Abstract
Wild cyclamen (Cyclamen purpurascens) is considered as a precious breeding material for the development of new cultivars. Malvidin 3,5-diglucoside is the main anthocyanin in the petals of C. purpurascens, whereas the F1 progeny of the C. persicum × C. purpurascens cultivars cross contains 3,5-diglucoside-type anthocyanins as the main pigment. The anthocyanin 5-O-glucosyltransferase (A5GT) enzyme is responsible for the glycosylation of the A ring of anthocyanin at the 5-O-position, which implies that the expression of A5GT is dominant in the petals of C. purpurascens × C. persicum cultivars. Here, we isolated the complete open reading frame of the A5GT gene from C. purpurascens (Cpur5GT). Results of qRT-PCR revealed that Cpur5GT shows tissue-specific expression, with strong expression in fully opened petals and weak expression in young petals. In vitro enzyme assay showed that when uridine diphosphate glucose was used as the sugar donor, recombinant Cpur5GT could catalyze the glycosylation of 3-glucoside-type anthocyanidins at the 5-O-position, but when uridine diphosphate galactose was served as glycosyl donor, the reaction could not be performed. These results demonstrate that Cpur5GT exhibits valid anthocyanin glucosylation activity and could be used to analyze the mechanism of A5GT-mediated flower coloration in cyclamen in future studies.
Keywords: anthocyanin 5-O-glucosyltransferase, cyclamen, flower coloration, pigmentation
Anthocyanin is one of the most important flavonoids involved in plant coloration, and the type and content of anthocyanins directly determine the tissue color. A wide variety of anthocyanins exist in nature, but only six anthocyanidins are commonly found in higher plants: pelargonidin (Pg), cyanidin (Cy), delphinidin (Dp), peonidin (Pn), petunidin (Pt), and malvidin (Mv) (Kong et al. 2003). The biosynthesis pathway of anthocyanins has been divided into two stages: anthocyanidin formation and anthocyanin modification such as glycosylation, methylation, and acylation (Yamazaki et al. 1999). Synergistic effects of these modifications facilitate the formation of more diverse kinds of anthocyanins. However, glycosylation always precedes other modifications and plays a crucial role in improving the solubility and chemical stability of plant anthocyanins as well as facilitating their storage and transportation in cells (Vogt and Jones 2000). Based on sequence similarity, motifs composition, and catalytic mechanism, glycosyltransferases (GTs) are classified into 111 subfamilies (http://www.cazy.org/fam/acc_GT.html). Uridine diphosphate (UDP) glycosyltransferase (UGTs) belong to the GT1 subfamily, a major class of enzymes that plays a vital part in plant secondary metabolism. UGTs have several receptor molecules, including alkaloids, flavonoids, isoflavonoids, and terpenoids (Akere et al. 2020). Flavonoid 3-O-glucosyltransferase (3GT/UFGT) is commonly considered as the first enzyme that catalyzes the glycosylation of anthocyanidins in most higher plants. To date, several 3GT genes involved in flavonoid glycosylation have been isolated and adequately characterized in diverse plant species, including Vitis vinifera (Ford et al. 1998), Petunia hybrida (Yamazaki et al. 2002), and Freesia hybrida (Meng et al. 2019; Sun et al. 2016). In most cases, the mono-glucosidic anthocyanins synthesized in plants will be transformed into di-glucosidic anthocyanins under the catalysis of anthocyanin 5-O-glucosyltransferase (A5GT), but reports on plant A5GT are relatively less. A5GT is responsible for the glycosylation of anthocyanins at the 5-O-position, leading to the formation of highly stable and soluble anthocyanidin 3,5-diglucoside, which plays an indispensable role in the development of flower color, particularly bright-purple flowers (Jonsson et al. 1984). Since the cloning of the first A5GT cDNA from Perilla frutescens by Yamazaki et al. (1999), several A5GTs have been cloned from different plant species, including Dahlia variabilis (Ogata et al. 2001), P. hybrida (Yamazaki et al. 2002), Gentiana triflora (Nakatsuka et al. 2008), and F. hybrida (Ju et al. 2018). Based on these studies, it can be concluded that the expression level of A5GT is correlated with the level of major pigments, and A5GTs exhibit a wide range of substrate specificity. However, to the best of our knowledge, the A5GT gene of wild cyclamen (Cyclamen purpurascens) has not been characterized in detail. Cyclamen is a popular ornamental plant grown either in pots or in the garden. Most of the commercial cyclamen cultivars have been obtained through the natural mutation and hybridization of wild cyclamen (Cyclamen persicum) and exhibit a range of color, shape, and size but without good fragrance (Grey-Wilson 2002). Ishizaka and Uematsu (1992) established the ovule cultivation system and solved the problem of strong cross-incompatibility between C. persicum and other species. The wild cyclamen C. purpurascens has a sweet fragrance and had been used as a breeding material. Interestingly, the F1 progeny of C. purpurascens × C. persicum contains 3,5-diglucoside-type anthocyanins in petals; this is similar to C. purpurascens, which contains malvidin 3,5-diglucoside (Mv3,5dG) as the major pigment (Takamura et al. 2005). The fragrant cyclamen cultivar “Kaori-no-mai” (KM), produced by crossing C. persicum cultivars with C. purpurascens, accumulates Mv3,5dG as a dominant anthocyanin and produces purple flowers. By contrast, the ion beam-irradiated mutant of KM, KMmv3 (commercial name: “Mayabi-no-mai” [MY]), accumulates malvidin 3-glucoside (Mv3G) and produces reddish-purple flowers (Ishizaka et al. 2012). It seems that A5GT contributes to the development of flower color in cyclamen. Previously, three 5GT-like genes (Cpur5GT1, Cpur5GT2 and Cpur5GT3) have been isolated partially from C. purpurascens. Among these, Cpur5GT2 most likely encodes a functional A5GT (Hase et al. 2012). On that basis, we amplified their complete ORFs and found that the nucleotide sequences of Cpur5GT1 and Cpur5GT3 were identical. The original differences in sequences of these two gene fragments were probably caused by mutations due to the Taq polymerase during PCR. Therefore, we assumed two candidate genes, Cpur5GT (previously named Cpur5GT2) and CpurGTL1 (previously named Cpur5GT1 and Cpur5GT3). The results of in vitro enzyme assay showed that recombinant Cpur5GT can catalyze the glycosylation of the tested anthocyanidin 3-glucoside. We also examined the relative activity of Cpur5GT on different substrates.
Cyclamen was grown in a greenhouse at Saitama Institute of Technology, Japan. The petals, leaves, immature anthers, and petioles of C. purpurascens were collected and immediately frozen in liquid nitrogen. The frozen samples were stored at −80°C until needed for further analysis. Total RNA was extracted from petals, according to the cetyltrimethylammonium bromide method (Chang et al. 1993) and then treated with DNase I (TaKaRa Bio, Japan) to eliminate genomic DNA contamination. First-strand cDNA was synthesized from 2.0 µg of total RNA using an oligo(dT)-anchor primer (5′-GAC TCG AGT CGA CAT CGA T17-3′), reverse transcriptase enzyme, and PrimeScript II 1st cDNA Synthesis Kit (TaKaRa Bio), according to the manufacturer’s instructions. Based on the obtained gene fragments (Cpur5GT1, Cpur5GT2, and Cpur5GT3) (Hase et al. 2012), 3′ rapid amplification of cDNA ends (RACE) and 5′ RACE were carried out using the 5′/3′-RACE 2nd Generation Kit (Merck, Germany), to cloned the full-length ORFs from C. purpurascens. The PCR products were gel extracted and cloned into the pTAC-2 Easy vector (BioDynamics Laboratory Inc., Japan). The resultant construct was transformed into Escherichia coli JM109 competent cells, and white (positive) colonies were picked for sequencing with a DNA sequencer (Model 3500, Applied Biosystems, USA) using the BigDye® Terminator ver. 3.1 Cycle Sequencing Kit (Applied Biosystems). Multi-alignment analysis was conducted using the Clustal W program (Thompson et al. 1994). Phylogenetic trees were constructed using the neighbor-joining method (Saitou and Nei 1987) with MEGA7 (Kumar et al. 2016). To investigate the expression levels of Cpur5GTs in different tissues, qRT-PCR was conducted on the Quant Studio™ 1 System using the standard cycling mode with Power Up™ SYBR® Green Master Mix (Thermo Fisher Scientific, USA). Each 20 µl reaction contained 100 ng of cDNA (template), 10 µM of primer, and 10 µl of 2×Master Mix. The elongation factor-1α (eEF1α) gene was used as an internal control. Each experiment was conducted in four replicates, and relative gene expression was determined using the 2−ΔΔCT method (Livak and Schmittgen 2001).
The Cpur5GT gene was cloned into the pET16b expression vector using NdeI and BamHI restriction endonucleases. The resulting pET16b-Cpur5GT vector was transformed into E. coli strain BL21 (DE3) competent cells (Novagen, Germany). A single E. coli BL21 (DE3) colony harboring the reconstructed plasmid was cultured in LB liquid medium containing 100 µg ml−1 ampicillin at 37°C until the optical density (OD600) of the culture reached 0.4–0.6. Then, the expression of Cpur5GT was induced by adding 5, 10, or 20 µM isopropyl-β-D-thiogalactopyranoside (IPTG), and the culture was grown at 28°C for 16 h. The cells were harvested and resuspended in Fast Break™ Cell Lysis Reagent (Promega, USA). The Cpur5GT fusion protein was purified using the HisLink™ Spin Protein Purification System (Promega), according to the manufacturer’s protocol. The purified proteins were separated on 15% acrylamide gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to detect the expression of the target protein; the uninduced recombinant expression vector was used as a control. The enzyme assay of recombinant Cpur5GT was tested in a reaction mixture (final volume: 200 µl) containing 37.5 µg of purified protein, 2 mM of UDP-glucose or UDP-galactose (donor), 0.2 mM of 3-glucoside-type anthocyanidins (acceptor) (such as Mv3G, delphinidin 3-glucoside [Dp3G], cyanidin 3-glucoside [Cy3G], peonidin 3-glucoside [Pn3G], or petunidin 3-glucoside [Pt3G]), 100 mM of Tris (pH 7.5). All the reaction mixtures were incubated at 28°C for 1 h. To determine the Km values, the concentrations of UDP-glucose and Mv3G were varied respectively; Km values were obtained by double-reciprocal plots. Every reaction was terminated by adding 10% phosphoric acid. The sample was centrifuged at 15,000 rpm for 5 min, and the supernatant was filtrated through a 0.22 mm filter (Shimadzu, Japan) and analyzed via high-performance liquid chromatography (HPLC). A Prodigy ODS-3 reversed-phase column (4.6 mm×100 mm, 3 µm, 100 Å; Phenomenex) was used to separate the metabolites at 30°C. The mobile phase comprised 1.5% (v/v) phosphoric acid (A), 1.5% (v/v) phosphoric acid, 20% (v/v) acetic acid, and 25% (v/v) acetonitrile solution (B). The elution was conducted for 60 min at a flow rate of 0.3 ml min−1. The reaction products were quantified by measuring the absorbance peak area at 525 nm.
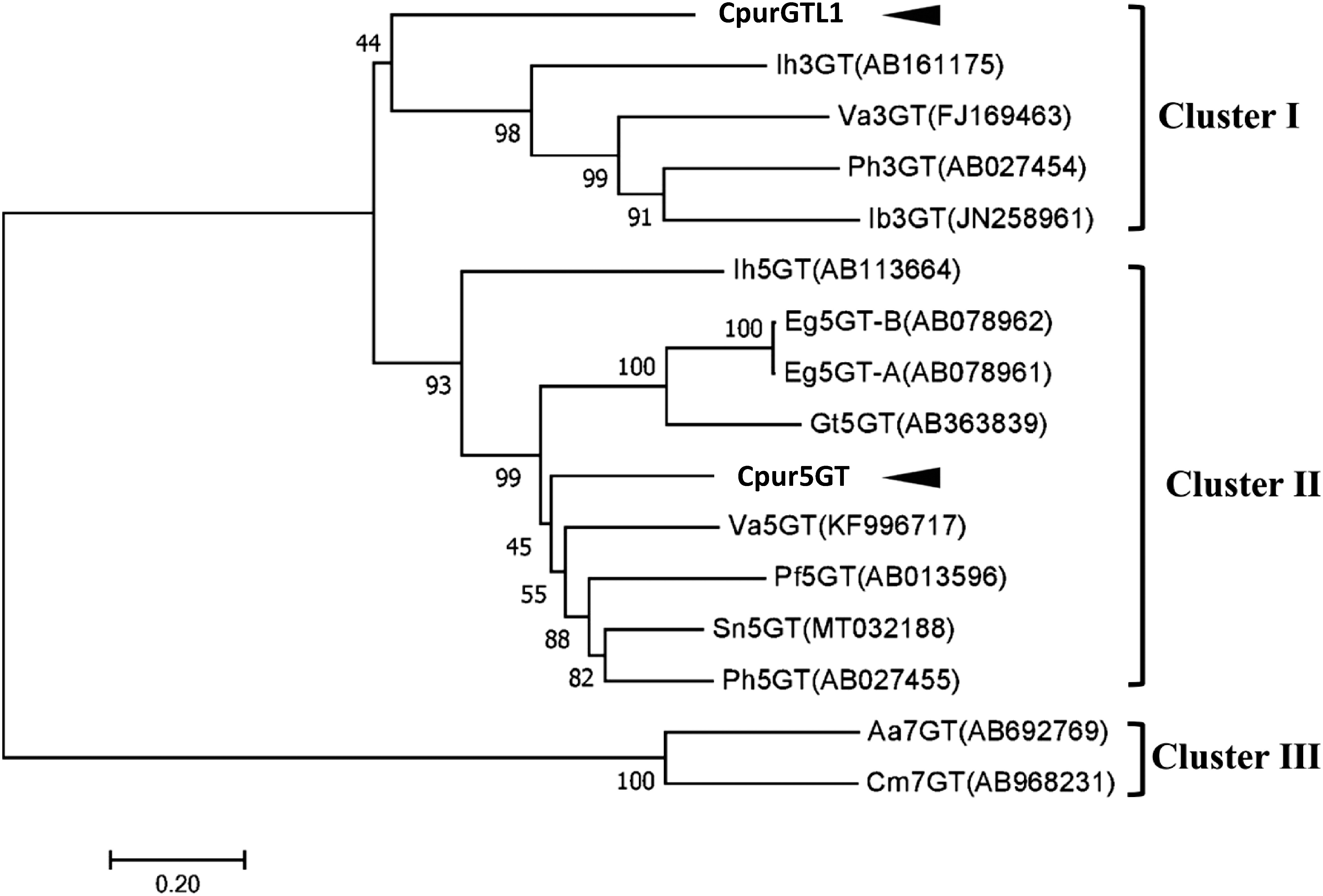
This time we have confirmed the full-length ORFs of Cpur5GT and CpurGTL1, the deduced amino acid sequences were presented in Supplementary Figure S1. The phylogenetic analysis of the putative Cpur5GT, CpurGTL1 and several functionally characterized plant UGTs (Figure 1) revealed three clusters, depending on the regiospecificity and regioselectivity of UGTs to sugar acceptors. Since Cpur5GT and CpurGTL1 were clustered in 5GT and 3GT clade, respectively, we considered that Cpur5GT is more likely to be the functional A5GT for anthocyanin glycosylation.
Figure 1. Phylogenetic analysis of Cpur5GT, CpurGLT1, and GT family members from other plant species. The phylogenetic tree was constructed using the neighbor-joining method with MEGA 7.0. The optimal tree (sum of branch length=6.59939181) is shown. Branch numbers represent the percentage of bootstrap values in 1,000 sampling replicates. Arrowheads indicate candidate 5GTs isolated from C. purpurascens in this study. EMBL/DDBJ/GenBank DNA database accession numbers and species names are shown.
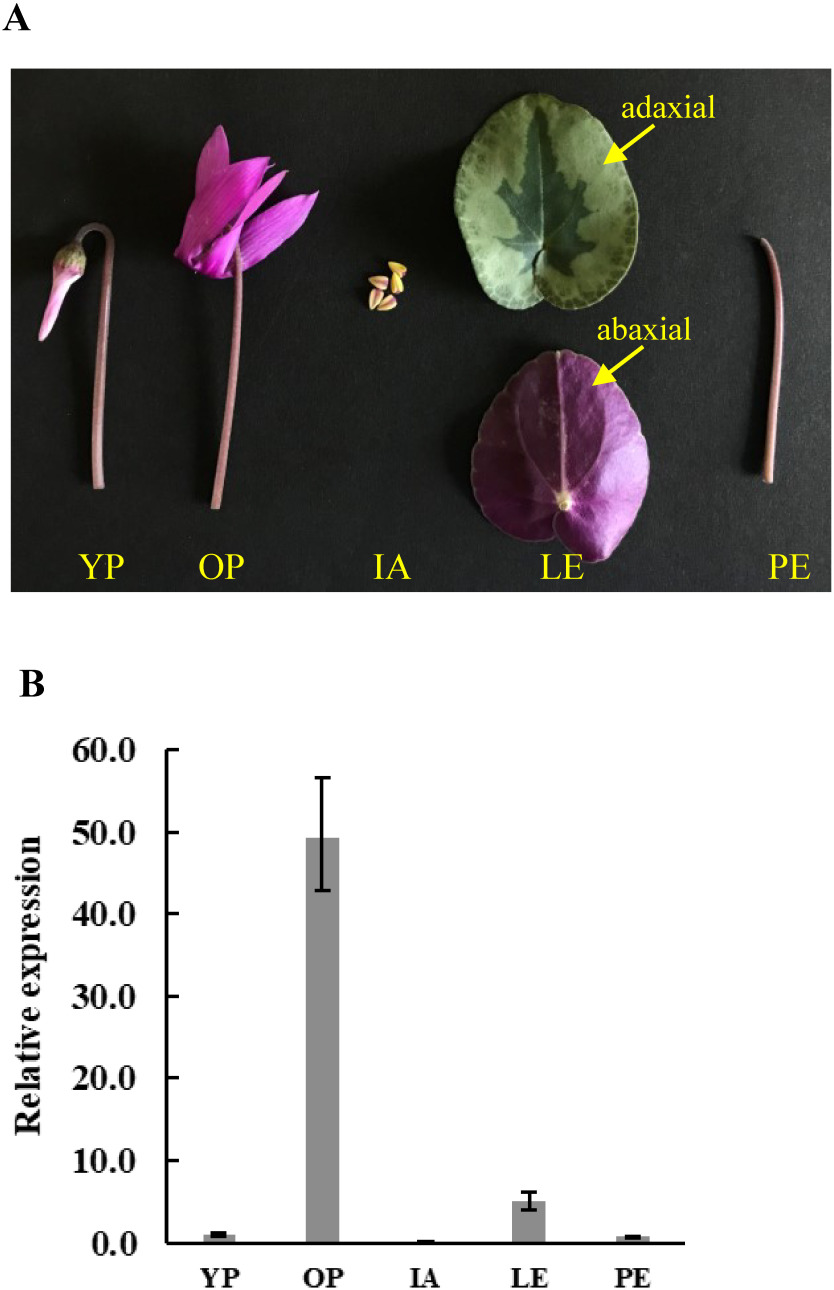
The expression profile of Cpur5GT was examined in young petals (pale pigmentation stage), fully opened petals, immature anthers, leaves, and petioles (Figure 2A) by qRT-PCR. It showed the lowest expression in immature anthers and the highest expression in fully opened petals (10- and 50- fold higher than that in leaves and young petals, respectively) (Figure 2B). The expression patterns of some genes that related to flower coloration change with the development of flowering. For example, Akita et al. (2018) have identified a functional co-pigmentation-related flavonol synthase gene from C. purpurascens (CpurFLS2). This gene strongly expressed at the early stage of flower development, and reduced expression in full opened flowers. While, Cpur5GT was observed strong expression at later stage and weak expression at early stage of flower development (Figure 2B). Previous studies showed that the expression of A5GT was closely related to the accumulation of bis-glycosidic anthocyanins; in berry skins of Vitis amurensis, the expression level of Va5GT increased with the accumulation of anthocyanins (He et al. 2015), in G. trifloral, the expression level of Gt5GT7 in petals coincided with the flower development (Nakatsuka et al. 2008), and in F. hybrida, Fh5GT1 and Fh5GT2 were expressed at all stages of flower development and showed the highest expression level in almost completely pigmented petals (Ju et al. 2018). In this study, Cpur5GT presented the similar expression profile to those functional A5GTs, which further suggested that Cpur5GT is more likely participate in color formation of cyclamen.
Figure 2. Relative expression levels of Cpur5GT in different tissues of C. purpurascens. (A) Photographs of the tested tissues of C. purpurascens. YP, young petals; OP, opened petals; IA, immature anthers; LE, leaves, (upper picture is adaxial and lower picture is abaxial); PE, petioles. (B) Expression analysis of Cpur5GT in different tissues of C. purpurascens. Data represent the means±standard deviation (SD) of four biological replicates.
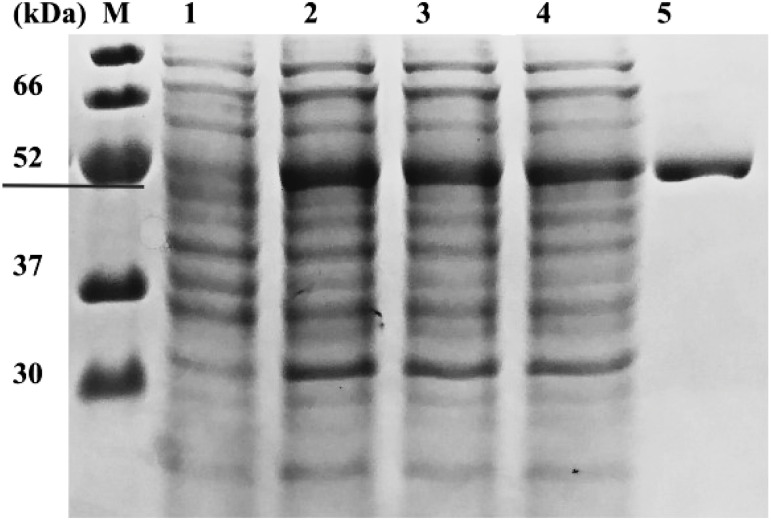
SDS-PAGE analysis showed that the molecular mass of the crude protein induced by adding IPTG was approximately equal to the expected molecular weight of the Cpur5GT protein (52 kDa), regardless of the concentration of IPTG (5, 10, or 20 µM) used for induction. Based on SDS-PAGE analysis, 5 µM IPTG was determined as the optimal concentration for the induction of the recombinant soluble Cpur5GT protein (Figure 3). Previous study mentioned that Cpur5GT could catalyze 5-O-position glycosylation of Mv3G (Hase et al. 2012). Here we determined the apparent Km value for Mv3G was 79.2 µM against UDP-glucose as the glycosyl donor, and the Vmax value for M3,5dG formation was 18.6 nM min−1 mg−1. This was comparable with Km value of Va5GT, 80.9 µM for Mv3G (He et al. 2015). For sugar donor specificity analyses, UDP-glucose and UDP-galactose were tested as glycosyl donors respectively, with Mv3G as acceptor. When tested with UDP-galactose, recombinant Cpur5GT could not further catalyze 5-O-glycosylation of Mv3G (Supplementary Figure S2), which was similar to the capability of Dv5GT and Va5GT (He et al. 2015; Ogata et al. 2001). The apparent Km value for UDP-glucose of 454 µM and a Vmax value of 18.1 nM min−1 mg−1 were obtained. And the result was similar to the Km value of Va5GT, 0.213 mM for UDP-glucose (He et al. 2015). To determine the acceptor specificity, the purified recombinant Cpur5GT protein was incubated with UDP-glucose and five potential substrates (acceptors), and the reaction products were examined via HPLC. When incubated with five kinds of anthocyanidin 3-glucoside as substrates, additional peaks were formed, which were identified as the corresponding anthocyanidin 3,5-diglucoside, based on the retention time and UV spectrum of standard samples (Figure 4). Cpur5GT showed a higher relative activity using Mv3G (Table 1). According to previous reports, A5GTs always exhibit the highest enzymatic activity when incubated with the precursors of primary anthocyanins. The Perilla frutescens 5GT enzyme showed the highest preference for Cy3G among all tested substrates, as cyanidin-derived anthocyanins mainly accumulated in its red leaves (Yamazaki et al. 1999). Since cyanidin- and pelargonidin-derived anthocyanins are the main pigments in D. variabilis, the Dv5GT protein shows a higher affinity for Cy3G and pelargonidin 3-glucoside (Pg3G) but very low relative activity on Dp3G (which has not yet been detected in Dahlia) (Ogata et al. 2001). Although cyanidin 3,5-diglucoside (Cy3,5dG), delphinidin 3,5-diglucoside (Dp3,5dG), peonidin 3,5-diglucoside (Pn3,5dG), and petunidin 3,5-diglucoside (Pt3,5dG) have not been identified in C. purpurascens, we showed that Cpur5GT could catalyze the glycosylation of the corresponding 3-O-glycoside precursors at the 5-O-position, with higher relative activity on Pn3G, Cy3G and Dp3G (Table 1). However, this is inconsistent with the results of substrate analysis of Va5GT extracted from grape cultivar “Zuoshanyi”; Va5GT preferentially glycosylated O-methoxylated anthocyanins (Mv3G, Pt3G, and Pn3G) rather than their hydroxylated counterparts (Dp3G and Cy3G) (He et al. 2015). Several scholars also argued that UGTs involved in plant secondary metabolism always exhibit higher regiospecificity or regioselectivity for specific positions of the hydroxyl group rather than the structure of sugar acceptors (Sun et al. 2016; Vogt and Jones 2000). However, anthocyanin biosynthesis is a complex process, and different enzymes do not act independently. The relative concentration of potential substrates may be one of the most crucial factors determining the enzyme activity in plants. The possibility that C. purpurascens contains other A5GTs, which exhibit different relative activities against various anthocyanin substrates, could not be excluded since UGTs involved in flavonoid biosynthesis are usually encoded by a multi-gene family in many plant species (Yonekura-Sakakibara and Hanada 2011). The above results indicate that Cpur5GT transfers the glucose moiety from UDP-glucose to the 5-O-position of anthocyanidin 3-glucoside and displays a broad range of substrate specificity. We plan to analyze the substrate specificity of Cpur5GT using other flavonoids such as flavonols in order to investigate the function of Cpur5GT in detail. Moreover, we will use the flower color mutant, such as MY (the major anthocyanin has been changed from Mv3,5dG to Mv3G), to reveal the possible role of Cpur5GT in cyclamen flower color formation. These studies will be highly valuable and will enhance our understanding of the molecular mechanism underlying flower coloration in cyclamen.
Figure 3. SDS-PAGE analysis of the recombinant Cpur5GT. Lane M, protein marker; lane 1, E. coli lysate without IPTG; lanes 2-4, E. coli lysate treated with 5, 10, or 20 µM IPTG; lane 5, purified recombinant protein.
Figure 4. HPLC analysis of Cpur5GT reaction products obtained from the in vitro enzyme assay. (A–J) Five potential substrates including Mv3G (A, B), Dp3G (C, D), Pt3G (E, F), Cy3G (G, H), and Pn3G (I, J) were incubated without (A, C, E, G, I) or with (B, D, F, H, J) the purified Cpur5GT. UDP-glucose was used as the sugar donor.
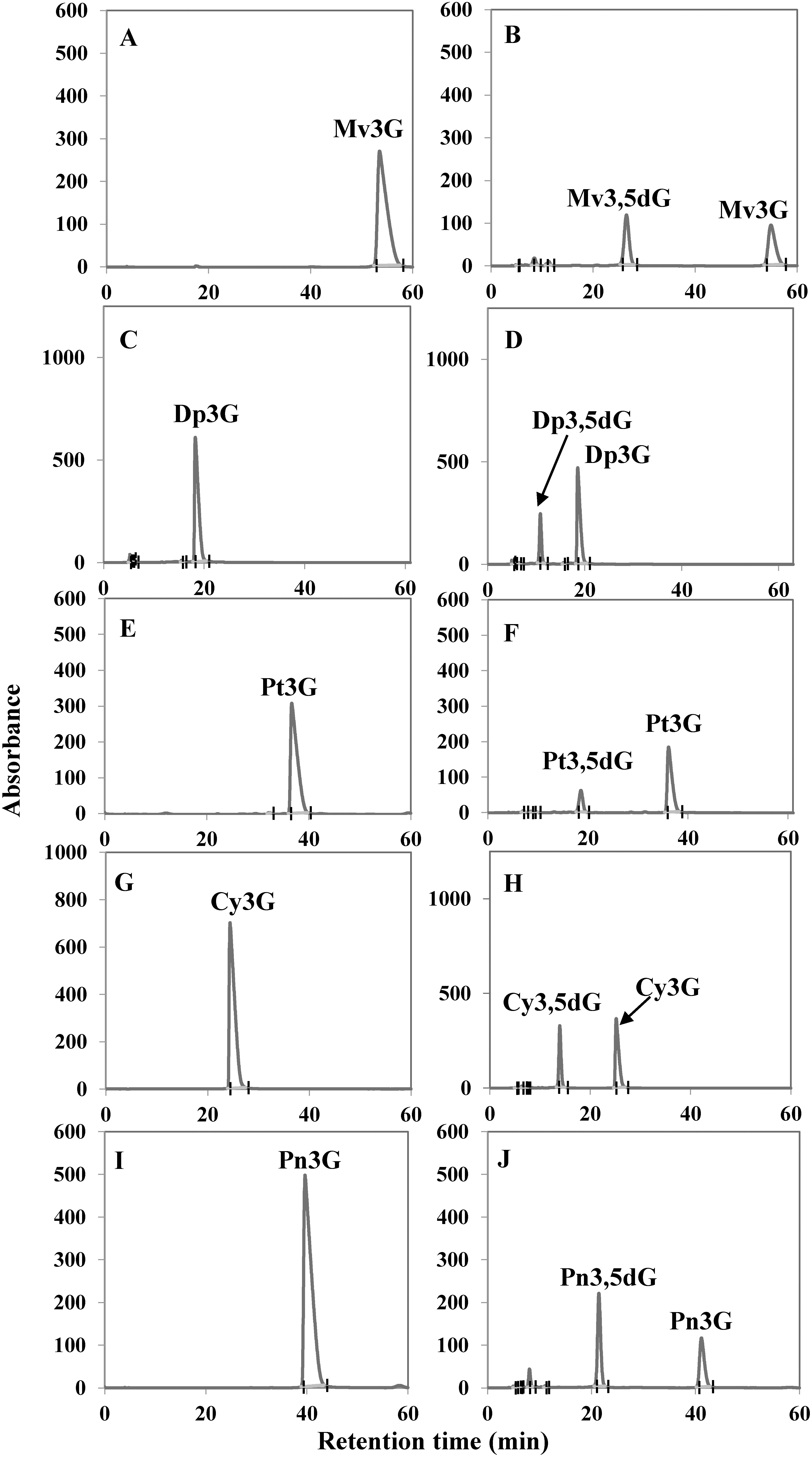
Table 1. Relative activity of Cpur5GT against several potential substrates.
| Substrate | Product | Relative activity (%) |
|---|---|---|
| Mv3G | Mv3,5dG | 100a±2.2 |
| Dp3G | Dp3,5dG | 86.8±3.3 |
| Pt3G | Pt3,5dG | 18.8±2.3 |
| Cy3G | Cy3,5dG | 78.3±12.4 |
| Pn3G | Pn3,5dG | 72.4±7.1 |
Relative activity analyses were performed with UDP-glucose as the donor. The amount of reaction productwas calculated from the area of the HPLC chromatogram recorded at 525 nm. The activity of Cpur5GTagainst malvidin 3-glucoside (Mv3G) was considered 100%, which was used to determine its relativeactivity against other substrates.
Abbreviations
- A5GT
anthocyanin 5-O-glucosyltransferase
- GT
glycosyltransferase
- UGT
uridine diphosphate (UDP) glycosyltransferase
- 3GT/UFGT
flavonoid 3-O-glucosyltransferase
- ORF
open reading frame
- Pg
pelargonidin
- Cy
cyanidin
- Dp
delphinidin
- Pn
peonidin
- Pt
petunidin
- Mv
malvidin
- Pg3G
pelargonidin 3-glucoside
- Cy3G
cyanidin 3-glucoside
- Dp3G
delphinidin 3-glucoside
- Pn3G
peonidin 3-glucoside
- Pt3G
petunidin 3-glucoside
- Mv3G
malvidin 3-glucoside
- Pg3,5dG
pelargonidin 3,5-diglucoside
- Cy3,5dG
cyanidin 3,5-diglucoside
- Dp3,5dG
delphinidin 3,5-diglucoside
- Pn3,5dG
peonidin 3,5-diglucoside
- Pt3,5dG
petunidin 3,5-diglucoside
- Mv3,5dG
malvidin 3,5-diglucoside
- EF1α
elongation factor-1α
- qRT-PCR
real-time quantitative reverse transcription
- HPLC
high-performance liquid chromatography
- IPTG
isopropyl-β-D-thiogalactopyranoside
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Data
References
- Akere A, Chen SH, Liu X, Chen Y, Dantu SC, Pandini A, Bhowmik D, Haider S (2020) Structure-based enzyme engineering improves donor-substrate recognition of Arabidopsis thaliana glycosyltransferases. Biochem J 477: 2791–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akita Y, Kitamura S, Mikami R, Ishizaka H (2018) Identification of functional flavonol synthase genes from fragrant wild cyclamen (Cyclamen purpurascens). J Plant Biochem Biotechnol 27: 147–155 [Google Scholar]
- Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11: 113–116 [Google Scholar]
- Ford CM, Høj PB (1998) Multiple glucosyltransferase activities in the grapevine Vitis vinifera L. Aust J Grape Wine Res 4: 48–58 [Google Scholar]
- Grey-Wilson C (2002) Cyclamen: A guide for gardeners, Horticulturists and Botanists. Timber Press, England
- Hase Y, Akita Y, Kitamura S, Narumi I, Tanaka A (2012) Development of an efficient mutagenesis technique using ion beams: Toward more controlled mutation breeding. Plant Biotechnol 29: 193–200 [Google Scholar]
- He F, Chen WK, Yu KJ, Ji XN, Duan CQ, Reeves MJ, Wang J (2015) Molecular and biochemical characterization of the UDP-glucose: Anthocyanin 5-O-glucosyltransferase from Vitis amurensis. Phytochemistry 117: 363–372 [DOI] [PubMed] [Google Scholar]
- Ishizaka H, Kondo E, Kameari N (2012) Production of novel flower color mutants from the fragrant cyclamen (Cyclamen persicum×C. purpurascens) by ion-beam irradiation. Plant Biotechnol 29: 201–208 [Google Scholar]
- Ishizaka H, Uematsu J (1992) Production of interspecific hybrids of Cyclamen persicum Mill. and C. hederifolium Aiton by ovule culture. Ikushugaku Zasshi 42: 353–366 (in Japanese) [Google Scholar]
- Jonsson LMV, Aarsman MEG, van Diepen J, de Vlaming P, Smit N, Schram AW (1984) Properties and genetic control of anthocyanin 5-O-glucosyltransferase in flowers of Petunia hybrida. Planta 160: 341–347 [DOI] [PubMed] [Google Scholar]
- Ju Z, Sun W, Meng X, Liang L, Li Y, Zhou T, Shen H, Gao X, Wang L (2018) Isolation and functional characterization of two 5-O-glucosyltransferases related to anthocyanin biosynthesis from Freesia hybrida. Plant Cell Tissue Organ Cult 135: 99–110 [Google Scholar]
- Kong JM, Chia LS, Goh NK, Chia TF, Brouillard R (2003) Analysis and biological activities of anthocyanins. Phytochemistry 64: 923–933 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Meng X, Li Y, Zhou T, Sun W, Shan X, Gao X, Wang L (2019) Functional differentiation of duplicated flavonoid 3-O-glycosyltransferases in the flavonol and anthocyanin biosynthesis of Freesia hybrida. Front Plant Sci 10: 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuka T, Sato K, Takahashi H, Yamamura S, Nishihara M (2008) Cloning and characterization of the UDP-glucose: Anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. J Exp Bot 59: 1241–1252 [DOI] [PubMed] [Google Scholar]
- Ogata J, Sakamoto T, Yamaguchi M, Kawanobu S, Yoshitama K (2001) Isolation and characterization of anthocyanin 5-O-glucosyltransferase from flowers of Dahlia variabilis. J Plant Physiol 158: 709–714 [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sun W, Liang L, Meng X, Li Y, Gao F, Liu X, Wang S, Gao X, Wang L (2016) Biochemical and molecular characterization of a flavonoid 3-O-glycosyltransferase responsible for anthocyanins and flavonols biosynthesis in Freesia hybrida. Front Plant Sci 7: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura T, Aizawa M, Kim SY, Nakayama M, Ishizaka H (2005) Inheritance of flower pigment in crosses between cyclamen cultivars and Cyclamen purpurascens. Acta Hortic 673: 437–441 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T, Jones P (2000) Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci 5: 380–386 [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Gong Z, Fukuchi-Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Saito K (1999) Molecular cloning and biochemical characterization of a novel anthocyanin 5-O-glucosyltransferase by mRNA differential display for plant forms regarding anthocyanin. J Biol Chem 274: 7405–7411 [DOI] [PubMed] [Google Scholar]
- Yamazaki M, Yamagishi E, Gong Z, Fukuchi-Mizutani M, Fukui Y, Tanaka Y, Kusumi T, Yamaguchi M, Saito K (2002) Two flavonoid glucosyltransferases from Petunia hybrida: Molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol Biol 48: 401–411 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K (2011) An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J 66: 182–193 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.