGGE assays (Figure 1f). The measured Tm was vastly higher than that under physiological conditions (37 °C), which protected the RNA particles from dissociation in vivo.
Assessment of Chemical and Serum Stability of Tritium-Labeled RNA Nanoparticles.
The ability of RNA nanoparticles to retain their intact structure is critical to maintaining desirable pharmacokinetic/pharmacodynamic profiles and reducing nonspecific toxicity. In addition to high thermodynamic stability, a 2′F chemical modification confers high enzymatic and chemical stability on 3WJ RNA nanoparticles. A serum stability assay of RNA nanoparticles demonstrated no detectable RNA degradation after 24 h of incubation in 20% serum, as shown in clear gel bands. Quantification of the gel imaging further revealed no significant degradation after 24 h (Figure 2a). Tritium/hydrogen exchange is a pseudo-monomolecular reaction with a highly temperature-dependent rate. Reverse equilibration of hydrogen occurs, but only slowly, at physiological temperatures. The in vivo tritium label stability of tritium and fluorescent dual-labeled RNA nanoparticles was further analyzed under physiological conditions. Colocalization between fluorescent and radioactive signal peaks was observed after size exclusion column chromatography, while a minimal shift appeared after 24 h of incubation. These results indicated that tritium markers were successfully introduced to RNA nanoparticles. Tritium labeling possesses strong chemical stability under physiological conditions, which allows a confident interpretation of the effects seen in in vivo studies (Figure 2b).
Figure 2.
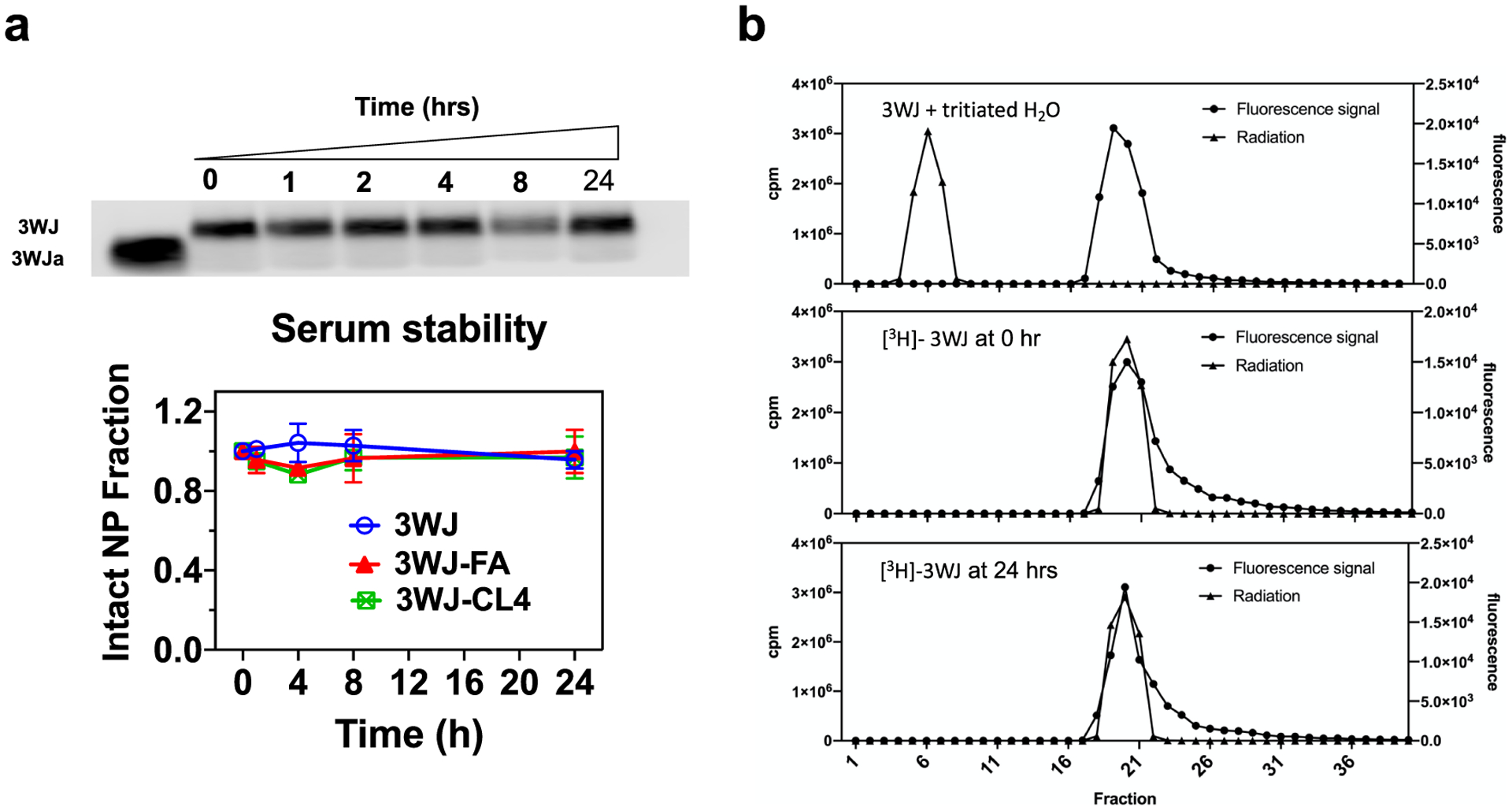
Stability assay of the RNA nanoparticles. (a) Serum stability assay by 2% agarose gel and quantification by ImageJ. (b) Elution profiles of the RNA nanoparticle on the Sephadex G-10 size exclusion column. From top to bottom: a mixture of unlabeled RNA nanoparticles and tritiated water, fractions obtained after purification of [3H]-3WJ nanoparticles, and fractions of [3H]-RNA nanoparticles after a 24 h incubation with 20% fetal bovine serum.
In Vitro Binding (Kd) and Uptake of RNA Nanoparticles.
To demonstrate that the binding affinity of the 3WJ nanoparticles is not affected by tritium labeling, the AFDye 647 fluorophore was conjugated to allow tracking of the RNA nanoparticles. A small-molecule DCL was conjugated to 3WJ nanoparticles as a negative control ligand to a folic acid ligand. A PSMA RNA aptamer of similar size to the CL4 aptamer was synthesized into 3WJ particles as a negative aptamer control.24 Two cell lines were used in the following experiment. KB cells with folate receptor overexpression were used to analyze the specific binding of the folic acid ligand. The TNBC cell line MDA-MB-231 with EGFR overexpression was cultured for CL4 RNA aptamer analysis. Flow cytometry data showed strong binding of 3WJ-FA to KB cells, while 3WJ and 3WJ-DCL displayed little binding (Figure 3a). A similar result was observed for 3WJ-CL4 in EGFR-overexpressing MDA-MB-231 cells (Figure 3a). Confocal microscopy imaging was also performed to evaluate the internalization of RNA NPs into cells. As shown in Figure 3b, confocal images scanned under three separate emission channels clearly showed more substantial cellular uptake of aptamer-conjugated RNA nanoparticles in receptor-overexpressing cell lines compared with the ligand-free and negative ligand 3WJ controls. We further investigated the apparent dissociation constant (Kd) for the 3WJ NPs by measuring the medium fluorescence intensity with a series of increasing concentrations of RNA particles. The Kd values of 3WJ-FA to KB cells and of 3WJ-CL4 to MDA-MB-231 cells were calculated to be 322.8 and 72.1 nM, respectively (Figure 3c).
Figure 3.
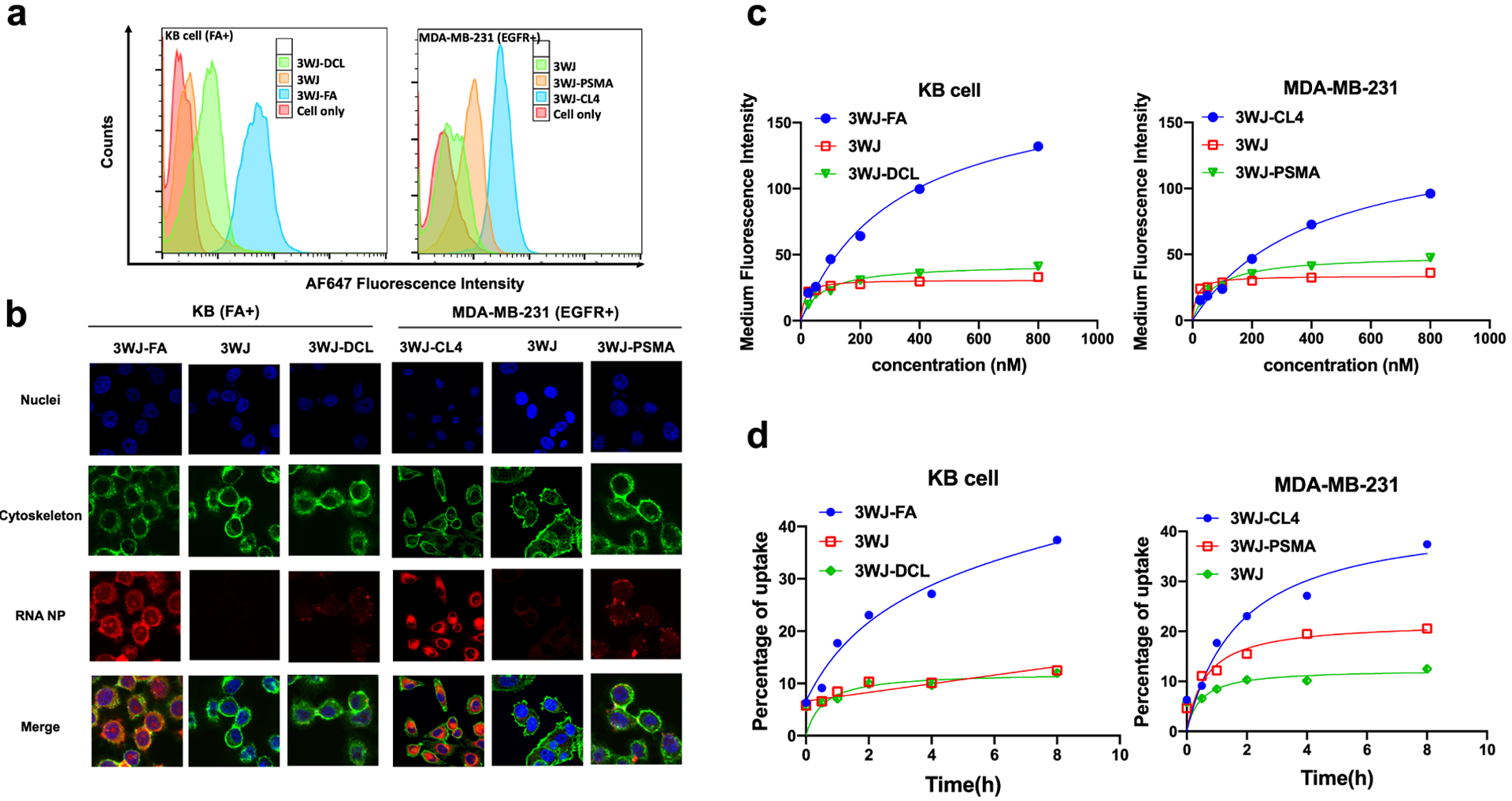
In vitro binding, internalization, and uptake of [3H]-labeled 3WJ RNA nanoparticles. (a) Binding affinity of RNA nanoparticles in KB and MDA-MB-231 cells. (b) Confocal microscopy imaging of cellular uptake of RNA nanoparticles (blue: nuclei; green: cytoskeleton; red: RNA nanoparticles). (c) Effect of concentration on the binding affinity of 3WJ nanoparticles (n = 3 biological replicates). (d) Cell uptake of 3WJ nanoparticles into KB and MDA-MB-231 cells quantified by LSC (n = 3 biological replicates).
Next, to further evaluate the validity of the [3H]-labeled RNA nanoparticles in cellular systems, the quantified uptake of [3H]-labeled RNA nanoparticles harboring ligands was examined in different cell lines. The cellular uptake of RNA nanoparticles was time-dependent (Figure 3d). In the folate receptor-positive cell line KB, approximately 30% of the total input radioactivity of 3WJ-FA nanoparticles was associated with the corresponding cells after 8 h, which was significantly higher than that of the 3WJ nanoparticles with the ligand control (10%). In the EGFR-overexpressing cell line, 3WJ-CL4 nanoparticles showed higher (35%) accumulation in cells than the negative ligand controls (19%).
Quantitative Biodistribution Analysis of [3H]-Labeled RNA Nanoparticles.
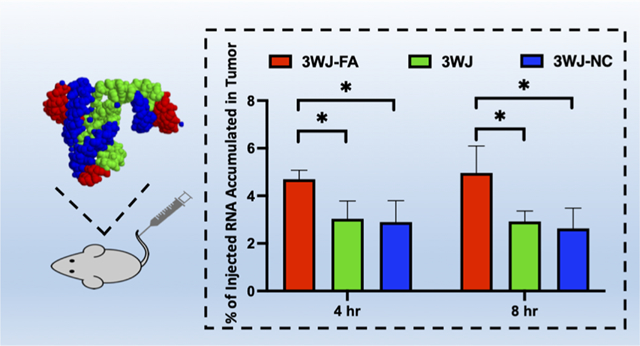
A quantitative biodistribution assay provided critical information for RNA nanoparticle design and optimization. First, the RNA nanoparticle plasma concentration curve was determined by quantifying the radioactivity in 100 μL of collected plasma. The clearance half-life was determined to be ~12 h for 3WJ-FA and ~10 h for 3WJ-CL4 nanoparticles. (Figures 4a and 5a). Pharmacokinetic parameters were calculated via WinNonlin 8.1 (Table 1).
Figure 4.
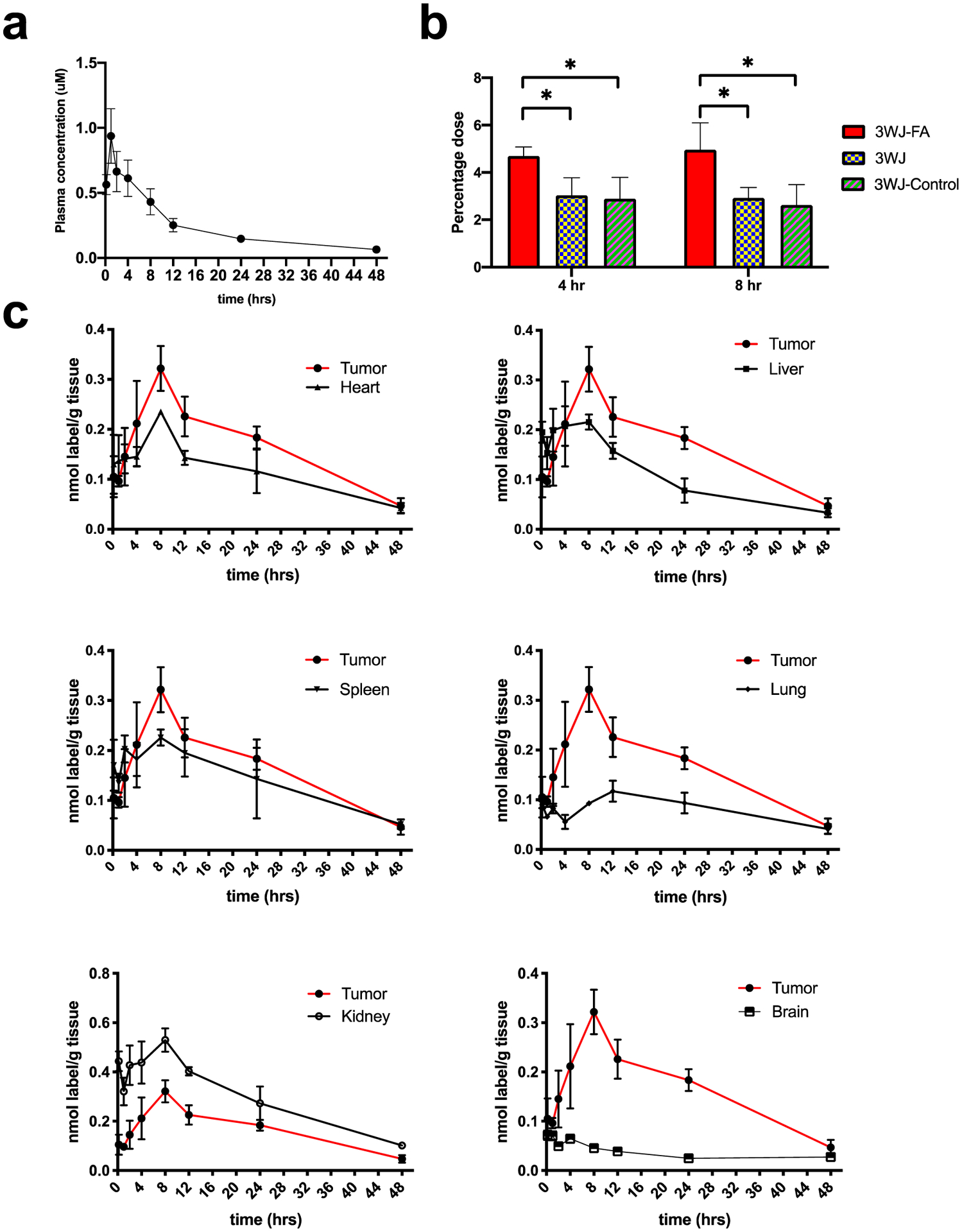
In vivo PK/biodistribution studies of [3H]-3WJ-FA RNA nanoparticles (n = 3 biologically independent animals). (a) [3H]-3WJ-FA plasma concentration curve (n = 3 biologically independent animals). (b) Oligonucleotide equivalent percentage (% of total injection) in tumor collected 4 and 8 h after intravenous injection (n = 3 biologically independent animals, statistical calculations were performed by the two-tailed unpaired t-test and presented as the mean ± SD, *p < 0.05). (c) Mass-normalized levels of [3H]-RNA nanoparticles in the organs or tissues expressed as the percentage of [3H] label per gram of tissue (n = 3 biologically independent animals).
Figure 5.
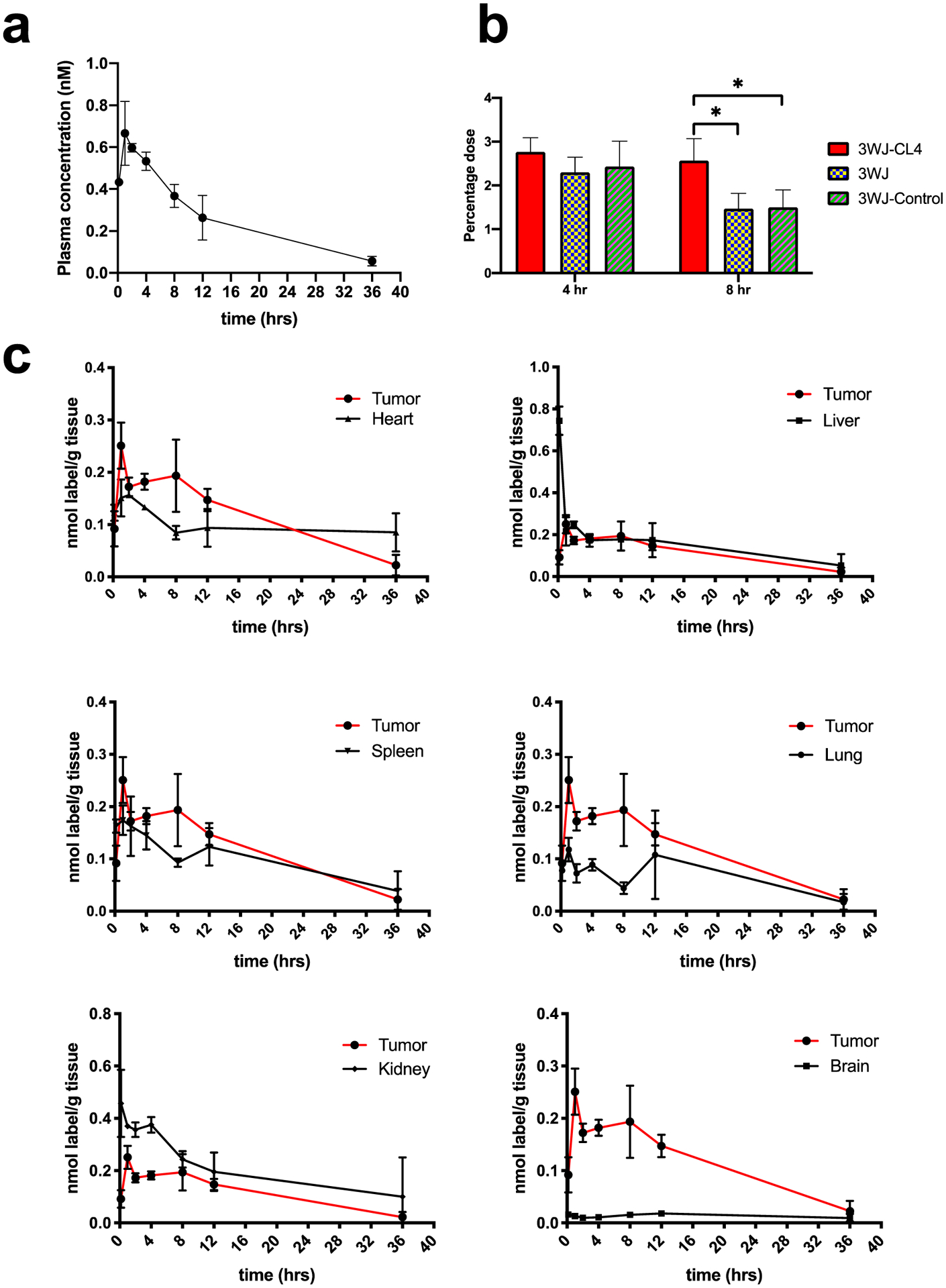
In vivo PK/biodistribution studies of [3H]-3WJ-CL4 RNA nanoparticles (n = 3 biologically independent animals). (a) [3H]-3WJ-CL4 plasma concentration curve (n = 3 biologically independent animals). (b) Oligonucleotide equivalent percentage (% of total injection) in tumors collected 4 and 8 h post intravenous injection (n = 5 biologically independent animals, statistics calculated by two-tailed unpaired t-test presented as the mean ± SD, *p < 0.05). (c) Mass-normalized levels of [3H]-RNA nanoparticles in the organs or tissues expressed as percentage of [3H] label per gram of tissue (n = 3 biologically independent animals).
Table 1.
PK Parameters of RNA Nanoparticlesa
| RNA NPs | T1/2 (h) | Tmax (h) | Cmax (nmol/L) | AUClast (h nmol/L) | Vd (L/kg) | Cl (1/h) |
|---|---|---|---|---|---|---|
| 3WJ-Cl4 | ||||||
| mean | 10.12 | 1.33 | 0.70 | 9.17 | 29.24 | 2.05 |
| SD | 2.08 | 0.58 | 0.11 | 1.70 | 2.84 | 0.41 |
| 3WJ-FA | ||||||
| mean | 18.03 | 1.33 | 0.97 | 11.17 | 41.47 | 1.55 |
| SD | 1.80 | 0.58 | 0.16 | 1.07 | 5.00 | 0.22 |
Abbreviations: AUC, area under the curve; Cl, clearance; PK, pharmacokinetic; pRNA, packaging RNA; siRNA, small interfering RNA; T, half-life; Vd, volume of distribution.
Based on the plasma concentration information and our previous studies, the highest tumor accumulation was achieved between 4 and 8 h We compared the 3WJ-FA and 3WJ-CL4 organs and tumor accumulation with the negative ligand and ligand-free 3WJ controls at 4 h and 8 h after injection in two tumor xenografts (Figures 4b and 5b) (Tables 2 and 3). Among the organs examined, both types of tritium-labeled RNA nanoparticles showed similar biodistribution profiles. The highest uptake of [3H]-labeled RNA nanoparticle equivalents (normalized to the administered dose) was found in the tumor, kidney, and liver (Figures 4b and 5b). In contrast, tritium molar activity was unmeasurable in the other healthy organs. More importantly, the tumor showed higher accumulation (5% of 3WJ-FA and 3% of 3WJ-CL4) at 4–8 h than the previously published average accumulation of nanoparticles (0.7%). The high tumor accumulation could contribute to the favorable tumor inhibition effect that we found in our previous publications.9,11,37 Compared with the negative ligand control, the RNA nanoparticles with specific ligands had significantly higher accumulation, demonstrating that the targeting specificity came from the ligand affinity rather than the increase in the background level. Notably, the negative ligand 3WJ controls also showed 2% of the total injection dose accumulation in the tumor. Although lower than the accumulation of the ligand displaying RNA particles, this number was still higher than the average. The results demonstrated higher tumor accumulation derived from both passive tumor accumulation mechanisms, such as the EPR effect, as well as the active targeting effect derived from the target ligand.
Table 2.
Oligonucleotide Equivalent Percentage (% of Total Injection) in KB Xenograft 4 and 8 h Post Injectiona
| heart | liver | spleen | lung | kidney | tumor | brain | |
|---|---|---|---|---|---|---|---|
| 4 h | |||||||
| 3WJ | 1.0 ± 0.2 | 13.6 ± 2.4 | 0.9 ± 0.3 | 0.5 ± 0.1 | 7.3 ± 0.5 | 3.0 ± 0.8 | 0.1 ± 0.1 |
| 3WJ-FA | 1.7 ± 0.1 | 14.4 ± 0.9 | 1.0 ± 0.2 | 0.8 ± 0.2 | 8.3 ± 0.5 | 4.7 ± 0.4 | 0.1 ± 0.1 |
| 3WJ-DCL | 1.8 ± 0.1 | 11.5 ± 2.1 | 1.0 ± 0.5 | 0.7 ± 0.2 | 7.5 ± 1.1 | 2.9 ± 0.9 | ND |
| 8 h | |||||||
| 3WJ | 1.0 ± 0.1 | 11.4 ± 1.7 | 1.2 ± 0.4 | 1.6 ± 0.4 | 6.6 ± 0.2 | 2.9 ± 0.4 | 0.1 ± 0.1 |
| 3WJ-FA | 1.6 ± 0.2 | 13.4 ± 1.6 | 1.0 ± 0.2 | 0.8 ± 0.2 | 8.3 ± 0.6 | 5.0 ± 1.1 | ND |
| 3WJ-DCL | 1.1 ± 0.1 | 11.7 ± 1.5 | 1.8 ± 0.2 | 0.5 ± 0.1 | 6.6 ± 0.2 | 2.6 ± 0.9 | ND |
ND: no detection.
Table 3.
Oligonucleotide Equivalent Percentage (% of Total Injection) in MDA-MB-231 Xenograft 4 and 8 h Post Injection
| heart | liver | spleen | lung | kidney | tumor | brain | |
|---|---|---|---|---|---|---|---|
| 4 h | |||||||
| 3WJ | 0.6 ± 0.1 | 12.9 ± 0.7 | 0.6 ± 0.3 | 0.5 ± 0.1 | 4.2 ± 0.6 | 2.3 ± 0.3 | 0.3 ± 0.1 |
| 3WJ-CL4 | 0.8 ± 0.1 | 15.5 ± 1.8 | 0.8 ± 0.2 | 0.9 ± 0.2 | 6.3 ± 0.1 | 2.8 ± 0.3 | 0.5 ± 0.3 |
| 3WJ-PSMA | 0.6 ± 0.2 | 12.5 ± 2.2 | 0.8 ± 0.2 | 0.5 ± 0.1 | 4.2 ± 0.6 | 2.3 ± 0.3 | 0.3 ± 0.2 |
| 8 h | |||||||
| 3WJ | 0.6 ± 0.2 | 11.0 ± 3.8 | 0.6 ± 0.2 | 0.9 ± 0.4 | 3.9 ± 1.2 | 1.5 ± 0.4 | 0.3 ± 0.1 |
| 3WJ-CL4 | 0.6 ± 0.1 | 11.7 ± 1.4 | 0.6 ± 0.3 | 0.5 ± 0.1 | 4.2 ± 0.6 | 2.6 ± 0.5 | 0.2 ± 0.1 |
| 3WJ-PSMA | 0.6 ± 0.2 | 11.5 ± 2.1 | 0.6 ± 0.3 | 0.5 ± 0.1 | 3.7 ± 1.0 | 2.5 ± 0.9 | 0.2 ± 0.1 |
The observed tissue distributions of the two types of [3H]-nanoparticles were generally similar; however, there was a notable difference. For 3WJ-FA, uptake by the kidneys accounted for approximately 10% of the administered dose at all sampling times, whereas for 3WJ-CL4, uptake by the kidneys accounted for only <5% of the administered dose. Overall, the levels of 3WJ-FA in the kidney were significantly higher than those of 3WJ-CL4 at all time points examined. The difference could be due to two mechanisms: (1) kidney cells have higher folate receptor expression;38 (2) smaller 3WJ-FA particles were eliminated faster through kidney filtration.19 Notably, the liver is expected to express high levels of folate receptor;39 however, 3WJ-FA nanoparticles exhibited a preferable tumor/liver accumulation ratio. We attribute this advantage to the elastic property of RNA nanoparticles. RNA particles can undergo conformational changes and squeeze out of the leaky tumor vasculature (EPR effect), resulting in higher accumulation in tumors.19
The distribution over time of [3H]-labeled ligands harboring RNA nanoparticles in essential organs (liver, kidney, lungs, heart, spleen, and brain) and tumors was also determined in vivo in mice receiving a single intravenous administration.
The biodistribution of [3H]-nanoparticles was analyzed with data normalized for organ or tumor weight (Figures 4c and 5c). From a qualitative standpoint, the RNA nanoparticle biodistribution profiles in healthy organs established after 1 h were largely maintained over the 24 h duration of the experiment. Although the highest mass-normalized concentrations of [3H]-nanoparticle equivalents were seen in highly perfused organs such as kidney and liver in general, more importantly, comparatively high levels of both nanoparticles were also detected in the tumor. Notably, for the 3WJ-FA nanoparticles, higher mass-normalized concentrations were measured in the tumor than in the liver. This research demonstrates that RNA nanoparticles harboring ligands have a tumor-specific targeting effect and supports the potential use of RNA nanoparticles as carriers to deliver therapeutic drugs to tumor sites. In addition to the desired therapeutic effect, the lower essential organ accumulation also explained the low side effects of RNA nanoparticles.11,13,37
In Vivo Fluorescence Biodistribution Study.
The whole-body fluorescence imaging technique used an in vivo imaging system (IVIS) to visualize the biodistribution of nanoparticles in live animals or in collected organs and tumors. This technique is widely used due to its fast image acquisition time and easy handling. We also studied the biodistribution based on fluorophores conjugated to RNA nanoparticles. The RNA nanoparticles harboring ligands exhibited relatively strong tumor accumulation compared with the ligand-free control particles, indicated by the higher fluorescence intensity in the tumor (Figure 6a). The quantification of organ and tumor accumulation was also analyzed by the IVIS (Figure 6b). The biodistribution profile was similar to that obtained using radioactive markers. RNA nanoparticles possessed a preferable tumor/organ ratio, demonstrating that the elastic property facilitates highly specific particle accumulation in the tumor. However, the fluorescence signal intensity decreased faster and was nondetectable after 12 h, whereas the nanoparticles were still detectable after 24 h using radiolabeling. The rapid elimination might be due to the cleavage of fluorescent dye from RNA nanoparticles followed by rapid excretion. Comparison of the results revealed the discrepancy between the radioactivity and the fluorescence signals, which called our attention to the quantification and interpretation of fluorescence imaging measurements.
Figure 6.
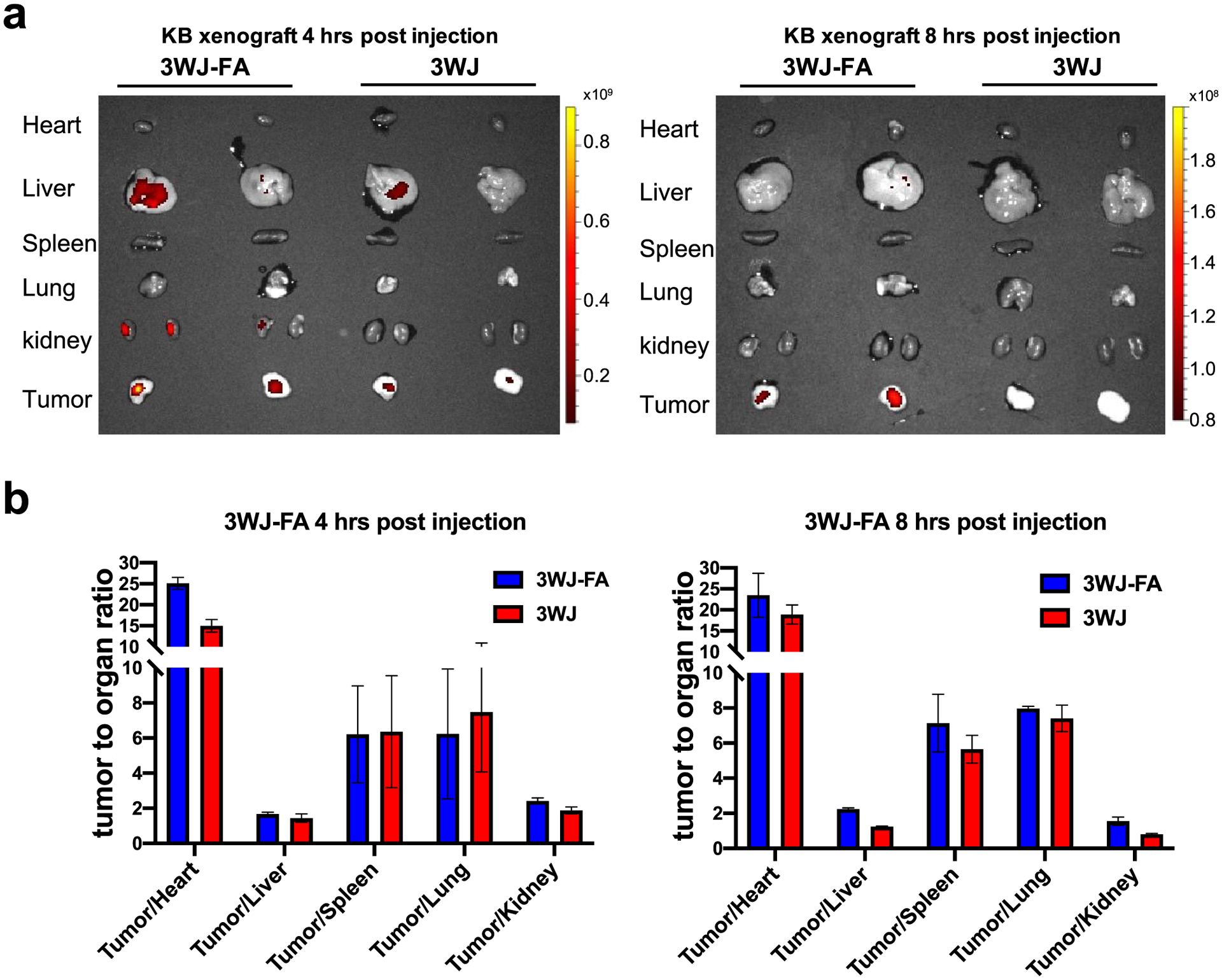
In vivo biodistribution studies of RNA nanoparticles via the IVIS. (a) Ex vivo fluorescent tumor and organ images of RNA nanoparticles. Representative images of organs and tumors treated with 3WJ-FA and 3WJ at 4 and 8 h post injection shown by the IVIS fluorescent imaging system. (b) Quantitative analysis of the biodistribution tumor to organ ratio using ex vivo fluorescence imaging (n = 3 biologically independent animals).
Urinary Excretion of RNA Nanoparticles.
Urine samples were collected and analyzed by scintillation counting. The mean total recovery of radiolabeled components up to 24 h after the administration of 3WJ-FA and 3WJ recovered in urine was approximately 55%, while the other 45% includes the radiative counts that cannot be recovered due to whole-body distribution and blood dilution after intravenous injection (Figure 7a). However, this does not mean that the other 45% of RNA nanoparticles remained in the body since this result may be due to the rate of radioactive material recovery. Therefore, the majority of the RNA particles may have been cleared from the body. The nontargeted RNA nanoparticles were excreted rapidly, as 40% of nanoparticles were detected 2 h after injection. Intact [3H]-RNA nanoparticles were observed in the urine up to 4 h after administration, which was consistent with the previous results (Figure 7b).19,40 3WJ-CL4 RNA nanoparticles, due to their large size, have a slower urine excretion rate than 3WJ alone. These results demonstrated that urinary excretion was the major excretion pathway of these RNA nanoparticles. The fast renal excretion contributed to lower off-target effects and lack of observable toxicity.
Figure 7.
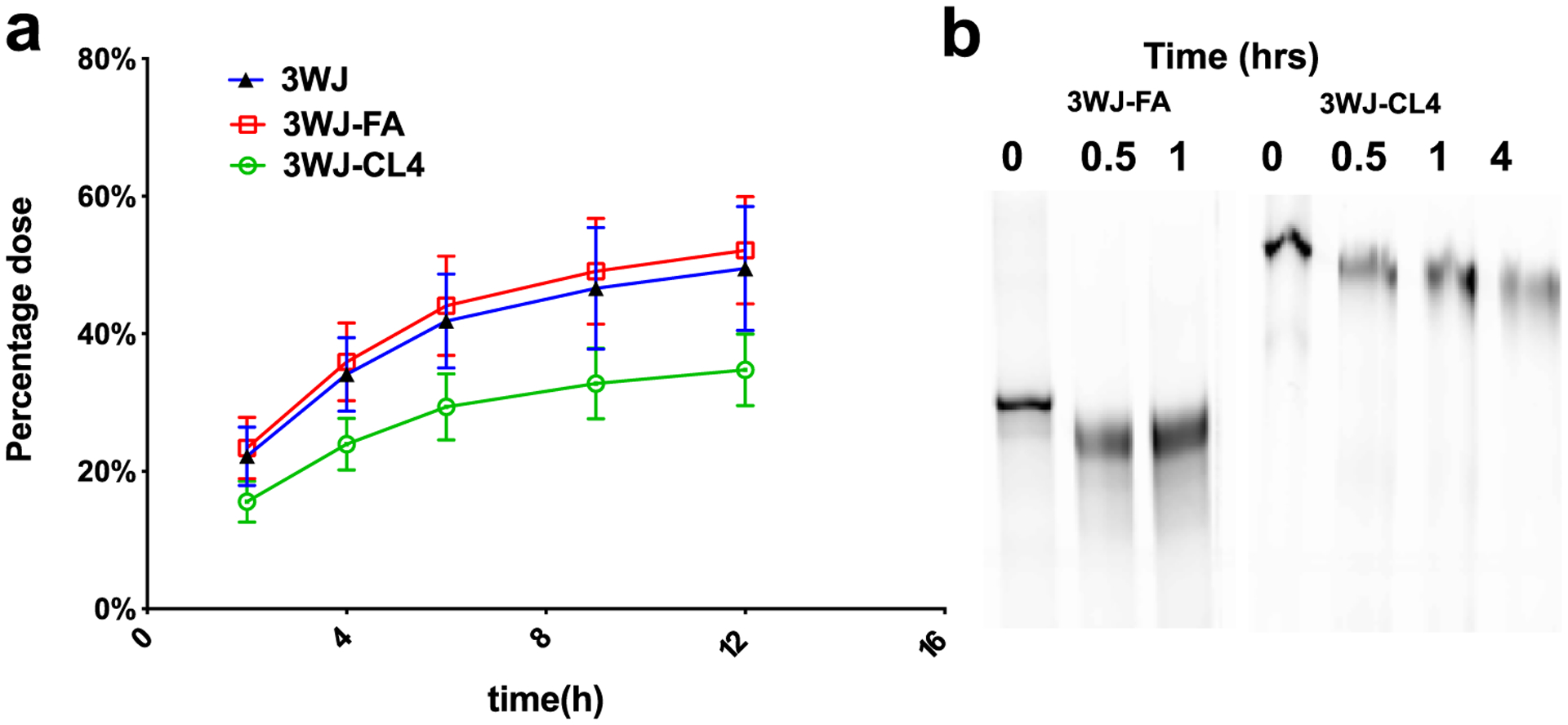
In vivo excretion studies of RNA nanoparticles. (a) Urinary elimination of [3H]-labeled RNA nanoparticles. Urine was collected at intervals from mice receiving a single intravenous dose of RNA nanoparticles. Dose-normalized [3H]-RNA nanoparticles in urine samples were measured by LSC (n = 3 biologically independent animals). (b) PAGE image of RNA nanoparticles in the urine sample harvested at different time points.
DISCUSSION
The quantitative PK profile of two RNA nanoparticles was investigated using radioactive markers. The results revealed that approximately 5% of 3WJ-FA and 3% of 3WJ-CL4 RNA nanoparticles accumulated in the tumor at 4 h after injection and retained at the tumor site for 12 h. The tumor accumulation rate was higher than the previously reported average of 0.7% of total injected nanoparticles accumulated in the tumor.18 RNA nanoparticles could also be excreted rapidly through kidney filtration, resulting in low organ accumulation and fast body clearance, which can lead to low side effects. We attribute these favorable pharmacokinetic parameters and high tumor retention to a combination of (1) passive targeting, resulting from the favorable physical properties of RNA nanoparticles, and (2) active targeting by the tumor-specific targeting ligand. We have reported that RNA nanoparticles display rubbery properties. The ability to undergo reversible conformational change allows RNA particles to squeeze through leaky tumor vasculature, increasing the EPR effect and consequently tumor accumulation. It also enables them to pass through kidney filtration, leading to fast excretion and a lack of observable toxicity. In addition to the passive accumulation, the active tumor targeting effect plays a vital role in drug delivery. Unlike the passive accumulation effect, which facilitates the accumulation of nanoparticles in the tumor vasculatures, the active targeting ligand promotes the particles to enter tumor cells after reaching the tumor vasculature. Both the passive tumor accumulation effect and the active targeting capability contribute to the rubbery and amoeba deformative property of RNA nanoparticles. These special physical and chemical properties also cause rapid excretion by the kidneys. The majority of the IV-injected radiolabeled RNA nanoparticles were cleared from the body in urine within 12 h. The fast body clearance phenomenon will lead to low toxicity and side effect in future clinical applications.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health Grant U01CA207946 and R01EB019036 to P. G. P.G.’s Sylvan G. Frank Endowed Chair position in Pharmaceutics and Drug Delivery from the CM Chen Foundation. The authors are grateful to Dr. Zhefeng Li and Dr. Hongran Yin for assistance in animal studies and Dr. Daniel Binzel, Nicolas Burns, Lora McBride, Xin Li, and Peter for assistance in the preparation of the manuscript. We thank the Analytical Cytometry Shared Resource, the University Laboratory Animal Resources Facilities, and OSU campus Microscopy and Imaging Facility (NIH Grant CA016058) at The Ohio State University Comprehensive Cancer Center.
Footnotes
The authors declare the following competing financial interest(s): P.G. is the consultant of Oxford Nanopore Technologies; the cofounder of Shenzhen P&Z Bio-medical Co. Ltd, as well as cofounder of ExonanoRNA, LLC and its subsidiary Weina Biomedical LLC. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.molpharmaceut.1c00035
REFERENCES
- (1).Guo P The emerging field of RNA nanotechnology. Nat. Nanotechnol 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Shu D; Shu Y; Haque F; Abdelmawla S; Guo P Thermodynamically stable RNA three-way junction for constructing multifuntional nanoparticles for delivery of therapeutics. Nat. Nanotechnol 2011, 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Jasinski D; Haque F; Binzel DW; Guo P Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano 2017, 11, 1142–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhang Y; Leonard M; Shu Y; Yang Y; Shu D; Guo P; Zhang X Overcoming Tamoxifen Resistance of Human Breast Cancer by Targeted Gene Silencing Using Multifunctional pRNA Nanoparticles. ACS Nano 2017, 11, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jaeger L; Chworos A The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol 2006, 16, 531–543. [DOI] [PubMed] [Google Scholar]
- (7).Afonin KA; Lindsay B; Shapiro BA Engineered RNA Nanodesigns for Applications in RNA Nanotechnology. DNA RNA Nanotechnol 2013, 1, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Jaeger L; Westhof E; Leontis NB TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res 2001, 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Shu D; Li H; Shu Y; Xiong G; Carson WE III; Haque F; Xu R; Guo P Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano 2015, 9, 9731–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yin HR; Xiong GF; Guo SJ; Xu CC; Xu R; Guo PX; Shu D Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133. Mol. Ther 2019, 27, 1252–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Piao X; Yin H; Guo S; Wang H; Guo P RNA Nanotechnology to Solubilize Hydrophobic Antitumor Drug for Targeted Delivery. Adv. Sci 2019, 6, 1900951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Xu Y; Pang L; Wang H; Xu C; Shah H; Guo P; Shu D; Qian SY Specific delivery of delta-5-desaturase siRNA via RNA nanoparticles supplemented with dihomo-γ-linolenic acid for colon cancer suppression. Redox Biol 2019, 21, 101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Guo S; Vieweger M; Zhang K; Yin H; Wang H; Li X; Li S; Dong Y; Chui W; Guo P Ultra-thermostable RNA nanoparticles for solubilizing and high yield loading of paclitaxel for breast cancer therapy with undetectable toxicity. Nat. Commun 2019, 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Xu C; Li H; Zhang K; Binzel DW; Yin H; Chiu W; Guo P Photo-controlled release of paclitaxel and model drugs from RNA pyramids. Nano Res 2019, 12, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Afonin KA; Kireeva M; Grabow WW; Kashlev M; Jaeger L; Shapiro BA Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano Lett 2012, 12, 5192–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Shapiro BA Computational Design Strategies for RNA Nanostructures. J. Biomol. Struct. Dyn 2009, 26, 820–820. [Google Scholar]
- (17).Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L; Heldman E; Reiser J; Chiu W; Freed EO; Shapiro BA Multifunctional RNA nanoparticles. Nano Lett 2014, 14, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wilhelm S; Tavares AJ; Dai Q; Ohta S; Audet J; Dvorak HF; Chan WCW Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater 2016, 1, 16014. [Google Scholar]
- (19).Ghimire C; Wang H; Li H; Vieweger M; Xu C; Guo P RNA Nanoparticles as Rubber for Compelling Vessel Extravasation to Enhance Tumor Targeting and for Fast Renal Excretion to Reduce Toxicity. ACS Nano 2020, 14, 13180–13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Maeda H The enhanced permeability and retention (EPR) effect in tumor vasculature: the key role of tumor-selective macromolecular drug targeting. Adv. Enzyme Regul 2001, 41, 189–207. [DOI] [PubMed] [Google Scholar]
- (21).Stylianopoulos T EPR-effect: utilizing size-dependent nanoparticle delivery to solid tumors. Ther Deliv 2013, 4, 421–423. [DOI] [PubMed] [Google Scholar]
- (22).Tanaka T; Shiramoto S; Miyashita M; Fujishima Y; Kaneo Y Tumor targeting based on the effect of enhanced permeability and retention (EPR) and the mechanism of receptor-mediated endocytosis (RME). Int. J. Pharm 2004, 277, 39–61. [DOI] [PubMed] [Google Scholar]
- (23).Haque F; Shu D; Shu Y; Shlyakhtenko LS; Rychahou PG; Evers BM; Guo P Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano Today 2012, 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Binzel DW; Shu Y; Li H; Sun M; Zhang Q; Shu D; Guo B; Guo P Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther 2016, 24, 1267–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lee TJ; Yoo JY; Shu D; Li H; Zhang JY; Yu JG; Jaime-Ramirez AC; Acunzo M; Romano G; Cui R; Sun HL; Luo ZH; Old M; Kaur B; Guo PX; Croce CM RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic miR-21. Mol. Ther 2017, 25, 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Cui DX; Zhang CL; Liu B; Shu Y; Du T; Shu D; Wang K; Dai FP; Liu YL; Li C; Pan F; Yang YM; Ni J; Li H; Brand-Saberi B; Guo PX Regression of Gastric Cancer by Systemic Injection of RNA Nanoparticles Carrying both Ligand and siRNA. Sci. Rep 2015, 5, 10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Abdelmawla S; Guo S; Zhang L; Pulukuri SM; Patankar P; Conley P; Trebley J; Guo P; Li QX Pharmacological characterization of chemically synthesized monomeric phi29 pRNA nanoparticles for systemic delivery. Mol. Ther 2011, 19, 1312–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Liu Y; Tseng YC; Huang L Biodistribution studies of nanoparticles using fluorescence imaging: a qualitative or quantitative method? Pharm. Res 2012, 29, 3273–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Arms L; Smith DW; Flynn J; Palmer W; Martin A; Woldu A; Hua S Advantages and Limitations of Current Techniques for Analyzing the Biodistribution of Nanoparticles. Front Pharmacol 2018, 9, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hilgenbrink AR; Low PS Folate receptor-mediated drug targeting: from therapeutics to diagnostics. J. Pharm. Sci 2005, 94, 2135–2146. [DOI] [PubMed] [Google Scholar]
- (31).Sabharanjak S; Mayor S Folate receptor endocytosis and trafficking. Adv. Drug Delivery Rev 2004, 56, 1099–1109. [DOI] [PubMed] [Google Scholar]
- (32).Sudimack J; Lee RJ Targeted drug delivery via the folate receptor. Adv. Drug Delivery Rev 2000, 41, 147–162. [DOI] [PubMed] [Google Scholar]
- (33).Esposito CL; Passaro D; Longobardo I; Condorelli G; Marotta P; Affuso A; de Franciscis V; Cerchia L A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS One 2011, 6, No. e24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Castillo L; Etienne-Grimaldi M; Fischel J; Formento P; Magne N; Milano G Pharmacological background of EGFR targeting. Ann. Oncol 2004, 15, 1007–1012. [DOI] [PubMed] [Google Scholar]
- (35).Dassonville O; Bozec A; Fischel JL; Milano G EGFR targeting therapies: Monoclonal antibodies versus tyrosine kinase inhibitors. Crit. Rev. Oncol. Hematol 2007, 62, 53–61. [DOI] [PubMed] [Google Scholar]
- (36).Rocha-Lima CM; Soares HP; Raez LE; Singal R EGFR targeting of solid tumors. Cancer Control 2007, 14, 295–304. [DOI] [PubMed] [Google Scholar]
- (37).Wang H; Ellipilli S; Lee WJ; Li X; Vieweger M; Ho YS; Guo P Multivalent rubber-like RNA nanoparticles for targeted co-delivery of paclitaxel and MiRNA to silence the drug efflux transporter and liver cancer drug resistance. J. Controlled Release 2021, 330, 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Muller C; Schibli R Prospects in folate receptor-targeted radionuclide therapy. Front. Oncol 2013, 3, 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Gates SB; Mendelsohn LG; Shackelford KA; Habeck LL; Kursar JD; Gossett LS; Worzalla JF; Shih C; Grindey GB Characterization of folate receptor from normal and neoplastic murine tissue: influence of dietary folate on folate receptor expression. Clin. Cancer Res 1996, 2, 1135–1141. [PubMed] [Google Scholar]
- (40).Piao X; Wang H; Binzel DW; Guo P Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2’-F RNA, and LNA in the context of Phi29 pRNA 3WJ. RNA 2018, 24, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]


