Abstract
Background
Blacks tend to have a stronger inflammatory immune response than Whites. We hypothesized that racial differences in host immunity also manifest in the tumor microenvironment, constituting part of a distinct aggressive tumor biology underlying higher mortality in Black women.
Methods
Pathological and gene expression profiling approaches were used for characterizing infiltrating immune cells in breast tumor microenvironment from 1315 patients from the Women’s Circle of Health Study. Racial differences in tumor immune phenotypes were compared, with results validated in a publicly accessible dataset. Prognostic associations of immune phenotypes were assessed in 3 independent cohorts.
Results
We found marked and consistent differences in tumor immune responses between Black and White patients. Not only did tumors from Blacks display a stronger overall immune presence but also the composition and quality of immune infiltrates differed, regardless of tumor subtypes. Black patients had a stronger CD4+ and B-cell response, and further, a more exhausted CD8+ T-cell profile. A signature indicating a higher ratio of exhausted CD8+ T cells to total CD8+ T cells (ExCD8-r) was consistently associated with poorer survival, particularly among hormone receptor–positive patients. Among hormone receptor–negative patients, combinations of the absolute fraction of CD8+ T cells and ExCD8-r signature identified the CD8lowExCD8-rhigh subgroup, the most prevalent among Blacks, with the worst survival.
Conclusions
Our findings of a distinct exhausted CD8+ T-cell signature in Black breast cancer patients indicate an immunobiological basis for their more aggressive disease and a rationale for the use of immune checkpoint inhibitors targeting the exhaustion phenotype.
In the United States, breast cancer mortality rates are the highest in African American or Black women (1). Findings from the last decade suggest that tumor biology may play a role in these racial disparities (2‐5). Whereas previous research has focused on the tumor itself, less attention was paid to immune infiltrates in the tumor microenvironment (TME), increasingly recognized for their roles in carcinogenesis and as a target of cancer immunotherapy (6).
Multiple lines of evidence indicate possible racial differences in immune responses to tumors. Systemic immune responses, measured by circulating cytokine levels (7) and transcriptomic profiles of cultured immune cells, differ across ancestral populations (8‐10). Further genetic analysis attributed these differences, in part, to immune adaptation to ancestral environments of early human populations (7,9,10). Two earlier studies also showed differential gene expression in immune-related pathways in prostate and breast cancers from Black and European American orWhite patients (11,12). Because the sample sizes in these studies were limited and methods predated the advent of advanced profiling of immune infiltrates, it remains to be established whether immune responses in breast cancer differ by race.
Two recent studies based on The Cancer Genome Atlas (TCGA) data showed only modest racial differences in tumor immune gene expression between Black and White patients after accounting for breast cancer subtypes (13,14). Despite being an invaluable public resource used for many important studies of immune infiltration in TME, TCGA tumor tissues were collected and processed primarily for sequencing of tumor cells but not necessarily immune cells, which might preside outside of tumor foci (15). Moreover, the design of TCGA, with no matching of patients by race, was not optimal for racial comparisons. To overcome these limitations and to systematically investigate breast TME between Black and White patients, we profiled immune infiltrates using pathological and molecular methods in tumors from the Women’s Circle of Health Study (WCHS), which was established specifically to investigate breast cancer racial disparities, with results validated in publicly accessible data.
Methods
Patient Population
Tumor tissues and data for this study were derived from WCHS as previously described (16) and in the Supplementary Methods (available online). Clinicopathological data were obtained from pathology reports, supplemented with data obtained from the New Jersey State Cancer Registry, which actively maintains updated information on deaths and causes of deaths through various follow-up sources. All study participants provided consent for the use of their data and specimens for research. The studies were approved by the institutional review boards at participating institutions.
Pathological Tumor-Infiltrating Lymphocytes (TIL) Assessment
As part of the protocol of receiving unstained slides and tumor blocks in WCHS, hematoxylin and eosin sections were prepared and reviewed for histopathologic scoring of stromal TILs by a board-certified breast pathologist (TK) blinded to patient characteristics, following the recommendations by the International TILs Working Group (17). TIL data were available from 1315 WCHS patients with invasive cancer (920 Blacks and 395 Whites). Descriptive characteristics of this patient population are summarized in Table 1.
Table 1.
Descriptive characteristics of patients used for pathological and molecular analysis of tumor-infiltrating lymphocytesa
| Patient characteristics | Patients with pathological TIL data (n = 1315) |
Patients with molecular TIL data (n = 367) |
||||
|---|---|---|---|---|---|---|
| Black (n = 920) | White (n = 395) | P | Black (n = 190) | White (n = 177) | P | |
| Age at diagnosis, mean (SD), y | 53.3 (10.6) | 52.6 (10.3) | .29 | 53.4 (10.5) | 54.8 (13.4) | .29 |
| Tumor grade, No. (%) | .63 | .63 | ||||
| I | 106 (12.1) | 98 (26.0) | 11 (5.9) | 15 (8.6) | ||
| II | 341 (38.9) | 165 (43.8) | 51 (27.6) | 46 (26.3) | ||
| III | 430 (49.0) | 114 (30.2) | 123 (66.5) | 114 (65.1) | ||
| ER status, No. (%) | <.001 | .34 | ||||
| Positive | 630 (68.7) | 322 (82.8) | 85 (44.7) | 88 (49.7) | ||
| Negative | 287 (31.3) | 67 (17.2) | 105 (55.3) | 89 (50.3) | ||
| PR status, No. (%) | <.001 | .29 | ||||
| Positive | 567 (63.5) | 280 (74.5) | 64 (33.7) | 69 (39.0) | ||
| Negative | 326 (36.5) | 96 (25.5) | 126 (66.3) | 108 (61.0) | ||
| HER2 status, No. (%) | .46 | .09 | ||||
| Positive | 170 (19.1) | 67 (17.3) | 61 (32.1) | 72 (40.7) | ||
| Negative | 722 (80.9) | 320 (82.7) | 129 (67.9) | 105 (59.3) | ||
| IHC subtype, No. (%) | <.001 | .16 | ||||
| Luminal (ER+ or PR+ and HER2-) | 526 (59.6) | 270 (72.8) | 54 (28.4) | 38 (21.5) | ||
| HER2+ | 166 (18.8) | 59 (15.9) | 61 (32.1) | 72 (40.7) | ||
| Triple-negative (ER-, PR- and HER2-) | 191 (21.6) | 42 (11.3) | 75 (39.5) | 67 (37.9) | ||
Pathologic TIL analysis was performed on all tumors from the Women’s Circle of Health Study (WCHS); molecular TIL analysis was performed on tumors from a subset of WCHS patients, supplemented with additional tumors from the Roswell Park Pathology Network Shared Resource. Patients were selected based on matching by race, age, and tumor subtype, oversampling patients with HER2+ and triple-negative breast cancer (TNBC) for Black–White comparisons. ER = estrogen receptor; PR = progesterone receptor; HER2 = human epidermal growth factor 2; TIL = tumor-infiltrating lymphocytes.
Gene Expression Profiling of Breast TME
The NanoString PanCancer Immune Panel was used to quantify immune cell subsets in the breast TME from WCHS patients, complemented with samples from the Roswell Park Pathology Network Shared Resource. Tissue samples from 190 Black breast cancer patients were frequency matched to 177 White patients by age at diagnosis and tumor subtypes. Women with HER2-positive and triple-negative breast cancer were oversampled to allow comparisons between White and Black women by subtype. Patient characteristics are summarized in Table 1. Two 10-micron curls were cut from a formalin-fixed parafin-embedded block for RNA extraction.
Normalized and transformed gene expression data were used to estimate the absolute fractions (ie, relative to all types of cells in the bulk tissue) of 10 major immune cell subsets derived from the literature where consensus could be reached across multiple algorithms (18) (total T cells, CD4+ T cells, CD8+ T cells, regulatory T cells, B cells, macrophages, neutrophils, mast cells, dendritic cells, and natural killer cells). The relative fractions of each immune cell type (ie, relative to all infiltrating immune cells) were calculated as the ratio of the absolute fraction to CD3+ cells. In a subset of patients with both pathological and molecular TIL scores, the correlations between the 2 were moderate to strong (Supplementary Figure 1, available online; r = 0.65; P < 2.2 x 10-16).
Publicly Accessible Datasets
We used 2 publicly accessible datasets: TCGA breast cancer subset (n = 1080, including 180 Blacks and 714 Whites) (19) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC; n = 1904 Whites only) (20). TCGA was used as a validation cohort for racial differences in immune infiltrates identified in WCHS, and TCGA and METABRIC were used for survival analysis of tumor immune phenotypes. In addition, genomic estimates of immune receptor repertoire were obtained from TCGA (21). Patient selection and inclusion of the studies are depicted in a CONSORT flow diagram (Supplementary Figure 2, available online).
Statistical Analysis
Comparisons of tumor-immune phenotypes between tumors from Black and White patients were conducted using Wilcoxon tests. Multivariable linear regression models were used to derive standardized residuals for race after controlling for hormone receptor status (estrogen receptor and progesterone receptor). Analyses of all-cause mortality with tumor immune phenotypes were conducted using Cox proportional hazards regression, with hazard ratios (HR) and 95% confidence intervals (CI) adjusted for clinical prognostic variables and the assumption of proportionality verified by time-independent Schoenfeld residuals. Analyses of disease-specific mortality were conducted by treating other causes of death as competing risk. The mean follow-up time (range) of TCGA, WCHS, and METABRIC was 27 (0-287) months, 72 (9-153) months, and 115 (0-355) months, respectively. All analyses were 2-sided and performed using R 3.6.1. Multiple comparison error was controlled for using the Bonferroni method, with a family-wise error rate of .05.
Results
Pathological TIL Scores
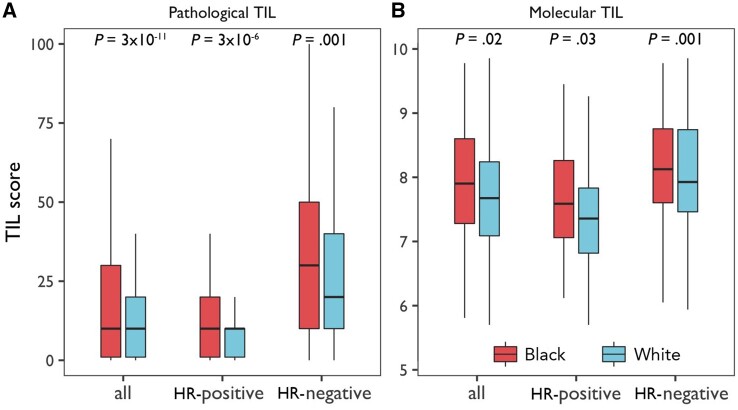
Among WCHS patients, TIL scores were higher in hormone receptor–negative than hormone receptor–positive tumors; no associations with HER2 status were found after adjusting for hormone receptor status (Supplementary Figure 3, available online). In comparisons by race overall and by hormone receptor subtype, tumors from Black women had higher pathological TIL scores than those from White women (all P ≤ .001) (Figure 1, A), with similar associations noted in molecular estimates of TILs in a subset of the patients (all P ≤ .03) (Figure 1, B).
Figure 1.
Pathological and molecular estimates of tumor-infiltrating lymphocytes in Black and White breast cancer patients in the Women’s Circle of Health Study. A) Boxplots of pathological tumor-infiltrating lymphocytes (TIL) scores by race, with and without stratification by tumor hormone receptor (HR) status. The bar in the middle of a box indicates the subgroup median, and the lower and upper edges indicate the first and third quartiles, respectively. The extended lines indicate the range in each subgroup. P values were derived from 2-sided Wilcoxon test between Black and White patients. B) Boxplots of molecular TIL scores estimated based on immune panel gene expression data, with and without stratification by tumor hormone receptor status.
Higher pathological TIL scores in Black patients in WCHS were associated with all-cause and disease-specific mortality (Supplementary Table 3, available online). Patients with lymphocyte-predominant breast cancer (TIL score ≥50%) had a statistically significantly lower hazard of all-cause mortality (HR = 0.38, 95% CI = 0.19 to 0.76), in comparison to those with lymphocyte-depleted breast cancer (TIL score <10%).
Molecular Estimates of Immune Infiltrates in Breast TME
Consistent with the results of pathological TIL assessment, no associations between HER2 status and the absolute or relative fractions of any immune cell subsets were found after adjusting for hormone receptor status (Supplementary Figure 4, available online). Thus, we relied on hormone receptor status to assess the tumor influences on immune estimates in TME, which is also supported by previous studies demonstrating 2 major etiological subtypes of breast cancer proximated by hormone receptor status (22). Hormone receptor–negative cancers had statistically significantly higher absolute fractions of all immune cell subsets than hormone receptor–positive cancers, except for mast cells, which differed in an opposite direction (Supplementary Figure 4, available online). In contrast, similar comparisons of the relative fractions of immune cells revealed no differences in any of the immune cells except mast cells. Because the relative fractions were not strongly confounded by tumor hormone receptor status, they were used in subsequent analyses unless otherwise specified.
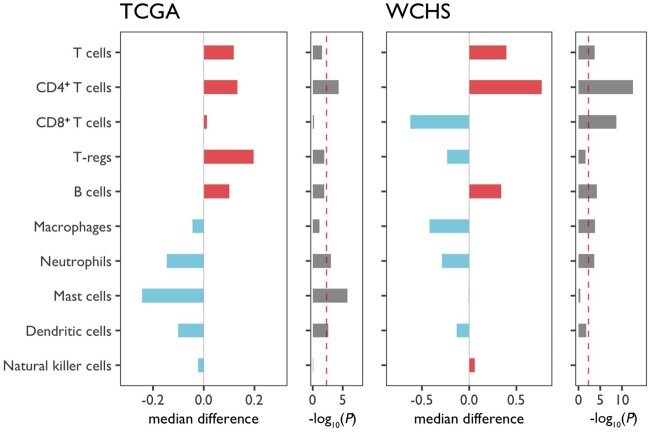
As shown in Figure 2 and Supplementary Figure 5 (available online), in contrast to lower fractions of neutrophils and dendritic cells in Blacks, the most statistically significant and consistent racial differences in immune infiltrates were higher fractions of CD4+ T cells in Black patients compared with White patients in both WCHS and TCGA. The racial differences in immune cell subsets remained after adjusting for or stratifying by tumor hormone receptor status (Supplementary Figures 6 and 7, available online). In a validation study of CD4+ T cells using immunohistochemical (IHC) staining and WCHS tissue microarrays, there was a moderate-to-strong correlation between IHC and molecular scores of CD4+ T cells (r = 0.59; P < 2.2 x 10-16); higher IHC scores of CD4 were also observed in Black than in White patients, although the differences were restricted to hormone receptor–negative cancer subtype (Supplementary Figure 8, available online).
Figure 2.
Racial differences in the relative fractions of immune cells between Black and White breast cancer patients. Relative fractions of the 10 major immune cell subsets plus CD4+ to CD8+ T-cell ratios were centered to an overall mean of 0 before subgroup medians were calculated for Whites and Blacks. The differences in subgroup medians are plotted in bar graphs, with red indicating higher immune cell estimates in Blacks, and blue indicating higher estimates in Whites. P values from 2-sided Wilcoxon test were log10-transformed, negated, and plotted as bar graphs along the median differences. The red reference line indicates a cutoff of statistical significance level after adjusting for multiple testing [-log10(0.05/11) = 2.3]. Patient populations from Women’s Circle of Health Study (WCHS) and The Cancer Genome Atlas (TCGA) were included for cross-validation.
Among CD4+ T-cell subsets, the fraction of T follicular helper (TFH) cells, as estimated by CIBERSORT algorithm (23), showed the most statistically significant difference between Black and White patients in WCHS and TCGA (Supplementary Figure 9, available online). In a recent pan-cancer TCGA analysis, it was reported that rs3366 in the 3’ untranslated region (UTR) of SIK1 was associated with the fraction of TFH cells present in bulk tumor tissues (24). We thus assessed the racial differences in the distribution of this variant. According to gnomAD data (25), the allele associated with a higher TFH fraction had a markedly higher frequency in Black than in White populations (0.45 vs 0.09). This corroborated our findings of higher fractions of TFH cells in Black than in White breast cancer patients.
Consistent with a stronger CD4+ T-cell response, B-cell fractions and B-cell receptor (BCR) clonality were also higher in Black than White patients in TCGA, but not T-cell receptor clonality (Supplementary Figure 10, available online). Moderate-to-strong correlations were noted between the fractions of CD4+ T cells and B cells and BCR diversity and richness (Supplementary Figure 11, available online).
T-Cell Exhaustion Markers and “ExCD8-r” Signature
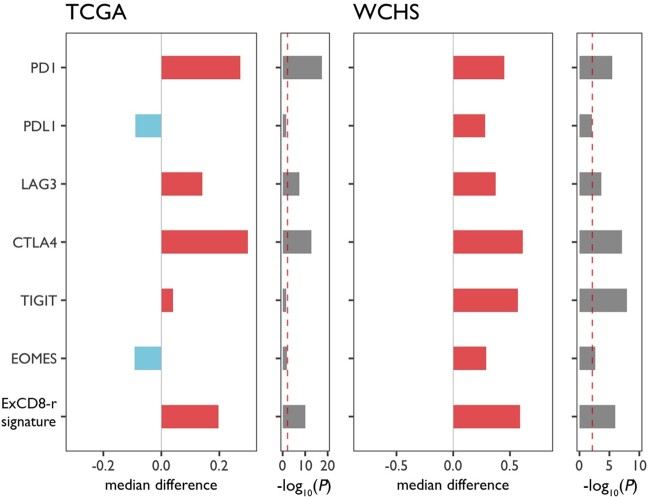
Because no consistent racial differences were seen in the CD8+ T-cell fractions (Figure 2), the major immune cell subtype with antineoplastic activity, we examined the expression of 4 major inhibitory receptors (IRs: PD-1, LAG-3, CTLA-4 and TIGIT) and the PD-1 ligand. The co-expression of these markers represents cardinal features of T-cell exhaustion and serves as targets of immune checkpoint inhibitors (26). As expected, these IRs and the PD-1 ligand displayed a strong co-expression pattern (Supplementary Figure 12, available online). When the expression levels were analyzed relative to the absolute fraction of CD8+ T cells, PD-1, LAG-3, and CTLA-4 were statistically significantly higher in breast tumors from Black patients in comparison with White patients in both WCHS and TCGA (all P < .001; Figure 3).
Figure 3.
Racial differences in T-cell exhaustion markers and ExCD8-r signature between Black and White breast cancer patients. The differences between Blacks and Whites in the expression levels of T-cell exhaustion markers relative to the absolute fractions of CD8+ T cells, as well as the ExCD8-r signatures, are plotted in bar graphs, with red indicating higher immune cell estimates in Blacks, and blue indicating higher estimates in Whites. P values from Wilcoxon test are log10-transformed, negated, and plotted as bar graphs along the median differences. The red reference line indicates log10(0.05/6) = 2.1. Patient populations from Women’s Circle of Health Study (WCHS) and The Cancer Genome Atlas (TCGA) were used for cross-validation. PD-1 = programmed cell death protein 1; PD-L1 = PD-1 ligand; LAG-3: lymphocyte-activation gene 3; CTLA-4: cytotoxic T-lymphocyte-associated protein 4; TIGIT = T Cell Immunoreceptor With Ig And ITIM Domains.
To develop an expression signature reflecting the balance between the cytotoxic and exhausted states of CD8+ T cells, we leveraged the co-expression pattern of PD-1 and LAG-3 as hallmarks of T-cell exhaustion, demonstrated in previous studies from our group and others (27‐29). Because Eomesodermin (Eomes) is a key transcription factor upregulated in terminally differentiated exhausted T cells (30), we defined the resultant signature “ExCD8-r” as the ratio of the aggregated expression of PD-1, LAG-3, and Eomes to the absolute CD8+ T-cell fraction. The signature scores were statistically significantly higher in tumors from Black patients (all P ≤ .002; Figure 3), regardless of hormone receptor status (Supplementary Figure 13, available online), suggesting that the CD8+ T-cell responses in breast tumors from Black women were more likely to assume an exhausted state than in Whites. In analysis of a recently developed T-cell exhaustion signature by Cai et al. (31), a strong correlation was noted with the signature we developed prior to adjusting for the absolute CD8+ T-cell fraction (r = 0.89; P < 2.2 x 10-16); the signature scores by Cai et al. were also statistically significantly higher in tumors from Black vs White patients in WCHS (Supplementary Figure 14, available online).
Prognostic Value of ExCD8-r Signature
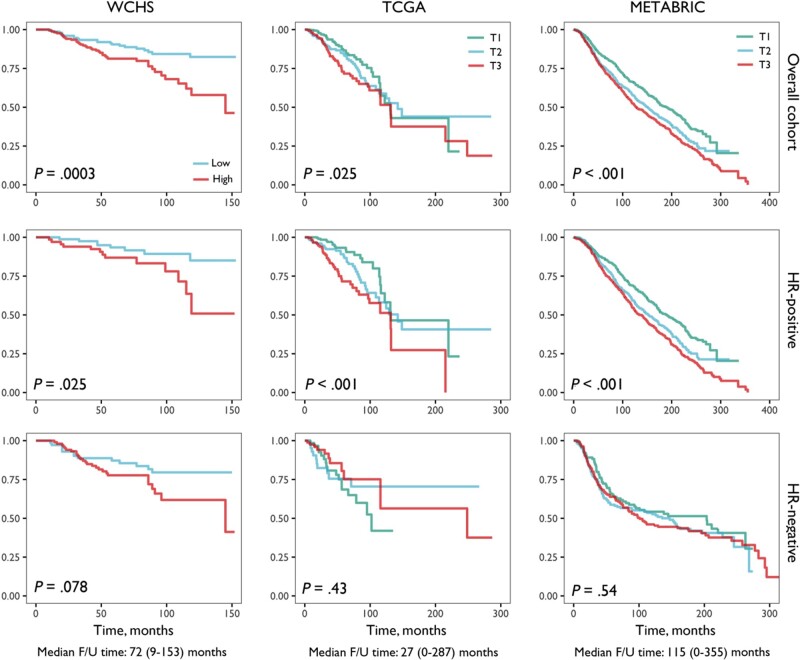
We next examined the relationships of the ExCD8-r signature with patient survival in WCHS, TCGA, and METABRIC with all races combined because of limited sample size for race-stratified analysis. A higher ExCD8-r signature was consistently associated with poorer all-cause mortality (Figure 4) and disease-specific mortality (Supplementary Figure 14, available online). The associations remained statistically significant after adjusting for age and clinical prognostic factors (Supplementary Table 2, available online, for all-cause mortality, and Supplementary Table 3, available online, for disease-specific mortality). Further adjusting for other immune cell subsets in the models had no substantial effects. Upon stratification by hormone receptor status, the prognostic value of ExCD8-r signature was limited to hormone receptor–positive breast cancer and not statistically significant in hormone receptor–negative cancer (Figure 4;Supplementary Tables 2 and 3, available online).
Figure 4.
Kaplan-Meir survival curves of all-cause mortality by the levels of ExCD8-r signature. Kaplan-Meier survival curves of all-cause mortality by the levels of ExCD8-r signature are plotted for Women’s Circle of Health Study (WCHS), The Cancer Genome Atlas (TCGA), and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) datasets, with and without stratification by tumor hormone receptor (HR) status. The signature levels in WCHS were categorized into binary based on the median because of limited sample size and categorized into thirds in TCGA and METABRIC datasets with large sample sizes. P values from log-rank test are displayed within the plots. The medians and ranges of the follow-up (F/U) time are shown at the bottom. The analyses were conducted with all races combined because of limited sample size of Black patients in the cohorts.
Because high absolute fractions of CD8+ T cells have been consistently associated with better prognosis among hormone receptor–negative breast cancer patients (32), we sought to determine the prognostic value of ExCD8-r signature in the context of CD8+ abundance. Notably, the absolute fractions of CD8+ T cells were statistically significant only in hormone receptor–negative patients in METABRIC but not in WCHS or TCGA, possibly because of limited sample size. In METABRIC, when CD8+ T-cell fractions were combined with the ExCD8-r signature, patients classified as CD8lowExCD8-rhigh had the highest all-cause mortality and disease-specific mortality; although unexpectedly, those classified as CD8highExCD8-rhigh, instead of CD8highExCD8-rlow, had the lowest mortality (Supplementary Figure 15 and Supplementary Table 4, available online).
Lastly, we compared the proportions of the 4 subgroups defined by the combination of the absolute fractions of CD8+ T cells and the ExCD8-r signature between Black and White patients. As shown in Figure 5, in both WCHS and TCGA, a CD8lowExCD8-rhigh profile was the most common subgroup in Black patients, in contrast to the CD8highExCD8-rlow profile being the most prevalent in White patients. Even among those with high CD8+ T-cell fractions, Black patients were still more likely than White patients to have tumors with CD8highExCD8-rhigh features. Similar findings were observed when stratified by tumor hormone receptor status (Supplementary Figure 16, available online).
Figure 5.
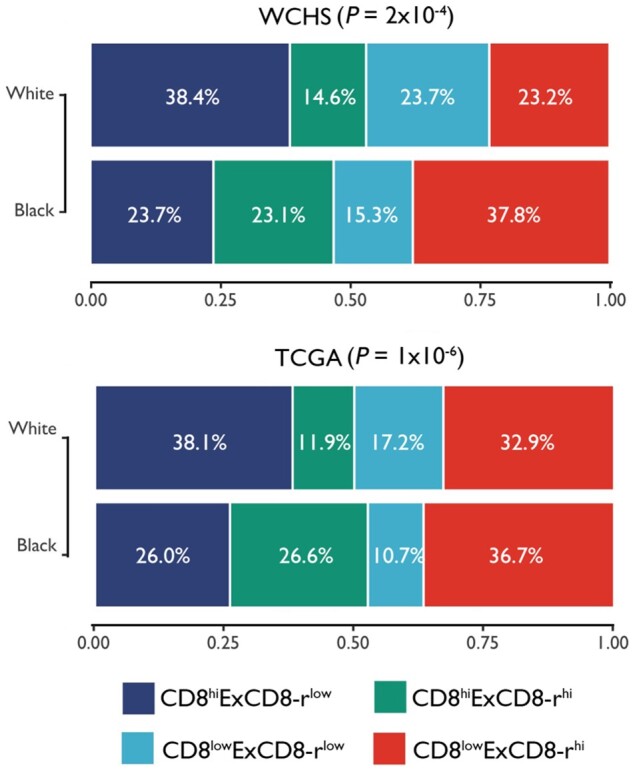
Breast cancer patient subgroups defined by the combination of the absolute CD8+ T-cell fractions and the ExCD8-r signature in Blacks and Whites. Stacked bar graphs of the proportions of the 4 patient subgroups were defined on the basis of the combination of the absolute fractions of CD8+ T cells and the ExCD8-r signature in Black and White breast cancer patients. The bar height of each subgroup corresponds to the percentage of that group within the racial group. P values from χ2 test between subgroup and race are displayed for Women’s Circle of Health Study (WCHS) and The Cancer Genome Atlas (TCGA) datasets.
Discussion
Our findings of stronger overall immune response in breast tumors from Black women compared with White women are consistent with results from a prior TCGA analysis showing that African ancestry was associated with a higher leukocyte fraction in breast tumors (21), as well as those from a study in colorectal cancer, showing a stronger lymphocytic reaction in Black patients (33). As Black individuals tend to exhibit stronger inflammation-related systemic immunity (8‐10), these data together suggest that it may give rise to stronger immune responses in the TME among Black women with breast cancer.
Our data support the notion that the composition and quality of immune infiltrates may be as important as their absolute presence in the TME. Black patients have tumor immune reactions shifted toward humoral immunity characterized by higher levels of CD4+ T cells, TFH cells, B cells, and BCR clonality. Although the antineoplastic activity of humoral immunity has been less well understood than cellular immunity, several recent studies have demonstrated the importance of TFH cells and B cells in immunotherapy (34‐37). Moreover, Black patients also exhibited a T-cell response of exhaustion features associated with poor survival, possibly because of diminished effector cytokine production and cytolytic activity (26). Thus, it may be the higher proportion of the T-cell exhaustion feature among Black patients that accounts for the seeming contradiction of poorer survival outcomes, despite the stronger overall immune presence of TILs in the TME.
The greater presence of exhausted immune phenotypes in tumors from Black women is consistent with the premise that a more active and prolonged inflammatory immune response in individuals of African descent, shaped over millennia in tropical Africa, could be related to more aggressive breast cancer (7,38). Host immunity is among the biologic mechanisms most frequently subject to natural selection (39,40) driven by infectious pathogens, a major environmental force in recent human evolutionary history (41). Because tropical regions in Africa are rich in pathogenic agents, early populations likely benefited from a more active immune defense, and genetic variants that enhance immune responses were likely positively selected. Indeed, allele frequencies of immune genes often display large, and sometimes extreme, racial differences (42,43). A potential approach to further investigate this hypothesis is to identify germline genetic variants in association with tumor immunophenotypes in racially diverse populations, ideally through genome-wide methods. Probably more than a coincidence, the only genome-wide significant association identified in a recent study of tumor immunophenotypes was between a variant in SIK and TFH cells (24), which happened to be the most statistically significantly differentially occurring CD4+ T-cell subset found in our study, and the variant has a much higher allele frequency in Blacks than in Whites. Given the limited sample size of our study, a genome-wide study of genetic variations is out of scope, but future studies in this area are warranted.
The literature indicates that hormone receptor–negative breast cancers are more immunologically “hot” than hormone receptor–positive tumors (44,45), and our data support that premise. Interestingly, the ExCD8-r signature was prognostic only among hormone receptor–positive but not hormone receptor–negative cancers. A new study using single cell proteomics also revealed CD8+ T-cell exhaustion in a subset of hormone receptor–positive breast cancers characterized by increased PD-1 and IR co-expression (46). In another study of patients with estrogen receptor–positive cancer, expression feature of CD8+ PD-1+/CTLA-4+ predicted response to immunotherapy (47). These data suggest that even among the immunologically “cold” hormone receptor–positive breast cancers, tumor immune response is relevant to patient survival and could potentially be targeted for immunotherapy, which is currently being evaluated in clinical trials. The finding may also have relevance to breast cancer health disparities, because some studies reported that the most persistent Black–White differences in breast cancer survival occurred within hormone receptor–positive cancers (48).
Our findings of distinct tumor immune responses in Black breast cancer patients may have clinical implications. Immune checkpoint inhibitors target the inhibitory receptors expressed on T cells, and their corresponding ligands on tumor or stromal populations such as myeloid cells, to reinvigorate the exhausted effector CD8+ T cells. The apparently stronger but more exhausted lymphocytic state in Black patients implies that a higher response rate to checkpoint inhibitors might be expected. Further, the stronger B-cell response in Black patients and the newly discovered role of tumor-infiltrating B cells in regulating response to immunotherapy provide further support this postulation. To our knowledge, however, few clinical trials on immune checkpoint inhibitors have reported race-specific outcome data. Enhanced recruitment of racial and ethnic minorities into clinical trials is warranted to close the gaps in cancer disparities and to ensure that all groups of patients will benefit from advances in cancer therapeutics (49).
In summary, we found statistically significant and consistent racial differences in immune infiltrates in the breast TME, in particular, a CD8+ T-cell response shifted toward an exhausted state in Black patients. These findings may explain the contradiction of a strong immune presence in the breast TME of Black patients, yet poorer survival, providing a new immunobiological perspective to underlying causes of breast cancer racial disparities.
Funding
This work was supported by the Breast Cancer Research Foundation (Principal Invesitgator: Ambrosone CB). It is also supported, in part, by the National Cancer Institute (NCI) grants P01CA151135 (Multiple Principal Invesitgators: Ambrosone CB, Palmer JR, Olshan A), R01CA100598 (Principal Invesitgator: Ambrosone CB), and R01 CA185623 (Multiple Principal Invesitgators: Bandera EV, Hong CC, Demissie K). Dr Cheng is a recipient of an NCI career development award (K07 CA201334). Roswell Park Comprehensive Cancer Center Data Bank and Biorepository, the Pathology Network Shared Resource, and Genomics Shared Resource are Cancer Center Support Grant (CCSG) Shared Resource are supported by the hormone receptor–grant P30CA16056 (Principal Investigator: Johnson CS).
Notes
Role of the funders: The study sponsors play no roles in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: The authors declare no potential conflicts of interest.
Acknowledgements: We thank participants and staff of the Women’s Circle of Health Study.
Author contributions: Conceptualization: SY, TK, CBA; Methodology: SY, TYC, AE, LY, WD, SL, TK, CBA; Validation: SY, AO, TK, CBA; Formal analysis: SY, TYC, AE, LY, TK, CBA; Investigation: SY, TY, AE, TK, CBA; Resources: SY, CBA; Data curation: SY, TYC, AE, KT, TK, CBA; Writing—original draft: SY, TYC, AE, CBA; Writing—review & editing: SY, TYC, AE, LY, AO, SIA, SE, CCH, QQ, WD, SL, EVB, KO, KT, TK, CBA; Visualization: SY, LY; Supervision: KT, TK, CBA; Funding acquisition: CBA; Project administration: SY, KT, CBA.
Data Availability
The tumor gene expression data from the NanoString panel were deposited into Gene Expression Omnibus (GEO) database for public access (GSE163418).
Supplementary Material
References
- 1. DeSantis CE, Miller KD, Goding Sauer A, et al. Cancer statistics for African Americans, 2019. CA A Cancer J Clin. 2019;69(3):211–233. [DOI] [PubMed] [Google Scholar]
- 2. Danforth DN Jr. Disparities in breast cancer outcomes between Caucasian and African American women: a model for describing the relationship of biological and nonbiological factors. Breast Cancer Res. 2013;15(3):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ademuyiwa FO, Edge SB, Erwin DO, et al. Breast cancer racial disparities: unanswered questions. Cancer Res. 2011;71(3):640–644. [DOI] [PubMed] [Google Scholar]
- 4. Amend K, Hicks D, Ambrosone CB.. Breast cancer in African-American women: differences in tumor biology from European-American women. Cancer Res. 2006;66(17):8327–8330. [DOI] [PubMed] [Google Scholar]
- 5. Albain KS, Unger JM, Crowley JJ, et al. Racial disparities in cancer survival among randomized clinical trials patients of the Southwest Oncology Group. J Natl Cancer Inst. 2009;101(14):984–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yao S, Hong CC, Ruiz-Narvaez EA, et al. Genetic ancestry and population differences in levels of inflammatory cytokines in women: role for evolutionary selection and environmental factors. PLoS Genet. 2018;14(6):e1007368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ye CJ, Feng T, Kwon HK, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345(6202):1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nedelec Y, Sanz J, Baharian G, et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016;167(3):657–669.e21. [DOI] [PubMed] [Google Scholar]
- 10. Quach H, Rotival M, Pothlichet J, et al. Genetic adaptation and neandertal admixture shaped the immune system of human populations. Cell. 2016;167(3):643–656.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin DN, Boersma BJ, Yi M, et al. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One. 2009;4(2):e4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wallace TA, Prueitt RL, Yi M, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68(3):927–936. [DOI] [PubMed] [Google Scholar]
- 13. O’Meara T, Safonov A, Casadevall D, et al. Immune microenvironment of triple-negative breast cancer in African-American and Caucasian women. Breast Cancer Res Treat. 2019;175(1):247–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pitt JJ, Riester M, Zheng Y, et al. Characterization of Nigerian breast cancer reveals prevalent homologous recombination deficiency and aggressive molecular features. Nat Commun. 2018;9(1):4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aran D, Sirota M, Butte AJ.. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6(1):8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ambrosone CB, Ciupak GL, Bandera EV, et al. Conducting molecular epidemiological research in the age of HIPAA: a multi-institutional case-control study of breast cancer in African-American and European-American women. J Oncol. 2009;2009:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salgado R, Denkert C, Demaria S, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26(2):259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Danaher P, Warren S, Dennis L, et al. Gene expression markers of tumor infiltrating leukocytes. J Immunother Cancer. 2017;5(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Curtis C, Shah SP, Chin SF, et al. ; METABRIC Group. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anderson WF, Rosenberg PS, Prat A, et al. How many etiological subtypes of breast cancer: two, three, four, or more? J Natl Cancer Inst. 2014;106(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shahamatdar S, He MX, Reyna MA, et al. Germline features associated with immune infiltration in solid tumors. Cell Rep. 2020;30(9):2900–2908.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wherry EJ, Kurachi M.. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, et al. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci USA. 2010;107(17):7875–7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grosso JF, Goldberg MV, Getnet D, et al. Functionally distinct LAG-3 and PD-1 subsets on activated and chronically stimulated CD8 T cells. J Immunol. 2009;182(11):6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blackburn SD, Shin H, Haining WN, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Intlekofer AM, Takemoto N, Wherry EJ, et al. Effector and memory CD8+ T cell fate coupled by T-bet and Eomesodermin. Nat Immunol. 2005;6(12):1236–1244. [DOI] [PubMed] [Google Scholar]
- 31. Cai MC, Zhao X, Cao M, et al. T-cell exhaustion interrelates with immune cytolytic activity to shape the inflamed tumor microenvironment. J Pathol. 2020;251(2):147–159. [DOI] [PubMed] [Google Scholar]
- 32. Ali HR, Provenzano E, Dawson SJ, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–1543. [DOI] [PubMed] [Google Scholar]
- 33. Wallace K, Lewin DN, Sun S, et al. Tumor-infiltrating lymphocytes and colorectal cancer survival in African American and Caucasian patients. Cancer Epidemiol Biomarkers Prev. 2018;27(7):755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hollern DP, Xu N, Thennavan A, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell. 2019;179(5):1191–1206.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Helmink BA, Reddy SM, Gao J, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petitprez F, de Reynies A, Keung EZ, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. [DOI] [PubMed] [Google Scholar]
- 37. Cabrita R, Lauss M, Sanna A, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. [DOI] [PubMed] [Google Scholar]
- 38. Gong Z, Quan L, Yao S, et al. Innate immunity pathways and breast cancer risk in African American and European-American women in the Women’s Circle of Health Study (WCHS). PLoS One. 2013;8(8):e72619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barreiro LB, Quintana-Murci L.. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11(1):17–30. [DOI] [PubMed] [Google Scholar]
- 40. Deschamps M, Laval G, Fagny M, et al. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet. 2016;98(1):5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fumagalli M, Sironi M, Pozzoli U, et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7(11):e1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quan L, Gong Z, Yao S, et al. Cytokine and cytokine receptor genes of the adaptive immune response are differentially associated with breast cancer risk in American women of African and European ancestry. Int J Cancer. 2014;134(6):1408–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hong CC, Sucheston-Campbell LE, Liu S, et al. Genetic variants in immune-related pathways and breast cancer risk in African American women in the AMBER consortium. Cancer Epidemiol Biomarkers Prev. 2018;27(3):321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stanton SE, Adams S, Disis ML.. Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes: a systematic review. JAMA Oncol. 2016;2(10):1354–1360. [DOI] [PubMed] [Google Scholar]
- 45. Ali HR, Chlon L, Pharoah PD, et al. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PLoS Med. 2016;13(12):e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wagner J, Rapsomaniki MA, Chevrier S, et al. A single-cell atlas of the tumor and immune ecosystem of human breast cancer. Cell. 2019;177(5):1330–1345.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Terranova-Barberio M, Pawlowska N, Dhawan M, et al. Exhausted T cell signature predicts immunotherapy response in ER-positive breast cancer. Nat Commun. 2020;11(1):3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tao L, Gomez SL, Keegan TH, et al. Breast cancer mortality in African-American and non-Hispanic White women by molecular subtype and stage at diagnosis: a population-based study. Cancer Epidemiol Biomarkers Prev. 2015;24(7):1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harris Y, Gorelick PB, Samuels P, et al. Why African Americans may not be participating in clinical trials. J Natl Med Assoc. 1996;88(10):630–634. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The tumor gene expression data from the NanoString panel were deposited into Gene Expression Omnibus (GEO) database for public access (GSE163418).