Abstract
IgA nephropathy is the most prevalent primary glomerulonephritis worldwide, with identical immunopathological characteristics caused by multiple etiologies as well as influenced by geographical and ethnical factors. To elucidate the role of immunologic and inflammatory mechanisms in the susceptibility to IgA nephropathy, we explored single nucleotide polymorphisms of related molecules in the immune pathways. We searched the PubMed database for studies that involved all gene variants of molecules in the 20 immunologic and inflammatory pathways selected from the Kyoto Encyclopedia of Genes and Genomes database. The odds ratios with their corresponding 95% confidence intervals in six genetic models (allele model, dominant model, homozygote model, heterozygote model, overdominant model, and recessive model) were summarized using fixed or random effect models. Subgroup analysis was conducted based on different ethnicities with generalized odds ratios. Heterogeneity was evaluated using the Q and I2 tests. Begg’s funnel plot and Egger’s linear regression test were used to evaluating possible publication bias among the included studies, and sensitivity analysis was used to test the stability of the overall results. A total of 45 studies met our selection criteria and eight related genetic association studies were retrieved, including 320 single-nucleotide polymorphisms from 20 candidate pathways, ranging from 2000 to 2021. A total of 28,994 healthy people versus 20,600 IgA nephropathy patients were enrolled. Upon meta-analyzed results that TGFB1 (rs1800469, rs1982073, rs1800471), IL-1B (rs1143627), IL-18 (rs1946518), and TLR1 (rs5743557) showed effect with or without ethnicity difference. And 10 variants presented stable and robust related to IgA nephropathy. This research showed that genetic variants are related to the immunologic and inflammatory effects of IgA nephropathy pathogenesis. The meta-analysis results supported the previous researches, and may help deepen the understanding of pathogenesis and explore new targets for IgA nephropathy-specific immunotherapy.
Keywords: IgA nephropathy, susceptibility, inflammatory molecules, immune pathway, single-nucleotide polymorphism, meta-analysis
Introduction
IgA nephropathy (IgAN) is the most prevalent primary glomerulonephritis in the world, and 30–40% of patients progress to the end-stage renal disease within 20–30 years after diagnosis, requiring kidney transplantation or renal replacement therapy (1). However, the recurrence of IgAN most likely occurs during kidney transplantation treatment, because the abnormalities in the immune system are still not corrected (2). The prevalence of IgAN shows great variations across the world; among patients undergoing renal biopsy, the incidence of IgAN is highest in Asian populations, occupying almost 45%; the prevalence of IgAN is moderate in European populations and is around 25%, and African populations show the lowest prevalence at just under 5% (3). IgAN also shows gender differences in prevalence: its prevalence in males is five times as that in females and male gender is a prognostic factor which indicates poor renal outcomes (4). Although the disease appears to be sporadic, there is an increasing number of reports showing familial clusters of IgAN. In addition, these familial cases have poorer renal outcomes and have fewer gender differences than sporadic cases (5). Gender as well as geographical factors hint at a possibility of genetic vulnerability. To date, the understanding of the pathogenic mechanisms of IgAN is still being explored but based on previous studies, immunologic regulation and inflammation contributes significantly to IgAN (6, 7). The characteristics of IgAN includes an IgA deposition in the glomerular mesangium area with complexes which activate mesangial cells to overproduce cytokines, chemokines, and complements (8). These mesangial-released inflammatory mediators cause local glomerular damage and may also aggravate podocytes and the renal tubular interstitium through humoral crosstalk (9). Recently, several susceptibility loci have been identified in different populations which involved in antigen processing and presentation, the mucosal defense system and the alternative complement pathway (10). Previous systematic review and meta-analysis usually focused on one gene and its single variant. However, the immunologic regulation is a huge network with interactions between various proteins which requires systematic studying of the immune pathways. The magnitude and significance of risk effects in genetic association studies is usually estimated by different genetic models with odds ratio (OR) and its 95% confidence interval (CI). However, the genetic contrasts are not independent and might cause confusion during interpretation of results and due to the use of inappropriate method without general justification. In reality, the true underlying genetic model is unknown and most studies have provided several potential models which may contribute to the loss of information resolution. The generalized linear odds ratio (ORG) analyzes the account of cases/healthy control pairs in the study to indicate the mutational load of disease susceptibility (11).
In past studies, the chosen of genetic models might not have been detailed enough to support results. In order to provide a strong support to existing evidence, we performed cluster analysis on six genetic models using SNPs in studies and calculated ORG. However, the large disparities in prevalence based on ethnicity that most studies have reported in cohorts of Asian populations has limited the overall review and meta-analysis of IgAN.
Materials and Methods
Search Strategy
To elucidate the role of the immunologic and inflammatory mechanisms in the susceptibility to IgA nephropathy, we selected 20 candidate pathways classified by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database ( Supplementary Box 1 ). In the meta-analysis, we searched the PubMed database for genetic association studies of all molecules in the aforementioned pathways involved in IgA nephropathy. The search strategy was based on the following combined MeSH terms: “IgA nephropathy”[Title/Abstract] AND (“genes”[MeSH Terms] OR “genes”[All Fields] OR “gene”[All Fields]) AND “related molecule name”[All Fields], (GWAS[Title]) AND (IgA nephropathy[Title]) (genome-wide association study[Title]) AND (IgA nephropathy[Title]). The eligible studies were all published in English and peer-reviewed, up to February 24, 2021, and we did not search gray literature (i.e., preprints, reports, or conference abstracts). Finally, references in the included articles were retrieved to identify further relevant studies.
Selection Criteria
The following inclusion criteria were met for the meta-analysis: (1) the IgA nephropathy diagnosis was confirmed by renal biopsy; (2) healthy individuals included in the study as a control group; (3) sufficient genotypic data, such as genotype count data or allele frequencies, were provided; and (4) disease susceptibility was investigated.
Studies meeting any one of the following criteria were excluded: (1) the research subjects were patients after kidney transplantation or recurrent patients; (2) no healthy subjects as a control group; (3) studies investigating progression, prognosis, severity, response to treatment; (4) comments, case reports, editorials, letters, reviews, and non-English articles; (5) animal studies and in vitro experiments; and (6) data unextractable studies.
Two reviewers independently screened the abstracts of the retrieved articles, and if necessary, the full text was also reviewed. Discrepancies regarding potentially eligible studies were finalized through discussion to reach a consensus.
Data Extraction
We used electronic data to extract the following information from each eligible study: first author, publication year, study country, PMID, ethnicity, disease type, sample size, age, and sex. For genotypic data, we extracted genotype count data in cases and controls, and, if available, the odds ratios (ORs) with their corresponding 95% confidence intervals (CIs) of five genetic models and evidence of Hardy-Weinberg equilibrium (HWE) in controls were also recorded.
Quality Assessment
We appraised the quality and risk of bias of the included studies according to the Newcastle–Ottawa Quality Assessment Scale (NOS). The total score of NOS is 9 points, including three aspects: selection, comparability, and exposure. According to the final score, the studies could be categorized into high quality (score >6), medium quality (score between 4 and 6), and low quality (score less than 4).
Statistical Analysis
STATA SE software (version 15.0) was used to conduct all the data processing. HWE was examined using Fisher’s exact test and whether controls were confronted with HWE according to P-value results, with 0.05 as the boundary. The association between the mutational load of a variant and susceptibility to IgAN was presented by ORs with 95% CIs. For each variant, the OR estimates were calculated using the fixed-effects model (Mantel-Haenszel method). Statistical significance was set at P < 0.05. If the heterogeneity test I2≥50% or p < 0.05 of the Q test, indicating that the homogeneity was insignificant, then the random-effects model (DerSimonian and Laird method) was considered. Since genetic models are not independent and there is no a priori biological justification for the choice, we performed genetic association analyses using six genetic models (allele model, dominant model, homozygote model, heterozygote model, overdominant model, and recessive model) for each molecule (12). Multiple genetic models could also reduce the probability of type I errors in genotype distribution in previous studies (13). Hence, ORG was calculated using ORGGASMA (http://biomath.med.uth.gr) software that was programmed by statist Elias Zintzaras (11).
Additionally, when the heterogeneity between studies was statistically significant, we used meta-regression analysis to identify potential sources of such heterogeneity. Begg’s funnel plot and Egger’s linear regression test were used to evaluating possible publication bias among the included studies. The stability of the combined results was examined by performing a sensitivity analysis.
Results
Characteristics of Studies
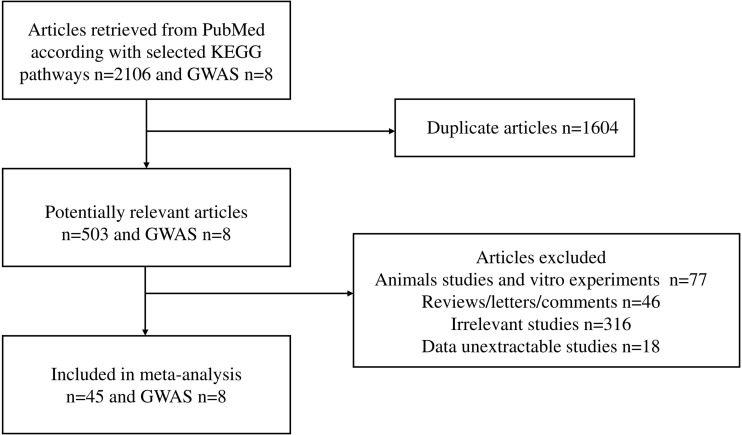
The workflow of the literature selected for meta-analysis is shown in Figure 1 . We obtained 45 studies, of which 8 studies involved Caucasians, and 37 studies recruited Asians (25 studies in Chinese, 1 study in Japanese, and 11 studies in Korean). Across these publications, nine studies included only pediatric patients, whereas most study subjects were adults. The participants ranged in age from 11.15 ± 5.11 to 48.1 ± 13.79, and 57.1% were male patients. Overall, 96 molecules from 20 candidate pathways and 320 polymorphisms were investigated in 45 studies published between 2000 and 2021. A total of 28,994 healthy people versus 20,600 patients had IgA nephropathy. The general characteristics of each included study and their references are shown in Supplementary Table 1 . Among a total of 320 gene polymorphisms, 48 were not confronted with HWE.
Figure 1.
Flow chart of studies selected for meta-analysis.
Meta-Analysis Results
Allele Model
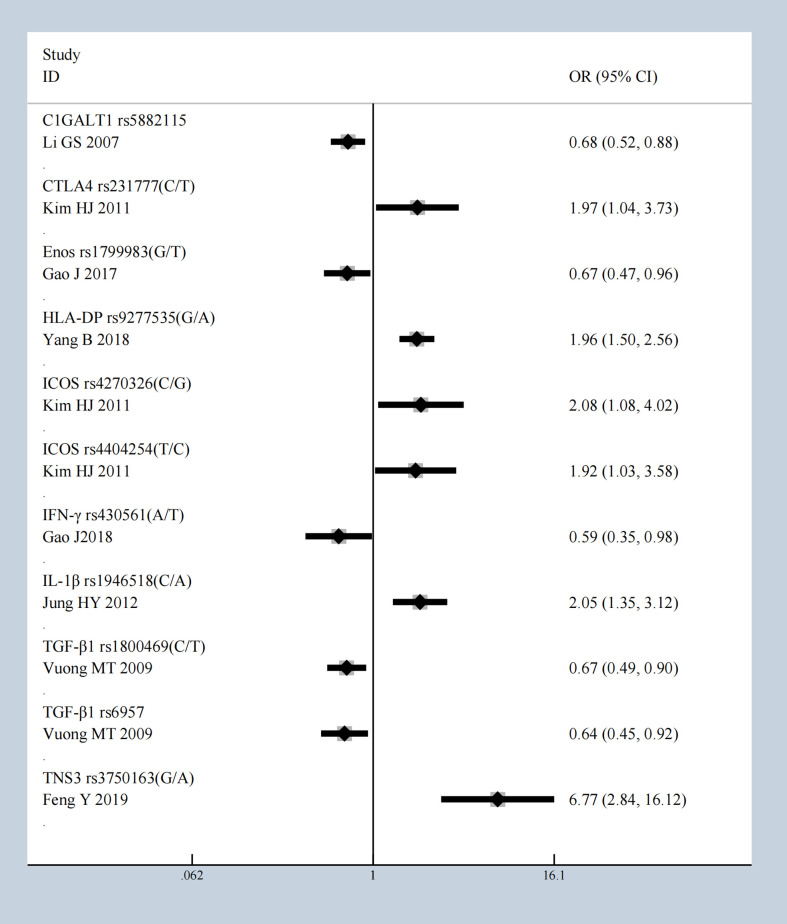
We conducted a meta-analysis of all molecules with gene polymorphisms that were confronted with HWE. The results show significant association with susceptivity to IgAN under the allele contrast model, as follows: core 1 synthase, glycoprotein-n-acetylgalactosamine 3-beta-galactosyltransferase 1 (C1GALT1), complement factor B (CFB), Cytotoxic T-Lymphocyte Associated Protein 4 (CTLA4), C-X-C Motif Chemokine Ligand 8 (CXCL8), endothelial nitric oxide synthase (Enos), Fc Receptor Like A (FCRLA), FCRLB, G Protein Subunit Gamma 2 (GNG2), Major Histocompatibility Complex, Class II, DP (HLA-DP), Inducible T Cell Costimulator (ICOS), Interferon-γ (IFN-γ), Interleukin 10 (IL-10), IL-1B, Interleukin 1 Receptor Antagonist (IL1RN), IL-6, Integrin Subunit Alpha X (ITGAX), LEM Domain Nuclear Envelope Protein 2 (LEMD2), Netrin 4 (NTN4), Pleckstrin Homology Like Domain Family B Member 1 (PHLDB1), Signal Transducer And Activator Of Transcription 4 (STAT4), Transforming Growth Factor-β (TGF‐β1), Toll Like Receptor 1 (TLR1), TLR10, Tumor Necrosis Factor Superfamily Member 13 (TNFSF13), Tensin 3 (TNS3), and Vascular Endothelial Growth Factor (VEGF); the details are shown in Supplementary Table 2 . The studies with the most statistically significant results, and the detailed information on per-single-nucleotide polymorphisms (SNPs) of significant OR with 95% CIs under the allele model are presented in Table 1 and Figure 2 .
Table 1.
SNPsof most significant OR with 95% CIs under the allele model.
| author | year | PMID | SNP | Ethnicity | Allele model OR | 0.95LCI | 0.95UCI | P |
|---|---|---|---|---|---|---|---|---|
| Li GS (14) | 2007 | 17228361 | C1GALT1 rs5882115 | Han Chinese | 0.679 | 0.522 | 0.884 | 0.004 |
| Kim HJ (15) | 2011 | 21677403 | CTLA4 rs231777 | Korean pediatric patients | 1.973 | 1.045 | 3.725 | 0.036 |
| Gao J (16) | 2017 | 28946141 | Enos rs1799983 | Han Chinese | 0.671 | 0.469 | 0.959 | 0.029 |
| Yang B (17) | 2018 | 29467950 | HLA-DP rs9277535 | Han Chinese | 1.958 | 1.497 | 2.560 | 0 |
| Kim HJ (15) | 2011 | 21677403 | ICOS rs4270326 | Korean pediatric patients | 2.082 | 1.079 | 4.018 | 0.029 |
| Kim HJ (15) | 2011 | 21677403 | ICOS rs4404254 | Korean pediatric patients | 1.919 | 1.029 | 3.579 | 0.04 |
| Gao J (18) | 2018 | 28391282 | IFN-γ rs430561 | Han Chinese | 0.589 | 0.353 | 0.982 | 0.042 |
| Jung HY (19) | 2012 | 26889427 | IL-1β rs1946518 | Korean | 2.055 | 1.353 | 3.120 | 0.001 |
| Vuong MT (20) | 2009 | 19258388 | TGF‐β1 rs1800469 | Sweden | 0.666 | 0.494 | 0.898 | 0.008 |
| Vuong MT (20) | 2009 | 19258388 | TGF‐β1 rs6957 | Sweden | 0.644 | 0.449 | 0.924 | 0.017 |
| Feng Y (21) | 2019 | 30928649 | TNS3 rs3750163 | Han Chinese | 6.768 | 2.841 | 16.123 | 0 |
Figure 2.
The most statistically significant SNPs under allele model.
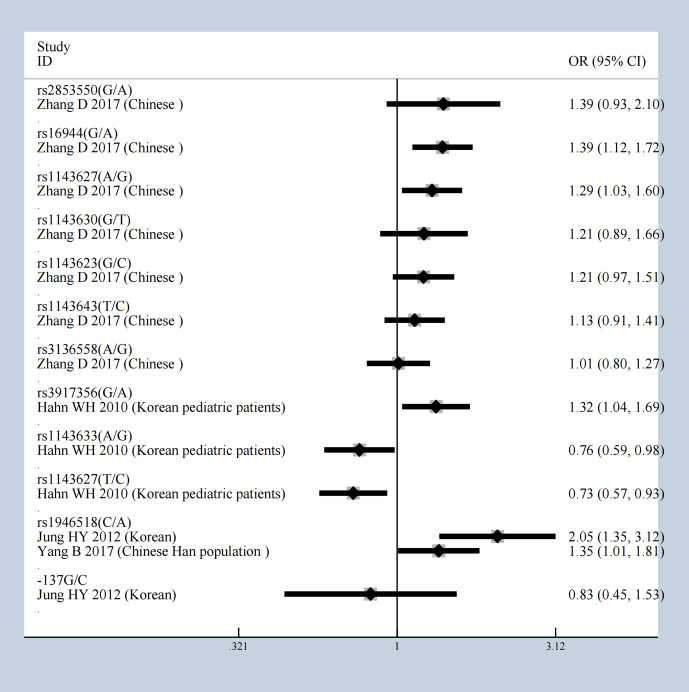
It is worth mentioning that the five gene variants of FCRLB (rs12079477, rs1891019, rs1891020, rs1417582, and rs4657093) have all been shown to have a statistical impact on IgAN susceptibility in the allele model Supplementary Figure 1 (22). The meta-analysis results of 13 gene variants of IL-1β are shown in Figure 3 , in which rs1946518 showed an increase in the IgAN susceptibility both in the Chinese Han population and Korean. CTLA4 has three SNPs (rs231777, rs5742909, and rs231779), all of which were significant. Both rs9277535 and rs3077 in HLA-DP showed an increased risk of developing IgAN. In addition, ICOS has eight gene variants, five of which have protective effects under the allele model (rs4270326, rs4404254, rs10183087, rs11571314, rs1559931) were associated with susceptibility to IgA in Supplementary Figure 2 .
Figure 3.
The results of 13 gene variants of IL-1b under allele model.
Dominant Model
To further validate the association of these significant SNPs with the risk of IgAN under allele model, we analyzed all gene polymorphisms in dominant contrast model. The genes significantly associated with IgAN occurrence and compounded with HWE were as follows: C1GALT1, CFB, CTLA4, CXCL8, FCRLA, FCRLB, HLA-DP, ICOS, IFN-γ, IL-10, IL-1β, IL1RN, IL4R, IL5RA, IL-6, ITGAM, ITGAX, Netrin 4 (NTN4), ST6GAL1, ST6GALNAC2, TGF‐β1, TLR1, TLR10, TNFRSF6B, TNFSF13, and TNS3. The general characteristics and details of each research on significant SNPs are described in Supplementary Table 3 . We identified ORs higher than 2 and lower than 0.5 as the most significant and the results are presented in Table 2 . Then, we noticed that ICOS has five SNPs (rs4270326, rs4404254, rs10183087, rs11571314, and rs1559931), all of which showed a robust significant risk of developing IgAN under dominant model which corresponded with the allele model results. Besides that, TNFRSF6B with the mutations of rs1291205 [OR = 0.45 (95% CI 0.31–0.54)], rs1291206 [OR = 0.46 (95% CI 0.31–0.67)], rs3208008 [OR = 0.47 (95% CI 0.32–0.69)] showed significant protective effect. And we obtained interesting results that did not show significance in allele model. The SNPs rs12054151, rs2239611, rs1990677, rs4686838, rs2284750, rs6784233, and rs7634389 in ST6GAL1 showed robust association with the occurrence of IgAN; rs3840858 in ST6GALNAC2 [OR = 3.68 95% CI (1.75–7.73)] showed a great correlation with IgAN risk in Chinese Uyghur population.
Table 2.
The most significant SNPs with ORs under dominant model higher than 2 and lower than 0.5.
| Author | Year | PMID | Ethnicity | SNP | Dominant model OR | 0.95LCI | 0.95UCI | P |
|---|---|---|---|---|---|---|---|---|
| Yang B (17) | 2018 | 29467950 | Han Chinese | HLA-DP rs9277535 | 2.05354 | 1.34 | 3.158399 | 0.001 |
| Kim HJ (15) | 2011 | 21677403 | Korean pediatric patients | ICOS rs4270326 | 2.23918 | 1.08 | 4.632189 | 0.03 |
| Kim HJ (15) | 2011 | 21677403 | Korean pediatric patients | ICOS rs4404254 | 2.21088 | 1.09 | 4.467172 | 0.027 |
| Kim HJ (15) | 2011 | 21677403 | Korean pediatric patients | ICOS rs10183087 | 2.17830 | 1.09 | 4.347564 | 0.027 |
| Kim HJ (15) | 2011 | 21677403 | Korean pediatric patients | ICOS rs11571314 | 2.17830 | 1.09 | 4.347564 | 0.027 |
| Kim HJ (15) | 2011 | 21677403 | Korean pediatric patients | ICOS rs1559931 | 2.17830 | 1.09 | 4.347564 | 0.027 |
| Liu XQ (23) | 2008 | 18256355 | Caucasians (St. Etienne) | IL4R rs1805015 | 0.43366 | 0.28 | 0.664227 | 0 |
| Fu D (24) | 2020 | 32747022 | Han Chinese | ST6GAL1 rs4686838 | 0.36516 | 0.30 | 0.443134 | 0 |
| Lu C (25) | 2015 | 26136946 | Uyghur Chinse | ST6GALNAC2 rs3840858 | 3.67568 | 1.75 | 7.73074 | 0.001 |
| Vuong MT (20) | 2009 | 19258388 | Sweden(male) | TGF‐β1 rs6957 | 0.14469 | 0.04 | 0.542827 | 0.004 |
| Vuong MT (20) | 2009 | 19258388 | Sweden(male) | TGF‐β1 rs180047 | 0.04851 | 0.00 | 0.907181 | 0.043 |
| Liu XQ (23) | 2008 | 18256355 | Caucasians (St. Etienne) | TNFRSF6B rs3208008 | 0.46880 | 0.32 | 0.684625 | 0 |
| Liu XQ (23) | 2008 | 18256355 | Caucasians (St. Etienne) | TNFRSF6B rs1291206 | 0.45731 | 0.31 | 0.667693 | 0 |
| Liu XQ (23) | 2008 | 18256355 | Caucasians (St. Etienne) | TNFRSF6B rs1291205 | 0.44846 | 0.31 | 0.654722 | 0 |
Overdominance Model
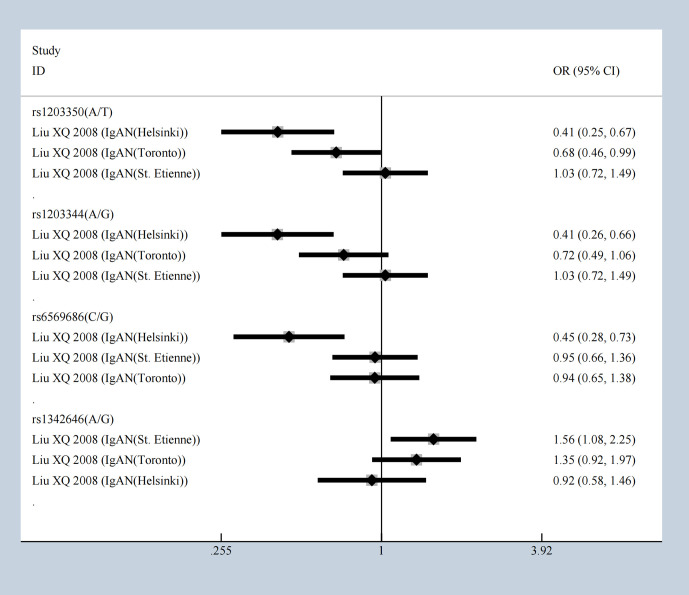
The SNPs rs3840858 and rs23840858 in ST6GALNAC2 showed protective effect respectively with [OR = 0.27 (95% CI 0.13–0.57)] and [OR = 0.64 (95% CI 0.41–0.99)] under the overdominance model which contradicted with dominance model result, but still hinted at IgAN occurrence in Uyghur Chinese population. The five SNPs of ICOS mentioned above were stably and significantly associated with IgAN but were associated with protective effects under overdominance model. Our meta-analysis results showed that rs1203350 and rs1203344 in IGAN1 decrease the susceptibility of IgAN in Helsinki and Toronto population, but not in St. Etienne patients. Besides that, according to the results of overdominance model, rs6569686 decreases the susceptibility of IgAN in Helsinki people ( Figure 4 ). The following showed a significant association including C1GALT1, CFB, CXCL8, FCRLA, FCRLB, HLA-DP, ICOS, IFN-γ, IL-10, IL-1β, IL4R, ITGAM, ITGAX, ST6GAL1, ST6GALNAC2, TGF‐β1. TNFRSF6B, TNS3, IGAN1, IL22RA1, and STAT4 emerged with significant statistic meaning from multitudinous immune molecules under overdominance model ( Supplementary Table 4 ) and most significant results were presented in Table 3 .
Figure 4.
The SNPs of IGAN1 in different population under overdominance model.
Table 3.
SNPs of most significant OR with 95% CIs under the overdominant model.
| Author | Year | Ethnicity | PMID | SNP | Overdominant OR | 0.95-LCI | 0.95-UCI | P |
|---|---|---|---|---|---|---|---|---|
| Yang B (17) | 2018 | Han Chinese | 29467950 | HLA-DP rs3077 | 4.62 | 2.809256 | 7.588952 | 0 |
| Liu XQ (23) | 2008 | Caucasians (St. Etienne) | 18256355 | IL4R rs1805015 | 2.45 | 1.58403 | 3.788273 | 0 |
| Yang B (17) | 2018 | Han Chinese | 29467950 | STAT4 rs7574865 | 2.38 | 1.553347 | 3.638879 | 0 |
| Fu D (24) | 2020 | Han Chinese | 32747022 | ST6GAL1 rs4686838 | 1.85 | 1.490156 | 2.274617 | 0 |
| Liu XQ (23) | 2008 | Caucasians (St. Etienne) | 18256355 | TNFRSF6B rs1291205 | 1.78 | 1.209218 | 2.614096 | 0.003 |
| Gao J (18) | 2018 | Han Chinese (St. Etienne) | 28391282 | IFN-γ rs430561 | 1.75 | 1.033877 | 2.949718 | 0.037 |
| Liu XQ (23) | 2008 | Caucasians | 18256355 | TNFRSF6B rs1291206 | 1.74 | 1.184463 | 2.562628 | 0.005 |
| Liu XQ (23) | 2008 | Caucasians (St. Etienne) | 18256355 | TNFRSF6B rs3208008 | 1.70 | 1.154023 | 2.499533 | 0.007 |
| Fu D (24) | 2020 | Han Chinese | 32747022 | ST6GAL1 rs2284750 | 1.58 | 1.274685 | 1.964356 | 0 |
| Liu XQ (23) | 2008 | Caucasians (St. Etienne) | 18256355 | IGAN1 rs1342646 | 1.56 | 1.080724 | 2.245152 | 0.017 |
| Yang B (26) | 2017 | Han Chinese | 27028244 | IL-1β rs1946518 | 1.53 | 1.00525 | 2.338833 | 0.047 |
| Hahn WH (27) | 2010 | Korean pediatric patients | 19280228 | IL-1β rs1143633 | 1.50 | 1.060556 | 2.127999 | 0.022 |
| Kim HJ (15) | 2011 | Korean pediatric patients | 21677403 | ICOS rs4270326 | 0.47 | 0.22301 | 0.992154 | 0.048 |
| Kim HJ (15) | 2011 | Korean pediatric patients | 21677403 | ICOS rs10183087 | 0.45 | 0.221188 | 0.92188 | 0.029 |
| Kim HJ (15) | 2011 | Korean pediatric patients | 21677403 | ICOS rs11571314 | 0.45 | 0.221188 | 0.92188 | 0.029 |
| Kim HJ (15) | 2011 | Korean pediatric patients | 21677403 | ICOS rs1559931 | 0.45 | 0.221188 | 0.92188 | 0.029 |
| Kim HJ (15) | 2011 | Korean pediatric patients | 21677403 | ICOS rs4404254 | 0.44 | 0.214075 | 0.91871 | 0.029 |
| Lu C (25) | 2015 | Uyghur Chinese | 26136946 | ST6GALNAC2 rs3840858 | 0.27 | 0.129354 | 0.572199 | 0.001 |
| Feng Y (21) | 2019 | Han Chinese | 30928649 | TNS3 rs3750163 | 0.06 | 0.013265 | 0.23499 | 0 |
Homozygote Model
It is generally recognized that homozygote of the mutation increases the risk for disease than the heterozygote, whereas homozygotes of the wild allele are not risky (11). C1GALT1 rs5882115 showed a significant effect under homozygote model [OR = 0.70 95% CI (0.55–0.91], allele model [OR = 0.68 95% CI (0.52–0.89)], dominant model [OR = 0.68 95% CI (0.51–0.90)], and overdominant model [OR = 1.39 95% CI (1.04–1.85)]. Several SNPs showed significance only in homozygote model compared to the other genetic models mentioned above including Activating Transcription Factor 6 (ATF6), Bone Morphogenetic Proteins 2 (BMP2), C Motif Chemokine Receptor 6 (CCR6), F-Box And Leucine Rich Repeat Protein 21 (FBXL21), LEM Domain Nuclear Envelope Protein 2 (LEMD2), Pecanex 3 (PCNXL3), RAS Guanyl Releasing Protein 1(RASGRP1),and Regulator Of G Protein Signaling 1 (RGS1). The details of the homozygote model with p<0.05 and HWE≥ 0.05 are shown in Supplementary Table 5 . Based on the homozygote model, compared to healthy controls, the OR value of IgA nephropathy was greater than 1.5, or less than 0.5 as shown in Supplementary Figure 3 . Twelve variants of IL-1B under homozygote model are shown in Supplementary Figure 4 , of which rs1946518 has a similar effect as that suggested by the allele model in Korean and Chinese Han population that increase the susceptibility of IgAN.
Heterozygote Model
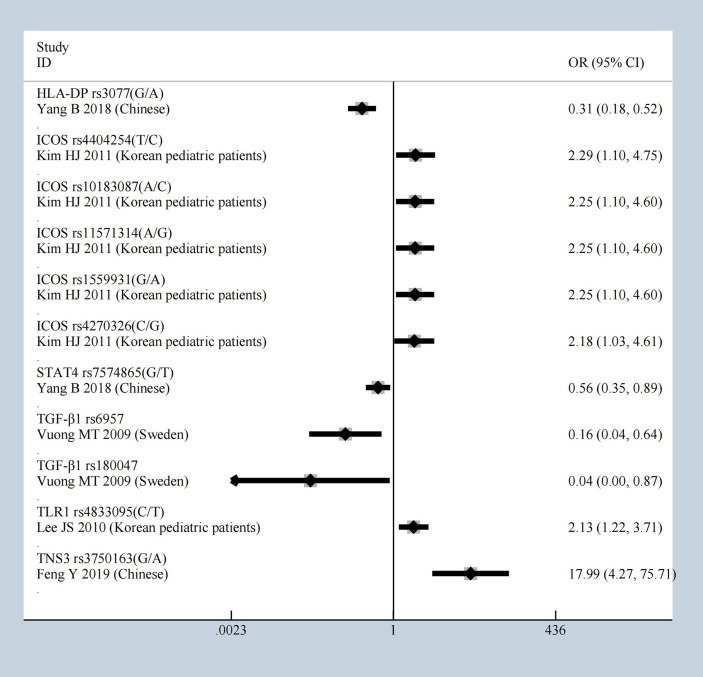
Under heterozygote model, compared to healthy controls, ICOS showed significant effect again with the mutations of rs4404254, rs10183087, rs11571314, rs1559931, rs4270326, and HLA-DP rs3077 [OR = 0.31 (95% CI 0.18–0.52)], STAT4 rs7574865 [OR = 0.56 (95% CI 0.35–0.89)], TGF‐β1 rs6957 [OR = 0.16 (95% CI 0.04–0.64)] and TGF‐β1 rs180047 [OR = 0.04 (95% CI 0–0.87)], TLR1 rs4833095 [OR = 2.13 (95% CI 1.22–3.71)], TNS3 rs3750163 [OR = 17.99 (95% CI 4.27–75.71)]; all had a significant effect which was identified as having an OR greater than 1.5 or less than 0.5, the results shown in Figure 5 . All results of the heterozygote model (p < 0.05 and HWE ≥ 0.05) were: CFB, C1GALT1, and CCR6, CD29, CTLA4, CXCL8, F-Box and Leucine Rich Repeat Protein 21 (FBXL21), Fc Fragment Of IgG Receptor IIb (FCGR2B), FCRLA, FCRLB, HLA-DP, ICOS, IFN-γ, IL-10, IL-1β, ITGAM, ITGAX, RGS1, STAT4, TGF‐β1, TLR1, TLR10, TNFSF13, and TNS3. General characteristics of each studies about molecules are described in Supplementary Table 6 .
Figure 5.
The most statistically significant SNPs under Heterozygote model.
Recessive Model and Cluster Analysis
All results of recessive model were described in Supplementary Table 7 . In order to find the SNPs which are stable and contribute to the predisposition of IgAN, we clustered the molecules with significant ORs under the six genetic models above, and the result was represented as a Venn diagram in Figure 6 . And five SNPs (IL-10 rs1800872, CXCL8 rs2227543, CXCL8 rs2227306, ITGAM rs4597342, and TGF‐β1 rs2241715) have a significant effect in dominant model, overdominant model, and heterozygote model. Among alleles, FCRLA rs1954174, IL1RN rs928940, IL1RN rs315951, LEMD2 rs751728, and VEGF 405C-G showed significance in homozygote and recessive models. Furthermore, HLA-DP rs3077 and STAT4 rs7574865 showed statistical significance in allele model, overdominant model, homozygote model, heterozygote model, and recessive model. In addition, TNFSF13 rs3803800 showed the most stable effect under five genetic models excepted the overdominant model; CFB rs549182 has a robust impact on susceptibility in all models except the homozygote one; there were two gene variants of CFB (rs549182 and rs4151657) that were significant in different patient groups with IgA nephropathy and we summarized it with ORG in Table 4 .
Figure 6.
The cluster analysis of the SNPs with significant ORs under the six genetic models.
Table 4.
Summary of the pooled ORs of two variants of CFB in patients with IgAN stage I or stage II.
| Allele model | Overdominant model | Dominant model | Heterozygote model | Homozygote model | Recessive model | ORG (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | PH | OR (95% CI) | PH | OR (95% CI) | PH | OR (95% CI) | PH | OR (95% CI) | PH | OR (95% CI) | PH | ||
| rs549182 | |||||||||||||
| IgAN Stage I | 1.33 (1.10,1.61) | 0.003 | 0.88 (0.66,0.99) | 0.040 | 1.30 (1.07,1.60) | 0.010 | 1.25 (1.02,1.53) | 0.032 | 4.03 (1.34,12.19) | 0.013 | 3.90 (1.29,11.77) | 0.016 | 1.31 (1.07,1.60) |
| IgAN stage II | 1.19 (1.02,1.40) | 0.031 | 1.0 (0.73,1.39) | 0.979 | 0.94 (0.69,1.29) | 0.716 | 0.99 (0.71,1.36) | 0.937 | 0.41 (0.11,1.59) | 0.197 | 0.41 (0.11,1.59) | 0.197 | 0.94 (0.68,1.28) |
| IgAN Stage I and stage II | 0.91 (0.68,1.22) | 0.513 | 0.86 (0.72,1.02) | 0.085 | 1.19 (1.00,1.41) | 0.050 | 1.17 (0.98,1.39) | 0.077 | 1.70 (0.80,3.61) | 0.167 | 1.66 (0.78,3.52) | 0.187 | 1.19 (1.01,1.41) |
| rs4151657 | |||||||||||||
| IgAN Stage I | 1.13 (1.01,1.27) | 0.031 | 0.80 (0.69,0.93) | 0.003 | 1.25 (1.07,1.44) | 0.004 | 1.28 (1.09,1.49) | 0.002 | 1.11 (0.86,1.44) | 0.417 | 0.99 (0.77,1.27) | 0.928 | 1.18 (1.04,1.35) |
| IgAN stage II | 1.21 (1.02,1.45) | 0.033 | 0.77 (0.61,0.97) | 0.029 | 1.35 (1.07,1.70) | 0.012 | 1.36 (1.06,1.73) | 0.014 | 1.30 (0.86,1.98) | 0.217 | 1.12 (0.75,1.67) | 0.584 | 1.28 (1.04,1.58) |
| IgAN Stage I and stage II | 1.15 (1.05,1.27) | 0.003 | 0.79 (0.70,0.90) | 0.000 | 1.27 (1.12,1.44) | 0.000 | 1.30 (1.14,1.48) | 0.000 | 1.16 (0.93,1.45) | 0.181 | 1.02 (0.83,1.26) | 0.839 | 1.21 (1.08,1.35) |
Genetic Association Studies
To date, eight GWASs have been performed that provide valuable information on the susceptibility of IgAN. Table 5 shows the information and significant molecules of the eight GWAS studies. Because none of them provided extractable data, we used their conclusions to prove our meta-analysis results. C1GALT1, HLA-DP, ITGAX, ITGAM, TNFSF13, and ST6GAL1 showed significant associations according to our meta-analysis results, further confirmed by GWAS studies. Recently, a large-scale genome-wide meta-analysis included four GWAS studies on people of Chinese and European descent to investigate genetic variants of IgAN susceptibility (34). This analysis revealed novel SNPs as susceptibility genes for IgAN: FCRL3 rs6427389, Dual Specificity Phosphatase 22 (DUSP22), interferon regulatory factor 4 (IRF4) rs6942325, and Peptidyl Arginine Deiminase 4 (PADI4) rs2240335.
Table 5.
The general characteristics and relevant molecules of genetic association studies.
| Number | Author | Year | PMID | Ethnicity | Case | Control | Molecule |
|---|---|---|---|---|---|---|---|
| 1 | John Feehally (28) | 2010 | 20595679 | British | 431 | 4,980 | HLA-A, HLA-B, HLA-C, DRB, DQA, DRB (9) |
| 2 | Xue-Qing Yu (29) | 2011 | 22197929 | Chinese | 4,137 | 7,734 | TNFSF13, DEFA, MHC, HLA-A, HLA-DR-DQ (10) |
| 3 | Krzysztof Kiryluk (10) | 2014 | 25305756 | European | 2,747 | 3,952 | VAV3, CARD9, ITGAM-ITGAX, HLA-DQ/DR, DEFA, TAP1/PSMB8, HLA-DP, CFHR3, TNFSF13, HORMAD2 (11) |
| East Asian | |||||||
| 4 | Krzysztof Kiryluk (30) | 2012 | 22737082 | Asian, European, African-American | 4,789 | 10,755 | HLA-DQB1/DRB1, PSMB9/TAP1, DPA1/DPB2, CFHR3/R1, HORMAD2 (12) |
| 5 | Ali G Gharavi (31) | 2011 | 21399633 | Chinese | 3,144 | 2,852 | CFHR1, CFHR3, HLA-DQB1, DQA1, DRB1, PSMB8, TAP2, TAP1, PSMB9, HLA-DPA1, -DPB1, -DPB2 (13) |
| European | |||||||
| 6 | Sanae Saka (32) | 2015 | 26202575 | Japanese | 915 | 481 | HLA-DRA, HLA-DRB1, HLA-DQA1, HLA-DQB1, TSPAN8 and PTPRR (22) |
| 7 | Yan-Na Wang (33) | 2021 | 33593824 | Chinese | 2,352 | 2,632 | GALNT12, C1GALT1 (34) |
| 8 | Ming Li (35) | 2015 | 26028593 | Chinese | 8,313 | 19,680 | ST6GAL1, ACCS, ODF1-KLF10, ITGAX-ITGAM, DEFA (20) |
Ethnicity Differences Analysis
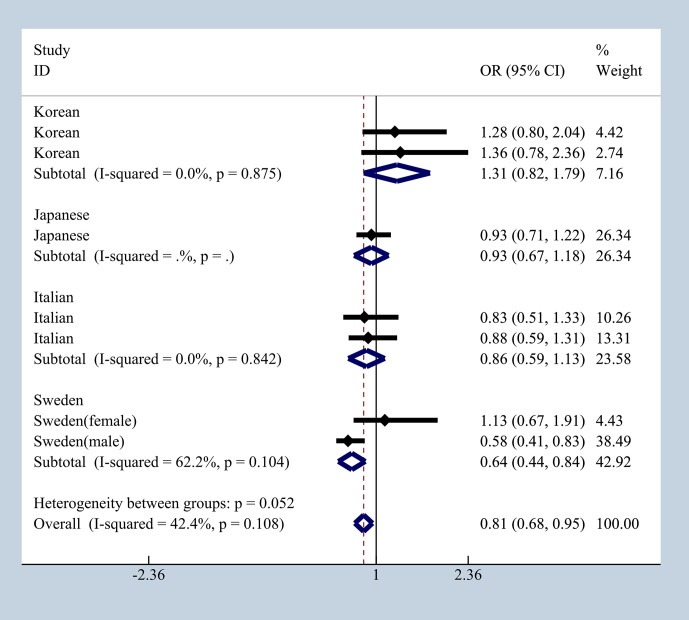
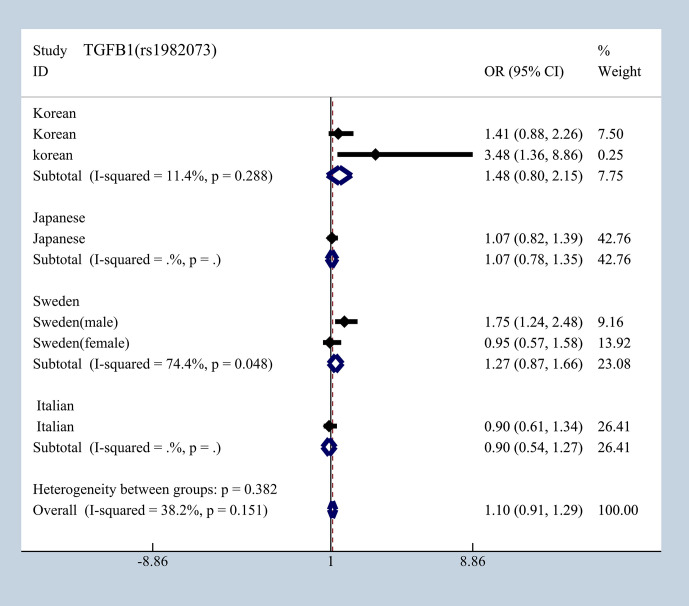
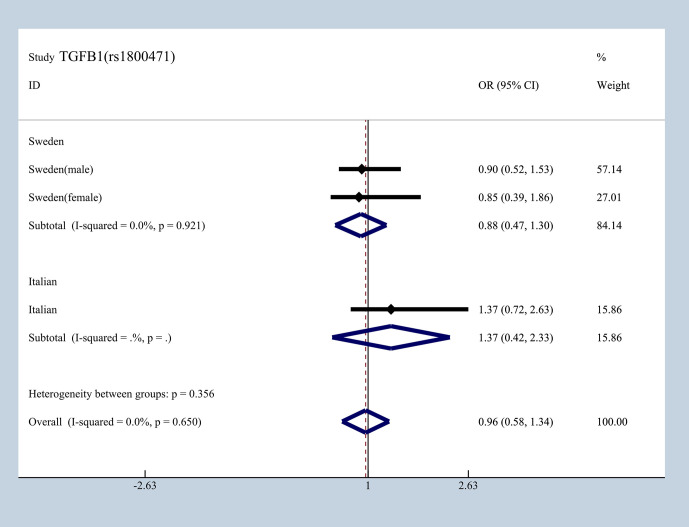
To elucidate the effect of different ethnicities on SNPs of immunologic process in IgAN, we investigated all available data to conduct a meta-analysis based on ORG and 95% CI. We summarized published studies related to rs1800469, rs1982073, and rs1800471 in TGFB1. Excluding those that did not meet our selection criteria, we finally enrolled five studies involving Korean, Japanese, Italian, and Swedish populations (19, 20, 36–39). In subgroup analyses by ethnicity and gender that we found in Japanese, Italian, and Sweden male group, we found that rs1800469 in TGFB1 has a moderate protective effect, whereas contradicting results was observed in Korean and Swedish female group with overall OR = 1.10, 95% CI 0.91–1.29, I2 = 38.2% ( Figure 7 ). The mutation in SNP 1982073 in TGFB1 presented a slightly increased susceptibility of IgAN among Korean, Japanese, and Swedish male group and showed a decreased possibility risk of disease ( Figure 8 ). The mutation in SNP rs1800471 has a similar moderate protective effect in Swedish population, both in males and females but showed an increased occurrence in Italian group ( Figure 9 ) (20, 38).
Figure 7.
The different effects in four different populations of rs1800469 in TGFB1.
Figure 8.
The different effects in four different populations of rs1982073 in TGFB1.
Figure 9.
Different effects between Sweden and Italian in rs1800471of TGFB1.
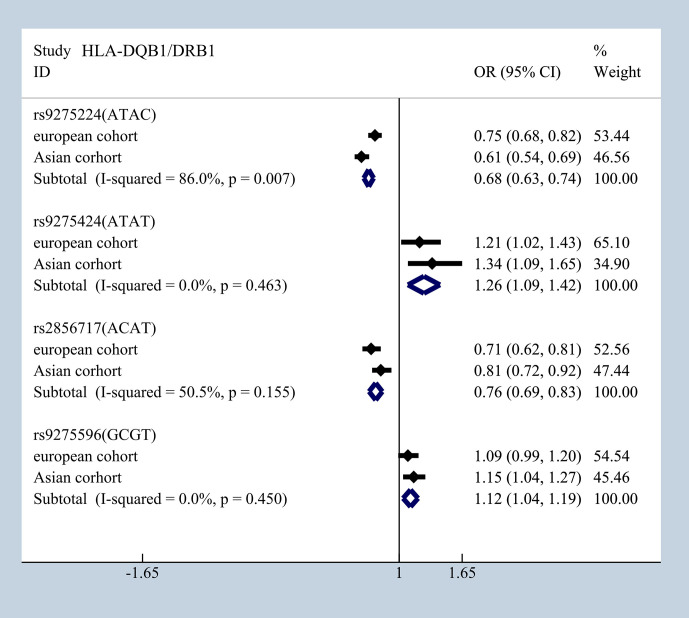
Furthermore, based on available data, interleukin family proteins were investigated in Korean and Chinese Han population that had an IL-18 (rs1946518) mutation; it showed a significant protective effect with overall ORG = 0.56, 95% CI (0.39–0.73) (19, 26) ( Supplementary Figure 5 ). However, the contradictory result was observed in Chinese Han population and Korean pediatric patients in IL-1B (rs1143627) (40, 41) ( Supplementary Figure 6 ). Chinese population and Korean pediatric patients shared the same predisposition of rs4833095 and rs5743557 in TLR1which presented in ( Supplementary Figure 7 ) (42, 43). Besides that, mutations of HLA-DQB1/DRB1, rs9275224 [ORG = 0.68 95% CI (0.63–0.74)], rs9275424 [ORG = 1.26 95% CI (1.09–1.42)]; rs2856717 [ORG = 0.76 95% CI (0.69–0.83)], and rs9275596 [ORG = 1.12 95% CI (1.04–1.19)] had consistent significant results in European and Asian cohorts ( Figure 10 ).
Figure 10.
Consistent results in European and Asian cohorts in mutations of HLA-DQB1/DRB1.
Sensitivity Analysis
In the sensitivity analysis, recalculated ORs after removing any single eligible study showed that no material alterations were detected implying that our results are reliable and robust ( Supplementary Figures 8–10 ).
Publication Bias
Potential publication bias among the included studies was assessed through the visual inspection of Begg’s funnel plot, shown in Figure 11 , that the funnel plot is symmetrical. The P-value of Egger’s test was 0.037, and the regression line crossed the zero line, which indicated the absence of significant publication bias ( Supplementary Figure 11 ). Additionally, most of enrolled studies got high scores of NOS quality assessment and details of it are shown in Supplementary Table 8 .
Figure 11.
Begg's funnel plot of publication bias.
Discussion
We analyzed 45 studies, 9 of which involved Caucasians, and 37 studies recruited Asians (25 studies in Chinese, 1 study in Japanese, and 11 studies in Korean). These studies were consistent with the global prevalence of IgAN. Although the prevalence in some Western countries has progressively increased, they still have a lower prevalence of IgAN, and Asian populations such as the Chinese, Japanese, and Korean are more vulnerable to IgAN (44).
The pathogenetic progress of IgAN are mainly characterized by four biochemical hits: (1) aberrant glycosylation of IgA1. (2) anti-glycan/glycopeptides antibodies against unnormal IgA1 synthesized (3) aberrant glycosylation of IgA1 combined with anti- glycan/glycopeptides antibodies so that immune complexes (ICS) are formed, (4) of which these ICS accumulated in the glomerular mesangium, induce cell proliferation, and secrete immune molecular to aggravate renal injuries (9, 45).
To avoid generating aberrant IgA1 with galactose-deficient O-glycans, it is necessary to maintain the normal activity and expression of core 1 β1,3galactosyltransferase 1 (encoded by C1GALT1), αNacetylgalactosaminide α2,6sialyltransferase 2 (encoded by ST6GALNAC2), and core 1 β1,3galactosyltransferase-specific chaperone 1 (encoded by C1GALT1C1) (46). Two studies enrolled Chinese Han and Chinese Uyghur populations both of which revealed that rs3840858 in ST6GALNAC2 is associated with susceptibility to IgAN; this was supported by our results. However, a larger patient size and different ethnic populations are required to further bolster the results. Moreover, the mutations of these three molecules are not only associated with the risk of developing IgAN but interactively contribute to the predisposition and severity of it (14, 25). According to a recent Chinese genetic association study and interactive protein network analysis, a possible interaction between C1GALT1 and GANT12 through O-glycan processing or the mucin-type O-glycan biosynthesis pathway exists, which is associated with susceptibility to IgAN (33).
Upon investigation of 20 immune pathways, we obtained results consistent with previous research that several significant SNPs were involved in the complement pathway especially the alternative pathway. CFB plays a pivotal role in the production of C3; it combines with C3b to form C3 convertase in the complement response (47). Products of CFH and CFH-related genes co-modulate the activation of alternative complement pathway (48). The SNP rs4151657 in CFB showed stable association with the risk of IgAN in Chinese Han population (under allele dominant overdominant and heterozygote models). It was also associated with higher serum CFB and decreased serum C3 level that indicated poor renal outcomes (49). A single deletion in both CFHR1 and CFHR3 confers a 30% reduction in the risk of IgA nephropathy, and a meta-analysis with risk-score model including Asian, European, and African population evaluated five susceptibility loci of three MHC, one CHF and one HORMAD2. These five loci moderately increased the risk of disease but together explained 4–5% of the predisposition after being examined (30). The region of IgG (FcγRs) and the complement system are engaged in renal injury caused by autoantibody/immune complex and IC clearance (50). C3 is the most common co-deposited molecule with IgA in up to 90% of patients, followed by properdin and complement factor H. Genes encoding low-affinity FcγRs FCGR2A, FCGR3A, FCGR2C, FCGR3B, and FCGR2B (rs12118043, FCRLA rs1954174, FCRLB rs4657093) showed a significant association with the predisposition of IgAN, but the linkage disequilibrium and conditional analysis suggest that these SNPs have a weak association with each other (22).
TGF-β1 encoded by TGFB1 is a multifunctional cytokine that contributes to cell proliferation, differentiation, expansion of mesangial matrix, the development of fibrosis, and various immunologic processes (51). We conducted a meta-analysis of rs1800469, rs1982073, and rs1800471 in TGFB1 among Korean, Japanese, Italian, and Swedish populations with ORG. The result showed that genotype shows a different effect regarding the susceptibility to IgAN according to ethnicity and gender. It is also noteworthy to mention that rs1800469, rs1982073, and rs1800471 shared high LD in the recombination block. Following haplotype analysis, we found that having TAC haplotype significantly increased the risk of IgAn by more than two times (39). Considering SNP effect on prognosis, no significant difference as observed in different genotype as contributing to either progressive or stable IgAN, but patients with homozygote mutation producing higher TGF-β1 levels show poorer survival due to renal failures (36).
Toll-like receptors (TLRs) are a family of innate immune receptors expressed in the membrane of leukocytes, renal tubular epithelial cells, and various cells which induce inflammatory cytokine expression through intracellular signaling pathway. The agonized TLRs can directly injure kidney and also overproduce antibodies via B lymphocytes (52). TLR1 is expressed in tubular epithelial cells and enhances the activation level of NFκB. Our meta-analysis found that rs4833095 and rs5743557 in TLR1 increases the risk of IgAN both in Chinese and Korean pediatric patients group; upon haplotype analysis, we found that Trs4833095 and Trs5743557 haplotype were associated with IgAN. In European population, the major allele of rs4833095 is T but in African American, sub-Saharan African, and Asian group, the major allele is C allele which corresponded with the prevalence of IgAN. The SNP rs4833095 was associated with the severity of pathology according to Lee’s grades (42).
Interleukin-1 receptors shared the same pathway with TLRs to produce cytokines. IL-1Ra is an anti-inflammatory contributor, while IL-1β as pro-inflammatory, which balances the immune response. In patients with IgAN with a lower IL-1Ra/IL-1βr ratio, more severe pathological changes were observed (53). IL-1B (rs1143627) in Korean pediatric patients show moderate protective effect but the Chinese Han group show a risk of IgAN. The reason for the difference may be that the pediatric patients were prone to have milder lesions in early stage of the disease. Haplotype analysis found CAT showed significant association with the disease (41).
IL-6, a common cytokine, elevated the activity of the GalNAc-specific sialyltransferase encoded by the ST6GALNAC2 gene and decreased the activity of core 1 β1,3galactosyltransferase 1 encoded by C1GALT1, leading to the accentuation of aberrant glycan-deficient IgA (54).
IL-18 is a cytokine that stimulates the production of several immune molecules such as tumor necrosis factor-α, IFN-γ, and IL-2 to regulate autoimmune conditions. In our meta-analysis which showed that rs1946518 in IL-18 has a robust effect in both Chinese Han and Korean populations. Although the IL-18 level hinted at development of glomerulonephritis and genotype of IL-18 rs1946518CC was related with a higher level of IL-18 mRNA, the current study revealed that rs1946518 may not be associated with clinical symptoms and pathology grade in IgAN (26, 55).
In the results of meta-analysis, in the T-cell co-stimulatory pathway, CTLA4 and ICOS have variant mutations and most of them presented different degree statistical meaning under different genetic models. CTLA4 is a negative regulator in CD28 signaling and plays a key role in the initiation and termination of immune responses, whereas ICOS regulates the activation and tolerance of T cells. The G allele in CTLA4 rs231779 significantly increased the risk and was related to the prevalence of proteinuria (15, 56). As for the correlation between SNPs and clinical symptoms, HLA-DQB1/DRB1 had a high association with a minor allele in rs2856717 that cannot be covered by adjustment of LD structure, protective haplotype (ACAT) and at-risk haplotypes (ATAT, GCGT). Additionally, rs12537 was related to severe proteinuria and rs2523946 indicated a higher incidence of gross hematuria (30). Besides the co-stimulating pathway of T cells, TNFSF13 has a key role in T cell-dependent antibody responses by encode proliferation-inducing ligand (APRIL) which is a powerful cytokine stimulated by Toll-like receptor 9 and other immune-associated factors (57). The minor allele A of TNFSF13(rs3803800) was observed to have a significant association with IgAN under additive and recessive model and was also associated with several clinical phenotypes such as severe proteinuria, low eGFR, and higher occurrence of mesangial hypercellularity (58).
There are several advantages of this meta-analysis. Since genetic comparisons are not independent, the true underlying genetic model is unknown, and the genetic contrasts are defined by merging information of the genotype distribution, we tested six kinds of genetic models to provide as much information as possible to reveal the SNPs in immunologic proteins with susceptibility to IgAN, which decreased the loss of information resolution. Furthermore, the subgroup analysis based on different ethnicities were performed with ORG. Genetic variants with a stable and robust risk of susceptibility to IgAN were found through cluster analysis. In addition, we provide a new perspective that combines genetic variability with the inflammatory effects of IgAN and attempts to further explore disease susceptibility and pathological mechanisms.
Our meta-analysis also has some limitations. IgAN is a complex disease, and SNPs without consideration of epistatic genes or gene interactions have reliable and conclusive inferences. We enrolled studies that only included healthy individuals as controls and excluded cohorts of secondary IgAN and comorbidity with other nephropathy diseases. We did not analyze the genetic risk of the subtypes of IgAN, and so we may conduct systematic review and meta-analysis in the future. Considering the large disparities of prevalence in ethnicity, most studies of the same SNP in different ethnicities did not provide sufficient data which limited the subgroup analysis performed. Several of our conclusions are based on individual studies and need to be interpreted with caution.
Conclusion
The overall meta-analysis results revealed most supportive SNPs in immunology or related pathways significantly associated with susceptibility to IgAN, and six of them were proved by genetic association studies. Furthermore, the subgroup analysis based on ethnicities revealed detailed information. In a polygenic complex disorder such as IgAN, the association of individual polymorphisms in genes may be limited compared with combinations of specific genotypes. However, multiple genetic contrasts may produce diverse results and positive associations resulted from pooling a small number of studies; therefore, these results must be interpreted with caution.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Author Contributions
Conceptualization: XD, YM, ZM, and HZ. Methodology: XD. Validation: XD, YM, and ZM. Formal analysis: XD. Investigation: XD and YM. Resources: XD and HZ. Data curation: XD, YM, and HZ. Writing—original draft preparation: XD. Writing—review and editing: XD, YM, and HZ. Supervision: YY and HZ. Project administration: YY and HZ. Funding acquisition: HZ. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by The National Natural Science Foundation of China (Nos. 61971441) and the National Key R&D Program of China (Nos. 2016YFC1305500).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.683913/full#supplementary-material
References
- 1. Kiryluk K, Novak J, Gharavi AG. Pathogenesis of Immunoglobulin A Nephropathy: Recent Insight From Genetic Studies. Annu Rev Med (2013) 64:339–56. 10.1146/annurev-med-041811-142014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Infante B, Rossini M, Di Lorenzo A, Coviello N, Giuseppe C, Gesualdo L, et al. Recurrence of Immunoglobulin A Nephropathy After Kidney Transplantation: A Narrative Review of the Incidence, Risk Factors, Pathophysiology and Management of Immunosuppressive Therapy. Clin Kidney J (2020) 13(5):758–67. 10.1093/ckj/sfaa060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jarrick S, Lundberg S, Welander A, Carrero JJ, Höijer J, Bottai M, et al. Mortality in IgA Nephropathy: A Nationwide Population-Based Cohort Stud. J Am Soc Nephrol (2019) 30(5):866–76. 10.1681/asn.2018101017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Donadio JV, Grande JP. IgA Nephropathy. N Engl J Med (2002) 347(10):738–48. 10.1056/NEJMra020109 [DOI] [PubMed] [Google Scholar]
- 5. Shi M, Yu S, Ouyang Y, Jin Y, Chen Z, Wei W, et al. Increased Lifetime Risk of ESRD in Familial IgA Nephropath. Kidney Int Rep (2021) 6(1):91–100. 10.1016/j.ekir.2020.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lai KN, Tang SC, Schena FP, Novak J, Tomino Y, Fogo B, et al. IgA Nephropathy. Nat Rev Dis Primers (2016) 2:16001. 10.1038/nrdp.2016.1 [DOI] [PubMed] [Google Scholar]
- 7. Wyatt RJ, Julian BA. IgA Nephropathy. N Engl J Med (2013) 368(25):2402–14. 10.1056/NEJMra1206793 [DOI] [PubMed] [Google Scholar]
- 8. Roberts I. Pathology of IgA Nephropathy. Nat Rev Nephrol (2014) 10(8):445–54. 10.1038/nrneph.2014.92 [DOI] [PubMed] [Google Scholar]
- 9. Novak J, Raskova Kafkova L, Suzuki H, Tomana M, Matousovic K, Brown R, et al. IgA1 Immune Complexes From Pediatric Patients With IgA Nephropathy Activate Cultured Human Mesangial Cells. Nephrol Dial Transplant (2011) 26(11):3451–7. 10.1093/ndt/gfr448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, et al. Discovery of New Risk Loci for IgA Nephropathy Implicates Genes Involved in Immunity Against Intestinal Pathogens. Nat Genet (2014) 46(11):1187–96. 10.1038/ng.3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zintzaras E. The Generalized Odds Ratio as a Measure of Genetic Risk Effect in the Analysis and Meta-Analysis of Association Studies. Stat Appl Genet Mol Biol (2010) 9:Article21. 10.2202/1544-6115.1542 [DOI] [PubMed] [Google Scholar]
- 12. Lewis CM, Knight J. Introduction to Genetic Association Studies. Cold Spring Harb Protoc (2012) 2012(3):297–306. 10.1101/pdb.top068163 [DOI] [PubMed] [Google Scholar]
- 13. Minelli C, Thompson JR, Abrams KR, Thakkinstian A, Attia J. The Choice of a Genetic Model in the Meta-Analysis of Molecular Association Studies. Int J Epidemiol (2005) 34(6):1319–28. 10.1093/ije/dyi169 [DOI] [PubMed] [Google Scholar]
- 14. Li GS, Zhu L, Zhang H, Lv JC, Ding JX, Zhao MH, et al. Variants of the ST6GALNAC2 Promoter Influence Transcriptional Activity and Contribute to Genetic Susceptibility to IgA Nephropathy. Hum Mutat (2007) 28(10):950–7. 10.1002/humu.20543 [DOI] [PubMed] [Google Scholar]
- 15. Kim HJ, Chung JH, Kang S, Kim SK, Cho BS, Kim SD, et al. Association of CTLA4, CD28 and ICOS Gene Polymorphisms With Clinicopathologic Characteristics of Childhood IgA Nephropathy in Korean Population. J Genet (2011) 90(1):151–5. 10.1007/s12041-011-0042-5 [DOI] [PubMed] [Google Scholar]
- 16. Gao J, Wei L, Fu R, Wei J, Niu D, Wang L, et al. Association of Interleukin-10 Polymorphisms (Rs1800872, Rs1800871, and Rs1800896) With Predisposition to IgA Nephropathy in a Chinese Han Population: A Case-Control Stud. Kidney Blood Press Res (2017) 42(1):89–98. 10.1159/000471899 [DOI] [PubMed] [Google Scholar]
- 17. Yang B, Zhang J, Liu X, Huang Z, Su Z, Liao Y, et al. Genetic Polymorphisms in HLA-DP and STAT4 are Associated With IgA Nephropathy in a Southwest Chinese Population. Oncotarget (2018) 9(6):7066–74. 10.18632/oncotarget.23829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao J, Wei L, Liu X, Wang L, Niu D, Jin T, et al. Association Between IFN-γ Gene Polymorphisms and IgA Nephropathy in a Chinese Han Population. Kidney Blood Press Res (2017) 42(1):136–44. 10.1159/000473889 [DOI] [PubMed] [Google Scholar]
- 19. Jung HY, Cho JH, Lim JH, Yu CH, Choi JY, Yoon SH, et al. Impact of Gene Polymorphisms of Interleukin-18, Transforming Growth Factor-β, and Vascular Endothelial Growth Factor on Development of IgA Nephropathy and Thin Glomerular Basement Membrane Disease. Kidney Res Clin Pract (2012) 31(4):234–41. 10.1016/j.krcp.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vuong MT, Lundberg S, Gunnarsson I, Wramner L, Seddighzadeh M, Hahn-Zoric M, et al. Genetic Variation in the Transforming Growth Factor-Beta1 Gene is Associated With Susceptibility to IgA Nephropathy. Nephrol Dial Transplant (2009) 24(10):3061–7. 10.1093/ndt/gfp079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng Y, Su Y, Ma C, Jing Z, Yang X, Zhang D, et al. 3'UTR Variants of TNS3, PHLDB1, NTN4, and GNG2 Genes are Associated With IgA Nephropathy Risk in Chinese Han Population. Int Immunopharmacol (2019) 71:295–300. 10.1016/j.intimp.2019.03.041 [DOI] [PubMed] [Google Scholar]
- 22. Zhou XJ, Cheng FJ, Qi YY, Zhao YF, Hou P, Zhu L, et al. FCGR2B and FCRLB Gene Polymorphisms Associated With IgA Nephropathy. PloS One (2013) 8(4):e61208. 10.1371/journal.pone.0061208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu XQ, Paterson AD, He N, St George-Hyslop P, Rauta V, Gronhagen-Riska C, et al. IL5RA and TNFRSF6B Gene Variants are Associated With Sporadic IgA Nephropathy. J Am Soc Nephrol (2008) 19(5):1025–33. 10.1681/asn.2007091013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu D, Zhong Z, Shi D, Peng Y, Li B, Wang D, et al. ST6GAL1 Polymorphisms Influence Susceptibility and Progression of IgA Nephropathy in a Chinese Han Population. Immunobiology (2020) 225(4):151973. 10.1016/j.imbio.2020.151973 [DOI] [PubMed] [Google Scholar]
- 25. Lu C, Li WL, Ma YR. Study of Correlation Between Polymorphism of ST6GALNAC2 and Susceptibility to IgA Nephropathy. Exp Ther Med (2015) 9(6):2127–32. 10.3892/etm.2015.2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang B, Feng W, Li Y, Shi Y, Cai B, Liao Y, et al. Interleukin 18 -607 A/C Gene Polymorphism is Associated With Susceptibility to IgA Nephropathy in a Chinese Han Populatio. Appl Immunohistochem Mol Morphol (2017) 25(10):725–30. 10.1097/pai.0000000000000364 [DOI] [PubMed] [Google Scholar]
- 27. Hahn WH, Cho BS, Kim SD, Kim SK, Kang S. Interleukin-1 Cluster Gene Polymorphisms in Childhood IgA Nephropathy. Pediatr Nephrol (2009) 24(7):1329–36. 10.1007/s00467-009-1146-5 [DOI] [PubMed] [Google Scholar]
- 28. Feehally J, Farrall M, Boland A, Gale DP, Gut I, Heath S, et al. HLA has Strongest Association With IgA Nephropathy in Genome-Wide Analysis. J Am Soc Nephrol (2010) 21(10):1791–7. 10.1681/asn.2010010076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ, et al. A Genome-Wide Association Study in Han Chinese Identifies Multiple Susceptibility Loci for IgA Nephropathy. Nat Genet (2011) 44(2):178–82. 10.1038/ng.1047 [DOI] [PubMed] [Google Scholar]
- 30. Kiryluk K, Li Y, Sanna-Cherchi S, Rohanizadegan M, Suzuki H, Eitner F, et al. Geographic Differences in Genetic Susceptibility to IgA Nephropathy: GWAS Replication Study and Geospatial Risk Analysis. PloS Genet (2012) 8(6):e1002765. 10.1371/journal.pgen.1002765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, et al. Genome-Wide Association Study Identifies Susceptibility Loci for IgA Nephropathy. Nat Genet (2011) 43(4):321–7. 10.1038/ng.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saka S, Hirawa N, Oka A, Yatsu K, Hirukawa T, Yamamoto R, et al. Genome-Wide Association Study of IgA Nephropathy Using 23 465 Microsatellite Markers in a Japanese Population. J Hum Genet (2015) 60(10):573–80. 10.1038/jhg.2015.88 [DOI] [PubMed] [Google Scholar]
- 33. Wang YN, Zhou XJ, Chen P, Yu GZ, Zhang X, Hou P, et al. Interaction Between G ALNT12 and C1GALT1 Associates With Galactose-Deficient IgA1 and IgA Nephropathy. J Am Soc Nephrol (2021) 32(3):545–52. 10.1681/asn.2020060823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li M, Wang L, Shi DC, Foo JN, Zhong Z, Khor CC, et al. Genome-Wide Meta-Analysis Identifies Three Novel Susceptibility Loci and Reveals Ethnic Heterogeneity of Genetic Susceptibility for IgA Nephropathy. J Am Soc Nephrol (2020) 31(12):2949–63. 10.1681/asn.2019080799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li M, Foo JN, Wang JQ, Low HQ, Tang XQ, Toh KY, et al. Identification of New Susceptibility Loci for IgA Nephropathy in Han Chines. Nat Commun (2015) 6:7270. 10.1038/ncomms8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lim CS, Kim YS, Chae DW, Ahn C, Han JS, Kim S, et al. Association of C-509T and T869C Polymorphisms of Transforming Growth Factor-Beta1 Gene With Susceptibility to and Progression of IgA Nephropathy. Clin Nephrol (2005) 63(2):61–7. 10.5414/cnp63061 [DOI] [PubMed] [Google Scholar]
- 37. Sato F, Narita I, Goto S, Kondo D, Saito N, Ajiro J, et al. Transforming Growth Factor-Beta1 Gene Polymorphism Modifies the Histological and Clinical Manifestations in Japanese Patients With IgA Nephropathy. Tissue Antigens (2004) 64(1):35–42. 10.1111/j.1399-0039.2004.00256.x [DOI] [PubMed] [Google Scholar]
- 38. Brezzi B, Del Prete D, Lupo A, Magistroni R, Gomez-Lira M, Bernich P, et al. Primary IgA Nephropathy is More Severe in TGF-Beta1 High Secretor Patients. J Nephrol (2009) 22(6):747–59. [PubMed] [Google Scholar]
- 39. Carturan S, Roccatello D, Menegatti E, Di Simone D, Davit A, Piazza A, et al. Association Between Transforming Growth Factor Beta1 Gene Polymorphisms and IgA Nephropathy. J Nephrol (2004) 17(6):786–93. [PubMed] [Google Scholar]
- 40. Zhang D, Xie M, Yang X, Zhang Y, Su Y, Wang Y, et al. Determination of IL-1B (rs16944) and IL-6 (rs1800796) Genetic Polymorphisms in IgA Nephropathy in a Northwest Chinese Han Population. Oncotarget (2017) 8(42):71750–8. 10.18632/oncotarget.17603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hahn WH, Cho BS, Kim SD, Kim SK, Kang S. Interleukin-1 Cluster Gene Polymorphisms in Childhood IgA Nephropathy. Pediatr Nephrol (2009) 24(7):1329–36. 10.1007/s00467-009-1146-5 [DOI] [PubMed] [Google Scholar]
- 42. Lee JS, Park HK, Suh JS, Hahn WH, Kang SW, Park HJ, et al. Toll-Like Receptor 1 Gene Polymorphisms in Childhood IgA Nephropathy: A Case-Control Study in the Korean Population. Int J Immunogenet (2011) 38(2):133–8. 10.1111/j.1744-313X.2010.00978.x [DOI] [PubMed] [Google Scholar]
- 43. Gao J, Wei L, Wei J, Yao G, Wang L, Wang M, et al. TLR1 Polymorphism Rs4833095 as a Risk Factor for IgA Nephropathy in a Chinese Han Population: A Case-Control Study. Oncotarget (2016) 7(50):83031–9. 10.18632/oncotarget.12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schena FP, Nistor I. Epidemiology of IgA Nephropathy: A Global Perspective. Semin Nephrol (2018) 38(5):435–42. 10.1016/j.semnephrol.2018.05.013 [DOI] [PubMed] [Google Scholar]
- 45. Suzuki H, Kiryluk K, Novak J, Moldoveanu Z, Herr AB, Renfrow MB, et al. The Pathophysiology of IgA Nephropathy. J Am Soc Nephrol (2011) 22(10):1795–803. 10.1681/asn.2011050464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hou L, Lin HH, Wu L, Luo XP. Clinical Characteristics of Wolfram Syndrome. Zhongguo Dang Dai Er Ke Za Zhi (2009) 11(2):113–5. [PubMed] [Google Scholar]
- 47. Gros P, Milder FJ, Janssen BJ. Complement Driven by Conformational Changes. Nat Rev Immunol (2008) 8(1):48–58. 10.1038/nri2231 [DOI] [PubMed] [Google Scholar]
- 48. Shi D, Zhong Z, Wang M, Cai L, Fu D, Peng Y, et al. Identification of Susceptibility Locus Shared by IgA Nephropathy and Inflammatory Bowel Disease in a Chinese Han Population. J Hum Genet (2020) 65(3):241–9. 10.1038/s10038-019-0699-9 [DOI] [PubMed] [Google Scholar]
- 49. Pan M, Zhang J, Li Z, Jin L, Zheng Y, Zhou Z, et al. Increased C4 and Decreased C3 Levels are Associated With a Poor Prognosis in Patients With Immunoglobulin A Nephropathy: A Retrospective Study. BMC Nephrol (2017) 18(1):231. 10.1186/s12882-017-0658-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jancar S, Sánchez Crespo M. Immune Complex-Mediated Tissue Injury: A Multistep Paradigm. Trends Immunol (2005) 26(1):48–55. 10.1016/j.it.2004.11.007 [DOI] [PubMed] [Google Scholar]
- 51. Park SR, Lee JH, Kim PH. Smad3 and Smad4 Mediate Transforming Growth Factor-Beta1-Induced IgA Expression in Murine B Lymphocytes. Eur J Immunol (2001) 31(6):1706–15. [DOI] [PubMed] [Google Scholar]
- 52. Smith KD. Toll-Like Receptors in Kidney Disease. Curr Opin Nephrol Hypertens (2009) 18(3):189–96. 10.1097/MNH.0b013e32832a1d5f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rauta V, Teppo AM, Törnroth T, Honkanen E, Grönhagen-Riska C. Lower Urinary-Interleukin-1 Receptor-Antagonist Excretion in IgA Nephropathy Than in Henoch-Schönlein Nephritis. Nephrol Dial Transplant (2003) 18(9):1785–91. 10.1093/ndt/gfg234 [DOI] [PubMed] [Google Scholar]
- 54. Suzuki H, Raska M, Yamada K, Moldoveanu Z, Julian BA, Wyatt RJ, et al. Cytokines Alter IgA1 O-Glycosylation by Dysregulating C1GalT1 and ST6GalNAc-II Enzymes. J Biol Chem (2014) 289(8):5330–9. 10.1074/jbc.M113.512277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Giedraitis V, He B, Huang WX, Hillert J. Cloning and Mutation Analysis of the Human IL-18 Promoter: A Possible Role of Polymorphisms in Expression Regulation. J Neuroimmunol (2001) 112(1-2):146–52. 10.1016/s0165-5728(00)00407-0 [DOI] [PubMed] [Google Scholar]
- 56. Jacob M, Ohl K, Goodarzi T, Harendza S, Eggermann T, Fitzner C, et al. CTLA-4 Polymorphisms in Patients With IgA Nephropathy Correlate With Proteinuria. Kidney Blood Press Res (2018) 43(2):360–6. 10.1159/000488069 [DOI] [PubMed] [Google Scholar]
- 57. He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, et al. Intestinal Bacteria Trigger T Cell-Independent Immunoglobulin A(2) Class Switching by Inducing Epithelial-Cell Secretion of the Cytokine APRIL. Immunity (2007) 26(6):812–26. 10.1016/j.immuni.2007.04.014 [DOI] [PubMed] [Google Scholar]
- 58. Zhong Z, Feng SZ, Xu RC, Li ZJ, Huang FX, Yin PR, et al. Association of TNFSF13 Polymorphisms With IgA Nephropathy in a Chinese Han Population. J Gene Med (2017) 19(6-7). 10.1002/jgm.2966 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.