Abstract
Long non-coding RNAs (lncRNAs) play crucial roles in many human diseases, particularly in tumorigenicity and progression. Although lncRNA research studies are increasing rapidly, our understanding of lncRNA mechanisms is still incomplete. The long intergenic non-protein coding RNA 662 (LINC00662) is a novel lncRNA, and accumulating evidence suggests that it is related to a variety of tumors in multiple systems, including the respiratory, reproductive, nervous, and digestive systems. LINC00662 has been shown to be upregulated in malignant tumors and has been confirmed to promote the development of malignant tumors. LINC00662 has also been reported to facilitate a variety of cellular events, such as tumor-cell proliferation, invasion, and migration, and its expression has been correlated to clinicopathological characteristics in patients with tumors. In terms of mechanisms, LINC00662 regulates gene expression by interacting with both proteins and with RNAs, so it may be a potential biomarker for cancer diagnosis, prognosis, and treatment. This article reviews the expression patterns, biological functions, and underlying molecular mechanisms of LINC00662 in tumors.
Keywords: LINC00662, oncogene, proliferation, molecular mechanism, competing endogenous RNA
Introduction
Cancer is a leading cause of death worldwide, second only to cardiovascular disease (Ferlay et al., 2018; Essa et al., 2020), and it represents a terrible threat to the health of mankind. In recent years, malignant-tumor incidence and mortality rates have increased in all countries at all income levels (Fidler et al., 2016; Torre et al., 2016). With the development of the Encyclopedia of DNA Elements project, most of the gene sequences in the genome have been clarified to be non-coding genome (Consortium, 2012; Consortium et al., 2020; Zhou et al., 2020). The non-coding RNAs (ncRNAs), which correspond to messenger RNAs (mRNAs), have traditionally been assumed to have many biological functions (Cech and Steitz, 2014; Matsui and Corey, 2017; Coker et al., 2019; Wang J. et al., 2019). In particular, current progress in sequencing technologies and large-scale genome sequencing projects have demonstrated that lncRNAs (>200 nucleotides) are crucial regulators of many human diseases, especially cancers (Xingming Jiang, 2015; Anastasiadou et al., 2018; Slack and Chinnaiyan, 2019).
Accumulating research has revealed that many lncRNAs play vital roles in cancer tumorigenicity (Yari et al., 2019; Feng et al., 2020) and progression (Zhu et al., 2017; Gong et al., 2020), are aberrantly expressed in a variety of cancers, and participate in many biological functions, such as cell proliferation (Ducoli et al., 2021; Shen et al., 2021), epithelial-to-mesenchymal transition (EMT) (Grelet et al., 2017; Zhao et al., 2017), invasion (Schmidt et al., 2016; Jin et al., 2019), migration (Liu L. et al., 2019; Zhao et al., 2018), angiogenesis (Zhao et al., 2020; Li et al., 2021), and differentiation (Luo et al., 2019; Tong et al., 2020). Moreover, lncRNAs can also function as competing endogenous RNAs (ceRNAs) (Wang et al., 2017; Zhou et al., 2019), transcription factors (Long et al., 2017; Ducoli et al., 2021), and in post-transcriptional processing (Zhang et al., 2018; Botti et al., 2019). All these findings suggest that lncRNAs may be able to serve as potential cancer biomarkers, ameliorating some of the current problems caused by cancer.
Several lines of evidence support an oncogenic role for LINC00662, the sequence for which is located on chromosome 19. Upregulation of LINC00662 expression has been detected in many cancers when compared to adjacent non-cancerous tissues. Research studies focusing on the oncogenic properties of LINC00662 have shown that it can be an oncogenic regulator in several tumor types in a variety of systems, including the respiratory, reproductive, nervous, and digestive systems.
In the present work, we have summarized the latest evidence concerning the abnormal expression of LINC00662, its associated clinical features, and molecular mechanisms, and we discuss both its prognostic and therapeutic values for malignant tumors.
The Expression Levels, Biological Processes, and Clinical Characteristics of LINC00662 in Human Cancers
In recent years, largely due to the rapid development of high-throughput technologies, many cancer-related lncRNAs have received widespread attention (Li et al., 2017; Yu et al., 2017; Wang J. et al., 2018; Li Y. et al., 2020), including LINC00662. However, a possible pan-cancer role for LINC00662 has not been clear, so a comprehensive review of its expression levels, biological functions, and clinical features is warranted (see Table 1).
TABLE 1.
Expression level and biological functions of LINC00662 in human cancers.
| Human body systems | Cancer type | Expression | Property | Prognosis | Function | References |
| Respiratory system | lung cancer | up-regulated | oncogene | poor | proliferation and colony formation | Xu et al. (2020) |
| lung cancer | up-regulated | oncogene | poor | invasion, migration and CSCs stemness | Gong et al. (2018) | |
| non-small cell lung cancer | up-regulated | oncogene | / | progression, proliferation, apoptosis, cell cycle arrest, migration and invasion | Lv et al. (2021) | |
| Reproductive system | breast cancer | up-regulated | oncogene | poor | proliferation and migration | Cheng L. et al. (2020) |
| breast cancer | up-regulated | oncogene | poor | / | Xiong et al. (2020) | |
| prostate cancer | up-regulated | oncogene | poor | tumorigenesis, proliferation, migration, invasion and apoptosis | Li et al. (2019) | |
| prostate cancer | up-regulated | oncogene | poor | proliferation, migration, invasion and apoptosis | Yao et al. (2020a) | |
| cervical cancer | up-regulated | oncogene | / | proliferation, metastasis, progression and the radioresistance | Wei et al. (2020) | |
| Nervous system | glioma | up-regulated | oncogene | / | proliferation and migration | Geng et al. (2020) |
| glioma | up-regulated | oncogene | poor | proliferation, and invasion | Wu et al. (2020) | |
| chordoma | up-regulated | oncogene | poor | malignant progression, proliferation, colony formation, invasion, migration, and glycolysis | Wang et al. (2020) | |
| Digestive system | colorectal cancer | up-regulated | oncogene | poor | tumorigenesis, metastasis, cell cycle arrest at G2/M phase, proliferation, apoptosis, migration and invasion | Wang H. et al. (2019) |
| colorectal cancer | up-regulated | oncogene | poor | proliferation, apoptosis, and invasion | Yao et al. (2020b) | |
| colon cancer | up-regulated | oncogene | poor | proliferation, migration and invasion, apoptosis | Cheng B. et al. (2020) | |
| hepatocellular carcinoma | up-regulated | oncogene | / | / | Tian et al. (2020) | |
| hepatocellular carcinoma | up-regulated | oncogene | poor | proliferation, migration, invasion and apoptosis | Guo et al. (2020) | |
| esophageal squamous cell carcinoma | up-regulated | oncogene | poor | proliferation, migration, invasion, viability and metastasis | Zhang et al. (2020) | |
| gastric cancer | up-regulated | oncogene | poor | proliferation, and chemo-sensitivity | Liu et al. (2018) | |
| oral squamous cell carcinoma | up-regulated | oncogene | poor | proliferation, apoptosis, migration and invasion | Xu et al. (2019) | |
| oral squamous cell carcinoma | up-regulated | oncogene | / | radioresistance | Chen et al. (2020) | |
| Other systems | melanoma | up-regulated | oncogene | poor | proliferation, migration and invasion | Xia et al. (2020) |
| acute myeloid leukemia | up-regulated | oncogene | / | malignant growth | Liu Y. et al. (2019) |
Respiratory-System Tumors
Globally, the leading cause of cancer deaths is lung cancer (LC) (Chen et al., 2016; Duma et al., 2019; Nasim et al., 2019). Numerous studies have confirmed that lncRNAs, including LINC00662, participate in LC progression (Yang et al., 2017; Wei et al., 2019; Pan et al., 2020; Zang et al., 2020). Xu et al. (2020) reported elevated LINC00662 expression in LC cells, which accelerated both their proliferation and colony formation. In addition, LINC00662 expression has been shown to promote invasion, metastases, and the stemness of LC stem cells as well (Gong et al., 2018). Moreover, Lv et al. (2021) reported that LINC00662 promoted the growth progression of non-small cell LC in vivo on the basis of enhanced proliferation in LC cells. LINC00662 may therefore be considered a promising diagnostic target for LC patients.
Reproductive-System Tumors
For women, breast cancer (BC) is one of the most lethal cancers worldwide, and BC morbidity in China is gradually increasing (Fahad Ullah, 2019; Ding et al., 2020). The expressions of lncRNAs have been reported to be closely associated with BC (Kansara et al., 2020). Cheng L. et al. (2020) showed that LINC00662 expression was elevated in both BC tissues and cells when compared to normal BC cell lines and tissue. Furthermore, the silencing of LINC00662 was reported to significantly inhibit both BC cell proliferation and motility. In addition, Xiong et al. (2020) found that LINC00662 expression levels in BC patients showed a significant positive correlation with overall survival.
Although overall survival for patients with prostate cancer (PCa) is already high, PCa remains the most common tumor type in men (Li et al., 2019), and metastatic PCa remains incurable at present (Wang G. et al., 2018). Therefore, the identification of new therapeutic targets for PCa patients remains necessary. Interestingly, Li et al. (2019) demonstrated that LINC00662 was highly expressed both in PCa cells and in tissue samples compared to levels in para-cancerous tissues and a normal prostate epithelial cell line. While LINC00662 overexpression was shown to be positively correlated to both distant metastases and to shorter overall survival, in vitro LINC00662 silencing inhibited PCa cell proliferation and motility and promoted cell apoptosis. Yao et al. (2020a) also revealed that LINC00662 overexpression was positively associated with TNM staging, primary lesion size, lymph-node metastases, and distant metastases.
Cervical cancer (CC), a common malignant tumor in females, accounts for 529,800 newly diagnosed cases annually, with radio-resistance significantly reducing its therapeutic effect in many patients. Therefore, exploring the possible mechanisms governing CC tumorigenesis and progression remains important for the diagnosis and treatment of CC patients (Wei et al., 2020). In this regard, Wei et al. (2020) showed that LINC00662 expression was significantly upregulated both in CC tissues and in CC cells and that this high LINC00662 expression facilitated CC cell proliferation, motility, and radio-resistance. In contrast, knockdown of LINC00662 expression inhibited all of the above-mentioned CC cellular events. We therefore suggest that LINC00662 expression may be of therapeutic value as a diagnostic biomarker for reproductive-system tumors in clinical practice.
Nervous-System Tumors
As the most lethal of all primary brain tumors, gliomas account for 80% of all central nervous system neoplasms (Geng et al., 2020). Accumulating evidence also indicates that the abnormal expression of lncRNAs participate in the occurrence and development of nervous-system tumors. For glioma patients, Wu et al. (2020) showed that LINC00662 overexpression was related to both unfavorable clinical characteristics and poor prognoses; however, the silencing of LINC00662 was shown to suppress glioma cell proliferation and invasiveness in vitro. Moreover, they also confirmed that knocking down LINC00662 expression in an in vivo nude-mouse model inhibited glioma growth.
Unlike gliomas, chordomas are uncommon malignant tumors and still merit the identification of novel biomarkers for diagnosis and treatment development. Both LINC00662 and RNF144B expressions have been shown to be aberrantly upregulated in chordoma tissues, and their knockdowns resulted in the attenuation of chordoma cell proliferation, colony formation, invasiveness, migration, EMT, and glycolysis (Wang et al., 2020).
Taken together, these studies suggest that LINC00662 serves as an oncogene in both glioma and chordoma and should be considered as a potential biomarker for treating these two tumor types that originate in the nervous system.
Digestive-System Tumors
Colorectal cancer (CRC) is the deadliest and highest-incidence cancer worldwide, characterized by high rates of both metastasis and reoccurrence (Wang L. et al., 2019). Studies have reported that many lncRNAs participate in CRC progression, including MALAT1 (Xu et al., 2018) and GAS5 (Ni et al., 2019). Wang H. et al. (2019) found that LINC00662 was significantly upregulated in CRC cells and positively correlated with the degree of tumor differentiation, tumor stage, and lymphatic metastasis. The overexpression of LINC00662 was also an indicator of poor overall survival. However, Yao et al. (2020b) reported that the down-regulation of LINC00662 was also correlated with good patient prognosis and with the significant repression of CRC cell proliferation and promotion of apoptosis. The overexpression of LINC00662 was reported by Cheng B. et al. (2020) to enhance CRC cell invasiveness and migration and to dramatically accelerate CRC growth.
As the third most common cause of cancer-related deaths globally (He et al., 2020), hepatocellular carcinoma (HCC) represents an immense burden to society, with high rates of morbidity and mortality, particularly in China. Numerous research reports have shown that the expressions of lncRNAs (e.g., MIAT, HULC, and PDPK2P) are closely correlated with HCC tumorigenesis and progression. For LINC00662, Tian et al. (2020) reported that it was overexpressed in transforming growth factor beta-exposed HCC cells using existing transcriptomic data, and (Guo et al., 2020) reported that LINC00662 was up-regulated in HCC, positively correlated with patient survival, promoted HCC cell proliferation and motility, and reduced apoptosis.
Esophageal cancer is the eighth-most common cancer worldwide, with a 5-year survival rate lower than 20% (Hirano and Kato, 2019; Reichenbach et al., 2019). Histologically, esophageal cancer can be classified into squamous cell carcinoma and adenocarcinoma, as well as other types. Esophageal squamous cell carcinoma (ESCC) is especially prevalent in Linxian County, Henan Province, China. Zhang et al. (2020) demonstrated that the level of LINC00662 expression was upregulated in ESCC, and this was correlated with the adverse clinical characteristics for ESCC patients. Moreover, the knockdown of LINC00662 was shown to reduce ESCC cell proliferation, migration, and invasiveness.
The mortality rate of gastric cancer (GC) remains high due to its frequent diagnosis only at advanced stages, making it the third-most frequent cause of cancer-related deaths (Smyth et al., 2020). Liu et al. (2018) reported that high LINC00662 expression in GC tissues and cells was associated with poor patient prognoses compared to patients with low LINC00662 expressions. Furthermore, the knockdown of LINC00662 was shown to both suppress GC cell proliferation and to enhance GC chemo-sensitivity.
Among head and neck cancers, oral squamous cell carcinoma (OSCC) is considered highly aggressive and represents the top cause of deaths globally (Xu et al., 2019; Chen et al., 2020). Notably, Chen et al. (2020) revealed that compared to normal tissues, LINC00662 expression was elevated in OSCC and that blocking its expression ameliorated OSCC radio-resistance. Similarly, Xu et al. (2019) also demonstrated that the overexpression of LINC00662 was closely correlated with the adverse clinical characteristics of tumor size, tumor stage, and lymph node metastasis ascribed to its influences on OSCC cell proliferation and motility. The above studies shed new light on the value of lncRNAs for both therapeutic and prognostic purposes for digestive-system tumors and suggest that LINC00662 may be a promising biomarker for many digestive-system tumors.
Tumors in Other Systems
In the United States, potentially fatal melanoma is the fifth-most prevalent cancer in males and the sixth-most prevalent cancer in females (Marco Rastrelli, 2014). Xia et al. (2020) showed that, in patients with melanoma, high levels of LINC00662 expression were associated with shorter survival times and that both melanoma tissues and cell lines showed high LINC00662 expression levels. In addition, the knockdown of LINC00662 was shown to restrain cell proliferation, migration, and invasiveness, and in in vivo experiments, LINC00662 expression was shown to facilitate tumor growth.
Acute myeloid leukemia (AML) is a malignant disease caused by myeloid hematopoietic progenitor cells. It is characterized by the aberrant proliferation of primitive and immature myeloid cells in both the bone marrow and peripheral blood (Ari Pelcovits, 2020). Even with the best current treatments, AML prognoses are poor, especially in patients 65 years or older (De Kouchkovsky and Abdul-Hay, 2016). Therefore, a better understanding of AML pathogenesis and its molecular mechanisms is necessary for the development of novel AML therapies. In AML, Liu Y. et al. (2019) demonstrated that LINC00662 was upregulated in AML cells and significantly promoted AML cell growth, while LINC00662 downregulation was reported to suppress AML cell growth and to accelerate apoptosis. Therefore, LINC00662 can be regarded as a promising prognostic and therapeutic target for patients with OSCC, melanoma, and AML.
LINC00662 Mechanisms in Human Tumors
Studies have determined that lncRNAs are primarily localized to the nucleus and chromatin, indicating that their significant influence may be on DNA (West Jason et al., 2014; Sun et al., 2018). However, even though lncRNAs lack protein-coding ability, they also play important roles in a variety of cellular processes via many molecular mechanisms as described below. Some lncRNAs have cis epigenetic roles, interacting with proteins to mediate protein-coding gene expression (Wang et al., 2008), while other lncRNAs can interact with transcription factors to competitively suppress them, in a capacity known as a molecular decoy (Geisler and Coller, 2013). Some lncRNAs may also function as miRNA sponges to suppress miRNA activity (Cesana et al., 2011), modify enhancer activities, and modulate active-chromatin states (Wang et al., 2011). In addition, lncRNAs can also regulate gene expression post-transcriptionally by binding to antisense mRNAs (Carrieri et al., 2012). In terms of the molecular mechanisms by which LINC00662 acts in cancers, studies have increasingly shown that LINC00662 mainly participates by regulating target miRNA by acting as a ceRNA (Shuai et al., 2020).
LINC00662 Can Act as a ceRNA in Post-transcriptional Regulation
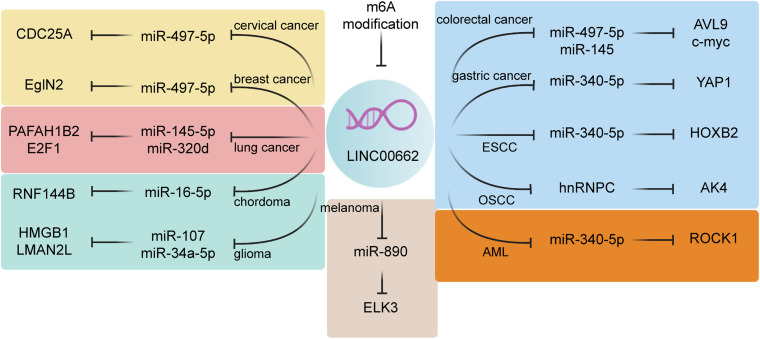
As long as RNA transcripts (e.g., lncRNAs, circular RNAs, and mRNAs) can combine with miRNA response elements (MREs), they are considered to have the ability to act as ceRNAs (Karreth and Pandolfi, 2013; Li X. et al., 2020). The examples mentioned above are entirely ncRNAs, so the ceRNA proposition suggests that miRNAs provide the link between ncRNAs and protein-coding RNAs. This idea of lncRNA/miRNA/mRNA network has previously been advocated as having an indispensable role in both tumorigenesis and the development of many human tumors. Many studies have confirmed that LINC00662 is an important lncRNA member and participates in this lncRNA/miRNA/mRNA network. The ceRNA functions attributed to LINC00662 are presented in Figure 1 and Table 2.
FIGURE 1.
The ceRNA network of LINC00662. LINC00662 is involved in a variety of tumors through the sponging of miRNAs, including miR-497-5p, miR-145-5p, miR-320d, miR-16-5p, miR-16-5p, miR-107, miR-34a-5p, miR-340-5p, hnRNPC, and miR-890. Yellow represents reproductive-system tumors; green represents nervous-system cancers, and blue represents digestive-system tumors. The level of LINC00662 expression is regulated by m6A-level modifications.
TABLE 2.
CeRNA networks related to LINC00662 in multiple cancers.
| CeRNA function of LINC00662 | Cancer type | References |
| miR-145-5p/PAFAH1B2 axis | lung cancer | Xu et al. (2020) |
| miR-320d/E2F1 axis | non-small cell lung cancer | Lv et al. (2021) |
| miR-497-5p/EglN2 axis | breast cancer | Cheng L. et al. (2020) |
| miR-497-5p/CDC25A axis | cervical cancer | Wei et al. (2020) |
| miR-34a-5p/LMAN2L axis | glioma | Geng et al. (2020) |
| miR-107/HMGB1 axis | glioma | Wu et al. (2020) |
| miR-16-5p/RNF144B axis | chordoma | Wang et al. (2020) |
| miR-497-5p/AVL9 axis | colorectal cancer | Wang H. et al. (2019) |
| miR-145/c-myc axis | colorectal cancer | Yao et al. (2020b) |
| miR-340-5p/HOXB2 axis | esophageal squamous cell carcinoma | Zhang et al. (2020) |
| miR-497-5p/YAP1 axis | gastric cancer | Liu et al. (2018) |
| hnRNPC/AK4 axis | oral squamous cell carcinoma | Chen et al. (2020) |
| miR-890/ELK3 axis | melanoma | Xia et al. (2020) |
| miR-340-5p/ROCK1 axis | acute myeloid leukemia | Liu Y. et al. (2019) |
LINC00662/miR-340-5p/mRNA for ROCK1/HOXB2
Liu Y. et al. (2019) demonstrated that LINC00662 could mediate miRNA-mRNA crosstalk interactions in AML by showing that LINC00662 combined directly with miR-340-5p and decreased its expression level. When LINC00662 was silenced, miR-340-5p levels were then significantly upregulated, AML cell proliferation was suppressed, and cell apoptosis increased. The restraint of miR-340-5p clearly attenuated the inhibitory effect of LINC00662 on proliferation suppression. In addition, decreased Rho-associated protein kinase 1 (ROCK1) expression was also shown to reverse the inhibitory effects of LINC00662 and miR-340-5p on proliferation. We therefore consider ROCK1 to be the target of miR-340-5p in AML cells. Currently, another important ESCC study has also reported a different ceRNA network related to the miRNA/mRNA axis. Zhang et al. (2020) showed that LINC00662 functioned as a ceRNA to sponge miR-340-5p and miR-340-5p led directly to the downregulation of homeobox B2 (HOXB2). Therefore, LINC00662 can upregulate HOXB2 expression and promote the development and progression of ESCC. In contrast, both the inhibition of HOXB2 and the upregulation of miR-340-5p slowed ESCC tumorigenesis. Taken together, these studies emphasize the vital roles of ceRNA interactions in mediating biological processes in both AML and ESCC.
LINC00662/miR-497-5p/mRNA EGLN2/CDC25A/AVL9/YAP1
Mechanistically, Cheng L. et al. (2020) found that LINC00662 promoted BC cell proliferation via the regulation of the miRNA/mRNA axis. Their bioinformatics analysis revealed that LINC00662 could be a sponge for miR-497-5p, and further BC studies showed that LINC00662 directly combined with it resulting in its downregulation. In addition, LINC00662 was shown to directly target Egl-9 family hypoxia inducible factor 2 (EGLN2), which was also considered to be a direct target for miR-497-5p. Therefore, this study revealed that LINC00662 overexpression accelerated BC cell growth via competitive binding with miR-497-5p and promoting EglN2 expression. Mechanistically in CC, LINC00662 was shown to also interact competitively with miR-497-5p, similar to its interaction in BC cells. In this study, Wei et al. (2020) showed that LINC00662, as a ceRNA, promoted CC progression and radio-resistance via the sponging of miR-497-5p, resulting in increased CDC25A levels. Therefore, this lncRNA/miRNA/mRNA axis in CC accelerated the malignant behavior of CC cells and the targeting of LINC00662 may enhance radio-resistance in these cells. In a CRC study, LINC00662 was found to exert a ceRNA function via the binding/decrease of miR-497-5p, leading to the overexpression of AVL9 (Wang H. et al., 2019). Collectively, LINC00662 appears to play a crucial role in development of CRC by modulating the miR-497-5p/AVL9 axis, shedding new light on CRC pathogenesis. Moreover, one of the vital regulatory mechanisms related to GC was the LINC00662/miR-497-5p/YAP1 ceRNA network proposed by Liu et al. (2018), where the knockdown of LINC00662 in GC cells was shown to suppress yes-associated protein 1 (YAP1) expression via the sponging of miR-497-5p. Furthermore, YAP1 has been shown to be a crucial downstream regulator of Hippo signaling and a mediator of GC-cell proliferation. Therefore, LINC00662 represents a potentially new biomarker for patients with BC, CC, CRC, and GC.
LINC00662/miR-16-5p/mRNA for RNF144B
Both LINC00662 and RNF144B have been shown to be aberrantly upregulated in chordoma tissues, and the knockdown of either LINC00662 or RNF144B impeded chordoma cell proliferation, colony formation, invasiveness, migration, EMT, and glycolysis. In addition, RNF144B overexpression was reported to reverse the effects of LINC00662 knockdown. Additionally, miR-16-5p was shown to target RNF144B and be a potential target for LINC00662. Thus, the relationship between LINC00662 and RNF144B was established through demonstrating their miR-16-5p interactions, and further studies also showed that the stable knockdown of LINC00662 inhibited tumor growth in vivo (Wang et al., 2020). These findings suggest that LINC00662 may also be an important therapeutic target for chordoma clinically.
LINC00662/miR-107/mRNA for HMGB1
Wu et al. (2020) showed that LINC00662 mechanistically regulated the miR-107/HMGB1 axis as a ceRNA. Moreover, the upregulation of high-mobility group box 1 protein (HMGB1) was reported to weaken the inhibition of glioma growth caused by the knockdown of LINC00662, suggesting that HMGB1 should be considered an miR-107 target. As a DNA-binding protein, HMGB1 is well-conserved, and many studies have shown that it accelerates the development of gliomas.
LINC00662/miR-34a-5p/mRNA for LMAN2L
Geng et al. (2020) reported that LINC00662 also acted as a ceRNA through miR-34a-5p binding and the resulting upregulation of lectin, mannose-binding 2-like protein (LMAN2L). For measures of functionality, the downregulation of LINC00662 has been shown to significantly suppress both cell proliferation and clonogenicity. Interestingly, inhibitors of miR-34a-5p (including LINC00662) have been reported to partially weaken the inhibitory effect on cell proliferation and migration induced by LINC00662 silencing in glioma cells. Therefore, these results suggest that the LINC00662/miR-34a-5p/LMAN2L axis can modulate glioma progression, bringing a new perspective to both glioma diagnosis and possible therapeutic methods for glioma patients.
LINC00662/miR-145-5p/mRNA for PAFAH1B2
Mechanistically, Xu et al. (2020) further revealed that LINC00662 functions as an LC oncogene (acting as a ceRNA) through the binding and downregulation of miR-145-5p. In addition, the knockdown of LINC00662 was shown to elevate miR-145-5p levels and result in the downregulation of platelet activating factor acetylhydrolase 1B catalytic subunit 2 (PAFAH1B2). Specifically, Xu et al. (2020) also found that miR-145-5p could combine with the 3′untranslated region (UTR) of PAFAH1B2 for negative regulation and suppressing miR-145-5p blocked the LC-repressive effect of LINC00662 silencing.
LINC00662/miR-890/mRNA for ELK3
LINC00662 has been reported to combine with miR-890, with ELK3 being the downstream gene target of miR-890. Furthermore, miR-890 has also been shown to negatively regulate ELK3 expression. Using rescue assays, the overexpression of ELK3 was shown to reverse the inhibitory effects of either LINC00662 knockdown or miR-890 mimics on the cell proliferative, migratory, and invasive abilities of melanoma (Xia et al., 2020).
LINC00662/miR-320d/mRNA for E2F1
LINC00662 has also been shown to act as an miR-320d sponge in non-small cell LC cells, and E2F transcription factor 1 (E2F1) is known to be a target for miR-320d in these cells. In addition, exosomal LINC00662 was reported to accelerate non-small cell LC progression via the sponging of miR-320d in vitro, and this exosomal LINC00662 also significantly enhanced non-small cell LC growth. Thus, this exosomal LINC00662 study showed that non-small cell LC progression was promoted through modulation of the miR-320d/E2F1 axis and expands our understanding of potential exosomal LINC00662 mechanisms. LINC00662, miR-320d, and E2F1 may therefore all serve as potential targets for non-small cell LC therapies (Lv et al., 2021).
LINC00662/hnRNPC/mRNA for AK4
Chen et al. (2020) reported that LINC00662 could interact with both heterogeneous nuclear ribonucleoprotein C (hnRNPC) and adenylate kinase 4 (AK4) in OSCC and that LINC00662 mainly interacted with hnRNPC protein to regulate AK4 mRNA stability, resulting in AK4 protein overexpression. Interestingly, LINC00662 was shown not to be able to interact with hnRNPC directly, and its modulation of OSCC cell radiosensitivity was shown to be through hnRNPC-modulated AK4. In fact, the knockdown of LINC00662 actually enhanced OSCC-cell radiosensitivity via the upregulation of AK4.
LINC00662/miR-145/mRNA for c-myc
The results of a bioinformatics analysis reported miR-145 to be a speculative miRNA target for LINC00662. In addition, LINC00662 was shown to directly interact with miR-145 and to reduce its expression in CRC. The upregulation of miR-145 was also reported to attenuate CRC cell growth and to accelerate apoptosis, while its suppression was shown to markedly reverse the inhibitory effect of LINC00662 knockdown on CRC cell growth. In addition, the recovery of c-myc expression was reported to partially reverse the inhibitory effects on CRC cell growth mediated either by LINC00662 low-expression or by miR-145 overexpression. Taken together, this study indicates that LINC00662 modulated CRC cell biology by raising c-myc levels via its binding with miR-145 and that the LINC00662/miR-145/c-myc axis plays a crucial role in regulating CRC cell growth (Yao et al., 2020b).
The Interaction Between LINC00662 and Protein
In addition to acting as a ceRNA, LINC00662 has also been shown to influence genomic methylation through its interactions not only with miRNA but also with protein (Guo et al., 2020). This research demonstrated that the effect of LINC00662 was mainly on the expressions of methionine adenosyltransferase 1A (MAT1A) and S-adenosylhomocysteine hydrolase (AHCY); key enzymes for the production of S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH). On the one hand, as previously described, LINC00662 exerted its ceRNA function via the MAT1A 3′UTR by direct sponging, thereby reducing MAT1A mRNA and downregulating MAT1A protein expression. On the other hand, and more importantly, LINC00662 was also shown to directly interact with AHCY protein, thereby facilitating AHCY instability via the upregulation of its ubiquitin-mediated degradation (Guo et al., 2020). Taken together, LINC00662 has also been shown to participate in the activation of multiple oncogenes through protein interactions, shedding additional light on possible LINC00662 mechanisms.
Pathway Related to LINC00662 Function in Cancers
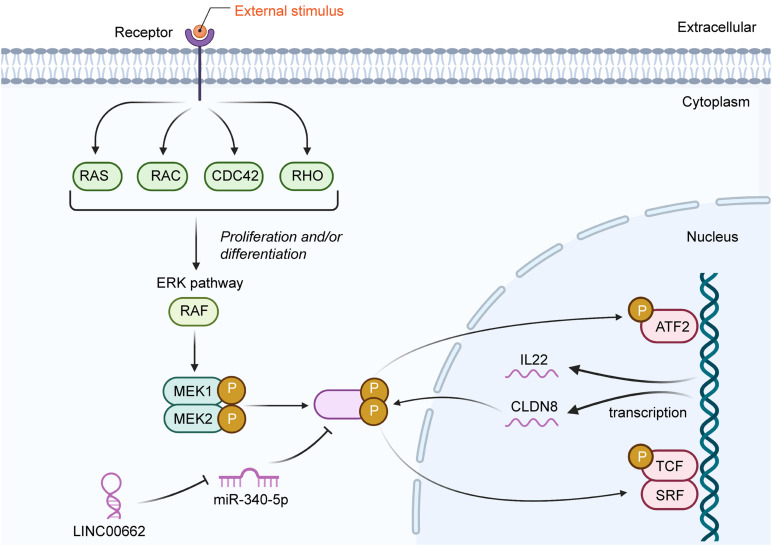
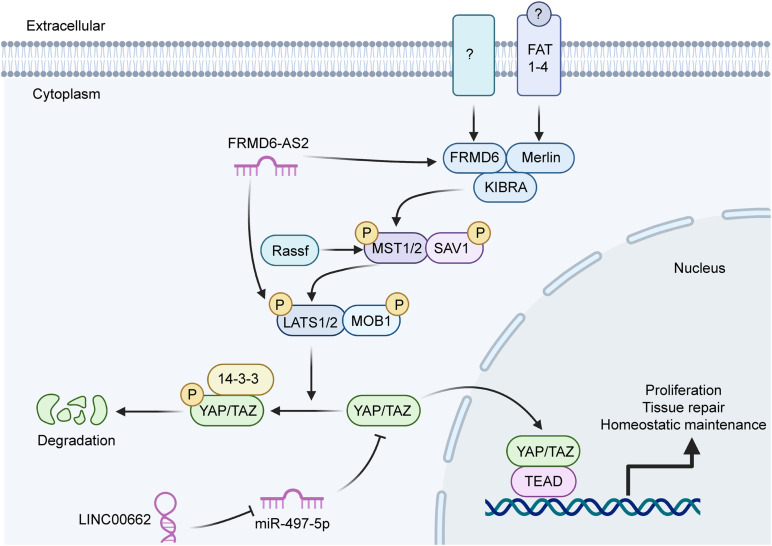
A variety of signaling pathways are active in cancer cells and are indispensable for their cellular processes. Recent evidence has indicated that lncRNAs, including LINC00662, are involved in many of these signaling pathways (Table 3). The extracellular signal-related kinase (ERK) signaling pathway is a vital contributor to many cellular processes and to the survival of cancer cells (Wen et al., 2019). The ERK family has five main members (ERK1–5); ERK1 and ERK2 are involved in regulating many biological processes, including meiosis and mitosis (Hu et al., 2019). LINC00662 has been shown to activate ERK signaling by raising the expression levels of both claudin 8 (CLDN8) and interleukin 22 (IL22) via the targeting of miR-340-5p (Cheng B. et al., 2020). These two genes play important roles in the pathogenesis of several intestinal diseases, including colon cancer, by promoting cell growth and metastasis (Figure 2). The Hippo signaling pathway has also become recognized as being important in GC. Liu et al. (2018) previously found that this pathway was linked to many cellular events via many biological molecules. Specifically, a decrease in LINC00662 expression has been shown to significantly reduce YAP1 expression, resulting in modulation of YAP1-mediated GC cell proliferation by the sponging of miR-497-5p (Figure 3). In addition, the Wnt/β-catenin signaling pathway also is of great importance for regulating cell proliferation in tumors. Xu et al. (2019) reported that the expression of LINC00662 promoted the activation of the Wnt/β-catenin signaling pathway. In their study, the upregulation of LINC00662 significantly increased Wnt3a and β-catenin proteins, while the knockdown of LINC00662 inhibited these proteins and blocked the overall activation of the Wnt/β-catenin signaling pathway. These studies indicate that LINC00662 may exert its oncogenic functions by regulating mitogen-activated protein kinase (MAPK)/ERK signaling, Hippo signaling, and the Wnt/β-catenin signaling pathway.
TABLE 3.
Signaling pathways related to LINC00662.
| Pathways related to LINC00662 | Cancer type | PMID |
| MAPK/ERK signaling pathway | colon cancer | Cheng B. et al. (2020) |
| Hippo signaling pathway | gastric cancer | Liu et al. (2018) |
| Wnt/β-catenin signaling pathway | oral squamous cell carcinoma | Xu et al. (2019) |
FIGURE 2.
A schematic diagram of LINC00662 participation in the ERK signaling pathway. LINC00662 promotes p-ERK via binding to (and suppressing) miR-340-5p. Both CLDN8 and IL22 are target genes of ERK, and CLDN8 promotes p-ERK simultaneously.
FIGURE 3.
A schematic representation of LINC00662 involved in the Hippo signaling pathway. LINC00662 regulates YAP1 through the sponging of miR-497-5p, similar to the ERK signaling-pathway mechanism. Activation of the Hippo signaling pathway promotes cell proliferation, tissue repair, and homeostatic maintenance.
Potential Clinical Application of LINC00662
As a Biomarker for Cancer Diagnosis
In recent years, accumulating evidence has indicated that lncRNAs could be detected in plasma and tissues as biomarkers for the diagnosis of cancers (Li et al., 2019). Interestingly, LINC00662 has been found to have a potential diagnostic value for patients with lung cancer (Gong et al., 2018), chordoma (Wang et al., 2020), colorectal cancer (Wang H. et al., 2019; Yao et al., 2020b), gastric cancer (Liu et al., 2018), and acute myelocytic leukemia (Liu Y. et al., 2019). Gong et al. (2018) observed that the LINC00662 expression level in lung cancer tissues was significantly higher than that in adjacent normal tissues. LINC00662 expression was also upregulated in colorectal cancer tissues, betokening potential application of LINC00662 in colorectal cancer diagnosis (Wang H. et al., 2019; Yao et al., 2020b).
As a Prognostic Biomarker for Cancer
Based on the recent studies, the overexpression of LINC00662 was significantly linked to poor prognosis in different cancer types. Guo et al. (2020) demonstrated that the increased expression of LINC00662 was associated with lower overall survival (P = 0.0071) and recurrence-free survival rate (P = 0.0064) in HCC. Gong et al. (2018) found that LINC00662 expression level in lung cancer was higher in advanced TNM stages. Therefore, overexpression of LINC00662 led to poor prognosis of lung cancer. Similarly, cervical cancer patients with high LINC00662 expression level had shorter overall survival time and relapse free survival time (Wei et al., 2020). Xu et al. (2019) found that in OSCC, LINC00662 expression was significantly correlated with tumor size, TNM stage, and lymph node metastasis. Many researches had reported that LINC00662 was a prognostic biomarker for patients with prostate cancer (Li et al., 2019), lung cancer (Wang et al., 2020), glioma (Geng et al., 2020; Wu et al., 2020), melanoma (Xia et al., 2020), colon cancer (Cheng B. et al., 2020), colorectal cancer (Wang H. et al., 2019), and gastric cancer (Liu et al., 2018).
As a Therapeutic Target for Cancer
Though not much study has been done on the therapeutic value of LINC00662 for cancer patients, studies on LINC00662 in tumor have provided the probability of LINC00662 as a therapeutic target for cancers. Xia et al. (2020) found that suppression of LINC00662 repressed cell proliferation, migration, and invasion of melanoma, which indicated that LINC00662 could be a promising therapeutic target for patients with melanoma. Xu et al. (2019) discovered that downregulation of LINC00662 inhibited the proliferation, migration, and invasion abilities of OSCC cell, and promoted apoptosis. In vivo, LINC00662-depleted U87 cells were injected into the BALB/C immunodeficiency mice, and the tumor volumes and weights of the LINC00662 knockdown group were significantly decreased. Therefore, silenced LINC00662 suppressed glioma proliferation in vivo, proving that LINC00662 could serve as therapeutic potential in glioma (Geng et al., 2020). Moreover, LINC00662 also could be a novel therapeutic target for treatment of chordoma (Wang et al., 2020), prostate cancer (Li et al., 2019), breast cancer (Cheng L. et al., 2020), lung cancer (Gong et al., 2018), AML (Liu Y. et al., 2019), gastric cancer (Liu et al., 2018), and colorectal cancer (Wang H. et al., 2019; Yao et al., 2020b).
Conclusion
Technological developments have provided efficient research tools for exploring lncRNAs and have led to the discovery of many lncRNAs in recent years (Ma et al., 2018; Xing et al., 2021). Accumulating research has identified crucial roles for lncRNAs in both tumor occurrence and progression (Peng et al., 2017), and the dysregulation of LINC00662 is among those identified as leading to a variety of cancers. The transcription site for LINC00662 is located on the 19th chromosome from site 28,281,401 to 28,284,848. Consistently, LINC00662 has been shown to be overexpressed in many different tumors, including lung cancer, breast cancer, cervical cancer, prostate cancer, chordoma, glioma, gastric cancer, and hepatocellular carcinoma. Mechanistically, when localized to the cytoplasm, LINC00662 has been shown to serve as a ceRNA for gene regulation and to influence RNA metabolism. In parallel, LINC00662 has also been shown to participate in regulating mRNA stability as a mediator of gene expression. Notably, LINC00662 has also been shown to interact with protein and RNA and to participate in a variety of vital signaling pathways, including the MAPK/ERK pathway, the Hippo pathway, and the Wnt/β-catenin signaling pathway.
Although the traditional treatment methods have been greatly developed, at present, the clinical treatment of cancer still faces with many problems. High recurrence rate and low treatment accuracy are still obstacles to further improve the prognosis of cancer patients. Therefore, LINC00662, which is a potential biomarker for early prediction of cancer initiation, is a novel promising target for cancer. However, the studies of LINC00662 are limited and mainly focus on investigating the vital role of LINC00662 in oncogenesis. We consider that the upstream molecular mechanism controlling the expression of LINC00662 remains to be revealed. Moreover, the interaction of ncRNA network and the clinical effectiveness of targeting LINC00662 are also unclear. Though further clinical studies are needed to assess the clinical value of LINC00662 in different tumors, LINC00662 still shows extraordinary promise.
Author Contributions
YH and WG designed the study. YX and ZS searched the articles and made figures. YH and XY wrote this manuscript. All authors worked collaboratively on the work presented here, read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (819028328), the Youth Talent Lifting Project of Henan Province (2021HYTP059), and Key Scientific Research Project of Henan Higher Education Institutions of China (21A320026).
References
- Anastasiadou E., Jacob L. S., Slack F. J. (2018). Non-coding RNA networks in cancer. Nat. Rev. Cancer 18 5–18. 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ari Pelcovits R. N. (2020). Acute myeloid leukemia: a review. R I Med. J. 103 38–40. [PubMed] [Google Scholar]
- Botti G., Scognamiglio G., Aquino G., Liguori G., Cantile M. (2019). LncRNA HOTAIR in tumor microenvironment: what role? Int. J. Mol. Sci. 20:2279. 10.3390/ijms20092279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491 454–457. 10.1038/nature11508 [DOI] [PubMed] [Google Scholar]
- Cech T. R., Steitz J. A. (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157 77–94. 10.1016/j.cell.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147 358–369. 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zheng R., Baade P. D., Zhang S., Zeng H., Bray F., et al. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66 115–132. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- Chen Y., Bao C., Zhang X., Lin X., Fu Y. (2020). Knockdown of LINC00662 represses AK4 and attenuates radioresistance of oral squamous cell carcinoma. Cancer Cell Int. 20:244. 10.1186/s12935-020-01286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B., Rong A., Zhou Q., Li W. (2020). LncRNA LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR-340-5p to regulate CLDN8/IL22 co-expression and activating ERK signaling pathway. J. Exp. Clin. Cancer Res. 39:5. 10.1186/s13046-019-1510-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Xing Z., Zhang P., Xu W. (2020). Long non-coding RNA LINC00662 promotes proliferation and migration of breast cancer cells via regulating the miR-497-5p/EglN2 axis. Acta Biochim. Pol. 67 229–237. 10.18388/abp.2020_5203 [DOI] [PubMed] [Google Scholar]
- Coker H., Wei G., Brockdorff N. (2019). m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim. Biophys. Acta Gene. Regul. Mech. 1862 310–318. 10.1016/j.bbagrm.2018.12.002 [DOI] [PubMed] [Google Scholar]
- Consortium E. P. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E. P., Moore J. E., Purcaro M. J., Pratt H. E., Epstein C. B., Shoresh N., et al. (2020). Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583 699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Kouchkovsky I., Abdul-Hay M. (2016). ‘Acute myeloid leukemia: a comprehensive review and 2016 update’. Blood Cancer J. 6:e441. 10.1038/bcj.2016.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Chen X., Zhang Q., Liu Q. (2020). Historical trends in breast Cancer among women in China from age-period-cohort modeling of the 1990-2015 breast Cancer mortality data. BMC Public Health 20:1280. 10.1186/s12889-020-09375-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducoli L., Agrawal S., Sibler E., Kouno T., Tacconi C., Hon C. C., et al. (2021). LETR1 is a lymphatic endothelial-specific lncRNA governing cell proliferation and migration through KLF4 and SEMA3C. Nat. Commun. 12:925. 10.1038/s41467-021-21217-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duma N., Santana-Davila R., Molina J. R. (2019). Non-Small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc. 94 1623–1640. 10.1016/j.mayocp.2019.01.013 [DOI] [PubMed] [Google Scholar]
- Essa H., Dobson R., Wright D., Lip G. Y. H. (2020). Hypertension management in cardio-oncology. J. Hum. Hypertens 34 673–681. 10.1038/s41371-020-0391-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahad Ullah M. (2019). Breast cancer: current perspectives on the disease status. Adv. Exp. Med. Biol. 1152 51–64. 10.1007/978-3-030-20301-6_4 [DOI] [PubMed] [Google Scholar]
- Feng Y. C., Liu X. Y., Teng L., Ji Q., Wu Y., Li J. M., et al. (2020). c-Myc inactivation of p53 through the pan-cancer lncRNA MILIP drives cancer pathogenesis. Nat. Commun. 11:4980. 10.1038/s41467-020-18735-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Colombet M., Soerjomataram I., Dyba T., Randi G., Bettio M., et al. (2018). Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur. J. Cancer 103 356–387. 10.1016/j.ejca.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Fidler M. M., Soerjomataram I., Bray F. (2016). A global view on cancer incidence and national levels of the human development index. Int. J. Cancer 139 2436–2446. 10.1002/ijc.30382 [DOI] [PubMed] [Google Scholar]
- Geisler S., Coller J. (2013). RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14 699–712. 10.1038/nrm3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y., Wu Y., Xu C., Li T., Zhang L. (2020). Long Non-Coding RNA LINC00662 regulated proliferation and migration by targeting miR-34a-5p/LMAN2L Axis in Glioma. Onco Targets Ther. 13 10161–10172. 10.2147/OTT.S272616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., Su Y., Liu Y., Sun P., Wang X. (2018). Long non-coding RNA Linc00662 promotes cell invasion and contributes to cancer stem cell-like phenotypes in lung cancer cells. J. Biochem. 164 461–469. 10.1093/jb/mvy078 [DOI] [PubMed] [Google Scholar]
- Gong X., Dong T., Niu M., Liang X., Sun S., Zhang Y., et al. (2020). lncRNA LCPAT1 upregulation promotes breast cancer progression via enhancing MFAP2 Transcription. Mol. Ther. Nucleic Acids 21 804–813. 10.1016/j.omtn.2020.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelet S., Link L. A., Howley B., Obellianne C., Palanisamy V., Gangaraju V. K., et al. (2017). A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 19 1105–1115. 10.1038/ncb3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Gong C., Wu P., Battaglia-Hsu S. F., Feng J., Liu P., et al. (2020). LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Differ. 27 2191–2205. 10.1038/s41418-020-0494-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Yu X., Li J., Zhang Q., Zheng Q., Guo W. (2020). Role of m 5 C-related regulatory genes in the diagnosis and prognosis of hepatocellular carcinoma. Am. J. Transl. Res. 12 912–922. [PMC free article] [PubMed] [Google Scholar]
- Hirano H., Kato K. (2019). Systemic treatment of advanced esophageal squamous cell carcinoma: chemotherapy, molecular-targeting therapy and immunotherapy. Jpn. J. Clin. Oncol. 49 412–420. 10.1093/jjco/hyz034 [DOI] [PubMed] [Google Scholar]
- Hu H., Dong Z., Wang X., Bai L., Lei Q., Yang J., et al. (2019). Dehydrocorydaline inhibits cell proliferation, migration and invasion via suppressing MEK1/2-ERK1/2 cascade in melanoma. Onco Targets Ther. 12 5163–5175. 10.2147/OTT.S183558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Xu X. E., Jiang Y. Z., Liu Y. R., Sun W., Guo Y. J., et al. (2019). The endogenous retrovirus-derived long noncoding RNA TROJAN promotes triple-negative breast cancer progression via ZMYND8 degradation. Sci. Adv. 5:eaat9820. 10.1126/sciadv.aat9820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansara S., Pandey V., Lobie P. E., Sethi G., Garg M., Pandey A. K. (2020). Mechanistic involvement of long non-coding RNAs in oncotherapeutics resistance in triple-negative breast cancer. Cells 9:1511. 10.3390/cells9061511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth F. A., Pandolfi P. P. (2013). ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 3 1113–1121. 10.1158/2159-8290.CD-13-0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li Z., Zheng W., Li X., Wang Z., Cui Y., et al. (2017). LncRNA-ATB: an indispensable cancer-related long noncoding RNA. Cell Prolif. 50 12–20. 10.1111/cpr.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Zhang L. Y., Qiao Y. H., Song R. J. (2019). Long noncoding RNA LINC00662 functions as miRNA sponge to promote the prostate cancer tumorigenesis through targeting miR-34a. Eur. Rev. Med. Pharmacol. Sci. 23 3688–3698. [DOI] [PubMed] [Google Scholar]
- Li X., Ding J., Wang X., Cheng Z., Zhu Q. (2020). NUDT21 regulates circRNA cyclization and ceRNA crosstalk in hepatocellular carcinoma. Oncogene 39 891–904. 10.1038/s41388-019-1030-0 [DOI] [PubMed] [Google Scholar]
- Li Y., Jiang T., Zhou W., Li J., Li X., Wang Q., et al. (2020). Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat. Commun. 11:1000. 10.1038/s41467-020-14802-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. J., Cheng J., Song Y., Li H. H., Zheng J. F. (2021). LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia. Lab Invest. 101 341–352. 10.1038/s41374-020-00519-9 [DOI] [PubMed] [Google Scholar]
- Liu L., Wang H. J., Meng T., Lei C., Yang X. H., Wang Q. S., et al. (2019). lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-Signaling Pathway in CRC. Mol. Ther. Nucleic Acids 17 644–656. 10.1016/j.omtn.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu Y., Gao X., Tian X. (2019). High expression of long intergenic non-coding RNA LINC00662 contributes to malignant growth of acute myeloid leukemia cells by upregulating ROCK1 via sponging microRNA-340-5p. Eur. J. Pharmacol. 859:172535. 10.1016/j.ejphar.2019.172535 [DOI] [PubMed] [Google Scholar]
- Liu Z., Yao Y., Huang S., Li L., Jiang B., Guo H., et al. (2018). LINC00662 promotes gastric cancer cell growth by modulating the Hippo-YAP1 pathway. Biochem. Biophys. Res. Commun. 505 843–849. 10.1016/j.bbrc.2018.09.191 [DOI] [PubMed] [Google Scholar]
- Long Y., Wang X., Youmans D. T., Cech T. R. (2017). How do lncRNAs regulate transcription? Sci. Adv. 3:eaao2110. 10.1126/sciadv.aao2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Wang K., Yeh S., Sun Y., Liang L., Xiao Y., et al. (2019). LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat. Commun. 10:2571. 10.1038/s41467-019-09784-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X., Lian Y., Liu Z., Xiao J., Zhang D., Yin X. (2021). Exosomal long non-coding RNA LINC00662 promotes non-small cell lung cancer progression by miR-320d/E2F1 axis. Aging (Albany NY) 13 6010–6024. 10.18632/aging.202522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Zhang J., Wen L., Lin A. (2018). Membrane-lipid associated lncRNA: a new regulator in cancer signaling. Cancer Lett. 419 27–29. 10.1016/j.canlet.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Marco Rastrelli S. T. (2014). Carlo Riccardo Rossi Melanoma: epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo 28 1005–1011. [PubMed] [Google Scholar]
- Matsui M., Corey D. R. (2017). Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 16 167–179. 10.1038/nrd.2016.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim F., Sabath B. F., Eapen G. A. (2019). Lung cancer. Med. Clin. North Am. 103 463–473. 10.1016/j.mcna.2018.12.006 [DOI] [PubMed] [Google Scholar]
- Ni W., Yao S., Zhou Y., Liu Y., Huang P., Zhou A., et al. (2019). Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer 18:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Fang S., Tian H., Zhou C., Zhao X., Tian H., et al. (2020). lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/beta-catenin signaling. Mol. Cancer 19:9. 10.1186/s12943-020-1133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W. X., Koirala P., Mo Y. Y. (2017). LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36 5661–5667. 10.1038/onc.2017.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach Z. W., Murray M. G., Saxena R., Farkas D., Karassik E. G., Klochkova A., et al. (2019). Clinical and translational advances in esophageal squamous cell carcinoma. Adv. Cancer Res. 144 95–135. [DOI] [PubMed] [Google Scholar]
- Schmidt K., Joyce C. E., Buquicchio F., Brown A., Ritz J., Distel R. J., et al. (2016). The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep. 15 2025–2037. 10.1016/j.celrep.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Feng X. P., Hu R. B., Wang H., Wang Y. L., Qian J. H., et al. (2021). N-methyladenosine reader YTHDF2-mediated long noncoding RNA FENDRR degradation promotes cell proliferation in endometrioid endometrial carcinoma. Lab Invest. 101 775–784. 10.1038/s41374-021-00543-3 [DOI] [PubMed] [Google Scholar]
- Shuai Y., Ma Z., Lu J., Feng J. (2020). LncRNA SNHG15: a new budding star in human cancers. Cell Prolif. 53:e12716. 10.1111/cpr.12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Chinnaiyan A. M. (2019). The role of non-coding RNAs in oncology. Cell 179 1033–1055. 10.1016/j.cell.2019.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth E. C., Nilsson M., Grabsch H. I., van Grieken N. C. T., Lordick F. (2020). Gastric cancer. Lancet 396 635–648. 10.1016/s0140-6736(20)31288-5 [DOI] [PubMed] [Google Scholar]
- Sun Q., Hao Q., Prasanth K. V. (2018). Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 34 142–157. 10.1016/j.tig.2017.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Wu Y., Yang Y., Wang J., Niu M., Gao S., et al. (2020). Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/β-catenin signaling. Mol. Oncol. 14 462–483. 10.1002/1878-0261.12606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Yang L., Yu C., Zhu W., Zhou X., Xiong Y., et al. (2020). Tumor-Secreted Exosomal lncRNA POU3F3 promotes cisplatin resistance in ESCC by inducing fibroblast differentiation into CAFs. Mol. Ther. Oncolytics 18 1–13. 10.1016/j.omto.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre L. A., Siegel R. L., Ward E. M., Jemal A. (2016). Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol. Biomarkers Prev. 25 16–27. 10.1158/1055-9965.epi-15-0578 [DOI] [PubMed] [Google Scholar]
- Wang C. B., Wang Y., Wang J. J., Guo X. L. (2020). LINC00662 triggers malignant progression of chordoma by the activation of RNF144B via targeting miR-16-5p. Eur. Rev. Med. Pharmacol. Sci. 24 1007–1022. [DOI] [PubMed] [Google Scholar]
- Wang G., Zhao D., Spring D. J., DePinho R. A. (2018). Genetics and biology of prostate cancer. Genes Dev. 32 1105–1140. 10.1101/gad.315739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Huo X., Yang X. R., He J., Cheng L., Wang N., et al. (2017). STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 16:136. 10.1186/s12943-017-0680-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yu M., Hu W., Chen X., Luo Y., Lin X., et al. (2019). Linc00662 promotes tumorigenesis and progression by regulating miR-497-5p/AVL9 axis in colorectal cancer. Front. Genet. 10:1385. 10.3389/fgene.2019.01385 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang J., Su Z., Lu S., Fu W., Liu Z., Jiang X., et al. (2018). LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin. Chim. Acta 485 229–233. 10.1016/j.cca.2018.07.004 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhu S., Meng N., He Y., Lu R., Yan G. R. (2019). ncRNA-encoded peptides or proteins and cancer. Mol. Ther. 27 1718–1725. 10.1016/j.ymthe.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C., Yang Y. W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472 120–124. 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Cho K. B., Li Y., Tao G., Xie Z., Guo B. (2019). Long Noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 20:5758. 10.3390/ijms20225758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., et al. (2008). Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454 126–130. 10.1038/nature06992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Wang L., Sun Y., Bao Y. (2020). LINC00662 contributes to the progression and the radioresistance of cervical cancer by regulating miR-497-5p and CDC25A. Cell Biochem. Funct. 38 1139–1151. 10.1002/cbf.3580 [DOI] [PubMed] [Google Scholar]
- Wei W., Zhao X., Zhu J., Zhang L., Chen Y., Zhang B., et al. (2019). lncRNAu50535 promotes the progression of lung cancer by activating CCL20/ERK signaling. Oncol. Rep. 42 1946–1956. 10.3892/or.2019.7302 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wen Z., Jiang R., Huang Y., Wen Z., Rui D., Liao X., et al. (2019). Inhibition of lung cancer cells and Ras/Raf/MEK/ERK signal transduction by ectonucleoside triphosphate phosphohydrolase-7 (ENTPD7). Respir. Res. 20:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West Jason A., Davis Christopher P., Sunwoo H., Simon Matthew D., Sadreyev Ruslan I., Wang Peggy I., et al. (2014). The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55 791–802. 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Guo X., Xu D., Zhang H. (2020). LINC00662 sponges miR-107 accelerating the invasiveness and proliferation of glioma cells. J. Cancer 11 5700–5712. 10.7150/jca.46381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. Q., Lu W. L., Ye Y. Y., Chen J. (2020). LINC00662 promotes cell proliferation, migration and invasion of melanoma by sponging miR-890 to upregulate ELK3. Eur. Rev. Med. Pharmacol. Sci. 24 8429–8438. [DOI] [PubMed] [Google Scholar]
- Xing C., Sun S. G., Yue Z. Q., Bai F. (2021). Role of lncRNA LUCAT1 in cancer. Biomed. Pharmacother 134:111158. 10.1016/j.biopha.2020.111158 [DOI] [PubMed] [Google Scholar]
- Xingming Jiang N. M. (2015). Dayong Wang Metformin inhibits tumor growth by regulating multiple miRNAs in human cholangiocarcinoma. Oncotarget 6 3178–3194. 10.18632/oncotarget.3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Chen Z., Chen W., Li Q., Lin B., Jia Y. (2020). FKBP-related ncRNA-mRNA axis in breast cancer. Genomics 112 4595–4607. 10.1016/j.ygeno.2020.08.017 [DOI] [PubMed] [Google Scholar]
- Xu D., Chen Y., Yuan C., Zhang S., Peng W. (2019). Long non-coding RNA LINC00662 promotes proliferation and migration in oral squamous cell carcinoma. Onco Targets Ther. 12 647–656. 10.2147/OTT.S188691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhang X., Hu X., Zhou W., Zhang P., Zhang J., et al. (2018). The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol. Med. 24:52. 10.1186/s10020-018-0050-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. Y., Peng J., Shi Z. Z., Chen X. L., Cheng H. Z., Wang H., et al. (2020). Silencing linc00662 inhibits cell proliferation and colony formation of lung cancer cells via regulating miR-145-5p/PAFAH1B2 axis. Biochem. Cell Biol. 99 330–338. 10.1139/bcb-2019-0396 [DOI] [PubMed] [Google Scholar]
- Yang B., Zhang L., Cao Y., Chen S., Cao J., Wu D., et al. (2017). Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol. Cancer 16:154. 10.1186/s12943-017-0722-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Xu D., Li Z., Yao J., Pan Z., Chen J. (2020b). Long intergenic non-coding RNA LINC00662 contributes to malignant growth of colorectal cancer cells by upregulating c-myc via sponging microRNA-145. Biosci. Rep. 10.1042/BSR20200011 Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Yao Z. F., Pan Z. Y., Yao Y. W., Chen J. F. (2020a). Long noncoding RNA Linc00662 promotes the tumorigenesis of prostate cancer cells. Zhonghua Nan Ke Xue 26 588–594. [PubMed] [Google Scholar]
- Yari H., Jin L., Teng L., Wang Y., Wu Y., Liu G. Z., et al. (2019). LncRNA REG1CP promotes tumorigenesis through an enhancer complex to recruit FANCJ helicase for REG3A transcription. Nat. Commun. 10:5334. 10.1038/s41467-019-13313-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li Z., Zheng H., Chan M. T., Wu W. K. (2017). NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif. 50:e12329. 10.1111/cpr.12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang X., Gu J., Zhang J., Shi H., Hou S., Xu X., et al. (2020). Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 11:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Li S., Lu J., Ge Y., Wang Q., Ma G., et al. (2018). LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer 17:87. 10.1186/s12943-018-0829-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Liang X., Ren L., Zhang S., Li S., Wan T., et al. (2020). LINC00662 promotes cell viability and metastasis in esophageal squamous cell carcinoma by sponging miR-340-5p and upregulating HOXB2. Thorac. Cancer 11 2306–2315. 10.1111/1759-7714.13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Sun H., Kong H., Chen Z., Chen B., Zhou M. (2017). The Lncrna-TUG1/EZH2 axis promotes pancreatic cancer cell proliferation, migration and EMT phenotype formation through sponging Mir-382. Cell Physiol. Biochem. 42 2145–2158. 10.1159/000479990 [DOI] [PubMed] [Google Scholar]
- Zhao W., Geng D., Li S., Chen Z., Sun M. (2018). LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 7 842–855. 10.1002/cam4.1353 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao Z., Sun W., Guo Z., Zhang J., Yu H., Liu B. (2020). Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 254:116900. 10.1016/j.lfs.2019.116900 [DOI] [PubMed] [Google Scholar]
- Zhou R. S., Zhang E. X., Sun Q. F., Ye Z. J., Liu J. W., Zhou D. H., et al. (2019). Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer 19:779. 10.1186/s12885-019-5983-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Sun Y., Huang D., Li M. J. (2020). epiCOLOC: integrating large-scale and context-dependent epigenomics features for comprehensive colocalization analysis. Front. Genet. 11:53. 10.3389/fgene.2020.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K. P., Ma X. L., Zhang C. L. (2017). LncRNA ODRUL contributes to osteosarcoma progression through the miR-3182/MMP2 Axis. Mol. Ther. 25 2383–2393. 10.1016/j.ymthe.2017.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]