Graphical abstract
Peptide-inhibitor hits were designed with small molecules published before used as the starting point through structure-based drug design and biochemically and structurally characterized the interaction modes between the FK1 domain of FKBP51 and hits.
Keywords: FKBP51, X-ray crystallography, Structure-based drug design, Molecular dynamics simulation, Peptide-inhibitor hit
Abstract
FKBP51 is well-known as a cochaperone of Hsp90 machinery and implicated in many human diseases including stress-related diseases, tau-mediated neurodegeneration and cancers, which makes FKBP51 an attractive drug target for the therapy of FKBP51-associated diseases. However, it has been reported that only nature product rapamycin, cyclosporine A, FK506 and its derivatives exhibit good binding affinities when bound to FKBP51 by now. Given the advantages of peptide-inhibitors, we designed and obtained 20 peptide-inhibitor hits through structure-based drug design. We further characterized the interaction modes of the peptide-inhibitor hits on the FK1 domain of FKBP51 by biochemical and structural biology methods. Structural analysis revealed that peptide-inhibitor hits form U-shaped conformations and occupy the FK506 binding pocket and share similar interaction modes with FK506. Using molecular dynamics simulations, we delved into the interaction dynamics and found that hits are anchored to the FK506 binding pocket in a quite stable conformation. Meanwhile, it was shown that interactions between FK1 and peptide-inhibitor hits are mainly attributed to the hydrogen bond networks comprising I87 and Y113 and FPF cores of peptide-inhibitors involved extensive hydrophobic interactions. We presumed that the peptide design strategy based on the small molecule structure probably shed new lights on the peptide-inhibitor discovery of other targets. The findings presented here could also serve as a structural basis and starting point facilitating the optimization and generation of FKBP51 peptide-inhibitors with better bio-activities.
1. Introduction
FK506 binding protein 51 (FKBP51), encoded by gene FKBP5 located at the chromosome 6 [1], [2], functions as a cochaperone in heat shock protein 90 (Hsp90) machinery. It has been revealed that FKBP51 comprises three distinct domains including FKBP12 like domain1 and 2 (named as FK1 and FK2) and C-terminal tetratricopeptide repeated (TPR) domain [3]. The FK1 domain conferring FKBP51 the peptidyl-prolyl isomerase activity displays high binding affinities to immunosuppressant compounds FK506 and rapamycin, which leads to FKBP51 being classified as an immunoglobulin protein while the function of FK2 domain is currently unclear [3], [4]. The TPR domain is mainly responsible for specifically binding to the extreme C-terminal of HSP90 [5], [6]. During the last decade, numerous studies have revealed that FKBP51 plays significant roles in steroid hormone receptor mature, hormone binding and nuclear translocation, especially for androgen, glucocorticoid and progesterone receptors, while itself could be induced by respective hormones [7], [8], [9], [10], [11]. Various studies also suggested that FKBP51 is involved in the NF-κB-mediated cancers and inflammation. For the effect of FKBP51 on the NF-κB signaling pathway, controversial results were obtained. Sun et al. reported that FKBP51 promotes the NF-κB activation in which both TPR and PPIase domains are required [12]. In contrast, Galigniana et al. determined that the inhibitory effect of FKBP51 on the NF-κB nuclear translocation rate and the transcription activity is independent on the FK1 domain of FKBP51 [13]. Meanwhile, it was reported that FKBP51 is associated to the development of stress-related diseases [14], [15], [16], [17] and certain cancers [18], [19], [20]. Even recently, Dickey et al. has reported that FKBP51, to a larger extent, is implicated in the Tau-mediated neurodegeneration diseases [21]. These studies all suggest that FKBP51 is relevant to the occurrence of many diseases and therefore would be an attractive drug target for the therapy of FKBP51-associated diseases.
To obtain FKBP51-targeted inhibitors of good quality, substantial works have been performed and the structure of the FK1 domain of FKBP51 in complex with the nature product FK506 was described. Many FK506 derivatives have been further rationally designed and evaluated by Hausch group [22], [23], [24]. Though these FK506 derivatives exhibit strong binding affinities to FKBP51, they hardly selectively recognize the FKBP51 from FKBPs yet, especially FKBP52 which is the FKBP51 closest paralog with 55% sequence identity and structural similarities but antagonistic functions. After extensive studies, the iFit series compounds which could discriminate the FKBP51 and FKBP52 were later discovered [25].
Compared with small molecule compounds and antibodies, peptide-inhibitors indeed represent a unique class of molecules because of their sizes and balance of the rigidity and flexibility. It was reported that peptides account for up to 40% protein–protein interactions which are found to be implicated in the development of several diseases [26]. Peptide-inhibitors sometimes could be a better choice to disrupt abnormal protein–protein interactions and further turns the peptide-based inhibitor design into a promising industry. Since insulin was first introduced into the clinical use for the therapy of type 1 diabetes in 1921, almost 80 peptide drugs have been approved by food and drug administration (FDA) and reached the market for a wide range of diseases. Over past decades, peptide-drugs experienced the revival of scientific interests because of their excellent complements to small molecules and biological macromolecules. It is known that the design of small molecules adopting FKBP51-selectivities is a daunting challenge, though iFit series compounds reported in 2015, because of presences of other subtypes of FKBPs. For this class of targets, the peptide-inhibitors exhibit much more potencies than small molecule compounds in view of the advantages of peptide-inhibitors including easy synthesis, lower toxicities, remarkable selectivities and larger surface areas. Therefore, we here designed and obtained 20 FKBP51-targeted peptide-inhibitor hits by combining structure-based drug design and Gaussian accelerated molecular dynamics (GaMD) simulations. Furthermore, peptide-inhibitor hits were synthesized and their binding affinities to FKBP51 were determined via fluorescence polarization (FP) and isothermal titration calorimetry (ITC) assays. Meanwhile, the binding modes and interaction dynamics were characterized by using X-ray crystallography and MD simulations. We expect that our findings would provide a new strategy for the design of peptide-inhibitors and a starting point for FKBP51-targeted peptide-inhibitor discoveries.
2. Results
2.1. Peptide-inhibitors design and the spontaneous folding study
Previously, many FKBP51-inhibitors have been reported. Amongst these inhibitors, the iFit series compounds were well-known not only for their high bio-activities, but also good FKBP51-selectivities. Therefore, it seemed that inhibitors designed based on iFit compounds would have great potencies to selectively leverage FKBP51. By analyzing structural characteristics of FK1 in complex with iFit series compounds (PDB ID: 5DIU, 5DIV, 4TW6, 4TW7 and 5DIT) [25], [27], we found that the middle part of iFit compounds, cyclohexyl moiety and pyridine, occupy the bottom of the FK506 binding pocket and tightly contact with FK1, indicating these moieties probably play crucial roles in the interactions between FK1 and compounds. Intriguingly, it was shown that both cyclohexyl moiety and pyridine are linked with amide moiety, which makes cyclohexyl moiety and pyridine structurally resemble proline and phenylalanine, respectively. Besides, it was reported that a hydrophobic loop from handle region of RYR1 is anchored into the FKBP12 pocket which shares 48% sequence identity and the similar structure with the FK1 domain of FKBP51 (PDB ID: 3J8H) [28]. Given the advantages of peptide-inhibitors over small molecules, we expected that we could design and obtain the peptide-inhibitors which harbored good FKBP51-targeted activities and selectivities by substituting iFit compounds with amino acids. To this end, we firstly divided iFit series compounds into five fragments and designated as A3′, A2′, A1, A2 and A3 (Fig. S1), respectively. As shown in the Fig. S1, A1 and A2 are very similar with proline and phenylalanine structurally. Therefore, we replaced A1 and A2 with proline and phenylalanine and yield a dipeptide “stub” and the molecular docking was employed to study the interaction between the stub and FK1 (Fig. S2A). It was revealed that pyrrole of proline occupied the pocket formed by Y57, F77, W90 and F130 while the side chain of phenylalanine inserted into the sub-pocket created by the sidechains of D68, K121, I122, K66 and L61, which was consistent with the interaction details of cyclohexyl moiety and pyridine in the FK506 binding pocket. By using the amino acids substitution, we next extended the stub at both terminuses and a small pool of 4-mer peptide-inhibitors were generated. It was found that A3 could be substituted with A, G, Q, S or N by docking scores and binding modes (Table S1). The structural analysis revealed that the interaction mode between peptide QFP and FK1 was quite identical to that between iFit compounds and FK1 with the residue Q being proximity to D68 and hydrogen bonding to residues Y57, D68, S69 and R72 (Fig. S2B). Furthermore, the docking results suggested that peptides had favorable docking scores (Table S2) and exhibited similar binding modes with iFit series compounds when A2’ was displaced with F, P, I or L (Fig. S3). Collaborating with results mentioned above, A3’ was replaced by random residues and the molecular docking was also carried out to decipher the interaction mode and evaluate the binding affinity. In silico experiments showed that the resulting peptides exhibited similar interaction patterns but different bio-activities. Finally, 20 peptide-inhibitor hits were selected and subjected to the further assessment.
Considering the flexibility of peptide-inhibitors in the solution, GaMD simulations [29] were performed to investigate the spontaneous folding of peptide-inhibitor QFPFV which was taken as a representative hit started from the extended conformation. As summarized in the Table 1, the GaMD potential boost was 11.700 kcal/mol with the standard deviation of 3.142 kcal/mol. All three simulations were combined to construct the free energy scale with the distance between terminal residues of the peptide-inhibitor and the root mean square deviation (RMSD) relative to the expected conformation used as reaction coordinates. The 2D PMF profile reweighting was performed using cumulant expansion to the second order since the anharmonicity of potential energy ΔV distribution was calculated to be 0.012 (Fig. 1A) [30], [31]. The results of reweighted 2D PMF (Fig. 1B) showed that three distinct conformational states, unfolded (U), folded (F) and intermediate (I), were captured during three combined GaMD simulations. The unfolded state corresponded to the global energy minimum which was centered at (3.0 Å, 11.8 Å) while the intermediate and folded state free energy wells were centered around (2.5 Å, 8.8 Å) and (2.0 Å, 4.3 Å), respectively. The structural clustering was carried out with CPPTRAJ [32] module to analyze the most probable conformation of peptide-inhibitor QFPFV in the solution. The top 5 ranked structural centroids were extracted and we found that the structural centroid of cluster Ⅳ formed the stable U-shaped conformation which was similar to the binding state of peptide-inhibitor hit QFPFV on the FK1, indicating that peptide-inhibitors we designed possibly adopted the FK1 binding-competent conformation (Fig. 1C).
Table 1.
Summary of GaMD simulations performed to capture the folding state of peptide-inhibitor hit.
| Systems | Natoms | ID | Length (ns) | ΔVavg (kcal/mol) | σΔV (kcal/mol) |
|---|---|---|---|---|---|
| Peptide-inhibitor: QFPFV | 6168 | Sim1 | 500 | 11.692 | 3.137 |
| Sim2 | 500 | 11.696 | 3.141 | ||
| Sim3 | 500 | 11.711 | 3.146 |
Natoms is the atoms number of the simulation systems. ΔVavg is the average of GaMD boost potential. σΔV is the standard error of GaMD boost potential.
Fig. 1.
The spontaneous folding of peptide-inhibitor hit QFPFV was monitored via GaMD. The distribution of boost potential ΔV with the anharmonicity of 0.012 (A). The 2D (RMSD to expected conformation, distance between terminal atoms) PMF (B) was built by reweighting three 500-ns GaMD simulations combined. Structural alignment of the expected FKBP51 binding-competent conformation with folded conformation obtained by GaMD (C).
2.2. The interactions between peptide-inhibitors and FK1 determined by using FP and ITC assays
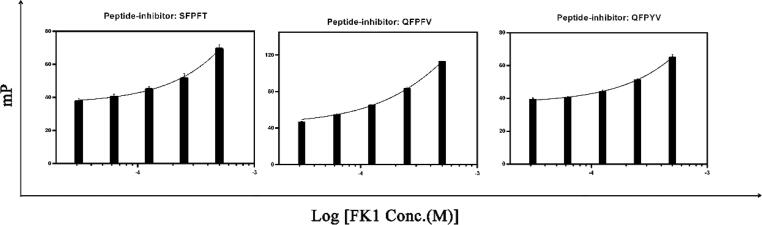
To characterize the binding affinities between peptide-inhibitor hits and FKBP51, the sequence encoding the FK1 domain of FKBP51 was cloned into pET28b vector. FK1 was over-expressed in E. coli BL21 (DE3) and further purified to homogeneity with the purity over 95%. Given the peptide-inhibitor sequence similarities and synthesis costs, 3 peptide-inhibitor hits with FITC-tag and 10 tag-free peptide-inhibitor hits were synthesized. The FP and ITC assays were further carried out to evaluate the interactions between FK1 and peptide-inhibitors. The peptide TAT was treated as a non-inhibitory control to rule out the non-specific binding (Fig. S4). It was revealed that peptide-inhibitor hits interacted with FK1 in a dose-dependent manner. The dissociation constants were not obtained because of the protein costs and weak binding affinities (Fig. 2,). Using ITC assays, the binding affinities were estimated and the thermodynamics parameters were calculated. The results showed that all 10 peptide-inhibitor hits could modestly bind to FK1 with dissociation constants higher than 400 μM (Fig. 3). Moreover, it was clearly revealed that the interactions between hits and FK1 were mainly enthalpy forced while the entropy was adverse for the complex formation. According to the rules concluded by Ross et al. [33], the negative ΔH0 and ΔS0 indicate that the hydrogen bonds and van der Waals’ introduced as the consequence of hydrophobic interactions were the main force contributing to the interaction. Notably, these results of FP and ITC implied that all peptide-inhibitors we designed probably shared similar interaction modes on the FK1 domain of FKBP51, which further favorably proved the results of protein-peptide dockings.
Fig. 2.
Interactions between peptide-inhibitor hits and FK1 were determined via fluorescence polarization assays. FITC-tagged peptide-inhibitors were incubated with different concentrations (ranging from 3.125 to 500 μM) of FK1. Millipolarization was measured and data were processed with GraphaPad Prism 6 and fitted with one-phase association model. Each panel was labelled with the peptide-inhibitor hit sequence on the top.
Fig. 3.
The isothermal titration calorimetry assays were employed to determinate the interactions between peptide-inhibitors and FK1. Data were processed with MicroCal PEAQ-ITC Analyze software and the dissociation constants and thermodynamic parameters were listed in each panel. Every figure was labelled with the peptide-inhibitor hit sequence on the top.
2.3. Structures of FK1 in complex with peptide-inhibitor hits determined using X-ray diffraction
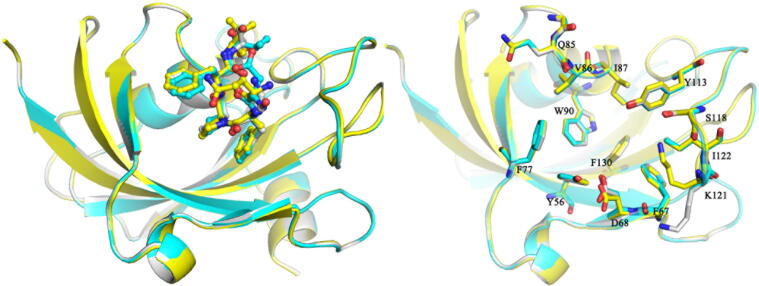
In order to investigate binding modes between hits and FK1, we solved the structures of FK1-SFPFT, FK1-QFPFV and FK1-DFPFV at resolutions of 1.39 Å, 1.50 Å and 1.31 Å, respectively. Three forms of crystals all belonged to P212121 space group with one molecule in the asymmetric unit. Three final structure models were well defined with clear electron density and consisted of residues from G16 to G139/E140/E140, 219/110/205 water molecules and one peptide-inhibitor molecule. Although the sidechains of terminal residues of peptide-inhibitor hits were not well defined because of the ambiguous density, the 2FO-FC electron-density maps clearly revealed that peptide-inhibitor hits SFPFT, DFPFV and QFPFV occupied the FK506 binding pocket with the P3 sitting in the bottom and the pyrrole faced with the side chain of W90 (Fig. 4A–C). It was found that peptide-inhibitor hits SFPFT, DFPFV and QFPFV formed U-shape conformations which were mainly stabilized by hydrogen bonds formed between S1/Q1/D1 and F4. The sidechain from P3 sandwiched between sidechains of F2 and F4 formed stacking interactions which further stabilized the U-shaped conformation (Fig. 4D–F). Structural alignment clearly showed that three structures could be superimposed pretty well with the RMSD less than 0.1 Å, which further confirmed our inference generated from the results of FP and ITC assays (Fig. 5). Unfortunately, structural studies revealed that the interaction modes of peptide-inhibitor hits with FK1 were entirely different from that we expected.
Fig. 4.
Overall structures of SFPFV-FK1 (A), QFPFT-FK1 (B) and DFPFV-FK1 (C) complexes. The FK1 was shown as surface and peptide-inhibitors were shown as sticks. The 2FO-FC electron maps of peptide-inhibitors SFPFT (D), QFPFV (E) and DFPFV (F) were contoured at 0.8 σ and shown in blue and intra-molecule hydrogen bonds of three peptide-inhibitors were depicted as yellow dash lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Superposition of SFPFT-, DFPFV- and QFPFV-FK1 complex structures. Peptide-inhibitor hits were shown as sticks and residues within 4 Å were displayed as lines. SFPFT- DFPFV- and QFPFV-FK1 complex structures were colored grey, cyan and yellow, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In the complex of FK1-QFPFV, F2 occupied the hydrophobic pocket and made hydrophobic contacts to a number of surrounding residues including F67, Y57, W90 and F130. It was also displayed that F4 and P3 engaged F77 in T-shape stacking and alkyl/π stacking interactions (Fig. 6D–F), respectively. Apart from hydrophobic interactions, two polar contacts were directly formed between the carbonyl oxygen from F4 and hydroxyl of Y113 and the main-chain nitrogen of V5 and carbonyl oxygen from Q85 at distances of 2.8 Å and 3.1 Å, respectively. Meanwhile, there was another water-mediated hydrogen bond formed between the nitrogen of I87 and carbonyl oxygen from P3 ((Fig. 6D–F). In comparison with the FK1-QFPFV complex, the sidechains of F2, P3 and F4 involved extensive hydrophobic stacking interactions and polar contacts mentioned above were maintained in FK1-SFPFT and FK1-DFPFV complexes (Fig. 6A–C, H, I). In addition, the water-bridged hydrogen bonds between F2 and S118, L119 were discovered in the FK1-SFPFT and FK1-DFPFV structures (Fig. 6A–C, H, I). Comparison of three peptide-inhibitor binding patterns demonstrated that hydrogen bond networks comprising the I87 backbone and Y113 sidechain and the F2-, P3- and F4-sidechain involved hydrophobic interactions were conserved in the structures we obtained, which further verified the conclusion drawn from FP and ITC assays.
Fig. 6.
Interaction details of peptide-inhibitors SFPFT, QFPFV and DFPFV with FK1. Top-view and side-view of the interactions between FK1 and peptide-inhibitors SFPFT (A, B), QFPFV (D, E) and DFPFV (G, H). Peptide-inhibitors were shown as sticks and residues contributing to the interaction were displayed as lines. Hydrogen bonds were depicted with yellow dash lines and water molecules were represented with red spheres. The 2D interaction diagrams of peptide-inhibitors SFPFT (C), QFPFV (F) and DFPFV (I) with FK1 were generated with Ligplot. The hydrogen bond interactions were displayed as dash lines and labelled with distances. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Superposition of peptide-inhibitor bound FK1 structures with FK1 native structure (PDB ID: 3O5P) [34] showed that bindings of peptide-inhibitor hits were quite rigid and barely induced marked conformational changes in the FK506 binding pocket (Fig. 7A, B). Furthermore, it was found that peptide-inhibitors replaced other 11 water molecules presented in the native FK1 structure but the water403, which was still present in complex structures we obtained and bridged the polar contact between I87 and P3. The displacement of water molecules might contribute an important entropy to the binding energy. Comparison of FK1-peptide structures with the FK1-iFit complex structure (PDB ID: 5DIU) disclosed that the bindings of peptide-inhibitor hits didn’t like iFit series compounds displacing the F67 sidechain and inducing conformation changes in the FK506 binding pocket, namely peptide-inhibitor hits probably not harboring FKBP51-selectivities. Nevertheless, detailed structural analysis revealed that peptide-inhibitors were surrounded by a few hydrophobic residues including Y57, F67, D68, F77, Q85, V86, I87, W90, Y113, S118 and F130, which are located on the rim of pocket and involved in the interactions between iFit series compounds (PDB ID: 5DIU) (Fig. 7E, F) [25] or FK506 (PDB ID: 3O5R) (Fig. 7C, D) [34] and FK1 as well, indicating our peptide-inhibitor hits probably leverage the FKBP51-involved biological processes which could be modulated by FK506 or iFit series compounds.
Fig. 7.
Comparison of FK1-peptide-inhibitor complex structures with native FK1 (A, B), FK1-FK506 complex (C, D) and FK1-iFit complex (E, F). Ligands were shown as sticks and residues within 4 Å were displayed as lines. All ligands were not shown for clarity in figure B, D and F. The native structure of FK1 was shown in magenta. The SFPFT-, QFPFV- and DFPFV-FK1 structures were colored white, yellow and green while the FK1-iFit and FK1-FK506 complex structures were colored cyan and light purple, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.4. MD simulations to monitor the interaction dynamics between FK1 and peptide-inhibitor hits
To inspect the interaction dynamics between FK1 and peptide-inhibitor hits, the 300-ns conventional MD simulations were carried out with structures obtained experimentally used as initial coordinates. By monitoring root mean square deviation (RMSD) of complex backbone, we found that RMSDs of three systems fluctuated around 1.0 Å throughout the MD simulations, implying three complex structures were fairly stable (Fig. 8A). To evaluate the binding stabilities of peptide-inhibitors SFPFT, DFPFPV and QFPFV, the RMSD evolutions of peptide-inhibitors were monitored and the results showed that RMSDs of peptide-inhibitors SFPFT and DFPFV fluctuated around 0.5 Å during the simulations while that of peptide-inhibitor QFPFV decreased from 1.5 Å to 0.8 Å rapidly and oscillated around 0.8 Å, which suggested that U-shaped conformations of three peptide-inhibitors were quite stable when bound to FK1 (Fig. 8B). The root mean square fluctuations (RMSFs) of three systems suggested that most parts of the FK1 domain were rather rigid, except some loop regions and terminal residues (Fig. 8C). As for peptide-inhibitor hits, it was shown that the terminal residues S/T, D/V and Q/V exhibited relative flexibilities compared to the FPF cores of peptide-inhibitors (Fig. 8D). The hydrogen bonds directly formed between peptide-inhibitors and FK1 were traced and we found that hydrogen bonds formed between residues Q85, Y113, I87 of FK1 and T5 (V5 for peptide-inhibitors QFPFV and DFPFV), F4, P3 from peptide-inhibitors were conserved and maintained in the last 20-ns simulations with occupancies higher than 60%. Additionally, one more hydrogen bond formed between F2 and L119 was discovered in the FK1-DFPFV simulation system (Fig. 9).
Fig. 8.
Peptide-inhibitor hits interact with FK1 in a quite stable U-shaped conformation. The RMSD evolutions of the complexes (A) and peptide-inhibitors (B) were monitored to evaluate the stabilities of MD simulation systems. The RMSFs of backbone atoms from FK1 (C) and peptide-inhibitors (D) were calculated to assess the flexibilities of the complexes.
Fig. 9.
The hydrogen bonds formed between I87 and P3, Y113 and F4, Q85 and T5/V5 of QFPFV-FK1 (A, B), SFPFT-FK1 (C, D) and DFPFV-FK1 (E, F) systems were monitored during the last 20-ns simulations and their occupancies were calculated and showed with histograms.
We next calculated the binding free energies of peptide-inhibitor hits with FK1 using MM-GBSA method. As shown in the Table 2, the peptide-inhibitors SFPFT, QFPFV and DFPFV favorably bound to FK1 with binding free energies of −7.97 kcal/mol, −5.75 kcal/mol and −8.10 kcal/mol, respectively. The results also revealed that the non-polar interaction (−51.23 kcal/mol for QFPFV, −53.49 kcal/mol for SFPFT, −53.60 kcal/mol for DFPFV) was the main contributor to the interactions between peptide-inhibitor hits and FK1 while the polar solvation free energy led to the polar interaction (20.05 kcal/mol for QFPFV, 20.32 kcal/mol for SFPFT, 18.72 kcal/mol for DFPFV) being adverse for the binding. Meanwhile, the results indicated that the interactions between FK1 and peptide-inhibitors were mainly enthalpy (−31.18 kcal/mol for QFPFV, −33.07 kcal/mol for SFPFT, −34.88 kcal/mol for DFPFV) forced, which was in agreement with the results of thermodynamic parameters generated via ITC.
Table 2.
The binding free energies for QFPFV-FK1, SFPFT-FK1 and DFPFV-FK1 systems calculated using MM-GBSA.
| Terms (kcal/mol) | QFPFV | SFPFT | DFPFV |
|---|---|---|---|
| ΔE vdw, gas | −44.60 | −47.02 | −46.93 |
| ΔE ele, gas | −19.86 | −24.59 | −74.53 |
| ΔE nonpl, sol | −6.63 | −6.47 | −6.67 |
| ΔE polar, sol | 39.90 | 44.91 | 93.24 |
| ΔE polar, total | 20.05 | 20.32 | 18.72 |
| ΔE nonpl, total | −51.23 | −53.39 | −53.60 |
| ΔH | −31.18 | −33.07 | −33.48 |
| -TΔS | −25.43 | −25.11 | −25.38 |
| ΔG | −5.75 | −7.97 | −8.10 |
ΔE vdw, gas: van der walls energy in gas state; ΔE ele, gas: electrostatic energy in gas state; ΔE nonpl, sol: nonpolar solvation energy; ΔE polar, sol: polar solvation energy; ΔE polar, total: polar free energy; ΔE nonpl, total: nonpolar free energy; ΔH: enthalpic contributions; -TΔS: entropic contributions; ΔG: binding free energy.
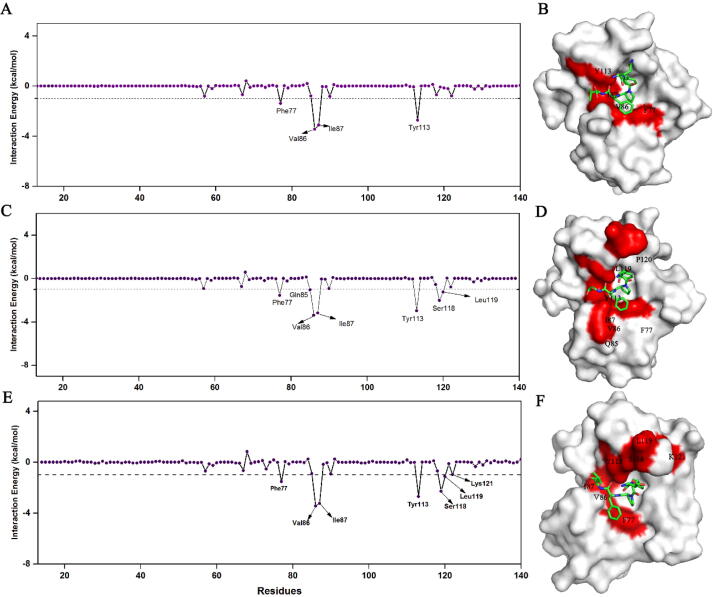
To further map out the hotspot residues contributing to the binding, the per-residue energy decomposition was performed and residues with energy contributions greater than 1 kcal/mol were indicated (Fig. 10). It was shown that all of hotspot residues including F77, V86, I87 and Y113 were conserved in three systems. Apart from residues listed above, four more residues, Q85, L119, K121 and P120, found in the FK1-SFPFT and FK1-DFPFV systems, also played significant roles in peptide-inhibitor bindings. The energy contributions of residues from peptide-inhibitors were characterized as well and the results unveiled that N-terminal residues of three peptides were adverse for the interactions between peptide-inhibitors and FK1 whereas other four residues were favorable for the bindings with the energy contributions greater than 1 kcal/mol (Fig. S5). Collectively, these findings from MD simulations clearly showed that three peptide-inhibitor hits could bind to the FK506 binding pocket on FK1 with a quite stable U-shaped conformation. The hydrogen bond networks presented in three complexes were conserved and the residues contributing to the binding were similar in three complexes, which further proved the results obtained from X-ray crystallography. In addition, it was indicated that FPF cores of peptide-inhibitor hits were the main contributors of the interactions between FK1 and hits.
Fig. 10.
The per-residue binding free energy spectrums of QFPFV-FK1 (A) SFPFT-FK1 (B) and DFPFV-FK1 (C) systems. Residues contributing less than −1 kcal/mol were indicated. The FK1 was shown as surface and peptide-inhibitor hits were displayed as sticks while the key residues were highlighted in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
FKBP51 has emerged as a key regulator of the steroid hormone receptor signaling including receptor maturation and nuclear translocation. Recently, it was shown that FKBP51 is implicated in many cellular signaling pathways and various FKBP51-interacting partners including Hsp90, steroid hormone receptors, NF-κB and Tau were reported. FKBP51 was thought as a contributor of many clinical diseases including neurodegeneration, stress-related diseases and cancers, which made FKBP51 an attractive drug target for the therapy of FKBP51-related diseases. Substantial works have been carried out to explore lead compounds leveraging FKBP51. Many FK506 derivatives were discovered and published by Hausch group. In this study, by combining structure-based drug design and peptide-protein docking, we designed 20 peptide-inhibitor hits and biochemically characterized interactions between FK1 and 10 peptide-inhibitors by using FP and ITC assays. The binding experiments showed that all 10 peptide-inhibitor hits could slightly bind to FK1, which was mainly enthalpy driven. We further determined structures of FK1 in complex with 3 peptide-inhibitor hits using X-ray crystallography. Structural studies revealed that peptide-inhibitor hits occupied the FK506 binding pocket on FK1 without inducing marked conformational changes. Structural alignment clearly revealed that three peptide-inhibitor hits shared identical binding modes when bound to FK1. Furthermore, MD simulations were employed to delve into the interaction dynamics and we found that interactions between FK1 and peptide-inhibitor hits were mainly attributed to FPF cores of peptide-inhibitors implicated extensive hydrophobic interactions and polar contacts while the terminal residues barely contributed to the binding. Meanwhile, the binding free energy and the entropic change were obtained by using MM-GBSA calculation and normal mode analysis, respectively. It was indicated that enthalpy was the main force driving the interactions between FK1 and peptide-inhibitor hits, which was in accordance with the results of FP and ITC. It was worthy to note that the effect of the entropic cost on the interaction was significant although the favorability of enthalpy outweighed entropy compensation. Based on that, we speculated that the modest binding affinity could probably result from the entropy compensation which was introduced as the consequence of high flexibilities of peptide-inhibitor hits. Collaborating with results of per-residue energy decomposition, we presumed that the cyclization and any other method increasing the propensities of peptide-inhibitors folding to FKBP51 binding-competent conformation with the FPF core treated as the starting point would facilitate the optimization in future.
It was reported that FK506 functioning as an inhibitor could downregulate the FKBP51-promoted NF-κB signaling activation and further reduce the apoptosis resistance of melanoma via suppressing the FKBP51 isomerase activity [12]. The finding that FK506 negatively regulates the proliferation of prostate cancer through targeting FKBP51 was reported by Periyasamy et al. [35]. Herein, structural analysis showed that structures of FK1 in complex with peptide-inhibitor hits and FK1-FK506 could be superposed pretty well with the RMSD of 0.161 Å over 141 atoms, indicating that peptide-inhibitor hits and FK506 bound to FKBP51 in a similar binding mode. Reasonably, we assumed that our peptide-inhibitors were likely to interfere with FK506-modulated biological processes as well through targeting the FK1 domain of FKBP51. We believed that our findings could provide a novel clue for the rational design of peptide-inhibitors targeting FKBP51 and a molecule probe for studies of FKBP51-invovled signaling pathways.
As a paralog of FKBP51, FKBP52 is also reported to be an important regulator in modulating steroid hormone receptor maturation, hormone binding and nuclear translocation and involved in the development of neurodegeneration diseases and a few cancers [36]. Even though the FK1 domain of FKBP52 and FKBP51 shared the 70% sequence identity and structural similarities, they acted frequently in an antagonistic manner. Therefore, the design and discovery of inhibitors harboring FKBP51-selectivities are urgently needed. To the best of our knowledge, iFit series compounds which could discriminate the FKBP51 and FKBP52 were reported, for the first time, in 2015 by Hausch group, but the number of these compounds was still too small. As peptide-inhibitors exhibit many advantages over small molecules when the target contains multifunctional subtypes, we here aimed at designing the FKBP51-targeted peptide-inhibitors by using structure-based drug design. Since the insulin was first medically used for the therapy of type 1 diabetes in 1921, 80 peptide-drugs have been approved, of which most were hormone-derived and others were mainly from the natural products of plant, bacteria, fungi and even venom [37]. In this work, a novel peptide-design method that peptide-inhibitor hits were derived from the reported small molecule compounds, iFit series compounds in this case, was described and applied. Originally, we expected that our peptide-inhibitors could harbor FKBP51-selectivities because of the qualities of iFit series compounds and work as a complement to the small molecule compounds in view of advantages of the peptide-inhibitors. Unfortunately, structural analysis clearly suggested that our peptide-inhibitors could not induce conformational changes in the FK506 binding pocket and further discriminate the FKBP51 from other FKBPs. We hope that our design strategy would be applicable to the design of peptide-inhibitors of other targets. Meanwhile, the results presented here showed that the interference of FKBP51-invovled signaling pathways by peptide-inhibitors was practicable and we expected that our peptide-inhibitor hits could provide a starting point and guideline for the discovery of FKBP51-selective peptide-inhibitors.
4. Conclusions
Herein, by combining computational modeling and structure-based drug design, we obtained 20 peptide-inhibitor hits targeting FKBP51. Further biochemical and structural studies were performed to characterize the interaction details between peptide-inhibitor hits and the FK1 domain of FKBP51. Using MD simulations, the interaction dynamics between FK1 and peptide-inhibitor hits were further explored. Our results unveiled the interaction modes and dynamics between FK1 and peptide-inhibitors. The binding affinities were demonstrated to be in the low micromolar regime. Guided by results of X-ray crystallography and MD simulations, we hope that our peptide-inhibitor hits could provide a good starting point for the generation of peptide-inhibitors which harbor better FKBP51 binding affinities and selectivities.
5. Materials and methods
5.1. Peptide synthesis
All peptides used in this work were synthesized via standard Fmoc-based solid-phase synthetic methods. Crude peptides were obtained and purified to homogeneity using preparative HPLC. The purities of peptides were further ascertained through analytical HPLC, and the structure assignment was performed by ESI-TOF MS. The purified peptides were dissolved into phosphate buffer saline and stocked at −20 °C for further use.
5.2. Docking-based peptide-inhibitors design
The iFit series inhibitors were divided into 5 segments according to their structural characteristics. A series of natural peptide structures were designed using bioelectronic isosteric and random amino acid substitution methods. For the characterization of the binding mode and binding affinity, the peptide-protein docking was performed by using the protein-peptide docking module of Schrödinger (Schrödinger, LLC, New York, USA). Finally, 20 pentapeptide-inhibitor hits were selected by docking scores and interaction modes and subjected to further evaluation.
5.3. Protein expression and purification
The tag-free FK1 (residues G16-E140 with A19T mutation) domain of FKBP51 was codon-optimized and synthesized at Genewiz (Genewiz, Suzhou, China) and cloned into pET28b expression vector. The plasmid was transformed into E. coli BL21 (DE3) and was cultured in 50 ml fresh Luria Broth (LB) broth supplemented with kanamycin at 37 °C overnight. The culture was inoculated into 1 L LB broth and further cultured to an optical density (OD600) of 0.6–0.8. The expression of FK1 was induced by adding 0.2 μM IPTG and cultured extra 20 h at 16 ℃. Cells were collected by centrifugation at 5000 rpm for 10 min and the pellet was resuspended with buffer A (20 mM Tris-HCl pH 6.0). Cells were lysed by sonication in ice-bath for 30 min with 1-s pulse and 3-s interval under the 40% output of the machine. The debris was removed by centrifuging at the speed of 12000 rpm at 4 °C for 50 min. The supernatant was loaded onto the pre-equilibrated SPFF column (Sunresin, Xian, China) with buffer A. The protein was eluted with buffer A supplemented with 300 mM NaCl and concentrated to a final volume of 5 ml. Furthermore, the protein was purified by gel filtration using superdex75 (GE Healthcare, Waukesha, USA) which was equilibrated with buffer B (20 mM Tris-HCl pH 8.0, 100 mM NaCl) at the flow rate of 1 ml/min. The fraction corresponding to the FK1 protein was pooled and concentrated to a final concentration of 50 mg/ml. The protein was flash-cooled with liquid nitrogen and stored at −80 ℃ for further use.
5.4. Isothermal titration calorimetry (ITC) assay
Isothermal Titration Calorimetry was carried out to determinate the interactions between peptide-inhibitor hits and FK1. The hits and FK1 were prepared by diluting into PBS buffer to final concentrations of 1.6/3.2 mM and 40/80 μM, respectively. Samples were centrifuged at 12,000g for 20 min to remove the precipitation. The titration was performed by titrating 40/80 μM protein in the cell with 1.6/3.2 mM peptide in the syringe. The peptide solution was titrated into PBS buffer alone, the resulting heat of dilution was used to subtract the experiment curve. Data analysis was carried out with MicroCal PEAQ-ITC Analyze software (Malvern, Malvern, UK) and one-binding site model was used to calculate the binding constants and thermodynamic parameters of the interactions between peptide-inhibitor hits and FK1.
5.5. Fluorescence polarization (FP) assay
Fluorescence polarization assays were performed to evaluate the binding affinity between FK1 and FITC-tagged peptide-inhibitors. In detail, 5 μM FITC-tagged peptide-inhibitors were incubated with serially diluted FK1 protein for 10 min at the room temperature. Then the FP in mP (millipolarization) unit was measured with microplate reader SpectraMax M5 (Molecular Device, San Jose, USA) and data were processed with GraphPad Prism 6 and fitted with one-phase association model.
5.6. Crystallization and structure determination
FK1-peptide-inhibitor complexes were prepared by mixing purified FK1 (30 mg/ml) with peptide-inhibitor hits in a ratio of 1:5 and incubated for 2–3 h at 4 °C. Crystallization was carried out at 16 °C using hanging drop vapor diffuse method with mixtures of 1 μl protein and 1 μl reservoir against 500 μl reservoir solution. Crystals appeared overnight and grew to full size in a week at conditions of 30–36% PEG3350, 0.1 M HEPES-NaOH (pH 7.5) and 0.2 M NH4-acetate. Diffraction-quality crystals were picked up with appropriate nylon loops and flash frozen with liquid nitrogen without cryoprotection. Diffraction data were collected at Shanghai Synchrotron Radiation Facility (SSRF) BL18U and BL17U and processed with HKL2000/3000 [38] and autoPROC [39]. Crystal structures were solved via molecular replacement using auto MR module of CCP4 software [40] and the native FK1 structure (PDB ID: 3O5R) [34] was used as the search model. The peptide-inhibitor hits in crystal models were built with Coot [41] manually. Iteration improvement and refinement of crystal models were carried out with Coot and REFMAC5 [42] and structure models were refined to the convergences of R-work and R-free. For the complex FK1-SFPFT, the structure was refined to R-work/R-free = 16.65%/19.40%, and 126 residues were in the Ramachandran preferred regions and 1 in the allowed region. For the complex FK1-QFPFV, the structure was refined to R-work/R-free = 18.48%/19.82%, and 127 residues were in the Ramachandran preferred regions and 1 in the allowed region. For the FK1-DFPFV complex, the final model structure was refined to 17.14%/18.97%, and all residues were in the allowed regions. The structure qualities were testified with MORPROBITY [43] and PROCHECK [44]. The final coordinates were deposited on the protein data bank (PDB) under the accession of 7ETT, 7ETU and 7ETV, respectively. The crystal diffraction data collection and structural refinement parameters were list in the Table 3. All structural figures were generated with PyMOL [45] and 2D interaction diagram was obtained by Ligplot [46].
Table 3.
Data collection and structure refinement statistics.
| QFPFV | DFPFV | SFPFT | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Wavelength(Å) | 0.9793 | 0.9793 | 0.9793 |
| Cell dimensions | |||
| 42.468 | 42.359 | 42.326 | |
| a, b, c (Å) | 54.283 | 54.377 | 54.425 |
| 56.334 | 56.466 | 56.319 | |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–1.50 (1.53–1.50) | 50–1.31 (1.35–1.31) | 50–1.39 (1.41–1.39) |
| Rpim (%) | 3.5 (6.0) | 1.6 (2.6) | 2.6 (5.5) |
| Mean I/σ(I) | 21.29 (16.2) | 14.9 (5.9) | 55.14 (25.44) |
| Completeness (%) | 100 (1 0 0) | 98.7 (81.3) | 99.2 (95.5) |
| Redundancy | 11.6 (11.9) | 10.7 (8.3) | 12 (11.5) |
| Unique reflections | 21,382 (1044) | 31,353 (2111) | 26,679 (1265) |
| Refinement | |||
| Resolution (Å) | 24.50–1.50 | 39.20–1.31 | 28.18–1.39 |
| Rwork/Rfree (%) | 18.48/19.82 | 17.14/18.97 | 16.65/19.40 |
| No. atoms | 1032 | 1035 | 1020 |
| Protein | 986 | 989 | 977 |
| Ligand | 46 | 46 | 43 |
| Average B factors (Å2) | 6.254 | 11.586 | 11.797 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.005 | 0.016 | 0.015 |
| Bond angles (°) | 0.879 | 1.865 | 1.797 |
| Ramachandran Plot (%) | |||
| Favored regions | 99.22 | 99.22 | 99.22 |
| Allowed regions | 0.78 | 0.78 | 0.78 |
| PDB entry | 7ETT | 7ETV | 7ETU |
Rfree = ΣT||Fobs| − |Fcalc||/ΣT|Fobs|, where T is a test data set of about 5% of the total reflections randomly chosen and set aside prior to refinement.
*Values in parentheses correspond to the highest resolution shell.
5.7. Molecule dynamic simulation
All MD simulations were carried out with Amber 18 package. Amber ff14SB force field [47] was used to parameterize the FK-peptide complexes which were further solvated into the periodic TIP3P [48] water model with the distance of solvate boundary to solute set to 12 Å. And negative charged Cl- ions were used to neutralize the system. The steepest descent and conjugated gradient method were used to eliminated the local collisions of the system under the restraint of force constant of 10, 5 and 0 kcal mol−1 Å−2, respectively. Then the system was gradually heated from 0 K to 300 K in the NVT ensemble with a weak constraint of 5 kcal mol−1 Å−2. Subsequently, the system was equilibrated for 100 ps under the restraint of 5, 2, 1, 0.5 kcal mol−1 Å−2, respectively, and further 100 ps equilibrium was carried out without any constraint. Finally, the 300-ns production MD simulation was carried out in the NPT ensemble. The particle mesh Ewald (PME) [49] and SHAKE algorithm [50] were used to compute long-distance electrostatics and constraint hydrogen involved bonds, respectively.
5.8. GaMD simulation system setup and simulation protocol
The GaMD simulations were carried out with AMBER 18. The initial structure of peptide-inhibitor hit QFPFV was built with the LEaP module and parameterized with ff14SB force field. Then it was immersed into explicit TIP3P water box and extended 10 Å from the solute surface. The system minimization was performed using 5000 cycles procedure (2500 cycles of steepest descent and 2500 cycles of conjugate gradient minimization). Then the system was gradually heated to 300 K in 50 ps in NVT ensemble and five steps equilibrium was employed to converge the water density in NPT ensemble. The GaMD simulation started with 10 ns cMD to collect potential statistics and 40 ns GaMD equilibration was performed, followed by three independent 500 ns production GaMD simulations.
5.9. Binding free energy calculation and per-residue energy decomposition
The binding free energy between ligand and receptor was calculated using MM-GBSA [51] method based on the equation below:
where G complex is the sum of free energy of complex in gas phase and its solvation free energy, G receptor and G ligand represent the solvation energy of receptor and ligand, respectively.
where ΔG bind consists of three components including ΔG gas, ΔG sol and entropy term (- TΔS). ΔG gas, ΔG sol can be calculated according to the equation below:
where ΔG gas was composed of ΔE int, ΔE ele and ΔE vdw, ΔE int is the internal energy come from bond, torsion and dihedral terms while the ΔE ele and ΔE vdw represent the electrostatic energy and van der Waals, respectively.
ΔG polar, sol can be obtained by solving the Generalized Born equation with dielectric constants of solvent and solute are set to 1.0 and 80.0, respectively. And ΔG nonp, sol can be estimated by solvent access surface area with water probe radius and surface tension constant set to 1.4 Å and 0.0072 kcal mol−1 Å−2 [52]. The MM-GBSA was carried out by extracting 250 snapshots from the last 20-ns trajectory using mmpbsa.pl module. The per-residue energy decomposition was performed to map out the most important residues contributing to the interaction between ligand and receptor. Given to the computational cost, only 100 out 1000 snapshots were extract to compute the entropy term.
CRediT authorship contribution statement
Jian-Ting Han: Conceptualization, Investigation, Formal analysis, Methodology, Validation, Visualization, Writing - original draft. Yongchang Zhu: Investigation, Methodology, Resources, Validation. Da-Bo Pan: Conceptualization, Methodology, Validation, Investigation. Hong-Xiang Xue: Investigation, Methodology, Resources. Shuang Wang: Investigation, Methodology. Yali Peng: Supervision, Resources. Huanxiang Liu: Supervision, Validation, Project administration, Resources, Methodology. Yong-Xing He: Supervision, Validation, Resources, Methodology. Xiaojun Yao: Conceptualization, Supervision, Validation, Project administration, Resources, Funding acquisition, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the grant from the National Natural Science Foundation of China (Grant No. 21775060). We thank the supercomputing center of Lanzhou University for providing high performance computing resource. We also thank the staff of beamlines BL17U and BL18U in the Shanghai Synchrotron Radiation Facility (SSRF) for the support and help of X-ray diffraction and data acquisition.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.07.015.
Contributor Information
Huanxiang Liu, Email: hxliu@lzu.edu.cn.
Yong-Xing He, Email: heyx@lzu.edu.cn.
Xiaojun Yao, Email: xjyao@lzu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Matosin N., Halldorsdottir T., Binder E.B. Understanding the molecular mechanisms underpinning gene by environment interactions in psychiatric disorders: the FKBP5 model. Biol Psychiat. 2018;83:821–830. doi: 10.1016/j.biopsych.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 2.Zannas A.S., Wiechmann T., Gassen N.C., Binder E.B. Gene–stress–epigenetic regulation of FKBP5: clinical and translational implications. Neuropsychopharmacology. 2016;41:261–274. doi: 10.1038/npp.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinars C.R., Cheung-Flynn J., Rimerman R.A., Scammell J.G., Smith D.F., Clardy J. Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc Natl Acad Sci USA. 2003;100:868–873. doi: 10.1073/pnas.0231020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozany C., März A., Kress C., Hausch F. Fluorescent probes to characterise FK506-binding proteins. ChemBioChem. 2009;10:1402–1410. doi: 10.1002/cbic.200800806. [DOI] [PubMed] [Google Scholar]
- 5.Pirkl F., Buchner J. Functional analysis of the hsp90-associated human peptidyl prolyl Cis/Trans isomerases FKBP51, FKBP52 and cyp4011Edited by R. Huber. J Mol Biol. 2001;308:795–806. doi: 10.1006/jmbi.2001.4595. [DOI] [PubMed] [Google Scholar]
- 6.Chen S., Sullivan W.P., Toft D.O., Smith D.F. Differential interactions of p23 and the TPR-containing proteins Hop, Cyp40, FKBP52 and FKBP51 with Hsp90 mutants. Cell Stress Chaperon. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barent R.L., Nair S.C., Carr D.C., Ruan Y., Rimerman R.A., Fulton J. Analysis of FKBP51/FKBP52 chimeras and mutants for Hsp90 binding and association with progesterone receptor complexes. Mol Endocrinol. 1998;12:342–354. doi: 10.1210/mend.12.3.0075. [DOI] [PubMed] [Google Scholar]
- 8.Ebong I.-O., Beilsten-Edmands V., Patel N.A., Morgner N., Robinson C.V. The interchange of immunophilins leads to parallel pathways and different intermediates in the assembly of Hsp90 glucocorticoid receptor complexes. Cell Discov. 2016;2:16002. doi: 10.1038/celldisc.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermeer H., Hendriks-Stegeman B.I., van der Burg B., van Buul-Offers S.C., Jansen M. Glucocorticoid-induced increase in lymphocytic FKBP51 messenger ribonucleic acid expression: a potential marker for glucocorticoid sensitivity, potency, and bioavailability. J Clin Endocrinol Metab. 2003;88:277–284. doi: 10.1210/jc.2002-020354. [DOI] [PubMed] [Google Scholar]
- 10.Hubler T.R., Denny W.B., Valentine D.L., Cheung-Flynn J., Smith D.F., Scammell J.G. The FK506-binding immunophilin FKBP51 is transcriptionally regulated by progestin and attenuates progestin responsiveness. Endocrinology. 2003;144:2380–2387. doi: 10.1210/en.2003-0092. [DOI] [PubMed] [Google Scholar]
- 11.Hubler T.R., Scammell J.G. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperon. 2004;9:243–252. doi: 10.1379/CSC-32R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romano S., Xiao Y., Nakaya M., D'Angelillo A., Chang M., Jin J. FKBP51 employs both scaffold and isomerase functions to promote NF-κB activation in melanoma. Nucleic Acids Res. 2015;43:6983–6993. doi: 10.1093/nar/gkv615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erlejman A.G., De Leo S.A., Mazaira G.I., Molinari A.M., Camisay M.F., Fontana V. NF-κB transcriptional activity is modulated by FK506-binding proteins FKBP51 and FKBP52: a role for peptidyl-prolyl isomerase activity. J Biol Chem. 2014;289:26263–26276. doi: 10.1074/jbc.M114.582882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Binder E.B., Salyakina D., Lichtner P., Wochnik G.M., Ising M., Pütz B. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36:1319–1325. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 15.Kirchheiner J., Lorch R., Lebedeva E., Seeringer A., Roots I., Sasse J. Genetic variants in FKBP5 affecting response to antidepressant drug treatment. Pharmacogenomics. 2008;9:841–846. doi: 10.2217/14622416.9.7.841. [DOI] [PubMed] [Google Scholar]
- 16.Zou Y.-F., Wang F., Feng X.-L., Li W.-F., Tao J.-H., Pan F.-M. Meta-analysis of FKBP5 gene polymorphisms association with treatment response in patients with mood disorders. Neurosci Lett. 2010;484:56–61. doi: 10.1016/j.neulet.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Roy A., Gorodetsky E., Yuan Q., Goldman D., Enoch M.-A. Interaction of FKBP5, a stress-related gene, with childhood trauma increases the risk for attempting suicide. Neuropsychopharmacology. 2010;35:1674–1683. doi: 10.1038/npp.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stechschulte L.A., Sanchez E.R. FKBP51—a selective modulator of glucocorticoid and androgen sensitivity. Curr Opin Pharmacol. 2011;11:332–337. doi: 10.1016/j.coph.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni L., Yang C.-S., Gioeli D., Frierson H., Toft D.O., Paschal B.M. FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells. Mol Cell Biol. 2010;30:1243–1253. doi: 10.1128/MCB.01891-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Periyasamy S., Hinds T., Shemshedini L., Shou W., Sanchez E.R. FKBP51 and Cyp40 are positive regulators of androgen-dependent prostate cancer cell growth and the targets of FK506 and cyclosporin A. Oncogene. 2010;29:1691–1701. doi: 10.1038/onc.2009.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jinwal U.K., Koren J., 3rd, Borysov S.I., Schmid A.B., Abisambra J.F., Blair L.J. The Hsp90 cochaperone, FKBP51, increases Tau stability and polymerizes microtubules. J Neurosci. 2010;30:591–599. doi: 10.1523/JNEUROSCI.4815-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopalakrishnan R., Kozany C., Gaali S., Kress C., Hoogeland B., Bracher A. Evaluation of synthetic FK506 analogues as ligands for the FK506-binding proteins 51 and 52. J Med Chem. 2012;55:4114–4122. doi: 10.1021/jm201746x. [DOI] [PubMed] [Google Scholar]
- 23.Gopalakrishnan R., Kozany C., Wang Y., Schneider S., Hoogeland B., Bracher A. Exploration of pipecolate sulfonamides as binders of the FK506-binding proteins 51 and 52. J Med Chem. 2012;55:4123–4131. doi: 10.1021/jm201747c. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Kirschner A., Fabian A.-K., Gopalakrishnan R., Kress C., Hoogeland B. Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control. J Med Chem. 2013;56:3922–3935. doi: 10.1021/jm400087k. [DOI] [PubMed] [Google Scholar]
- 25.Gaali S., Kirschner A., Cuboni S., Hartmann J., Kozany C., Balsevich G. Selective inhibitors of the FK506-binding protein 51 by induced fit. Nat Chem Biol. 2015;11:33–37. doi: 10.1038/nchembio.1699. [DOI] [PubMed] [Google Scholar]
- 26.Petsalaki E., Russell R.B. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr Opin Biotechnol. 2008;19:344–350. doi: 10.1016/j.copbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Gaali S., Feng X., Hähle A., Sippel C., Bracher A., Hausch F. Rapid, structure-based exploration of pipecolic acid amides as novel selective antagonists of the FK506-binding protein 51. J Med Chem. 2016;59:2410–2422. doi: 10.1021/acs.jmedchem.5b01355. [DOI] [PubMed] [Google Scholar]
- 28.Yan Z., Bai X.-C., Yan C., Wu J., Li Z., Xie T. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miao Y., Feher V.A., McCammon J.A. Gaussian accelerated molecular dynamics: unconstrained enhanced sampling and free energy calculation. J Chem Theory Comput. 2015;11:3584–3595. doi: 10.1021/acs.jctc.5b00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miao Y., Sinko W., Pierce L., Bucher D., Walker R.C., McCammon J.A. Improved reweighting of accelerated molecular dynamics simulations for free energy calculation. J Chem Theory Comput. 2014;10:2677–2689. doi: 10.1021/ct500090q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miao, Y., Bhattarai, A. and Wang, J.Ligand Gaussian accelerated molecular dynamics (LiGaMD): Characterization of ligand binding thermodynamics and kinetics. bioRxiv (2020),pp. 2020.2004.2020.051979. [DOI] [PMC free article] [PubMed]
- 32.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 33.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20:3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 34.Bracher A., Kozany C., Thost A.K., Hausch F. Structural characterization of the PPIase domain of FKBP51, a cochaperone of human Hsp90. Acta Crystallogr D Biol Crystallogr. 2011;67:549–559. doi: 10.1107/S0907444911013862. [DOI] [PubMed] [Google Scholar]
- 35.Periyasamy S., Warrier M., Tillekeratne M.P.M., Shou W., Sanchez E.R. The immunophilin ligands cyclosporin A and FK506 suppress prostate cancer cell growth by androgen receptor-dependent and -independent mechanisms. Endocrinology. 2007;148:4716–4726. doi: 10.1210/en.2007-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storer C.L., Dickey C.A., Galigniana M.D., Rein T., Cox M.B. FKBP51 and FKBP52 in signaling and disease. Trends Endocrinol Metab. 2011;22:481–490. doi: 10.1016/j.tem.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muttenthaler Markus, King Glenn F., Adams David J., Alewood Paul F. Trends in peptide drug discovery. Nat Rev Drug Discov. 2021;20(4):309–325. doi: 10.1038/s41573-020-00135-8. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski, R., Macarthur, M.W., Moss, D.S. and Thornton, J.PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr, 26 (1993), pp. 283-291.
- 45.DeLano W.L. DeLano Scientific LLC; San Carlos, CA, USA: 2015. The PyMOL Molecular Graphics System. [Google Scholar]
- 46.Wallace, A.C., Laskowski, R.A. and Thornton, J.M.LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng., 8 (1995),pp. 127-134. [DOI] [PubMed]
- 47.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 49.Darden T., York D., Pedersen L. Particle mesh ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 50.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 51.Case D.A., Cheatham T.E., 3rd, Darden T., Gohlke H., Luo R., Merz K.M., Jr. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sitkoff D., Sharp K.A., Honig B. Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem. 1994;98:1978–1988. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.