Abstract
The RAS-associated domain family 9 (RASSF9), a RAS-associated domain family gene, is expressed in a variety of tissues. However, its roles in tumorigenesis, particularly in non-small cell lung cancer (NSCLC), are still not understood well. In the present study, we aimed to examine the potential roles of RASSF9 in NSCLC and the underlying mechanisms. Our data showed that RASSF9 expression was upregulated in NSCLC tissues and cell lines. Increased expression of RASSF9 promotes NSCLC cell proliferation. On the contrary, knockdown of RASSF9 represses cell proliferation. Moreover, the effects of RASSF9 on NSCLC cell proliferation were further confirmed in vivo by using a subcutaneous tumor model. Mechanistically, pharmacological intervention studies revealed that the MEK/ERK axis is targeted by RASSF9 for transducing its regulatory roles on NSCLC cell proliferation. Collectively, our data indicate that RASSF9 plays a key role in tumorigenesis of NSCLC by stimulating tumor cell proliferation, which relies on activation of the MEK/ERK axis. Thus, RASSF9 might be a druggable target for developing novel agents for treating NSCLC.
Subject terms: Cell growth, Non-small-cell lung cancer
Introduction
Lung cancer is the most common cancer and a major cause of cancer-related death worldwide according to the newly report from the Global Cancer Statistics 2018 [1]. Non-small cell lung cancer (NSCLC) is the most common type, accounting for 85% of all lung cancer cases [2]. NSCLC can be further subdivided into three major pathologic subtypes, including lung adenocarcinoma, squamous cell carcinoma, and large cell carcinoma [3]. Lung adenocarcinoma is the most common subtype of NSCLC comprising ~40% of NSCLC cases, followed by squamous cell carcinoma 25–30% and large cell carcinoma 5–10% [4]. In recent decades, the clinical diagnosis and treatment of tumors have made great progresses due to the underlying molecular mechanisms for lung cancer development have been deeply studied. However, the long-term prognosis for NSCLC patients remains poor, with overall 5-year survival rates of <15% [5]. Hence, it is urgent to elucidate the detailed molecular pathogenesis of cell proliferation in NSCLC, so as to develop more effective therapeutic targets.
RASSF9 is a RAS-association domain family gene and is known to be expressed in multiple organs. The RASSF family includes 10 genes from RASSF1 to RASSF10 which contains a consensus RAS-association (RA) domain and bound to activated Ras [6, 7]. The RASSF family can be further subdivided into C-terminally (RASSF1-6) and N-terminally (RASSF7-10) based on whether there is a coiled-coil motif called the Salvador/RASSF/Hippo (SARAH) domain near the C-terminal or N-terminal RA domain [8, 9]. The N-terminal RASSF genes participate in numerous vital biological progresses, such as cell survival, apoptosis, tumor growth, and metastasis [7]. In addition, previous reports in the literature show RASSF7 expression is increased in NSCLC [10] and hepatocellular carcinoma [11] and plays a role in stimulating tumor proliferation. On the contrary, RASSF8 and RASSF10 have been shown to be reduced in multiple cancers, including lung cancer, gastric cancer, cervical cancer, breast cancer, colorectal esophageal squamous cell carcinoma, and hepatocarcinoma, and thus are considered as tumor suppressors [12–21]. However, the exact roles of RASSF9 in tumorigenesis in a variety of cancers, especially in NSCLC, are not fully understood.
In the present study, we observed RASSF9 expression is increased in NSCLC tissues and cell lines. Increased expression of RASSF9 promotes NSCLC cell proliferation, whereas knockdown of RASSF9 suppresses tumor cell proliferation. We further revealed that, by activating the MEK/ERK axis, RASSF9 plays a positive role in NSCLC cell proliferation. Our data suggest that RASSF9 might be a druggable target for developing novel agents for treating NSCLC.
Results
RASSF9 shows increased expression in NSCLC tissues and cell lines
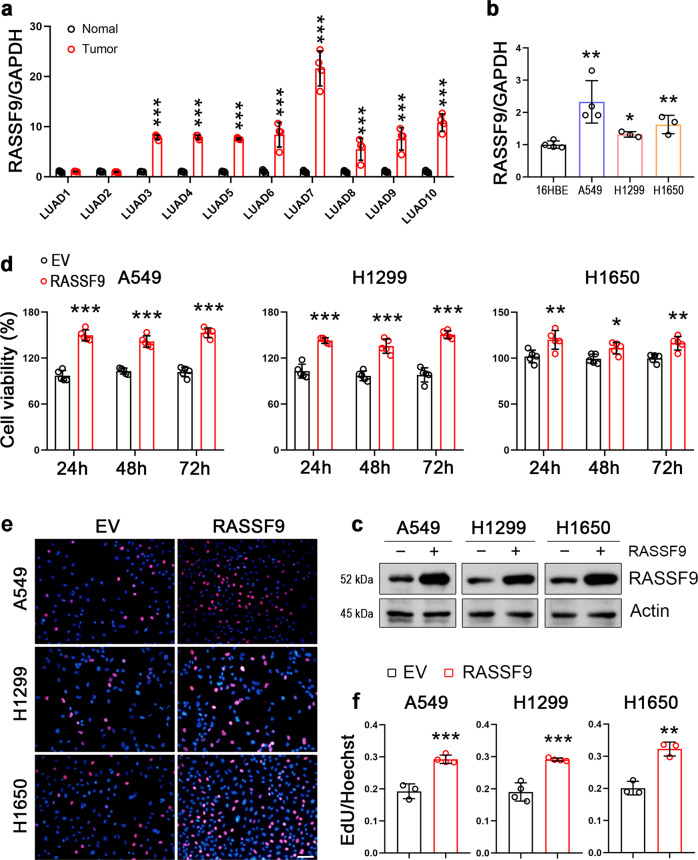
To investigate the potential role of RASSF9 in the development and progression of NSCLC, we first detected the mRNA levels of RASSF9 in clinical samples from NSCLC patients. Our results showed that, in eight paired tissues, RASSF9 was significantly increased in NSCLC tissues with the comparison of the adjacent normal tissues (Fig. 1a). In addition, we also measured RASSF9 expression in several non-small cell lung cancer cell lines, including A549, H1299, and H1650. As shown in Fig. 1b, we found that RASSF9 was markedly increased in A549, H1299, and H1650 as compared to 16HBE cells. Taken together, these data indicated that RASSF9 expression is increased in NSCLC, suggesting RASSF9 might play a crucial role in the progression of NSCLC.
Fig. 1. RASSF9 is highly expressed in NSCLC and plays a role in stimulating tumor cell proliferation.
a The mRNA levels of RASSF9 in normal and NSCLC tissues. n = 10. b The mRNA levels of RASSF9 in human bronchial epithelial cells (16HBE) and NSCLC tumor cell lines (A549, H1299, and H1650). c RASSF9 was increased by the transfection with the plasmid carrying Rassf9. The control cells were transfected with the empty vector pcDNA3.1. Twenty-four hours post-transfection, cells were harvested for analyzing RASSF9 by western blot. Actin was used as a loading control. d RASSF9 improves cell viability. A549, H1299, and H1650 cells were transfected with plasmid expressing Rassf9 and cell viability was analyzed by the method of MTT at different time points as indicated. The control cells were transfected with the empty vector pcDNA3.1 (EV). e RASSF9 promotes cell proliferation. A549, H1299, and H1650 cells were transfected with the plasmid expressing Rassf9. Twenty-four hours post-transfection, cell proliferation was analyzed by EdU incorporation assay. The control cells were transfected with the empty vector pcDNA3.1 (EV). Scale bar = 500 μm. f Quantitative analysis of EdU incorporation. Data are presented as means ± SD (error bars). *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t-test.
RASSF9 facilitates NSCLC cell viability and proliferation
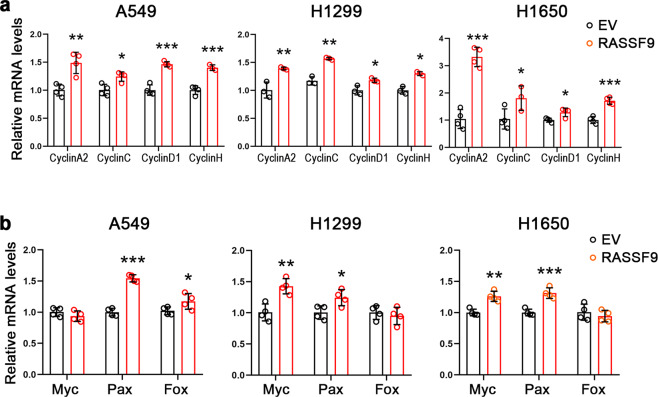
We next examined the potential roles of RASSF9 on the progression of NSCLC such as cell proliferation. To this aim, cells were transfected with the plasmid carrying Rassf9 to increase expression of RASSF9 (Fig. 1c). First, we observed that elevated RASSF9 expression caused appreciable growth in cell viability in these NSCLC cells (Fig. 1d). The EdU incorporation results showed that RASSF9 increased the cell proliferation in all three tested NSCLC cell lines including A549, H1299, and H1650 (Fig. 1e and f). Moreover, we also detected the expression of genes related to cell cycle and cell viability. The results showed that the mRNA levels of cyclins were markedly enhanced by RASSF9 in A549, H1299, and H1650 cells (Fig. 2a). We also detected the expression of three oncogenes (c-Myc, Pax, and Fos) and found that at least two genes were enhanced by RASSF9 in these NSCLC cells (Fig. 2b). These data indicate that RASSF9 has a positive regulatory role in cell proliferation in NSCLC.
Fig. 2. RASSF9 increases the expression of cyclins and oncogenes.
a Elevated expression of RASSF9 increases the mRNA levels of cyclins in A549, H1299, and H1650 cells. b Effects of RASSF9 on the expression of c-Myc, Pax, and Fos. Cells were transfected with the plasmid expressing Rassf9. Control cells were transfected with the empty vector pcDNA3.1 (EV). Twenty-four hours post-transfection, total RNA was extracted for gene expression analysis by qRT-PCR, and GAPDH was used as a house-keeping gene. Data are presented as means ± SD (error bars). *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t-test.
Knockdown of RASSF9 inhibits NSCLC cell viability and proliferation
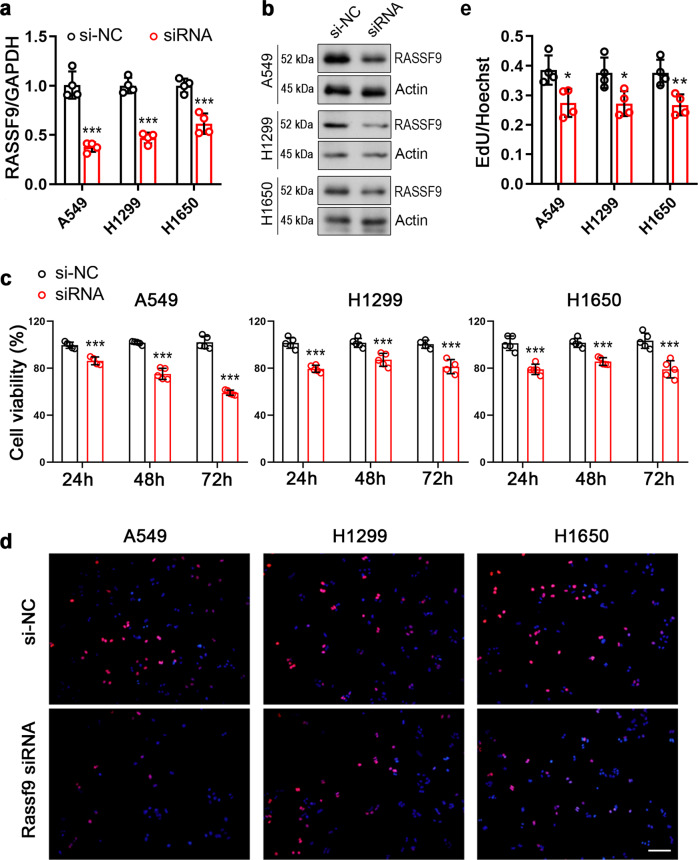
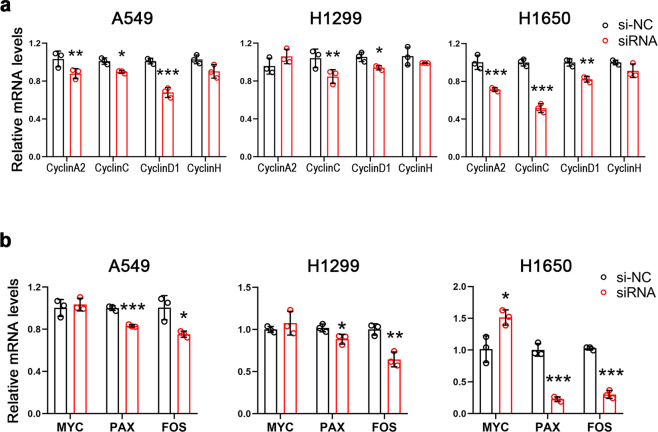
To further evaluate the significance of RASSF9 in the NSCLC progression, we downregulated RASSF9 expression in these NSCLC cells via small interference RNA (siRNA) (Fig. 3a, b). We found that siRNA-induced RASSF9 knockdown dramatically weakened cell viability (Fig. 3c). Furthermore, the results of EdU incorporation showed that decreased expression of RASSF9 markedly suppressed cell proliferation in A549, H1299, and H1650 cells (Fig. 3d, e). In addition, we also examined the mRNA levels of cyclins and the results showed that at least two cyclins were decreased by knockdown of RASSF9 in these NSCLC cells (Fig. 4a). Moreover, upon RASSF9 knockdown in these cells, the mRNA level of Myc, Pax, or Fos was significantly reduced (Fig. 4b). Collectively, these findings further indicate that RASSF9 plays a positive regulatory role on cell proliferation in NSCLC.
Fig. 3. Knockdown of Rassf9 suppresses NSCLC cell proliferation.
a Knockdown of Rassf9 by siRNA. A549, H1299, and H1650 cells were transfected with Rassf9 siRNA or negative control (si-NC). Forty-eight hours post-transfection, total RNA was extracted for gene expression analysis by qRT-PCR and GAPDH was used as a house-keeping gene. b The protein levels of RASSF9 were reduced by siRNA. Cell treatments were described in (a). Protein levels were analyzed by western blot and Actin was used a loading control. c Knockdown of Rassf9 decreases NSCLC cell viability. A549, H1299, and H1650 cells were transfected with Rassf9 siRNA or si-NC. Cell viability was analyzed by the method of MTT at 24 h, 48 h, and 72 h post-transfection. d Knockdown of Rassf9 reduces EdU incorporation. A549, H1299, and H1650 cells were transfected with Rassf9 siRNA or negative control (si-NC). Forty-eight hours post-transfection, cell proliferation was analyzed by EdU immunofluorescence assay. Scale bar = 500 μm. e Quantitative analysis of EdU incorporation. Data are presented as means ± SD (error bars). *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t-test.
Fig. 4. Knockdown of Rassf9 decreases the expression of cyclins and oncogenes.
a Knockdown of Rassf9 inhibits the mRNA levels of cyclins. b Effects of Rassf9 knockdown on the expression of c-Myc, Pax, and Fos. A549, H1299, and H1650 cells were transfected with Rassf9 siRNA. Forty-eight hours post-transfection, total RNA was extracted for gene expression analysis by qRT-PCR and GAPDH was used as a house-keeping gene. Data are presented as means ± SD (error bars). *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t-test.
RASSF9 stimulates cell proliferation through the MEK/ERK axis
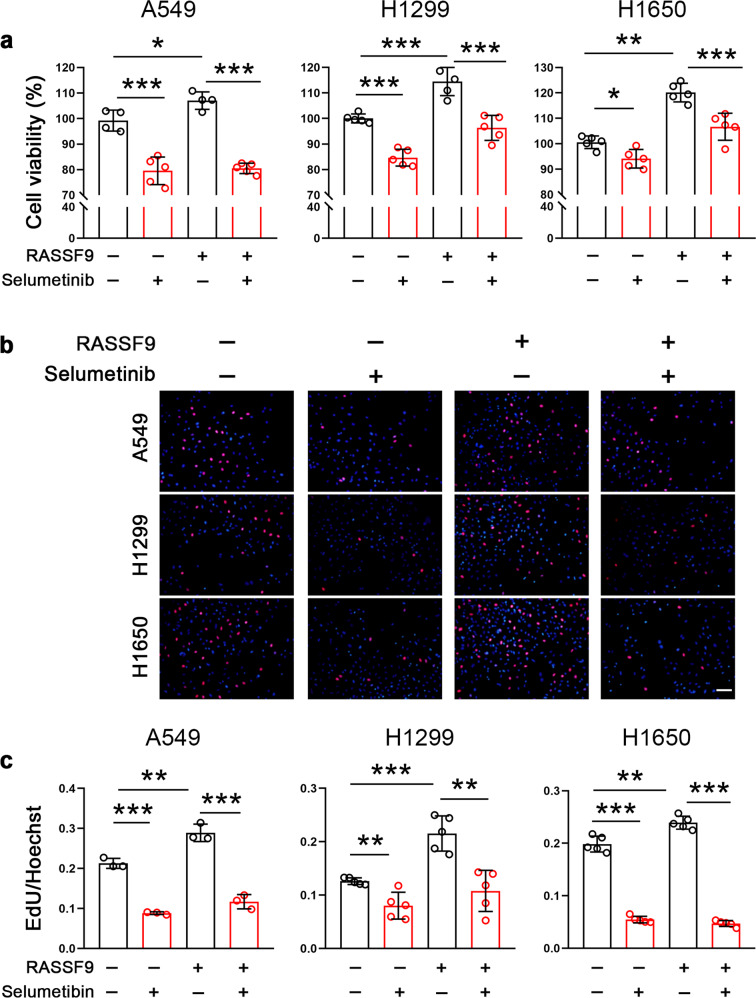
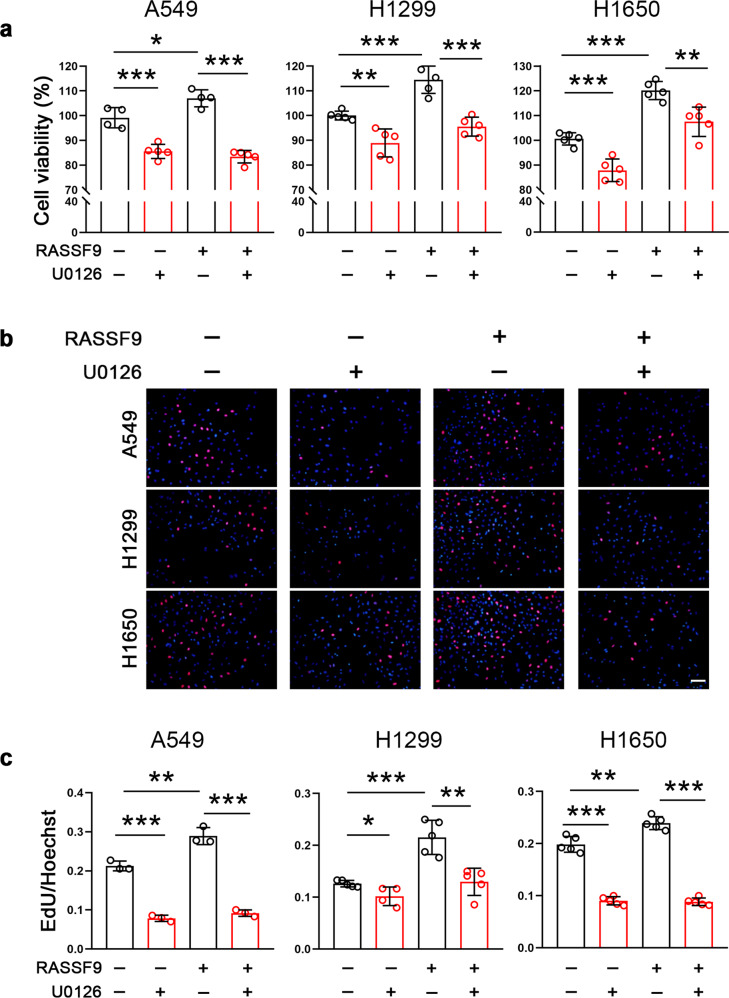
RASSF9 has been shown to positively regulate RAS signaling by inducing RAS dimerization, which activates downstream signal pathway including MEK and ERK to drive tumor cell proliferation [22]. Therefore, we next chose a pharmacological intervention strategy to examine the effects of RASSF9 on MEK/REK signal transduction. The results showed that selumetinib, a potent inhibitor of MEK, suppressed cell viability both in the presence or absence of RASSF9 in these NSCLC cells (Fig. 5a). Selumetinib also significantly reduced the EdU incorporation in A549, H1299, and H1650 cells (Fig. 5b, c). The increases in EdU incorporation induced by RASSF9 were also markedly blocked by selumetinib (Fig. 5b, c). Furthermore, we also chose another MEK inhibitor U1026 to confirm the above findings. As expected, the cell viability and the EdU incorporation were largely blocked by U0126 in A549, H1299, and H1650 cells (Fig. 6a–c).
Fig. 5. MEK inhibition by selumetinib reduces RASSF9-induced NSCLC cell viability and proliferation.
a Selumetinib reduces RASSF9-induced upregulation of cell viability. A549, H1299, and H1650 cells were transfected with the plasmid expressing Rassf9. Twelve hours post-transfection, cells were treated with 10 μM of selumetinib for additional 24 h and then subjected for cell viability assay by the method of MTT. b Selumetinib decreases RASSF9-induced EdU incorporation. Cell treatments were described in (a). Scale bar = 500 μm. c Quantitative analysis for EdU incorporation. Data are presented as means ± SD (error bars). *P < 0.05, **P < 0.01, and ***P < 0.001. One-way ANOVA test.
Fig. 6. MEK inhibition by U0126 abolishes RASSF9-induced NSCLC cell viability and proliferation.
a U0126 reduces the increased cell viability induced by RASSF9. A549, H1299, and H1650 cells were transfected with the plasmid expressing Rassf9. Twelve hours post-transfection, cells were treated with 10 μM of U0126 for additional 24 h and then subjected for cell viability assay by the method of MTT. b U0126 decreases RASSF9-induced EdU incorporation. Cell treatments were described in (a). Scale bar = 500 μm. c Quantitative analysis of EdU incorporation. Data are presented as means ± SD (error bars). *P < 0.05, **P < 0.01, and ***P < 0.001. One-way ANOVA test.
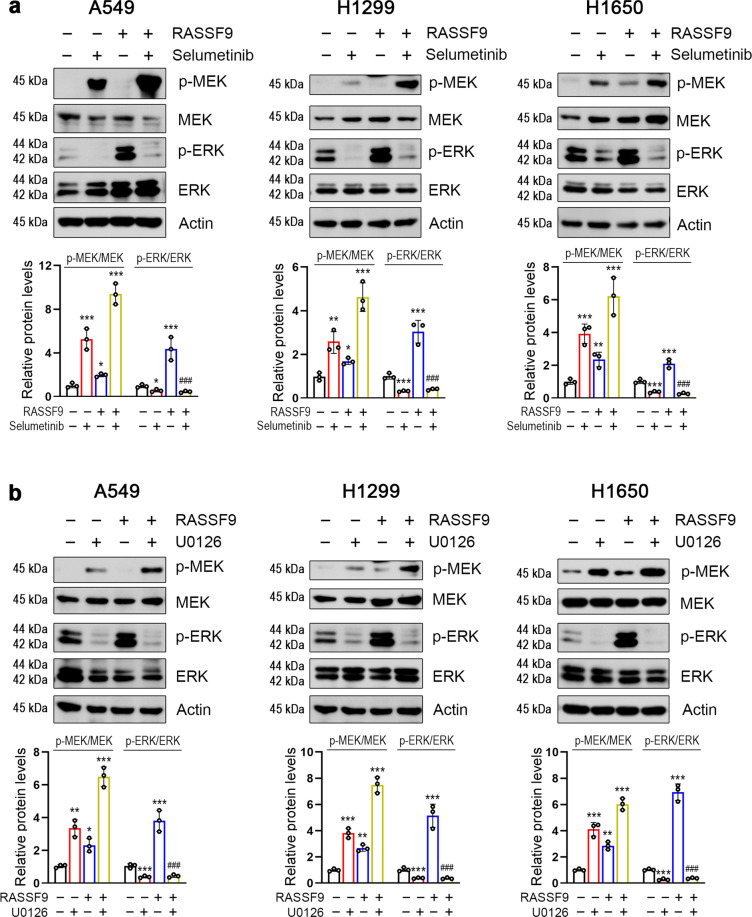
Moreover, we also examined the protein levels involved in the axis of MEK/ERK to ascertain the inhibitory effects of selumetinib and U0126. As shown in Fig. 7a, the phospho-ERK (p-ERK) was robustly stimulated by RASSF9. In the presence of selumetinib, however, this stimulation was almost completely counteracted in all three tested NSCLC cells (Fig. 7a). The phospho-MEK (p-MEK) was also increased by RASSF9 (Fig. 7a). Selumetinib resulted in remarkable increases in p-MEK, similar findings were also reported previously [22, 23]. Similar to selumetinib, U0126 treatments also greatly blocked RASSF9 stimulated p-ERK in A549, H1299, and H1650 cells (Fig. 7b). Meanwhile, p-MEK was increased by U1026 (Fig. 7b). These data clearly indicated that the treatments of selumetinib and U0126 are effective strategies for blocking the axis of MEK/ERK induced by RASSF9 in NSCLC cells.
Fig. 7. MEK inhibitors neutralize RASSF9-induced ERK activation.
a Inhibition of MEK using selumetinib abolishes RASSF9-induced ERK activation. b Inhibition of MEK by U0126 reduces RASSF9-induced ERK activation. A549, H1299, and H1650 cells were transfected with the plasmid expressing Rassf9. Twelve hours post-transfection, cells were treated with 10 μM of U0126 or 10 μM of selumetinib for additional 24 h. Total cell lysates were prepared and subjected to western blot analysis. Actin was used as a loading control. Data are presented as means ± SD (error bars). *P < 0.05, **P < 0.01, and ***P < 0.001 versus the control cells. ###P < 0.001 versus the cells transfected with the plasmid expressing Rassf9. One-way ANOVA test.
RASSF9 stimulates NSCLC cell proliferation in vivo
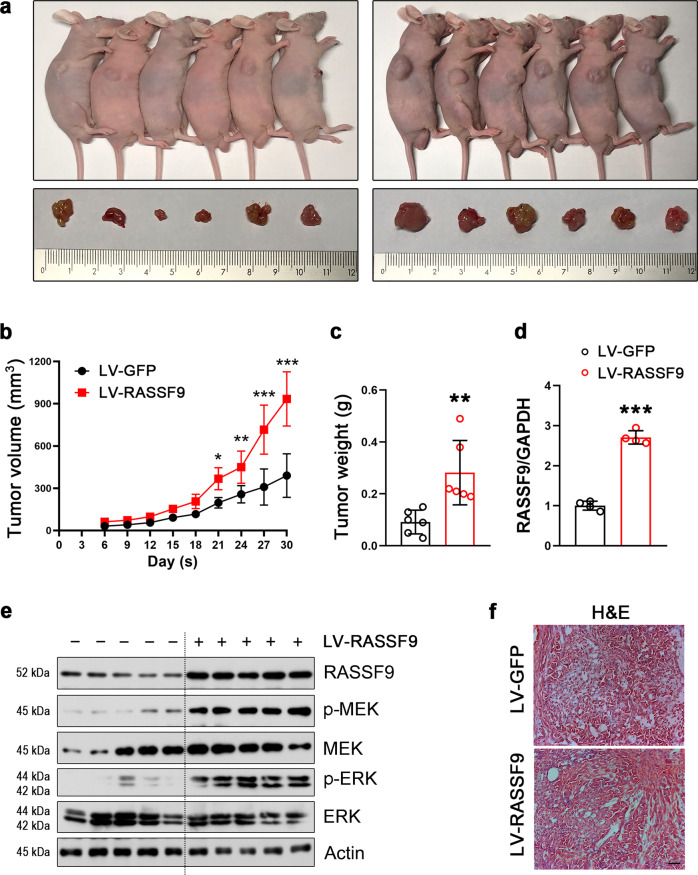
We next attempted to determine whether RASSF9 positively regulates NSCLC cell proliferation in vivo. Therefore, we first established a stable cell line expressing RASSF9. To this aim, A549 cells were transduced with lentivirus carrying Rassf9 (LV-RASSF9) and then subjected to puromycin screening. The resulted cells were injected subcutaneously into nude mice. Our results revealed that elevated expression of RASSF9 induced by LV-RASSF9 greatly stimulated cell growth, as evidenced by increased tumor mass, volume, and weight (Fig. 8a–c). Subsequently, RASSF9 expression in the LV-RASSF9 transduced tumors was confirmed by qRT-PCR (Fig. 8d). As expected, LV-RASSF9 induced an increase in RASSF9 protein levels, thereby stimulating an increase in p-MEK and p-ERK (Fig. 8e). Moreover, the hematoxylin/eosin (H&E) staining showed that tumor cells in the RASSF9 group were presented in nest-like distribution, and the nuclei were deeply stained, suggesting these cells grew vigorously (Fig. 8f). These in vivo data further confirmed the positive regulatory roles of RASSF9 on NSCLC cell proliferation.
Fig. 8. RASSF9 promotes NSCLC tumorigenesis in vivo.
A549 cells were transduced with lentivirus expressing Rassf9 (LV-RASSF9) or Gfp (LV-GFP). After screening with puromycin, the cells stably expressed RASSF9 or GFP were implanted into nude mice. a The representative images of gross subcutaneous tumors in nude mice and images of xenograft tumors dissected from nude mice (n = 6). b RASSF9 stimulates tumor volume (n = 6). c RASSF9 increases tumor weight (n = 6). d RASSF9 expression in the transplanted tumors was increased by LV-RASSF9 (n = 4). Gene expression was analyzed by qRT-PCR and GAPDH was used as house-keeping gene. e The protein levels of RASSF9, p-MEK, MEK, p-ERK, ERK in the transplanted tumors (n = 5). Protein levels were analyzed by western blot and Actin was used as loading control. f H&E staining. Scale bar = 200 μm. Data are presented as means ± SD (error bars). *P < 0.05, **P < 0.01, and ***P < 0.001. Student’s t-test.
Discussion
RASSF9 is a member of RAS-association domain family. There is growing evidence that the dysregulation of RASSF9 expression is closely associated with many human diseases, including cancer [7, 24–26]. RASSF9 was initially identified as a protein based on interaction with the cytoplasmic domain of PAM (peptide acylglycine α-amino monooxygenase) through a yeast two-hybrid screen [27]. The follow-up study has shown that the protein encoded by this gene is located in the perinuclear endosomes [28]. This protein associates with PAM, and may be involved with the trafficking of this enzyme through secretory or endosomal pathways [28].
RASSF family is divided into two categories: C-terminal RASSFs (RASSF1-6) and N-terminal RASSFs (RASSF7-10) based on the location of the RA domain. Members of the C-terminal RASSF proteins are commonly considered as cancer suppressors and are involved in many important biological processes including regulation of apoptosis and proliferation, cell cycle, control of cell migration and adhesion, nuclear transport, and microtubule stability [29, 30]. RASSF7-10 is a newly identified family member that may also have important biological functions, some of which may be different from previously studied Ras effector molecules. Among them, RASSF7 regulates microtubule cytoskeleton and spindle formation, and plays a decisive role in mitosis and cell growth [31]. As a potential tumor suppressor gene, RASSF8 interacts with adherens junction component and adhesion-related β-catenin/E-cadherin to regulate cell adhesion function [12]. Moreover, lack of RASSF8 expression leads to increased cell migration and tumor aggression [32]. Likewise, RASSF10 also inhibits cell growth and colony formation, and induces apoptosis in various cell lines [33, 34]. RASSF9, a member of the N-terminal RASSF protein, has been shown to bind to Ras protein The RASSF9 and RAS family GTPases pull-down experiments indicate that RASSF9 binds to N-Ras, K-Ras, and R-Ras [34]. A previous study showed that RASSF9 induces dimerization of RAS, and eventually promotes cell growth in esophageal squamous cancer [22]. In line with this, our results suggest that RASSF9 expression is elevated in NSCLC tissues and cell lines. Meanwhile, the upregulation of RASSF9 expression enhanced the proliferation of tumor cells, while downregulation of RASSF9 resulted in the opposite outcome. In contrast, two recent studies have described that RASSF9 is decreased in breast and gastric cancer, and that RASSF9 suppresses the proliferation of these cancer cells [35, 36]. We speculated that the discrepancy is most likely due to different tumor cell types.
The RAS/RAF/MEK/ERK signaling cascade is a mitogen-activated protein kinase (MAPK) pathway that can be widely activated. It can transmit extracellular signals into the nucleus and cause changes in the expression profile of specific proteins in the cell, thereby affecting cell proliferation, differentiation, survival, and apoptosis [37, 38]. Once this pathway is activated, RAS combines with RAF family kinases to phosphorylate and activate RAF, MEK, and ERK kinases in turn. In addition, the latest evidence from current reports revealed elevated expression of RASSF9 lead to RAS dimer formation and RAF, MEK, and ERK activation [22]. In line with this view, in our study, we detected the expression of MEK, p-MEK, ERK, and p-ERK, and found that RASSF9 stimulated the p-MEK and p-ERK. Meanwhile, inhibition of MEK by selumetinib or U0126 eliminated the effects of RASSF9 on cell proliferation. These results suggest that RASSF9 may stimulate the proliferation of NSCLC cells by activating the MEK/ERK signaling pathway.
Taken together, our study revealed that expression of RASSF9 is upregulated in NSCLC tissues and cell lines. RASSF9 promotes the proliferation of NSCLC cells, while knockdown of RASSF9 inhibits the growth of tumor cells. In addition, we further confirm the roles of RASSF9 on the proliferation of NSCLC cells by using xenograft model. Pharmacological interventions revealed that activation of the MEK/ERK axis is responsible for RASSF9-induced tumor cell proliferation. Moreover, RASSF9 might be a druggable target for developing agents for treating NSCLC.
Materials and methods
Human specimens
To examine Rassf9 expression, a cohort of 10 patients with lung adenocarcinoma patients were enrolled from the Affiliated Hospital of Nantong University (Nantong, China). The patients are varied from 60 to 75 years of age, male, and Chinese Han nationality. All diagnoses were pathologically confirmed. None of the patients had received any anti-tumor treatments such as radiotherapy, chemotherapy, and immunotherapy prior to the surgery. Following surgical excision, all the fresh tissues including lung adenocarcinoma and matched adjacent tissues were immediately frozen in liquid nitrogen and placed in −80 °C freezer for long-term storage. All patients received written informed consent. This study was approved by the Ethics Committee of Affiliated Hospital of Nantong University.
Cell culture
All human NSCLC cell lines including A549, H1299, and H1650 were obtained from the National Collection of Authenticated Cell Cultures (Shanghai, China). 16HBE cells were from Sigma-Aldrich (City of Saint Louis, USA). Cells were cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and incubated at 37 °C in a humidified atmosphere with 5% CO2.
Cell treatments
Cell transfection was achieved by using Lipofectamine RNAiMAX (Invitrogen) for siRNAs, or Lipofectamine 2000 (Invitrogen) for plasmids according to the manufacturer’s protocols. The Rassf9 siRNA sequence and the plasmid expressing Rassf9 were described previously [22]. Twenty-four hours post-transfection with plasmids or 48 h post-transfection with siRNAs, cells were then subjected to various experiments such as MTT, cell colony formation, or EdU incorporation assays. For pharmacological interventions, cells were treated with U0126 (10 μM) or selumetinib (10 μM) for additional 24 h.
MTT assay
NSCLC cells (5000 cells/well in 100 µl DMEM) were seeded into 96-well plates and cultured for 24 h, followed by transfection with the plasmid expressing Rassf9 or Rassf9 siRNA. Cell viability was then examined by MTT assay with the manufacturer’s instructions. Cell growth was evaluated by the absorbance at 570 nM in a Microplate Reader (BioTek, Winooski, VT).
EdU assay
Ten micromolar of EdU (5-ethynyl-2′-deoxyuridine) was added to cells and then cultured for 2 h at 37 °C. After fixation, cells were incubated in the Click reaction mixture, in which Alexa Fluor 594 was contained, for 30 min at room temperature in the dark, and then cells were washed with PBS thrice and stained with Hoechst (diluted 1:1000 with PBS) at room temperature for 10 min to label the nucleus. Finally, EdU incorporation was observed with a fluorescent microscope (Zeiss, Germany).
Lentivirus transduction
The production of lentivirus expressing Rassf9 (LV-RASSF9) was described previously [22]. Briefly, the coding sequences of Rassf9 were synthesized and incorporated into pHBLV-CMV-MCS-3FLAG-EF1-ZsGreen-T2A-PURO at the sites of EcoR I and BamH I. The prepared plasmid was then transfected into 293T cells for producing lentivirus expressing Rassf9 (LV-RASSF9). The plasmid construction and lentivirus generation were performed at HANBIO (Shanghai, China). A549 cells were transduced with LV-RASSF9 to overexpress RASSF9 (MOI = 10). The control cells were transduced with LV-GFP at the same dosage. Meanwhile, 5 µg/ml polybrene was added to improve the transduction efficiency. Transfected A549 cells were selected with 2.5 μg/ml puromycin for 1 week. The efficiency for lentivirus-mediated Rassf9 expression in A549 was verified by qRT-PCR and western blot analysis. The cells stably expressed RASSF9 or GFP were used for tumor cell transplantation experiments.
Animal experiments
Six-week-old male BALB/c nude mice (18–22 g) were purchased from Shanghai Slake Laboratory Animal Co. Ltd (Shanghai, China). Mice were bred and maintained in a specific pathogen-free (SPF) environment (group housing up to 5 mice per cage). Mice were randomly divided into two groups: LV-GFP group (n = 6) and LV-RASSF9 group (n = 6). No statistical methods were used to pre-determine sample size. The tumor model was established by hypodermic injection of 2 × 106 A549 cells under the right axilla of BALB/C nude mice. Tumor growth was monitored every 3 days and measured using a vernier calipers. The volume of tumor based on caliper measurements was estimated by the formula: tumor volume = 1/2 length × width × width. After 1 month, all mice were sacrificed and tumor tissues were excised for further analysis. All experiments involving animals were approved by the Institutional Animal Care and Use Committees of the Nantong University (Approval ID: SYXK [SU] 2017-0046).
Quantitative real time-PCR (qRT-PCR)
Total RNA was extracted from the lung cancer cells using TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol. Then RNA quantity and quality were determined with a spectrophotometer (ND-1000; NanoDrop Technologies). cDNA was generated using a Prime Script RT Master Mix (Bio-Rad). The primer sequences are Rassf9, 5′-GAG GAC CTG AGC GAA AGT GAT-3′ (forward) and 5′-TCT GGA TGC CAC TCA AAT GAG A-3′ (reverse); Fos, 5′-GAT ACA CTC CAA GCG GAG AC-3′ (forward) and 5′-CCC AGT CAG ATC AAG GGA AG-3′ (reverse); Myc, 5′-CGA CTC TGA GGA GGA ACA AG-3′ (forward) and 5′-CGT AGT TGT GCT GAT GTG TG-3′ (reverse); Pax, 5′-ACC AAT CAG CAT AGG AAT CTG-3′ (forward) and 5′-TGC TGT TGT TGC TTG AAG AC-3′ (reverse); Ccna2 (Cyclin A2), 5′-CGC TGG CGG TAC TGA AGT C-3′ (forward) and 5′-GAG GAA CGG TGA CAT GCT CAT-3′ (reverse); Ccnc (Cyclin C), 5′-CCT TGC ATG GAG GAT AGT GAA TG-3′ (forward) and 5′-AAG GAG GAT ACA GTA GGC AAA GA-3′ (reverse); Ccnd1 (Cyclin D1), 5′-GCT GCG AAG TGG AAA CCA TC-3′ (forward) and 5′-CCT CCT TCT GCA CAC ATT TGA A-3′ (reverse); Ccnh (Cyclin H), 5′-TGT TCG GTG TTT AAG CCA GCA-3′ (forward) and 5′-TCC TGG GGT GAT ATT CCA TTA CT-3′ (reverse). Gapdh, 5′-GAC CTG ACC TGC CGT CTA-3′ (forward) and 5′-AGG AGT GGG TGT CGC TGT-3′ (reverse). qRT-PCR was performed using SYBR Premix Ex Taq II on a Lightcycler 96 system (Roche). Data were analyzed using the 2−△△CT method.
Protein extraction and western blot analysis
Cells or tissues were lysed with ice-cold lysis buffer (25 mM Tris-HCl, pH 7.4; 100 mM NaF; 50 mM Na4P2O7; 10 mM Na3VO4; 10 mM EGTA; 10 mM EDTA; 1% NP-40; 10 μg/ml Leupeptin; 10 μg/ml Aprotinin; 2 mM PMSF and 20 nM Okadaic acid). Protein concentrations were determined by BCA assay (Thermo Scientific) according to the manufacturer’s directions. The membranes were blocked for 1 h at room temperature in TBS-T (Tris-buffered saline + 0.05% Tween-20) containing 5% BSA and incubated overnight at 4 °C with the following primary antibody: rabbit anti-RASSF9 (Invitrogen; PA5-58785), anti-MEK1/2 (Cell Signal Technology; #9126), anti-phospho-MEK1/2 (Cell Signal Technology; #9154), anti-ERK1/2 (Cell Signal Technology; #9102), anti-phospho-ERK1/2 (Cell Signal Technology; #4370), and mouse anti-β-actin (Cell Signal Technology; #3700) was used as an internal control. Then followed by incubation with the secondary anti-rabbit (Santa Cruz Biotechnology; sc-2357) or anti-mouse (Santa Cruz Biotechnology; sc-2005) HRP-conjugated antibodies at room temperature for 1 h. Finally, protein signals were visualized using an enhanced chemilluminescense detection reagent (Roche). The western blot data were quantified by using the ImageJ software.
H&E staining
H&E staining was performed with the following standard procedures. Briefly, the collected tumors from nude mice were fixed with 4% paraformaldehyde and dehydrated with successive sucrose gradient for 24–48 h. Then, the tissues were cut into 10-μm-thick frozen sections and processed for H&E staining. After washes, the slides were counterstained with haematoxylin for 5 min, rinsed with a continuous flow of running water and then immerged in hydrochloric-alcohol solution for 15 s. Thereafter, the slides were stained with 0.5% eosin for 30 s. Finally, the slides were dehydrated with ethanol, transparentized with xylene, and eventually sealed with neutral gum. After staining, the slides were analyzed and photographed under a Zeiss microscope.
Statistical analysis
The investigators were not blind to the group allocation during the experiments. However, data collection and analysis were performed blindly. All results are expressed as the mean ± standard deviation (SD) of three independent experiments. Statistical analysis was conducted by GraphPad Prism version 8.0 (GraphPad Software, CA). Similar variances between the groups are being statistically compared. The data were analyzed by one-way ANOVA test or Student’s t-test. A value of P < 0.05 was accepted as statistical significance. No data were excluded in the analyses. No statistical methods were used to pre-determine sample size but our sample sizes are similar to those reported in our previous publications [39, 40].
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2017YFA0701304); the National Natural Science Foundation of China (81770841, 81970747, 81770266, and 81701222); and the “Six-one” Project for High-level Health Talents (LGY2016037) in Jiangsu, China.
Author contributions
J.Y. and Q.J. performed the experiments and analyzed the data. J.Z., Y.J., X.Y., X.L. and J.M. analyzed the data. J.Y. and C.S. wrote the manuscript. C.S. and J.S. conceived the idea and supervised the study.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by Silvia von Karstedt
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cheng Sun, Email: suncheng1975@ntu.edu.cn.
Jiahai Shi, Email: sjh@ntu.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–94. doi: 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberg AJ, Brock MV, Samet JM. Epidemiology of lung cancer: looking to the future. J Clin Oncol. 2005;23:3175–85. doi: 10.1200/JCO.2005.10.462. [DOI] [PubMed] [Google Scholar]
- 4.Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5:288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosell R, Karachaliou N. Lung cancer: maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol. 2013;10:549–50. doi: 10.1038/nrclinonc.2013.152. [DOI] [PubMed] [Google Scholar]
- 6.Donninger H, Schmidt ML, Mezzanotte J, Barnoud T, Clark GJ. Ras signaling through RASSF proteins. Semin Cell DevBiol. 2016;58:86–95. doi: 10.1016/j.semcdb.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: a new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochem J. 2009;425:303–11. doi: 10.1042/BJ20091318. [DOI] [PubMed] [Google Scholar]
- 8.Iwasa H, Hossain S, Hata Y. Tumor suppressor C-RASSF proteins. Cell Mol Life Sci. 2018;75:1773–87. doi: 10.1007/s00018-018-2756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Underhill-Day N, Hill V, Latif F. N-terminal RASSF family: RASSF7-RASSF10. Epigenetics. 2011;6:284–92. doi: 10.4161/epi.6.3.14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Dong Q, Zhang X, Han Q, Han X, Han Y, et al. The coiled-coil domain of oncogene RASSF 7 inhibits hippo signaling and promotes non-small cell lung cancer. Oncotarget. 2017;8:78734–48. doi: 10.18632/oncotarget.20223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M, Li Q, Zhang L, Wang Y, Wang L, Li Q, et al. RASSF7 promotes cell proliferation through activating MEK1/2-ERK1/2 signaling pathway in hepatocellular carcinoma. Cell Mol Biol. 2018;64:73–79. doi: 10.14715/cmb/2018.64.5.12. [DOI] [PubMed] [Google Scholar]
- 12.Lock FE, Underhill-Day N, Dunwell T, Matallanas D, Cooper W, Hesson L, et al. The RASSF8 candidate tumor suppressor inhibits cell growth and regulates the Wnt and NF-kappaB signaling pathways. Oncogene. 2010;29:4307–16. doi: 10.1038/onc.2010.192. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Liu W, Zhang YP, Huang XR. The miR-224 promotes non-small cell lung cancer cell proliferation by directly targeting RASSF8. Eur Rev Med Pharm Sci. 2017;21:3223–31. [PubMed] [Google Scholar]
- 14.He C, Wang L, Zhang J, Xu H. Hypoxia-inducible microRNA-224 promotes the cell growth, migration and invasion by directly targeting RASSF8 in gastric cancer. Mol Cancer. 2017;16:35. doi: 10.1186/s12943-017-0603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Li Y, Wang FF, Lv W, Xie X, Cheng X. Over-expressed miR-224 promotes the progression of cervical cancer via targeting RASSF8. PLoS ONE. 2016;11:e0162378. doi: 10.1371/journal.pone.0162378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hesson LB, Dunwell TL, Cooper WN, Catchpoole D, Brini AT, Chiaramonte R, et al. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer. 2009;8:42. doi: 10.1186/1476-4598-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schagdarsurengin U, Richter AM, Wohler C, Dammann RH. Frequent epigenetic inactivation of RASSF10 in thyroid cancer. Epigenetics. 2009;4:571–6. doi: 10.4161/epi.4.8.10056. [DOI] [PubMed] [Google Scholar]
- 18.Liu W, Wang J, Wang L, Qian C, Qian Y, Xuan H, et al. Ras-association domain family 10 acts as a novel tumor suppressor through modulating MMP2 in hepatocarcinoma. Oncogenesis. 2016;5:e237. doi: 10.1038/oncsis.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter AM, Walesch SK, Dammann RH. Aberrant promoter methylation of the tumour suppressor RASSF10 and its growth inhibitory function in breast cancer. Cancers. 2016;8:26. [DOI] [PMC free article] [PubMed]
- 20.Li X, Liang Q, Liu W, Zhang N, Xu L, Zhang X, et al. Ras association domain family member 10 suppresses gastric cancer growth by cooperating with GSTP1 to regulate JNK/c-Jun/AP-1 pathway. Oncogene. 2016;35:2453–64. doi: 10.1038/onc.2015.300. [DOI] [PubMed] [Google Scholar]
- 21.Lu D, Ma J, Zhan Q, Li Y, Qin J, Guo M. Epigenetic silencing of RASSF10 promotes tumor growth in esophageal squamous cell carcinoma. Disco Med. 2014;17:169–78. [PubMed] [Google Scholar]
- 22.Shi H, Ju Q, Mao Y, Wang Y, Ding J, Liu X, et al. TAK1 phosphorylates RASSF9 and inhibits esophageal squamous tumor cell proliferation by targeting the RAS/MEK/ERK axis. Adv. Sci. 2021;8:2001575. doi: 10.1002/advs.202001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ambrogio C, Kohler J, Zhou ZW, Wang H, Paranal R, Li J, et al. KRAS dimerization impacts MEK inhibitor sensitivity and oncogenic activity of mutant KRAS. Cell. 2018;172:857–868 e815. doi: 10.1016/j.cell.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 24.Donner I, Katainen R, Kaasinen E, Aavikko M, Sipila LJ, Pukkala E, et al. Candidate susceptibility variants in angioimmunoblastic T-cell lymphoma. Fam Cancer. 2019;18:113–9. doi: 10.1007/s10689-018-0099-x. [DOI] [PubMed] [Google Scholar]
- 25.Kita R, Fraser HB. Local adaptation of sun-exposure-dependent gene expression regulation in human skin. PLoS Genet. 2016;12:e1006382. doi: 10.1371/journal.pgen.1006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nejadtaghi M, Jafari H, Farrokhi E, Samani KG. Familial colorectal cancer type X (FCCTX) and the correlation with various genes—A systematic review. Curr Probl Cancer. 2017;41:388–97. doi: 10.1016/j.currproblcancer.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Alam MR, Caldwell BD, Johnson RC, Darlington DN, Mains RE, Eipper BA. Novel proteins that interact with the COOH-terminal cytosolic routing determinants of an integral membrane peptide-processing enzyme. J Biol Chem. 1996;271:28636–40. doi: 10.1074/jbc.271.45.28636. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Johnson RC, Milgram SL. P-CIP1, a novel protein that interacts with the cytosolic domain of peptidylglycine alpha-amidating monooxygenase, is associated with endosomes. J Biol Chem. 1998;273:33524–32. doi: 10.1074/jbc.273.50.33524. [DOI] [PubMed] [Google Scholar]
- 29.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–28. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Volodko N, Gordon M, Salla M, Ghazaleh HA, Baksh S. RASSF tumor suppressor gene family: biological functions and regulation. FEBS Lett. 2014;588:2671–84. doi: 10.1016/j.febslet.2014.02.041. [DOI] [PubMed] [Google Scholar]
- 31.Recino A, Sherwood V, Flaxman A, Cooper WN, Latif F, Ward A, et al. Human RASSF7 regulates the microtubule cytoskeleton and is required for spindle formation, Aurora B activation and chromosomal congression during mitosis. Biochem J. 2010;430:207–13. doi: 10.1042/BJ20100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richter AM, Walesch SK, Wurl P, Taubert H, Dammann RH. The tumor suppressor RASSF10 is upregulated upon contact inhibition and frequently epigenetically silenced in cancer. Oncogenesis. 2012;1:e18. doi: 10.1038/oncsis.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei Z, Chen X, Chen J, Wang W, Xu X, Cai Q. RASSF10 is epigenetically silenced and functions as a tumor suppressor in gastric cancer. Biochem Biophys Res Commun. 2013;432:632–7. doi: 10.1016/j.bbrc.2013.02.033. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Viciana P, Sabatier C, McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol. 2004;24:4943–54. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B, Chen P, Wang J, Wang L, Ren M, Zhang R, et al. MicroRNA-1254 exerts oncogenic effects by directly targeting RASSF9 in human breast cancer. Int J Oncol. 2018;53:2145–56. doi: 10.3892/ijo.2018.4530. [DOI] [PubMed] [Google Scholar]
- 36.Liu WL, Wang HX, Shi CX, Shi FY, Zhao LY, Zhao W, et al. MicroRNA-1269 promotes cell proliferation via the AKT signaling pathway by targeting RASSF9 in human gastric cancer. Cancer Cell Int. 2019;19:308. doi: 10.1186/s12935-019-1026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuen HF, Chan KK, Grills C, Murray JT, Platt-Higgins A, Eldin OS, et al. Ran is a potential therapeutic target for cancer cells with molecular changes associated with activation of the PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Clin Cancer Res. 2012;18:380–91. doi: 10.1158/1078-0432.CCR-11-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michael JV, Wurtzel JG, Goldfinger LE. Regulation of H-Ras-driven MAPK signaling, transformation and tumorigenesis, but not PI3K signaling and tumor progression, by plasma membrane microdomains. Oncogenesis. 2016;5:e228. doi: 10.1038/oncsis.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun C, Wang M, Liu X, Luo L, Li K, Zhang S, et al. PCAF improves glucose homeostasis by suppressing the gluconeogenic activity of PGC-1alpha. Cell Rep. 2014;9:2250–62. doi: 10.1016/j.celrep.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 40.Lee J, Sun C, Zhou Y, Lee J, Gokalp D, Herrema H, et al. p38 MAPK-mediated regulation of Xbp1s is crucial for glucose homeostasis. Nat. Med. 2011;17:1251–60. doi: 10.1038/nm.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]