Abstract
The CRISPR/Cas9 system has been used for genome editing in several organisms, including higher plants. This system induces site-specific mutations in the genome based on the nucleotide sequence of engineered guide RNAs. The complex genomes of C4 grasses makes genome editing a challenge in key grass crops like maize (Zea mays), sorghum (Sorghum bicolor), Brachiaria spp., switchgrass (Panicum virgatum), and sugarcane (Saccharum spp.). Setaria viridis is a diploid C4 grass widely used as a model for these C4 crop plants. Here, an optimized CRISPR/Cas9 binary vector that exploits the non-homologous end joining (NHEJ) system was used to knockout a green fluorescent protein (gfp) transgene in S. viridis accession A10.1. Transformation of embryogenic callus by A. tumefaciens generated ten glufosinate-ammonium resistant transgenic events. In the T0 generation, 60% of the events were biallelic mutants in the gfp transgene with no detectable accumulation of GFP protein and without insertions or deletions in predicted off-target sites. The gfp mutations generated by CRISPR/Cas9 were stable and displayed Mendelian segregation in the T1 generation. Altogether, the system described here is a highly efficient genome editing system for S. viridis, an important model plant for functional genomics studies in C4 grasses. Also, this system is a potential tool for improvement of agronomic traits in C4 crop plants with complex genomes.
Keywords: CRISPR/Cas9, genome editing, non-homologous end-joining, panicoid grasses, Setaria viridis, stable genetic transformation
Introduction
Genome editing is a powerful tool to introduce deletions, insertions, or sequence modifications into organismal genomes. Genome editing with the Clustered Regulatory Interspaced Short Palindromic Repeat-associated endonuclease protein 9 (CRISPR/Cas9) system has been optimized for a range of organisms, replacing other site-specific nucleases (e.g., Meganucleases, ZNFs, and TALENs) due to its greater simplicity, specificity, and efficiency (Cong et al. 2013; Jinek et al. 2012; Makarova et al. 2015; Mali et al. 2013; May et al. 2013; Zhang 2013). The CRISPR/Cas9 system creates double-stranded DNA breaks (DSBs) at a specific target sequence specified by the sequence of a guide RNA (gRNA). The break is repaired by either the non-homologous end joining (NHEJ) pathway, which generates nucleotide insertions or deletions (indels) at the target site, or the homology-directed repair (HDR) pathway, which can generate single and double knock-ins of a desired target gene or promoter sequence (Li et al. 2016; Sun et al. 2016; Zhao et al. 2016). The NHEJ pathway is more efficient than the HDR pathway (Li et al. 2016; Liang et al. 2017; Sun et al. 2016; Tang et al. 2017; Woo et al. 2015; Zhang et al. 2016). However, the efficiency of either CRISPR/Cas9 system varies between phylogenetically distant plant species. Thus, it is necessary to optimize the elements of the CRISPR/Cas9 system for the target genome (Basso et al. 2020; Li et al. 2016; Yin et al. 2017).
Several features of the C4 grass Setaria viridis make it an experimental model plant for C4 grass crops, including a short life cycle, high seed production, and established protocols for genetic transformation and plant regeneration (Brutnell et al. 2010; Li and Brutnell 2011; Martins et al. 2015), as well as a small diploid genome that is fully sequenced (Bennetzen et al. 2012). This model plant is widely used to study C4 grass functional genomics and for proof-of-concept experiments for panicoid bioenergy feedstocks and food crops with highly complex genomes like maize (Zea mays), sorghum (Sorghum bicolor), Brachiaria spp., switchgrass (Panicum virgatum), and sugarcane (Saccharum spp.).
Huang et al. (2019) edited the genome of S. viridis with a CRISPR/Cas9 genome system to introduce mutations in the SvLes1 (Sevir.5G085400) gene. More recently, Weiss et al. (2020) demonstrated efficient genome editing in S. viridis with a combination of a CRISPR/Cas9 and expression of exonuclease Trex2 (CRISPR/Cas9_Trex2), which promoted repair by either NHEJ or microhomology-mediated end joining (MMEJ). These authors showed that this multiplex CRISPR/Cas9_Trex2 system induced targeted indels in both the svDrm1a and svDrm1b genes of T0 transgenic events at a frequency of 73% to 100%. These indels were transmitted to at least 60% of the transgene-free T1 plants, with 33% of them containing biallelic or homozygous indels in both genes.
Here, we optimized a CRISPR/Cas9 binary vector that promotes the activity of the NHEJ pathway and this vector was used to edit a transgenic copy of the green fluorescent protein (gfp) gene in S. viridis accession A10.1. This study generated ten biallelic mutants (homozygous) at the T0 generation. These indels in the gfp transgene were stable and showed Mendelian segregation in the T1 generation. These lines carried no off-target indels in predicted off-target sites, indicating highly site-specific targeting by this combination of CRISPR/Cas9 binary vector and gRNA.
Materials and methods
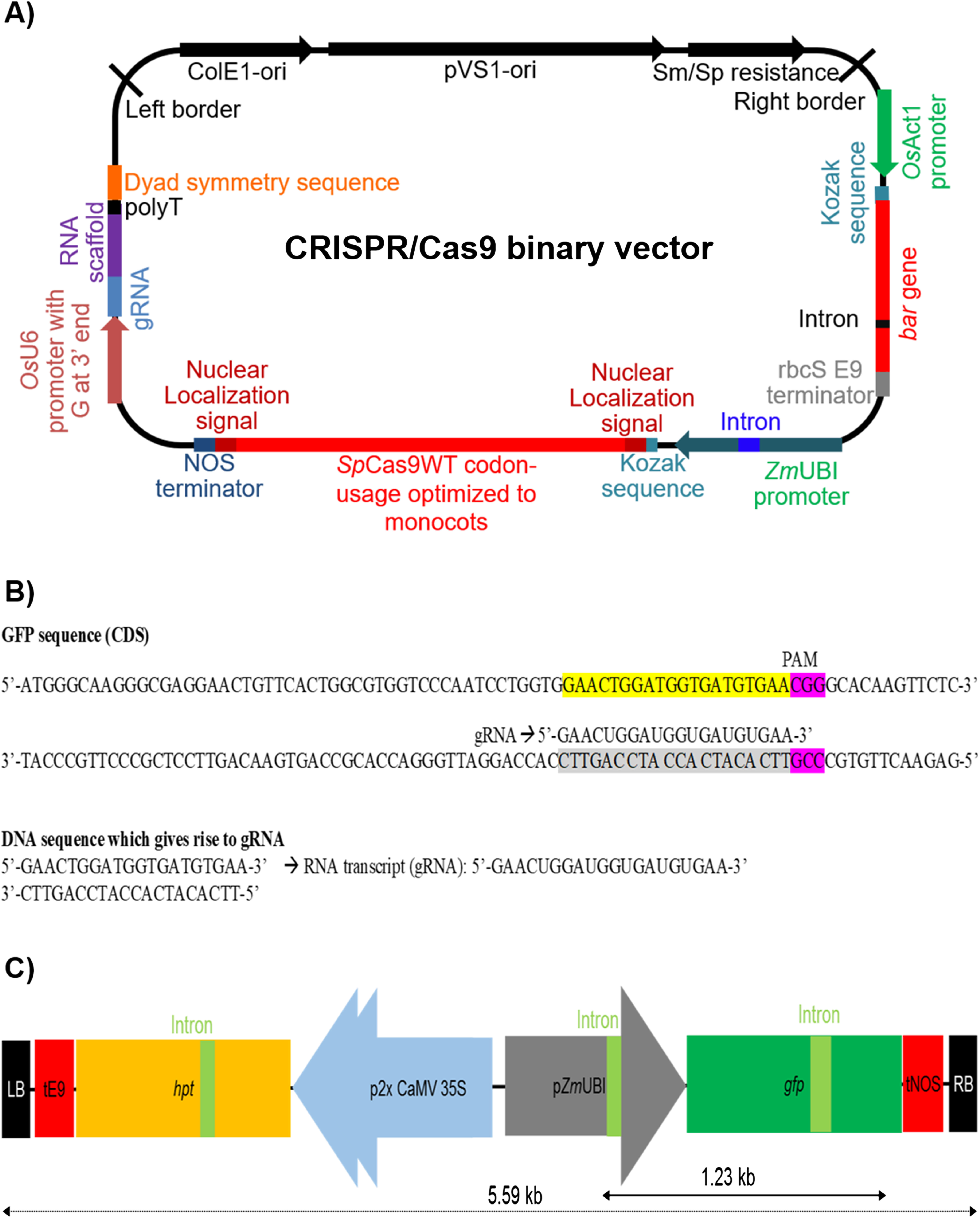
Construction of a CRISPR/Cas9 binary vector to enhance genome editing through NHEJ pathway
The T-DNA of the binary vector is composed of (i) selection marker gene (bar gene from Streptomyces hygroscopicus for resistance to glufosinate-ammonium herbicide), with an intron sequence, controlled by Oryza sativa constitutive promoter actin 1 (OsAct-1), and 3′rbcsE9 transcription terminator; (ii) Cas9 nuclease from Streptococcus pyogenes (SpCas9WT), with codon-usage optimized for monocots and fused in-frame with two SV40 nuclear localization signal (NLS) at the 5′ and 3′ end of the gene. This fragment was engineered under control of the constitutive ubiquitin-1 promoter from Zea mays (ZmUBI-1), with an intron sequence and linked with the nopaline synthase (NOS) transcription terminator sequence. The Kozak sequence CCG AA (optimized Kozak sequence for monocots) was introduced in front of the translation initiation site (ATG); (iii) DNA sequence that corresponds to the gRNA sequence under control of the constitutive U6-2 small nuclear RNA promoter (coding for RNA polymerase III) from Oryza sativa (OsU6), with extra guanine (G) nucleotide at 3′ end fused in-frame at the 3′ end of a synthetic RNA sequence (scaffold RNA used for Cas9 coupling). The dyad symmetry sequence was added after the transcription stop signal (poly A7). The gRNA was designed for targeting the 5′ end of the gfp (green fluorescent protein) gene and direct the cleavage by Cas9 from the sense strand of target DNA using NGG sequence as the Protospacer Adjacent Motif (PAM). Thus, the gfp transgene was used as target DNA to optimize the CRISPR/Cas9 NHEJ system in S. viridis and the GFP protein as a reporter of the knockout efficiency. The absence of off-target mutations was analyzed by scanning the Phytozome 12 S. viridis genome (Setaria viridis v2.1, DOE-JGI, (http://phytozome.jgi.doe.gov/) using GT-Scan (O’Brien and Bailey 2014) and CCTop (Stemmer et al. 2015) software. The binary vector also contained the CoIE1 and pVS1 origin of replication from Escherichia coli and Agrobacterium tumefaciens, respectively, and resistance marker gene to streptomycin/spectinomycin (Sm/Sp). The CRISPR/Cas9 binary vector was synthesized, and assembled by DNA Cloning Service (www.dna-cloning.com; Hamburg, Germany).
Stable transformation and regeneration of Setaria viridis gene editing events from embryogenic callus
S. viridis (accession A10.1) was co-transformed by a high-efficiency Agrobacterium-mediated transformation protocol (Martins et al. 2015). After removing the lemmas and paleas, mature seeds were disinfested, blotted onto sterile filter paper, and transferred to callus induction medium (CIM2) for embryogenic callus induction. After 22 days of incubation in the dark at 25±2°C, embryogenic calli were isolated and transferred onto a fresh CIM2 medium. After a week, selected embryogenic calli were transformed by A. tumefaciens EHA105. Embryogenic calli from a homozygous transgenic S. viridis plants constitutively expressing GFP and resistant to hygromycin B (30 mg/l) (Martins et al. 2015) were co-transformed with the CRISPR/Cas9 NHEJ binary vector. Selection with glufosinate-ammonium herbicide (3 mg/l; Liberty®-Bayer CropScience) identified ten independent CRISPR/Cas9 NHEJ events after. Transgenic seedlings were planted into pots containing 250 g of latosoil, substrate (Plantmax®), and vermiculite (Agrifloc, Brasil Minérios) mixture (3 : 1 : 0.5; w/w/w). The plants were maintained in a growth chamber under 16/8-h photoperiod at 500 µmol m−2 s−1 light intensity, 26±2°C temperature, and 65% relative humidity until seed establishment (seeds from T1 generation).
Genotyping of the gene editing events
Genomic DNA was isolated from co-transforming T0 lines and control (GFP positive, but not co-transformed with our CRISPR/Cas9 binary vector) plants using the CTAB method (Doyle and Doyle 1987). PCR assays were carried out using specific primers targeting bar gene and ZmUBI-1:GFP to confirm the insertion of ZmUBI-1:GFP and CRISPR/Cas9 transgene in the regenerated plants from selection with glufosinate-ammonium herbicide. PCR-positive events were also confirmed for BAR protein accumulation with a quick test strip (QuickStix™ Kit for PAT/bar, EnviroLogix, Inc., USA), according to the manufacturer’s instructions. The ZmUBI-1:GFP PCR products (1.23 kb length) from ten co-transforming events and two control plants were directly purified and dual sequenced (forward and reverse) using the Sanger method by Macrogen Service (Geumcheon-gu, Seoul, South Korea). Chromatograms were analyzed using Geneious R10 software (Kearse et al. 2012). Mono (heterozygous) and biallelic (homozygous) indels in the DNA sequence targeted by the gRNA were decoded using standard degenerate symbols (IUPAC/IUB), based on the color peaks of the chromatograms, according to the Degenerate Sequence Decoding (DSD) method (Ma et al. 2015). Chromatograms from both forward and reverse sequencing confirmed the indels and their exact positions in the target sequence. The inheritance and stability of indels induced by the CRISPR/Cas9 system were confirmed by PCR, and subsequent Sanger sequencing of the ZmUBI-1:GFP transgene from EC1 to EC3, EC5, and EC7 events from T1 generation. Wild-type plants (ECWT) were used as control. The T-DNA copy number in each event was estimated using seeds from T1 co-transforming events. One hundred seeds were grown in a selective MS medium containing 5 mg/l glufosinate-ammonium. Ten-day-old resistant and sensitive plants were counted and analyzed statistically using the χ-Squared test. Events with a 3 : 1 Mendelian segregation ratio were considered as single T-DNA insertion. The occurrence of putative off-targets was screened by Sanger sequencing of PCR product from the top two predicted off-target sites, which have respectively 1 and 2 nucleotide mismatches in the core sequence, besides 3 and 2 nucleotide mismatches farther from the PAM sequence, compared with the sequence of gRNA. Highly pure DNA samples from co-transforming T0 transgenic events named EC1 to EC5 and control plant ECWT1 were amplified by PCR using primers flanking the predicted off-target sites. PCR products were previously checked using agarose gel electrophoresis, purified using a commercial kit, and sequenced by the Sanger method in a forward direction using the same primers.
Relative expression of gfp mRNA in gene editing events
Total RNA was isolated from leaf tissue of S. viridis adult plants (30 days after germination), with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. RNA concentration was estimated using a spectrophotometer (NanoDrop 2000, Thermo Scientific, Massachusetts, USA). RNA integrity was analyzed in 1% agarose gel electrophoresis. RNA samples were treated with RNase-free RQ1 DNase I (Promega), according to the manufacturer’s instructions. Then, 2 µg of DNase-treated RNA were used as the template for cDNA synthesis using oligo(dT)18 primer and SuperScript III RT (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. The cDNA produced was quantified by spectrophotometry and diluted with nuclease-free water to 200 ng/µl. Quantitative reverse transcription PCR (qRT-PCR) assays were carried out in ABI StepOne Plus Real-Time PCR System (Applied Biosystems, USA). The reactions were performed using 300 ng cDNA, 0.2 µM of each gene-specific primer, and Platinum™ SYBR™ Green qPCR SuperMix-UDG w/ROX (Invitrogen). Statistical difference between co-transforming EC1 to EC10 events and S. viridis A10.1 (wild-type) was confirmed by Tukey’s HSD test (p-value<0.01) using the SASM-Agri statistical package (Canteri et al. 2001). All reactions were performed with three technical replicates for each sample and conditions, according to Martins et al. (2016). The relative expression was calculated using the 2−∆Ct method (Schmittgen and Livak 2008) using SvSUI (Sevir.2G348300) as an endogenous reference gene. All primers used in the present work are listed in Supplementary Table S2.
GFP protein detection in gene editing events
GFP-specific fluorescence in leaves of the co-transforming events was visualized in Leica M205 FA stereomicroscope equipped with either a long pass filter with a 395–455 nm excitation filter and a 480 nm emission filter or a GFP filter with a 450–490 nm excitation filter and a 500–550 nm emission filter. Detection of GFP protein in the crude protein extracts isolated from leaf tissue of adult co-transforming and transgenic plants was carried out using indirect ELISA (Clark 1981). Initially, the protein amount in the total protein extracts was quantified using Bradford’s method (Bradford 1976). Serological assays were conducted using the monoclonal IgG1 Anti-GFP (G1546, Sigma-Aldrich) as a primary antibody produced in mice, which were immunized with a synthetic peptide corresponding to amino acids 132–144 of GFP. Polyclonal anti-mouse IgG produced in goat and alkaline phosphatase-conjugated (A3688, Sigma-Aldrich) was used as a secondary antibody. Samples consisting of leaves from S. viridis A10.1 non-transgenic plants (wild-type plant used as a negative control for GFP in the indirect ELISA), transgenic ECWT1 and ECWT2 plants (GFP-positive control for the indirect ELISA), and co-transforming EC1 to EC10 events were grounded to a fine powder with pestle and mortar in liquid nitrogen and homogenized in 1 : 2 (w/v) extraction buffer (10 mM Na2CO3, 10 mM NaHCO3, 15 mM NaN3, 5 mM Na-DIECA, 0.2% bovine serum albumin, 2% PVP40, pH 9.6). Plant samples were considered GFP-positive when absorbance at 405 nm was at least twice the average value of the negative control.
Results
We constructed a new CRISPR/Cas9 binary vector engineered with several genetic elements to enhance the efficiency and specificity of genome editing in S. viridis, as described in Material and Methods and shown in Figure 1a. We also designed a gRNA to target the 5′-end of a gfp transgene (Figure 1b) previously inserted into the genome of S. viridis (Figure 1c), directing Cas9 cleavage of the sense strand of the target site using NGG nucleotides as the PAM sequence.
Figure 1. Genome editing and gene knock-out strategy using the CRISPR/Cas9 system exploring the non-homologous end joining (NHEJ) pathway in S. viridis accession A10.1. (a) Optimized CRISPR/Cas9 NHEJ binary vector for genome editing in S. viridis; (b) schematic diagram of the guide RNA targeting the gfp gene. (c) Schematic T-DNA region of the binary vector used to transform Setaria viridis for constitutive expression of GFP protein. The generated Setaria viridis events were edited in co-transforming events by CRISPR/Cas9 system.
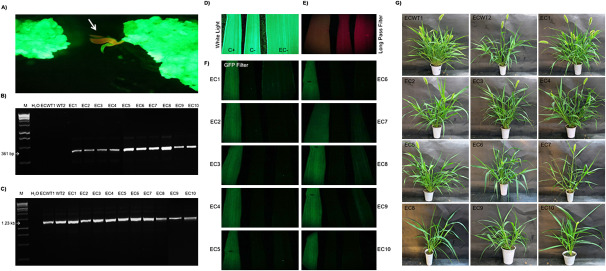
Stable genetic transformation and genotyping of recovered genome editing events
Embryogenic calli from transgenic plants overexpressing gfp were transformed with the engineered CRISPR/Cas9 binary vector to generate indels that disrupt the gfp transgene (Figure 2a). Ten independent transgenic events obtained following Agrobacterium-mediated transformation were sequentially named as EC1 to EC10. PCR analysis confirmed these lines carried the CRISPR/Cas9 minimal expression cassette (Figure 2b and 2c), which was confirmed by the presence of the BAR protein from the bar gene (Table 2). Plants from these events were compared to two independent gfp overexpressing plants (GFP-positive control plants, named ECWT1 and ECWT2) and two non-transgenic wild-type plants (GFP-negative control plants). GFP accumulation in leaves of young gene edited plants, detected by fluorescence microscopy, presented three accumulation patterns: (i) absence, (ii) low GFP accumulation, and (iii) GFP accumulation equivalent to ECWT1 or ECWT2 control plants (Figure 2c). The results indicate different levels of allele disruption in each event. The morphology of adult T0 gene edited plants appeared similar to control plants, indicating transformation with the CRISPR/Cas9 binary vector did not cause pleiotropic alterations of growth and development (Figure 2d). The CRISPR/Cas9 minimal expression cassette segregated at a 3 : 1 ratio in the T1 progeny of events EC1 to EC10, indicating a single copy insertion for each event, as expected for A. tumefaciens-mediated transformation.
Figure 2. Genetic transformation of Setaria viridis with CRISPR/Cas9 NHEJ binary vector and plant evaluation. (a) Embryogenic callus observed under GFP long-pass filter (excitation and emission wavelength 395–455 nm and 480 nm, respectively). Arrowhead indicates a transgenic shoot regenerated under glufosinate-ammonium selection. (b) Agarose gel electrophoresis of PCR products from CRISPR-edited plants using primers specific for bar gene (top) and ZmUBI-1:GFP (bottom). MWM=1 kb ladder DNA molecular weight marker; ECWT1 and ECWT2 are transgenic plants positive for ZmUBI-1:GFP and negative for CRISPR/Cas9 NHEJ minimal expression cassette; EC1 to EC10 are independent co-transforming and edited events. (c) Imaging using GFP Filter (excitation and emission wavelength 450–490 nm and 500–550 nm, respectively). Leaf from S. viridis A10.1 constitutively expressing GFP protein (positive control: C+); leaf from non-transgenic plants (negative control: C−); leaves from S. viridis A10.1 constitutively expressing GFP protein and co-transformed with CRISPR/Cas9 NHEJ binary vector (EC1 to EC10). (d) Phenotype of mature edited T0 event (EC1) compared with a non-edited plant (ECWT1 and ECWT2), illustrating the normal development of edited plants.
Table 2. Results of indirect ELISA assays (absorbance values at 405 nm) and quick test strip to serological detection, respectively, of GFP and BAR proteins in leaf samples from co-transforming events of Setaria viridis. Primary antibody against the C-terminal portion was used for GFP detection, while for BAR protein detection was used the QuickStix™ Kit for PAT/bar (EnviroLogix, Inc., USA).
| Sample | Absorbance values at 405 nm | ELISA for GFP | Quick test strip for BAR |
|---|---|---|---|
| Buffer | 0.101 | Negative | Negative |
| SvA10.1 (negative control; wild-type plants)a | 0.332 | Negative | Negative |
| ECWT1 (GFP-positive control 1)b | 1.372 | Positive | Negative |
| ECWT2 (GFP positive control 2)c | 1.624 | Positive | Negative |
| EC1 | 0.762 | Positive | Positive |
| EC2 | 0.412 | Negative | Positive |
| EC3 | 0.358 | Negative | Positive |
| EC4 | 0.421 | Negative | Positive |
| EC5 | 0.456 | Negative | Positive |
| EC6 | 0.309 | Negative | Positive |
| EC7 | 0.695 | Positive | Positive |
| EC8 | 0.869 | Positive | Positive |
| EC9 | 0.987 | Positive | Positive |
| EC10 | 0.379 | Negative | Positive |
a=wild-type Setaria viridis A10.1 (non-transgenic; negative control for GFP in the indirect ELISA); b and c=transgenic plants positive control for ZmUBI-1:GFP and negative control for CRISPR/Cas9 NHEJ (used as positive controls for GFP protein in the indirect ELISA); EC1 to EC10=co-transforming events PCR-positive to ZmUBI-1:GFP and CRISPR/Cas9 NHEJ minimal expression cassette.
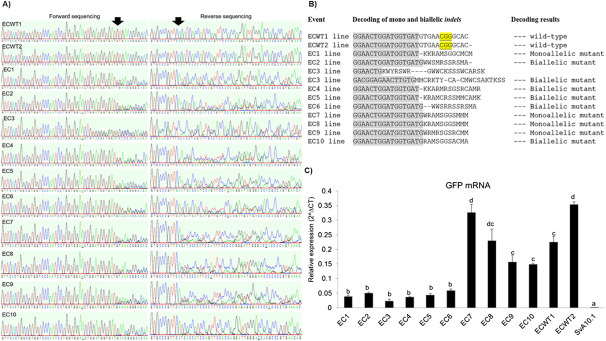
To identify the nature of the mutation introduced by gene editing in each event, purified PCR products amplified from the targeted genomic region in the T0 EC1-EC10 and GFP-positive control plants (ECWT1 or ECWT2) were sequenced by Sanger sequencing (Figure 3a). Sequencing revealed the presence of indels at the gRNA-targeted site in the gfp transgene of plants from each of the ten events and these changes appeared as either mono and biallelic indels (Table 1, Figure 3B). Finally, the effect of these mutations on gfp expression and GFP protein accumulation in the adult T0 plants was determined with RT-qPCR and indirect ELISA assays, respectively. mRNA from the gfp transgene was observed in all plants from all events (Figure 3C), while GFP protein levels were different amongst the lines, ranging from low accumulation in plants with monoallelic mutations to no accumulation in plants with biallelic mutations (Table 2). These results indicate the mutations introduced by gene editing modified the mRNA sequence to encode either a truncated protein (resulting from a premature stop codon) or a protein with an amino acid sequence different from GFP (resulting from a frameshift mutation) (Supplementary File S1).
Figure 3. Gene editing and gene expression. (a) Sanger sequencing of PCR products from target sequence of the CRISPR/Cas9 NHEJ system. Forward and reverse sequencing were carried out using ZmUBI(F) and GFP(R) primers (Supplemental Table S2). ECWT1 and ECWT2 are transgenic plants positive for ZmUBI:GFP plants and negative for CRISPR/Cas9 NHEJ, while EC1 to EC10 are transgenic positive. Arrows above the sequencing chromatograms indicate the location of indels in the target sequence, resulting in the overlap of peaks in the chromatograms. (b) Decoding of the mono and biallelic indels in gfp transgene of the Setaria viridis co-transforming EC1 to EC10 events compared with ECWT1 and ECWT2 plants (negative controls for edition, gfp gene wild-type). (c) Relative expression of gfp mRNA by qRT-PCR from EC1 to EC10 co-transforming events (T0 lines), ECWT1 and ECWT2 plants (GFP-positive control), and SvA10.1 wild-type plants (non-transgenic; negative control). Statistical difference between each independent event was confirmed using Tukey’s HSD test (p-value<0.01).
Table 1. Sequences of the mono and biallelic indels in gfp transgene induced by CRISPR/Cas9 NHEJ system, gfp mRNA expression level, GFP protein accumulation, and segregation behavior of the CRISPR/Cas9 transgene in gene edited plants from events EC1 to EC10 compared to parental unedited gfp transgenic plants (ECWT1 and ECWT2).
| Events | Decoding of mono and biallelic mutations | Decoding results | gfp mRNA accumulation | GFP protein accumulation | Segregation ratio in T1 |
|---|---|---|---|---|---|
| ECWT1 | GGATGGTGATGTGAACGGGCAC | Non-edited control lineage 1 | Positive | Positive | — |
| ECWT2 | GGATGGTGATGTGAACGGGCAC | Non-edited control lineage 2 | Positive | Positive | — |
| EC1 | ECWT line GGAACTGGATGGTGATGTGAACGGGCAC- EC1 line GGAACTGGATGGTGAT-KKRAMSGGCMCM Allele 1 GGAACTGGATGGTGAT TGAACGGGCACA ****************-************ Allele 2 GGAACTGGATGGTGATGTGAACGGGCAC **************************** | Allele 1: deletion of G nucleotide; Allele 2: no indels; Monoallelic mutant. | Positive | Positive | 3 : 1 |
| EC2 | ECWT line GGAACTGGATGGTGATG-TGAACGGGCAC EC2 line GGAACTGGATGGTGATGWWSMRSSRSMA- Allele 1 GGAACTGGATGGTGATGATGAACGGGCA- *****************-**********- Allele 2 GGAACTGGATGGTGATGT--ACGGGCACAA ******************--********** | Allele 1: insertion of A nucleotide; Allele 2: deletion of GA nucleotides; Biallelic mutant. | Positive | Negative | 3 : 1 |
| EC3 | Chromatogram from forward sequencing ECWT line GGAACTGGATGGTGATGTGAACGGGCACAAGT EC3 line GGAACTGKWYRSWR----GWWCKSSSWCARSK Allele 1 GGAACTGGATGGTG----GAACGGGCACAAGT **************----**********???? Allele 2 GGAACTG-TT-CACAAGTTCTCCGTCACAA *******--*----*-**---*-*-***** | Allele 1: deletion of ATGT nucleotides; Allele 2: two deletions of G nucleotide; Biallelic mutant. | Positive | Negative | 3 : 1 |
| Chromatogram from reverse sequencing ECWT line GACGGAGAACTTGTGCCCGTTCACATC-ACCA-T-CC EC3 line GACGGAGAACTTGTGMMCRKTY-CA-CMWCSAKTKSS Allele 1 GACGGAGAACTTGTGCCCGTTC-CA-CCACCATTGCC **********************-**-*-****-*-** Allele 2 GACGGAGAACTTGTGAA--CAGTTCACATCGA--GTT-CAGTTCC ***************----*-*********-*----*-******* | Allele 1: deletions of A and T nucleotides and insertions of C/A, T and G nucleotides; Allele 2: insertions of AA, A, G, and G nucleotides and two deletions of CC nucleotides; Biallelic mutant. | ||||
| EC4 | ECWT line GGAACTGGATGGTGATGTGAACGGGCACAAG EC4 line GGAACTGGATGGTGAT-KKARMRSGSRCAMR Allele 1 GGAACTGGATGGTGAT-TGAACGGGCACAAG ****************-************** Allele 2 GGAACTGGATGGTGATGTAGAACGGGCACA ******************-*--********* | Allele 1: deletion of G nucleotide; Allele 2: insertion of A nucleotide; Biallelic mutant. | Positive | Negative | 3 : 1 |
| EC5 | ECWT line GGAACTGGATGGTGATGTGAACGGGCACAAG EC5 line GGAACTGGATGGTGAT-KRAMCRSSMMCAMK Allele 1 GGAACTGGATGGTGAT-TGAACGGGAACAAG ****************-************** Allele 2 GGAACTGGATGGTGAT--GAACGGGCACA ****************--*********** | Allele 1: deletion of G nucleotide; Allele 2: deletions of GT nucleotides; Biallelic mutant. | Positive | Negative | 3 : 1 |
| EC6 | ECWT line GGAACTGGATGGTGATGTGAACGGGCACAA EC6 line GGAACTGGATGGTGATG--WWSRRSSRSMA Allele 1 GGAACTGGATGGTGATG--AACGGGCACAA *****************--*********** Allele 2 GGAACTGGATGGTGATGTTGAACGGGCA *****************-********** | Allele 1: deletion of TG nucleotides; Allele 2: one insertion of T nucleotide; Biallelic mutant. | Positive | Negative | 3 : 1 |
| EC7 | ECWT line GGAACTGGATGGTGATGTGAACGGGCAC EC7 line GGAACTGGATGGTGATGWRAMSGGSMMM Allele 1 GGAACTGGATGGTGATGTGAACGGGCAC **************************** Allele 2 GGAACTGGATGGTGATG--AACGGGCACA *****************--*********** | Allele 1: no indels; Allele 2: deletion of TG nucleotides; Monoallelic mutant. | Positive | Positive | 3 : 1 |
| EC8 | ECWT line GGAACTGGATGGTGATGTGAACGGGCACA EC8 line GGAACTGGATGGTGATGKRAMSGGSMMM Allele 1 GGAACTGGATGGTGATG-GAACGGGCACA *****************-*********** Allele 2 GGAACTGGATGGTGATGTGAACGGGCAC **************************** | Allele 1: deletion of T nucleotide; Allele 2: no indels; Monoallelic mutant. | Positive | Positive | 3 : 1 |
| EC9 | ECWT line GGAACTGGATGGTGATGTGAACGGGCACAC EC9 line GGAACTGGATGGTGATGWRMRSGSRCMM Allele 1 GGAACTGGATGGTGATGTGAACGGGCACAC ****************************** Allele 2 GGAACTGGATGGTGATG--AACGGGCACAC *****************--*********** | Allele 1: no indels; Allele 2: deletion of TG nucleotides Monoallelic mutant. | Positive | Positive | 3 : 1 |
| EC10 | ECWT line GGAACTGGATGGTGATGTGAACGGGCACAC EC10 line GGAACTGGATGGTGATGRAMSGGSACMA Allele 1 GGAACTGGATGGTGATG--AACGGGCACAC *****************--*********** Allele 2 GGAACTGGATGGTGATG-GAACGGGACCAC *****************-************ | Allele 1: deletions of TG nucleotides; Allele 2: deletion of T nucleotide; Biallelic mutant. | Positive | Negative | 3 : 1 |
Inheritance of introduced indels and the absence of off-target mutations in recovered events
Previous studies in other plant species show that the mutations induced by the CRISPR/Cas9 system are stable and heritable as classical Mendelian alleles. Similarly, the mutations present in the T0 edited plants where heritable in the T1 generation and segregated as expected (Supplementary Figure S1A). Because CRISPR/Cas9 can target off-target sequences, we evaluated the specificity of our CRISPR/Cas9 system by sequencing the two genome locations that best matched the gRNA sequence according to in silico predictions (Supplementary Table S1). Sanger sequencing of PCR fragments amplified from these genomic regions showed no mutations in each region, demonstrating the specificity of gRNA targeting to the gfp gene sequence within transgene (Supplementary Figure S1B).
Discussion
The development of genome editing technology over the last 20 years has employed several types of nucleases (Boch et al. 2009; Christian et al. 2010; Durai et al. 2005; Joung and Sander 2013; Kim et al. 1996; Paques and Duchateau 2007; Smith et al. 2006; Streubel et al. 2012), but CRISPR/Cas9 has revolutionized genome editing because of its simplicity, versatility, efficiency, and specificity (Gil-Humanes et al. 2017; Li et al. 2016; Liang et al. 2017; Shan et al. 2014; Yin et al. 2017; Zhang et al. 2016). CRISPR/Cas9 has been used successfully in numerous organisms, including several plant species (Chang et al. 2016; Gao et al. 2015; Gerasimova et al. 2017; Jacobs et al. 2015; Jiang et al. 2013, 2014; Li et al. 2016; Lowder et al. 2015; Michno et al. 2015; Yin et al. 2015; Zhang et al. 2016). However, C4 grasses like maize, sorghum, Brachiaria, switchgrass, and sugarcane have highly complex genomes, which contributes to low efficiency for genome editing (Mohan 2016). Despite the economic importance of C4 crop plants as food crops or biomass for biofuel production, there are still few studies reporting successful genome editing by CRISPR/Cas9 in these plant species (Gerasimova et al. 2017). Also, genetic manipulation of these crops is laborious and time-consuming, as long periods are usually necessary to advance generations, making functional genomic studies difficult (Basso et al. 2019; Basso et al. 2020). The use of model plants is a common alternative for functional genomics studies, as work in these translates to the target C4 crop species (Nguyen et al. 2020; Santos et al. 2020). In this context, S. viridis serves as a model plant for important phylogenetically related C4 crops (Brutnell et al. 2015). Therefore, new resources and investigative tool development for S. viridis represent important advances. Huang et al. (2019) showed that CRISPR/Cas9 efficiently induced mutations in S. viridis to create new mutant alleles.
Here we describe a successful pipeline for S. viridis genome editing. We engineered a CRISPR/Cas9 binary vector to promote NHEJ. This vector coupled with a high-efficiency transformation protocol (Martins et al. 2015) generated ten independent gene editing events, all containing indels in the gfp target gene, in which 60% were biallelic. The efficiency achieved with this system was higher than reported with other approaches like transient expression of CRISPR/Cas9 components (Zhang et al. 2016), viral vectors (Yin et al. 2015), or ribonucleoprotein complexes (Liang et al. 2017; Woo et al. 2015). Also, the efficiency was similar to or higher than DNA-integration-based genome-editing methods used in other monocots (Svitashev et al. 2016). Further, the pipeline describe here was simple, accurate, and highly efficient with indels equivalent to the CRISPR/Cas9_Trex2 system (Weiss et al. 2020). A contributor to the efficiency of editing observed here may be the use of strong constitutive promoters driving Cas9 nuclease and the gRNA, which improves the frequency of DSB due to high expression in transgenic cells at the early stages of dedifferentiation and differentiation (Gil-Humanes et al. 2017; Yang et al. 2017).
No indels occurred in potential off-target sites in the T0 events obtained here, confirming the specificity of the Cas9-gRNA combination, which is in part due to the low level of off-target cleavage when gRNAs are designed to be specific to target sequences (Young et al. 2019). The occurrence of indels in off-target sites and toxicity of strong and prolonged Cas9-gRNA expression in plant and bacteria are major concerns (Johnson et al. 2015; Lowder et al. 2015, 2016; Peng et al. 2016). However, highly target-specific gRNAs might eliminate the need for strong Cas9-gRNA expression (Osakabe et al. 2016). In addition, long term Cas9 cytotoxic effects have not been observed in most plant species. The use of the CRISPR/Cas9_Trex2 system to enhance indels in human cells indicated that Trex2 exonuclease increases frequency of off-target mutations (Chari et al. 2015). Weiss et al. (2020) demonstrated that CRISPR/Cas9_Trex2 system improves indels in S. viridis compared to the canonical CRISPR/Cas9 system, along with enhanced off-target mutations.
The results here showed codon optimized Cas9 nuclease was not cytotoxic for E. coli, A. tumefaciens, and S. viridis cells. Furthermore, indels introduced in S. viridis by this CRISPR/Cas9 system at single or multiple sites were stably inherited from the T0 to T1 generations, indicating Mendelian inheritance similar to that observed in other plant species (Yang et al. 2017; Zhang et al. 2016; Zhao et al. 2016) or even in S. viridis (Weiss et al. 2020). Transgene-free mutant plants can be obtained by self-crossing or backcrossing in the T1 generation from events carrying either homozygous or heterozygous for the CRISPR/Cas9 T-DNA (Zhang et al. 2014). Recently, transgene-free plants were obtained in the T0 generation with new strategies based on transient expression of CRISPR/Cas9 DNA or RNA (Zhang et al. 2016), viral vectors (Gil-Humanes et al. 2017; Yin et al. 2015), and ribonucleoprotein complexes (Liang et al. 2017; Svitashev et al. 2016; Woo et al. 2015).
In summary, we developed an optimized CRISPR/Cas9 binary vector, and this study confirms the efficiency of a pipeline for site-directed editing using this CRISPR/Cas9 system in S. viridis, similar to that shown by Weiss et al. (2020). This work shows that promoting NHEJ pathway activity leads to accurate gene editing in S. viridis, with 60% of the transgenic events in a biallelic state at the T0 generation. This demonstrates the CRISPR/Cas9 NHEJ system is highly efficient in S. viridis. Furthermore, our results showed that a binary vector containing Cas9 nuclease codon-optimized for monocots and flanked by two NLS driven by strong constitutive promoters, and an optimized gRNA sequence driven by specific promoters were highly efficient in generating site-specific and biallelic indels in the S. viridis genome. Currently, our group is using this pipeline for genome editing in the sugarcane genome and we expect it to be effective in other important C4 grass and monocot crops.
Conclusion
In this work, we have successfully established a pipeline using the CRISPR/Cas9 NHEJ system for efficient editing and knock-out of target genes in S. viridis, which utilizes a new CRISPR/Cas9 binary vector, which exploits the NHEJ pathway, that is optimized and engineered for C4 monocots. This pipeline is expected to be useful for reverse genetics studies or to introduce desirable agronomic traits into economically important C4 crops such as maize, Brachiaria, wheat, rice, sorghum, and sugarcane.
Acknowledgments
We are grateful to Embrapa and FAPDF for their financial support. HBCM is grateful to FAPDF from Edital Universal 03/2016, Embrapa from Macroprograma number 03/2017 (13.16.05.026.00.00), and number 02/2017 (02.16.05.022.00.00). MFB is grateful to CNPq for the postdoctoral research fellowship (PDJ 150936/2018-4). WRS is thankful to Fapesp (2019/04878-7) for the financial support. We also thank Dr. Frank G. Harmon to help improve the English writing of the revised manuscript.
Abbreviations
- ATG
Translation initiation site
- cDNA
Complementary DNA
- CIM2
Callus induction medium
- CRISPR/Cas9
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9
- DNA
Deoxyribonucleic acid
- DSD
Degenerate Sequence Decoding
- GFP or gfp
Green fluorescent protein
- gRNA
Guide RNA
- HDR
homology-directed repair
- NHEJ
Non-homologous end joining
- NLS
nuclear localization signal
- NOS
Nopaline synthase
- OsAct-1
Oryza sativa actin 1
- OsU6
Oryza sativa
- PAM
protospacer adjacent motif
- RNA
Ribonucleic acid
- qRT-PCR
Quantitative reverse transcription PCR
- S. viridis
Setaria viridis
- SpCas9WT
Cas9 nuclease from Streptococcus pyogenes
- SV40
Simian vacuolating virus 40
- T-DNA
transfer DNA
- ZmUBI-1
Zea mays
Authors’ contributions
MFB, TRS, and HBCM planned and designed CRISPR/Cas9 NHEJ strategy for gfp transgene editing and knockout in S. viridis accession A10.1; KED and BDBC carried out the S. viridis gfp transformation; MFB carried out the co-transformation of S. viridis using the CRISPR/Cas9 NHEJ binary vector, conducted biological and molecular analyses, wrote the manuscript, and prepared the figures and tables; WRS, BOG, and AKK provided inputs in the manuscript. All authors revised and approved the final version of the manuscript.
Competing interests
All authors declare that the research was carried out in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Consent for publication or Ethics approval and consent to participate
Not applicable.
Supplementary Data
References
- Basso MF, Arraes FBM, Grossi-de-Sa M, Moreira VJV, Alves-Ferreira M, Grossi-de-Sa MF (2020) Insights into genetic and molecular elements for transgenic crop development. Front Plant Sci 11: 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso MF, Ferreira PCG, Kobayashi AK, Harmon FG, Nepomuceno AL, Molinari HBC, Grossi-de-Sa MF (2019) MicroRNAs and new biotechnological tools for its modulation and improving stress tolerance in plants. Plant Biotechnol J 17: 1482–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J, et al. (2012) Reference genome sequence of the model plant Setaria. Nat Biotechnol 30: 555–561 [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brutnell TP, Bennetzen JL, Vogel JP (2015) Brachypodium distachyon and Setaria viridis: Model Genetic Systems for the Grasses. Annu Rev Plant Biol 66: 465–485 [DOI] [PubMed] [Google Scholar]
- Brutnell TP, Wang L, Swartwood K, Goldschmidt A, Jackson D, Zhu X-G, Kellogg E, Van Eck J (2010) Setaria viridis: A model for C4 photosynthesis. Plant Cell 22: 2537–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canteri MG, Althaus RA, Virgens Filho JS, Giglioti EA, Godoy CV (2001) SASM-Agri: Sistema para análise e separação de médias em experimentos agrícolas pelos métodos Scoft-Knott, Tukey e Duncan. Rev Bras Agrocomputação 1: 18–24 [Google Scholar]
- Chang H, Yi B, Ma R, Zhang X, Zhao H, Xi Y (2016) CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep 6: 22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R, Mali P, Moosburner M, Church GM (2015) Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods 12: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF (2010) Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MF (1981) Immunosorbent assays in plant pathology. Annu Rev Phytopathol 19: 83–106 [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19: 11–15 [Google Scholar]
- Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S (2005) Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res 33: 5978–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87: 99–110 [DOI] [PubMed] [Google Scholar]
- Gerasimova SV, Khlestkina EK, Kochetov AV, Shumny VK (2017) Genome editing system CRISPR/Cas9 and peculiarities of its application in monocots. Russ J Plant Physiol 64: 141–155 [Google Scholar]
- Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sánchez-León S, Baltes NJ, Starker C, Barro F, Gao C, et al. (2017) High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J 89: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Mamidi S, Healey A, Grimwood J, Jenkins J, Barry K, Sreedasyam A, Shu S, Feldman M, Wu J, et al. (2019) The Setaria viridis genome and diversity panel enables discovery of a novel domestication gene. /bioRxiv/ doi: https://doi.org/10.1101/744557
- Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Yang B, Weeks DP (2014) Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS One 9: e99225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RA, Gurevich V, Filler S, Samach A, Levy AA (2015) Comparative assessments of CRISPR-Cas nucleases cleavage efficiency in planta. Plant Mol Biol 87: 143–156 [DOI] [PubMed] [Google Scholar]
- Joung JK, Sander JD (2013) TALENs: A widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: Zinc finger fusions to FokI cleavage domain. Proc Natl Acad Sci USA 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C (2016) Gene replacements and insertions in rice by intron targeting using CRISPR/Cas9. Nat Plants 2: 16139. [DOI] [PubMed] [Google Scholar]
- Li P, Brutnell TP (2011) Setaria viridis and Setaria italica, model genetic systems for the Panicoid grasses. J Exp Bot 62: 3031–3037 [DOI] [PubMed] [Google Scholar]
- Liang Z, Chen K, Li T, Zhang Y, Wang Y, Zhao Q, Liu J, Zhang H, Liu C, Ran Y, et al. (2017) Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun 8: 14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder L, Malzahn A, Qi Y (2016) Rapid evolution of manifold CRISPR systems for plant genome editing. Front Plant Sci 7: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, Paul JW 3rd, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169: 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Chen L, Zhu Q, Chen Y, Liu Y-G (2015) Rapid decoding of sequence-specific nuclease-induced heterozygous and biallelic mutations by direct sequencing of PCR products. Mol Plant 8: 1285–1287 [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, et al. (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13: 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins PK, Mafra V, de Souza WR, Ribeiro AP, Vinecky F, Basso MF, Cunha BADB, Kobayashi AK, Molinari HBC (2016) Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci Rep 6: 28348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins PK, Ribeiro AP, da Cunha BADB, Kobayashi AK, Molinari HBC (2015) A simple and highly efficient Agrobacterium-mediated transformation protocol for Setaria viridis. Biotechnol Rep (Amst) 6: 41–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May AP, Haurwitz RE, Doudna JA, Berger JM, Carter MM, Donohoue P (2013) Compositions and methods of nucleic acid-targeting nucleic acids. Patent WO2014150624A1.
- Michno JM, Wang X, Liu J, Curtin SJ, Kono TJ, Stupar RM (2015) CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6: 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan C (2016) Genome editing in sugarcane: Challenges ahead. Front Plant Sci 7: 1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DQ, Van Eck J, Eamens AL, Grof CPL (2020) Robust and reproducible Agrobacterium-mediated transformation system of the C4 genetic model species Setaria viridis. Front Plant Sci 11: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien A, Bailey TL (2014) GT-Scan: Identifying unique genomic targets. Bioinformatics 30: 2673–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Watanabe T, Sugano SS, Ueta R, Ishihara R, Shinozaki K, Osakabe K (2016) Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci Rep 6: 26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Duchateau P (2007) Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr Gene Ther 7: 49–66 [DOI] [PubMed] [Google Scholar]
- Peng R, Lin G, Li J (2016) Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J 283: 1218–1231 [DOI] [PubMed] [Google Scholar]
- Santos CM, Romeiro D, Silva JP, Basso MF, Molinari HBC, Centeno DC (2020) An improved protocol for efficient transformation and regeneration of Setaria italica. Plant Cell Rep 39: 501–510 [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9: 2395–2410 [DOI] [PubMed] [Google Scholar]
- Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. (2006) A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res 34: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL (2015) CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10: e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel J, Blucher C, Landgraf A, Boch J (2012) TAL effector RVD specificities and efficiencies. Nat Biotechnol 30: 593–595 [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia L (2016) Engineering Herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol Plant 9: 628–631 [DOI] [PubMed] [Google Scholar]
- Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016) Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7: 13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, et al. (2017) A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3: 17018. [DOI] [PubMed] [Google Scholar]
- Weiss T, Wang C, Kang X, Zhao H, Elena Gamo M, Starker CG, Crisp PA, Zhou P, Springer NM, Voytas DF, et al. (2020) Optimization of multiplexed CRISPR/Cas9 system for highly efficient genome editing in Setaria viridis. Plant J 104: 828–838 [DOI] [PubMed] [Google Scholar]
- Woo JW, Kim J, Kwon SI, Corvalán C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Yang H, Wu J-J, Tang T, Liu K-D, Dai C (2017) CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci Rep 7: 7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K, Gao C, Qiu J-L (2017) Progress and prospects in plant genome editing. Nat Plants 3: 17107. [DOI] [PubMed] [Google Scholar]
- Yin K, Han T, Liu G, Chen T, Wang Y, Yu AYL, Liu Y (2015) A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5: 14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Zastrow-Hayes G, Deschamps S, Svitashev S, Zaremba M, Acharya A, Paulraj S, Peterson-Burch B, Schwartz C, Djukanovic V, et al. (2019) CRISPR-Cas9 editing in maize: Systematic evaluation of off-target activity and its relevance in crop improvement. Sci Rep 9: 6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F (2013) CRISPR-Cas systems and methods for altering expression of gene products. Patent: US8697359B1.
- Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, et al. (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12: 797–807 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liang Z, Zong Y, Wang Y, Liu J, Chen K, Qiu J-L, Gao C (2016) Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun 7: 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang C, Liu W, Gao W, Liu W, Song G, Li WX, Mao L, Chen B, Xu Y, et al. (2016) An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci Rep 6: 23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.