Abstract
Background
The function of major histocompatibility complex (MHC) molecules is to bind peptide fragments derived from genomic mutations or pathogens and display them on the cell surface for recognition by cognate T cells to initiate an immune response.
Methods
In this study, we provide a comprehensive investigation of HLA gene expression in a pan-cancer manner involving 33 cancer types. We utilised gene expression data from several databases and immune checkpoint blockade-treated patient cohorts.
Results
We show that MHC expression varies strongly among cancer types and is associated with several genomic and immunological features. While immune cell infiltration was generally higher in tumours with higher HLA gene expression, CD4+ T cells showed significantly different correlations among cancer types, separating them into two clusters. Furthermore, we show that increased HLA gene expression is associated with prolonged survival in the majority of cancer types. Lastly, HLA gene expression is associated with patient response to immune checkpoint blockade, which is especially prominent for HLA class II expression in tumour biopsies taken during treatment.
Conclusion
We show that HLA gene expression is an important feature of tumour biology that has significant impact on patient prognosis.
Subject terms: Tumour immunology, MHC
Background
Major histocompatibility complex (MHC) molecules are cell-surface molecules that present antigens to T cells. These antigens allow T cells to differentiate healthy cells from tumour or infected cells. Different classes of MHC molecules with specific functions exist. MHC I, encoded by class I human leukocyte antigen (HLA) genes HLA-A, HLA-B and HLA-C, presents protein fragments that originate from inside of the cell to CD8+ T cells.1 Together with the β-2 microglobulin (B2M) protein, which forms the shorter MHC chain, the HLA class I proteins form the MHC I complex.2 MHC I complexes are expressed on the surface of most nucleated cells.3 MHC II complexes, encoded by HLA-DR, HLA-DQ and HLA-DP, present endosomal protein fragments to CD4 T cells.1 MHC II complexes have limited expression, and are typically expressed on professional antigen-presenting cells (APCs), including dendritic cells, macrophages and B cells.1 MHC II can also act as signal receptors, and can induce apoptosis of APCs.1 In addition to the classical antigen-presenting MHCs, a number of non-classical HLA genes are expressed—these include class I genes HLA-E, HLA-F and HLA-G and class II genes HLA-DOA, HLA-DOB and HLA-DM. These non-classical MHC complexes have a broad range of functions. For example, HLA-DM assists in the processing of antigens for loading onto HLA class II complexes,4 whereas HLA-E presents antigens to NK cells.5
Tumour cells have evolved several means to escape T-cell recognition through HLA-related mechanisms. They can alter HLA expression to downregulate the production of MHC complexes and therefore display fewer identifying antigens.6 HLA class I gene mutations have been reported in many cancers, including in head and neck cancer, squamous cell lung cancer, stomach adenocarcinoma, diffuse large B-cell lymphoma and colon cancer.7 Rates of mutation were the highest on exon 4, which encodes the a3 domain to which the CD8 co-receptor binds, and the next highest on exons 3 and 2, which encode the a1 and a2 domains that bind the protein and present antigens.7 Loss-of-function mutations often occurred at the N-terminal end of the protein, which likely caused the protein to be non-functional.7 Loss of heterozygosity (LOH) of HLA alleles may allow cancer cells to escape immune recognition.8 In non-small-cell lung cancers, HLA LOH occurs in 40% of early-stage cancers.8 Additionally, the HLA homozygosity was preferentially selected for at metastatic cancer sites.8 Advanced cancer patients that were heterozygous at all HLA class I loci had increased survival as compared to patients who were homozygous at any one locus.9
While it is understood that tumours utilise various strategies that result in altered MHC expression on the cell surface, the impact of HLA gene expression changes in clinical outcomes across different cancer types has not been systematically investigated for all cancer types. For example, plasma levels of HLA-G proteins in thyroid cancer have been investigated in multiple studies,10,11 but no studies have evaluated gene expression of all HLA genes in primary thyroid tumours. While downregulation of classical HLA class I genes leads to poorer prognosis in many cancer types,12–14 some discrepancies between prognostic associations within cancer types have been reported. For example, studies in oesophageal cancer have shown that HLA class I genes are associated with both good and poor prognosis. Lastly, several studies have performed in-depth analyses of HLA gene expression in particular cancer types, but few studies have evaluated HLA gene expression differences among cancer types. Investigation of HLA gene expression differences might provide insights into varying prognostic associations involving HLA gene expression between cancer types or show some rationale for different levels of clinical responses to immune checkpoint blockade therapy (ICBT).
In this study, we have investigated these gaps in knowledge using a pan-cancer approach that included 33 different tumour types. We found that HLA gene expression is significantly associated with distinct genomic and immunologic tumour characteristics. We also show that HLA gene expression has prognostic value in both standard treatment and ICBT settings. Our analyses provide a comprehensive investigation of HLA gene expression across cancer types.
Methods
TCGA data
All datasets used in this study are publicly available. RNAseq data generated by The Cancer Genome Atlas (TCGA) for KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; KICH, kidney chromophobe; LGG, brain lower-grade glioma; GBM, glioblastoma multiforme; BRCA, breast cancer; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma; READ, rectum adenocarcinoma; COAD, colon adenocarcinoma; UCS, uterine carcinosarcoma; UCEC, uterine corpus endometrial carcinoma; OV, ovarian serous cystadenocarcinoma; HNSC, head and neck squamous carcinoma; THCA, thyroid carcinoma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; SKCM, skin cutaneous melanoma; BLCA, bladder urothelial carcinoma; LIHC, liver hepatocellular carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; ACC, adrenocortical carcinoma; PCPG, pheochromocytoma and paraganglioma; SARC, sarcoma; LAML, acute myeloid leukaemia; PAAD, pancreatic adenocarcinoma; ESCA, oesophageal carcinoma; TGCT, testicular germ cell tumours; THYM, thymoma; MESO, mesothelioma; UVM, uveal melanoma; DLBC, lymphoid neoplasm diffuse large b-cell lymphoma; CHOL, cholangiocarcinoma, were obtained from TCGA on FireBrowse (http://firebrowse.org). These datasets included 13,717 Level 3 RNAseq samples of which 12,015 were obtained from tumour tissue and 1702 samples were obtained from tumour adjacent normal tissue. RSEM-normalised gene expression for 20,502 genes for each sample was provided (see Supplementary Table 1).
TCGA DNA methylation profiles were downloaded from FireBrowse (http://firebrowse.org). Based on the annotation file for Infinium HumanMethylation450K BeadChip platform, CpGs located in the promoter of each HLA gene were identified. Methylation levels of CpGs were then averaged to obtain promoter methylation scores for each gene. Chromosome-level aneuploidy scores were downloaded as a supplemental file from prior work.15 Chromosome 6p (Chr6p) was considered to be lost if chr6p or chr6 was deleted (‘−1’) and gained if chr6p or chr6 was amplified (‘+1’). Samples were excluded for chr6p copy number variation (CNV) analysis if no CNV call had been made (‘NA’). HLA mutation information was obtained from Castro et al.16 HLA genes were considered to be mutated if any amino acid change occurred as a result of a mutation, i.e. any mutation other than “silent” was considered a mutation. A list of hypermutated samples was obtained from prior work.17
PRECOG data
Meta-z-scores for prognostic analyses were obtained from PREdiction of Clinical Outcomes from Genomic profiles (PRECOG, https://precog.stanford.edu/precog_data.php,18). For the Principal Component Analysis on PRECOG data, gene expression data and the associated survival data from 166 datasets were obtained from https://precog.stanford.edu/. All datasets with less than 40 samples or with a death event percentage <20% were excluded, resulting in the inclusion of 141 datasets.
Immunotherapy data
Raw gene expression data and clinical information for the Van Allen dataset (anti-cytotoxic T-lymphocyte-associated protein 4 (CTLA4)) were obtained from the Database of Genotypes and Phenotypes (dbGaP) under accession number phs000452. Reads were aligned to the GRCh37 human genome assembly using TopHat (v. 2.1.0).19 Transcript counts were obtained through Cufflinks (v. 2.2.1).19 Processed gene expression data for the Snyder dataset (anti-Programmed death-ligand 1 (PD-L1)) were acquired from the GitHub repository (https://github.com/hammerlab/multi-omic-urothelial-anti-pdl1) that accompanied the original publication.20 Processed gene expression data for the Hugo and Riaz datasets (anti-Programmed cell death protein 1 (PD-1) and anti-PD-1, respectively) were obtained from Gene Expression Omnibus (GEO) under accession numbers GSE78220 and GSE91061, respectively. Clinical data for these datasets were obtained from supplementary files in the original publications21,22 or from GEO. In all datasets, clinical benefit designations were defined based on response annotations by the original authors. Clinical benefit was assumed for patients with complete or partial response, whereas no clinical benefit was designated to patients with progressive disease. Patients with stable disease were excluded from the analysis due to classification uncertainty; patients with stable disease are not considered responders when evaluating objective response rates (ORRs),23,24 but might gain some long-term clinical benefit from ICBT,25 which precluded a clear classification of patients with stable disease as experiencing clinical or no clinical benefit.
Calculation of immune cell infiltration scores
Immune infiltration scores of specific immune cell types were calculated using our established framework detailed in refs. 26,27 In short, immune cell-specific weight profiles for naive B cells, memory B cells, CD4+ T cells, CD8+ T cells, NK cells and monocytes were utilised. The enrichment of these immune cell signatures in patient samples was determined by BASE,28 a rank-based gene set enrichment method, for each immune cell type. Immune inference scores were normalised by 1000-fold randomisation of patient profiles, resulting in patient-specific inference scores for each immune cell type.
Gene set variation analysis
Gene set variant analysis (GSVA)29 was performed on all TCGA samples using the C2 pathway database from the molecular signatures database (MSigDB).30 Pathways highly related to the immune system were selected for evaluation.
Principal component analysis
Principal component analysis (PCA) was performed using the prcomp R function for each individual cancer type. Principal component coordinates for each sample were extracted using the factoextra R package (https://github.com/kassambara/factoextra). Principal component 1 (PC1) was used to represent meta-HLA expression. Since the sign (+ or −) of PC1 can be reversed without impacting the information contained within PC1, we correlated PC1 with HLA/B2M expression and reversed all PC1 signs within a cancer type if PC1 and HLA/B2M expression were negatively correlated. This was done to make sure that the meta-HLA gene accurately represented overall HLA gene expression.
Survival analysis
Survival analyses were performed using the R survival package (version 3.1-8). Log-rank tests were performed to evaluate overall or relapse-free survival probabilities between two groups, using the survdiff function. Kaplan–Meier (KM) plots were generated using the survfit function. For all reported KM plots and accompanying log-rank tests, samples were separated into two groups based on median HLA gene expression or as indicated otherwise.
Statistical analysis
HLA gene expression comparisons between normal and corresponding tumour tissue were performed using Student’s t-tests. All comparisons of chr6p CNV and mRNA between were performed using Student’s t-tests. The Spearman correlation coefficient (SCC) was reported for all correlation analyses. Total HLA gene expression was calculated for each cancer type by counting the total number of reads that were aligned to HLA/B2M genes in all samples and dividing the total count by the number of samples within each cancer type. Correlations were calculated using the R function cor and significance was assessed using cor.test. P-values < 0.05 were considered significant. All analyses were conducted in R (version 3.6.1).
Results
HLA genes are deregulated in tumours with expression varying dramatically among tumour types
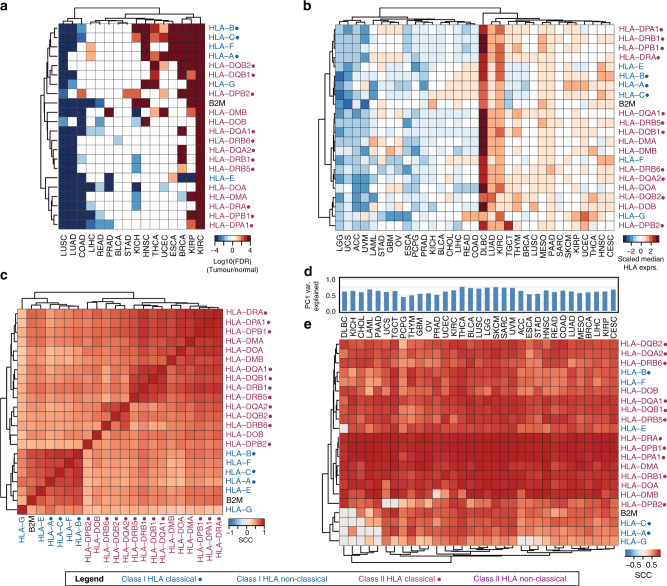
We first explored the differential expression of HLA/B2M genes in tumours with respect to normal samples across 16 cancer types from the TCGA with at least ten normal controls (Supplementary Table 1). We found that HLA/B2M genes tended to be significantly downregulated in Lung squamous cell carcinoma (LUSC) and Lung adenocarcinoma (LUAD), but upregulated in Kidney renal clear cell carcinoma (KIRC), Kidney chromophobe (KICH) and Head and neck squamous carcinoma (HNSC) (adjusted p < 0.05). Several of these findings are consistent with previous literature in which downregulation of HLA class I has been reported in LUAD31 and upregulation of HLA class I genes has been reported in renal cell carcinoma32 as compared to normal tissue. The variation in HLA gene expression across cancer types was much larger compared to variation between HLA genes and hierarchical clustering revealed that HLA genes from similar HLA classes clustered together (Fig. 1a). Next, we compared the median HLA/B2M expression in the 33 TCGA cancer types (Supplementary Table 1). Again, expression of HLA/B2M genes was highly correlated but varied substantially among different cancer types (Fig. 1b). Diffuse large B-cell lymphoma (DLBC) showed the highest HLA/B2M gene expression levels, consistent with the known high expression of HLA genes in B cells.33,34 Low-grade glioma (LGG) showed the lowest expression of HLA class I genes across all tumour types.
Fig. 1. HLA mRNA expression comparison across cancer types and HLA genes.
a Heatmap comparing the expression between all HLA/B2M between normal and tumour tissue. Displayed are −log10(p-value) values of t-tests comparing HLA/B2M expression in normal and tumour tissue. P-values were adjusted by the Bonferroni procedure. All adjusted p-values > 0.05 were considered non-significant and set to −log10(p) = 0. P-values received a negative sign if the log2FC between normal and tumour was negative (i.e. higher expression in normal tissue). Values greater than |4| were set to 4. b Median HLA/B2M gene expression by cancer type and HLA gene. c Correlation matrix of HLA/B2M gene expression across all cancer types. d Percentage of variance captured by principal component 1 (PC1) in each cancer type. e Correlation matrix of the meta-HLA gene and gene expression of HLA/B2M. P-values were adjusted by the Bonferroni procedure. All correlations with adjusted p-values > 0.05 were considered non-significant and set to 0. Classical HLA genes indicated with a dot (●), HLA class I genes indicated in blue, HLA class II genes in purple.
At the pan-cancer level, expression of all HLA/B2M genes was highly correlated based on correlation coefficients (Fig. 1c). This correlation pattern was also further confirmed by analysis in individual cancer types (Supplementary Fig. 1). In fact, PCA indicated that over 60% of the expression variance in HLA/B2M genes could be explained by the first principal component (PC1) (Fig. 1d, Supplementary Table 2). As such, we denoted PC1 as the meta-HLA gene to approximate the overall gene expression level of the HLA/B2M gene cluster. As shown, almost all HLA/B2M genes were significantly correlated with the meta-HLA gene (Fig. 1e).
Association between HLA gene expression and genomic features
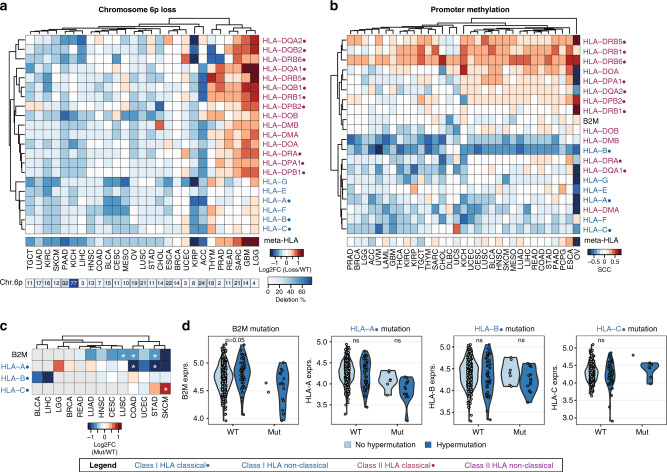
Having observed the high variation of HLA/B2M expression among different cancer types, we examined whether this variation could be explained by several major genomic features. All of the HLA genes are located on chr6p, which has been reported to be associated with high-frequency gain/loss in cancers.15 Thus, we first analysed the relationship between chr6p CNV and HLA gene expression (Supplementary Table 3). As expected, chr6p loss was associated with downregulation of HLA gene expression in the majority of cancer types (Fig. 2a). Increased HLA gene expression upon chr6p loss was observed in a few cancer types, especially, in two brain cancer types, glioblastoma multiforme (GBM) and LGG. In contrast, the correlation between chr6p gain and HLA gene expression was moderate. Although a number of cancer types showed overall increased or decreased HLA gene expression upon chr6p gain, the majority of genes displayed minimal changes (Supplementary Fig. 2A). These results suggest that chr6p CNV only partially explains the expression changes of HLA genes in certain cancer types and that additional HLA-regulating mechanisms might be at play.
Fig. 2. Association between HLA gene expression and genomic features.
a Heatmap of the log2 fold change (log2FC) comparing HLA and B2M expression between patients with chr6p loss and patients with diploid chr6p (WT). b Heatmap of Spearman correlation coefficients (SCCs) comparing HLA/B2M promoter methylation and mRNA expression in samples with diploid chr6p. c Heatmap the log2FC comparing HLA and B2M mRNA expression between patients with and without mutations in HLA genes. Only cancer types with at least five samples displaying HLA mutations were included. Grey boxes indicate <5 samples with HLA mutations available. Stars indicate significant comparisons at p < 0.05, two-sided Student’s t-test. d Comparison of HLA gene expression in STAD between patients without (WT) and with HLA mutations (Mut). WT and Mut samples were stratified by hypermutation status (light blue = no hypermutation, dark blue = hypermutation). Significance indicated by two-sided Student’s t-tests for comparisons in which both groups had at least five samples. Classical HLA genes indicated with a dot (●), HLA class I genes indicated in blue, HLA class II genes in purple.
We next investigated the relationship between HLA/B2M promoter methylation and mRNA expression in tumour samples with diploid chr6p. Approximately half of the HLA/B2M genes showed a negative correlation between promoter methylation and HLA gene expression, whereas a small number of HLA genes (e.g. HLA-DBP1 and HLA-DRB6) showed strong positive correlations (Fig. 2b). Lastly, we investigated the relationship between mutations and HLA gene expression. Due to the low incidence of HLA mutations, especially in HLA class II genes, we focused our efforts on mutations in HLA class I genes. In general, mutations in B2M and HLA genes were associated with decreased HLA gene expression of the concordant mRNA transcript (Fig. 2c). This is consistent with previous studies that evaluated the relationship between B2M/HLA mutations and transcript abundance.7,35
It has been reported that mutations in HLA genes are associated with high mutation burden or somatic hypermutation in several cancer types7,36 (Supplementary Table 3). Indeed, we observed that HLA/B2M-mutated samples were more likely to display a hypermutation phenotype. For example, in Stomach adenocarcinoma (STAD), the cancer type with the highest percentage of hypermutated samples, 15 out of 17 (88%) B2M-mutated samples showed hypermutation, compared to 47 out of 386 (12%) B2M wild-type samples (p = 1E−11, Fisher’s exact test). Similarly, samples with HLA-A, HLA-B and HLA-C mutations were enriched for hypermutation with p-values of 6E−7, 1E−8 and 1E−5, respectively (Fisher’s exact test). However, only weak or no correlation was observed between HLA/B2M gene expression and HLA/B2M mutations, irrespective of hypermutation (Fig. 2d). Taking together, the expression of HLA genes in tumours seems affected by many different genomic features, but their contributions vary among different cancer types.
HLA gene expression is associated with patient prognosis in many cancer types
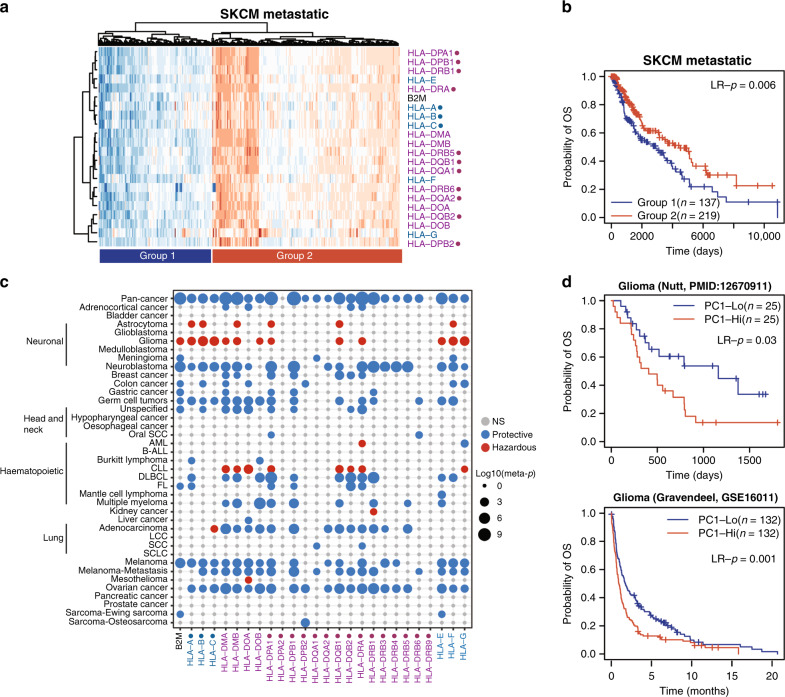
Given critical roles played by HLA genes in cancer immunology, we investigated the association between HLA gene expression and patient survival. First, we used the TCGA metastatic skin cutaneous melanoma (SKCM) data as an example to show the expression pattern of HLA/B2M genes in tumour samples. We performed hierarchical clustering analysis using all HLA/B2M genes and observed two groups of patients (Fig. 3a): patients in group 1 (n = 137) had high expression of almost all HLA genes, whereas patients in group 2 (n = 219) had low expression of HLA genes. Survival analysis indicated that patients with high HLA gene expression had significantly longer survival compared to patients with low HLA gene expression (p = 0.006, Log-rank test, Fig. 3b).
Fig. 3. HLA gene expression associated with prognosis.
a Heatmap showing clustering of metastatic melanoma patients based on HLA gene expression. b KM plot showing a difference in survival between metastatic melanoma patients in group 1 or group 2 from (a). Significance indicated by Log-rank tests. c Heatmap displaying the −log10(meta-p-value) of univariate CoxPH regression between HLA/B2M gene expression and overall survival. Blue dots indicate a protective role for HLA gene expression, whereas red dots indicate a hazardous role. Dot sizes indicate −log10(meta-p-value). d KM plot showing the difference in survival between glioma patients with low and high meta-HLA scores in two independent datasets. Significance indicated by Log-rank tests. Classical HLA genes indicated with a dot (●), HLA class I genes indicated in blue, HLA class II genes in purple.
Next, we extended our analysis to all cancer types. We utilised the PRECOG dataset,18 which has a large number of microarray datasets from different cancer types with extensive follow-up time. We found that all HLA/B2M genes, with the exceptions of HLA-DRB9, were associated with overall survival of patients in at least one cancer type (Fig. 3c). In almost all cancer types, the association between HLA gene expression and prognosis was consistent between different HLA/B2M genes. In contrast, the association between HLA/B2M gene expression and patient survival varied between different cancer types. For the majority of cancer types, high levels of HLA/B2M gene expression were associated with good prognosis. However, in glioma, astrocytoma and CLL, we observed consistent negative correlations between HLA/B2M expression and prognosis. The meta-HLA gene was also associated with survival. For example, the meta-HLA gene showed a negative relationship between high HLA gene expression and poor prognosis in glioma (Fig. 3d), consistent with the negative association between individual HLA genes and prognosis in this cancer type. Thus, HLA gene expression is associated with patient prognosis in a large number of cancer types, high HLA gene expression being protective in the majority of cancer types but not in glioma, astrocytoma and CLL.
High HLA gene expression is associated with a hot tumour microenvironment
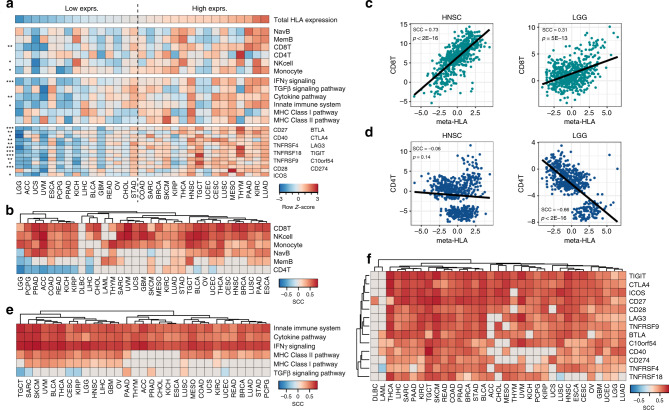
The tumour microenvironment (TME) is an important contributor to HLA gene expression in the tumour, since HLA genes are highly expressed on certain immune cell types.1 We thus investigate the relationship between HLA/B2M expression and the TME in greater depth. Since most of the HLA/B2M genes are highly correlated, we used the meta-HLA gene to represent their expression to simplify the presentation of the results. We ordered cancer types based on total HLA gene expression and displayed the total infiltration of different immune cell types, immune-associated pathways and the expression of immune checkpoint proteins accordingly (Fig. 4a). As shown, cancer types with higher HLA gene expression displayed the presence of a more immunologically active TME, also referred to as an immune hot TME, showing significantly higher immune cell infiltration and the enrichment of pathways associated with the immune system (Fig. 4a).
Fig. 4. Association between meta-HLA gene expression and the tumour microenvironment.
a Overview of HLA gene expression and the TME characteristics in all cancer types. Heatmap displaying z-transformed values of total HLA gene expression, immune cell infiltration, pathway activity and immune checkpoint expression. Cancer types ordered based on total HLA gene expression. Stars indicate significance between cancer types with low and high overall HLA expression (separated by dotted line) assessed by Student t-tests. *** = <0.001, ** = <0.01, * = <0.05. b Spearman correlation coefficients (SCCs) between meta-HLA gene expression and the infiltration of six major immune cell types. c Correlation between CD8+ T-cell infiltration and meta-HLA expression in HNSC and LGG. d Correlation between CD4+ T-cell infiltration and meta-HLA expression in HNSC and LGG. e SCCs between meta-HLA gene expression and the activity of six immune pathways. f SCCs between meta-HLA gene expression and the mRNA expression of immune checkpoint proteins. All non-significant correlations (Bonferroni-corrected p > 0.05) are shown in grey.
Next, we assessed the relationship between HLA gene expression and the infiltration of different immune cell types within each cancer type (Fig. 4b). Based on the correlation patterns between immune cell infiltration and meta-HLA expression, we observed two clusters of cancer types. These two clusters displayed similar patterns of CD8+ T cells, NK cells and monocytes, but were distinguished by memory B and CD4+ T-cell infiltration. As shown, almost all cancer types showed a significant positive association between HLA gene expression and the infiltration of cytotoxic effector cells CD8+ T and NK cells, indicating high HLA gene expression is associated with a hot TME. For example, HNSC and even the least immunologically active cancer type, LGG, showed highly significant positive relationships between HLA gene expression and CD8+ T-cell infiltration (Fig. 4c). In contrast, CD4+ T cells had a significant negative correlation with HLA gene expression in the minor cluster consisting of LGG, PCPG, PRAD, ACC, COAD, READ, KICH and KIRP, but weak or no correlation in the other major cluster (Fig. 4b). For example, no association was found for HNSC but LGG showed a strong negative association between HLA gene expression and CD4+ T-cell infiltration (SCC = −0.66, p = 7E−69, Fig. 4d). Immune-related pathways active within the TME were also strongly associated with HLA gene expression (Fig. 4e). The innate immune system, cytokine and IFNγ signalling pathways were positively correlated with HLA gene expression in all cancer types (Fig. 4e), further reaffirming that samples with a hot TME are associated with increased HLA gene expression.
We observed that IFNγ signalling was positively correlated with the meta-HLA gene in almost all cancer types (Fig. 4e). Generally, IFNγ activity is associated with immune activation and an effective antitumour immune response.37,38 CD8+ T and NK cells were concordantly positively correlated with HLA gene expression (Fig. 4b). However, since these patients still presented to the clinic with a tumour, we reasoned that immunosuppressive features might present in the tumour to prevent an effective immune response from occurring. We thus looked at the correlation between HLA gene expression and the expression of immune checkpoints to further investigate this possibility. Indeed, we found that HLA gene expression correlated positively with the vast majority of immune checkpoint proteins across the majority of cancer types (Fig. 4f). Both hematopoietic cancer types, Diffuse large b-cell lymphoma (DLBC) and acute myeloid leukaemia (LAML), showed minimal correlations, suggesting an overt difference in immune checkpoint expression between solid and liquid cancers. In conclusion, we have shown that increased HLA gene expression within a tumour is associated with a hot TME, both on the cance- type level and within cancer types.
HLA gene expression is associated with patient response to immunotherapy
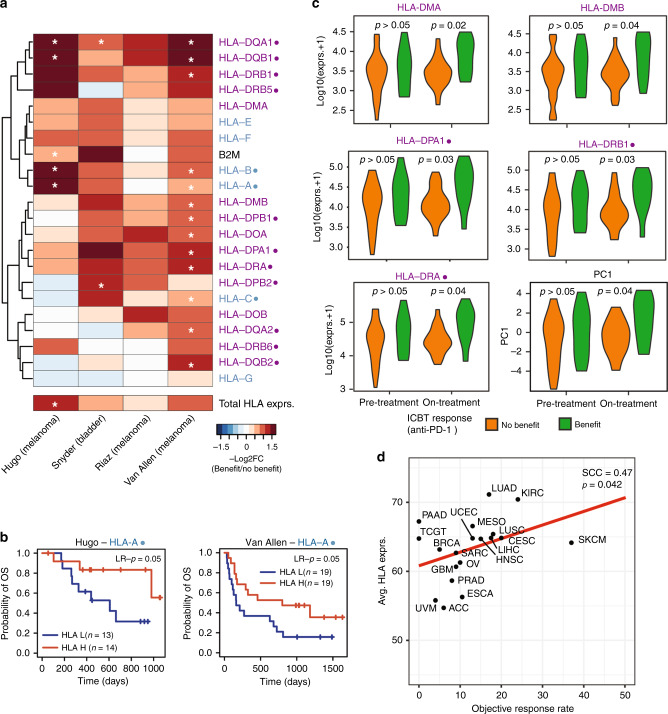
During our investigation of TME characteristics associated with HLA gene expression, we observed strong positive correlations between immune checkpoint expression and HLA gene expression. In addition, ICBT is standard-of-care in several cancer types.39,40 We thus focused on HLA gene expression in patients treated with ICBT. Four datasets with pre-treatment samples and ICBT response information were collected and patients were stratified based on clinical benefit (complete response (CR) or partial response (PR)) or no clinical benefit (progressive disease (PD)). We compared the expression of all HLA genes between these two response groups (Fig. 5a). Both class I and II HLA genes showed higher baseline expression levels in patients who experienced clinical benefit from ICBT compared to those who did not benefit. This suggests that HLA gene expression in pre-treatment samples was predictive of patient response to ICBT. Next, we investigated whether HLA gene expression was associated with survival in these datasets. HLA-A and HLA-F were consistently significantly (p < 0.05) associated with survival in two melanoma datasets (Fig. 5b for HLA-A (p = 0.05 and p = 0.05) and Supplementary Fig. 3A for HLA-F (p = 0.05 and p = 9E−4)).
Fig. 5. HLA Class I genes associated with response to immune checkpoint blockade.
a Heatmap showing the -log10(p-values) comparing HLA gene expression in pre-treatment samples from patients with clinical benefit (complete response (CR) and partial response (PR)) to patients without the clinical benefit (progressive disease (PD)). Due to the small sample sizes, significance was indicated by a one-sided t-test hypothesising that patients with clinical benefit had higher HLA gene expression compared to patients without clinical benefit. White stars indicate significance at p < 0.05. b KM plot comparing overall survival between patients with low and high HLA-A gene expression in the Hugo and Van Allen datasets. Significance calculated by Log-rank tests. c Violin plots comparing gene expression of 5 HLA class II genes and the meta-HLA gene between patients who benefitted or did not benefit from anti-PD-1 treatment in pre- or on-treatment biopsies. Significance calculated by two-sided Student’s t-tests. d Correlation between -log10(total HLA gene expression) and ICBT objective response rate based on ref. 41 Classical HLA genes indicated with a dot (●), HLA class I genes indicated in blue, HLA class II genes in purple.
To investigate if differences in HLA gene expression were also present during treatment, we compared the expression levels of HLA/B2M genes between pre- and on-treatment biopsies using a dataset from Riaz et al.22 Importantly, we observed a much more significant increase in an HLA class II gene expression when comparing patients with and without clinical benefit in on-treatment samples as compared to pre-treatment samples (Fig. 5c), suggesting that on-treatment samples are more informative of clinical benefit when using HLA class II gene expression as an indicator of response. This was not the case for HLA class I genes, which showed much less significance in on-treatment samples compared to HLA class II genes (Supplementary. Fig. 3B). This suggests that HLA class II genes might be more important for an effective response to ICBT. The meta-HLA gene could also much more significantly stratify patients with and without clinical benefit in on-treatment samples compared to pre-treatment samples (Fig. 5c). Extending our studies to additional cancer types, we investigated if there was a relationship between HLA gene expression and overall objective response to ICBT. We correlated total HLA gene expression for each cancer type with the objective response rate presented by Yarchoan et al. (Fig. 5d).41 We indeed observed a positive relationship between response and HLA gene expression (SCC = 0.47, p = 0.042, Fig. 5D), suggesting that tumour types with a hotter TME tend to be more responsive to ICBT. In conclusion, we showed that several HLA genes are associated with response to ICBT and that this association is much stronger in samples obtained during ongoing ICBT compared to pre-treatment samples in HLA class II genes.
Discussion
Proper function of MHC is essential to an effective antitumour response.1 Tumour cells devise numerous ways to escape the immune system and a number of these strategies involve alteration of HLA genes.6 In this study, we have comprehensively investigated the relationship between HLA gene expression and genomic alterations, TME characteristics and prognosis. Although our study confirms many findings from smaller studies that either focus on a single cancer type or a set of HLA genes, we show that a systematic analysis across cancer types and HLA genes provides additional valuable information. When comparing patterns of HLA gene expression between different HLA genes, we consistently found that expression patterns are often similar within HLA classes. Specifically, HLA class I genes, HLA-A, HLA-B and HLA-C, clustered tightly together in the majority of analyses. Non-classical HLA class I gene HLA-G was often separated from the other HLA class I genes. HLA-G has restricted expression in healthy tissues and is commonly only expressed in placental tissue during pregnancy to induce immune tolerance.42 Pathological expression of HLA-G has been observed in cancer and other diseases43 and we hypothesise that due to its distinctly different function, HLA-G expression is often distinct from other HLA class I genes. Although some variation in gene expression exists between HLA genes, as exemplified by HLA-G, we observed that the majority of HLA gene expression could be approximated using the meta-HLA gene. The meta-HLA expression reflects the overall HLA abundance at the mRNA level, which should capture changes that impact HLA gene expression (e.g. transcription rate and mRNA stability), including epigenetic changes. However, it does not capture potentially important other HLA-related mechanisms, including regulation at the protein level, alteration in HLA trafficking or antigen- processing machinery components. Nevertheless, the use of a meta-HLA gene instead of all HLA genes can significantly simply downstream analyses and we have shown in multiple instances that the meta-HLA gene provides an excellent summary of associations seen on the gene level.
We investigated several genomic mechanisms that could affect HLA gene expression, including HLA CNV, HLA promoter methylation, HLA mutations and tumour type. We have shown that these alterations are associated with HLA gene expression to a different degree in different cancer types, suggesting the presence of some cancer-type-specific evasion mechanisms. Previous studies have shown that both increased and decreased promoter methylation are associated with altered HLA gene expression.44–46 For example, increased promoter methylation of HLA class I genes and HLA-DQB1 led to decreased mRNA expression in oesophageal cancer,45,46 while hypermethylation of HLA-DRA and HLA-DQ has been associated with higher mRNA levels in oesophageal cancer and melanoma.44,46 We indeed confirmed the positive correlation between HLA-DRA/HLA-DQA1 and mRNA expression and the negative correlation between promoter methylation of HLA Class I genes and mRNA expression reported in oesophageal cancer (Fig. 2b),45,46 providing some validation that our results parallel previous findings. A previous study has shown that mutations in HLA class I genes tend to occur in ‘hotspot’ sites and that mutations likely alter HLA protein functionality.7 Based on our analyses, we did not observe any alterations in HLA gene expression upon HLA mutation, suggesting that the structural consequences of mutations are more prominent than effects on mRNA stability or mRNA expression regulation. Investigations into additional HLA-related mechanisms that tumour cells exploit to evade immune recognition would provide us with an even better understanding of tumour-type-specific and HLA gene-specific mechanisms that are at play. These could include alterations in HLA mRNA splicing,47,48 altered HLA protein stability49 and deficits in the antigen-presenting machinery.50
The TME is an important consideration when investigating HLA gene expression in cancer. The infiltration of myeloid, CD8+ T and NK cells seemed especially important, as these immune cell types were almost uniformly positively associated with HLA gene expression in all cancer types. In many cancers, HLA class I downregulation on tumour cells (‘missing-self’) leads to NK cell activation and tumour cell destruction,51,52 which is seemingly in contrast with our findings of increased NK cell infiltration in tumours expressing high levels of HLA mRNA. However, it seems that the context in which HLA/NK engagement takes place is an important aspect to consider.53 For example, NK cell interactions with tumour cell HLA-E in the presence of inhibitory NK cell receptor NKG2A indeed inhibit NK cell function,54 whereas the presence of NKG2C activates NK cells when bound to HLA-E.55,56 In addition, cytokines secreted by NK cells, most notably IFNγ, stimulate the expression of HLA class I molecules in tumour cells,57 leading to heightened HLA expression in tumours with high NK cell infiltration. Since macrophages express HLA class II genes,1 these positive correlations were not unexpected. A more intricate association between HLA gene expression and other major immune cell types was observed. For example, CD4+ T cells were negatively associated with HLA gene expression in a number of cancer types, whereas no associated was observed in other cancer types. This negative association was especially prominent in LGG, which also showed a strong negative association between HLA gene expression and patient prognosis. This suggests that CD4+ T cells play an important role in LGG. In addition, we found that the mRNA expression of immune checkpoints is strongly positively correlated with HLA gene expression, suggesting that immunosuppression through immune checkpoint proteins might counteract increased HLA gene expression to prevent immune activation. Lastly, we hypothesise that the observation between immune ‘cold’ tumours and low HLA gene expression is related to the transition from HLA-positive tumours cells to HLA-negative tumour cells due to immunoselection. Several studies have reported that HLA expression is gradually lost as tumours progress and HLA expression is often absent at metastatic sites.58,59 HLA gene expression in a ‘hot’ TME could represent early stages of T-cell-mediated immune activation that is followed by a ‘cold’ TME phase in which most tumour cells with HLA gene expression have been eliminated.60
Lastly, we investigated the association between HLA gene expression and prognosis in the settings of ICBT and standard-of-care chemotherapy. We confirmed that metastatic melanoma patients with increased HLA gene expression tended to respond better to ICBT and survive longer. This observation was especially clear in biopsies taken during the treatment period since multiple HLA class II genes were much more strongly associated with ICBT response during treatment as compared to before treatment initiation. The importance of HLA class II genes has been shown in the context of ICBT in a number of studies.61,62 It is proposed that the activation of CD4+ T cells by HLA class II expression helps to initiate CD8+ T cells that consequently mount a successful antitumour immune response during ICBT.63 We also found that increased HLA gene expression is associated with good prognosis in the majority of cancer types under chemotherapy regimens. In contrast to other studies that implicated specific genes in prognosis,12–14 we found that often many HLA genes are associated with prognosis within a given cancer type. Co-expression between HLA genes is a likely explanation for this observation.
Although our study provides novel insights into HLA gene expression in cancers, we note a few limitations of our study. First, the normal tissue samples in the TCGA dataset were collected directly adjacent to the tumour and likely resemble but not represent truly normal tissue. Comparison of tumour tissue to distant normal tissues would provide a more accurate evaluation of normal/tumour HLA gene expression differences. Some of the discrepancies between the established downregulation of HLA genes in tumour cells and our observation of significant upregulation of HLA genes in some cancer types could be due to this. Second, we have only investigated a small number of potential alterations that can affect HLA gene expression. Other HLA-related alterations that would be of considerable interest are HLA mRNA splicing,47,48 altered HLA protein stability49 and deficits in the antigen-presenting machinery.50 Third, the majority of our prognostic analyses were performed using univariate models due to the differential availability of clinical variables for different datasets. To prevent the comparison of associations that were based on different multivariate models, we thus choose to limit our analyses to univariate analyses. Consequently, some of our results could have had different results if additional variables would have been adjusted for. Fourth, our study focused solely on gene expression studies from bulk tumour resections without taking differences in HLA gene expression between tumour and stromal cells into consideration. Studies involving microdissected tumour sections or single-cell RNAseq approaches would provide a more comprehensive image of HLA gene expression in tumours. Lastly, as our study is mostly based on HLA gene expression, we are unable to provide insights into how, for example, HLA CNV or HLA mutations are related to functional MHC impairments within tumour samples.
In conclusion, we provide a comprehensive overview of HLA gene expression in a variety of tumour types in which we contrasted both HLA genes and tumour types. While several studies investigated individual HLA genes across cancer types or all HLA genes within a specific cancer type, we have systematically investigated the impact of all HLA gene expression changes in clinical outcomes across a broad range of cancer types. We show pan-cancer patterns as well as tumour-type-specific patterns that can spark additional studies into the role of HLA genes in cancer progression and prognosis. HLA gene expression is significantly associated with distinct genomic and immunologic tumour characteristics and HLA gene expression has prognostic value in both standard treatment and immune checkpoint blockade therapy (ICBT) settings.
Supplementary information
Acknowledgements
We would like to thank all members of the Cheng lab for their suggestions and critical feedback.
Author contributions
C.C. conceived the project. E.S., C.M.F. and C.C. performed computational analyses. E.S., C.M.F., X.W. and C.C. wrote the paper. E.S., C.M., X.W. and C.C. interpreted the results. X.W. and C.C. supervised the project. C.C., E.S., CF. and X.W. critically reviewed the content. C.C., E.S., C.F. and X.W. read and approved the final paper.
Ethics approval and consent to participate
Not applicable. All data utilised in this study are publicly available.
Consent to publish
Not applicable.
Data availability
All data utilised in this study are publicly available. See ‘Methods’ for data sources.
Competing interests
The authors declare no competing interests.
Funding information
This work is supported by the Cancer Prevention Research Institute of Texas (CPRIT) (RR180061 to C.C.), the National Cancer Institute of the National Institutes of Health (1R21CA227996 to C.C.) and the T32 training grant of the National Institutes of Health (T32 AI007363 to ES). C.C. is a CPRIT Scholar in Cancer Research.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Evelien Schaafsma, Chloe M. Fugle
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01400-2.
References
- 1.Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 2.Albitar M, Johnson M, Do KA, Day A, Jilani I, Pierce S, et al. Levels of soluble HLA-I and beta2M in patients with acute myeloid leukemia and advanced myelodysplastic syndrome: association with clinical behavior and outcome of induction therapy. Leukemia. 2007;21:480–488. doi: 10.1038/sj.leu.2404506. [DOI] [PubMed] [Google Scholar]
- 3.Shiina T, Inoko H, Kulski JK. An update of the HLA genomic region, locus information and disease associations: 2004. Tissue Antigens. 2004;64:631–649. doi: 10.1111/j.1399-0039.2004.00327.x. [DOI] [PubMed] [Google Scholar]
- 4.Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015;15:203–216. doi: 10.1038/nri3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braud VM, Allan DS, O’Callaghan CA, Söderström K, D’Andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 6.Garner H, de Visser KE. Immune crosstalk in cancer progression and metastatic spread: a complex conversation. Nat. Rev. Immunol. 2020;20:483–497. doi: 10.1038/s41577-019-0271-z. [DOI] [PubMed] [Google Scholar]
- 7.Shukla SA, Rooney MS, Rajasagi M, Tiao G, Dixon PM, Lawrence MS, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA Loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–1271.e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science. 2018;359:582–587. doi: 10.1126/science.aao4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertol BC, de Araújo JNG, Sadissou IA, Sonon P, Dias FC, Bortolin RH, et al. Plasma levels of soluble HLA-G and cytokines in papillary thyroid carcinoma before and after thyroidectomy. Int J. Clin. Pract. 2020;74:e13585. doi: 10.1111/ijcp.13585. [DOI] [PubMed] [Google Scholar]
- 11.Akın M, Aral LA, Yavuz A, Karabacak H, Dikmen K, Bostancı H. Plasma human leukocyte antigen-G (HLA-G) in patients with thyroid cancer. Turk. J. Med Sci. 2017;47:1263–1266. doi: 10.3906/sag-1611-25. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhuri S, Cariappa A, Tang M, Bell D, Haber DA, Isselbacher KJ, et al. Genetic susceptibility to breast cancer: HLA DQB*03032 and HLA DRB1*11 may represent protective alleles. Proc. Natl Acad. Sci. USA. 2000;97:11451–11454. doi: 10.1073/pnas.97.21.11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ECM Zeestraten, Reimers MS, Saadatmand S, Goossens-Beumer IJ, Dekker J-WT, Liefers GJ, et al. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br. J. Cancer. 2014;110:459–468. doi: 10.1038/bjc.2013.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Najafimehr H, Hajizadeh N, Nazemalhosseini-Mojarad E, Pourhoseingholi MA, Abdollahpour-Alitappeh M, Ashtari S, et al. The role of Human leukocyte antigen class I on patient survival in Gastrointestinal cancers: a systematic review and meta-analysis. Sci. Rep. 2020;10:728. doi: 10.1038/s41598-020-57582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–689.e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro A, Ozturk K, Pyke RM, Xian S, Zanetti M, Carter H. Elevated neoantigen levels in tumors with somatic mutations in the HLA-A, HLA-B, HLA-C and B2M genes. BMC Medical. Genomics. 2019;12:107. doi: 10.1186/s12920-019-0544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385.e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder A, Nathanson T, Funt SA, Ahuja A, Novik JB, Hellmann MD, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: an exploratory multi-omic analysis. PLOS Med. 2017;14:e1002309. doi: 10.1371/journal.pmed.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anagnostou V, Yarchoan M, Hansen AR, Wang H, Verde F, Sharon E, et al. Immuno-oncology trial endpoints: capturing clinically meaningful activity. Clin. Cancer Res. 2017;23:4959–4969. doi: 10.1158/1078-0432.CCR-16-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villaruz LC, Socinski MA. The clinical viewpoint: definitions, limitations of RECIST, practical considerations of measurement. Clin. Cancer Res. 2013;19:2629–2636. doi: 10.1158/1078-0432.CCR-12-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Queirolo P, Spagnolo F. Atypical responses in patients with advanced melanoma, lung cancer, renal-cell carcinoma and other solid tumors treated with anti-PD-1 drugs: a systematic review. Cancer Treat. Rev. 2017;59:71–78. doi: 10.1016/j.ctrv.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Varn FS, Andrews EH, Mullins DW, Cheng C. Integrative analysis of breast cancer reveals prognostic haematopoietic activity and patient-specific immune response profiles. Nat. Commun. 2016;7:10248. doi: 10.1038/ncomms10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varn FS, Wang Y, Mullins DW, Fiering S, Cheng C. Systematic pan-cancer analysis reveals immune cell interactions in the tumor microenvironment. Cancer Res. 2017;77:1271–1282. doi: 10.1158/0008-5472.CAN-16-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng C, Yan X, Sun F, Li LM. Inferring activity changes of transcription factors by binding association with sorted expression profiles. BMC Bioinforma. 2007;8:452. doi: 10.1186/1471-2105-8-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinforma. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perea F, Bernal M, Sánchez‐Palencia A, Carretero J, Torres C, Bayarri C, et al. The absence of HLA class I expression in non-small cell lung cancer correlates with the tumor tissue structure and the pattern of T cell infiltration. Int. J. Cancer. 2017;140:888–899. doi: 10.1002/ijc.30489. [DOI] [PubMed] [Google Scholar]
- 32.Sáenz‐López P, Gouttefangeas C, Hennenlotter J, Concha A, Maleno I, Ruiz‐Cabello F, et al. Higher HLA class I expression in renal cell carcinoma than in autologous normal tissue. Tissue Antigens. 2010;75:110–118. doi: 10.1111/j.1399-0039.2009.01409.x. [DOI] [PubMed] [Google Scholar]
- 33.Zajacova M, Kotrbova‐Kozak A, Cerna M. Expression of HLA-DQA1 and HLA-DQB1 genes in B lymphocytes, monocytes and whole blood. Int. J. Immunogenetics. 2018;45:128–137. doi: 10.1111/iji.12367. [DOI] [PubMed] [Google Scholar]
- 34.Heng TSP, Painter MW. Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 35.Middha S., Yaeger R., Shia J., Stadler Z. K., King S., Guercio S., et al. Majority of B2M-mutant and -deficient colorectal carcinomas achieve clinical benefit from immune checkpoint inhibitor therapy and are microsatellite instability-high. JCO Precis. Oncol.3, 1–14 (2019). [DOI] [PMC free article] [PubMed]
- 36.Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, et al. Genetic mechanisms of immune evasion in colorectal. Cancer Cancer Discov. 2018;8:730–749. doi: 10.1158/2159-8290.CD-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 38.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat. Rev. Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 39.Dummer R, Hauschild A, Lindenblatt N, Pentheroudakis G, Keilholz U, ESMO Guidelines Committee. Cutaneous melanoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015;26:v126–132. doi: 10.1093/annonc/mdv297. [DOI] [PubMed] [Google Scholar]
- 40.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarchoan M., Hopkins A., Jaffee E. M. Tumor mutational burden and response rate to PD-1 inhibition. 10.1056/NEJMc1713444. (Massachusetts Medical Society, 2017). [DOI] [PMC free article] [PubMed]
- 42.McMaster M., Librach C., Zhou Y., Lim K., Janatpour M., DeMars R. et al. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J. immunol. 154, 3771–3778 (1995). [PubMed]
- 43.Carosella ED, Moreau P, Lemaoult J, Rouas-Freiss N. HLA-G: from biology to clinical benefits. Trends Immunol. 2008;29:125–132. doi: 10.1016/j.it.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Lee JS, O’Neill L. Methylation of the HLA-DR alpha gene is positively correlated with expression. Immunogenetics. 1987;26:92–98. doi: 10.1007/BF00345460. [DOI] [PubMed] [Google Scholar]
- 45.Nie Y, Yang G, Song Y, Zhao X, So C, Liao J, et al. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. 2001;22:1615–1623. doi: 10.1093/carcin/22.10.1615. [DOI] [PubMed] [Google Scholar]
- 46.Hu JM, Li L, Chen YZ, Liu C, Cui X, Yin L, et al. HLA-DRB1 and HLA-DQB1 methylation changes promote the occurrence and progression of Kazakh ESCC. Epigenetics. 2014;9:1366–1373. doi: 10.4161/15592294.2014.969625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sangrouber D, Marcou C, Le Discorde M, Chang C-C, Carosella ED, Moreau P. Cellular co-localization of intron-4 containing mRNA and HLA-G soluble protein in melanoma analyzed by fluorescence in situ hybridization. J. Immunol. Methods. 2007;326:54–62. doi: 10.1016/j.jim.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Goodson-Gregg FJ, Rothbard B, Zhang A, Wright PW, Li H, Walker-Sperling VE, et al. Tuning of NK-Specific HLA-C Expression by Alternative mRNA Splicing. Front Immunol. 2019;10:3034. doi: 10.3389/fimmu.2019.03034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Z. & Jensen P. E. Structural characteristics of HLA-DQ that may impact DM editing and susceptibility to type-1 diabetes. Front Immunol.4, 262 (2013). [DOI] [PMC free article] [PubMed]
- 50.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ljunggren HG, Kärre K. In search of the “missing self”: MHC molecules and NK cell recognition. Immunol. Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-S. [DOI] [PubMed] [Google Scholar]
- 52.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev. Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López-Botet M, Llano M, Navarro F, Bellón T. NK cell recognition of non-classical HLA class I molecules. Semin Immunol. 2000;12:109–119. doi: 10.1006/smim.2000.0213. [DOI] [PubMed] [Google Scholar]
- 55.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/S1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 56.Hammer Q, Rückert T, Borst EM, Dunst J, Haubner A, Durek P, et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 2018;19:453–463. doi: 10.1038/s41590-018-0082-6. [DOI] [PubMed] [Google Scholar]
- 57.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat. Rev. Cancer. 2016;16:7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 58.Ryschich E, Nötzel T, Hinz U, Autschbach F, Ferguson J, Simon I, et al. Control of T-cell-mediated immune response by HLA class I in human pancreatic carcinoma. Clin. Cancer Res. 2005;11:498–504. [PubMed] [Google Scholar]
- 59.So T, Takenoyama M, Mizukami M, Ichiki Y, Sugaya M, Hanagiri T, et al. Haplotype loss of HLA class I antigen as an escape mechanism from immune attack in lung cancer. Cancer Res. 2005;65:5945–5952. doi: 10.1158/0008-5472.CAN-04-3787. [DOI] [PubMed] [Google Scholar]
- 60.Aptsiauri N, Ruiz-Cabello F, Garrido F. The transition from HLA-I positive to HLA-I negative primary tumors: the road to escape from T-cell responses. Curr. Opin. Immunol. 2018;51:123–132. doi: 10.1016/j.coi.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 61.Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alspach E, Lussier DM, Miceli AP, Kizhvatov I, DuPage M, Luoma AM, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature. 2019;574:696–701. doi: 10.1038/s41586-019-1671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Borst J, Ahrends T, Bąbała N, Melief CJM, Kastenmüller W. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018;18:635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data utilised in this study are publicly available. See ‘Methods’ for data sources.