Abstract
Inflammation increases the risk of cardiometabolic disease. Delineating specific inflammatory pathways and biomarkers of their activity could identify the mechanistic underpinnings of the increased risk. Plasma levels of kynurenine, a metabolite involved in inflammation, associates with cardiometabolic disease risk. We used genetic approaches to identify inflammatory mechanisms associated with kynurenine variability and their relationship to cardiometabolic disease. We identified single-nucleotide polymorphisms (SNPs) previously associated with plasma kynurenine, including a missense-variant (rs3184504) in the inflammatory gene SH2B3/LNK. We examined the association between rs3184504 and plasma kynurenine in independent human samples, and measured kynurenine levels in SH2B3-knock-out mice and during human LPS-evoked endotoxemia. We conducted phenome scanning to identify clinical phenotypes associated with each kynurenine-related SNP and with a kynurenine polygenic score using the UK-Biobank (n = 456,422), BioVU (n = 62,303), and Electronic Medical Records and Genetics (n = 32,324) databases. The SH2B3 missense variant associated with plasma kynurenine levels and SH2B3−/− mice had significant tissue-specific differences in kynurenine levels.LPS, an acute inflammatory stimulus, increased plasma kynurenine in humans. Mendelian randomization showed increased waist-circumference, a marker of central obesity, associated with increased kynurenine, and increased kynurenine associated with C-reactive protein (CRP). We found 30 diagnoses associated (FDR q < 0.05) with the SH2B3 variant, but not with SNPs mapping to genes known to regulate tryptophan-kynurenine metabolism. Plasma kynurenine may be a biomarker of acute and chronic inflammation involving the SH2B3 pathways. Its regulation lies upstream of CRP, suggesting that kynurenine may be a biomarker of one inflammatory mechanism contributing to increased cardiometabolic disease risk.
Subject terms: Prognostic markers, Cardiovascular diseases, Disease genetics, Gene regulation
Introduction
Epidemiological studies have identified numerous biomarkers associated with clinical disease1. However, the etiological relationship between the biomarker and disease, and the molecular mechanisms contributing to their association often cannot be determined by traditional association approaches. This knowledge gap limits the opportunities to identify clinical applications for a putative biomarker. Genetic approaches, which link molecular mechanisms with disease outcomes, can ascertain for possible causal relationships between a biomarker and a disease, and can identify the discrete disease-associated mechanisms captured by the biomarker2. Here, we leverage such genetic approaches to more fully characterize the plasma metabolite kynurenine.
Kynurenine is a modulatory biomolecule synthesized from the essential dietary amino acid tryptophan, in part, by the inducible enzyme indoleamine 2,3-dioxygenase (IDO)3. Plasma kynurenine has been associated with a range of phenotypes including cardiovascular diseases (CVD) [heart disease, atherosclerosis, and endothelial dysfunction], hypertension, diabetes, obesity and neuropsychiatric disorders3,4. Animal and cell models suggest that kynurenine pathway metabolites are linked to disease through modulation of inflammation5,6. Therapies that decrease inflammation lower levels of non-specific inflammatory biomarkers such as c-reactive protein (CRP)7, reduce cardiovascular disease risk8 and improve cardiometabolic indices including levels of glucose and insulin which would subsequently lead to disease reduction9. However, findings relating the kynurenine pathway to inflammation and cardiometabolic disease are inconsistent, and the mechanistic relationship between plasma kynurenine levels and cardiometabolic disease risk is not well-defined.
We hypothesized that characterizing the genetic determinants underlying plasma kynurenine levels would define its role in health and disease risk. We used single nucleotide polymorphisms (SNPs) associated with plasma kynurenine levels as instrumental variables to determine how genetic regulation of kynurenine contributes to disease risk. We leveraged data from observational and interventional human studies and mouse models and showed that plasma kynurenine is a biomarker associated with multiple cellular mechanisms, and that only one of these mechanisms appears to associate with clinical disease.
Results
SNPs in four independent genomic regions associate with plasma kynurenine
Of all SNPs reaching genome-wide significance (p < 5 × 10−8), we identified six lead SNPs associated with plasma kynurenine in the Cooperative Health Research in the Region of Augsburg (KORA) and TwinsUK GWAS meta-analysis at (Table 1). Based on Linkage disequilibrium (LD) and genomic location, we concluded that these 6 lead SNPs represent 4 independent loci. Three SNPs are cis-expression quantitative trait loci (eQTLs) for genes involved in tryptophan/kynurenine metabolism: the IDO 1/2 genes (rs10085935, Chr8), which are the rate limiting enzymes in the kynurenine extrahepatic pathway3,10; and SLC7A5 (rs750950 and rs8051149, Chr16), which participates in tryptophan transport across the cell membrane11. Two SNPs mapped to a region on Chr 12. The strongest association was with rs3184504, a missense variant in a known inflammatory gene, SH2B3, and has been associated with multiple phenotypes including myocardial infarction and hypertension12. The final SNP (rs16924894, Chr10) is an intergenic variant located near the KIAA1217 and ARHGAP21 genes. Tryptophan 2,3-dioxygenase, encoded by the TDO2 gene, is another key regulatory enzyme in the kynurenine pathway13. We assessed the SNPs located in or near the TDO2 genes and none were significantly associated with plasma kynurenine levels. Of 6 downstream metabolites (3-hydroxy anthranilic acid (3HAA), and quinolinic acid (QA), NAM (nicotinamide), NA (nicotinic acid), KA (kynurenic acid), HK (hydroxykynurenine)) in the kynurenine pathway, GWAS data of QA and NAM levels14,15 showed no significant associations with the SH2B3 rs3184504 variant. (Supplementary Table 1).
Table 1.
Annotations for top GWAS loci associated with circulating kynurenine levels derived from the KORA-TwinsUK meta-analysis GWAS summary statistics.
| SNP | Chromosome | Position | Reference allele | Alternative allele | Reference allele frequency | Beta | SE | P value | Closest gene (s) | Genomic location | eQTL (s)a | Possible relationship to kynurenine |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs10085935 | 8 | 39,806,267 | T | C | 0.37 | − 0.010 | 0.002 | 3.326E−09 | IDO2 | intronic | IDO1 | ✓ |
| rs16924894 | 10 | 24,845,525 | A | T | 0.02 | 0.081 | 0.015 | 2.329E−08 | KIAA1217 (dist = 8748), ARHGAP21 (dist = 27,013) | intergenic | No significant eQTLs were found | |
| rs3184504 | 12 | 111,884,608 | T | C | 0.48 | 0.015 | 0.002 | 6.046E−18 | SH2B3 | exonic-Missense (R262W) | ALDH2, LINC01405, TMEM116, ADAM1B, MAPKAPK5-AS1 | ✓ |
| rs11066320 | 12 | 112,906,415 | A | G | 0.43 | 0.012 | 0.002 | 3.101E−11 | PTPN11 | intronic | ALDH2, ADAM1B, MAPKAPK5, NAA25, BRAP | |
| rs750950 | 16 | 87,873,247 | A | C | 0.64 | 0.014 | 0.002 | 2.342E−16 | SLC7A5 | intronic | SLC7A5 | ✓ |
| rs8051149 | 16 | 87,878,822 | A | G | 0.21 | 0.026 | 0.003 | 9.073E−26 | SLC7A5 | intronic | SLC7A5, RP4-536B24.2 | ✓ |
Data was obtained from http://metabolomics.helmholtz-muenchen.de/gwas/. Four shading rows indicate SNPs belonging to 4 independent regions.
SNP single nucleotide polymorphism, SE standard error.
aData was obtained from https://gtexportal.org/home/
SH2B3 and rs3184504 associate with plasma kynurenine
To confirm the SH2B3 rs3184504 SNP association with kynurenine levels, we examined the genetic association with plasma kynurenine using data from the ABO Glycoproteomics in Platelets and Endothelial Cells (ABO) Study (N = 66 young healthy volunteers). The pattern of association was consistent with a recessive effect (Fig. 1), with a significant association between homozygosity for the T allele and increased kynurenine (β = 0.17, p = 0.04, recessive genetic model).
Figure 1.
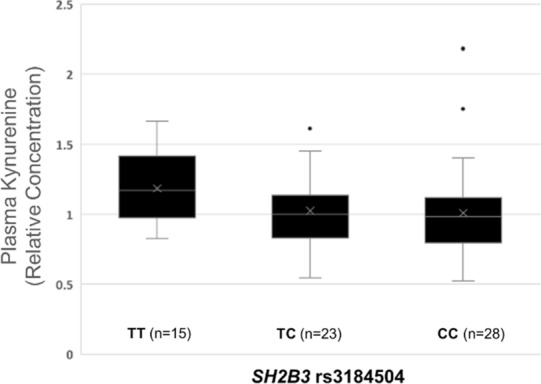
The association of SH2B3 rs3184504 genotype with plasma kynurenine was confirmed in a sample of 66 healthy men and women. This figure was created in IBM SPSS Statistics 27.
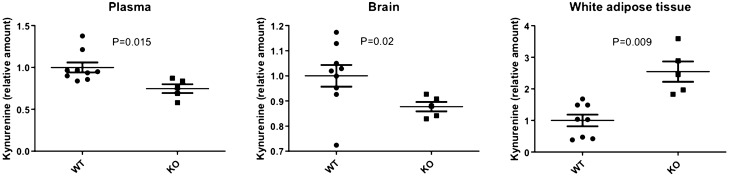
We further characterized regulation of kynurenine by SH2B3 by measuring kynurenine levels in an Sh2b3−/− mouse, as compared to wild-type controls. The most marked differences in kynurenine levels were observed in white adipose tissue where there were more than 2.5-fold (p = 0.009) higher kynurenine levels in Sh2b3−/− compared to wild type mice (Fig. 2). In contrast, significantly lower levels of kynurenine were observed in plasma (≈ 0.25-fold, p = 0.02) and brain (≈ 0.13-fold, p = 0.02) of Sh2b3−/− mice. There was no difference in kynurenine levels in the spleen and modestly lower, but non-significant, levels in the liver in KO mice compared to controls. (Supplementary Fig. 1). These data suggest that SH2B3 is a regulator of kynurenine metabolism, but that the effects on the pathway may differ by tissue.
Figure 2.
Kynurenine levels in selected tissues in wild type (WT, N = 8) and SH2B3 knockout (KO, N = 5) mice. Kynurenine levels were measured by ELISA assay. Values are expressed as relative concentrations, normalized to WT. Differences between WT and KO animals were analyzed by unpaired t test. This figure was created in GraphPad Prism v.9.1.1.
Plasma Kynurenine and inflammation
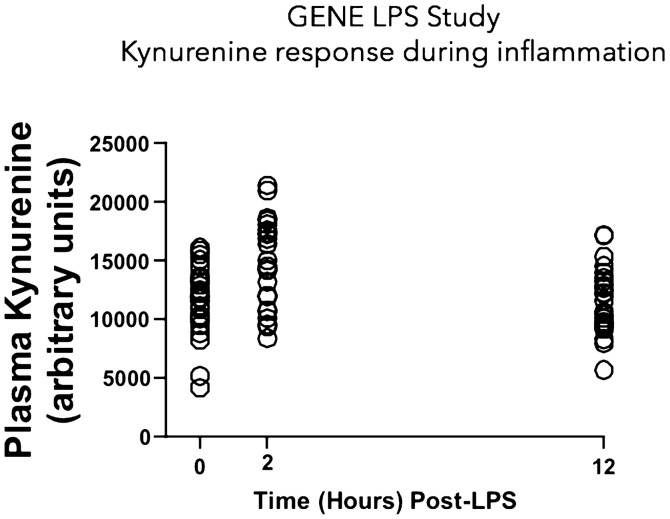
Inflammation is a risk factor for cardiometabolic disease. To understand a possible genetic relationship between kynurenine and CRP, an inflammatory biomarker associated with cardiometabolic disease risk, we conducted bi-directional Mendelian randomization analyses. Genetically determined kynurenine levels were associated with increased CRP (inverse-variance weighted average meta-analysis [IVWA] β = 0.21 (0.10), p = 0.04), but the reverse association was not significant (p = 0.71), suggesting that kynurenine may increase CRP levels. We also examined the effects of lipopolysaccharide (LPS) challenge (an acute inflammatory stimulus) on kynurenine levels in healthy human subjects (N = 24). In the Genetics of Evoked responses to Niacin and Endotoxemia (GENE) study, plasma kynurenine significantly increased by 25% (p = 0.0008) 2 h post-LPS challenge (Fig. 3), confirming that an inflammatory stimulus causes rapid increases in plasma kynurenine levels. This timepoint is concurrent with peak cytokine responses to LPS (e.g. TNFα, IL-6), but precedes an increase in CRP, which peaked 24 h post-LPS16, highlighting kynurenine as an early marker of acute inflammation.
Figure 3.
Plasma Kynurenine response to evoked endotoxemia (LPS, 1 ng/kg) in healthy individuals (N = 24). Mean kynurenine levels significantly increased 2 h post-LPS (p = 0.0008) and returned to baseline levels by 12 h post-LPS. This figure was created in GraphPad Prism v.9.1.1.
Kynurenine and body composition
Obesity is an inflammatory state, and is associated with elevated CRP levels17. We tested whether genetically determined central adiposity, measured by waist circumference is associated with kynurenine levels, and found a significant positive association (IVWA β = 0.04 (0.01), p = 0.0002). Similarly, in the ABO Study, measured waist circumference was positively correlated with plasma kynurenine levels (r = 0.279, p = 0.02). Collectively, these results suggest that central adiposity is associated with higher kynurenine levels.
The kynurenine association with cardiometabolic diseases risk is specific to the SH2B3 pathway
The individual SNPs associated with plasma kynurenine represent distinct mechanisms of kynurenine regulation and may have distinct patterns of disease associations. We conducted PheWAS for each of the six SNPs individually. The SNPs near the IDO1/2 and SLC7A5 genes, which directly contribute to either kynurenine synthesis or metabolism, had no significant associations with disease. In contrast, 30 phenotypes significantly associated with rs3184504 in SH2B3 at FDR q < 0.05 (Supplementary Table 2). Among these associations were three distinct collections of diagnoses related to: hypothyroidism, hypertension and heart diseases including myocardial infarction, all of which were associated with higher kynurenine levels. The associations were similar in the Electronic Medical Records and Genetics (eMERGE)/BioVU (Fig. 4) and the UK Biobank data sets (Supplementary Fig. 2). In sum, the SH2B3 rs3184504 variant associates with a range of diseases, and that previously reported epidemiological disease associations with plasma kynurenine may be driven by genetic mechanisms associated with SH2B3.
Figure 4.
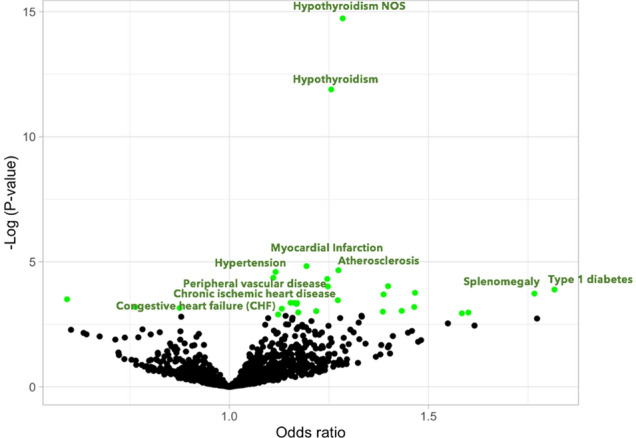
Volcano plot summarizing pheWAS associations for the SH2B3 rs3184504 variant in the BioVU and eMERGE data sets. Each point indicates a phenotype association with T allele, from a logistic regression association analysis assuming an additive genetic model and are adjusted for age, gender and principal components. Odd-ratios greater than 1 indicated increased risk associated with increased kynurenine levels. Only some points are annotated for clarity, and points in green have FDR q < 0.05.
We performed phenome-wide association testing between the 6-SNP kynurenine polygenic risk score and 894 phenotypes in the BioVU and eMERGE datasets to explore whether genetically-predicted plasma kynurenine associated with clinical disease risk. We did not identify any significant associations (FDR q < 0.05). (Supplementary Fig. 3). Thus, plasma kynurenine is likely not a causal mediator of cardiometabolic diseases.
The SH2B3 rs3184504 variant may modulate disease though modulation of metabolic rate
We hypothesized that the mechanism linking SH2B3 to disease may be through effects on energy metabolism. In the ABO study the rs3184504 risk variant was associated with significantly lower heart rate (β = −4.98, p = 0.0004, additive genetic model and β = −4.98, p = 0.017, recessive genetic model) and body temperature (β = −0.002, p = 0.0007, additive genetic model and β = −0.003, p = 0.004, recessive genetic model), suggestive of physiological differences. In the UK biobank (UKBB), we observed significant inverse relationship between the rs3184504 variant and whole body fat-free mass (β = −0.91, FDR p = 1.39E−23), and basal metabolic rate (β = −0.91, FDR p = 3.38E−21). However, this variant was not associated with body fat percentage (β = −0.05, FDR p = 0.89), suggesting a relationship with mitochondrial energy metabolism rather than a direct effect on adiposity.
Figure 5 summarizes the findings of the current study.
Figure 5.
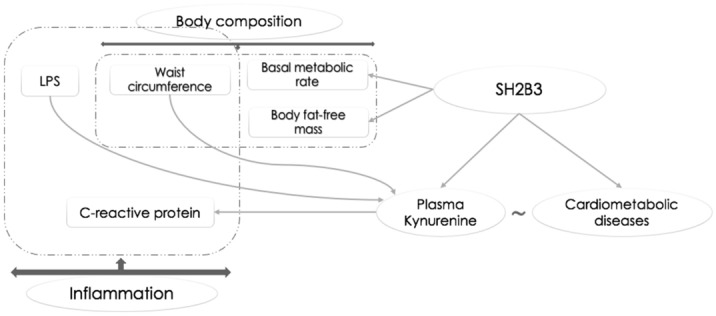
An overview of the study results.
Discussion
We leveraged genetic information to characterize the relationship between plasma levels of kynurenine and cardiometabolic diseases. Plasma kynurenine levels are determined by genetic variation associated with genes involved in tryptophan metabolism as well as genes without a clear previous link to kynurenine biosynthesis or catabolism. We confirmed a role for SH2B3 regulation of kynurenine in a knock-out mouse model. Additional human studies (the GENE evoked endotoxemia study) also confirmed that kynurenine is induced by inflammatory stimuli and is likely up-stream of the inflammatory biomarker CRP, which is also associated, but not causally related, with multiple inflammatory diseases. A polygenic predictor that assimilated the genetics from all of the kynurenine-modulating mechanisms was not associated with clinical phenotypes, suggesting that plasma kynurenine itself does not cause disease. Indeed, the individual SNPs near genes with well-characterized roles in kynurenine metabolism (IDO, SLC7A5) were not associated with disease. However, we observed significant associations linking a kynurenine-modulating missense variant in SH2B3 to a range of diseases including atherosclerosis, hypertension and hypothyroidism, as well as with white blood cell and platelet counts.
Genetic predictors of plasma kynurenine included some which impact established regulatory pathways; kynurenine is generated by tryptophan degradation by IDO, the first and rate-limiting enzyme in tryptophan catabolism, whose activity is stimulated by inflammation3,10. SLC5A7 encodes a protein involved in tryptophan transport across the cell membrane11. IDO and SLC5A7 are thought to predominately regulate kynurenine production. However, neither IDO or SLC5A7 individually, or polygenic risk score analyses, showed genetic associations with cardiometabolic disorders or other disease. These data suggest that plasma kynurenine levels per se do not have a causal relationship to these diseases.
The SH2B3 rs3184504 variant was associated with many diseases, including cardiometabolic and thyroid disease. This variant is a nonsynonymous SNP located in exon 3 of the SH2B3 gene which leads to a R262W amino acid change and has been shown to be involved in controlling immune responses. SH2B3 (LNK) is important in hematopoiesis, regulates the expansion of dendritic cells in lymph nodes, acts as a negative regulator of cytokine signaling, and has been associated with increased susceptibility to aortic dissection18–20. It is thought that the rs3184504 variant in SH2B3 approximates a loss of function21. Thus, to confirm the relationship between SH2B3 and kynurenine, we measured kynurenine levels in SH2B3−/−mice, and observed significant plasma and tissue-specific changes in kynurenine.
Up to now, no mechanistic relationship between SH2B3 and kynurenine metabolism had been reported. SH2B3 may regulate kynurenine levels indirectly through modulation of inflammatory signaling, and subsequent activation of IDO22. However, our data supported an association between rs3184504 and kynurenine in healthy individuals, without concurrent inflammation, suggesting SH2B3 may directly modulate the kynurenine pathway.
The kynurenine pathway controls production of NAD + , with important consequences for energy metabolism, and a known relationship to acute kidney injury23. Our data support a link between the SH2B3-kynurenine axis and energy homeostasis. We found that the SH2B3 rs3184504 variant associates with lower whole body fat-free mass and basal metabolic rate, in addition to lower body temperature and heart rate. There may be an important distinction between kynurenine that is elevated due to increased synthesis from tryptophan (controlled by IDO and SLC7A5), versus kynurenine that is elevated due to reduced breakdown of kynurenine or altered downstream pathway flux (potentially controlled by SH2B3). This hypothesis remains to be further interrogated, and there may be other mechanisms linking SH2B3 to kynurenine, which remain to be explored.
Epidemiological studies have shown that higher plasma kynurenine levels associate with increased inflammation and CVD prevalence in patients with end-stage renal disease. Individuals with higher kynurenic acid levels were more likely to have higher risk of coronary artery disease mortality and myocardial infarction24. Furthermore, kynurenine has been associated with the risk of all-cause mortality, particularly death from CVD25. We found that plasma kynurenine was acutely regulated during evoked endotoxemia, suggestive of a role in acute inflammatory responses. Kynurenine has recently been reported to be upregulated during COVID-19 infection, further highlighting a potential role for kynurenine as a biomarker of inflammatory activation26.
Waist circumference, a marker of central obesity, was positively associated with kynurenine. We also observed an inverse association between the rs3184504 variant and whole body fat-free mass and basal metabolic rate. PheWAS analyses revealed that the rs3184504 variant was directly associated with some obesity associated co-morbidities including hypertension and heart-related diseases, but not some other obesity-related comorbidities including obstructive sleep apnea and type 2 diabetes. Consistent with these findings, previous research demonstrated that SH2B3-related genetic alterations contribute to the development of hypertension and hematological disorders27,28. Consistently, the SH2B3 rs3184504 variant is associated with increased risk of CAD29, increased platelet counts and leukocytosis30, diastolic blood pressure31, atherosclerosis and thrombosis21. Thus, these data suggest that this SH2B3-related mechanism, which regulates both disease risk and kynurenine levels, may account for the observed associations between plasma kynurenine and the risk of cardiometabolic diseases. Traditional prospective studies seeking to link a metabolomic biomarker to a disease are limited based on sample sizes, follow-up times and do not provide insights into the molecular mechanisms account for candidate associations. We circumvented these limitations by using an approach which associated a putative biomarker with clinical traits based on their shared genetic structures. Thus, we were able probe for kynurenine associations in large, deeply phenotyped populations. Along with other strengths (increasing sample sizes and the number of diseases), this approach can also identify molecular mechanisms underlying an association. Another strength of our study is that we replicated findings in multiple populations (BioVU, eMERGE and the UKBB).
Our study had considerable strengths, but also some limitations. Using EHR billing codes rather than a systematic ascertainment of a diagnosis for classifying PheWAS phenotypes in BioVU and eMERGE databases might lead to both false-positive and false-negative associations. To address this, we replicated the study in other databases including UKBB and published data from large disease GWAS. Negative associations, such as the absence of an association between the kynurenine polygenic score (PRS) and disease, leave open the possibility of a false negative due to lack of power. However, we obtained very strong associations for the SNP in SH2B3 in the same datasets, indicating that the kynurenine genetic instruments should have been suitably powered. While experimental evidence is provided for showing kynurenine increases associated with acute inflammatory stimuli, additional studies are needed to confirm sustained elevations associated with inflammatory states. Furthermore, tissues in the heart, lungs and cardiopulmonary vasculature were not available for analyses and differences in levels in KO mice could not be assessed.
In conclusion: in this virtual biomarker study, we explored the association between kynurenine genetic predictors and clinical diagnoses derived from large datasets. Our findings suggest diverse molecular mechanisms regulate plasma kynurenine. The SH2B3-rs3184504 variant associates with both plasma kynurenine and diseases; our data suggest this is independent of kynurenine production. Plasma kynurenine, upregulated during inflammation, is upstream of the inflammatory biomarker CRP. The SH2B3 rs3184504 variant, which regulates kynurenine levels, associates with increased cardiometabolic disorders, potentially in a kynurenine-independent manner. In sum, although targeting plasma kynurenine directly is unlikely to be effective in disease treatment, interrogation of the SH2B3 pathways during inflammation may identify novel causal disease mechanisms.
Materials and methods
We identified SNPs associated with plasma kynurenine from published data32, and tested their associations with disease phenotypes. Identified associations were validated and probed using animal models and in independent populations.
Kynurenine GWAS summary statistics
SNPs associated with plasma kynurenine were identified from a GWAS meta-analysis of the KORA-TwinsUK studies32. Analyses were based on 7824 adult individuals of European descent33,34. Summary statistics were obtained from the Metabolomics GWAS server (http://metabolomics.helmholtz-muenchen.de/gwas/)32. An independent (r2 < 0.05 within 1000 kilobases) set of SNPs significantly associated (p < 5 × 10−8) with circulating kynurenine levels were selected from the KORA-TwinsUK meta-analysis GWAS summary statistics.
Electronic health record-linked genetic datasets
BioVU
BioVU is Vanderbilt University Medical Center’s DNA biobank, which is linked to de-identified EHR phenotype data35. A subset of BioVU (n = 62,303) participants of European Ancestry have SNP genotype data acquired on the Illumina MEGAEX platform. Quality control steps for the BioVU population have been previously described36. Genotypes were imputed with IMPUTE437, version 2.3.0 (University of Oxford), using the 10/2014 release of the 1000 Genomes cosmopolitan reference haplotypes and variants imputation quality scores less than 0.3 were excluded. One participant from each related pair (pi-hat > 0.2) was randomly excluded. Analyses were restricted to subjects of European ancestry, defined by principal components analyses in conjunction with HapMap reference populations. Quality control analyses used PLINK v1.938. The use of BioVU and other de-identified data presented in these analyses was approved by the VUMC Institutional Review Board (IRB), in accordance with the informed consent guidelines.
eMERGE
The eMERGE Phase I, II and III Network, a consortium of medical centers using EHRs as a tool for genomic research, included39 participants (n = 32,324) who were born prior to 1990 and were recruited from Geisinger Health System, Marshfield Clinic, Northwestern University, Mayo Clinic, and Kaiser Permanente/University of Washington. Consent was collected based on each site’s IRB protocols. eMERGE data were genotyped on multiple SNP arrays. QC procedures and imputation protocols for these data were conducted based on the established protocols developed by the eMERGE Genomics Working Group41. Use of de-identified eMERGE data was approved by the IRB at each site39, in accordance with the site-specific informed consent guidelines.
The UKBB study
UKBB is a British population-based self-reported study which is composed of approximately 0.5 million participants aged 37–73 at recruitment42. GWAS summary statistics for 2173 UKBB phenotypes43 were downloaded from the study by Bycroft et al.44. PheWAS results for individual kynurenine-associated SNPs were obtained from http://geneatlas.roslin.ed.ac.uk/43.
Disease genome-wide association study datasets
Summary statistics for CRP were obtained from a GWAS meta-analysis of 204,402 European individuals45. Additional summary statistics were downloaded from published GWAS of waist circumference20.
Phenome-wide Association Analysis (PheWAS)
Single SNP and multi SNP PheWAS analyses were conducted by testing associations with either individual SNPs (single SNP) associated with kynurenine or all kynurenine SNPs (a PRS comprising multi SNPs) PheWAS phenotypes. Analyses used the R PheWAS package46.
PheWAS were performed in BioVU and eMERGE using clinical phecode phenotypes (https://phewas.mc.vanderbilt.edu/) based on ICD-9-CM and ICD-10 diagnosis codes47,48. Individuals with two or more instances of a PheWAS diagnosis existing in their medical documents were considered as cases49. Clinical phenotypes with ≥ 300 cases were included and those affecting a single sex (like uterine prolapse and prostate cancer) were excluded. After exclusions, there were 894 phenotypes. Controls were individuals without any closely related PheWAS codes, and were matched to the age (BioVU) or decade of birth (eMERGE) ranges among the cases.
eQTL analysis
eQTL data for selected SNPs were obtained from the Genotype Tissue Expression (GTEx) portal, https://gtexportal.org.
Animal models
SH2B3−/− mouse
Plasma and tissues (white adipose tissue, brain, liver, spleen) were obtained from 14–15 week-old C57BL/6 J mice (wild type [WT, N = 9 plasma, N = 8 tissues)]) and from Lnk−/− (Sh2b3 knock out [KO], N = 5), as previously described50. Samples were frozen immediately following collection, and stored at − 80° C prior to analysis. Tissue samples were homogenized (Tissue Lyser LT) and stored at − 80° C prior to metabolite measurement. Mice were housed and taken care of in accordance with the Guide for the Care and Use of Laboratory Animals, US Department of Health and Human Services. All animal procedures were approved by the Vanderbilt Institutional Animal Care and Use Committee.
Enzyme-linked immunosorbent assay (ELISA) of kynurenine
Kynurenine in mouse plasma and tissue extracts was analyzed by ELISA (Cod. LS-F56401, LifeSpan BioScience Inc., Seattle, WA, USA), according to manufacturer’s instructions. All samples were run in duplicate, with an even distribution of samples from KO and WT animals across the three plates used. The intra-assay coefficient of variance was 9.8%. Due to relatively high inter-assay variability, likely attributable to differences by lot in the three ELISA kits used, we analyzed data as fold difference between WT and KO mice for each plate, rather than absolute values.
Clinical studies
GENE study
Healthy volunteers (294 non-pregnant/lactating women and men, age 18–45, BMI 18–30 kg/m2) were recruited to an evoked endotoxemia study (LPS, 1 ng/kg) at the University of Pennsylvania, as previously described16. Plasma metabolites, including kynurenine, were measured in a subset of individuals (n = 24) at baseline, and two hours post LPS-challenge, by mass spectrometry in positive and negative ion modes using well-established protocols51,52.
ABO study
The ABO Study recruited healthy volunteers (non-pregnant/lactating women and men, age 18–50) to a single study visit at the University of Pennsylvania from 2012 to 2014, as described53. Plasma metabolomics profiling, including measurement of kynurenine, was carried out at Metabolon (Metabolon Inc, Morrisville, NC; global metabolomics platform). Genotyping was performed using the Exome chip (Illumina, CoreExome, N > 540,000 variants, including rs3184504) at the Center for Applied Genomics at the Children's Hospital of Philadelphia. We analyzed data for a subset of individuals with overlapping metabolite and genetic data (N = 66).
The GENE and ABO studies were approved by the IRB of the University of Pennsylvania and Vanderbilt University. All participants provided written informed consent.
Statistical analyses
Genotyped populations (UKBB, BioVU, eMERGE and other large disease GWAS)
SingleSNP
PheWAS analyses for each kynurenine-associated SNP were performed in the BioVU and eMERGE populations. Associations were tested assuming an additive genetic model and used multivariable logistic regression models adjusting for 5 PCs, sex and either birth decade (eMERGE) or age (BioVU). BioVU and eMERGE data were combined using meta-analyses encoded by the METAL package (default settings were used)54.
MultiSNP
A PRS for plasma kynurenine levels was computed for each individual in the BioVU and eMERGE populations by summing their (Allele dosage x change in kynurenine levels per allele) for each kynurenine-associated SNPs. Association testing between the PRS and each phenotype was then tested and combined, as described above. Odds-ratios (ORs) represent the risk of disease per standard deviation (SD) increase in the PRS.
For UKBB phenotypes, and additional phenotypes based on GWAS summary statistics, mutliSNP association tests were conducted. Associations were tested using the IVWA, MR-Egger and Weighted Median methods, as implemented in the Mendelian Randomization R package55. Heterogeneity p values are based on the Cochran’s Q statistic, and a low p value may indicate horizontal pleiotropy. Association estimates represent the change in the log odds-ratio per standard deviation change in plasma kynurenine.
Multiple testing corrections
We applied a strict Benjamini–Hochberg (B–H) false discovery rate (FDR)56 to adjust for multiple testing, and associations with a q-value < 0.05 were considered significant40. All analyses were performed in accordance with relevant guidelines and regulations. The study was carried out in compliance with the ARRIVE guidelines.
Supplementary Information
Acknowledgements
The authors are grateful to the staff and participants of the BioVU, eMERGE, UK biobank and all GWAS studies used in this paper. The research is solely the responsibility of the authors and do not necessarily represent the views of Vanderbilt University Medical Center.
Abbreviations
- B–H
Benjamini–Hochberg
- CRP
C-reactive protein
- CVD
Cardiovascular diseases
- ELISA
Enzyme-linked immunosorbent assay
- eMERGE
Electronic medical records and genetics consortium
- eQTL
Expression quantitative trait loci
- FDR
False discovery rate
- GENE
Genetics of evoked responses to niacin and endotoxemia
- GTEx
Genotype tissue expression
- 3HAA
3-Hydroxy anthranilic acid
- HK
Hydroxykynurenine
- IDO
Indoleamine 2,3-dioxygenase
- IRB
Institutional Review Board
- IVWA
Inverse-variance weighted average
- KORA
Cooperative Health Research in the Region of Augsburg
- KA
Kynurenic acid
- LD
Linkage disequilibrium
- LPS
Lipopolysaccharide
- NA
Nicotinic acid
- NAM
Nicotinamide
- OR
Odds-ratio
- PheWAS
Phenome-wide association analysis
- PRS
Polygenic score
- QA
Quinolinic acid
- SD
Standard deviation
- SNP
Single nucleotide polymorphism
Author contributions
All authors reviewed, commented on and approved the paper for the submission. M.B., J.F.F., J.D.M. designed the study. M.B., J.F.F., J.D.M., C.W., M.S., A.M., K.S. conducted research. M.B., J.F.F., J.D.M., C.W., M.S., C.M.S., K.S. performed the statistical analysis. M.B., J.F.F., J.D.M. interpreted the results. M.B., J.F.F., J.D.M. wrote the paper.
Funding
This research was supported by the NIH, R01 HL142856 (Ferguson), and the American Heart Association; 16FTF30130005 (Mosley). Datasets used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding, the 1S10RR025141-01 instrumentation award, and by the CTSA grant UL1TR000445 from NCATS/NIH. The eMERGE Network was initiated and funded by NHGRI through the following grants: U01HG006828 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital); U01HG006830 (Children’s Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative/University of Washington); U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG004438 (CIDR) and U01HG004424 (the Broad Institute) serving as Genotyping Centers.
Data and code availability
eMERGE data are available through dbGaP (phs000360.v3.p1). Upon acceptance, the complete findings from the PheWAS analyses will be made available thought a publicly-available website.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jonathan D. Mosley and Jane F. Ferguson.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95154-9.
References
- 1.Vasan RS. Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation. 2006;113:2335–2362. doi: 10.1161/CIRCULATIONAHA.104.482570. [DOI] [PubMed] [Google Scholar]
- 2.Maher BS. Polygenic scores in epidemiology: risk prediction, etiology, and clinical utility. Curr. Epidemiol. Rep. 2015;2:239–244. doi: 10.1007/s40471-015-0055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michelhaugh SK, Guastella AR, Mittal S. Overview of the kynurenine pathway of tryptophan metabolism. In: Mittal S, editor. Targeting the Broadly Pathogenic Kynurenine Pathway. Berlin: Springer; 2015. pp. 3–9. [Google Scholar]
- 4.Song P, Ramprasath T, Wang H, Zou M-H. Abnormal kynurenine pathway of tryptophan catabolism in cardiovascular diseases. Cell Mol. Life Sci. 2017;74:2899–2916. doi: 10.1007/s00018-017-2504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawlak K, Domaniewski T, Mysliwiec M, Pawlak D. The kynurenines are associated with oxidative stress, inflammation and the prevalence of cardiovascular disease in patients with end-stage renal disease. Atherosclerosis. 2009;204:309–314. doi: 10.1016/j.atherosclerosis.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 6.Mangge H, et al. Disturbed tryptophan metabolism in cardiovascular disease. Curr. Med. Chem. 2014;21:1931–1937. doi: 10.2174/0929867321666140304105526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cozlea DL, et al. The impact of C reactive protein on global cardiovascular risk on patients with coronary artery disease. Curr. Health Sci. J. 2013;39:225–231. [PMC free article] [PubMed] [Google Scholar]
- 8.Aimo A, et al. Colchicine for the treatment of coronary artery disease. Trends Cardiovasc. Med. 2020 doi: 10.1016/j.tcm.2020.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Esser N, Paquot N, Scheen AJ. Inflammatory markers and cardiometabolic diseases. Acta Clin. Belg. 2015;70:193–199. doi: 10.1179/2295333715Y.0000000004. [DOI] [PubMed] [Google Scholar]
- 10.Badawy AA-B. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int. J. Tryptophan. Res. 2017;10:1178646917691938–1178646917691938. doi: 10.1177/1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scalise M, Galluccio M, Console L, Pochini L, Indiveri C. The human SLC7A5 (LAT1): the intriguing histidine/large neutral amino acid transporter and its relevance to human health. Front. Chem. 2018;6:243. doi: 10.3389/fchem.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao C, et al. Integromic analysis of genetic variation and gene expression identifies networks for cardiovascular disease phenotypes. Circulation. 2015;131:536–549. doi: 10.1161/CIRCULATIONAHA.114.010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breda C, et al. Tryptophan-2,3-dioxygenase (TDO) inhibition ameliorates neurodegeneration by modulation of kynurenine pathway metabolites. Proc. Natl. Acad. Sci. USA. 2016;113:5435. doi: 10.1073/pnas.1604453113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long T, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017;49:568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- 15.Rhee EP, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18:130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson JF, et al. Race and gender variation in response to evoked inflammation. J. Transl. Med. 2013;11:63. doi: 10.1186/1479-5876-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi J, Joseph L, Pilote L. Obesity and C-reactive protein in various populations: a systematic review and meta-analysis: obesity and CRP in various populations Obes. Rev. 2013;14:232–244. doi: 10.1111/obr.12003. [DOI] [PubMed] [Google Scholar]
- 18.Mori T, et al. Lnk/Sh2b3 controls the production and function of dendritic cells and regulates the induction of IFN-γ–producing T cells. J. Immunol. 2014;193:1728. doi: 10.4049/jimmunol.1303243. [DOI] [PubMed] [Google Scholar]
- 19.Laroumanie F, et al. LNK deficiency promotes acute aortic dissection and rupture. JCI Insight. 2018;3:e122558. doi: 10.1172/jci.insight.122558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, et al. LNK/SH2B3 loss of function promotes atherosclerosis and thrombosis. Circ. Res. 2016;119:e91–e103. doi: 10.1161/CIRCRESAHA.116.303577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang X, Karlsen TH. Genetics of primary sclerosing cholangitis and pathophysiological implications. Nat. Rev. Gastroenterol. Hepatol. 2017;14:279–295. doi: 10.1038/nrgastro.2016.154. [DOI] [PubMed] [Google Scholar]
- 23.Poyan Mehr A, et al. De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat. Med. 2018;24:1351–1359. doi: 10.1038/s41591-018-0138-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu E, et al. Increases in plasma tryptophan are inversely associated with incident cardiovascular disease in the Prevención con Dieta Mediterránea (PREDIMED) study. J Nutr. 2017;147:314–322. doi: 10.3945/jn.116.241711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo H, et al. Plasma biomarkers of inflammation, the kynurenine pathway, and risks of all-cause, cancer, and cardiovascular disease mortality: the hordaland health study. Am. J. Epidemiol. 2016;183:249–258. doi: 10.1093/aje/kwv242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas T, et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight. 2020 doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dale BL, Madhur MS. Linking inflammation and hypertension via LNK/SH2B3. Curr. Opin. Nephrol. Hypertens. 2016;25:87–93. doi: 10.1097/MNH.0000000000000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maslah N, Cassinat B, Verger E, Kiladjian J-J, Velazquez L. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia. 2017;31:1661–1670. doi: 10.1038/leu.2017.139. [DOI] [PubMed] [Google Scholar]
- 29.CARDIoGRAMplusC4D Consortium et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet.45, 25–33 (2013). [DOI] [PMC free article] [PubMed]
- 30.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 31.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin S-Y, et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moayyeri A, Hammond CJ, Hart DJ, Spector TD. The UK adult twin registry (TwinsUK Resource) Twin Res. Hum. Genet. 2013;16:144–149. doi: 10.1017/thg.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wichmann H-E, Gieger C, Illig T. KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67:26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 35.Roden D, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruderfer DM, et al. Significant shared heritability underlies suicide attempt and clinically predicted probability of attempting suicide. Mol. Psychiatry. 2019 doi: 10.1038/s41380-018-0326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gottesman O, et al. The electronic medical records and genomics (eMERGE) network: past, present, and future. Genet. Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosley JD, et al. A study paradigm integrating prospective epidemiologic cohorts and electronic health records to identify disease biomarkers. Nat. Commun. 2018;9:3522. doi: 10.1038/s41467-018-05624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuvich RL, et al. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality: pitfalls of Merging GWAS data: lessons learned. Genet. Epidemiol. 2011;35:887–898. doi: 10.1002/gepi.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sudlow C, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang L, et al. A resource-efficient tool for mixed model association analysis of large-scale data. Nat. Genet. 2019;51:1749–1755. doi: 10.1038/s41588-019-0530-8. [DOI] [PubMed] [Google Scholar]
- 44.Bycroft C, et al. The UK biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ligthart S, et al. Genome analyses of >200,000 individuals identify 58 loci for chronic inflammation and highlight pathways that link inflammation and complex disorders. Am. J. Hum. Genet. 2018;103:691–706. doi: 10.1016/j.ajhg.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–2376. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013;31:1102–1111. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei W-Q, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS ONE. 2017;12:e0175508. doi: 10.1371/journal.pone.0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saleh MA, et al. Lymphocyte adaptor protein LNK deficiency exacerbates hypertension and end-organ inflammation. J. Clin. Invest. 2015;125:1189–1202. doi: 10.1172/JCI76327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimberly WT, et al. Metabolite profiling identifies anandamide as a biomarker of nonalcoholic steatohepatitis. JCI Insight. 2017;2:e92989. doi: 10.1172/jci.insight.92989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang Z-Z, et al. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front. Genet. 2019;10:454. doi: 10.3389/fgene.2019.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
eMERGE data are available through dbGaP (phs000360.v3.p1). Upon acceptance, the complete findings from the PheWAS analyses will be made available thought a publicly-available website.