Abstract
Computer aided detection (CADe) and computer aided diagnostic (CADx) systems are ongoing research areas for identifying lesions among complex inner structures with different pixel intensities, and for medical image classification. There are several techniques available for breast cancer detection and diagnosis using CADe and CADx systems. However, some of these systems are not accurate enough or suffer from lack of sufficient data. For example, mammography is the most commonly used breast cancer detection technique, and there are several CADe and CADx systems based on mammography, because of the huge dataset that is publicly available. But, the number of cancers escaping detection with mammography is substantial, particularly in dense-breasted women. On the other hand, digital breast tomosynthesis (DBT) is a new imaging technique, which alleviates the limitations of the mammography technique. However, the collections of huge amounts of the DBT images are difficult as it is not publicly available. In such cases, the concept of transfer learning can be employed. The knowledge learned from a trained source domain task, whose dataset is readily available, is transferred to improve the learning in the target domain task, whose dataset may be scarce. In this paper, a two-level framework is developed for the classification of the DBT datasets. A basic multilevel transfer learning (MLTL) based framework is proposed to use the knowledge learned from general non-medical image datasets and the mammography dataset, to train and classify the target DBT dataset. A feature extraction based transfer learning (FETL) framework is proposed to further improve the classification performance of the MLTL based framework. The FETL framework looks at three different feature extraction techniques to augment the MLTL based framework performance. The area under receiver operating characteristic (ROC) curve of value 0.89 is obtained, with just 2.08% of the source domain (non-medical) dataset, 5.09% of the intermediate domain (mammography) dataset, and 3.94% of the target domain (DBT) dataset, when compared to the dataset reported in literature.
Keywords: Digital breast tomosynthesis, Transfer learning, Deep learning, Feature fusion, GLCM
Introduction
Medical image classification is an important research area, as it helps doctors in classifying the medical images into different categories, thus helping them in disease diagnosis. Recently, computer aided detection (CADe) and computer aided diagnostic (CADx) systems are gaining momentum in the medical image classification field, due to their improved performance in such detection and diagnosis tasks. Such systems are able to analyze medical images and identify suspicious areas, which are relevant to the radiologist findings. But the drawback with such CADe and CADx systems are in the domains, where data is insufficient and not publicly available. In such situations, the concept of transfer learning, where the knowledge learned by training with huge datasets in a source domain can be transferred and utilized for classification in the target domain with insufficient data, can be applied.
Breast cancer is a problem due to its high prevalence and mortality rate [1]. The survival rate of breast cancer depends on the stage at which it is diagnosed. Ultrasound, mammography, and digital breast tomosynthesis (DBT) are the predominant breast cancer imaging techniques used for the detection and diagnosis of breast cancer. Among these imaging techniques, mammography is the prevalent breast cancer screening technique, due to its wide spread availability. However, it has limitations in detecting lesions for women with dense breasts [2, 3]. DBT is a new imaging modality, which is widely used for breast cancer screening nowadays, and it is the most effective method for detection of early breast cancer. But, building CADe and CADx systems using this modality is challenging, since the DBT dataset is limited and is not publicly available. In order to overcome this problem, the concept of transfer learning can be applied to classify the DBT dataset, by transferring the knowledge learned from the mammography dataset and also non-medical image datasets.
In this paper, a two-level framework is developed for breast cancer classification. The first level builds a basic multilevel transfer learning (MLTL) framework based on the work proposed by [4]. This MLTL framework consists of the source, intermediate, and target domains. Widely available non-medical images dataset is used as the source domain, and the basic common features are transferred to the intermediate domain, consisting of the mammography dataset. Thereafter, the features of the mammography dataset are transferred to the target domain, consisting of the DBT dataset and the classification of the DBT dataset is performed. The proposed second-level framework, feature extraction based transfer learning (FETL), improves the performance of the MLTL framework using the following feature extraction techniques:
CNCF fusion algorithm to combine high-level and low-level features of the target domain images.
GLCM based feature extraction technique to extract a set of texture features from the target domain images.
Multi-input perceptron algorithm to extract features from the patient reports along with the target domain images.
These techniques retrieve the significant features from the images and play an important role in analyzing the medical images for identifying lesions, cysts, and calculi, which help in classification. The resultant features from the above three proposed techniques are used for classification separately and their results are analyzed.
Material and Methods
Related Work
In recent years, the concept of transfer learning has shown that the problems associated with the classification of the target domains with insufficient training data can be alleviated [5]. This can be done by transferring the knowledge from the source domain network, which is trained on a large-scale dataset. The concept of transfer learning has been successfully applied in the classification tasks of various domains.
Siyu Jiang et al. [6] have introduced multilabel metric transfer learning (MLMTL) for the classification of scene, emotion, BibTeX, and Enron multilabel datasets, by adjusting the instance-level, as well as label-level distributions, with the help of distance metrics, such as maximum mean discrepancy, whereas other algorithms consider only the differences between instance-level distributions.
In the manufacturing field, the concept of transfer learning has been used for fault diagnosis of a machine. Yan Xu et al. [7] have proposed two phase fault diagnosis transfer learning model to overcome the problem of insufficient training data and different data distributions, by simulating various manufacturing strategies virtually and have obtained a balanced dataset. Deep neural network-based diagnosis models have been trained with virtually generated data. Using transfer learning, the pretrained model is migrated to the physical space for real-time fault diagnosis in the car body-side production.
The transfer learning concept has also been used in text classification [8] and sentiment analysis [9] to improve the performance. To overcome the problem of imbalanced dataset in the real-world Chinese sentiment classification datasets, Xiao et al. [10] have used the pretrained network on hotel service reviews, which is a class balanced dataset. By using an under sampling technique and transferring the pretrained model, the classification performance of the tourist attraction dataset has been greatly improved.
Catherine Sandoval et al. [11] have proposed a two-stage classification approach for the classification of publicly available fine-art paintings. Five patches of image data have been trained using six standard pretrained convolutional neural networks (CNNs) such as AlexNet, VGG-16 in the first stage. The class probability vectors generated from the first stage classifier have been used to make the final decision more accurate in the second stage.
In recent times, transfer learning concepts have been used for applications dealing with online target domain data [12–14] as well as with heterogeneous source domains for the classification of target domain data [15, 16].
Transfer learning techniques have been used in the medical imaging field and have shown significant improvements in lung disease classification [17], EEG signal recognition [18], and telemonitoring of Parkinson’s disease [19]. Veronika Cheplygina et al. [17] have proposed an instance-transfer approach for the classification of chronic obstructive pulmonary disease from the chest CT scans, to generalize the existing methods across domains such as different scanners or scanning protocols. The joint-knowledge transfer approach with domain adaptation has been proposed by Zhaohong Deng et al. [18], for EEG signal recognition. Labeled dataset has been used for training, and unlabeled Bonn and CHBMIT datasets have been used as test sets. Iterative transfer learning has been applied to get higher accuracy. An important issue in transfer learning is negative transfer [19], which degrades the performance, when compared to techniques without using transfer learning. Hyunsoo Yoon and Jing Li [19] have proposed a positive transfer learning approach for telemonitoring of Parkinson’s disease by transferring the patient-specific model parameters, but not the patient’s data.
To reduce the domain difference between the source and the target domains, an intermediate domain can be introduced. Ravi K. Samala et al. [4] have proposed multistage transfer learning for the classification of the DBT dataset with AlexNet as source domain and the mammography dataset as intermediate domain.
With the advancements in image processing, machine learning, and deep learning techniques, many methodologies have been used for the classification of breast cancer images. Chenchen Wu et al. [20] have proposed transfer learning models, combined with an effective adaptive sampling method to identify tumors in whole-slide images of breast cancer that notably improves the classification accuracy.
Transfer learning based dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) method has been used for classifying fibroadenoma and invasive ductal carcinoma in breast tumors. Leilei Zhou et al. [21] have introduced two lesion-level models based on InceptionV3 and VGG19 networks, which are pretrained with the ImageNet dataset and the effects of different depths of transfer learning have been examined.
Xinfeng Zhang et al. [22] have developed an advanced ensemble classification based on deep learning approach, to boost the prediction efficiency of cancer prognosis. An autoencoder (AE) neural network has been used to classify different features within the gene expressions, and many microarray studies have been analyzed to assess the clinical outcome of breast cancer.
Mohammad Alkhaleefah et al. [23] have shown the importance of image augmentation for the classification of breast cancer using transfer learning techniques. Data augmentation techniques such as rotation, flipping, and zooming are applied to generate new training samples based on the existing breast x-ray images, to overcome the lack of sufficient datasets, and also to improve the classification accuracy.
Deep learning assisted efficient AdaBoost algorithm (DLA-EABA) for breast cancer detection has been mathematically proposed by Jing Zheng [24], with advanced computational techniques. In addition to traditional computer vision approaches, tumor classification methods have also been actively developed using deep convolutional neural networks.
Jonathan de Matos et al. [25] have proposed a classification approach for breast cancer histopathologic images (HI) using transfer learning technique. Irrelevant patches have been removed before training a support vector machine (SVM) classifier, to improve the classification accuracy. Hafiz Mughees Ahmad et al. [26] have proposed transfer learning based on AlexNet, GoogleNet, and ResNet to classify breast cancer histology images at multiple cellular and nuclei configurations.
Constance Fourcade et al. [27] have presented a deep learning-based approach with a super pixel segmentation method to segment organs of PET images that helps in finding the evolution of breast cancer metastasis. Mor Yemini et al. [28] have addressed the problem of mass detection in mammograms using CNN and transfer learning techniques. Mohammad Alkhaleefah and Chao-Cheng Wu [29] have used the concept of transfer learning to classify breast cancer images, where the convolutional neural network has been utilized as a feature extractor. In addition to this, an SVM classifier, based on radial basis function (RBF), has been adapted for its flexibility in fitting the data dimension space adequately by tuning the kernel width. This hybridization between CNN and RBF-based SVM has shown robust results.
Ensemble based machine learning algorithms [30, 31], extreme machine learning algorithms, and genetic algorithms [32] have also been used to effectively classify breast cancer images. Sanaz Mojrian et al. [33] have introduced an innovative method for detection and diagnosis of breast cancer, based on a multilayer fuzzy expert system that uses an extreme learning machines (ELM) classifier, combined with RBF, to give better performance compared to the linear SVM model.
In the image processing domain, several feature extraction techniques have been widely used for enhancing the performance of the classifier. Particularly, in the medical domain, various feature extraction techniques [34, 35] have been used to identify the lesion areas and to classify them more accurately. The shape, texture, and intensity features [36] and CNN based image features [37] have been extracted from the brain MRI images and then classified using various machine learning classifiers. The intrinsic pattern extractions from the breast and brain images have been done using hybrid genetic algorithm [35] and gray level co-occurrence matrix (GLCM) embedded with CNN [38], respectively.
Thus, it can be observed that transfer learning can be applied in the medical imaging field, where sufficient data is not available or is expensive. Also, the performance of such systems can be improved further by using appropriate feature extraction methods. This work proposes a two-level, multitransfer learning-based framework, with feature extraction techniques for breast cancer detection. The details of the proposed work are presented below.
Proposed Work
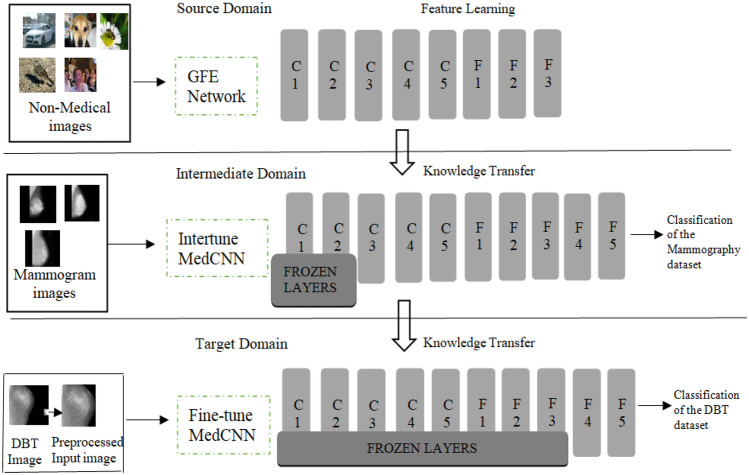
The architecture of the proposed basic MLTL framework is shown in Fig. 1. The generalized feature extraction (GFE) network corresponds to the source domain, which consists of easily available non-medical images. The fine-tune MedCNN corresponds to the target DBT domain. Since these two are totally unrelated domains, an intermediate Intertune MedCNN is introduced as proposed in [4]. The generalized features learned from the GFE network are transferred to the intermediate domain, and the specific features of the medical images learned from the intermediate domain are transferred to the target domain to give good classification results.
Fig. 1.
MLTL framework
The GFE network consists of five convolutional layers, with each convolutional layer followed by max-pooling layers, and two dense layers, followed by a softmax classifier layer. The input images are resized and the features are extracted by the convolutional layers, and the feature map is produced as the output. The dimensionality reduction is done using the max-pooling layer, and the sub feature map is generated as the output. The features are learned from the non-medical images, and the model is saved.
The intertune MedCNN acts as an intermediate domain. This is designed by restoring the saved model and fine-tuning the GFE network and adding new layers according to the mammography dataset. In the CNN, the layers near the input are generic and the layers towards the output are specific to the input. Hence, the layers near the input are frozen, and the other layers are fine-tuned according to the mammography dataset. Two dense layers are added, which correlate the high-level features more strongly to a particular class. The regularization is done using drop out strategy, which introduces randomness in the network. This is done by randomly selecting some nodes and removing some nodes for the incoming and outgoing connections, in order to avoid over fitting. The classifier layer is modified as the number of output classes is different in the source and intermediate domains. The specific features of the mammography dataset are learned, and the model is saved.
The target fine-tune MedCNN is designed from the intertune MedCNN by restoring the saved model and fine-tuning according to the DBT dataset. The learned knowledge from the intertune MedCNN, with the mammography dataset, is transferred to train the fine-tune MedCNN, which classifies the DBT dataset into three classes, namely, normal, benign, and malignant. In order to further improve the performance of the target domain dataset, well established feature extraction techniques are used for the target domain dataset, before feeding to the fine-tune MedCNN. This is the proposed second-level FETL framework.
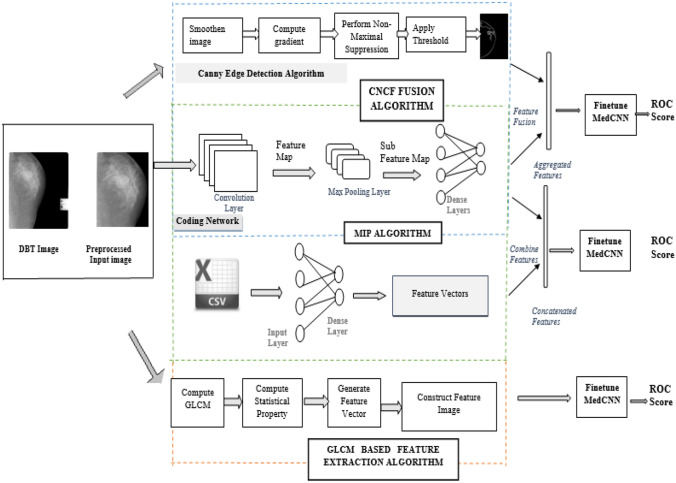
As shown in Fig. 2, the proposed second-level FETL framework consists of three different feature extraction techniques. Each technique has its unique way of extracting the most important features of the target domain medical instances. The extracted features are fed into the fine-tune MedCNN of the MLTL based framework and then classified as benign, malignant, and normal instances. The detailed explanation of the three different feature extraction techniques is given below.
Fig. 2.
FETL framework
CNCF Fusion Algorithm
A feature is a piece of information, which is relevant for solving the computation task related to a certain application. Features of images are of generally two types—low-level basic features and high-level domain-specific features. Both the features are extracted separately using different methods and then they are fused together, which is highly informative.
The color, shape, and texture are the low-level features of the image. Among these, the shape feature is extracted using the well known Canny edge detection algorithm. Edges define the boundaries between regions in an image, which help in segmentation and object recognition. The edge detection of an image significantly filters out the unimportant information, while preserving the important properties of an image and reduces the redundancy in data [39]. The Canny edge detection algorithm is the most popular optimal edge detector. The steps involved in Canny edge detection are (i) image smoothing with Gaussian filter, (ii) finding gradient through convolution operation, (iii) applying non-maximal suppression, and (iv) applying threshold. Thus, the low-level features of the target domain dataset are extracted using the Canny edge detection algorithm.
High-level domain-specific features are extracted using a coding network, which efficiently learns the representation of data. It consists of several convolutional and pooling layers for extracting the domain-specific features from the target domain dataset.
The structure of the coding network is shown in Table 1. The input to the coding network is the gray scale images of size 150 × 150. The coding network consists of six convolutional layers, followed by the pooling layers, the flatten layer, and the dense layer.
Table 1.
Coding network specification
| Layer type | Size/stride | Output dimension |
|---|---|---|
| Convolution | 11 × 11/1 | 140 × 140 × 32 |
| Convolution | 11 × 11/1 | 130 × 130 × 32 |
| Max pool | 5 × 5/2 | 63 × 63 × 32 |
| Convolution | 9 × 9/ 1 | 55 × 55 × 64 |
| Max pool | 5 × 5/2 | 26 × 26 × 64 |
| Convolution | 8 × 8 /1 | 19 × 19 × 128 |
| Convolution | 9 × 9/1 | 11 × 11 × 256 |
| Convolution | 8 × 8/1 | 4 × 4 × 256 |
| Flatten | 4096 | |
| Dense | 256 | |
| Dense | 128 |
The convolutional layer function is described with the help of the following equation (Eq. 1):
| 1 |
where,
n is the number of layers in the coding network.
h is the activation function.
is the input feature map.
is the output feature map.
is the weight kernel for convolutional layers.
× denotes the convolution operation.
is the bias of the coding network.
Max-pooling operation is carried out in a window size of 5 × 5 dimensions with 2 strides.
The pooling operation is described b (Eq. 2)
| 2 |
where is the output feature map obtained by the 5 × 5 dimension of pooling layers. There are two activation functions widely used, the sigmoid function and the tanh activation function. Both the functions suffer from gradient diffusion problem and low convergence rates. Therefore, to make the coding network efficient, this work uses rectified linear unit, ReLU. Thus, high-level features are extracted using the above-structured coding network for the target domain dataset. The low-level and high-level features need to be fused together, before feeding into the fine-tune MedCNN. Both the features are fused together using the single layer perceptron network, and the aggregated feature map is obtained as the output. This aggregated feature map is given as input to the pretrained fine-tune MedCNN for classification.
GLCM Based Feature Extraction
In an image, the texture is a repeated pattern of information or arrangement of the structure with regular intervals, and the texture feature describes the innate properties of an image surface. Therefore, texture analysis is one of the most important techniques, which characterizes the tissue, to define changes in functional characteristics of organs, in order to identify the disease in several organs like liver [40], skin, and breast. There are various texture feature extraction algorithms such as GLCM, GLDM, GLRLM, LBP, and SGLDM. Among these, GLCM is widely used for medical images [38, 44].
GLCM is a tabulation of how often different combinations of gray levels co-occur in an image. GLCM is a matrix, where the number of rows and columns is equal to the number of gray levels in the image. Each entry (x, y) in the GLCM corresponds to the number of occurrences of the pair of gray levels x and y, which are at distance d apart in the original image. It is constructed by calculating how often a pixel with intensity value x occurs in a specific spatial relationship to a pixel with value y.
Texture feature calculations are done from the contents of the GLCM, to obtain a measure of the variation in intensity at the pixel of interest. Various statistical properties of the image can be calculated from the GLCM. Among these, contrast, which is a measure of how the data in the image is distributed, is calculated, and the feature vector is obtained. The feature image is constructed from the feature vector for the target domain dataset. This feature image is given as input to the pretrained fine-tune MedCNN for classification.
Multi-Input Perceptron Algorithm
Along with the patient images, the report from the radiologist is used to extract more informative features. The patient report consists of quantitative readings of observation by the radiologist. The reports are analyzed and additional information such as mass, calcification, density, and lymph nodes, and impression of the respective patient images are extracted and converted into csv files. The patient reports are processed using the multilayer perceptron algorithm, and features are extracted from the csv files. At the same time, high-level features are also extracted from the images using the coding network discussed in the CNCF algorithm.
The features extracted from the report and the high-level features are concatenated using a single layer perceptron network. The concatenated features are given as input to the pretrained fine-tune MedCNN and classified.
Experimental Results
Dataset
The source domain dataset consists of 25,000 images from five different classes such as birds, cars, dogs, flowers, and humans. These images are a part of the ImageNet dataset [41]. The intermediate domain dataset consists of 1000 mammogram images with three different classes—benign, malignant, and normal. These images are from the standard dataset MIAS [42] and BCDR [43]. The target domain dataset consists of 500 DBT images, belonging to the three classes—normal, benign, and malignant. These are real-time images collected from Saveetha Institute of Medical and Technical Sciences of India. The classification labels of the DBT dataset are established using the corresponding radiologist reports, which form the ground truth.
EvaluationMetrics
The metrics chosen for evaluating the proposed framework using real-time DBT images are the “area under receiver operating characteristic (ROC) curve.” This paper deals with a medical dataset, which is extremely imbalanced. Hence, the area under the ROC curve is best applicable, compared to other metrics such as accuracy rate, precision, recall, and f-1 score. Since it is the medical dataset, the main goal is to clearly differentiate the patients, as positive or negative cases. The output from the softmax layer of the fine-tune MedCNN is used as the input for ROC curve analysis. The higher the area under the ROC curve, the better is the model in distinguishing between the presence and absence of a disease.
ROC curve depicts the true positive rate on the Y axis, and false positive rate on the X axis.
| 3 |
| 4 |
Using Eqs. 3 and 4, true positive rate gives the measure of the ratio of the images correctly classified as positive with that of the total predicted positive images. False positive rate gives the measure of the ratio of the images wrongly classified as positive with that of the total actual negative images.
Results
All the three networks discussed in the previous section are trained with mini-batch stochastic gradient descent optimization using a batch size of 64 on a GeForce GTX TITAN X GPU, with an initial learning rate of 0.01 and 150 epochs. Three different proportions are used for training and testing—85% for training and 15% for testing, 80% for training and 20% for testing, and 75% for training and 25% for testing. The training and testing data’s are randomly chosen. The proposed model is trained on each of the partitions, and the results are reported in terms of area under the ROC curve. The results obtained with all the proportions remain consistent. The results shown are calculated based on the ground-truth obtained using the radiologist reports for the DBT dataset.
First-Level MLTL Framework
The 25,000 non-medical images are used for feature learning in the source domain. The source domain GFE network is designed with two different network structures and their performances are evaluated. The first network structure consists of two convolutional layers each, followed by max-pooling layer and two dense layers. This network yields a training accuracy of 0.75. The second network structure consists of five convolutional layers each, followed by the max-pooling layer and three dense layers. This network, after adequate training, gives 0.90 training accuracy. Since the second network structure provides higher training accuracy, this network structure is selected to perform multilevel transfer learning.
The mammography dataset with 1000 images is used for intermediate domain feature learning and classification. The pretrained GFE network using non-medical images is used as the basis for the development of the intertune MedCNN.
Four different network structures are evolved by freezing all the layers (C1-F3), freezing the first convolutional layer (C1), freezing the first two convolutional layers (C1–C2), and freezing the first three convolutional layers (C1–C3), respectively. The area under the ROC curve obtained from all these network structures are 0.44, 0.63, 0.66, and 0.46, respectively.
The network structure having comparatively higher area under the ROC curve of 0.66, obtained by freezing the first two convolutional layers, is selected to perform transfer learning and to design the target fine-tune MedCNN. The area under the ROC curve of the intertune MedCNN is depicted in Fig. 3. Even though this intertune MedCNN structure is the best among the evolved structures, it is observed that the area under the ROC curve is still less, since the source and intermediate domains are unrelated.
Fig. 3.
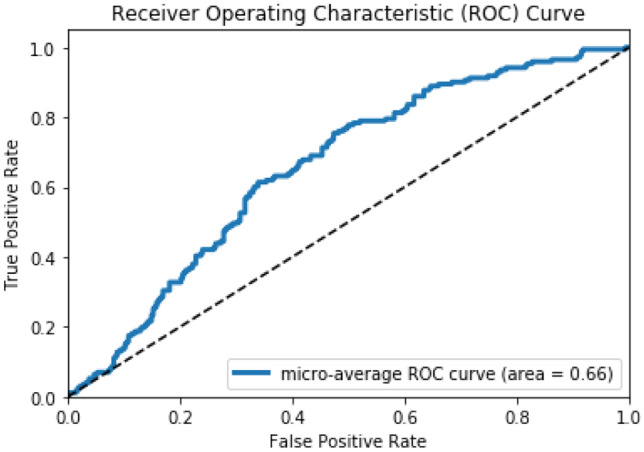
Area under the ROC curve of the intertune MedCNN (intermediate mammography domain)
The 500 DBT images are used to perform the target domain classification. The pretrained intertune MedCNN is used as the basis for developing the fine-tune MedCNN. The dense layer of this network alone is fine-tuned according to the DBT images. The area under the ROC curve for this fine-tune MedCNN is observed to be 0.81 as shown in Fig. 4. It is observed that the area under the ROC curve of the target domain dataset has improved significantly. This is because of using related auxiliary information from the intermediate domain, consisting of the mammography dataset.
Fig. 4.
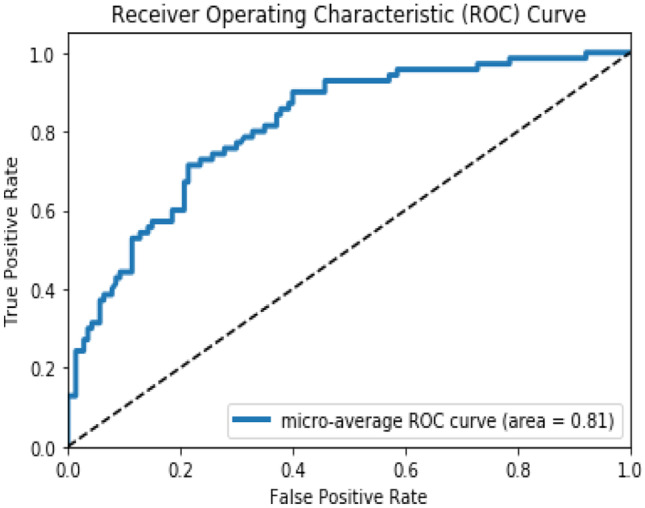
Area under the ROC curve of the Fine-tune MedCNN (target DBT domain)
Second-Level FETL Framework
In order to further improve the target domain classification performance, a second-level FETL framework with three different feature extraction techniques is implemented. The output images, after Canny edge detection, for a sample from each of the three different classes, are shown in Fig. 5.
Fig. 5.
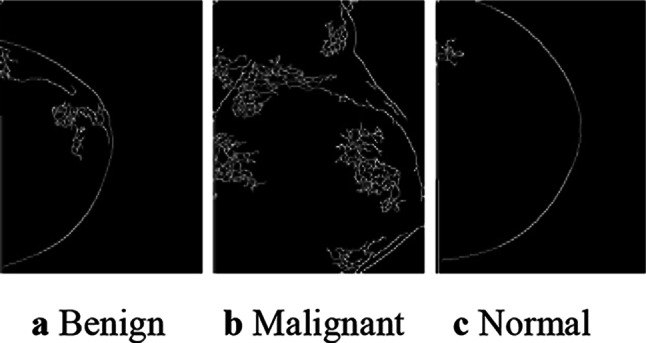
Canny edge detection output of the DBT images
The area under the ROC curve of the fine-tune MedCNN obtained using the CNCF fusion algorithm is 0.89, which is shown in Fig. 6. It is observed that there is a significant improvement in area under the ROC curve, when low-level and high-level features are fused and given as input for classification, rather than giving the raw image.
Fig. 6.
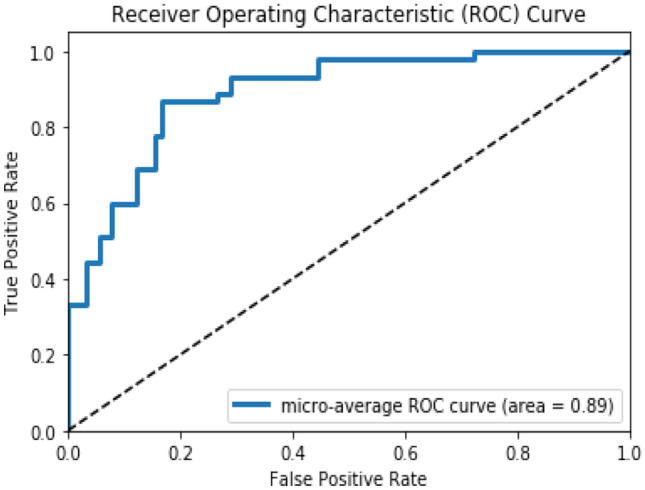
Area under the ROC curve of the fine-tune MedCNN using CNCF algorithm
The second technique, namely, GLCM based feature extraction algorithm, extracts the texture features of the DBT images. A sample of the output feature image for each of the three classes is shown in Fig. 7.
Fig. 7.
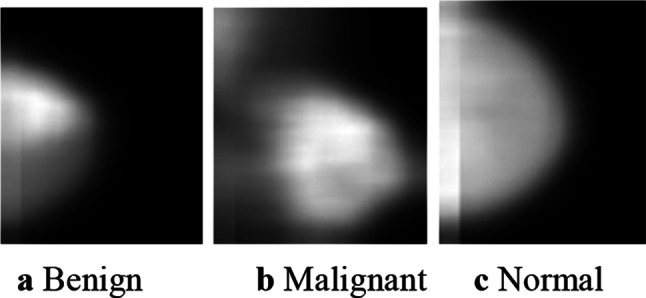
GLCM output feature images of the DBT images
The fine-tune MedCNN with GLCM based feature extraction algorithm gives 0.88 area under the ROC curve, as shown in Fig. 8. It is observed that the texture feature image produces improvement in the classification performance than the raw image.
Fig. 8.
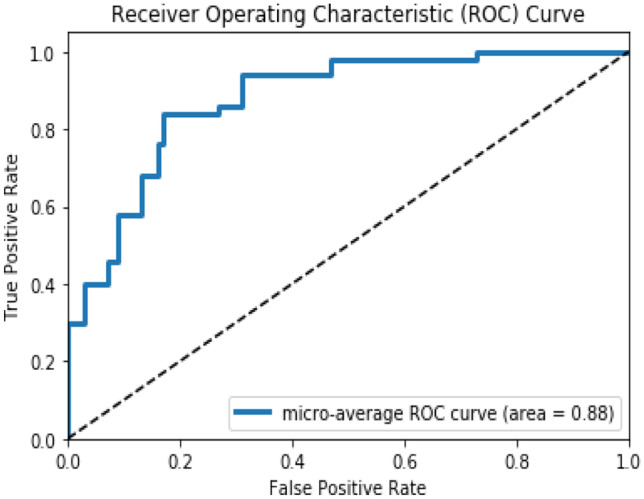
Area under the ROC curve of the fine-tune MedCNN using GLCM algorithm
The third feature extraction technique uses the patient report information along with the image features. The fine-tune MedCNN with the MIP algorithm gives 0.89 area under the ROC curve, which is depicted in Fig. 9. It is observed that the use of the patient report along with images improves the classification performance of the target domain dataset.
Fig. 9.
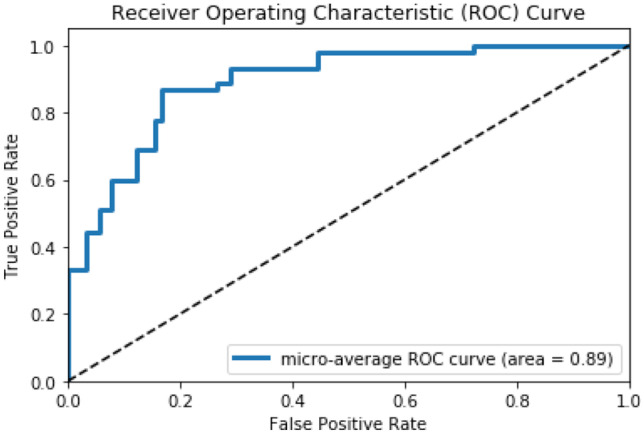
Area under the ROC curve of the fine-tune MedCNN using MIP algorithm
Thus, the experimental results clearly indicate that the proposed FETL framework, which uses various feature extraction techniques, augments the performance of the MLTL framework. Since the target domain dataset has only less number of images and it is different from the source domain distribution, the various feature extraction techniques have contributed to the improved performance of the fine-tune MedCNN.
Discussion
Table 2 provides a comparison between the existing work [4] and the proposed work. The existing work [4] reports an area under the ROC curve of 0.91 using AlexNet, which is trained with the ImageNet dataset consisting of 1.2 million non-medical images from 1000 classes. The intermediate domain consisted of 19,632 mammography images, and the target domain consisted of 12,680 DBT images. The proposed FETL based framework gives almost the same performance with the area under the ROC curve of 0.89 for the classification of the DBT dataset, with just 25,000 images in the source domain, 1000 images in the intermediate domain, and 500 images in the target domain. This, when compared to the datasets used in [4], is 2.08% of the source domain dataset, 5.09% of the intermediate domain dataset, and 3.94% of the target domain dataset.
Table 2.
Comparison with the earlier reported work
| Source domain | Intermediate domain | Target domain | Performance (area under the ROC curve) |
||||
|---|---|---|---|---|---|---|---|
| No. of instances |
No. of classes |
No. of instances |
No. of classes |
No. of instances |
No. of classes |
||
| Existing work [4] | 1.2 million | 1000 | 19,632 | 2 | 12,680 | 2 | 0.91 |
|
Proposed work |
25,000 (2.08%) | 5 |
1000 (5.09%) |
3 |
500 (3.94%) |
3 | 0.89 |
The proposed work has also been compared with three other state-of-the-art methods for classification of breast cancer. Table 3 presents this comparison. It can be seen that the proposed two level transfer learning framework provides an area under the ROC of 0.89. Mohammad et al. [23] have reported an area under the ROC as 96.1. However, it is to be noted that they have used 1.2 million images in the source domain dataset and 104,795 in the target domain dataset, whereas the proposed work uses only 2.08% of the source domain dataset and 0.477% of the target domain dataset. Thus, the results confirm that the proposed two level frameworks can be effectively utilized even in domains, where there is sparsity of data. The feature extraction carried out on the target domain has improved the performance, thus throwing open more avenues for multitransfer learning.
Table 3.
Comparison with recent three state of the art methods
| Work reported in literature | Target domain classification | Target domain dataset | Method followed | Performance (area under the ROC curve) |
|---|---|---|---|---|
|
Leilei Zhou et al. [21] Source domain: ImageNet (1.2 M) 1000 classes Target domain: MRI breast images |
Fibroadenoma, invasive ductal carcinoma (IDC) |
207 images (before augmentation) |
Inception V3 VGG 19 |
0.89 (after augmentation) 0.87 (after augmentation) |
|
Mohammad et al. [23] Source domain: ImageNet (1.2 M) 1000 classes Target domain: mammography breast images |
Normal, benign, and malignant |
57,430 (before augmentation) |
Fine-tuned VGG-19 | 81.3 |
|
104,795 (after augmentation) |
Fine-tuned VGG-19 | 96.1 | ||
|
Mor Yemini et al. [28] Source domain: ImageNet (1.2 M) 1000 classes Target domain: mammography breast images |
Normal and malignant |
410 images (before augmentation) |
Google Inception-V3 |
0.78 (after augmentation method 1) 0.86 (after augmentation method 2) |
|
Proposed two-level framework Source Domain: ImageNet (25,000) 5 classes Intermediate domain: mammography breast images Target domain: DBT images |
Normal, benign, malignant | 500 images |
CNN based MLTL and FETL frameworks |
0.81 (without feature extraction) 0.89 (with feature extraction) |
Conclusion
This work has successfully implemented a two-level MLTL and FETL based framework for classifying DBT images, with the information obtained from general images and mammography images. The significant contribution of this work is the use of the various feature extraction techniques, which have clearly proved that it is possible to apply this framework to even domains where there is a sparsity of data. The future work will focus on exploring the possibility of extending this framework to other medical domains, where data is not easily available.
Funding
This work was supported by the Department of Radiology, Saveetha Medical College & Hospital (SMCH), and Saveetha Institute of Medical and Technical Sciences of India.
Data Availability
The work was supported by the Department of Radiology, Saveetha Medical College & Hospital (SMCH). The real-time images are collected from Saveetha Institute of Medical and Technical Sciences of India.
Code Availability
Custom code.
Declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Aswiga R V, Email: aswiga91@gmail.com.
Aishwarya R, Email: aishwarya.rsv@gmail.com.
Shanthi A P, Email: shanthiap@gmail.com.
References
- 1.Nasrindokht Azamjah, Yasaman Soltan-Zadeh, Farid Zayeri.: Global trend of breast cancer mortality rate: A 25-year study. Asian Pacific Journal of Cancer Prevention, 20(7): 2015–2020, 2019. 10.31557/APJCP.2019.20.7.2015, https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6745227/ [DOI] [PMC free article] [PubMed]
- 2.Dr. Kamal Kant Koli: Fast MRI bests 3D mammograms for screening cancer in women with dense breasts. Medical Dialogues, 2020. https://medicaldialogues.in/radiology/news/fast-mri-bests-3d-mammograms-for-screening-cancer-in-women-with-dense-breasts-70882
- 3.Limitations of Mammograms. American Cancer Society. https://www.cancer.org/cancer/breast-cancer/screening-tests-and-early-detection/mammograms/breast-density-and-your-mammogram-report.html
- 4.Ravi K. Samala, Heang-Ping Chan, Lubomir Hadjiiski, Mark A. Helvie, Caleb D. Richter, Kenny H. Cha.: Breast cancer diagnosis in digital breast tomosynthesis-effects of training sample size on multi-stage transfer learning using deep neural nets. IEEE Transactions on Medical Imaging, vol. 38, no. 3: 686–696, 2019 [DOI] [PMC free article] [PubMed]
- 5.Ben Tan, Yangqiu Song, Erheng Zhong, Qiang Yang.: Transitive transfer learning. International Conference on Knowledge Discovery and Data Mining, ACM SIGKDD, ISBN: 978–1–4503–3664:1155–1164, Aug 2015
- 6.Jiang Siyu, Yonghui Xu, Wang Tengyun, Yang Haizhi, Qiu Shaojian, Han Yu, Song Hengjie. Multi-label metric transfer learning jointly considering instance space and label space distribution divergence. IEEE Access. 2019;7:10362–10373. doi: 10.1109/ACCESS.2018.2889572. [DOI] [Google Scholar]
- 7.Yan Xu, Yanming Sun, Xiaolong Liu, Yonghua Zhang. : A digital-twin-assisted fault diagnosis using deep transfer learning. Advances in Prognostics and System Health Management. IEEE Access, vol.7: 19990–19999, 2019. 10.1109/ACCESS.2018.2890566
- 8.Chongyu Pan, Jian Huang, Jianxing Gong, Xingsheng Yuan.: Few-shot transfer learning for text classification with light weight word embedding based models. IEEE Access, vol.7: 53296–53304, 2019. 10.1109/ACCESS.2019.2911850
- 9.Zhou Junhao, Yue Lu, Dai Hong-Ning, Wang Hao, Xiao Hong. Sentiment analysis of Chinese microblog based on stacked bidirectional LSTM. IEEE Access. 2019;7:38856–38866. doi: 10.1109/ACCESS.2019.2905048. [DOI] [Google Scholar]
- 10.Xiao, Wang, Du.: Improving the performance of sentiment classification on imbalanced datasets with transfer learning. IEEE Access, vol.7: 28281–28290, 2019. 10.1109/ACCESS.2019.2892094
- 11.Catherine Sandoval, Elena Pirogova, Margaret Lech.: Two-stage deep learning approach to the classification of fine-art paintings. IEEE Access, vol.7: 41770–41781, 2019. 10.1109/ACCESS.2019.2907986
- 12.Qingyao Wu, Hanrui Wu, Xiaoming Zhou, Mingkui Tan, Yonghui Xu, Yuguang Yan, and Tianyong Hao.: Online transfer learning with multiple homogeneous or heterogeneous source. IEEE Transactions on Knowledge and Data Engineering, vol.29, no. 7: 1494–1507, 2017
- 13.Yun-tao Du, Qian Chen, Heng-yang Lu, Chong-jun Wang.: Online single homogeneous source transfer learning based on AdaBoost. In: IEEE 30th International Conference on Tools with Artificial Intelligence, ISBN :2375–0197:344–349,2018. 10.1109/ICTAI.2018.00061
- 14.Yuguang Yan, Qingyao Wu, Mingkui Tan, Michael K. Ng, Huaqing Min, Ivor W. Tsang.: Online heterogeneous transfer by hedge ensemble of offline and online decisions. IEEE Transactions on Neural Networks and Learning Systems, vol. 29, no. 7: 3252–3263, 2018 [DOI] [PubMed]
- 15.Fuzhen Zhuang, Xiaohu Cheng, Sinno JialinPan.: Transfer learning with multiple sources via consensus regularized auto encoders. In: 2014th European Conference on Machine Learning and Knowledge Discovery in Databases, vol- III, LNCS 8726: 417–431, Sep 2014
- 16.Yong Luo, Yonggang Wen, Tongliang Liu, Dacheng Tao.: Transferring knowledge fragments for learning distance metric from a heterogeneous domain. IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 41, no. 4: 1013–1026, 2019 [DOI] [PubMed]
- 17.Veronika Cheplygina, Isabel Pino Pena, Jesper Holst Pedersen, David A. Lynch, Lauge Sorensen, and Marleen de Bruijne. Transfer learning for multicenter classification of chronic obstructive pulmonary Disease. IEEE Journal of Biomedical and Health Informatics, vol. 22, no. 5:1486–1496, 2018 [DOI] [PMC free article] [PubMed]
- 18.Zhaohong Deng, Peng Xu, LixiaoXie, Kup-Sze Choi and Shitong Wang.: Transductive joint-knowledge-transfer TSK FS for recognition of epileptic EEG signals. IEEE Transactions on Neural Systems and Rehabilitation Engineering, vol. 26, no. 8:1481–1494, 2018 [DOI] [PubMed]
- 19.Hyunsoo Yoon, Jing Li.: A novel positive transfer learning approach for telemonitoring of Parkinson’s disease. IEEE Transactions on Automation Science and Engineering, vol. 16, no. 1:180–191, 2019
- 20.Chenchen Wu, Jun Ruan, Guanglu Ye, Jingfan Zhou, Simin He, Jianlian Wang, Zhikui Zhu, Junqiu Yue: Identifying tumor in whole-slide images of breast cancer using transfer learning and adaptive sampling. Eleventh International Conference On Advanced Computational Intelligence (ICACI), 7–9, 2019. 10.1109/Icaci.2019.8778616
- 21.Leilei Zhou , Zuoheng Zhang , Xindao Yin , Hong-Bing Jiang , Jie Wang , Guan Gui : Transfer learning-based DCE-MRI method for identifying differentiation between benign and malignant breast tumors. IEEE Access, vol 8: 17527–17534, 2020. 10.1109/Access.2020.2967820
- 22.Zhang Xinfeng, He Dianning, Zheng Yue. HuaibiHuo, Simiao Li, Ruimei Chai, And Ting Liu: Deep learning based analysis of breast cancer using advanced ensemble classifier and linear discriminant analysis. Special Section on Deep Learning Algorithms for Internet of Medical Things, IEEE Access. 2020;8:120208–120217. doi: 10.1109/Access.2020.3005228. [DOI] [Google Scholar]
- 23.Mohammad Alkhaleefa, Praveen Kumar Chittem, Vishnu Priya Achhannagari, Shang-Chih Ma, Yang-Lang Chang.: The influence of image augmentation on breast lesion classification using transfer learning. International Conference on Artificial Intelligence and Signal Processing (AISP), 2020. 10.1109/Aisp48273.2020.9073516
- 24.Jing Zheng.: Deep learning assisted efficient AdaBoost algorithm for breast cancer detection and early diagnosis. Special Section on Deep Learning Algorithms for Internet of Medical Things, IEEE Access, 2020. 10.1109/Access.2020.2993536
- 25.Jonathan De Matos, Alceu De S. Britto Jr, Luiz E. S. Oliveira, And Alessandro L. Koerich.: Double transfer learning for breast cancer histopathologic image classification. IJCNN International Joint Conference on Neural Networks, 2019. 10.1109/IJCNN.2019.8852092
- 26.Hafiz Mughees Ahmad, Sajid Ghuffar, Khurram Khurshid.: Classification of breast cancer histology images using transfer learning. 16th International Bhurban Conference on Applied Sciences & Technology (IBCAST): 328–332. 2019. 10.1109/IBCAST.2019.8667221
- 27.Constance Fourcade, Ludovic Ferrer, Gianmarco Santini, Noemie Moreau Caroline Rousseau, Marie Lacombe, Camille Guillerminet, Mathilde Colombie, Mario Campone, Diana Mateus, Mathieu Rubeaux.: Combining super pixels and deep learning approaches to segment active organs in metastatic breast cancer pet images. Annu Int Conf IEEE Eng Med Biol Soc, 2020 Jul;2020:1536–1539. 10.1109/EMBC44109.2020.9175683 [DOI] [PubMed]
- 28.Mor Yemini, Yaniv Zigel, Dror Lederman.:Detecting masses in mammograms using convolutional neural networks and transfer learning. ICSEE International Conference on the Science of Electrical Engineering, 2018. 10.1109/ICSEE.2018.8646252
- 29.Mohammad Alkhaleefah, Chao-Cheng Wu.: A hybrid CNN and RBF-based SVM approach for breast cancer classification in mammograms. IEEE International Conference on Systems, Man, and Cybernetics, 2018. 10.1109/Smc.2018.00159
- 30.Naresh Khuriwal, Nidhi Mishra.:Breast cancer diagnosis using adaptive voting ensemble machine learning algorithm. IEEMA Engineer Infinite Conference (eTechNxT), 2018. 10.1109/ETECHNXT.2018.8385355
- 31.Naveen, R. K. Sharma, Anil Ramachandran Nair.:Efficient breast cancer prediction using ensemble machine learning models. International Conference on Recent Trends on Electronics, Information, Communication & Technology RTEICT, 2019. 10.1109/RTEICT46194.2019.9016968
- 32.Mohamed Nemissi, Halima Salah, Hamid Seridi.:Breast cancer diagnosis using an enhanced extreme learning machine based-neural network. International Conference on Signal, Image, Vision and their Applications (SIVA), 2018. 10.1109/Siva.2018.8661149
- 33.Sanaz Mojrian, Gergo Pinter, Javad Hassannataj Joloudari, ImreFelde, Akos Szabo-Gali, Laszlo Nadai, Amir Mosavi. : Hybrid machine learning model of extreme learning machine radial basis function for breast cancer detection and diagnosis; a multilayer fuzzy expert system. International Conference on Computing and Communication Technologies (RIVF), 2020. 10.1109/Rivf48685.2020.9140744
- 34.Veeramuthu, A., Meenakshi. S, Kameshwaran. A.: A plug-in feature extraction and feature subset selection algorithm for classification of medicinal brain image data. International Conference on Communication and Signal Processing, 2014. 10.1109/ICCSP.2014.6950108
- 35.Nagarajan G, Minu RI, Muthukumar B, Vedanarayanan V, Sundarsingh SD. Hybrid genetic algorithm for medical image feature extraction and selection. Procedia Computer Science, Elsevier. 2016;85:455–462. doi: 10.1016/j.procs.2016.05.192. [DOI] [Google Scholar]
- 36.Kailash D. Kharat, VikulJ.Pawar, Suraj R. Pardeshi.: Feature extraction and selection from MRI images for the brain tumor classification. International Conference on Communication and Electronics Systems ICCES, 2016. 10.1109/CESYS.2016.7889969
- 37.Aimin Yang, Xiaolei Yang, Wenrui Wu, Huixiang Liu, Yunxi Zhuansun.:Research on feature extraction of tumor image based on convolutional neural network. IEEE Access, vol 7: 24204–24213, 2019. 10.1109/ACCESS.2019.2897131
- 38.Yifan Hu, Yefeng Zheng.: A GLCM Embedded CNN strategy for computer-aided diagnosis in intracerebral hemorrhage. Computer Vision and Pattern Recognition:1–9, 2019. arXiv:1906.02040v1
- 39.Ruba Anas, Hadeel, A., Elhadi, Elmustafa Sayed Ali.: Impact of edge detection algorithms in medical image processing. International Scientific Journal of World Scientific News, vol.118:129–143, 2019
- 40.Aborisade, Ojo, Amole, Durodola.: Comparative analysis of textural features derived from GLCM for ultrasound liver image classification. International Journal of Computer Trends and Technology (IJCTT), vol-11, no.6 : 239–244, 2014
- 41.ImageNet online dataset http://www.image-net.org/
- 42.Mammography online dataset https://www.kaggle.com/kmader/mias-mammography
- 43.Mammography online dataset https://bcdr.eu/patient/list
- 44.Jiaxing Tan, Yongfeng Gao, Weiguo Cao, Marc Pomeroy, Shu Zhang, Yumei Huo, Lihong Li, Zhengrong Liang.: GLCM-CNN Gray level co-occurrence matrix based CNN model for Polyp diagnosis. IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), 2019. 10.1109/BHI.2019.8834585
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The work was supported by the Department of Radiology, Saveetha Medical College & Hospital (SMCH). The real-time images are collected from Saveetha Institute of Medical and Technical Sciences of India.
Custom code.