Abstract
Uridine 5′-diphosphate (UDP)-glucose dehydrogenase (UGD) produces UDP-glucuronic acid from UDP-glucose as a precursor of plant cell wall polysaccharides. UDP-glucuronic acid is also a sugar donor for the glycosylation of various plant specialized metabolites. Nevertheless, the roles of UGDs in plant specialized metabolism remain poorly understood. Glycyrrhiza species (licorice), which are medicinal legumes, biosynthesize triterpenoid saponins, soyasaponins and glycyrrhizin, commonly glucuronosylated at the C-3 position of the triterpenoid scaffold. Often, several different UGD isoforms are present in plants. To gain insight into potential functional differences among UGD isoforms in triterpenoid saponin biosynthesis in relation to cell wall component biosynthesis, we identified and characterized Glycyrrhiza uralensis UGDs (GuUGDs), which were discovered to comprise five isoforms, four of which (GuUGD1–4) showed UGD activity in vitro. GuUGD1–4 had different biochemical properties, including their affinity for UDP-glucose, catalytic constant, and sensitivity to feedback inhibitors. GuUGD2 had the highest catalytic constant and highest gene expression level among the GuUGDs, suggesting that it is the major isoform contributing to the transition from UDP-glucose to UDP-glucuronic acid in planta. To evaluate the contribution of GuUGD isoforms to saponin biosynthesis, we compared the expression patterns of GuUGDs with those of saponin biosynthetic genes in methyl jasmonate (MeJA)-treated cultured stolons. GuUGD1–4 showed delayed responses to MeJA compared to those of saponin biosynthetic genes, suggesting that MeJA-responsive expression of GuUGDs compensates for the decreased UDP-glucuronic acid pool due to consumption during saponin biosynthesis.
Keywords: Glycyrrhiza uralensis, triterpenoid saponin, UDP-glucose dehydrogenase, UDP-glucuronic acid
Introduction
Plant cells have cell walls that consist mainly of cellulose, hemicellulose, and pectin. Unlike cellulose, which consists only of glucose (Delmer and Amor 1995), hemicellulose and pectin contain various sugar components, including glucose, galactose, rhamnose, glucuronic acid, galacturonic acid, xylose, arabinose, apiose, mannose, and fucose (Caffall and Mohnen 2009; Ebringerová et al. 2005; Mohnen 2008). The monomeric precursors of the sugars in these polysaccharides are two sugar nucleotides: uridine 5′-diphosphate (UDP) sugars and guanosine 5′-diphosphate (GDP) sugars. UDP sugars are commonly synthesized from UDP-glucose, which is available from photosynthesis assimilates. UDP-xylose, UDP-arabinose, UDP-apiose, and UDP-galacturonic acid are derived from UDP-glucuronic acid (Feingold 1982; Seifert 2004), which is synthesized from UDP-glucose via irreversible two-step oxidation with no release of intermediates by UDP-glucose dehydrogenase (UGD; Ge et al. 2004). UGD activity is often the lowest in extracts containing pathway enzymes and is inferred as rate limiting in the synthesis of cell wall precursors (Amino et al. 1985; Dalessandro and Northcote 1977). Many studies have examined the biological roles of UGD in the biosynthesis of cell wall polysaccharides. One biological study suggested the importance of UGDs in the sugar nucleotide oxidation pathway and polysaccharide synthesis based on the reductions in arabinose, xylose, and apiose in the cell wall of ugd2,3 double mutant Arabidopsis thaliana plants, which exhibit a dwarf phenotype (Reboul et al. 2011). Moreover, a reduced sugar composition was detected only in UGD-A mutants, not in UGD-B mutants, in Zea mays, suggesting that UGD-A has a more important role than UGD-B in UDP-glucuronic acid synthesis (Kärkönen et al. 2005). Several UGD isoforms are present in plants, although comparative biochemical and biological studies of these isoforms have been limited to Z. mays and A. thaliana (Kärkönen et al. 2005; Klinghammer and Tenhaken 2007; Reboul et al. 2011). Plants have an alternative pathway for the biosynthesis of UDP-glucuronic acid that involves inositol oxygenase (Kanter et al. 2005; Kotake et al. 2007; Loewus et al. 1962; Seitz et al. 2000). In this pathway, UDP-glucuronic acid is synthesized from α-D-glucuronic acid 1-phosphate by UDP-sugar pyrophosphorylase (USP). Unlike UGDs, generally one USP isoform is present in plants.
Apart from the biosynthesis of cell wall polysaccharides, UDP-glucuronic acid is also used as a sugar donor for the glycosylation of various plant specialized metabolites, including flavonoids and triterpenoids. Nevertheless, the contribution of UGDs in plant specialized metabolisms has not been well studied.
Legumes, the third largest family of angiosperms, biosynthesize various glucuronosylated metabolites. Soyasaponins, which are representative plant specialized metabolites of legumes, are classified as oleanane-type triterpenoid saponins and are commonly glucuronosylated at the C-3 position of the triterpenoid scaffold. Moreover, Glycyrrhiza species (licorice), which are major medicinal legumes, characteristically accumulate glycyrrhizin, another oleanane-type triterpenoid saponin with a characteristic sugar chain composed of two glucuronic acids, in their roots and stolons (Hayashi and Sudo 2009). This unique sugar moiety of glycyrrhizin, which is 150 times sweeter than sucrose (Kitagawa et al. 1993), contributes sweetness (Esaki et al. 1978; Kitagawa 2002). Hence, licorice seems to be a good target for studying the contribution of UGD to the supply of UDP-glucuronic acid for triterpenoid saponin biosynthesis.
Most enzymes that catalyze the biosynthesis of soyasaponins or glycyrrhizin have been characterized in licorice. Both soyasaponins and glycyrrhizin are derived from β-amyrin, one of the most commonly occurring triterpenes in plants, which is biosynthesized from 2,3-oxidosqualene by β-amyrin synthase (bAS; Augustin et al. 2011; Hayashi et al. 2001; Thimmappa et al. 2014). In soyasaponin biosynthesis, the two cytochrome P450 monooxygenases (P450) CYP93E3 and CYP72A566 catalyze oxidation reactions at the C-24 and C-22β positions of β-amyrin, respectively, to produce soyasapogenol B as an aglycone part of soyasaponin I and II (Seki et al. 2008; Tamura et al. 2018). Soyasaponin I and II are produced via three glycosylation steps, in which first glucuronic acid, then galactose or arabinose, and finally rhamnose are attached. In glycyrrhizin biosynthesis, the P450s CYP88D6 and CYP72A154 catalyze oxidation reactions at the C-11 and C-30 positions of β-amyrin, respectively, producing glycyrrhetinic acid (Seki et al. 2008, 2011). Finally, two-step glucuronosylation of glycyrrhetinic acid at the C-3 hydroxy group by two glucuronosyltransferases, a cellulose synthase-like enzyme GuCSyGT (GuCsl) and UGT73P12, produces glycyrrhizin via glycyrrhetinic acid 3-O-monoglucuronide (Chung et al. 2020; Jozwiak et al. 2020; Nomura et al. 2019). However, no enzyme producing UDP-glucuronic acid has been identified in licorice.
To investigate the possible functional differentiation between UGD isoforms in specialized metabolisms in relation to cell wall component biosynthesis, UGDs should be identified and characterized individually. Therefore, in this study, we identified and characterized Glycyrrhiza uralensis UGDs (GuUGDs), which were discovered to have five isoforms. We analyzed the biochemical properties of each GuUGD isoform and examined their individual roles in the transition from UDP-glucose to UDP-glucuronic acid. We also explored the possible contribution of each GuUGD isoform to the sugar donor supply for triterpenoid saponin biosynthesis by comparing gene expression patterns between GuUGDs and known saponin biosynthetic genes.
Materials and methods
Plant materials
Roots of G. uralensis strain 308-19 harvested in June 2011 (Ramilowski et al. 2013) were used for RNA extraction to isolate UGD genes. Tissue-cultured stolons of G. uralensis (Hokkaido-iryodai strain) were maintained in Murashige and Skoog (MS) medium (Duchefa Biochemie, Haarlem, The Netherlands) supplemented with 6% sucrose and 0.01 mM 1-naphthaleneacetic acid (NAA), as reported previously (Kojoma et al. 2010). RNA was extracted from the roots of 3-week-old Medicago truncatula accession R108 grown in plant chambers under a 16-h-light/8-h-dark photoperiod at 24°C.
Database searches and isolation of UGD genes
We identified GuUGD unigene sequences with blastx similarity search against 32,840 protein coding sequences derived from G. uralensis transcriptome data (Ramilowski et al. 2013, http://ngs-data-archive.psc.riken.jp/Gur/index.pl) using the A. thaliana UGD1 gene sequence, previously isolated and characterized by Oka and Jigami (2006), as a query. To obtain the full-length coding sequences (CDSs) of six unigenes containing partial CDSs, we performed rapid amplification of cDNA ends polymerase chain reaction (RACE-PCR) using the SMARTer RACE cDNA Amplification Kit (Clontech/Takara Bio, Shiga, Japan). The full-length CDSs of GuUGDs were PCR-amplified from the first-strand cDNA library prepared from roots of G. uralensis strain 308-19 with primers 1–10 (Supplementary Table S1) and PrimeSTAR Max DNA Polymerase (Takara Bio), then cloned into pENTR/DTOPO (Thermo Fisher Scientific, Waltham, MA, USA) to produce an entry clone. After sequencing, each gene was assigned a G. uralensis gene ID (Supplementary Table S2) through a similarity search against the G. uralensis genome database (http://ngs-data-archive.psc.riken.jp/Gur-genome/index.pl). The nucleotide sequences isolated in this study have been submitted to the DNA Data Bank of Japan (DDBJ) under accession numbers LC528155 (GuUGD1), LC528156 (GuUGD2), LC528157 (GuUGD3), LC528158 (GuUGD4), and LC528159 (GuUGD5).
We identified putative M. truncatula UGD (MtUGD) protein sequences in a blastp similarity search against GenBank (http://www.ncbi.nlm.nih.gov/genbank/) using the isolated GuUGD1–5 sequences as queries. The corresponding genes (MtUGD1–3) were isolated from cDNA derived from M. truncatula root RNA using primers 22–33 (Supplementary Table S3).
Recombinant protein expression in Escherichia coli
Full-length CDSs encoding the GuUGD and MtUGD isoforms and A. thaliana UGD2 (used as a positive control) were cloned from entry clones into the SacI and SalI sites of the His-tag expression vector pCold I DNA (TaKaRa Bio) with the In-Fusion HD cloning kit (TaKaRa Bio) and primers 11–21 (Supplementary Table S1). The coding sequence of Arabidopsis UGD2 was amplified from the cDNA clone RAFL09-33-I02 provided by the RIKEN BRC through the national Bio-Resource Project of the MEXT, Japan. The plasmid was introduced into E. coli strain BL21 Star (DE3; Invitrogen, Carlsbad, CA, USA) with the chaperone plasmid pGro7 (TaKaRa Bio). To obtain recombinant proteins, we grew bacterial cultures at 37°C to an optical density at 600 nm of approximately 0.4–0.8 in LB medium supplemented with 0.5 mg ml−1 L-arabinose, 20 µg ml−1 chloramphenicol, and 100 µg ml−1 ampicillin. After cooling the cultures for 30 min at 15°C, we induced protein expression by adding 500 µM isopropyl β-D-thiogalactopyranoside (IPTG). After 1 day of cultivation at 15°C, cells were cooled by being shaken on ice, collected by centrifugation (4°C, 10 min, 4,500×g), and frozen in liquid nitrogen.
Purification of the enzyme
The frozen cells were resuspended in chilled disruption buffer (10 mM Tris, 50 mM sodium phosphate, 2 mM MgCl2, 10% [v/v] glycerol, 1 mM NAD+, 2 mM 2-mercaptoethanol, and 1× cOmplete Protease Inhibitor Cocktail [Roche, Basel, Switzerland]; pH 8.0) at 10 ml g−1 fresh weight (FW). Lysozyme (200 µg ml−1) and Nonidet P-40 (1%, v/v) were added to the suspension, and the mixture was shaken slowly at 4°C for 30 min. Then 2.4 U ml−1 benzonase nuclease (Sigma-Aldrich, St. Louis, MO, USA) was added, followed by incubation for 15 min with slow shaking at 4°C. Bacterial debris was removed by centrifugation (10 min, 9,500×g), and the clear supernatant was applied to a TALON metal affinity resin column (Clontech) equilibrated with purification buffer (10 mM Tris, 50 mM sodium phosphate, 300 mM NaCl, and 10% glycerol; pH 8.0) after the addition of 250 mM NaCl. Extracts were purified with the batch/gravity-flow method. The column was washed twice with 10 volumes of purification buffer to wash out all unbound proteins. Weakly bound proteins were eluted from the TALON column with five volumes of purification buffer containing 10 mM imidazole. The UGD enzyme was eluted in purification buffer containing 100 mM imidazole and immediately concentrated in storage buffer (20 mM Tris, 50 mM KCl, 0.1 mM EDTA, 10% [v/v] glycerol, and 0.5 mM NAD+; pH 8.0) by ultrafiltration on an Amicon Ultra 100 k device (Millipore, Burlington, MA, USA). The recombinant enzyme was stored at −80°C. His-tagged UGD was detected by western blotting with Anti-His-tag mAb-HRP-DirecT (Medical & Biological Laboratories, Aichi, Japan) and Chemi-Lumi One Super (Nacalai Tesque, Kyoto, Japan). The quality of the purified UGDs was verified by Coomassie Brilliant Blue (CBB) staining and UV detection after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The concentrations of the purified UGDs were calculated from the intensity of the corresponding band in the CBB-stained gel.
Enzyme assays
Enzyme activity of UGDs was analyzed by two approaches: analysis of reaction products with ultra-performance liquid chromatography–mass spectrometry (UPLC–MS); or analysis of the initial velocity by photometrical monitoring of the time-dependent increase in NADH converted from the cofactor NAD+, monitored at 340 nm with the Infinite 200 PRO multimode microplate reader (Tecan, Männedorf, Switzerland). All reactions were performed at 25°C in 100 µl of assay buffer (20 mM Tris, 1 mM EDTA, and 10% [v/v] glycerol; pH 8.7) following the addition of enzyme that had been preincubated for 90 min in storage buffer at room temperature. UDP-glucuronic acid production was confirmed by UPLC–MS using 1 µg of each isoform in a reaction solution containing 2 mM UDP-glucose and 600 µM NAD+ after determination of the optimal condition. To determine the optimal pH, 1 µg of GuUGD1–4 was analyzed at pH 4.0, 6.0, 8.0, and 10.0 to roughly determine the optimal pH range; then, GuUGD4 was analyzed using smaller increments between pH 7.1 and 10.6. We used glycine buffer for pH range 9.0–10.6 instead of the Tris buffer.
We evaluated the substrate specificity of each isoform by comparing the initial velocity with a standard enzyme assay containing 500 µM UDP-glucose and NAD+ as substrate and cofactor, respectively. The amounts of the enzymes were adjusted so that the conversion rate was in the range of 0.01 to 0.1 µM s−1 in the standard enzyme assay. GuUGD5, for which the conversion rate could not be calculated, was used in this assay in the same amount as GuUGD4 (20-fold larger volumes than GuUGD4 solution). All sugar nucleotides and cofactors were used at a concentration of 500 µM.
To determine kinetic parameters, we used sufficient concentrations of NAD+ (1 mM) and various concentrations of UDP-glucose (0–2,000 µM), or various NAD+ concentrations (5–1,305 µM) and a constant UDP-glucose concentration (2 mM). The reaction was performed three times using 1 µg of GuUGD1–4, and we calculated kinetic parameters from the nonlinear least squares based on the Gauss-Newton method using Python.
We evaluated the inhibitor sensitivity by comparing the initial velocity to the standard enzyme assay. The standard enzyme assay for inhibitor sensitivity was performed without an inhibitor, using UDP-glucose at twice the concentration of each Km calculated in the kinetic analyses. The amounts of the enzymes were adjusted so that the conversion rate was in the range of 0.01 to 0.1 µM s−1 without inhibitors.
Reaction product analyses using UPLC–MS
Reaction products were analyzed with an ACQUITY Ultra-Performance LC system with a tandem quadruple detector (Waters, Milford, MA, USA) using hydrophilic interaction chromatography on an ACQUITY UPLC BEH Amide column (1.7 µm, 2.1×150 mm; Waters) maintained at 75°C; 5 µl of each sample was injected for analyses. The mobile phase consisted of 50 : 50 acetonitrile/water with 10 mM triethylamine acetate. The flow rate was set at 0.4 ml min−1. Compounds were ionized by electrospray ionization in negative ion mode. Mass spectra were recorded in the range of 100–750 m/z. The settings of the mass spectrometer were as follows: capillary voltage, +3.5 keV; cone voltage, 45 V; source temperature, 150°C; desolvation temperature, 450°C; cone gas flow, 50 l h−1; and desolvation gas flow, 450 l h−1. MassLynx (ver. 4.1; Waters) was used for data acquisition and analyses. We identified peaks by comparing their retention times with that of authentic standard UDP-glucuronic acid (Nacalai Tesque).
3D Modeling
The human UGD homo-hexamer structure (Beattie et al. 2018) was used as a template. 3D-structure models of GuUGDs were constructed using the automated SWISS-MODEL server at ExPASy (https://swissmodel.expasy.org/; Benkert et al. 2011; Bertoni et al. 2017; Bienert et al. 2017; Guex et al. 2009; Waterhouse et al. 2018) based on protein homology. The predicted 3D structures of the GuUGDs were analyzed via superimposition on the X-ray crystal structure of human UGD with RasMol ver. 2.7.5.2.
Treatment of tissue-cultured stolons with abscisic acid
Tissue-cultured stolons were cultured for 2 weeks in MS medium supplemented with 6% sucrose without NAA before treatment. For abiotic stress treatments in time-course quantitative real-time PCR (qPCR) analyses, we diluted abscisic acid (ABA; BD Biosciences, San Jose, CA, USA) with ethanol to make 10 mM stock solutions, and then added 1 ml of stock solutions to 100 ml of medium (final concentration, 100 µM). Ethanol (1 ml) was added to 100 ml of medium as a mock treatment.
RNA extraction and first-strand cDNA synthesis
Total RNA was extracted from frozen tissue-cultured stolons with PureLink Plant RNA Reagent (Thermo Fisher Scientific), then treated with recombinant DNase I (RNase-free; Takara Bio) and purified with the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol. First-strand cDNA was synthesized with PrimeScript RT Master Mix (Perfect Real Time; Takara Bio) from 2.5 µg total RNA in a 50-µl reaction.
qPCR
We mixed 1× FastStart Essential DNA Green Master (Roche), 500 nM primers, and 0.5 µl of cDNA, and the reaction volume was brought to 10 µl with PCR-grade water. cDNA for the MeJA treatment analyses was synthesized from total RNA extracted from tissue-cultured stolons treated with 100 µM MeJA or 0.1% ethanol (mock for MeJA), as described previously (Tamura et al. 2017). Reactions were performed with a LightCycler Nano (Roche) at 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 15 s. The data were analyzed with LightCycler Nano ver. 1.1.0 (Roche). We calculated the relative transcript levels of each target gene using β-tubulin (Seki et al. 2008; GenBank accession number LC318135) as a reference gene. Each sample was amplified three times with primers 34–57 (Supplementary Table S4) designed using the Primer3 website (http://bioinfo.ut.ee/primer3-0.4.0/; Koressaar and Remm 2007; Untergasser et al. 2012).
Bioinformatics and phylogenetic analyses
We retrieved homologous plant UGD protein sequences from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) by blastp search using the isolated GuUGD1–5 sequences as queries. Sequences containing irregularly long N- or C- termini were removed. Phylogenetic trees were generated by the maximum likelihood method based on the JTT matrix-based model (Jones et al. 1992) after alignment by ClustalW in MEGA X (Kumar et al. 2018). Homologous UGD genes were retrieved as reference sequences of each homologous protein. Genes located upstream and downstream of each UGD homolog in G. uralensis and other plants were retrieved from the G. uralensis genome database (http://ngs-data-archive.psc.riken.jp/Gur-genome/index.pl) and GenBank, respectively.
Gene expression analyses of Fabales UGDs
Gene expression data for UGD based on fragments per kilobase of exon per million mapped reads (FPKM; G. uralensis), reads per kilobase of exon per million mapped sequence reads (RPKM; Glycine max), or normalized expression in microarrays (M. truncatula) were retrieved from public transcriptome data for each putative UGD gene. Normalized data derived from RNA extracted from roots and leaves of G. uralensis collected in summer (G. uralensis database; http://ngs-data-archive.psc.riken.jp/Gur/index.pl, Ramilowski et al. 2013), from roots and young leaves of G. max (SoyBase; https://www.soybase.org/, Severin et al. 2010), and from roots and leaves of M. truncatula (Mt Gene Expression Atlas; https://mtgea.noble.org/v3/, Benedito et al. 2008; He et al. 2009) were analyzed for UGD expression. Because GuUGD3, GuUGD4, and GuUGD5 each corresponded to two partial unigene sequences (Supplementary Table S2), their FPKM values were recalculated as follows: expression of GuUGD=(raw reads of unigene I+raw reads of unigene II)/(raw reads of unigene I/FPKM of unigene I+raw reads of unigene II/FPKM of unigene II). The number correspondence between GenBank and each database is shown in Supplementary Table S5.
Results
Identification of UDP-glucose dehydrogenases in G. uralensis
To identify UGDs in G. uralensis, we performed a blastx search against 32,840 protein coding sequences derived from G. uralensis transcriptome data (Ramilowski et al. 2013) using the A. thaliana UGD1 gene sequence as a query. We found eight unigene sequences, including two complete and six partial open reading frames (ORFs), encoding putative GuUGDs. RACE-PCR amplification revealed that the six partial sequences were derived from three genes. Thus, complete ORFs of five putative GuUGDs (referred to as GuUGD1–5) were obtained and assigned to five independent gene IDs, located on different scaffolds, in the G. uralensis genome database (Mochida et al. 2017; Supplementary Table S2). The deduced amino acid sequences of GuUGD1–5 showed high amino acid sequence identity (79–86%) to A. thaliana UGD1 (Table 1). Furthermore, very high identities were found among GuUGD1–5 (81–93%; Table 1).
Table 1. Amino acid sequence identity among GuUGD1–5.
| GuUGD2 | GuUGD3 | GuUGD4 | GuUGD5 | AtUGD1 | |
|---|---|---|---|---|---|
| GuUGD1 | 89% | 88% | 90% | 81% | 86% |
| GuUGD2 | 93% | 85% | 83% | 84% | |
| GuUGD3 | 86% | 83% | 83% | ||
| GuUGD4 | 81% | 83% | |||
| GuUGD5 | 79% |
In vitro enzyme activity of recombinant UGDs
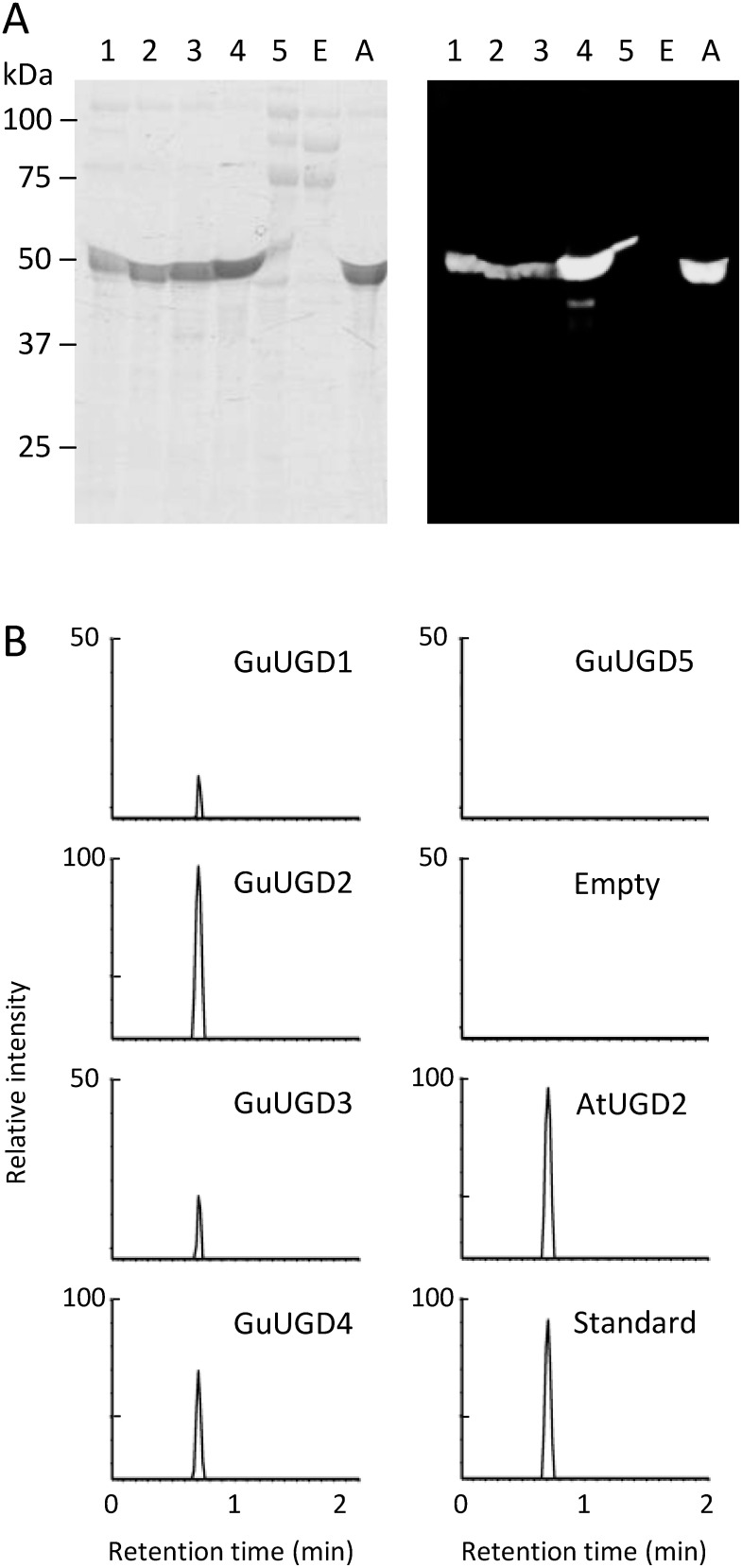
We successfully expressed GuUGDs in E. coli by cloning the ORF of each GuUGD into a cold-shock expression vector together with GroES/EL chaperones. The purified GuUGDs showed a protein band at approximately 50 kDa in a UV-illuminated SDS-PAGE gel and by His-tag detection in a western blotting gel (Figure 1A). The expression of recombinant GuUGD5 was much lower than that of the others, so GuUGD5 was purified at a low concentration and low purity (Figure 1A). The concentration of each purified GuUGD was calculated from the intensity of the corresponding band in CBB-stained gels, and UGD activity was analyzed using the same amount of purified GuUGD. Enzyme activity was detected using two approaches: detecting reaction products by UPLC–MS and photometrical monitoring of the time-dependent increases in NADH that was converted from the cofactor NAD+. Generally, the optimal pH of plant UGDs is 8.0–9.0 (Davies and Dickinson 1972; Hinterberg et al. 2002; Stewart and Copeland 1998; Strominger and Mapson 1957; Turner and Botha 2002). Consistently, increases in NADH were detected at pH 8.0 and 10.0, but not at pH 4.0 or 6.0 using GuUGD1–4 in preliminary experiments. Further analysis revealed that the UGD enzyme activity level was high at pH 8.5–9.0 (Supplementary Figure S1). Because of the difficulty in obtaining large amounts of purified GuUGD5, we could not perform optimization for GuUGD5; thus, GuUGD5 was analyzed under the optimal conditions for the other GuUGDs.
Figure 1. UGD activity of purified UGD recombinant proteins. (A) Electrophoresis of purified recombinant proteins. Purified His-tagged recombinant proteins were detected in a UV-stained SDS-PAGE gel (left panel) and by anti-His detection in a western blotting membrane transferred from an identical gel (right panel). UGDs were detected as ∼50 kDa. Lanes 1–5: purified recombinant GuUGD1–5 proteins (10, 10, 7.5, 5, and 15 µl of eluted GuUGD1, GuUGD2, GuUGD3, GuUGD4, and GuUGD5 solution were loaded, respectively); E: protein solution removed non-interactive protein for the TALON resin, extracted from IPTG-induced E. coli transformed with empty vector (10 µl of protein solution was loaded); A: purified recombinant AtUGD2 protein (7.5 µl of protein solution was loaded). (B) UPLC–MS chromatograms at m/z 579.1 for the in vitro reaction products. All reaction products were diluted 10 times after 1 day of incubation using 1 µg of each purified UGD (protein solution extracted from E. coli transformed with empty vector was used at the same volume used for UGD-containing protein solutions). 100% corresponds to the intensity indicated in products catalyzed by AtUGD2, which was used as a positive control. UDP-glucuronic acid was used as an authentic standard.
As shown in Figure 1B, with the coexistence of UDP-glucose as a substrate and NAD+ as a cofactor, GuUGD1–4 and AtUGD2 (positive control) yielded a single reaction product that showed the same m/z (579.1) and retention time (0.74 min) as the authentic UDP-glucuronic acid standard. This peak was not detected in GuUGD5 products or the empty vector control (Figure 1B). Consistent with this result, an increase in NADH was detected in the reactions containing GuUGD1–4 in the photometrical analyses, whereas it was not detected in the GuUGD5-containing reaction (Table 2). Some plant UGDs accept other sugar nucleotides as minor substrates and NADP+ as a minor cofactor with weak catalytic activity (0.5–23% of the main activity; Klinghammer and Tenhaken 2007; Stewart and Copeland 1998; Turner and Botha 2002). Hence, we analyzed enzyme activity for other substrates (UDP-galactose, UDP-N-acetylglucosamine, TDP-glucose, ADP-glucose, and GDP-glucose) in the presence of either NAD+ or NADP+. In the presence of NAD+, only GuUGD4 accepted UDP-galactose and UDP-N-acetylglucosamine as minor substrates with 15% and 10% activity compared to UDP-glucose, respectively (Table 2). By contrast, GuUGD5 did not show enzyme activity for any substrate. None of the GuUGDs used NADP+ as a cofactor instead of NAD+.
Table 2. Substrate specificity of GuUGDs.
| Nucleotide-sugar | Relative enzyme activity of isoforms (%) | ||||
|---|---|---|---|---|---|
| GuUGD1 | GuUGD2 | GuUGD3 | GuUGD4 | GuUGD5 | |
| UDP-glucose | 100 | 100 | 100 | 100 | n.d. |
| UDP-galactose | n.d. | n.d. | n.d. | 15 | n.d. |
| UDP-N-acetylglucosamine | n.d. | n.d. | n.d. | 10 | n.d. |
We calculated enzyme activity based on initial velocity within 10 min measured by monitoring the conversion of NAD(P)+ to NAD(P)H, detected by the absorbance at 340 nm. 100% corresponds to the activity indicated in the standard assay with UDP-glucose as a substrate and NAD+ as a cofactor. n.d., not detected.
To obtain deeper insight into the biochemical properties of each active GuUGD isoform, we analyzed kinetic parameters for UDP-glucose and NAD+. GuUGD1–4 exhibited typical hyperbolic reaction kinetics and were not inhibited by higher substrate concentrations (Supplementary Figure S2). Affinity for the substrate UDP-glucose and the cofactor NAD+ differed among the GuUGD isoforms (Table 3). GuUGD4 showed the highest affinity for UDP-glucose, with approximately less than one-fifth of the Km of the other active GuUGD isoforms. The highest catalytic constant was detected in GuUGD2, with more than twice the kcat of the other active GuUGD isoforms.
Table 3. Kinetic parameters of GuUGDs.
| Isoform | Kinetics for UDP-glucose | Kinetics for NAD+ | ||||
|---|---|---|---|---|---|---|
| Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | Km (µM) | kcat (s−1) | kcat/Km (s−1 µM−1) | |
| GuUGD1 | 261±84 | 0.09±0.01 | 0.0003 | 31±5 | 0.10±0.01 | 0.0031 |
| GuUGD2 | 285±54 | 0.89±0.09 | 0.0031 | 153±29 | 1.01±0.12 | 0.0066 |
| GuUGD3 | 570±46 | 0.39±0.01 | 0.0007 | 62±7 | 0.40±0.02 | 0.0064 |
| GuUGD4 | 55±5 | 0.25±0.02 | 0.0045 | 122±15 | 0.36±0.01 | 0.0029 |
We calculated enzyme activity based on initial velocity within 5 min measured by monitoring the conversion of NAD+ to NADH, detected by the absorbance at 340 nm. Values represent the mean±SE of three measurements.
Fine tuning of UGD activity is mediated by feedback inhibition of the enzyme by UDP-xylose, a product obtained from UDP-glucuronic acid after decarboxylation by UDP-xylose synthase (Hinterberg et al. 2002; Neufeld and Hall 1965; Turner and Botha 2002). The enzyme activity of all GuUGD isoforms was strongly inhibited and could scarcely be detected in the presence of 500 µM UDP-xylose (Table 4). The inhibitory effect differed at low UDP-xylose concentrations. GuUGD4 was the most sensitively inhibited by UDP-xylose at 20 µM, while GuUGD3 was not inhibited. Unlike UDP-xylose, UDP-glucuronic acid showed very weak inhibitory effects, as seen in other plants (Davies and Dickinson 1972; Oka and Jigami 2006; Turner and Botha 2002). Only GuUGD4 was sensitive to UDP-glucuronic acid. GuUGD1, GuUGD2, and GuUGD3 were not inhibited, even by high concentrations of UDP-glucuronic acid.
Table 4. Inhibitory effects of UDP-xylose and UDP-glucuronic acid on GuUGD1–4.
| Inhibitor | Relative enzyme activity of isoforms (%) | |||
|---|---|---|---|---|
| GuUGD1 | GuUGD2 | GuUGD3 | GuUGD4 | |
| None | 100 | 100 | 100 | 100 |
| UDP-xylose (20 µM) | 80 | 106 | 92 | 63 |
| UDP-xylose (500 µM) | n.d. | n.d. | n.d. | 7 |
| UDP-glucuronic acid (20 µM) | 104 | 111 | 109 | 89 |
| UDP-glucuronic acid (1250 µM) | 95 | 103 | 109 | 34 |
Enzyme activity was calculated based on initial velocity within 10 min by monitoring the conversion of NAD+ to NADH, detected by the absorbance at 340 nm. All inhibitors were added just before the reaction was initiated. The concentration of the substrate UDP-glucose was twice the concentration of each Km calculated in the kinetic analyses. 100% corresponds to the activity indicated in the standard assay performed without an inhibitor. n.d., not detected.
Molecular modeling and structural analyses of UGDs
In general, UGD amino acid sequences share striking identity among plants, animals, and microorganisms: amino acid sequences of A. thaliana UGD1 have 85% identity to G. max UGD, 58% identity to human UGD, and 22% identity to Streptococcus pyogenes UGD. As shown in Table 1, the five GuUGD isoforms showed very high amino acid sequence identities (81–93%) to one another; however, GuUGD5 showed no enzyme activity in this study. To explore the key amino acid residues affecting the activity of GuUGDs, we performed amino acid sequence comparison of the GuUGD isoforms. The five GuUGDs, comprising 480–482 amino acids, contained the full-length functional catalytic motifs (NAD+ binding motif, UDP binding motif, and central motif) and several residues were found to be specific to GuUGD5 among the five GuUGDs (Figure 2A). Several residues have been proposed as important residues for catalysis (shown in asterisks); some residues were proposed based on bond distances in native and mutant S. pyogenes UGD (Campbell et al. 2000), and some residues were proposed based on residue occupancy within the binding site and comparative kinetic analyses of wild-type and mutated human UGD (Egger et al. 2011). Several residues in human UGD were also identified as key residues that have hydrophobic interactions, hydrogen bonds, salt bridges, and water bridges with UDP-glucose (shown in circles) and NAD+ (shown in squares) in SWISS-MODEL. Among the residues specific to GuUGD5, Lys257 replaced arginine, which positions a key residue with a hydrogen bond in human UGD, with UDP-glucose in SWISS-MODEL (red box, Figure 2A). To confirm that this residue is located near UDP-glucose in the GuUGDs, we constructed homology-based structural models of the GuUGDs using human UGD (Beattie et al. 2018) as a template. The 3D structural models of GuUGD2 and GuUGD5, which displayed high sequence identity (59.4–61.5%) to human UGD (Supplementary Table S6), were predicted by SWISS-MODEL using human UGD as template. The 3D structural models of GuUGD1, GuUGD3, and GuUGD4 could not be predicted with this crystal structure in SWISS-MODEL and thus were predicted with the constructed 3D structure of GuUGD2 as a template. Superimposition analyses of the crystal structure of human UGD with the GuUGD models showed a highly conserved structure in all enzymes (Figure 2B). Compared to GuUGD1–4, there were fewer nitrogen atoms of Lys257 in GuUGD5 and they were farther from the oxygen atom of UDP-glucose, suggesting that this variation decreased the attraction between GuUGD5 and UDP-glucose and thus the affinity for UDP-glucose. Among the residues specific to GuUGD5, no other residue was located near the substrate and cofactor.
Figure 2. Amino acid sequences and predicted molecular structures of GuUGDs. (A) Alignment of the amino acid sequences deduced from cDNA of cloned GuUGDs. We made the alignment using BioEdit with ClustalW. The NAD+ binding motif, UDP binding motif, and central motif were annotated based on GenBank. Asterisks indicate key residues for catalysis proposed in previous studies (Campbell et al. 2000; Egger et al. 2011); circles and squares indicate amino acids shown as key residues interacting with UDP-glucose and NAD+, respectively, by SWISS-MODEL. Amino acid residues shown in yellow background were specific to GuUGD5. The amino acid residue framed in red was near UDP-glucose in the 3D protein models. (B) The 3D protein model of GuUGDs built with human UGD homo-hexamer structure as a template in SWISS-MODEL. Squares indicate the residue position that likely prevented UGD activity in GuUGD5; this position is indicated by a yellow circle in the enlarged panel (right). Atoms of oxygen, nitrogen, carbon, and phosphorous are indicated in red, blue, gray, and orange, respectively. Distances between substrates and the crystal structure of human UGD are displayed in the enlarged panel.
Expression patterns of GuUGD genes
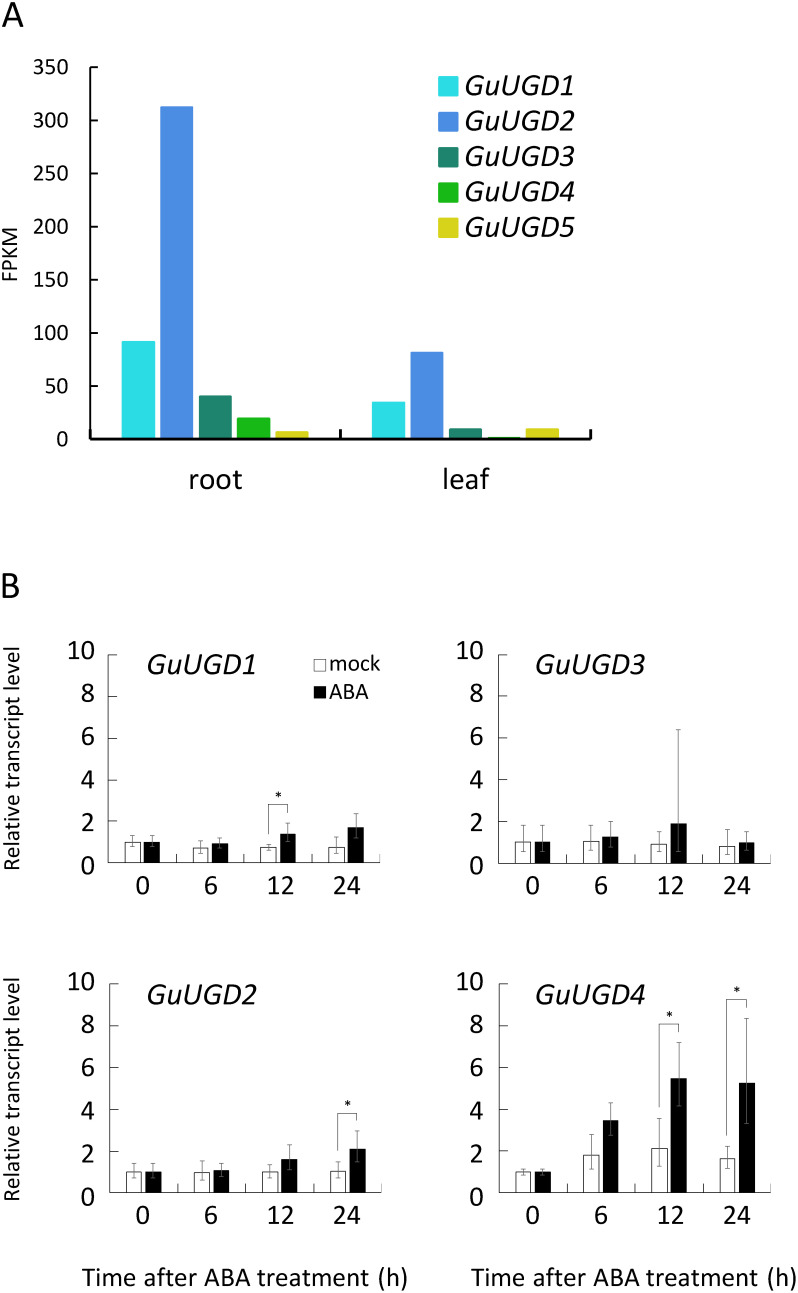
The gene expression of each GuUGD isoform was compared based on FPKM values obtained in RNA-Seq analyses of G. uralensis plants (Ramilowski et al. 2013). As shown in Figure 3A, GuUGD2 had the highest expression among all GuUGD genes in both roots and leaves. GuUGD1 showed moderate expression, whereas GuUGD3 and GuUGD4 showed weaker expression. GuUGD5 was expressed weakly in both roots and leaves.
Figure 3. Expression patterns of GuUGD genes. (A) Tissue-specific expression of GuUGDs. Expression levels of GuUGDs are based on FPKM values obtained in RNA sequencing (RNA-Seq) analyses of G. uralensis plants (Ramilowski et al. 2013). (B) Quantitative polymerase chain reaction (qPCR) analyses of GuUGDs in abscisic acid (ABA)-treated cultured stolons. Transcript levels in tissue-cultured stolons treated with ABA are indicated by filled bars; outlined bars indicate the mock treatment. Relative transcript levels 0 h after treatment were set equal to 1. Error bars indicate the standard deviation (SD) of three biological replicates. Asterisks indicate significant differences in the gene expression level between the ABA and mock treatments at each time point. Student’s t test, *** p<0.001, ** p<0.01, * p<0.05.
UGD expression is upregulated in leaves of Populus tomentosa under various stressful conditions, including ABA treatment (Tian et al. 2014). In G. uralensis, the accumulation of glycyrrhizin was promoted by exogenous ABA (Qiao et al. 2017). To obtain additional insight into the biological roles of each active GuUGD isoform, we analyzed the gene expression responses to ABA treatment in tissue-cultured stolons. Only GuUGD4 showed a strong expression response to ABA treatment (Figure 3B). GuUGD1 and GuUGD2 showed quite small responses to ABA treatment, while GuUGD3 was barely affected (Figure 3B). GuUGD5 did not differ significantly between ABA and mock treatments (data not shown).
Transcriptional response of GuUGD genes to MeJA compared to saponin biosynthetic genes
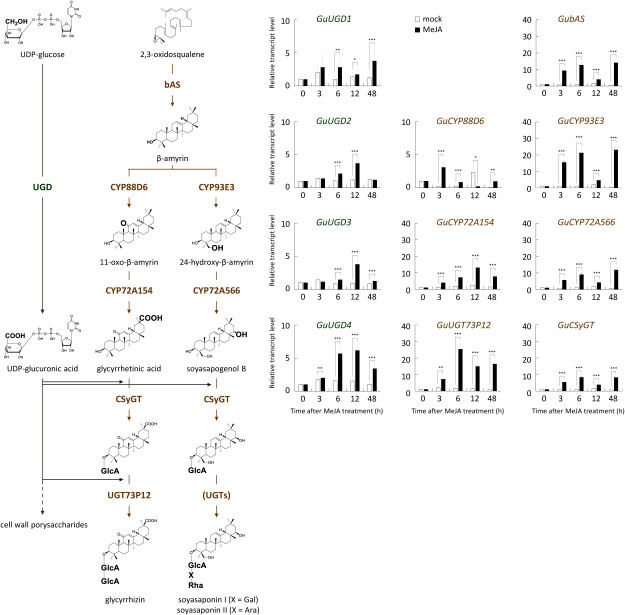
The biosynthesis of glycyrrhizin in the roots and hairy roots of Glycyrrhiza species is upregulated following the application of exogenous MeJA (Shabani et al. 2009; Wongwicha et al. 2011). Similarly, the biosynthesis of soyasaponin responds to MeJA treatment in cultured cells and tissue-cultured stolons (Hayashi et al. 2003; Tamura et al. 2018). To evaluate the possible involvement of each active GuUGD isoform in glycyrrhizin and soyasaponin biosynthesis, we analyzed the effects of MeJA on the expression of GuUGD1–4, as well as on known saponin biosynthetic genes, in tissue-cultured stolons of G. uralensis. GuUGD1–4 expression was induced after MeJA treatment (Figure 4). The expression of GuUGD genes was strongly enhanced after 6 h (peak times 12 or 48 h), with the greatest enhancement seen in GuUGD4. However, the upregulation of GuUGD gene expression was late compared to that of the glucuronosyltransferase genes CSyGT and UGT73P12. CSyGT and UGT73P12 expression were already upregulated at 3 h (peak time 6 h). Soyasaponin biosynthetic genes (bAS, CYP93E3, CYP72A566 and CSyGT) showed very similar expression responses to one another, with strong upregulation after 3 h. By contrast, glycyrrhizin biosynthetic genes (bAS, CYP88D6, CYP72A154, CSyGT, and UGT73P12) showed different response times to MeJA treatment. Upregulation of CYP88D6 expression immediately subsided at 6 h, although the enhanced bAS gene expression continued to strengthen, and the enhancement of CYP72A154 gene expression did not reach a peak by this time.
Figure 4. MeJA-responsive expression of GuUGDs and saponin biosynthetic genes. Biosynthetic pathways of glycyrrhizin and soyasaponin were predicted in previous studies (Seki et al. 2008, 2011). Transcript levels were analyzed by qPCR. Transcript levels in the mock treatment tissue-cultured stolons are indicated by outlined bars; MeJA treatments are indicated by filled bars. Relative transcript levels 0 h after treatment were set equal to 1. Error bars indicate the SD of three technical replicates. Asterisks indicate significant differences in gene expression level between the MeJA and mock treatments at each time point. Student’s t test, *** p<0.001, ** p<0.01, * p<0.05. Part of this figure was reproduced and modified from Plant and Cell Physiology 59(4) with permission from Oxford University Press.
Discussion
We identified four active UGD isoforms (GuUGD1–4) in G. uralensis. Another isoform (GuUGD5) was also analyzed; however, it showed no enzyme activity for any substrate tested in this study. The affinity of GuUGD1–4 for UDP-glucose and NAD+ was profiled as Km values of 55–570 µM and 31–153 µM, respectively, which is consistent with UGDs characterized in other plants: the affinity of recombinant UGDs has been reported as Km values of 15–335 µM and 67–70 µM for UDP-glucose and NAD+, respectively, in soybean (Hinterberg et al. 2002), A. thaliana (Klinghammer and Tenhaken 2007; Oka and Jigami 2006), and Eucalyptus grandis (Labate et al. 2010), and the affinity of UGDs purified from plant tissues has been reported as Km values of 19–950 µM and 72–400 µM for UDP-glucose and NAD+, respectively, in pea (Strominger and Mapson 1957), lily (Davies and Dickinson 1972), wheat (Stewart and Copeland 1998), canola (Stewart and Copeland 1998), soybean (Stewart and Copeland 1998), sugarcane (Turner and Botha 2002), and maize (Kärkönen et al. 2005). The calculated catalytic constants of GuUGD1–4 were 0.1–1.0 s−1, which is lower than those of other plants: catalytic activity of UGD has been reported as kcat values of 1.17–2.52 s−1 in A. thaliana recombinant UGDs (Klinghammer and Tenhaken 2007) or Vmax values of 2.17 and 68–172 µmol min−1 mg−1 in sugarcane UGD purified from plant tissues (Turner and Botha 2002) and E. grandis recombinant UGD (Labate et al. 2010; it can be calculated as 1.8–143 s−1 because UGDs were reported as ∼50 kDa), respectively. GuUGD2 had the highest catalytic constant (1.0 s−1), similar to that of AtUGDs (1.17–2.52 s−1), with 2- to 10-fold higher kcat values than the other active GuUGD isoforms (Table 3). GuUGD2 also had the highest gene expression level among the GuUGDs (Figure 3A), which suggests that GuUGD2 is the major isoform contributing to the transition from UDP-glucose to UDP-glucuronic acid in planta.
GuUGD4 showed the highest affinity for UDP-glucose, which suggests that GuUGD4 can produce UDP-glucuronic acid under various conditions, including UDP-glucose deficiency. Meanwhile, GuUGD4 showed the highest sensitivity for feedback inhibition to 20 µM UDP-xylose and UDP-glucuronic acid, which suggests its inactivation under conditions in which abundant products are present. Due to the properties of GuUGD4, we would expect a minimum transition from UDP-glucose into UDP-glucuronic acid to be maintained by continuous catalysis by GuUGD4 unless it is inhibited by accumulated products. The exact concentration of UDP-glucose in G. uralensis is unknown; however, the concentration of UDP-glucose can be estimated to range from 15 µM to 3.5 mM for the following reasons. UDP-glucose has been detected in amounts of 0.007–1.6 nmol mg−1 FW in several plants (Hayashi and Matsuda 1981; Macrae et al. 1992; Morrell and Rees 1986; Rees et al. 1984; Schlüpmann et al. 1994). In addition, the concentration and amount of UDP-glucose have been calculated as 3.5 mM and 1.6 nmol mg−1, respectively, in pollen tubes of Nicotiana alata (Schlüpmann et al. 1994). If we assume that both the amount and correspondence between the concentration and amount of UDP-glucose are almost the same in all cells, 0.007 nmol mg−1 and 1.6 nmol mg−1 of UDP-glucose can be calculated as 15 µM and 3.5 mM, respectively. Based on the concentration of UDP-glucose, the concentration of UDP-xylose can be assumed as 0.3–70 µM, because the amount of UDP-xylose was calculated as approximately one fiftieth of the amount of UDP-glucose in soybean cells (Hayashi and Matsuda 1981). Based on these observations, GuUGD4 is possibly inhibited by UDP-xylose in cells, while other GuUGDs are likely inhibited weakly. The ranges of the assumed concentrations of UDP-glucose and UDP-xylose cover the kinetic parameters of GuUGDs; thus, it can be inferred that total UGD activity can be regulated by UDP-glucose and UDP-xylose in planta via differences in the concentration-dependent activity of each GuUGD isoform.
Aside from its enzymatic properties, the total UGD activity can also be regulated by the differential expression of UGD genes. Among the GuUGDs, GuUGD4 was most strongly upregulated by ABA and MeJA, whereas the other GuUGDs, including the putative major contributing isoform GuUGD2, responded weakly to ABA or MeJA treatment compared to GuUGD4. This strong response to stress hormones suggests a large contribution by GuUGD4 to the transition from UDP-glucose to UDP-glucuronic acid under these stressful conditions.
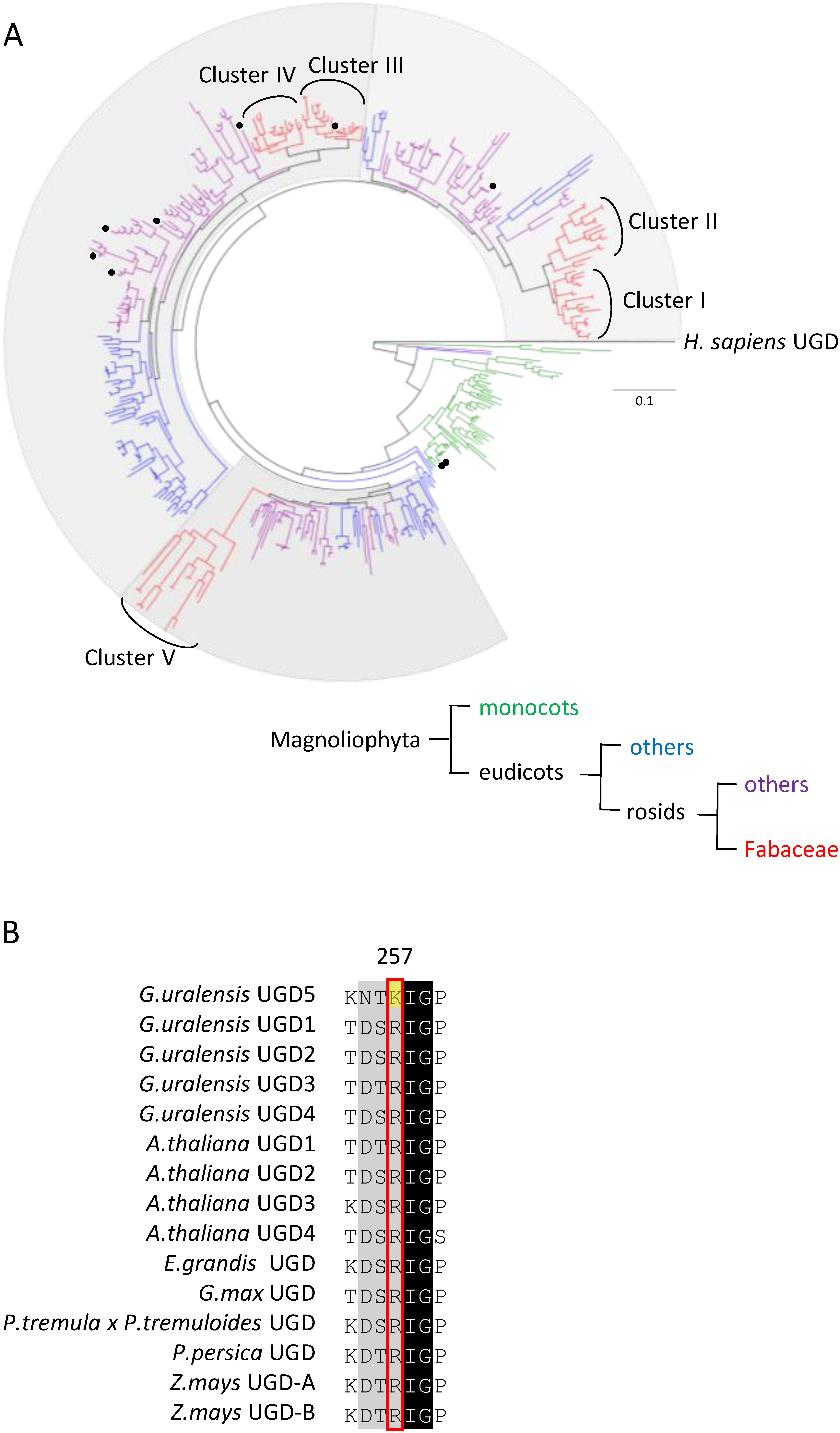
Plants often contain several UGD isoforms, probably generated via gene duplication. To reveal the molecular evolution of plant UGDs, we constructed a phylogenetic tree with various plant UGDs. As shown in Figure 5A, the phylogenetic tree contains three clades (gray backgrounds) composed of evenly assigned eudicot UGDs. No monocot UGDs belonged to these clades. This phylogenetic tree suggests that a gene duplication event occurred in the common ancestor of eudicots. In addition, sister clades of Fabales UGDs (clusters I & II and clusters III & IV) suggest an additional gene duplication in the common ancestor of Fabales. Consistently, many legumes have five types of UGDs with conserved synteny (Supplementary Figure S3). Additionally, similar tissue-specific gene expression was observed for UGDs belonging to the same clusters (Figure 3A, Supplementary Figure S4). These observations suggest that characteristic biological functions are maintained within each cluster.
Figure 5. Phylogenetic tree and alignment of amino acid sequences of GuUGDs and characterized plant UGDs. (A) Maximum likelihood phylogenetic tree of UGD homologous proteins from various plants, rooted on Homo sapiens UGD as the outgroup. The scale measures evolutionary distance in substitutions per amino acid. Protein sequences were retrieved from GenBank by blastp search with the GuUGD sequences as queries. Black circles on the tree indicate previously characterized UGDs. Monocots are shown in green. Eudicots except for rosids are shown in blue. Rosids except for Fabaceae are shown in purple. Fabaceae are shown in red. Fabaceae UGD homologous proteins are separated into five clusters. (B) Alignment of amino acid sequences of GuUGDs and characterized plant UGDs. We made the alignment using BioEdit with ClustalW. The amino acid residue shown with the yellow background is specific to GuUGD5.
In the enzyme assays, GuUGD5 did not show UGD activity. Because of the low concentration and low purity of recombinant GuUGD5 due to difficulties expressing GuUGD5 in E. coli at levels as high as with GuUGD1–4 (Figure 1A), we used a large volume of protein solution to add the same amount of GuUGD5 as the other GuUGDs to the reaction mixture. Therefore, GuUGD5 might not have shown UGD activity because contaminating endogenous E. coli proteins prevented GuUGD5 activity. Other possible reasons are that GuUGD5 has no or very weak UGD activity lower than the detection limit. To obtain deeper insight into the biochemical properties of legume UGD cluster I–V, we analyzed MtUGDs. Four MtUGD sequences were found in a blastp search, and three of them were isolated (Supplementary Tables S5, S7). MtUGD1, MtUGD2, and MtUGD3 belong to clusters II, III, and V, respectively (Supplementary Figure S3). The MtUGDs were successfully expressed in E. coli and purified (Supplementary Figure S5A). In the enzyme assays, the highest peak intensity of UDP-glucuronic acid was seen with MtUGD2, which belongs to cluster III with GuUGD2 (Supplementary Figure S5B). On the other hand, the very small UDP-glucuronic acid peak detected in the MtUGD3 product suggests that UGDs belonging to cluster V have the weakest UGD activity in the five Fabales UGD clusters.
Among five GuUGDs, the amino acid substitution from Arg257 to Lys257, which likely decreases attraction to UDP-glucose, was found only in GuUGD5, while Arg257 was conserved in all characterized UGDs (Figure 5B). Interestingly, Lys257 was conserved in all cluster V UGDs (Supplementary Figure S6). Considered with the results of the enzyme assay, UGD activity might be weakened by the substitution from Arg257 to Lys257. Likewise, several amino acids were conserved in GuUGD and MtUGD belonging to the same cluster (Supplementary Figure S7). Cluster-specific biochemical properties are likely derived from these cluster-specific amino acids. To reveal the relation between biochemical properties and amino acid residues, further analysis is needed.
Soyasaponin biosynthetic genes, which are collectively regulated by the transcription factor GubHLH3 (Tamura et al. 2018), showed very similar expression patterns under MeJA treatment in tissue-cultured stolons. Expression of the GuUGD1–4 genes was enhanced under MeJA treatment, although the enhancement of GuUGD expression was delayed compared to that of soyasaponin and glycyrrhizin biosynthetic genes. These expression patterns seem to reflect the fact that GuUGD gene expression is regulated separately from that of triterpenoid saponin biosynthetic genes. We infer that MeJA-responsive expression of GuUGDs compensates for the decreased UDP-glucuronic acid pool due to consumption in saponin biosynthesis rather than prior or concomitant supply of UDP-glucuronic acid. Expression of a gene encoding a protein homologous to USP, which produces UDP-glucuronic acid from α-D-glucuronic acid 1-phosphate in the myoinositol pathway, was not upregulated ahead of the enhancement of glucuronosyltransferase gene expression (Supplementary Figure S8), which suggests that the alternative pathway was not used to supply UDP-glucuronic acid for saponin biosynthesis.
Overall, the results of this study suggest that no GuUGD isoforms exhibit functional differentiation that is specific to saponin biosynthesis. Genes in triterpene pathways are organized in operon-like clusters in some plant genomes to facilitate co-regulation of biosynthetic genes (Field and Osbourn 2008; Field et al. 2011; Kliebenstein and Osbourn 2012; Krokida et al. 2013; Nützmann and Osbourn 2014; Qi et al. 2004). However, no genes related to triterpenoid saponin biosynthesis were found around legume UGD genes (Supplementary Figure S3), which supports the idea that a UGD isoform specific to saponin biosynthesis did not form.
Acknowledgments
We are very grateful to the Takeda Garden for Medicinal Plant Conservation, Kyoto, Japan (Takeda Pharmaceutical Company Ltd.) for providing G. uralensis 308-19 strain plants. We are also very grateful to Dr. Kojoma (Health Sciences University of Hokkaido) for providing tissue-cultured stolons of G. uralensis Hokkaido-iryodai strain. We thank Mr. Naoki Chiyo for technical advice and critical reading of the manuscript, and Ms. Keiko Fukamoto (Osaka University) for technical assistance.
Abbreviations
- ABA
abscisic acid
- ADP
adenosine 5′-diphosphate
- bAS
β-amyrin synthase
- CDSs
coding sequences
- FPKM
fragments per kilobase of exon per million mapped reads
- GDP
guanosine 5′-diphosphate
- MeJA
methyl jasmonate
- NAD+
nicotinamide adenine dinucleotide
- NADP+
nicotinamide adenine dinucleotide phosphate
- ORFs
open reading frames
- qPCR
quantitative real-time PCR
- RPKM
reads per kilobase of exon per million mapped sequence reads
- TDP
thymidine 5′-diphosphate
- UDP
uridine 5′-diphosphate
- UGD
UDP-glucose dehydrogenase
- USP
UDP-sugar pyrophosphorylase
Supplementary Data
References
- Amino S, Takeuchi Y, Komamine A (1985) Changes in enzyme activities involved in formation and interconversion of UDP-segars during the cell cycle in a synchronous culture of Catharanthus roseus. Physiol Plant 64: 111–117 [Google Scholar]
- Augustin JM, Kuzina V, Andersen SB, Bak S (2011) Molecular activities, biosynthesis and evolution of triterpenoid saponins. Phytochemistry 72: 435–457 [DOI] [PubMed] [Google Scholar]
- Beattie NR, Pioso BJ, Sidlo AM, Keul ND, Wood ZA (2018) Hysteresis and allostery in human UDP-glucose dehydrogenase require a flexible protein core. Biochemistry 57: 6848–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27: 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T (2017) Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci Rep 7: 10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert S, Waterhouse A, de Beer TAP, Tauriello G, Studer G, Bordoli L, Schwede T (2017) The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res 45(D1): D313–D319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900 [DOI] [PubMed] [Google Scholar]
- Campbell RE, Mosimann SC, Rijn IVD, Tanner ME, Strynadka NCJ (2000) The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation. Biochemistry 39: 7012–7023 [PubMed] [Google Scholar]
- Chung SY, Seki H, Fujisawa Y, Shimoda Y, Hiraga S, Nomura Y, Saito K, Ishimoto M, Muranaka T (2020) A cellulose synthase-derived enzyme catalyses 3-O-glucuronosylation in saponin biosynthesis. Nat Commun 11: 5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalessandro G, Northcote DH (1977) Possible control sites of polysaccharide synthesis during cell growth and wall expansion of pea seedlings (Pisum sativum L.). Planta 134: 39–44 [DOI] [PubMed] [Google Scholar]
- Davies MD, Dickinson DB (1972) Properties of uridine diphosphoglucose dehydrogenase from pollen of Lilium longiflorum. Arch Biochem Biophys 152: 53–61 [DOI] [PubMed] [Google Scholar]
- Delmer DP, Amor Y (1995) Cellulose biosynthesis. Plant Cell 7: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebringerová A, Hromádková Z, Heinze T (2005) Hemicellulose. In: Heinze T (ed) Polysaccharides I. Advances in Polymer Science, vol 186. Springer, Berlin, Heidelberg, pp 1–67
- Egger S, Chaikuad A, Kavanagh KL, Oppermann U, Nidetzky B (2011) Structure and mechanism of human UDP-glucose 6-dehydrogenase. J Biol Chem 286: 23877–23887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki S, Konishi F, Kamiya S (1978) Synthesis and taste of some glycosides of glycyrrhetic acid. Agric Biol Chem 42: 1599–1600 [Google Scholar]
- Feingold DS (1982) Aldo (and keto) hexoses and uronic acids. In: Loewus FA, Tanner W (eds) Plant Carbohydrates I. Encyclopedia of Plant Physiology (New Series), vol 13/A. Springer, Berlin, Heidelberg, pp 3–76
- Field B, Fiston-Lavier A-S, Kemen A, Geisler K, Quesneville H, Osbourn AE (2011) Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc Natl Acad Sci USA 108: 16116–16121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Osbourn AE (2008) Metabolic diversification: Independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Ge X, Penney LC, Rijn IVD, Tanner ME (2004) Active site residues and mechanism of UDP-glucose dehydrogenase. Eur J Biochem 271: 14–22 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 30(Suppl 1): S162–S173 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Huang P, Inoue K (2003) Up-regulation of soyasaponin biosynthesis by methyl jasmonate in cultured cells of Glycyrrhiza glabra. Plant Cell Physiol 44: 404–411 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Huang P, Kirakosyan A, Inoue K, Hiraoka N, Ikeshiro Y, Kushiro T, Shibuya M, Ebizuka Y (2001) Cloning and characterization of a cDNA encoding β-amyrin synthase involved in glycyrrhizin and soyasaponin biosyntheses in licorice. Biol Pharm Bull 24: 912–916 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Sudo H (2009) Economic importance of licorice. Plant Biotechnol 26: 101–104 [Google Scholar]
- Hayashi T, Matsuda K (1981) Sugar nucleotides from suspension-cultures soybean cells. Agric Biol Chem 45: 2907–2908 [Google Scholar]
- He J, Benedito VA, Wang M, Murray JD, Zhao PX, Tang Y, Udvardi MK (2009) The Medicago truncatula gene expression atlas web server. BMC Bioinformatics 10: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinterberg B, Klos C, Tenhaken R (2002) Recombinant UDP-glucose dehydrogenase from soybean. Plant Physiol Biochem 40: 1011–1017 [Google Scholar]
- Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Jozwiak A, Sonawane PD, Panda S, Garagounis C, Papadopoulou KK, Abebie B, Massalha H, Almekias-Siegl E, Scherf T, Aharoni A (2020) Plant terpenoid metabolism co-opts a component of the cell wall biosynthesis machinery. Nat Chem Biol 16: 740–748 [DOI] [PubMed] [Google Scholar]
- Kanter U, Usadel B, Guerineau F, Li Y, Pauly M, Tenhaken R (2005) The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 221: 243–254 [DOI] [PubMed] [Google Scholar]
- Kärkönen A, Murigneux A, Martinant J-P, Pepey E, Tatout C, Dudley BJ, Fry SC (2005) UDP-glucose dehydrogenases of maize: A role in cell wall pentose biosynthesis. Biochem J 391: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa I (2002) Licorice root. A natural sweetener and an important ingredient in Chinese medicine. Pure Appl Chem 74: 1189–1198 [Google Scholar]
- Kitagawa I, Hori K, Sakagami M, Hashiuchi F, Yoshikawa M, Ren J (1993) Saponin and sapogenol. Xlix. On the constitutents of the roots of Glycyrrhiza inflata BATALIN from Xinjiang, China. Characterization of two sweet oleanane-type triterpene oligoglycosides, apioglycyrrhizin and araboglycyrrhizin. Chem Pharm Bull 41: 1350–1357 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Osbourn A (2012) Making new molecules—evolution of pathways for novel metabolites in plants. Curr Opin Plant Biol 15: 415–423 [DOI] [PubMed] [Google Scholar]
- Klinghammer M, Tenhaken R (2007) Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J Exp Bot 58: 3609–3621 [DOI] [PubMed] [Google Scholar]
- Kojoma M, Ohyama K, Seki H, Hiraoka Y, Asazu SN, Sawa S, Sekizaki H, Yoshida S, Muranaka T (2010) In vitro proliferation and triterpenoid characteristics of licorice (Glycyrrhiza uralensis Fischer, Leguminosae) stolons. Plant Biotechnol 27: 59–66 [Google Scholar]
- Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23: 1289–1291 [DOI] [PubMed] [Google Scholar]
- Kotake T, Hojo S, Yamaguchi D, Aohara T, Konishi T, Tsumuraya Y (2007) Properties and physiological functions of UDP-sugar pyrophosphorylase in Arabidopsis. Biosci Biotechnol Biochem 71: 761–771 [DOI] [PubMed] [Google Scholar]
- Krokida A, Delis C, Geisler K, Garagounis C, Tsikou D, Peña-Rodríguez LM, Katsarou D, Field B, Osbourn AE, Papadopoulou KK (2013) A metabolic gene cluster in Lotus japonicus discloses novel enzyme functions and products in triterpene biosynthesis. New Phytol 200: 675–690 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labate MTV, Bertolo ALF, do Nascumento DD, Gutmanis G, De Andrade A, Rodrigues MJC, Camargo ELO, Boaretto LF, Moon DH, Bragatto J, et al. (2010) Cloning and endogenous expression of a Eucalyptus grandis UDP-glucose dehydrogenase cDNA. Genet Mol Biol 33: 686–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA, Kelly S, Neufeld EF (1962) Metabolism of myo-inositol in plants: Conversion to pectin, hemicellulose, D-xylose, and sugar acids. Proc Natl Acad Sci USA 48: 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae E, Quick WP, Benker C, Stitt M (1992) Carbohydrate metabolism during postharvest ripening in kiwifruit. Planta 188: 314–323 [DOI] [PubMed] [Google Scholar]
- Mochida K, Sakurai T, Seki H, Yoshida T, Takahagi K, Sawai S, Uchiyama H, Muranaka T, Saito K (2017) Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J 89: 181–194 [DOI] [PubMed] [Google Scholar]
- Mohnen D (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277 [DOI] [PubMed] [Google Scholar]
- Morrell S, Rees T (1986) Sugar metabolism in developing tubers of Solanum tuberosum. Phytochemistry 25: 1579–1585 [Google Scholar]
- Neufeld EF, Hall CW (1965) Inhibition of UDP-D-glucose dehydrogenase by UDP-D-xylose: A possible regulatory mechanism. Biochem Biophys Res Commun 19: 456–461 [DOI] [PubMed] [Google Scholar]
- Nomura Y, Seki H, Suzuki T, Ohyama K, Mizutani M, Kaku T, Tamura K, Ono E, Horikawa M, Sudo H, et al. (2019) Functional specialization of UDP-glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J 99: 1127–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H-W, Osbourn A (2014) Gene clustering in plant specialized metabolism. Curr Opin Biotechnol 26: 91–99 [DOI] [PubMed] [Google Scholar]
- Oka T, Jigami Y (2006) Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae. FEBS J 273: 2645–2657 [DOI] [PubMed] [Google Scholar]
- Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A (2004) A gene cluster for secondary metabolism in oat : Implications for the evolution of metabolic diversity in plants. Proc Natl Acad Sci USA 101: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao J, Luo Z, Li Y, Ren G, Liu C, Ma X (2017) Effect of abscisic acid on accumulation of five active components in root of Glycyrrhiza uralensis. Molecules 22: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Sawai S, Seki H, Mochida K, Yoshida T, Sakurai T, Muranaka T, Saito K, Daub CO (2013) Glycyrrhiza uralensis transcriptome landscape and study of phytochemicals. Plant Cell Physiol 54: 697–710 [DOI] [PubMed] [Google Scholar]
- Reboul R, Geserick C, Pabst M, Frey B, Wittmann D, Lütz-Meindl U, Léonard R, Tenhaken R (2011) Down-regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. J Biol Chem 286: 39982–39992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees TA, Leja M, Macdonald FD, Green JH (1984) Nucleotide sugars and starch synthesis in spadix of Arum macula tum and suspension cultures of Glycine max. Phytochemistry 23: 2463–2468 [Google Scholar]
- Schlüpmann H, Bacic A, Read SM (1994) Uridine diphosphate clucose metabolism and callose synthesis in cultured pollen tubes of Nicotiana alata Link et Otto. Plant Physiol 105: 659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ (2004) Nucleotide sugar interconversions and cell wall biosynthesis: How to bring the inside to the outside. Curr Opin Plant Biol 7: 277–284 [DOI] [PubMed] [Google Scholar]
- Seitz B, Klos C, Wurm M, Tenhaken R (2000) Matrix polysaccharide precursors in Arabidopsis cell walls are synthesized by alternate pathways with organ-specific expression patterns. Plant J 21: 537–546 [DOI] [PubMed] [Google Scholar]
- Seki H, Ohyama K, Sawai S, Mizutani M, Ohnishi T, Sudo H, Akashi T, Aoki T, Saito K, Muranaka T (2008) Licorice β-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc Natl Acad Sci USA 105: 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki H, Sawai S, Ohyama K, Mizutani M, Ohnishi T, Sudo H, Fukushima EO, Akashi T, Aoki T, Saito K, et al. (2011) Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 23: 4112–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin AJ, Woody JL, Bolon Y-T, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. (2010) RNA-Seq atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol 10: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabani L, Ehsanpour AA, Asghari G, Emami J (2009) Glycyrrhizin production by in vitro cultured Glycyrrhiza glabra elicited by methyl jasmonate and salicylic acid. Russ J Plant Physiol 56: 621–626 [Google Scholar]
- Stewart DC, Copeland L (1998) Uridine 5′-diphosphate-glucose dehydrogenase from soybean nodules. Plant Physiol 116: 349–355 [Google Scholar]
- Strominger JL, Mapson LW (1957) Uridine diphosphoglucose dehydrogenase of pea seedlings. Biochem J 66: 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Seki H, Suzuki H, Kojoma M, Saito K, Muranaka T (2017) CYP716A179 functions as a triterpene C-28 oxidase in tissue-cultured stolons of Glycyrrhiza uralensis. Plant Cell Rep 36: 437–445 [DOI] [PubMed] [Google Scholar]
- Tamura K, Yoshida K, Hiraoka Y, Sakaguchi D, Chikugo A, Mochida K, Kojoma M, Mitsuda N, Saito K, Muranaka T, et al. (2018) The basic helix-loop-helix transcription factor GubHLH3 positively regulates soyasaponin biosynthetic genes in Glycyrrhiza uralensis. Plant Cell Physiol 59: 783–796 [DOI] [PubMed] [Google Scholar]
- Thimmappa R, Geisler K, Louveau T, Maille PO, Osbourn A (2014) Triterpene biosynthesis in plants. Annu Rev Plant Biol 65: 225–257 [DOI] [PubMed] [Google Scholar]
- Tian J, Du Q, Li B, Zhang D (2014) Single-nucleotide polymorphisms in the 5′UTR of UDP-glucose dehydrogenase (PtUGDH) associate with wood properties in Populus tomentosa. Tree Genet Genomes 10: 339–354 [Google Scholar]
- Turner W, Botha FC (2002) Purification and kinetic properties of UDP-glucose dehydrogenase from sugarcane. Arch Biochem Biophys 407: 209–216 [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3: New capabilities and interfaces. Nucleic Acids Res 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018) SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1): W296–W303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongwicha W, Tanaka H, Shoyama Y, Putalun W (2011) Methyl jasmonate elicitation enhances glycyrrhizin production in Glycyrrhiza inflata hairy roots cultures. Z. Naturforsch 66: 423–428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.