Abstract
Dermatologists stand at the gateway of individualization of classification, treatment, and outcomes of acral melanoma patients. The acral melanoma genetic landscape differs in vital ways from that of other cutaneous melanomas. These differences have important implications in understanding pathogenesis, treatment, and prognosis. The selection of molecularly targeted therapy must be adapted for acral melanoma. It is also critical to recognize that tumor development is far more complex than an isolated event, reliably treated by a medication acting on a single target. Tumors exhibit intratumor genetic heterogeneity, metastasis may have different genetic or epigenetic features than primary tumors, and tumor resistance may develop because of the activation of alternative genetic pathways. Microenvironmental, immune, and epigenetic events contribute and sustain tumors in complex ways. Treatment strategies with multiple targets are required to effectively disrupt the tumor ecosystem. This review attempts to translate the current molecular understanding of acral melanoma into digestible concepts relevant to the practice of dermatology. The focus is tumor genetics defining potentially treatable cancer pathways, contextualized within the relevant pathologic and molecular features.
Key words: acral melanoma, dermatology, genetics, melanoma, molecular, oncology, tumor genetics
Graphical abstract
Capsule Summary.
-
•
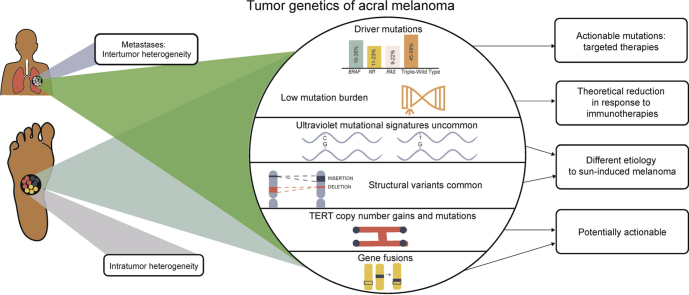
The tumor genetics of acral melanoma are very different from those of ultraviolet radiation–induced melanoma, having different significantly mutated genes, lower mutational burdens, more structural variants, and fewer ultraviolet radiation–induced mutations.
-
•
Clarification of acral melanoma subgroups on a clinical, pathologic, and molecular basis is necessary, but not yet complete.
Introduction
Acral melanoma is a unique tumor within the melanoma spectrum. Its molecular features are gradually being unraveled. Molecularly targeted therapies have revolutionized melanoma management by offering unprecedented responses in some patients. As such treatments become increasingly available, dermatologists should have a working understanding of these concepts. Melanoma classification will increasingly include, and likely be determined by, molecular findings. The molecular characteristics of acral melanoma will contribute to understanding its pathogenesis, enabling potentially preventive actions and therapies.
Vagaries and misconceptions have impaired our understanding of acral melanoma. Acral melanoma here refers specifically to melanoma on the palms, soles, and nail unit (sun-protected sites). Melanoma on the dorsal surface of the hands and feet should be grouped with other more common forms of cutaneous melanoma, frequently driven by ultraviolet (UV) radiation exposure.1 Traditional classification systems of melanoma have resulted in the misconception that all acral melanomas are acral lentiginous melanomas. Although the most common histologic subtype, not all acral melanomas take the lentiginous form.2,3 Many studies focus on acral lentiginous melanoma, omitting an important subgroup of acral melanoma. Whether subungual melanoma is a separate entity from other acral melanoma variants remains uncertain. A differentiation between melanomas arising from the nail matrix (initially linear melanonychia) and those arising on dorsal, sun-exposed skin of nail folds may be necessary.
A clear approach to the clinical, pathologic, and molecular subgrouping of acral melanomas will identify acral melanoma patients eligible for targeted therapies based on pharmacogenomic markers. Clarification of these acral melanoma subgroups is incomplete. This review presents an update on acral melanoma tumor genetics as it applies to the practice of dermatology. Clinical applications of this knowledge are presented together with a brief contextualization of tumor genetics within the larger tumor ecosystem. This overview first presents data to facilitate cursory reading about acral melanoma; second, more specific detail for specialized readers.
Tumor genetics of acral melanoma
Relevant pathways in melanoma development
Tumorigenesis is a result of complex interactions between genetic changes in the tumor and the patient and environmental (including microenvironmental) factors.4 Table I summarizes important cellular pathways in acral melanoma. Separation of these pathways gives the impression they are discrete, parallel processes; however, there are many interactions between them.
Table I.
Cellular pathways with a pathogenetic role in acral melanoma
| Pathway | Cellular activity | Role in acral melanoma |
|---|---|---|
| MAPK5 | Cellular proliferation, differentiation, and survival6 | Activated in more than 90% of melanomas7 Stimulated by activating BRAF and NRAS mutations and inactivating NF1 mutations5 Stimulated by upstream receptor tyrosine kinases (eg, KIT)8 Collateral effects allow tumor to evade immune system5 Constitutive activation of this pathway demonstrated in ALM (in situ and invasive) and the AM group as a whole5,9 |
| PI3K/AKT/PTEN5 | Permits cellular survival (antiapoptotic)10 |
PTEN antagonizes the PI3K/AKT/mTOR pathway, acts as a tumor suppressor10 Stimulated by upstream receptor tyrosine kinases (eg, KIT)8 |
| JAK/STAT35 | Regulates cellular proliferation, differentiation, migration, and survival (context dependent)11 | Regulates the PD-1 immune checkpoint, a mechanism of immune escape for melanomas12 May correlate with more advanced AM5 |
| TERT5 | Regulates telomere maintenance13 |
TERT activation essential in tumor development Confers immortality to melanoma cells by maintaining telomere length13 Abnormally activated in 37% of AM patients in 1 study14 |
| CDK4/CDKN2A5 | Directs the cell cycle CDK4 = an oncogene CDKN2A = a tumor suppressor gene that encodes p16 RB1 and p53 also involved in this pathway Also stimulated by the MAPK pathway via cyclin D115 |
CDK4 exhibits rare germline mutations leading to melanoma, whereas germline CDKN2A mutations detected in 10%–25% of melanoma-prone families16 Nongermline aberrations also critical: Activation in AM, especially in subungual/interdigital melanomas17,18 Pathway abnormally activated in 51% of AM patients in 1 study14 |
| MDM2/TP535 | Determines senescence and apoptosis14 P53 inhibited by MDM, supporting oncogenesis MDM and p53 interact in complex ways19 |
Abnormal activation identified in 17% of AM in 1 study14 |
| WNT signaling4 | Determines cellular proliferation, migration, polarity, and fate4 Interacts with MITF20 |
Exact role in melanoma unclear21 Mutated CTNNB1 in an AM raises the possibility21 |
| MCR1-MITF | Interaction between cellular activities such as melanin synthesis and the oncogene MITF, as well as the CDK4/CDKN2A pathway | Germline MCR1 variants associated with increased melanoma risk in general8 A subtype of AM shows MITF aberrations22 |
ALM, Acral lentiginous melanoma; AM, acral melanoma; MAPK, mitogen-activated protein kinase.
In acral melanoma, cancer pathways are activated in different ways from UV radiation–induced cutaneous melanoma. They interact with one another in complex ways. Inhibiting one pathway may result in deviation to and tumor stimulation via another. How aberration in specific pathways is more conducive to development of different acral melanoma subgroups, such as acral lentiginous melanoma, nodular acral melanoma, or nail melanoma, is unclear.
Genetic changes observed in acral melanoma tumors
Driver mutations
Driver mutations vary significantly among melanoma subtypes.23 Whole exome sequencing (analysis of the entire coding portion) of tumor DNA often leads to the identification of many mutations.24 The identification of driver mutations leading to tumorigenesis (as opposed to passenger mutations) is challenging.25 This is particularly problematic in cutaneous melanoma, in which the mutagenic effects of UV radiation result in a high mutational burden.26,27 Many driver mutations are current or potential targets of molecular therapies. Acral melanomas have significantly lower mutational burdens than cutaneous melanomas.26 It is unknown whether this applies to different subgroups and stages of acral melanoma. Driver mutations are more easily identified in acral melanoma because of their lower rate of somatic mutations.28
Intratumoral and intertumoral heterogeneity must also be considered. Analysis of a single clone of tumor cells may disregard important driver mutations in other tumor clones.7,29,30 Molecular alterations leading to tumor progression and metastases are poorly understood.
The genomic classification of melanomas by the Cancer Genome Atlas identifies 4 subtypes of melanomas based on their driver mutations: BRAF, RAS, NF1, and the triple wild type.31 These typically result in dysfunction of the mitogen-activated protein kinase pathway. Each group has distinct clinical and genetic features.32 Approximately half (42%-55%) of acral melanomas studied to date have BRAF, RAS, or NF1 mutations; the others fall into the triple-wild-type group.22 BRAF, NRAS, and NF1 alterations were mutually exclusive in 1 acral lentiginous melanoma study.14 Subungual and interdigital melanomas display the most diverse driver mutations.17
Absence of BRAF, RAS, or NF1 hot-spot mutations defines the triple-wild-type group (ie, all 3 “typical” melanoma mutations are wild type or normal).31 Mitogen-activated protein kinase pathway activation still occurs in most triple-wild-type melanomas.33 Triple-wild-type driver mutations are observed in 45% to 58% of acral melanoma cases.22 These include genetic alterations in a variety of genes, including KIT, CCND1, CTNNB1, KDR (VEGFR2), MDM2, BCL2, AKT3, IDH1, GNAQ (uveal melanoma), GNAS, CDK4, CDKN2A, MITF, PTEN, RB1, TP53, APC, ERBB2, ERBB3, NUAK2, ABCB5, and TERT.3,22,26,31,34, 35, 36, 37, 38, 39
Deleterious KIT mutations or amplifications are frequently an early event in acral melanoma development (3%-36%), specifically directing lentiginous growth.3,22,40 KIT mutations activate both the mitogen-activated protein kinase and PI3K/AKT pathways.41 Most KIT mutations described in acral melanoma are in exons 9, 11, 13, 17, and 18.42,43 PDGFRA is often coamplified with KIT, although the converse is also reported.3,36 It appears to be a critical event in acral melanoma.3 NF1 and SPRED1 losses may occur with or without KIT driver mutations.44
BRAF variants play a smaller role in acral melanoma (10%-35% of cases).12,22 Both typical V600E and other mutations (eg, V600L) are reported.45 Alternative BRAF mutations to V600E occurred in 5% of cases in a recent study.44 A molecular subclassification of acral melanoma based on BRAF mutation was proposed: acral melanoma with typical BRAF mutations observed in cutaneous melanoma, potentially responsive to BRAF inhibitors; and acral melanoma with non-BRAF mutations with other potentially actionable targets. This study also found that acral melanomas with BRAF mutations were less likely to be of the acral lentiginous melanoma subtype.
NF1 driver mutations are observed in 11% to 23% of acral melanomas.22 NF1 is a tumor suppressor; therefore, alterations leading to loss of function of this gene need to arise on both chromosomes. NF1 mutant tumors are generally associated with poor prognosis.31,32 Homozygous NF1 deletions were more common than point mutations in an acral lentiginous melanoma study.14
RAS genetic alterations are observed in 9% to 22% of cases.12,22 NRAS mutations are at the same nucleotide positions as observed in cutaneous melanoma.5
Melanoma genetics studies tend to originate from a few centers, and it is uncertain whether their findings apply to other geographic regions and understudied population groups.22 Lower prevalences of KIT, NRAS, and BRAF are reported in acral melanoma in some populations.21 The main findings of acral melanoma cohorts sequenced to date are summarized in Table II.
Table II.
Reported genetically sequenced acral melanoma cohorts (adapted from Chen et al5)
| Study | Cases, no. | KIT, % | BRAF, % | NF1, % | RAS, % | Other |
|---|---|---|---|---|---|---|
| Curtin et al46 2005 (targeted sequencing) |
36 | Not sequenced | 23 | Not sequenced | 10 | Structural changes, amplifications in 89%; CDK4 amplifications, CDKN2A losses more common than other CM |
| Krauthammer et al28 2012 |
17 (9 metastases) | 29.4 | 0 | 0 | 11.8 | 1 RAC1 mutation Copy gains in 5p13, 11q13 and 12q14 more common than other CM 3 DYNCC1I1 mutations |
| Zebary et al47 2013 (targeted sequencing) |
88 ALMs | 15 | 17 | Not sequenced | 15 | 4% PTEN mutations (25 tumors sequenced) |
| Furney et al26 2014 |
5 (all metastases) | 40 | 40 | 0 | 0 | 0% TERT promoter mutations |
| Puntervoll et al43 2014 (targeted sequencing) |
36 (24 Tanzanian) | 11.1 | 11.1 | Not sequenced | 11.1 NRAS | — |
| De Lima Vazquez et al48 2016 (targeted sequencing) |
61 ALM | 20.7 | 10.3 | 0 | 7.5 NRAS | 9.3% TERT promoter mutations PDGFRA mutation in 14.8% |
| Liang et al14 2017 |
34 ALM (17 metastases) | 2.6 | 18.4 (and 2.6 homozygous deletions) | 7.9 (loss/homozygous deletions) | 10.5 NRAS, 5.3 KRAS (and 2.6 amplifications) | 2.6% TERT promoter mutations 10.5% TERT amplifications PAK1 copy gains in 15% |
| Shim et al21 2017 |
Composite of Asian cases reported | 10.2 (51/498) | 10.4 (40/383) | — | 6.6 NRAS (18/273) | — |
| Hayward et al23 2017 |
35 | 8.6 | 22.8 | 25.7 | 17.1 11 NRAS |
Substantially more complex structural rearrangements CCND1 rearrangements |
| Kong et al18 2017 |
514 | — | — | — | — | 39.5% CDK4 gain 26.7% CCND1 gain 60.3% P16INK4a loss |
| Roh et al49 2017 |
46 | — | — | — | — | 10.9% TERT promoter mutations |
| Moon et al35 2018 |
64 | 10.9 | 34.4 | 17.2 | 21.9 NRAS | 17.2% GNAQ |
| Haugh et al17 2018 |
22 (9 volar, 13 nail unit/interdigital) | 4.5 | 13.6 | 4.5 loss | 22.7 NRAS | 9.1% TERT gains 22.7% CCND1 gains 13.6% CDK4 gains 13.6% PAK1 gains 18.2% GAB2 gains |
| Gao et al45 2018 (targeted sequencing) |
40 | Not sequenced | 30 | Not sequenced | 10 NRAS | 7.5% PTEN mutations |
| Yeh et al44 2019 (targeted sequencing) |
122 | 11.5 (and 2.5 fusions) | 21.3 | 14.8 (inactivation) | 27.9 NRAS | 5.3% TERT promoter mutations 10.7% TERT amplifications Amplifications in PAK1 and GAB2 (22.1%), CDK4 (22.1%), and CCND1 (19.7%), among others |
| Zaremba et al1 2019 |
50 (including dorsal lesions) | 6 (not clear whether it includes dorsal lesions) | 21.7 | 17.3 | 39.1 | 8.6% TERT promoter mutation |
| Sheen et al37 2019 |
45 | 24.4 | 8.9 | 11.0 | 26.7 NRAS and KRAS | 68.9% with cell cycle aberrations (CDK4/6, CCND1/2, CDKN2A) 35.6% with other receptor tyrosine kinase gains (eg, EGFR, PDGFRA) 33.3% with antiapoptosis gains (eg, BIRC 2, 3, 5) |
| Shi et al50 2019 |
29 | 13.8 | 27.6, 3.4 gains | 10.3 mutations | 10.3 NRAS | 10.3% CDKN2A mutations 17.2% CDKN2A losses 6.9% TERT promoter mutations 13.8% TERT copy number variants 6.9% fusions |
ALM, Acral lentiginous melanoma; CM, cutaneous melanoma; —, not available.
Mutational burden
Because acral melanomas are not typically UV radiation induced, they have lower mutational burdens.23,26 High tumor mutational burden theoretically improves responses to immunotherapies.51 In practice, case series show similar efficacy in cutaneous melanoma and acral melanoma.22
Mutational signatures
In cutaneous melanoma, high levels of cytosine to thymine mutations are observed. These frequently show UV radiation mutational signatures not observed in acral melanoma, even if cytosine to thymine mutations are observed.26 Mutational signatures in acral melanoma are different and are reported in other cancers, but not cutaneous melanoma.23 Rates of non–cytosine to thymine mutations (guanine to adenine) are also lower in acral melanoma.26 The presence of specific mutational signatures has prognostic implications in cutaneous melanoma.52 Whether this extends to non-UV radiation signatures in acral melanoma is uncertain.
UV radiation signatures are identified in only a small proportion of acral melanoma.14,50,53,54 In accordance with the relatively more frequent occurrence and worse prognosis of acral melanoma in people with darker Fitzpatrick skin types compared with that for other types of melanoma, authorities have justified the use of aggressive sun protection in this group. This approach is questionable because acral melanoma is not commonly associated with UV radiation–induced mutations.
UV radiation signatures in subungual melanomas are reported, whereas it has been shown that the nail plate blocks the majority of UV radiation.22,53,55
Structural variants
Structural variants represent genetic variation larger than 50 base pairs.56 Acral melanoma shows a higher frequency of structural variants than cutaneous melanoma23,57 because of entirely different mutational processes that occur in acral melanoma.23,31 Subungual and interdigital melanomas have more copy number aberrations compared with both volar and dorsal melanomas of acral skin (CDK4 and cyclin D1).17 Copy number gains in BIRC2, BIRC3, and BIRC5 (antiapoptosis genes) correlate with poor melanoma-specific survival.37 Amplifications in PAK1, GAB2, and IL7R are identified in acral melanoma.14,44,58 ALK break points occurred in 6.9% of acral melanomas in 1 study.59 CDKN2A deletions are common (15.8%-35%).14,23,35
Telomere length and pathway alterations
Alterations in telomere length do not correlate with melanoma subtype.23 An association was detected between short telomeres (and TERT aberrations) and poor melanoma survival.22,60 Telomerase pathway alterations are reported in 9% to 45% of acral melanoma.22 TERT promoter variants are less common in acral melanoma (9%-41%) than in cutaneous melanoma (more than 50%).22 However, in 45% of acral melanoma with TERT aberrations, TERT copy number gains are noted (as opposed to point mutations typically observed in cutaneous melanoma).22,61 A high frequency of TERT promoter mutations was reported in acral melanomas involving the digit and nail (38.8%).1
Gene fusions
Gene fusions occur when 2 previously independent genes are joined. When an upstream gene is turned on in the tumor cell, this can activate a downstream gene.62 Kinase fusions activate the mitogen-activated protein kinase pathway in acral melanoma through mechanisms other than BRAF, RAS, or KIT mutations.44 Kinase fusion genes seem to play a particular role in melanomas lacking common coding mutations (pan-negative melanoma, lacking BRAF, NRAS, KRAS, HRAS, NF1, KIT, GNAQ, and GNA11).62 Kinase gene fusions reported in acral melanoma include PAK1, DGKB, RET, and NTRK1.23,62 Receptor tyrosine kinase fusions were found in 4%, fusions of BRAF in 3%, and protein kinase C fusions in 1% of acral melanomas in a recent study.44 These gene fusions present potentially actionable targets.62
Intratumor heterogeneity
Intratumor heterogeneity is the morphologic and genetic variation of different clones of tumor cells within a single tumor.30 This is a possible mechanism for failure of targeted therapy, including the development of recurrences.7 It was detected in acral melanoma during assessment of BRAF and KIT mutations.7,63 It is an extensive and early event in acral melanoma development.64 Thus, the concept of “treating a single mutation” is an oversimplification.
Mutations in metastases and recurrences
Discordance has been reported between mutations detected in primary acral melanomas, their metastases, and recurrences (intrapatient intertumoral heterogeneity).29,30 This phenomenon has important implications when targeted therapies are used and advocates for multimodal therapy in patients with advanced disease.
Clinical applications of tumor genetics
Diagnosis
Fluorescence in situ hybridization can detect acral melanoma–specific genetic alterations to assist diagnosis of early or equivocal lesions. The analysis of genes such as RREB1, CEP6, MYB, and CCND1 provides valuable ancillary information.36,65,66 Single-nucleotide polymorphism arrays also provide a mechanism by detecting copy number changes in areas characteristic of melanoma.67 Mutational analyses may identify the primary tumor site in metastatic disease with uncertain primary, as described in ocular melanoma, in which distinguishing between conjunctival, uveal, and cutaneous primaries is relevant to treatment and follow-up.68
Excision margin assessment
CCND1 amplifications are described in otherwise apparently normal melanocytes surrounding acral melanoma.36 Isolated melanocytes showing gene amplifications may occur up to 3 mm from the histologic tumor-free surgical excision margin.69
Classification: correlation between genotype and phenotype in acral melanoma
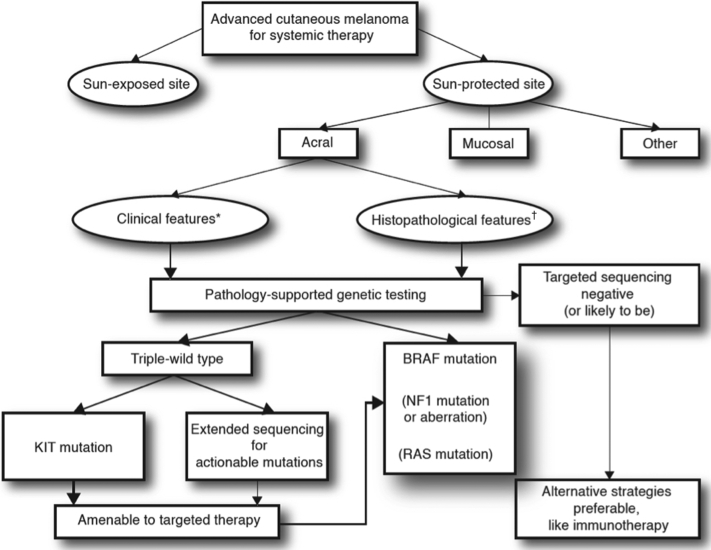
Clinical and pathologic features will be integrated to inform targeted sequencing of melanomas and identify suitable therapies. Extensive sequencing of tumors (whole exome or whole genome sequencing) is not practical, cost-effective, or easily interpreted on the clinical front line. An algorithm incorporating relevant variables such as age, UV radiation exposure status, and histologic subtype will direct genetic investigations in a way similar to that of pathology-supported genetic testing in breast cancer (Fig 1).70,71 Table III shows examples of clinical and pathologic factors associated with specific genotypes. The incorporation of genomics could refine traditional clinicopathologic classifications of melanoma.75
Fig 1.
Acral melanoma: Projected algorithm for pathology-supported genetic testing, composing targeted sequencing based on clinical∗ and histopathologic† features referred to in Table III.
Table III.
Clinical and histopathologic features (referred to in Fig 1) potentially helpful in predicting mutational status
| Clinical∗ and histopathologic features† | Associated mutation/alteration |
|---|---|
| Young (possibly <50 y) with ALM∗ | BRAF72 |
| Female sex∗ | BRAF47 |
| Nevus-associated AMs∗† | BRAF73 |
| ALM in general† | Non-BRAF44 |
| Lentiginous growth pattern† | KIT3 |
| Epithelioid cells† | BRAF mutation35 |
| Bizarre cells† | NRAS, NF1 mutations35 |
| Spindle cells with prominent dendrites† | NF1, GNAQ mutations35 |
| Advanced Clark level (≈Breslow thickness)† | KIT48 |
| Ulceration† | CDK4/6, CCND1/2, CDKN2A and receptor tyrosine kinase aberrations18,37 |
| ALM with low mitotic rate† | TERT promoter mutations48 |
| Amelanotic AM† | KIT aberrations35,74 |
ALM, Acral lentiginous melanoma; AM, acral melanoma.
Clinical.
Histopathologic.
Treatment: targeted therapies
In 1 large study of melanoma, including acral melanoma, the majority of tumors contained a potentially actionable target with currently available therapies.23 At present, treatment of BRAF-mutated melanoma with a combination of BRAF and MEK inhibitors offers rapid, albeit partial, responses in many cases.12 This pathway is less relevant to the treatment of acral melanoma. Less is known about the role of therapies targeted against other melanoma mutations, although studies show some efficacy (eg, KIT-mutated melanoma treated with tyrosine kinase inhibitors).12 Although these therapies are currently prohibitively expensive in many settings, patients may access them through clinical trials, and they are likely to become standard of care. Table IV refers to current and potential treatment strategies for melanoma. A combination of agents will better address the complexity of tumor and host biology. Agents currently under development may have a place in acral melanoma treatment.18,22,31,44,82,89
Table IV.
Correlations between molecular lesions or activity, potential metastatic acral melanoma treatments, responses, and resistance mechanisms∗
| Target | Treatments | Agents† | Responses‡ | Known resistance mechanisms‡ |
|---|---|---|---|---|
| Genetic pathway | ||||
| MAPK | BRAF inhibitors§ (BRAF mutation) | Vemurafenib∗ Dabrafenib mesylate∗ Encorafenib∗ |
ORR alone in AM 61.5% Combination BRAF/MEK inhibition in AM up to 79%5 |
Intrinsic and acquired resistance Development of new mutations, epigenetic and transcriptome alterations Paradoxic reactivation of MAPK pathway Upregulation of PI3K and Ral pathways76 |
| MEK inhibitors§ (BRAF mutation) | Trametinib∗ Cobimetinib∗ Binimetinib∗ |
BRAF/MEK inhibition in AM up to 79%5 | Upregulation of PI3K and Ral pathways76 | |
| RTK inhibitors (KIT mutation) | Imatinib Dasatinib Nilotinib Sunitinib |
Response rates up to 27%77 | Rapid development of resistance Downstream activation of various kinases Upregulation of receptor tyrosine kinases78 |
|
| ERK inhibitors | Ulixertinib Ravoxertinib |
Clinical trials76 | ||
| PI3K/AKT/PTEN | PI3K/Akt/mTOR inhibitors | Sirolimus (rapamycin) Everolimus AZD5363 LY294002 |
Investigational79 | |
| RTK inhibitors | As above | |||
| JAK/STAT3 | RTK inhibitors | May inhibit this pathway too80 | ||
| TERT | TERT or telomerase inhibitors | EGCG | Investigational, no clinical trials on melanoma to date81 | |
| CDK4/CDKN2A | CDK inhibitors | Abemaciclib Palbociclib Dinaciclib |
Clinical trials82 | |
| MDM2/TP53 | MDM2/p53 Interaction inhibitors | AMG232 Actinomycin D |
Clinical trials83 | |
| WNT signaling | WNT modulators | LGK974 | Clinical trial84 | |
| MCR1-MITF | P300/CBP inhibitors | A-485 | Investigational85 | |
| Immune system | ||||
| Immunotherapies | Immune checkpoint inhibitors | Ipilimumab∗ Nivolumab∗ Pembrolizumab∗ |
ORR with nivolumab or pembrolizumab in AM up to 32%5 | Reduced TILs in AM Reduced PD-L1 expression in AM Lower somatic mutation rate in AM5 |
| Vaccine | Talimogene laherparepvec (t-vec)∗ | Durable RR 16.3%86 | — | |
| Cytokine | IL-2 (high dose)∗ | ORR 16%87 | — | |
AM, Acral melanoma; EGCG, epigallocatechin-3-gallate; IL-2, interleukin-2; MAPK, mitogen-activated protein kinase; ORR, overall response rate; RR, response rate; TIL, tumor-invading lymphocyte.
Food and Drug Administration approved for clinical use in melanoma.88
Not an exhaustive list.
All melanoma subtypes, not exclusively acral melanoma (unless otherwise stated).
Existing and proposed combination therapies may overcome resistance (eg, BRAF and MEK inhibitors, BRAF/MEK inhibitors and immune checkpoint inhibitors, MEK inhibitors, CDK inhibitors, PI3-Akt-mTOR inhibitors).
Prognosis
Individualized prognostication in melanoma will incorporate genetic analysis.90 At present, certain genetic lesions found in melanoma confer worse prognosis; for example, NF1 mutations, TERT amplifications and mutations, AURKA copy gain, and some mutational signatures.22,32,52,91
Prevention
Specific recommendations for the prevention of acral melanoma remain elusive. No major mutagenic driver is confirmed, as in UV-induced cutaneous melanoma.3 Predispositions such as a history of penetrating injury or physical strain are proposed.5,92 Multiple melanocytic nevi on the foot was identified as a risk factor; however, it is unusual for acral melanomas to arise in existing melanocytic nevi.3,92
Other molecular and microenvironmental features
Although not the focus of this review, microenvironmental factors have an important role in explaining acral melanoma pathogenesis, treatment response, and prognosis. This ecosystem includes epigenetics, proteomics, metabolomics, the tumor microenvironment, and the host's immunologic response, germline mutations, and even microbiome.4,12,15,31,67,93, 94, 95, 96, 97, 98, 99, 100, 101, 102 Of particular importance are immunology and germline mutations.
A critical step in melanoma development is the ability of the tumor to evade detection by the immune system. This pathway is exploited by immune checkpoint inhibitors in the treatment of melanoma.12 Coordination of the immune response with therapies targeting oncogenes is a possible treatment strategy.12,89
CDKN2A, CDK4, MCR1, BAP1, and TERT promoter germline mutations increase melanoma risk within families.15,96,97 No specific germline mutation is associated with acral melanoma yet; in fact, MCR1 variants were less common in acral lentiginous melanoma patients in a Swedish study.103 An increased risk of melanoma in general (but not acral lentiginous melanoma specifically) was observed in families of acral lentiginous melanoma patients.104 One study showed a similar prevalence of acral lentiginous melanoma in a cohort of melanoma patients with familial CDKN2A mutations compared with a group without.3,105 Patients with multiple primary acral melanomas (but no family history) have been reported.106 An uncommon association of melanoma and some forms of inherited palmoplantar keratoderma has been reported (Mal de Meleda, Papillon-Lefèvre syndrome, Nagashima-type disease, and Greither disease).106, 107, 108, 109, 110 Mutations identified include SLURP-1 (Mal de Meleda), CTSC (Papillon-Lefèvre syndrome), and SERPINB7 (Nagashima-type disease). It is not clear whether these cases were a result of inherited predisposition to melanoma or another intrinsic susceptibility.
Conclusion
Critical knowledge gaps in the tumor genetics of acral melanoma remain. Complete molecular characterization of acral melanoma subsets, including different anatomic and histologic subgroups, is necessary. The development of pathology-supported genetic testing algorithms will herald the age of precision medicine in the treatment of acral melanoma. Clarity about the molecular events leading to progression and metastases, and whether these are unique in acral melanoma or even in acral melanoma subsets, will be helpful to explain treatment success and failure. Understanding how intratumoral and intertumoral heterogeneity leads to the biological behavior of acral melanoma is critical. The ultimate goal is unraveling the molecular events leading to acral melanoma development in the first place and to apply this knowledge to prevention and treatment.
Footnotes
Funding sources: Funding was received from the NIH/NCI (CA161870). Research reported in this review provides the background for development of an adaptive pathology-supported genetic testing framework for research translation supported by the Cancer Association of South Africa (S006385).
The content and findings reported are the sole deduction, view, and responsibility of the researchers and do not reflect the official position and sentiments of these funding agencies.
Conflicts of interest: None disclosed.
References
- 1.Zaremba A., Murali R., Jansen P. Clinical and genetic analysis of melanomas arising in acral sites. Eur J Cancer. 2019;119:66–76. doi: 10.1016/j.ejca.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ravaioli G., Dika E., Lambertini M., Chessa M., Fanti P., Patrizi A. Acral melanoma: correlating the clinical presentation to the mutational status. G Ital Dermatol Venereol. 2019;154(5):567–572. doi: 10.23736/S0392-0488.18.05791-7. [DOI] [PubMed] [Google Scholar]
- 3.Merkel E.A., Gerami P. Malignant melanoma of sun-protected sites: a review of clinical, histological, and molecular features. Lab Invest. 2017;97(6):630–635. doi: 10.1038/labinvest.2016.147. [DOI] [PubMed] [Google Scholar]
- 4.Paluncic J., Kovacevic Z., Jansson P.J. Roads to melanoma: key pathways and emerging players in melanoma progression and oncogenic signaling. Biochim Biophys Acta Mol Cell Res. 2016;1863(4):770–784. doi: 10.1016/j.bbamcr.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y.A., Teer J.K., Eroglu Z. Translational pathology, genomics and the development of systemic therapies for acral melanoma. Semin Cancer Biol. 2020;61:149–157. doi: 10.1016/j.semcancer.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison D.K. MAP kinase pathways. Cold Spring Harb Perspect Biol. 2012;4(11):a011254. doi: 10.1101/cshperspect.a011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandes M., Barcelos D., Comodo A.N. Acral lentiginous melanomas harbour intratumor heterogeneity in BRAF exon 15, with mutations distinct from V600E/V600K. Am J Dermatopathol. 2019;41(10):733–740. doi: 10.1097/DAD.0000000000001418. [DOI] [PubMed] [Google Scholar]
- 8.Bolognia J., Schaffer J., Cerroni L. 4th ed. Elsevier; London, UK: 2018. Dermatology. [Google Scholar]
- 9.Fernandes J.D., Hsieh R., De Freitas L.A.R. MAP kinase pathways: molecular roads to primary acral lentiginous melanoma. Am J Dermatopathol. 2015;37(12):892–897. doi: 10.1097/DAD.0000000000000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgescu M.M. PTEN tumor suppressor network in PI3K-Akt pathway control. Genes Cancer. 2010;1(12):1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison D. The JAK/STAT pathway:fact sheet. Cold Spring Harb Perspect Biol. 2012;4(3):a011205. doi: 10.1101/cshperspect.a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schadendorf D., van Akkooi A.C.J., Berking C. Melanoma. Lancet. 2018;392(10151):971–984. doi: 10.1016/S0140-6736(18)31559-9. [DOI] [PubMed] [Google Scholar]
- 13.Bell R.J.A., Rube H.T., Xavier-Magalhães A. Understanding TERT promoter mutations: a common path to immortality. Mol Cancer Res. 2016;14(4):315–323. doi: 10.1158/1541-7786.MCR-16-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang W.S., Hendricks W., Kiefer J. Integrated genomic analyses reveal frequent TERT aberrations in acral melanoma. Genome Res. 2017;27(4):524–532. doi: 10.1101/gr.213348.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheppard K.E., McArthur G.A. The cell-cycle regulator CDK4: an emerging therapeutic target in melanoma. Clin Cancer Res. 2013;19(19):5320–5328. doi: 10.1158/1078-0432.CCR-13-0259. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein A.M., Struewing J.P., Chidambaram A., Fraser M.C., Tucker M.A. Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J Natl Cancer Inst. 2000;92(12):1006–1010. doi: 10.1093/jnci/92.12.1006. [DOI] [PubMed] [Google Scholar]
- 17.Haugh A.M., Zhang B., Quan V.L. Distinct patterns of acral melanoma based on site and relative sun exposure. J Invest Dermatol. 2018;138(2):384–393. doi: 10.1016/j.jid.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 18.Kong Y., Sheng X., Wu X. Frequent genetic aberrations in the CDK4 pathway in acral melanoma indicate the potential for CDK4/6 inhibitors in targeted therapy. Clin Cancer Res. 2017;23(22):6946–6957. doi: 10.1158/1078-0432.CCR-17-0070. [DOI] [PubMed] [Google Scholar]
- 19.Nag S., Qin J., Srivenugopal K.S., Wang M., Zhang R. The MDM2-p53 pathway revisited. J Biomed Res. 2013;27(4):254–271. doi: 10.7555/JBR.27.20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ploper D., Taelman V.F., Robert L. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci U S A. 2015;112(5):E420–E429. doi: 10.1073/pnas.1424576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shim J.H., Shin H.T., Park J. Mutational profiling of acral melanomas in Korean populations. Exp Dermatol. 2017;26(10):883–888. doi: 10.1111/exd.13321. [DOI] [PubMed] [Google Scholar]
- 22.Rabbie R., Ferguson P., Molina-Aguilar C., Adams D.J., Robles-Espinoza C.D. Melanoma subtypes: genomic profiles, prognostic molecular markers and therapeutic possibilities. J Pathol. 2019;247(5):539–551. doi: 10.1002/path.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward N.K., Wilmott J.S., Waddell N. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 24.Pon J., Marra M. Driver and passenger mutations in cancer. Annu Rev Pathol. 2015;10:25–50. doi: 10.1146/annurev-pathol-012414-040312. [DOI] [PubMed] [Google Scholar]
- 25.Reddy B., Miller D., Tsao H. Somatic driver mutations in melanoma. Cancer. 2017;123:2104–2117. doi: 10.1002/cncr.30593. [DOI] [PubMed] [Google Scholar]
- 26.Furney S.J., Turajlic S., Stamp G. The mutational burden of acral melanoma revealed by whole-genome sequencing and comparative analysis. Pigment Cell Melanoma Res. 2014;27(5):835–838. doi: 10.1111/pcmr.12279. [DOI] [PubMed] [Google Scholar]
- 27.Hodis E., Watson I.R., Kryukov G.V. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krauthammer M., Kong Y., Ha B.H. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44(9):1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai B., Cai X., Kong Y.Y. Analysis of KIT expression and gene mutation in human acral melanoma: with a comparison between primary tumors and corresponding metastases/recurrences. Hum Pathol. 2013;44(8):1472–1478. doi: 10.1016/j.humpath.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Grzywa T.M., Paskal W., Włodarski P.K. Intratumor and intertumor heterogeneity in melanoma. Transl Oncol. 2017;10(6):956–975. doi: 10.1016/j.tranon.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akbani R., Akdemir K.C., Aksoy B.A. Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cirenajwis H., Lauss M., Ekedahl H. NF1-mutated melanoma tumors harbor distinct clinical and biological characteristics. Mol Oncol. 2017;11(4):438–451. doi: 10.1002/1878-0261.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sosman J. Melanoma: what every physician needs to know. 2017. https://www.cancertherapyadvisor.com/home/decision-support-in-medicine/imaging/melanoma/ Available at:
- 34.Leichsenring J., Stögbauer F., Volckmar A.L. Genetic profiling of melanoma in routine diagnostics: assay performance and molecular characteristics in a consecutive series of 274 cases. Pathology. 2018;50(7):703–710. doi: 10.1016/j.pathol.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Moon K.R., Choi Y.D., Kim J.M. Genetic alterations in primary acral melanoma and acral melanocytic nevus in Korea: common mutated genes show distinct cytomorphological features. J Invest Dermatol. 2018;138(4):933–945. doi: 10.1016/j.jid.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 36.Darmawan C.C.C., Gwanghyun J., Montenegro S.S.E. Early detection of acral melanoma: a review of clinical, dermoscopic, histopathologic, and molecular characteristics. J Am Acad Dermatol. 2019;81(3):805–812. doi: 10.1016/j.jaad.2019.01.081. [DOI] [PubMed] [Google Scholar]
- 37.Sheen Y., Tan K., Tse K. Genetic alterations in primary melanoma in Taiwan. Br J Dermatol. 2020;182(5):1205–1213. doi: 10.1111/bjd.18425. [DOI] [PubMed] [Google Scholar]
- 38.Namiki T., Coelho S.G., Hearing V.J. NUAK2: an emerging acral melanoma oncogene. Oncotarget. 2011;2(9):695–704. doi: 10.18632/oncotarget.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vásquez-Moctezuma I., Meraz-Ríos M., Villanueva-López C. ATP-binding cassette transporter ABCB5 gene is expressed with variability in malignant melanoma. Actas Dermosifiliogr. 2010;101(4):341–348. doi: 10.1016/j.ad.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 40.Yun J., Lee J., Jang J. KIT amplification and gene mutations in acral/mucosal melanoma in Korea. APMIS. 2011;119(6):330–335. doi: 10.1111/j.1600-0463.2011.02737.x. [DOI] [PubMed] [Google Scholar]
- 41.Desai A., Ugorji R., Khachemoune A. Acral melanoma foot lesions. Part 1: epidemiology, aetiology, and molecular pathology. Clin Exp Dermatol. 2017;42(8):845–848. doi: 10.1111/ced.13243. [DOI] [PubMed] [Google Scholar]
- 42.Slipicevic A., Herlyn M. KIT in melanoma: many shades of gray. J Invest Dermatol. 2015;135(2):337–338. doi: 10.1038/jid.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puntervoll H.E., Molven A., Akslen L.A. Frequencies of KIT and GNAQ mutations in acral melanoma. J Cutan Pathol. 2014;41(11):893–894. doi: 10.1111/cup.12382. [DOI] [PubMed] [Google Scholar]
- 44.Yeh I., Jorgenson E., Shen L. Targeted genomic profiling of acral melanoma. J Natl Cancer Inst. 2019;111:1–10. doi: 10.1093/jnci/djz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao H.W., Tsai W.C., Perng C.L., Wang W.M., Chiang C.P. Distinct MAPK and PI3K pathway mutations in different melanoma types in Taiwanese individuals. Eur J Dermatol. 2018;28(4):509–518. doi: 10.1684/ejd.2018.3359. [DOI] [PubMed] [Google Scholar]
- 46.Curtin J.A., Fridlyand J., Kageshita T. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 47.Zebary A., Omholt K., Vassilaki I. KIT, NRAS, BRAF and PTEN mutations in a sample of Swedish patients with acral lentiginous melanoma. J Dermatol Sci. 2013;72(3):284–289. doi: 10.1016/j.jdermsci.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 48.De Lima Vazquez V., Vicente A.L., Carloni A. Molecular profiling, including TERT promoter mutations, of acral lentiginous melanomas. Melanoma Res. 2016;26(2):93–99. doi: 10.1097/CMR.0000000000000222. [DOI] [PubMed] [Google Scholar]
- 49.Roh M.R., Park K.H., Chung K.Y., Shin S.J., Rha S.Y., Tsao H. Telomerase reverse transcriptase (TERT) promoter mutations in Korean melanoma patients. Am J Cancer Res. 2017;7(1):134–138. [PMC free article] [PubMed] [Google Scholar]
- 50.Shi K., Zhang B., Kong B.Y. Distinct genomic features in a retrospective cohort of mucosal, acral and vulvovaginal melanomas. J Am Acad Dermatol. 2019 doi: 10.1016/j.jaad.2019.07.017. [DOI] [PubMed] [Google Scholar]
- 51.Goodman A.M., Kato S., Bazhenova L. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trucco L.D., Mundra P.A., Hogan K. Ultraviolet radiation–induced DNA damage is prognostic for outcome in melanoma. Nat Med. 2019;25(2):221–224. doi: 10.1038/s41591-018-0265-6. [DOI] [PubMed] [Google Scholar]
- 53.Rawson R.V., Johansson P.A., Hayward N.K. Unexpected UVR and non-UVR mutation burden in some acral and cutaneous melanomas. Lab Invest. 2017;97(2):130–145. doi: 10.1038/labinvest.2016.143. [DOI] [PubMed] [Google Scholar]
- 54.Liu L., Zhang W., Gao T., Li C. Is UV an etiological factor of acral melanoma? J Expo Sci Environ Epidemiol. 2016;26(6):539–545. doi: 10.1038/jes.2015.60. [DOI] [PubMed] [Google Scholar]
- 55.Stern D.K., Creasey A.A., Quijije J., Lebwohl M.G. UV-A and UV-B penetration of normal human cadaveric fingernail plate. Arch Dermatol. 2011;147(4):439–441. doi: 10.1001/archdermatol.2010.375. [DOI] [PubMed] [Google Scholar]
- 56.Yi K., Ju Y.S. Patterns and mechanisms of structural variations in human cancer. Exp Mol Med. 2018;50(8):98. doi: 10.1038/s12276-018-0112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng P.F. Medical bioinformatics in melanoma. Curr Opin Oncol. 2018;30(2):113–117. doi: 10.1097/CCO.0000000000000428. [DOI] [PubMed] [Google Scholar]
- 58.Chernoff K.A., Bordone L., Horst B. GAB2 amplifications refine molecular classification of melanoma. Clin Cancer Res. 2009;15(13):4288–4291. doi: 10.1158/1078-0432.CCR-09-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niu H.T., Zhou Q.M., Wang F. Identification of anaplastic lymphoma kinase break points and oncogenic mutation profiles in acral/mucosal melanomas. Pigment Cell Melanoma Res. 2013;26(5):646–653. doi: 10.1111/pcmr.12129. [DOI] [PubMed] [Google Scholar]
- 60.Rachakonda S., Kong H., Srinivas N. Telomere length, telomerase reverse transcriptase promoter mutations, and melanoma risk. Genes Chromosomes Cancer. 2018;57(11):564–572. doi: 10.1002/gcc.22669. [DOI] [PubMed] [Google Scholar]
- 61.Carrera C., Puig-Butille J.A. Clinical, epidemiological, and molecular heterogeneity in acral melanoma. J Invest Dermatol. 2018;138(2):254–255. doi: 10.1016/j.jid.2017.09.027. [DOI] [PubMed] [Google Scholar]
- 62.Turner J., Couts K., Sheren J. Kinase gene fusions in defined subsets of melanoma. Pigment Cell Melanoma Res. 2017;30(1):53–62. doi: 10.1111/pcmr.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Comodo-Navarro A.N., Fernandes M., Barcelos D. Intratumor heterogeneity of KIT gene mutations in acral lentiginous melanoma. Am J Dermatopathol. 2020;42(4):265–271. doi: 10.1097/DAD.0000000000001475. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X., Peng Y., Li C. Genomic heterogeneity and branched evolution of early stage primary acral melanoma shown by multiregional microdissection sequencing. J Invest Dermatol. 2019;139(7):1526–1534. doi: 10.1016/j.jid.2019.01.019. [DOI] [PubMed] [Google Scholar]
- 65.Su J., Yu W., Liu J. Fluorescence in situ hybridisation as an ancillary tool in the diagnosis of acral melanoma: a review of 44 cases. Pathology. 2017;49(7):740–749. doi: 10.1016/j.pathol.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Diaz A., Puig-Butillé J.A., Valera A. TERT and AURKA gene copy number gains enhance the detection of acral lentiginous melanomas by fluorescence in situ hybridization. J Mol Diagn. 2014;16(2):198–206. doi: 10.1016/j.jmoldx.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Donnelly D., Aung P.P., Jour G. The “-omics” facet of melanoma: heterogeneity of genomic, proteomic and metabolomic biomarkers. Semin Cancer Biol. 2019;59:165–174. doi: 10.1016/j.semcancer.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 68.Griewank K.G., Schilling B., Scholz S.L. Oncogene status as a diagnostic tool in ocular and cutaneous melanoma. Eur J Cancer. 2016;57:112–117. doi: 10.1016/j.ejca.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 69.Bastian B.C., Kashani-Sabet M., Hamm H. Gene amplifications characterize acral melanoma and permit the detection of occult tumor cells in the surrounding skin. Cancer Res. 2000;60(7):1968–1973. [PubMed] [Google Scholar]
- 70.Grant K.A., Apffelstaedt J.P., Wright C. Mammaprint Pre-screen Algorithm (MPA) reduces chemotherapy in patients with early-stage breast cancer. S Afr Med J. 2013;103(8):522–526. doi: 10.7196/samj.7223. [DOI] [PubMed] [Google Scholar]
- 71.Grant K.A., Myburgh E.J., Murray E. Reclassification of early stage breast cancer into treatment groups by combining the use of immunohistochemistry and microarray analysis. S Afr J Sci. 2019;115(3/4):2–7. [Google Scholar]
- 72.Hong J.W., Lee S., Kim D.C., Kim K.H., Song K.H. Prognostic and clinicopathologic associations of BRAF mutation in primary acral lentiginous melanoma in Korean patients: a preliminary study. Ann Dermatol. 2014;26(2):195–202. doi: 10.5021/ad.2014.26.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shitara D., Tell-Martí G., Badenas C. Mutational status of naevus-associated melanomas. Br J Dermatol. 2015;173(3):671–680. doi: 10.1111/bjd.13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi Y.D., Chun S.M., Jin S.A., Lee J.B., Yun S.J. Amelanotic acral melanomas: clinicopathological, BRAF mutation, and KIT aberration analyses. J Am Acad Dermatol. 2013;69(5):700–707. doi: 10.1016/j.jaad.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 75.Scolyer R.A., Long G.V., Thompson J.F. Evolving concepts in melanoma classification and their relevance to multidisciplinary melanoma patient care. Mol Oncol. 2011;5(2):124–136. doi: 10.1016/j.molonc.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Savoia P., Fava P., Casoni F., Cremona O. Targeting the ERK signaling pathway in melanoma. Int J Mol Sci. 2019;20:1–37. doi: 10.3390/ijms20061483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cosgarea I., Ritter C., Becker J.C., Schadendorf D., Ugurel S. Update on the clinical use of kinase inhibitors in melanoma. J Dtsch Dermatol Ges. 2017;15(9):887–893. doi: 10.1111/ddg.13321. [DOI] [PubMed] [Google Scholar]
- 78.Domingues B., Lopes J., Soares P., Populo H. Melanoma treatment in review. Immunotargets Ther. 2018;7:35–49. doi: 10.2147/ITT.S134842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu X., Yu J., Yan J. PI3K/AKT/mTOR pathway inhibitors inhibit the growth of melanoma cells with mTOR H2189Y mutations in vitro. Cancer Biol Ther. 2018;19(7):584–589. doi: 10.1080/15384047.2018.1435221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delyon J., Chevret S., Jouary T. STAT3 mediates nilotinib response in KIT-altered melanoma: a phase II multicenter trial of the French Skin Cancer Network. J Invest Dermatol. 2018;138(1):58–67. doi: 10.1016/j.jid.2017.07.839. [DOI] [PubMed] [Google Scholar]
- 81.Yu J., Yu J., Wu X. The TERT copy number gain is sensitive to telomerase inhibitors in human melanoma. Clin Sci. 2020;134(2):193–205. doi: 10.1042/CS20190890. [DOI] [PubMed] [Google Scholar]
- 82.Lee B., McArthur G.A. CDK4 inhibitors an emerging strategy for the treatment of melanoma. Melanoma Manag. 2015;2(3):255–266. doi: 10.2217/mmt.15.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanz G., Singh M., Peuget S., Selivanova G. Inhibition of p53 inhibitors: progress, challenges and perspectives. J Mol Cell Biol. 2019;11(7):586–599. doi: 10.1093/jmcb/mjz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goldsberry W.N., Londoño A., Randall T.D., Norian L.A., Arend R.C. A review of the role of wnt in cancer immunomodulation. Cancers (Basel) 2019;11(6):1–19. doi: 10.3390/cancers11060771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang R., He Y., Robinson V. Targeting lineage-specific MITF pathway in human melanoma cell lines by A-485, the selective small-molecule inhibitor of p300/CBP. Mol Cancer Ther. 2018;17(12):2543–2550. doi: 10.1158/1535-7163.MCT-18-0511. [DOI] [PubMed] [Google Scholar]
- 86.Andtbacka R.H.I., Kaufman H.L., Collichio F. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33(25):2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 87.Davar D., Ding F., Saul M. High-dose interleukin-2 (HD IL-2) for advanced melanoma: a single center experience from the University of Pittsburgh Cancer Institute. J Immunother Cancer. 2017;5(1):1–10. doi: 10.1186/s40425-017-0279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.National Cancer Institute Drugs approved for melanoma. 2019. https://www.cancer.gov/about-cancer/treatment/drugs/melanoma Available at:
- 89.Veatch J.R., Lee S.M., Fitzgibbon M. Tumor-infiltrating BRAFV600E-specific CD4+ T cells correlated with complete clinical response in melanoma. J Clin Invest. 2018;128(4):1563–1568. doi: 10.1172/JCI98689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gershenwald J.E., Scolyer R.A., Hess K.R., Faries M.B., Kirkwood J.M., McArthur G.A. Melanoma staging: evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(6):472–492. doi: 10.3322/caac.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yan J., Yu J., Wu X. Increased AURKA gene copy number correlates with poor prognosis and predicts the efficacy of high-dose interferon therapy in acral melanoma. J Cancer. 2018;9(7):1267–1276. doi: 10.7150/jca.24013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Durbec F., Martin L., Derancourt C., Grange F. Melanoma of the hand and foot: epidemiological, prognostic and genetic features. A systematic review. Br J Dermatol. 2012;166(4):727–739. doi: 10.1111/j.1365-2133.2011.10772.x. [DOI] [PubMed] [Google Scholar]
- 93.Clish C.B. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud. 2015;1(1):a000588. doi: 10.1101/mcs.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim H.Y., Lee H., Kim S.H., Jin H., Bae J., Choi H.K. Discovery of potential biomarkers in human melanoma cells with different metastatic potential by metabolic and lipidomic profiling. Sci Rep. 2017;7(1):1–14. doi: 10.1038/s41598-017-08433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gopalakrishnan V., Spencer C.N., Nezi L. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Betti M., Aspesi A., Biasi A. CDKN2A and BAP1 germline mutations predispose to melanoma and mesothelioma. Cancer Lett. 2016;378(2):120–130. doi: 10.1016/j.canlet.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 97.Rojek N.W., Korcheva V., Leachman S.A. A large skin-colored nodule on the plantar foot: a quiz. Acta Derm Venereol. 2017;97(10):1265–1266. doi: 10.2340/00015555-2726. [DOI] [PubMed] [Google Scholar]
- 98.Moran B., Silva R., Perry A.S., Gallagher W.M. Epigenetics of malignant melanoma. Semin Cancer Biol. 2018;51:80–88. doi: 10.1016/j.semcancer.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 99.Hammerlindl H., Schaider H. Epigenetics in melanoma development and drug resistance. In: Blumenberg M., editor. Human Skin Cancers - Pathways, Mechanisms, Targets and Treatments. IntechOpen; 2017. pp. 3–24. [DOI] [Google Scholar]
- 100.Chan E., Patel R., Nallur S. MicroRNA signatures differentiate melanoma subtypes. Cell Cycle. 2011;10(11):1845–1852. doi: 10.4161/cc.10.11.15777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shruthi B., Vinodhkumar P., Selvamani M. Proteomics: a new perspective for cancer. Adv Biomed Res. 2016;5(1):67. doi: 10.4103/2277-9175.180636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sengupta D., Tackett A.J., Rock L., Rock L. Proteomic findings in melanoma. J Proteomics Bioinform. 2016;9(4):1–7. doi: 10.4172/jpb.1000e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Höiom V., Tuominen R., Käller M. MC1R variation and melanoma risk in the Swedish population in relation to clinical and pathological parameters. Pigment Cell Melanoma Res. 2009;22(2):196–204. doi: 10.1111/j.1755-148X.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 104.Fallah M., Pukkala E., Sundquist K. Familial melanoma by histology and age: joint data from five Nordic countries. Eur J Cancer. 2014;50(6):1176–1183. doi: 10.1016/j.ejca.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 105.Van Der Rhee J.I., Krijnen P., Gruis N.A. Clinical and histologic characteristics of malignant melanoma in families with a germline mutation in CDKN2A. J Am Acad Dermatol. 2011;65(2):281–288. doi: 10.1016/j.jaad.2010.06.044. [DOI] [PubMed] [Google Scholar]
- 106.Lacruz G., Cárdenas I., Carrera C. Multiple primary acral melanomas in two young Caucasian patients. Dermatology. 2014;228(4):307–310. doi: 10.1159/000362207. [DOI] [PubMed] [Google Scholar]
- 107.Bergqvist C., Kadara H., Hamie L. SLURP-1 is mutated in Mal de Meleda, a potential molecular signature for melanoma and a putative squamous lineage tumor suppressor gene. Int J Dermatol. 2017;57(2):162–170. doi: 10.1111/ijd.13850. [DOI] [PubMed] [Google Scholar]
- 108.Nakajima K., Nakano H., Takiyoshi N. Papillon-Lefèvre syndrome and malignant melanoma: a high incidence of melanoma development in Japanese palmoplantar keratoderma patients. Dermatology. 2008;217(1):58–62. doi: 10.1159/000124340. [DOI] [PubMed] [Google Scholar]
- 109.Kogame T., Kaku Y., Endo Y. A follow-up report of acral melanoma in a patient with Nagashima-type palmoplantar keratosis: validation of SERPINB7 mutation and local recurrence. Eur J Dermatol. 2018;28(4):519–520. doi: 10.1684/ejd.2018.3317. [DOI] [PubMed] [Google Scholar]
- 110.Seike T., Nakanishi H., Urano Y., Arase S. Malignant melanoma developing in an area of palmoplantar keratoderma (Greither's disease) J Dermatol. 1995;22(1):55–61. doi: 10.1111/j.1346-8138.1995.tb03342.x. [DOI] [PubMed] [Google Scholar]