Following myocardial infarction, tissue ischemia leads to activation of hypoxia-inducible factors. DeBerge et al. demonstrate that HIF-1α and HIF-2α activation in myeloid cells antagonizes cardiac repair pathways through cleavage of cardioprotective MerTK and suppression of anti-inflammatory mitochondrial metabolism, respectively.
Abstract
Hypoxia-inducible factors (HIFs) are activated in parenchymal cells in response to low oxygen and as such have been proposed as therapeutic targets during hypoxic insult, including myocardial infarction (MI). HIFs are also activated within macrophages, which orchestrate the tissue repair response. Although isoform-specific therapeutics are in development for cardiac ischemic injury, surprisingly, the unique role of myeloid HIFs, and particularly HIF-2α, is unknown. Using a murine model of myocardial infarction and mice with conditional genetic loss and gain of function, we uncovered unique proinflammatory roles for myeloid cell expression of HIF-1α and HIF-2α during MI. We found that HIF-2α suppressed anti-inflammatory macrophage mitochondrial metabolism, while HIF-1α promoted cleavage of cardioprotective MerTK through glycolytic reprogramming of macrophages. Unexpectedly, combinatorial loss of both myeloid HIF-1α and HIF-2α was catastrophic and led to macrophage necroptosis, impaired fibrogenesis, and cardiac rupture. These findings support a strategy for selective inhibition of macrophage HIF isoforms and promotion of anti-inflammatory mitochondrial metabolism during ischemic tissue repair.
Graphical Abstract
Introduction
During myocardial infarction (MI), reduced tissue perfusion and depleted metabolite availability lead to severe tissue ischemia. Cellular responses to low oxygen tension and nutrient deprivation lead to the stabilization of hypoxia-inducible factors (HIFs), a family of transcription factors that includes the two main isoforms, HIF-1α and HIF-2α (Ivan et al., 2001; Maxwell et al., 1999; Palazon et al., 2014; Semenza, 2012; Walmsley et al., 2011). HIF activation in cardiac parenchymal cells is thought to activate prosurvival pathways, with previous studies describing cardioprotective roles for HIF-1α (Kido et al., 2005) and HIF-2α (Koeppen et al., 2018) in cardiomyocytes after MI. In addition, hypoxic pathway targeting has improved tissue salvage in clinical trials of select MI patients (Bøtker et al., 2010); however, current strategies have failed to improve clinical outcomes (Eitel et al., 2015). The latter is consistent with the premise that HIF isoforms enact nonoverlapping functions, including in disparate cell types. Thus, a deeper understanding of the cell- and isoform-specific functions of critically important HIFs holds the potential to improve therapeutic strategies in MI and other scenarios of hypoxic injury.
Relative to most studies of parenchymal HIFs in ischemic injury, the functional significance of HIF isoforms in immune populations responsible for cardiac tissue repair is less understood. Ischemic cardiac injury mobilizes macrophages, both tissue resident (Dick et al., 2019) and recruited monocyte derived (Hilgendorf et al., 2014), which are central protagonists of tissue repair in hypoxic tissue. In both inflammation and limiting oxygen tensions, HIF-1α promotes a switch to glycolytic metabolism to fuel inflammatory cytokine production by macrophages (Cramer et al., 2003). While elegant initial studies found that knockdown of HIF-1α in hematopoietic stem cells improved post-MI cardiac function in association with reduced infiltration of myeloid cells (Dong et al., 2010), this approach also affected HIF-1α expression in nonmyeloid hematopoietic lineages, including lymphocytes, which positively (Rieckmann et al., 2019) or negatively (Santos-Zas et al., 2021) regulate cardiac repair after MI. HIF-2α has also been implicated in macrophage production of proinflammatory cytokines, but this is independent of changes in glycolytic metabolism, as HIF-2α is dispensable for ATP synthesis during hypoxia (Imtiyaz et al., 2010). In contrast to HIF-1α, the role of macrophage HIF-2α during cardiac ischemia is largely unknown, despite its potential clinical significance.
To ascertain the roles of myeloid HIF-1α and HIF-2α during ischemic tissue injury, we examined cardiac repair under experimental conditions of myeloid cell–specific HIF isoform loss and gain of function. Our findings reveal that although myeloid HIF isoforms act together to afford critical cardioprotection, HIF-1α and HIF-2α function uniquely and independently antagonize optimal cardiac repair by impeding anti-inflammatory macrophage metabolic reprogramming and promoting cleavage of cardioprotective MerTK (myeloid-epithelial-reproductive receptor tyrosine kinase), respectively.
Results
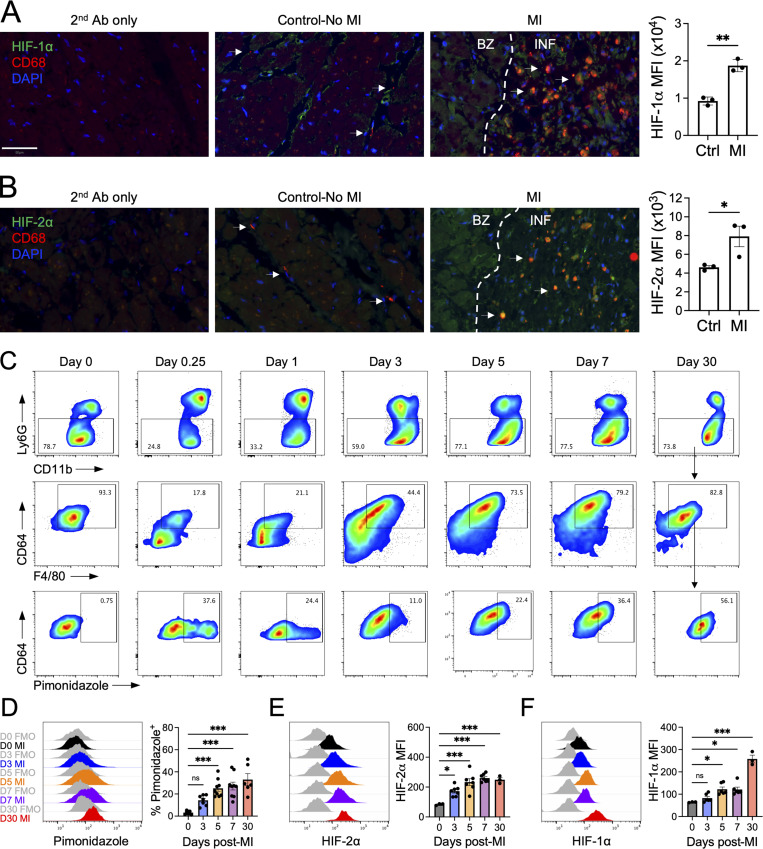
To discover the unknown role of HIF-2α in myeloid cells after MI, we first assessed myeloid cell expression of HIFs in cardiac autopsies of patients who died within 5–14 d after MI or patients with non–MI-related deaths. We found that both HIF-1α and HIF-2α levels were significantly increased in CD68+ macrophages after acute MI in humans compared with non-MI controls (Fig. 1, A and B). To determine the kinetics of macrophage hypoxia and HIF isoform protein expression after MI, we next subjected mice to an experimental model of MI by coronary artery ligation. Injection of the hypoxia tracer pimonidazole revealed progressive accumulation of hypoxic macrophages in the infarcted myocardium during the acute phase after MI (Fig. 1, C and D). Consistent with exposure to tissue hypoxia, induction of both HIF-1α and HIF-2α protein in infarct-associated macrophages was evident by flow cytometry after MI (Fig. 1, E and F). Interestingly, increases in HIF-2α were more acutely responsive to ischemic insult, which preceded subsequent escalations of HIF-1α. Furthermore, both pimonidazole staining and protein expression of HIF-1α and HIF-2α was sustained in infarct-associated macrophages and remained significantly increased 30 d after MI, indicating that hypoxic macrophages persist in ischemic myocardium through the progression to heart failure. To this latter point, flow cytometry (Fig. S1, A and B) and single-cell RNA sequencing (Fig. S1, D–G; and Table S1) of infarct/scar–associated macrophages from human patients with ischemic cardiomyopathy revealed HIF activation and signaling as measured by increases in HIF-1α and HIF-2α protein relative to remote tissue (Fig. S1 C), up-regulation of pathways involved in the response to hypoxia (Fig. S1 H), and enrichment of HIFs and HIF-dependent genes (Fig. S1 I).
Figure 1.
Sequential stabilization of HIF-2⍺ followed by HIF-1⍺ in cardiac macrophages after MI.(A and B) Histological analyses of HIF-1α (A) or HIF-2α (B) in CD68+ macrophages (arrows) in cardiac autopsy samples collected 5–14 d after MI with quantification of HIF mean fluorescence intensity in CD68+ macrophages. Patients with non-MI deaths were used as controls. Scale bar, 50 µm. n = 3 patients/group. *, P < 0.05; **, P < 0.01 by two-tailed, unpaired t test. (C) Gating strategy of cardiac macrophages after MI. Cells were first gated on live, single cells, and macrophages were identified as CD11b+Ly6G−F4/80+CD64+ cells. (D) Accumulation of infarct-associated cardiac macrophages within hypoxic myocardium using the hypoxia tracer pimonidazole after permanent occlusion MI in mice. n = 6–9 mice/group pooled from more than three independent experiments. ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. (E and F) Expression of HIF-2⍺ (E) or HIF-1⍺ (F) in infarct-associated cardiac macrophages after MI in mice. For E and F, n = 3–7 mice/group pooled from three independent experiments. *, P < 0.05; ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. All data presented as mean ± SEM. Gray histograms represent FMO staining controls. Ab, antibody; BZ, border zone; INF, infarct.
Figure S1.
HIF expression and signaling in human cardiac macrophages during ischemic cardiomyopathy.(A) Gross appearance of remote (RT) and peri-infarct (INF) regions sampled for flow cytometry. (B) Gating strategy for cardiac macrophages and peripheral blood mononuclear cells (PBMCs). Cardiac macrophages and PBMCs were first gated on live, single cells and identified as CD14+CD64+ cells. (C) Expression of HIF-1⍺ and HIF-2⍺ in cardiac macrophages and PBMCs. FMO was used as a negative staining control. (D) Gross appearance of INF region sampled for single-cell RNA-sequencing analyses. Transcriptomic analysis was performed on 1,149 cells using the 10x Genomics platform. (E) Identification of eight unique clusters by t-distributed stochastic neighbor embedding dimensionality reduction analysis. EC, endothelial cell; SMC, smooth muscle cell. (F) Heatmap of the 20 most differentially expressed genes within each cluster. (G) Feature plots representing single-cell gene expression of canonical macrophage markers (Cd68, C1qa, Lyve1, Mertk, and Csfr1) identified cluster 3 as a macrophage cluster. (H) Pathway enrichment of differentially expressed genes in the macrophage cluster (cluster 3) was determined using gProfiler. Enrichment is expressed as the −log[P] and is adjusted for multiple comparisons. (I) Violin plots of Hif1 and Epas1 (Hif2) expression and inflammatory cytokine and chemokine gene expression shown to be HIF dependent in primary human macrophages during hypoxia treatment. RT, remote tissue; FSC-A, forward scatter area; FSC-W, forward scatter width; SSC-A, side scatter area; SSC-W, side scatter width; EC, endothelial cell; NK, natural killer; UMAP, Uniform Manifold Approximation and Projection for Dimension Reduction.
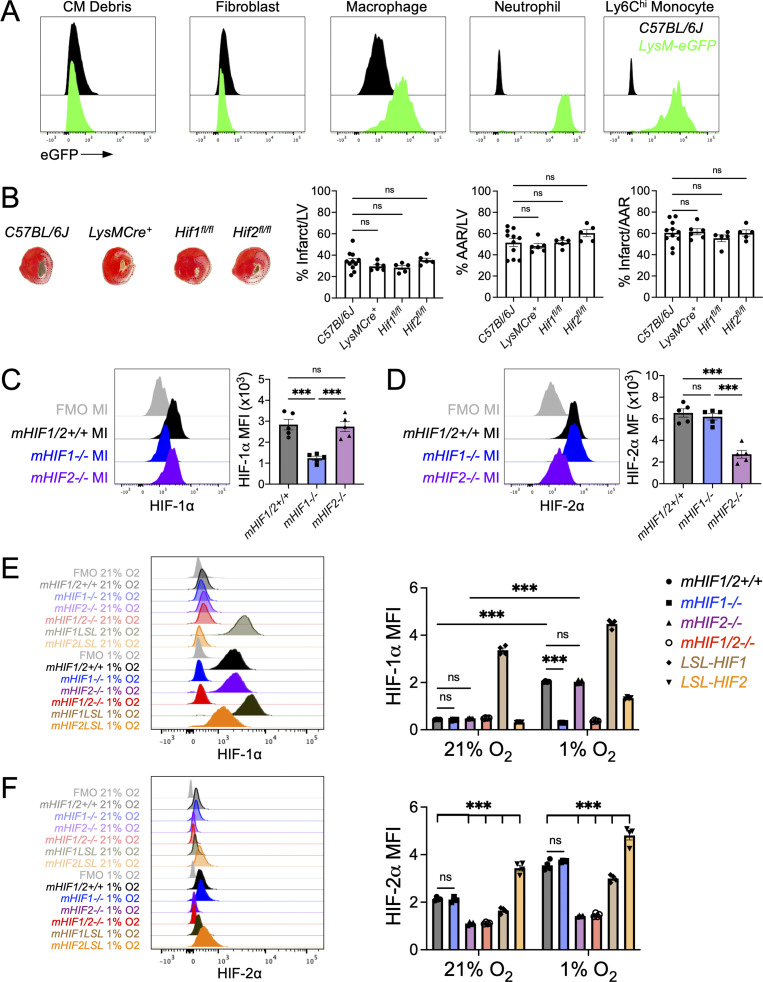
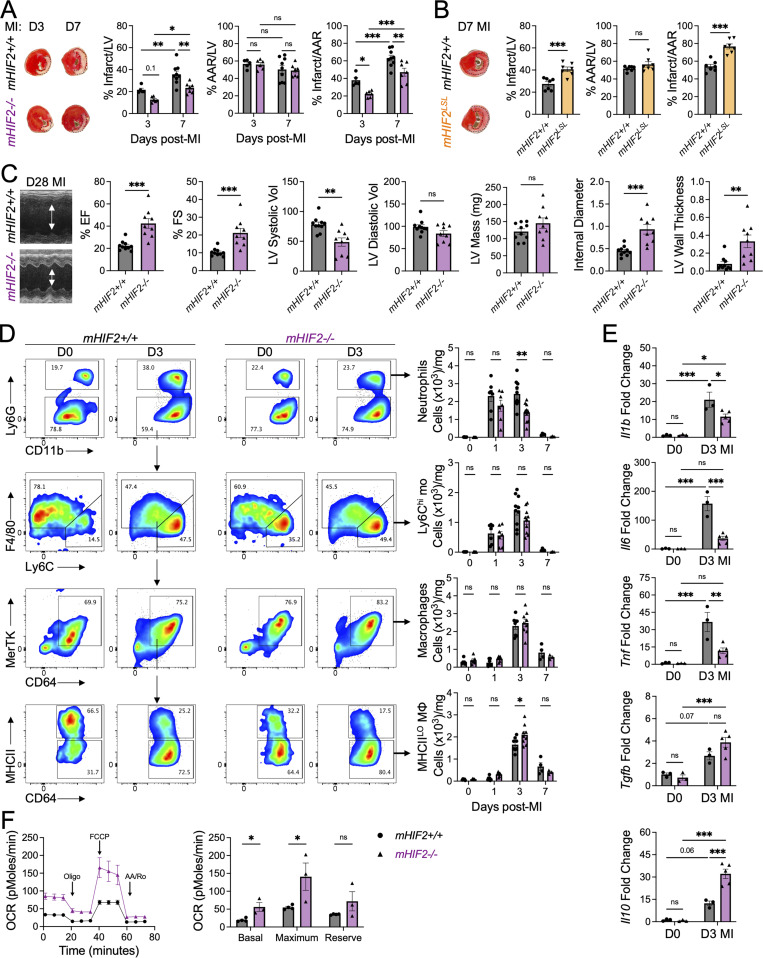
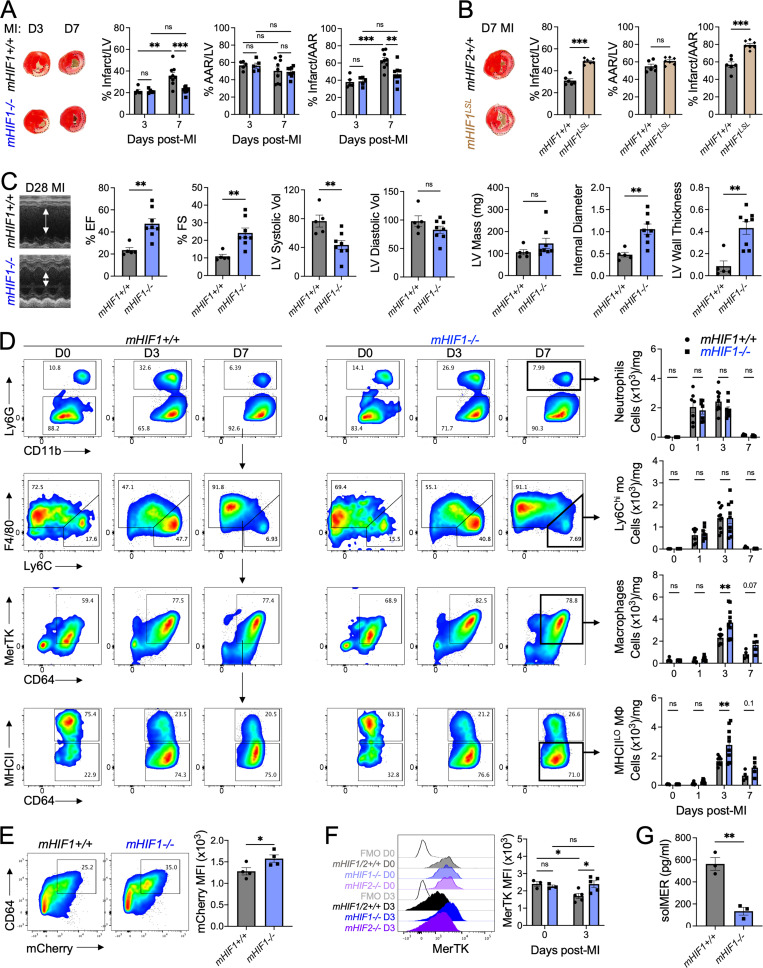
We next sought to determine the functional significance of HIF-2α expression in cardiac myeloid cells in the acute phase after MI. We selectively deleted HIF-2α in myeloid cells (mHIF2−/−) by crossing transgenic mice with a floxed Hif2 (Epas1) gene with mice expressing Cre recombinase under the control of the lysozyme M (Lyz2) promotor (Fig. S2 A). We compared left ventricle (LV) remodeling and contractile function after MI between mHIF2+/+ and mHIF2−/− mice. Despite comparable levels of ischemic injury (area at risk [AAR]), pathological measurement of infarct size at 7 d after MI revealed smaller sized infarcts in mHIF2−/− mice compared with controls (Figs. 2 A and S2 B). Smaller infarcts were also observed earlier at 3 d after MI in mHIF2−/− mice compared with controls, which was consistent with the early activation of HIF-2α that we observed in infarct-associated macrophages. Echocardiography performed 28 d after MI revealed improved LV systolic function, reduced LV dilatation, and increased LV wall thickness in mHIF2−/− mice compared with controls (Fig. 2 C). Importantly, loss of HIF-2α did not affect the levels of HIF-1α in cardiac macrophages (Fig. S2, C and D), suggesting that the effects of myeloid cell Hif2 deletion on cardiac remodeling and function were not due to compensatory effects of HIF-1α. To further explore the role of myeloid cell expression of HIF-2α in LV remodeling, we crossed mice that specifically express a Cre-dependent, prolyl hydroxylation-resistant HIF-2α (Kim et al., 2006b), with LysM-Cre mice to generate mice that express constitutively increased levels of HIF-2α in myeloid cells (mHIF2LSL). Relative to controls, infarct sizes in mHIF2LSL mice were increased 7 d after MI (Fig. 2 B), supporting a pathological role for HIF-2α activation in myeloid cells after MI.
Figure S2.
Loss of either HIF-1⍺ or HIF-2⍺ does not result in a compensatory increase in the other isoform in macrophages.(A) Flow cytometric analysis of lysozyme M expression in heart (cardiomyocytes [CM], fibroblasts, and macrophages) and peripheral blood (neutrophils, Ly6Chi monocytes) using LysM-eGFP mice, which express EGFP under the control of the LysM promoter region. C57BL/6J mice served as a negative staining control. n = 1/group from a single experiment. (B) Percent infarct/LV, percent AAR/LV, and percent infarct/AAR measured 7 d after MI in C57BL/6J, LysM-Cre+, Hif1fl/fl, or Hif2fl/fl mice. n = 5–11 mice/group pooled from more than three independent experiments. ns, one-way ANOVA followed by Tukey’s test. (C and D) Expression of HIF-1⍺ (C) or HIF-2⍺ (D) in cardiac macrophages from mice with myeloid-specific deletion of Hif1 (mHIF1−/−) or Hif2 (mHIF2−/−) compared with controls 5 d after MI. n = 5 mice/group pooled from two independent experiments. ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. (E and F) Expression of HIF-1⍺ (E) or HIF-2⍺ (F) in BMDMs from mice with myeloid-specific deletion (mHIF1−/−, mHIF2−/−, and mHIF1/2−/−) or overexpression (mHIF1LSL and mHIF2LSL) of HIFs cultured under normoxia (21% O2) or hypoxia (1% O2) for 3 h. n = 4 sets of cells/group. Data are representative of two independent experiments. ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. All data presented as mean ± SEM.
Figure 2.
Myeloid HIF-2⍺ promotes inflammation and worsens cardiac repair after MI.(A) Percent infarct/LV, percent AAR/LV, and percent infarct/AAR measured 3 or 7 d after MI in mice with myeloid-specific deletion of Hif2 (mHIF2−/−) or controls. n = 5–9 mice/group pooled from more than three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-way ANOVA followed by Tukey’s test. (B) Infarct measurement 7 d after MI in mice with overexpression of HIF-2⍺ (mHIF2LSL) or controls. n = 7 mice/group pooled from two independent experiments. ***, P < 0.001 by two-tailed, unpaired t test. (C) M-mode echocardiography measurements 28 d after MI with quantification of percent ejection fraction (EF), percent fractional shortening (FS), LV systolic and diastolic volume (microliter), LV mass, internal diameter (millimeters), and LV wall thickness (millimeters). n = 9–10 mice/group pooled from two independent experiments. **, P < 0.01, ***, P < 0.001 by two-tailed, unpaired t test. (D) Infarct-associated cellular responses in mHIF2−/− mice or controls after MI. n = 3–10 mice/group pooled from more than three independent experiments. *, P < 0.05; **, P < 0.01 by two-way ANOVA followed by Tukey’s test. (E) Gene expression of inflammatory mediators in whole cardiac extracts. n = 3–5 mice/group pooled from two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-way ANOVA followed by Tukey’s test. (F) OCR of infarct-associated cardiac macrophages 3 d after MI. n = 3–5 mice/group pooled from two independent experiments. *, P < 0.05 by two-tailed, unpaired t test. All data presented as mean ± SEM. D, day.
We next used flow cytometry to investigate whether myeloid cell Hif2 deficiency impacted the extent and quality of immune cell infiltrate in the heart after MI. Although no differences were observed in the total number of macrophages in the infarcted myocardium, there was an increase in macrophage polarization toward a reparative phenotype in mHIF2−/− mice compared with mHIF2+/+ mice, as measured by an increase in the total number of MHCIILO macrophages 3 d after MI (Fig. 2 D). MHCIILO cardiac macrophages secrete higher levels of IL-10 compared with MHCIIHI cardiac macrophages (Chakarov et al., 2019; DeBerge et al., 2017), and this was evident as increased Il10 levels in cardiac extracts from mHIF2−/− mice compared with mHIF2+/+ mice despite similar baseline levels (Fig. 2 E). HIF-2α has been shown to be dispensable for in vitro MHCIIHI macrophage polarization following LPS and IFN-γ treatment. However, HIF-2α directly regulates macrophage expression of proinflammatory genes, including Il6 (Imtiyaz et al., 2010), and we found decreased levels of the proinflammatory genes Il1b, Il6, and Tnf in cardiac extracts from mHIF2−/− mice compared with mHIF2+/+ mice (Fig. 2 E). These data support a proinflammatory role for HIF-2α in infarct-associated macrophages, yet, it is important to note that LysM-Cre is also active in neutrophils, Ly6Chi monocytes, and some dendritic cell subsets. While myeloid cell Hif2 deficiency did not affect initial neutrophil or Ly6Chi monocyte recruitment to the infarcted myocardium, the total number of neutrophils was reduced 3 d after MI in mHIF2−/− mice compared with mHIF2+/+ mice (Fig. 2 D). Hif2-deficient neutrophils exhibit increased apoptosis during inflammation (Thompson et al., 2014), so we next measured neutrophil apoptosis after MI. Annexin staining of infarct-associated neutrophils 1 d after MI revealed increased neutrophil apoptosis in mHIF2−/− mice compared with mHIF2+/+ mice (Fig. S3 A), which may have contributed to the enhanced resolution of neutrophilic inflammation in mHIF2−/− mice on day 3 after MI. Taken together, these data show that myeloid cell HIF-2α promotes inflammatory responses after MI to worsen cardiac repair.
Figure S3.
Effects of HIF deficiency on neutrophil apoptosis and macrophage proliferation. (A) Neutrophil apoptosis as measured by Annexin staining 1 d after MI in mice with myeloid-specific deletion of Hif2 (mHIF2−/−) or controls. Black contours represent FMO stained controls. n = 5 mice/group pooled from two independent experiments. *, P < 0.05 by two-tailed, unpaired t test. (B) Macrophage proliferation as determined by BrdU incorporation at day 3 after MI in mice with myeloid-specific deletion of Hif1 (mHIF1−/−) or controls. n = 4 mice/group pooled from two independent experiments. ns, two-tailed, unpaired t test. All data are presented as mean ± SEM.
Phagocytosis of apoptotic neutrophils (efferocytosis) by macrophages imprints anti-inflammatory cellular reprogramming through mitochondrial respiration (A-Gonzalez et al., 2017; Zhang et al., 2019) and is necessary for inflammation resolution and cardiac repair after MI (Wan et al., 2013). Since myeloid cell Hif2 deficiency was associated with increased reparative macrophages and improved inflammation resolution, we asked whether HIF-2α suppressed phagocytosis-associated metabolism in macrophages to impair inflammation resolution after MI. To measure cellular metabolism in cardiac macrophages, we isolated infarct-associated macrophages 3 d after MI and measured oxygen consumption rate (OCR) using a Seahorse Analyzer. Interestingly, HIF-2α suppressed mitochondrial respiration as basal respiration and maximum respiratory capacity were significantly increased in infarct-associated cardiac macrophages from mHIF2−/− mice compared with mHIF2+/+ mice after MI (Fig. 2 F). To determine how HIF-2ɑ suppressed mitochondrial respiration, we modeled efferocytosis in vitro by coculturing bone marrow–derived macrophages (BMDMs) with apoptotic Jurkat T cells. Apoptotic cells (ACs) reproducibly induced BMDMs to produce the canonical anti-inflammatory cytokine IL-10 (Fadok et al., 1998), and this response was also validated with elicited primary peritoneal macrophages (Fig. 3 A). Loss of Hif2 enhanced AC-induced macrophage production of IL-10, which was specific to HIF-2α, as macrophages deficient in both Hif2 and Hif1, but not Hif1 alone, also exhibited increased IL-10 production (Fig. 3 A). In contrast, overexpression of HIF-2α suppressed AC-induced IL-10 production by BMDMs (Fig. 3 B). Consistent with our in vivo data, loss or gain of HIF-2α was associated with increased or decreased mitochondrial respiration by efferocytic BMDMs, respectively (Fig. 3 D). Importantly, loss or gain of HIF-2α did not affect the levels of HIF-1α in BMDMs (Fig. S2, E and F) and did not affect BMDM uptake of ACs (Fig. 3 C), thereby implicating a HIF-2α–specific postengulfment signaling pathway.
Figure 3.
HIF-2⍺ suppresses anti-inflammatory macrophage mitochondrial metabolism. (A) IL-10 production by BMDMs or peritoneal elicited macrophages (PMac) from mice with myeloid-specific deletion of Hif1 (mHIF1−/−), Hif2 (mHIF2−/−), both Hif1 and Hif2 (mHIF1/2−/−), or controls after efferocytosis of ACs. Nonengulfed cells were removed from adherent phagocytes. n = 3 sets of cells/group. Data are representative of more than three independent experiments. *, P < 0.05; ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. (B) IL-10 production by BMDMs from mice with myeloid-specific overexpression of HIF-2⍺ (mHIF2LSL) or controls after efferocytosis of ACs. n = 5 sets of cells/group. Data are representative of two independent experiments. *, P < 0.05 by two-tailed, unpaired t test. (C) Efferocytosis of calcein-labeled ACs (green) by MitoTracker-labeled BMDMs (red). Scale bar, 10 µm. n = 3 sets of cells/group. Data are representative of three independent experiments. ns, two-tailed, unpaired t test. (D) OCR with quantification of mitochondrial respiration by untreated (⍉) or efferocytic (AC) BMDMs. n = 6–8 sets of cells/group. Data are representative of three independent experiments. ***, P < 0.001 by two-way ANOVA followed by Tukey’s test. AA/Ro, antimycin A/rotenone. (E) Expression of genes involved in fatty acid storage, synthesis, and oxidation in untreated mHIF2−/− BMDMs or controls. n = 3 sets of cells/group. Data are representative of three independent experiments. *, P < 0.05; **, P < 0.01 by two-tailed, unpaired t test. (F) Expression of Hilpda in untreated BMDMs. n = 5 sets of cells/group. Data represent three independent experiments. *, P < 0.05; ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. (G) IL-10 levels in cell culture medium after cocultivation of ACs and mHIF2−/− BMDMs treated with control or Cpt1a siRNA. n = 3 sets of cells/group. Data are representative of two independent experiments. **, P < 0.01 by two-tailed, unpaired t test. (H) OCR with quantification of mitochondrial respiration by vehicle or TOFA-treated efferocytic BMDMs. n = 8 sets of cells/group. Data are representative of three independent experiments. *, P < 0.05 by two-tailed, unpaired t test. (I) IL-10 production by vehicle (Veh) or TOFA-treated efferocytic BMDMs. n = 5 sets of cells/group. Data are representative of three independent experiments. ***, P < 0.001 by two-tailed, unpaired t test. (J) BODIPY levels in BMDMs after cocultivation of BMDMs with BODIPY FL C16-labeled ACs. n = 3 sets of cells/group. Data are representative of two independent experiments. ***, P < 0.001 by two-way ANOVA followed by Tukey’s test. (K) BODIPY staining in BMDMs after cocultivation of BMDMs with BODIPY FL C16-labeled ACs with quantification of lipid droplets (LD) per cell. Arrows indicate lipid droplets in BMDM. Scale bar, 10 µm. n = 4 sets of cells/group. Data are representative of three independent experiments. *, P < 0.05 by two-tailed, unpaired t test. (L) OCR with quantification of mitochondrial respiration by control or Hilpda siRNA-treated efferocytic BMDMs. n = 7–11 sets of cells/group. Data are representative of two independent experiments. **, P < 0.01; ***, P < 0.001 by two-tailed, unpaired t test. (M) IL-10 production by control or Hilpda siRNA-treated efferocytic BMDMs. n = 6 sets of cells/group. Data are representative of two independent experiments. ***, P < 0.001 by two-tailed, unpaired t test.
We have previously shown that efferocytic IL-10 production is dependent on mitochondrial β-oxidation of AC-derived fatty acids to fuel mitochondrial metabolism (Zhang et al., 2019). Separately, HIF-2α has been linked to suppression of fatty acid β-oxidation and increased lipid storage in hepatocytes (Rankin et al., 2009). Since HIF-2α is stably expressed by BMDMs during normoxia (Fig. S2 F; Imtiyaz et al., 2010; Dehn et al., 2016), we sought to determine whether HIF-2α suppressed postengulfment β-oxidation of AC-derived fatty acids by first assessing expression of genes involved in fatty acid synthesis, storage, and oxidation in untreated mHIF2+/+ and mHIF2−/− BMDMs. We found that loss of HIF-2α was associated with decreases in genes involved in fatty acid synthesis and storage with a concomitant increase in fatty acid oxidation (FAO) genes, including Cpt1a (Fig. 3 E), which encodes carnitine palmitoyltransferase 1 (CPT1), a rate-limiting enzyme of FAO. To determine whether the increase in Cpt1a with Hif2-deficiency was linked to the increased IL-10 production by efferocytic mHIF2−/− BMDMs, we specifically targeted Cpt1a gene expression with siRNAs. Targeted knockdown of Cpt1a in mHIF2−/− BMDMs significantly reduced efferocytosis-induced IL-10 production (Fig. 3 G), linking increased FAO with loss of HIF-2α. As a complementary approach, we used a small-molecule inhibitor to block acetyl–coenzyme A carboxylase (ACC), which is encoded by the gene Acaca and produces malonyl–coenzyme A, an endogenous inhibitor of CPT1. Loss of HIF-2α was associated with decreased Acaca expression (Fig. 3 E), and treatment of wild-type efferocytic BMDMs with the ACC-specific inhibitor 5-tetradecyloxy-2-furoic acid (TOFA) enhanced mitochondrial respiration (Fig. 3 H) and IL-10 production (Fig. 3 I), recapitulating the effects of Hif2 deficiency. Taken together, these data support a role for HIF-2α in suppression of anti-inflammatory mitochondrial metabolism in efferocytic macrophages.
While HIFs have been shown to directly repress gene transcription (Cavadas et al., 2016), including Cpt1a within tumor cells (Du et al., 2017), they are widely believed to function as transcriptional activators. This led us to hypothesize that HIF-2α may activate genes involved in directing AC-derived fatty acids away from the mitochondria instead of directly repressing fatty acid β-oxidation. To track AC-derived fatty acids during efferocytosis, we loaded ACs with a boron-dipyrromethene (BODIPY)–labeled, long-chain fatty acid and examined uptake and localization of BODIPY in BMDMs after efferocytosis. Consistent with our earlier observation that HIF-2α was dispensable for macrophage uptake of ACs (Fig. 3 C), we observed similar total levels of BODIPY-labeled fatty acids in mHIF2+/+ and mHIF2−/− BMDMs (Fig. 3 J). Despite similar levels, when we examined distribution of AC-derived fatty acids, we found increased localization of BODIPY-labeled fatty acids within lipid droplets of mHIF2+/+ BMDMs compared with mHIF2−/− BMDMs (Fig. 3 K). Since lipid droplets were reduced, but not completely eliminated, in efferocytic mHIF2−/− BMDMs, we wondered whether HIF-2α promoted lipid droplet stability. Recent studies of macrophages have identified a role for the hypoxia-inducible lipid droplet–associated protein (HILPDA) in promoting retention of lipids within lipid droplets (Maier et al., 2017; van Dierendonck et al., 2020). We found that loss or gain of HIF-2α in BMDMs was associated with decreased and increased expression of Hilpda, respectively (Fig. 3 F), suggesting that HIF-2α regulated Hilpda expression in macrophages. To determine whether HILPDA antagonized anti-inflammatory mitochondrial metabolism in efferocytic macrophages, we specifically targeted Hilpda gene expression with siRNAs. Targeted knockdown of Hilpda in wild-type BMDMs enhanced mitochondrial respiration (Fig. 3 L) and IL-10 production (Fig. 3 M), linking HIF-2α to storage of AC-derived fatty acids through HILPDA. Thus, HIF-2α activation leads to sequestration of AC-derived fatty acids in lipid droplets to deprive the macrophage of the fuel needed for anti-inflammatory mitochondrial metabolism.
To determine whether our findings were unique to HIF-2α, we next compared cardiac remodeling and function after MI between controls and mice with myeloid-specific loss (mHIF1−/−) or gain (mHIF1LSL) of HIF-1α. Similarly to mHIF2−/− mice, yet in contrast to previously published data (Dong et al., 2010), mHIF1−/− mice presented with smaller sized infarcts compared with controls 7 d after MI (Fig. 4 A). Alternatively, constitutive activation of HIF-1α in myeloid cells of mHIF1LSL mice led to increased infarct sizes compared with controls 7 d after MI (Fig. 4 B). Echocardiography performed 28 d after MI revealed improved LV systolic function, reduced LV dilatation and increased LV wall thickness in mHIF1−/− mice compared with controls (Fig. 4 C), linking HIF-1α activation in myeloid cells to adverse remodeling after MI. In contrast to mHIF2−/− mice, earlier infarct measurements at 3 d after MI revealed no differences in infarct sizes between mHIF1−/− mice and controls (Fig. 4 A), consistent with later activation of HIF-1α that we observed in infarct-associated macrophages (Fig. 1 D). Importantly, loss of HIF-1α did not affect the levels of HIF-2α in cardiac macrophages (Fig. S2, C and D), indicating that the effects of myeloid cell Hif1 deletion on cardiac remodeling and function were not due to compensatory effects of HIF-2α. Taken together, these data suggest that HIF-1α functioned independent of HIF-2α in myeloid cells to promote infarct expansion after MI.
Figure 4.
Myeloid HIF-1⍺ antagonizes cardiac repair through MerTK cleavage after MI.(A) Percent infarct/LV, percent AAR/LV, and percent infarct/AAR measured 3 or 7 d after MI in mice with myeloid-specific deletion of Hif1 (mHIF1−/−) or controls. n = 5–9 mice/group pooled from more than three independent experiments. **, P < 0.01, ***, P < 0.001 by two-way ANOVA followed by Tukey’s test. (B) Infarct measurement 7 d after MI in mice with overexpression of HIF-1⍺ (mHIF1LSL) or controls. n = 6–7 mice/group pooled from two independent experiments. ***, P < 0.001 by two-tailed, unpaired t test. (C) M-mode echocardiography measurements 28 d after MI with quantification of percent ejection fraction (EF), percent fractional shortening (FS), LV systolic and diastolic volume (microliters), LV mass, internal diameter (millimeters), and LV wall thickness (millimeters). n = 5–8 mice/group pooled from two independent experiments. **, P < 0.01 by two-tailed, unpaired t test. (D) Infarct-associated cellular responses in mHIF1−/− mice or controls after MI. n = 4–10 mice/group pooled from more than three independent experiments. **, P < 0.01 by two-way ANOVA followed by Tukey’s test. (E) Phagocytosis of apoptotic mCherry-expressing cardiomyocytes by cardiac macrophages 5 d after MI. n = 4 mice/group pooled from two independent experiments. *, P < 0.05 by two-tailed, unpaired t test. (F) MerTK expression on cardiac macrophages after MI. Unfilled histograms represent FMO staining controls. n = 3–5 mice/group pooled from two independent experiments. *, P < 0.05 by two-way ANOVA followed by Tukey’s test. (G) Serum levels of solMER 3 d after MI. n = 3 mice/group from three independent experiments. **, P < 0.01 by two-tailed, unpaired t test. All data are presented as mean ± SEM.
We next used flow cytometry to investigate whether changes in the extent and quality of cardiac immune cell infiltrate contributed to HIF-1α–dependent infarct expansion. In contrast to prior reports (Dong et al., 2010), Hif1 deficiency did not reduce myeloid cell infiltration into the heart, as we observed similar total numbers of Ly6Chi monocytes and neutrophils in mHIF1−/− mice compared with controls after MI (Fig. 4 D). However, there was a significant increase in MerTK+ macrophages in mHIF1−/− mice compared with controls after MI (Fig. 4 D). This was specific to HIF-1α, as numbers of MerTK+ macrophages in mHIF2−/−mice were similar to controls (Fig. 2 D). Differences in proliferation could not explain the increased macrophages in mHIF1−/− mice, as in vivo administration of BrdU labeled similar percentages of macrophages in mHIF1+/+ and mHIF1−/− mice after MI (Fig. S3 B). We address the effect of Hif1-deficiency on macrophage apoptosis in a section below. MerTK mediates the clearance of apoptotic cardiomyocytes to prevent infarct expansion (Howangyin et al., 2016; Wan et al., 2013) and promotes proreparative MHCIILO macrophage polarization (DeBerge et al., 2017). Accordingly, we observed increased macrophage efferocytosis of apoptotic cardiomyocytes (Fig. 4 E) and increased total number of MHCIILO macrophages (Fig. 4 D) in mHIF1−/− mice compared with controls after MI. During inflammation, MerTK levels are naturally reduced by its proteolytic cleavage (Cai et al., 2016; DeBerge et al., 2017), so we next examined MerTK levels on macrophages before and after MI to determine whether HIF-1α activation compromised inflammation resolution through MerTK cleavage. Despite similar levels of MerTK on cardiac macrophages at steady state, cell surface MerTK expression was significantly reduced in controls and mHIF2−/− mice after MI, but this effect was largely absent in mHIF1−/− mice (Fig. 4 F). This was associated with a reduction in serum levels of the cleavage product, soluble MerTK (solMER) in mHIF1−/− mice compared with controls after MI (Fig. 4 G). Together, these data show that myeloid cell HIF-1α promotes MerTK cleavage after MI to worsen cardiac repair.
To dig deeper into the cell-intrinsic relationship between HIF-1α and MerTK, we examined cell surface MerTK and solMER levels of BMDMs from mice with loss or gain of HIF-1α after stimulation of TLR4, which recognizes damage-associated molecular patterns released by dying cardiomyocytes (Rohde et al., 2014) and triggers MerTK cleavage (Thorp et al., 2011). TLR4 stimulation decreased cell surface MerTK and increased solMER in controls and mHIF2−/− BMDMs (Fig. 5, A and B). However, these effects were largely absent with mHIF1−/− BMDMs (Fig. 5, D and E), and led to preservation of efferocytosis of apoptotic Jurkat T cells after TLR4 stimulation compared with controls (Fig. 5 C). Constitutive HIF-1α activation in mHIF1LSL BMDMs led to even greater reductions or elevations of cell surface MerTK and solMER, respectively, both at steady state and after TLR4 stimulation (Fig. 5, D and E), implicating HIF-1α in MerTK cleavage on macrophages. A disintegrin and metalloproteinase 17 (ADAM17) mediates the proteolytic cleavage of MerTK to block efferocytosis (Thorp et al., 2011), so we next assessed ADAM17 levels. Loss (mHIF1−/−) or gain (mHIF1LSL) of HIF-1α was associated with decreased and increased expression of Adam17, respectively (Fig. 5, F and G). HIF-1α increases Adam17 expression under conditions of high glucose (Li et al., 2015) and is required for the TLR4-stimulated switch to glycolysis (Cramer et al., 2003), so we next determined the requirement for glycolytic metabolism in MerTK cleavage. Despite similar basal levels of glycolytic metabolism, TLR4 stimulation increased glycolysis and glycolytic capacity in mHIF1+/+ BMDMs but this effect was attenuated in mHIF1−/− BMDMs (Fig. 5 H). Constitutive HIF-1α activation in mHIF1LSL BMDMs increased both basal and TLR4-stimulated glycolysis (Fig. 5 H), linking HIF-1α to macrophage glycolytic metabolism. To block the HIF-1α–mediated increase in glycolysis, we stimulated TLR4 on BMDMs cultured in glucose-free media or in the presence of 2-deoxy-D-glucose (2-DG), an inhibitory glucose analogue. Both glucose-free media and 2-DG abrogated TLR4-stimulated MerTK cleavage (Fig. 5, I and J), thereby implicating HIF-1α–mediated glycolytic reprogramming of macrophages in MerTK cleavage.
Figure 5.
HIF-1⍺ promotes MerTK cleavage through glycolytic reprogramming of macrophages. (A and B) Cell surface expression of MerTK (A) or solMER (B) in culture media of untreated (⍉) or LPS-treated BMDMs from controls or mice with myeloid-specific deletion of Hif2 (mHIF2−/−). (C) Efferocytosis of calcein-labeled apoptotic Jurkat cells (green) by BMDMs from mice with myeloid-specific deletion of Hif1 (mHIF1−/−) or controls treated with LPS. Scale bar, 10 µm. n = 3 sets of cells/group. Data are representative of two independent experiments. *, P < 0.05 by two-way ANOVA followed by Tukey’s test. (D and E) Cell surface expression of MerTK (D) or solMER (E) in culture media of untreated (⍉) or LPS-treated BMDMs from controls, mHIF1−/− mice, or mice with overexpression of HIF-1⍺ (mHIF1LSL). For B–E, n = 3–4 sets of cells/group, and data are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-way ANOVA followed by Tukey’s test. (F) Expression of Adam17 in untreated (⍉) or LPS-treated mHIF1−/− BMDMs or controls. n = 3 sets of cells/group and data are representative of three independent experiments. *, P < 0.05 by two-way ANOVA followed by Tukey’s test. (G) Expression of Adam17 in untreated mHIF1LSL BMDMs or controls. n = 3 sets of cells/group, and data are representative of two independent experiments. *, P < 0.05 by two-tailed, unpaired t test. (H) ECAR with quantification of glycolytic function in untreated (⍉) or LPS-treated BMDMs. n = 5–7 sets of cells/group. Data are representative of more than three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-way ANOVA followed by Tukey’s test. (I and J) Cell surface expression of MerTK (I) or solMER (J) in culture media of untreated or LPS-treated BMDMs cultured in 0 mM glucose (Glu), 5 mM glucose, or 5 mM glucose with 25 mM 2-DG. n = 4 sets of cells/group. Data are representative of two independent experiments. **, P < 0.01; ***, P < 0.001 by two-way ANOVA followed by Tukey’s test. Unfilled histograms represent FMO staining controls. All data presented as mean ± SEM.
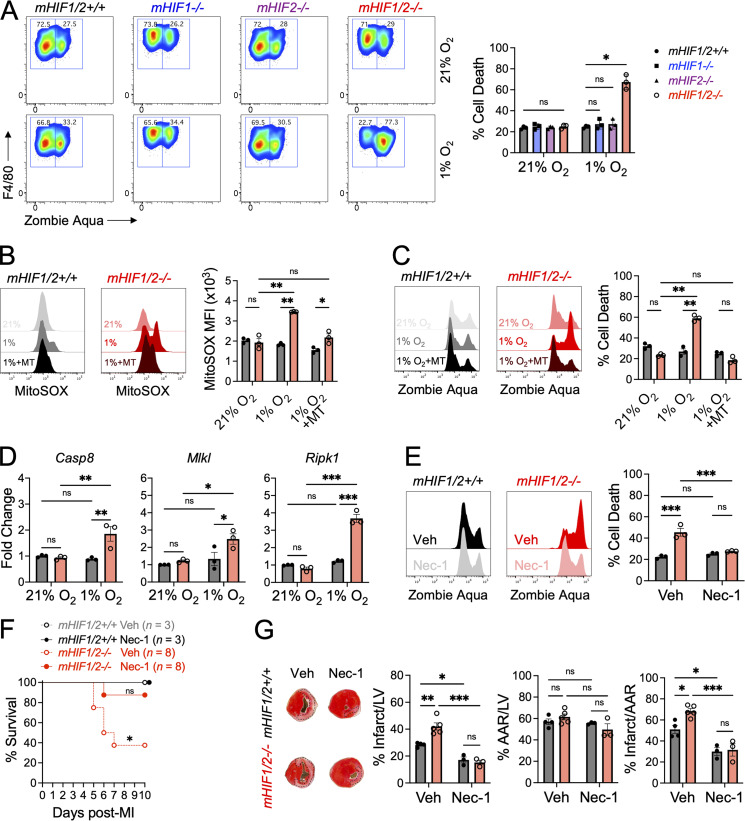
Given that HIF-2α and HIF-1α both antagonized cardiac repair, we hypothesized that loss of both isoforms in myeloid cells would maximize cardiac repair. In contrast to our expectations, loss of both HIF-2α and HIF-1α in myeloid cells (mHIF1/2−/−) increased death due to cardiac rupture in mHIF1/2−/− mice compared with controls after MI (Fig. 6, A and B). To determine the cause of increased ventricular wall rupture, we examined the extent and quality of cells involved in stable scar formation and tissue remodeling, including myofibroblasts and myeloid cells. While myofibroblast accumulation was reduced in mHIF1/2−/− mice compared with controls after MI, this was not specific to combined Hif1 and Hif2deficiency as myofibroblasts were also reduced in mHIF1−/− and mHIF2−/− mice (Fig. 6 C). Since we observed increased cardiac macrophages in mHIF1−/− mice without changes in proliferation (Fig. 4 D, Fig. S3 B), we next assessed whether HIFs affected macrophage survival using Annexin staining after MI. While Hif2 deficiency did not affect macrophage apoptosis after MI, loss of HIF-1α decreased macrophage apoptosis compared with controls (Fig. 6 D), which may have contributed to the increase in cardiac macrophages that we observed in mHIF1−/− mice. In contrast, combined Hif1 and Hif2 deficiency significantly increased macrophage apoptosis compared with controls (Fig. 6 D). Despite the increase in macrophage apoptosis, mHIF1/2−/− mice had increased total number of macrophages compared with controls 5 d after MI (Fig. 6 E). However, we also observed hyperinflammatory responses in mHIF1/2−/− mice, measured as increased neutrophils and Ly6Chi monocytes on days after MI when these responses were declining in controls (Fig. 6 E). These results demonstrate that clearance of apoptotic macrophages, inflammation resolution, and myofibroblast activation after MI were impaired by combined Hif1 and Hif2 deficiency in myeloid cells.
Figure 6.
Myeloid HIFs prevent cardiac rupture after MI. (A) Survival of mice with myeloid-specific deletion of both Hif1 and Hif2 (mHIF1/2−/−) or controls after MI. n = 6–7 mice/group pooled from two independent experiments. **, P < 0.01 by log-rank (Mantel–Cox) test. (B) Evidence of cardiac rupture in mHIF1/2−/− mice compared with controls after MI. Scale bars represent 100 µm (left panel) and 50 µm (right panel). n = 6–7 mice/group pooled from two independent experiments. ***, P < 0.001 by two-tailed, unpaired t test. (C) Absolute number of infarct-associated ⍺SMA+ myofibroblasts 3 d after MI in mice with myeloid-specific deletion of Hif1 (mHIF1−/−), Hif2 (mHIF2−/−), both Hif1 and Hif2 (mHIF1/2−/−), or controls. n = 4–5 mice/group pooled from two independent experiments. *, P < 0.05 by one-way ANOVA followed by Tukey’s test. (D) Cardiac macrophage apoptosis as measured by Annexin staining after MI. Black contour plots represent FMO staining controls. n = 8 mice/group pooled from three independent experiments. ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. (E) Infarct-associated cellular responses in mHIF1/2−/− mice or controls after MI. n = 4–9 mice/group pooled from more than three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. All data are presented as mean ± SEM.
To investigate the requirement for HIF-1α and HIF-2α in macrophage survival following ischemic injury, we measured survival of BMDMs from mHIF1/2−/− mice or controls following exposure to hypoxia (1% O2). Despite similar levels of cell death during normoxia, hypoxia exposure increased cell death in BMDMs from mHIF1/2−/− mice compared with controls (Fig. 7 A). However, this effect was not observed with BMDMs deficient in either Hif1 or Hif2 alone (Fig. 7 A), so we performed additional experiments to determine how loss of both HIFs increased hypoxic BMDM cell death. Hypoxia increases production of mitochondrial reactive oxygen species (mROS; Chandel et al., 1998), which can trigger necrotic cell death in the ischemic myocardium (Luedde et al., 2014). Following hypoxia exposure, increased mROS levels were observed in BMDMs from mHIF1/2−/− mice, but not controls, and elimination of mROS using a superoxide scavenger attenuated hypoxic cell death in BMDMs (Fig. 7, B and C), linking increased mROS to hypoxic cell death in mHIF1/2−/− BMDMs. mROS promote necroptotic signaling leading to necroptosis (Schenk and Fulda, 2015), and this was evident as increased gene expression of necroptosis effectors, such as receptor-interacting protein kinase 1 (Ripk1), in hypoxic mHIF1/2−/− BMDMs compared with controls (Fig. 7 D). To determine whether hypoxia-induced necroptosis was responsible for BMDM cell death, we blocked necroptosis using necrostatin-1, a specific inhibitor of RIPK1. Inhibition of necroptosis abrogated hypoxic cell death in mHIF1/2−/− BMDMs compared with controls (Fig. 7 E). To test whether hypoxia-induced necroptosis of macrophages contributed to cardiac rupture in mHIF1/2−/− mice, we administered necrostatin-1 or vehicle and measured survival and LV remodeling after MI between mHIF1/2+/+ and mHIF1/2−/− mice. Compared with vehicle-treated mHIF1/2−/− mice, administration of necrostatin-1 to mHIF1/2−/− mice significantly improved survival (Fig. 7 F) and reduced infarcts sizes to levels comparable with mHIF1/2+/+ mice (Fig. 7 G). Thus, in contrast to their isoform-specific roles, HIF-1α and HIF-2α work together to suppress mROS and inhibit macrophage necroptosis. This may have contributed to the cardioprotective response mediated by myeloid cell–derived HIF isoforms, which was necessary to prevent cardiac rupture.
Figure 7.
HIFs temper hypoxic production of mROS to inhibit macrophage necroptosis.(A) Cell death as measured by Zombie Aqua positivity of BMDMs from mice with myeloid-specific deletion of Hif1 (mHIF1−/−), Hif2 (mHIF2−/−), both Hif1 and Hif2 (mHIF1/2−/−), or controls cultured under normoxia (21% O2) or hypoxia (1% O2). n = 3 sets of cells/group. Data are representative of three independent experiments. *, P < 0.05 by one-way ANOVA followed by Tukey’s test. (B) Levels of mROS as measured by MitoSOX in BMDMs cultured under normoxia, hypoxia, or hypoxia with MitoTEMPO (MT). (C) Cell death of MitoTEMPO-treated BMDMs cultured under hypoxia. For B and C, n = 3 sets of cells/group and are representative of two independent experiments. *, P < 0.05; **, P < 0.01 by one-way ANOVA followed by Tukey’s test. (D) Necroptosis gene expression in BMDMs cultured under normoxia or hypoxia. n = 3 sets of cells/group. Data represent two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. (E) Cell death of vehicle (Veh)– or necrostatin-1 (Nec-1)–treated BMDMs cultured under hypoxia. n = 3 sets of cells/group. Data represent two independent experiments. ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. (F) Survival of mice treated with vehicle or necrostatin-1 after MI. n = 3–8 mice/group pooled from two independent experiments. *, P < 0.05 by log-rank (Mantel–Cox) test. (G) Percent infarct/LV, percent AAR/LV, percent infarct/AAR measured 7 d after MI in mice treated with vehicle or necrostatin-1. n = 3–5 mice/group pooled from two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA followed by Tukey’s test. All data are presented as mean ± SEM.
Discussion
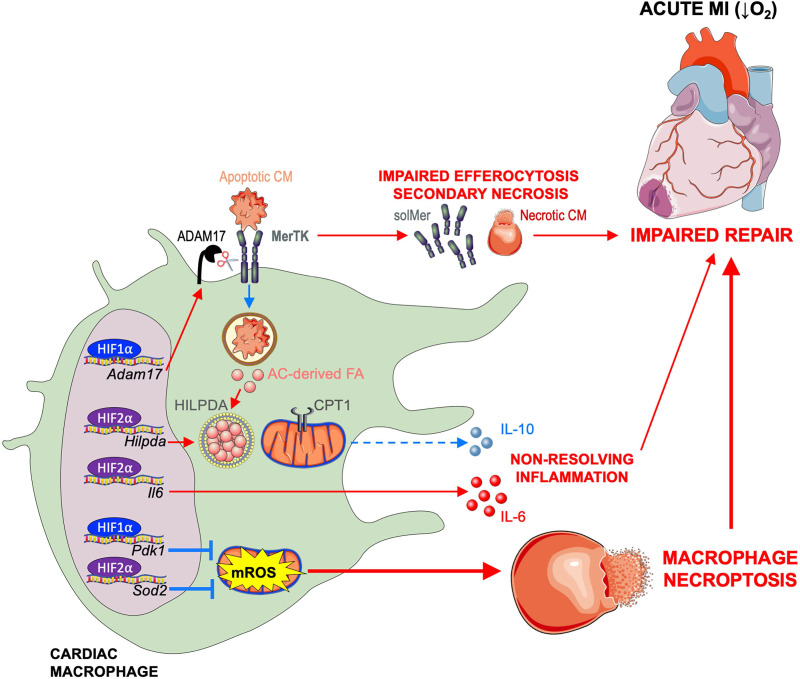
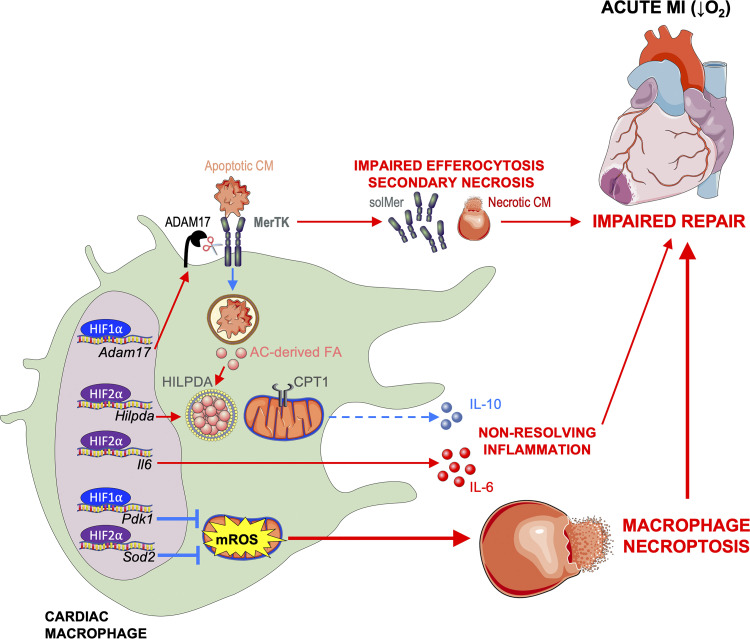
Taken together, our data reveal a new role for HIF-2α in cardiac repair. This occurs through anti-inflammatory metabolic reprogramming and through a mechanism that is distinct from HIF-1α. HIF-2α activated genes involved in fatty acid synthesis and storage, including Hilpda, to antagonize CPT1-dependent FAO of AC-derived fatty acids and impair anti-inflammatory production of IL-10 by efferocytic macrophages. In contrast to HIF-2α, HIF-1α mediated glycolytic reprogramming in macrophages to promote proteolytic cleavage of phagocytic receptor MerTK and impair clearance of dying cells and the initiation of inflammation resolution. Despite their distinct proinflammatory roles, these HIFs together were required for cardiac macrophage survival after MI, as loss of both HIF-1α and HIF-2α increased macrophage cell death, leading to cardiac rupture and impaired survival. These isoform-independent roles counteracted a combinatorial HIF-1α and HIF-2α cardioprotective response, as depicted in our working model (Fig. S4).
Figure S4.
Working model. MI causes ischemia and release of danger signals from dying cells, leading to sequential activation of HIF-2α and then HIF-1α in cardiac macrophages. HIF-2α activates genes involved in fatty acid (FA) synthesis and storage, including HILPDA protein, to antagonize mitochondrial oxidation of FA derived from ACs and anti-inflammatory production of IL-10 by macrophages. HIF-2α also activates inflammatory genes, including Il6, to worsen cardiac repair. In contrast, HIF-1α functions independent of HIF-2α to promote ADAM17-mediated cleavage of MerTK. This results in impaired efferocytosis of apoptotic cardiomyocytes (CM), leading to secondary necrosis and infarct expansion. The cumulative effect is adverse ventricular remodeling and progression to heart failure. Despite their distinct proinflammatory roles, HIFs are also required for cardiac macrophage survival after MI. Loss of both HIF-1α and HIF-2α increases mROS, likely due to reduced expression of antioxidant genes (Pdk1 and Sod2), leading to macrophage necroptosis, impaired scar formation, cardiac rupture, and ultimately, death.
HIF expression was first detectable in cardiac macrophages within days after the induction of ischemic injury in both humans and mice. HIF-2α levels increased before HIF-1α in cardiac macrophages, which may be of significance to early interventional strategies. This was somewhat surprising given an expectation that HIF-1α would be stabilized first as macrophage metabolism shifted to ischemia-associated glycolysis (Kim et al., 2006a). However, prolyl hydroxylation of HIF-2α is less efficient than HIF-1α, which may lead to its earlier stabilization and activation (Koivunen et al., 2004). Interestingly, expression of both HIF-1α and HIF-2α was still detectable in infarct-associated macrophages 30 d after MI, a time point that is considered equivalent to human heart failure. Similarly, HIF expression and signaling was detectable in cardiac macrophages from the explanted hearts in patients with end-stage heart failure, raising the possibility that macrophage expression of HIFs may drive the progression to heart failure. Pathway analyses of human cardiac macrophages revealed up-regulation of lipid storage and cellular responses to ROS, consistent with our findings that HIF-2α mediates retention of AC-derived fatty acids in lipid droplets and that both HIFs confer protection from the cytotoxic effects of hypoxic mROS, respectively. While our findings reveal how myeloid HIFs regulate the initial inflammatory and reparative macrophage responses after the acute ischemic injury, this only sets the trajectory for disease progression (Dick and Epelman, 2016), and the role for myeloid HIFs after chronic ischemic cardiomyopathy is established, which represents the majority of patients, will be an important area of future investigation.
Metabolic reprogramming is linked to macrophage polarization and is instrumental to their function during homeostasis, tissue injury, and host defense (Thorp, 2021). Similar to previous observations during infection (Tannahill et al., 2013), we found that TLR4 signaling activated a HIF-1α–dependent switch to glycolytic metabolism to facilitate the increased energetic demands of macrophage inflammatory activation. In contrast, we previously demonstrated that tissue reparative macrophages require mitochondrial metabolism for anti-inflammatory IL-10 production after MI (Zhang et al., 2019), Here, we found that HIF-2α suppressed this efferocytosis-induced FAO and mitochondrial metabolism to block the anti-inflammatory potential of macrophages after MI. By suppressing mitochondrial metabolism, HIF-2α indirectly cooperated with HIF-1α to promote a switch to glycolytic metabolism to fuel a proinflammatory, maladaptive response within the infarcted heart. Mechanistically, HIF-2α positively regulated macrophage expression of Hilpda, which promoted retention of AC-derived fatty acids within lipid droplets and prevented CPT1-dependent mitochondrial β-oxidation. Deficiency of Hilpda alone does not alter inflammatory gene expression in macrophages (van Dierendonck et al., 2020), suggesting that additional signals are required for anti-inflammatory IL-10 production by efferocytic macrophages. We found that HIF-2α was also associated with decreased expression of genes involved in FA synthesis and storage and inhibition of ACC, which catalyzes the first committed step of FA synthesis, enhanced IL-10 production by efferocytic macrophages. This necessitates future studies using targeted lipidomics to track the fate of AC-derived FAs within the efferocyte. Separately, HIF-2α suppresses FAO and mitochondrial metabolism in hepatocytes (Rankin et al., 2009) and intestinal HIF-2α contributes to systemic lipid synthesis and storage (Xie et al., 2017), suggesting that HIF-2ɑ regulation of metabolic programming is a conserved mechanism that will be translatable to other inflammatory conditions as well.
HIF-1α has been implicated in macrophage host defense against pathogens (Cheng et al., 2014), which may explain the myeloid isoform-specific maladaptive roles we observed for HIFs in sterile ischemic injury following MI. Recognition of pathogen-associated molecular patterns, including LPS, by germline-encoded pattern recognition receptors, triggers HIF-1α and HIF-2α signaling to initiate proinflammatory macrophage responses (Tannahill et al., 2013). Similar to pathogen-associated molecular patterns, damage-associated molecular patterns released by necrotic cardiomyocytes are sensed by pattern recognition receptors, leading to macrophage inflammatory responses after MI. While both sterile ischemic injury and infection trigger inflammation, the physiological purpose of the coordinated response by HIF-1α and HIF-2α to suppress pre- and postengulfment efferocytic signaling, respectively, and prevent inflammation resolution may have evolved to ensure pathogen clearance before the onset of tissue reparative processes. This could be explained by the cost of collateral tissue damage being outweighed by pathogen persistence and, ultimately, host death. During infection, impaired clearance of dying cells leads to secondary necrosis, which may limit pathogen replication in infected cells (Nailwal and Chan, 2019). In contrast, impaired clearance of dying cardiomyocytes following HIF-1α activation in myeloid cells contributed to infarct expansion after MI. HIF-1α was found to positively regulate Adam17 expression in macrophages, which supports recent observations of Adam17 as a direct transcriptional target gene of HIF-1α (Lian et al., 2019). This was associated with HIF-1α–dependent glycolytic reprogramming in macrophages leading to MerTK cleavage and impaired efferocytosis. Impaired clearance of dying cells has also been linked to autoimmune disease (Rothlin and Lemke, 2010), revealing a potential link between macrophage HIF-1α activation and autoimmunity. For example, hyperglycemia in type 1 diabetes patients is associated with an inflammatory state and increased cardiac autoimmunity (Sousa et al., 2020), which is initiated in part through myeloid cell function (DeBerge et al., 2020). Additional studies are needed to determine whether poor glycemic control favors HIF-1α–dependent inflammatory activation in cardiac macrophages, impairing clearance of apoptotic cardiomyocytes and leading to processing and presentation of cardiac self-antigens for the initiation of postinfarction autoimmunity.
Despite their distinct proinflammatory roles, HIF-1α and HIF-2α together were required to inhibit necroptotic cell death of macrophages during hypoxic stress, which may have contributed to the increase in infarct size, ventricular wall rupture, and premature death after MI in mice with combined loss of HIF-1α and HIF-2α in myeloid cells. Combined loss of HIF-1α and HIF-2α in macrophages was associated with elevated mROS during hypoxic stress, which impaired macrophage survival. Isoform-specific HIF regulation of antioxidant genes, such as pyruvate dehydrogenase kinase by HIF-1α (Kim et al., 2006a) and superoxide dismutase 2 by HIF-2α (Lian et al., 2019), likely explain the absence of increased macrophage death with loss of either HIF-1α or HIF-2α alone. Interestingly, Hif1 deficiency was associated with increased cardiac macrophage survival after MI, consistent with the proapoptotic role observed for HIF-1α activation in macrophages during atherosclerosis (Aarup et al., 2016). Preservation of cell surface MerTK may have contributed to enhanced survival of Hif1-deficient macrophages, as MerTK activates a protective response to oxidative stress in efferocytic macrophages (Lantz et al., 2020). In macrophages with combined loss of HIF-1α and HIF-2α, the increase in mROS during hypoxic stress led to increased necroptotic cell death, which was abrogated using necrostatin-1, a RIPK1-specific inhibitor of necroptosis. Systemic administration of necrostatin-1 to mice with combined loss of HIF-1α and HIF-2α in myeloid cells rescued the survival deficit after MI. Treatment of mice with necrostatin-1 also reduced infarct sizes, which was expected as necrostatin-1 inhibits both macrophage and nonmacrophage cell death, including cultured cardiomyocytes (Smith et al., 2007), leading to reduced infarct sizes after MI (Koudstaal et al., 2015). Importantly, necrostatin-1 blocked the increase in infarct sizes observed in mice with combined loss of HIF-1α and HIF-2α in myeloid cells and reduced infarcts to levels indistinguishable from controls, consistent with the premise that myeloid cell death was a significant contributor to infarct size and survival after MI. Necroptosis proteins, including RIP1K and activated MLKL, are increased in the hearts of patients with ischemic cardiomyopathy compared with controls (Szobi et al., 2017), suggesting that targeted inhibition of necroptosis during heart failure may improve outcomes in part through preserving cardiac macrophage survival.
In a therapeutic context, our findings suggest that strategies that nonspecifically increase HIF activation in the ischemic heart may fail to achieve full efficacy. Isoform-specific HIF antagonists are in clinical development for the treatment of cancer and have demonstrated favorable safety profiles and activity in patients (Courtney et al., 2018), supporting the therapeutic potential of inhibiting HIFs after MI. Targeted delivery of therapeutic cargo to cardiac macrophages would circumvent the differential role of HIFs in different myocardial cell populations. Selective targeting of either HIF-1α or HIF-2α would block isoform-specific proinflammatory activation of macrophages while also preserving isoform-specific antioxidant pathways that are necessary to prevent accumulation of hypoxic mROS and macrophage necroptotic death. This could reduce inflammation and adverse ventricular remodeling after MI to limit the progression to heart failure.
A limitation of our findings in the heart is that LysM-Cre is expressed in other myeloid cells, including neutrophils and Ly6Chi monocytes. Similar to macrophages, some subsets of neutrophils also constitutively express HIF-2α (Thompson et al., 2014). LysM-Cre deletion of Hif1 or Hif2 in myeloid cells did not affect initial cardiac Ly6Chi monocyte or neutrophil accumulation, consistent with a recent report that HIFs are dispensable for myeloid cell migration during sterile inflammation (Gardner et al., 2017). However, we found that neutrophil abundance was significantly reduced by 3 d after MI with Hif2 deficiency, and this was associated with an increase in neutrophil apoptosis. Neutrophil levels of HIF-2α have been shown to be unaffected by hypoxia or inflammatory stimuli, and loss of HIF-2α does not affect neutrophil function or expression of apoptosis regulators (Thompson et al., 2014), suggesting that the changes we observed in neutrophil survival may be secondary to the cell-intrinsic effects of HIF-2α that we found in efferocytic macrophages. Efferocytosis of apoptotic neutrophils by macrophages and resolution of neutrophilic inflammation limit the extent of tissue injury, so these findings are in line with a pathological role for myeloid cell HIF-2α in cardiac repair after MI. Additional studies using Cre driver mice that specifically target Hif2 expression in either neutrophils or macrophages are needed to dissect their precise roles in inflammation resolution after MI. HIFs also regulate glycolysis in neutrophils (Sadiku et al., 2017), while mitochondrial metabolism has been implicated in neutrophil differentiation (Riffelmacher et al., 2017), warranting future studies of HIFs in neutrophil metabolism.
In conclusion, our findings newly inform that myeloid HIF-2α aggravates cardiac repair by suppressing anti-inflammatory metabolic reprogramming and that the mechanism is distinct from HIF-1α; the latter suppresses apoptotic cell receptors that are necessary for dead cell clearance during tissue injury. Although deficiency of either macrophage HIF-1α or HIF-2α in isolation improves cardiac repair, blockade of both isoforms in combination contributes to the opposite and catastrophic result of cardiac rupture. This is because both isoforms are required together in order to protect from hypoxia-induced necroptosis. These findings support the contention that while macrophage HIF activation is critical to tissue repair, optimal therapeutic leveraging of HIFs requires strategies that employ isoform-specific targeting.
Materials and methods
Human ischemic cardiomyopathy specimens
This study was approved by the institutional review board at Northwestern University (#STU00012288) and performed in accordance with the Helsinki Doctrine on Human Experimentation. Written consent was obtained from all study participation. Cardiac tissue specimens were obtained from the explanted hearts of adult patients with ischemic cardiomyopathy undergoing cardiac transplantation at Northwestern Memorial Hospital. For single-cell RNA-sequencing analyses, the patient was a 57-yr-old male with no LV assist device and heart failure duration of 8.75 yr. For flow cytometry analyses, the patient was a 69-yr-old male with an LV assist device and heart failure duration of 1.5 yr. Explanted hearts were immediately immersed and rinsed with cold cardioplegia solution. Tissue specimens from infarct scar or viable remote (absence of fibrosis) myocardium were obtained from the lateral wall of the LV. Specimens were maintained in cold cardioplegia solution to preserve tissue integrity. Within 1 h of procurement, specimens were digested with collagenase type II (600 U/ml; Worthington) and DNase I (0.1 mg/ml; Sigma-Aldrich) in HBSS at 37°C for 30 min with agitation. Specimens were subsequently triturated through a 40-µm cell strainer to prepare a single-cell suspension for downstream analyses by single-cell RNA sequencing or flow cytometry.
Single-cell RNA-sequencing analysis
Single-cell suspensions were processed using the 10x Genomics Chromium System. To determine the quality and quantity of the single-cell 5′ gene expression library, the Illumina library was run on the Agilent Bioanalyzer High Sensitivity Chip and Kapa DNA Quantification kit. In collaboration with the Northwestern University Sequencing Core, the library was sequenced on the Illumina HiSeq 4000 with the following parameters: read 1, 26 cycles; index 1, 8 cycles; read 2, 98 cycles. The sequenced data were processed into expression matrices via the 10x Genomics Cell Ranger Single Cell software suite v2.2. The raw base call files were demultiplexed into FASTQ files and then aligned to the human transcriptome (GRCh38-3.0.0). Further filtering of cell barcodes and unique molecular identifiers was performed to build a transcript count table.
Filtering and normalization
Downstream analyses were performed in R using the packages Seurat (v4.0.0; Butler et al., 2018) and sctransform (v0.3.2; Hafemeister and Satija, 2019). To filter out cells with low-quality data and probable doublets, cells that expressed <400 features per 1,000 unique molecular identifiers and with >5,000 features were removed, respectively. Furthermore, features that were not detected in at least three single cells were filtered. To remove probable ACs, cells with greater than 40% mitochondrial gene content were also excluded. With these criteria in place, our dataset consisted of 1,150 single cells with 15,716 features. To reduce the effect of technical variation on confounding biological heterogeneity, normalization was performed using a modeling framework in sctransform.
Dimensionality reduction and clustering
The expression matrix underwent dimensionality reduction using principal-component analysis, which was performed on features selected from the FindVariableFeatures function. Using dimensional heatmaps, a jackstraw plot, and an elbow plot, the number of significant principal components was determined for downstream clustering and analyses. Clusters were determined by the FindCluster function in Seurat, which applies a graph-based clustering approach. This approach embeds cells into a K-nearest neighbor graph based on the Euclidean distance with edges drawn between cells with similar feature expression. Then, to cluster the cells, the Louvain algorithm (Blondel et al., 2008) was applied to iteratively group the cells together based on optimizing the modularity function. Finally, Uniform Manifold Approximation and Projection for Dimension Reduction was used to reduce the dimensionality of the dataset and for visualization of the data.
Cluster identification and pathway enrichment analysis
Differentially expressed features were determined using the FindAllMarkers function in Seurat. A comparison of these features with known markers for fibroblasts, vascular smooth muscle cells, neurons, myocytes, endothelial cells, pericytes, and lymphocytes determined the identification of each cluster. Consequently, further downstream analysis was completed with merged clusters to compare the myeloid cluster. To determine differentially expressed features in the myeloid cluster, the FindMarkers function was used. Features that had an adjusted P value less than 0.05 and had at least a twofold change were determined to be differentially expressed. gProfiler (Reimand et al., 2016) was employed to determine enriched pathways based on differentially expressed genes between myeloid cells and nonmyeloid cells. Gene sets from the gene ontology molecular function and gene ontology biological process were used. Processed single-cell RNA-sequencing data have been uploaded to the Gene Expression Omnibus (accession no. GSE180678).
Immunofluorescence of human acute MI
Formalin-fixed and paraffin-embedded sections were prepared from LV myocardial tissue specimens obtained from patients who died 5–14 d after acute MI or patients with non–MI-related deaths as previously described (van der Laan et al., 2014). Sections were deparaffinized and rehydrated and then incubated in methanol with 0.3% H2O2 for 10 min to block endogenous peroxidases. Antigen retrieval was performed by heating sections for 20 min at 125°C and 22 psi in citrate buffer, pH 6.0 (for HIF-1α), or Tris-EDTA buffer, pH 9.0 (for HIF-2α), using a Decloaking Chamber (Biocare Medical). Aldehyde reduction was then performed by washing sections in deionized water with 1% NaBH4. Sections were blocked for 1 h at room temperature in TBS with 5% normal goat serum and 0.3% Triton X-100. Primary antibodies for CD68 (1:100; Agilent), HIF-1α (1:100; Novus), and HIF-2α (1:100; Novus) were diluted in blocking buffer and incubated overnight at 4°C in a humidified stain tray (IHC World). Sections were washed extensively in TBS-Tween and incubated with Alexa Fluor 488 goat anti-rabbit (1:100) and Alexa Fluor 647 goat anti-mouse (1:100) secondary antibodies diluted in TBS with 1% BSA and 0.3% Triton X-100 for 1 h at room temperature in a dark, humidified stain tray. Sections were washed extensively in TBS-Tween and autofluorescence was quenched by incubating the sections with 1X TrueBlack in 70% ethanol for 1 min. Sections were rinsed with TBS, and sections were mounted with a coverslip using VECTASHIELD antifade mounting medium with DAPI. Sections were imaged at 20× magnification using a SLIDEVIEW VS200 (Olympus). Brightness and contrast were equally adjusted on all representative images using QuPath. Quantification of HIF-1α or HIF-2α mean fluorescence intensity of CD68+ macrophages was performed using ImageJ (National Institutes of Health). The fluorescence signal in the CD68 (red) channel was subjected to a threshold to remove background fluorescent and segment individual particles. Only signal within the CD68+ macrophage cell body was scored. Data represent the analysis of at least three fields per patient sample (more than 100 cells per patient sample).
Mice
LysM-Cre (B6.129P2-Lyz2tm1(cre)Ifo/J, stock no. 004781), HIF1flox/flox (B6.129-Hif1atm3Rsjo/J, stock no. 007561), HIF2flox/flox (Epas1tm1Mcs/J, stock no. 008407), and αMHC-mCherry (Tg(Myh6*-mCherry)2Mik, stock no. 021577) mice were purchased from The Jackson Laboratory and backcrossed to wild-type C57BL/6J mice for 6–12 generations. To generate mice with myeloid lineage–specific knockout of HIF-1α (mHIF1−/−) or HIF-2α (mHIF2−/−), LysM-Cre mice were bred with HIF1flox/flox or HIF2flox/flox mice, respectively. mHIF1−/− and mHIF2−/− mice were crossed to generate mice with combined loss of HIF-1α and HIF-2α in myeloid cells (mHIF1/2−/−). C57BL/6J mice were bred in our animal facility before use. A priori experiments revealed no difference in infarct sizes among LysM-Cre, HIF1flox/flox, HIF2flox/flox, and C57BL/6J mice (Fig. S2 B), so C57BL/6J mice were used as wild-type controls. Mice with myeloid-specific overexpression of HIF-1α (mHIF1LSL) or HIF-2α (mHIF2LSL) were generously provided by Yatrik Shah (University of Michigan, Ann Arbor, MI; Xie et al., 2014). Mice were housed in temperature- and humidity-controlled, pathogen-free environments and kept on a 14:10-h day/night cycle with access to standard mouse chow and water ad libitum. 2–4-mo-old female mice were used for experiments. Animal studies were conducted in accordance with guidelines using a protocol approved by the Institutional Animal Care and Use Committee at Northwestern University.
MI surgery
Permanent occlusion MI surgeries were performed on female mice aged 2–4 mo as previously described (Wan et al., 2013). In brief, mice were anesthetized with avertin (2,2,2-tribromoethanol, 99%, 0.1 mg/kg i.p.; Alfa Aesar) and received sustained-release buprenorphine (0.1 mg/kg s.c.; ZooPharm) before the first incision. Puralube Vet Ointment (Dechra) was applied to the eyes, and mice were secured in a supine position, endotracheal intubated, and ventilated with an Inspira Advanced Safety Single Animal Pressure/Volume Controlled Ventilator (Harvard Apparatus). Animals were maintained at 37°C using a far-infrared warming pad (Kent Scientific). The chest wall was shaved, depilated with Nair, and sterilized with povidone iodide and alcohol prep pads. Using a Leica S4E dissecting microscope and ACE Light Source (Schott), a left thoracotomy was performed with the aid of a Geiger Thermal Cautery Unit (Delasco) to maintain normal hemostasis. The LV was visualized and the proximal left anterior descending coronary artery was temporarily ligated with Surgipro II 7–0 monofilament polypropylene sutures (Covidien) ∼2 mm distal to the site of its emergence from under the left atrium. Blanching/pale discoloration and hypokinesis of the anterior wall verified ligation. Using Surgipro II 6–0 monofilament polypropylene sutures (Covidien), the surgical site was closed in layers starting with the chest wall followed by the pectoral muscle and finally skin and subcutaneous tissue. Animals were allowed to recover on a heating pad (Sunbeam) before being returned to cages. For necroptosis inhibition, mice received necrostatin-1 (5 mg/kg s.c.) or vehicle control daily beginning 24 h after MI. Mice dying within 48 h of MI surgery were treated as technical errors and excluded from analyses.
LV infarct and AAR measurements
Mice were anesthetized with avertin (0.1 mg/kg i.p.), secured in a supine position, endotracheal intubated, and ventilated with an Inspira Advanced Safety Single Animal Pressure/Volume Controlled Ventilator (Harvard Apparatus). With the aid of a dissecting microscope, the thoracic cavity was carefully opened to maintain normal hemostasis and expose the heart. Using an insulin syringe with 30-G needle, 100 µl FluoSpheres Polystyrene Microspheres (10 µm, red fluorescent 580/605) was injected into the LV. Hearts were excised 1 min later and sectioned into 1-mm coronal slices using a Mouse Heart Slicer Matrix (Zivic Instruments). Infarct and viable myocardium were visualized by staining the slices in 1.5-ml microfuge tubes with 1% 2,3,5-triphenyltetrazolium chloride in saline for 5 min at 37°C and then fixing in 10% buffered formalin phosphate for 1 h at 4°C. Slices were placed directly on an Epson Perfection V600 Photo scanner and scanned to generate digital infarct images. Brightness and contrast were equally adjusted on all images using Adobe Photoshop CC 2018 to aid infarct visualization. AAR was visualized by placing the slices under an Olympus IX51 fluorescent scope and imaging the slices with Olympus cellSens Imaging Software. Both infarct and AAR were measured as a percentage of the LV using ImageJ. Infarct size, expressed as a percentage of AAR, was calculated by dividing the sum of infarct areas from all sections by the sum of AAR from all sections and multiplying by 100.
Echocardiography
Cardiac function was assessed by transthoracic echocardiography on mice anesthetized with isoflurane before and 3–4 wk after MI using a Vevo 770 equipped with a 25-MHz probe (VisualSonics). Heart rate was maintained above 400 beats/min to ensure physiologically relevant measurements in all anesthetized animals. Parasternal short-axis images were acquired using M-mode 1 mm before, at, and after the papillary muscles. Image analysis was performed using Vevo LAB software (VisualSonics). Measurements of LV internal dimension at both end systole and end diastole were made in three consecutive cardiac cycles and averaged for analysis.
Hypoxyprobe and HIF staining
To detect hypoxia and HIF expression in mouse cardiac macrophages, mice were injected with pimonidazole (60 mg/kg i.v.; Hypoxyprobe) in PBS. Pimonidazole forms stable protein adducts under hypoxic conditions that are readily detectable using a fluorescently conjugated antibody. Mice were euthanized by CO2 asphyxiation 90 min later, and hearts were flushed with 20 ml ice-cold PBS. Cells were processed into single-cell suspensions and stained with surface antibodies as described below in tissue isolation and flow cytometry sections, respectively. For intracellular staining, cells were fixed and permeabilized using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences). After fixation/permeabilization, cells were first incubated with rabbit anti-mouse HIF-1α or HIF-2α antibodies (1:100) in BD Perm/Wash buffer for 30 min on ice. Cells were washed twice with BD Perm/Wash buffer and then incubated with Alexa Fluor 647 donkey anti-rabbit IgG (1:200) and FITC anti-pimonidazole IgG1 (1:100; Hypoxyprobe) antibodies in BD Perm/Wash buffer for 30 min on ice. Cells were washed twice in FACS buffer and stored in the dark at 4°C until flow cytometric analyses. Fluorescence minus one (FMO) was used as a negative staining control.
Tissue isolation
Mice were euthanized by CO2 asphyxiation, and hearts were extensively flushed through the LV with a volume of 20 ml ice-cold PBS to remove peripheral cells. Infarcted myocardium distal to the ligature was then excised, weighed, and transferred to a 1.5-ml microfuge tube. Heart tissue was then minced in 1 ml DMEM with 600 U/ml collagenase type 2, 60 U/ml hyaluronidase, and 60 U/ml DNase I and rotated on a LabQuake rotisserie shaker (ThermoFisher) at 37°C for 30 min. After digestion, the sample was carefully pipetted five times with a P1000 pipette to disrupt any remaining tissue and transferred through a 40-µm cell strainer to prepare a single-cell suspension. Cardiac cells were collected by centrifuging at 2,000 rpm for 5 min at 4°C, followed by red blood cell lysis and resuspension in ice-cold FACS buffer (1× PBS supplemented with 2% FBS and 2 mM EDTA). Viable cells were counted on a hemocytometer with trypan blue exclusion to determine the total number of viable cells for heart (cells/mg of tissue).
Flow cytometry
Single cells were resuspended in a volume of 200 µl FACS buffer and transferred to either 1.5-ml microfuge tubes or 96-well plates. Dead cells were labeled by incubating samples with Zombie Aqua Fixable Dye (1:1,000; BioLegend) in PBS for 15 min at room temperature in the dark. Cells were washed in FACS buffer, and then Fc receptors were blocked using TruStain FcX antibody (1:100) in FACS buffer for 15 min on ice. Cells were incubated with primary antibodies (1:200 dilution for mouse or the manufacturer’s recommended volume for human) in 100 µl FACS buffer for 20 min on ice in the dark. For secondary antibody staining of MerTK, cells were first labeled with a biotin-conjugated primary antibody (1:200), washed twice in FACS buffer, and then incubated with Alexa Fluor 647–streptavidin (1:200) in FACS buffer for 20 min on ice in the dark. For mouse myofibroblast staining, cells were surface stained with PerCP/Cy5.5-CD45 and APC-PDGFRα antibody, fixed and permeabilized using BD’s Cytofix/Cytoperm kit, and then incubated with Alexa Fluor 488–α-smooth muscle actin (αSMA) antibody in BD Perm/Wash buffer for 30 min on ice. After staining, cells were washed twice in FACS buffer, fixed with 1% paraformaldehyde in FACS buffer for 1 h, washed twice in FACS buffer, and stored in the dark at 4°C before analyses. All flow cytometric analyses were performed on either a FACS Canto II or LSRFortessa X-20 Cell Analyzer (BD Biosciences). BD Compbeads (BD Biosciences) were used to optimize fluorescence compensation settings to enable multicolor flow cytometric analyses. FMO and gene-deleted mice or cells were used as staining controls. All cells were pregated on live (live-dead exclusion), single cells (forward scatter area versus forward scatter height and side scatter area versus side scatter width). For human, cardiac macrophages and peripheral blood monocytes were identified as CD14+CD64+ cells. For mouse, neutrophils were identified as CD11b+Ly6G+, Ly6Chi; monocytes were identified as CD11b+Ly6G−F4/80loLy6Chi; macrophages were identified as CD11b+Ly6G−F4/80hiLy6CloCD64+MerTK+ and distinguished by MHCII expression; and myofibroblasts were identified as CD45−αSMA+PDGFRα+ cells. Data were analyzed on FlowJo software (Tree Star), and full gating strategies are depicted in figures.
In vivo efferocytosis assay
4–6-week-old αMHC-mCherry recipient mice, which specifically express mCherry protein in cardiomyocytes, were lethally irradiated with 1,000 rads from a cesium source before transplantation. Bone marrow was harvested from the tibia of mHIF1+/+ or mHIF1−/− mice by cutting both ends at the metaphysis and flushing the bone with a volume of 5 ml ice-cold PBS. Bone marrow cells were centrifuged at 1,500 rpm for 5 min at 4°C, and red blood cells were lysed using RBC lysis buffer (BioLegend). A total of 5 × 106 bone marrow cells from mHIF1+/+ or mHIF1−/− mice were injected into irradiated recipients by tail vein injection. After transplantation, mice were maintained on acidified drinking water containing neomycin. 6 wk after transplantation, mice were subjected to MI surgery. Heart tissue and cells were processed 5 d after injury as described above, and flow cytometric analyses were performed to identify CD64+ macrophages that were also positive for cardiomyocyte-derived mCherry protein.
Ex vivo respiratory analyses
To measure OCR in cardiac macrophages after MI, CD11b+ cells were magnetically selected from single-cell suspensions of enzymatically digested infarct/border zone tissue 3 d after MI using an EasySep Mouse CD11b positive selection kit II and EasyEights EasySep magnet (STEMCELL Technologies). Macrophages were subsequently enriched by removing nonadherent cells with sequential washes after 1 h of adherence to a Petri dish. Macrophages (5 × 105 cells/well) were seeded to a Cell-Tak–coated, XFe96 cell culture microplate. Experiments were conducted in prewarmed DMEM medium without phenol red, glucose, and sodium pyruvate and containing 2 mM L-glutamine, pH 7.4, using a Seahorse XFe96 analyzer (Agilent). After baseline measurements in triplicate, the following chemicals (all from Sigma-Aldrich) were injected in this order: (1) glucose (25 mM), (2) ATP-synthesis inhibitor oligomycin (1.5 mM), (3) sodium pyruvate (1 mM) and carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (1.5 mM) to uncouple ATP synthesis, and (4) rotenone (100 nM) to block complex I and antimycin A (1 mM) to block complex III. Measurements were taken in triplicate after the addition of each chemical. OCR analyses were generated by Wave Desktop software (Agilent).
Apoptosis detection
Cardiac single-cell suspensions were stained with Zombie Aqua Fixable Viability Dye (BioLegend) and surface antibodies as described above. Cells were washed with FACS buffer, resuspended in Annexin V binding buffer (BioLegend), and 106 cells in a volume of 100 µl were transferred to a 5-ml round-bottom polystyrene tube (Falcon). Cells were stained with 5 µl FITC–Annexin V for 15 min in the dark at room temperature. After staining, an additional 400 µl Annexin V binding buffer was added to the tube and cells were analyzed by flow cytometry. FMO (no FITC–Annexin V) controls are depicted as black contour plots and were used to set the Annexin V–positive gate.
BrdU proliferation
For macrophage proliferation after MI, mice were injected with 200 µl BrdU solution (2 mg/mouse, i.p.; BD Biosciences). Mice were euthanized 2 h later, and hearts were processed into single-cell suspensions. Cells were stained with surface antibodies and then fixed and permeabilized using buffers from the BD Pharmigen BrdU flow kit. Permeabilized cells were treated with DNase (300 µg/ml) in DPBS for 1 h at 37°C. Cells were washed and stained with APC-conjugated anti-BrdU antibody in BD Perm/Wash for 30 min at room temperature. Cells were washed and resuspended in FACS buffer for flow cytometric analyses. FMO (no APC-BrdU) were used to set the BrdU-positive gate.
BMDM culture
Bone marrow cells were harvested from tibia and femurs of mice by cutting the bone at both ends of the metaphysis and flushing cells from the bone with a volume of 5 ml ice-cold PBS. Red blood cells were removed by lysis and cells were filtered through a 40 µm cell strainer. Bone marrow cells were cultured in non–tissue culture Petri dishes (Fisherbrand) for 7 d with DMEM containing 20% L929-cell conditioned media, 10% fetal bovine serum, 1% penicillin-streptomycin, 1% sodium pyruvate, and 1% L-glutamine. A complete media change was performed on day 3 or 4 of culture. On day 7 of culture, media was removed, BMDMs were washed with PBS, and 1 ml of cold Cellstripper was added to the dish. After 5 min of incubation at room temperature, BMDMs were removed by gentle pipetting, washed with DMEM medium with 10% FBS, 1% penicillin/streptomycin, counted, and seeded to tissue culture–treated plates for experiments.
Peritoneal macrophage culture
To elicit peritoneal macrophages, mice received 1 ml sterile 3% Brewer’s thioglycolate medium by peritoneal injection. On day 3 after injection, mice were euthanized by CO2 asphyxiation, and macrophages were recovered from the peritoneal cavity by lavage with 5 ml ice-cold PBS. Peritoneal macrophages were subsequently enriched by removing nonadherent cells with sequential washes after 1 h of adherence to a Petri dish. Peritoneal macrophages were removed using Cellstripper, washed, and seeded to tissue culture–treated plates for experiments.
Hypoxia/ischemia treatment
BMDMs were seeded in a 24-well plate (Falcon) at a density of 5 × 105 cells/well in 500 µl DMEM containing 10% FBS and 1% penicillin-streptomycin and allowed to adhere overnight. Prior to hypoxia treatment, culture media was equilibrated to hypoxic conditions (1% O2, 94% N2, and 5% CO2) for 90 min in a temperature- and humidity-controlled Coy O2 Control In Vitro Glove Box (Coy Laboratory Products). BMDMs were then transferred into the hypoxic chamber and hypoxic culture media was added to the cells. BMDMs were exposed to hypoxia for 3 to 24 h. All assays on hypoxic BMDMs were performed within the hypoxic Coy O2 Control In Vitro Glove Box or immediately upon removal to minimize reoxygenation effects. To measure HIF activation, BMDMs were fixed and permeabilized using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences) and stained with antibodies for HIF-1α and HIF-2α as described above. To measure cell death, BMDMs were labeled with Zombie Aqua Fixable Viability Dye in the hypoxic chamber, fixed, and analyzed by flow cytometry. To detect mROS, BMDMs were loaded with 2.5 µM MitoSOX reagent (Invitrogen) in HBSS during the last 10 min of hypoxia exposure. BMDMs were then washed three times with HBSS and analyzed by flow cytometry. For reactive oxygen species scavenging, BMDMs were treated with 1 µM MitoTEMPO (Sigma-Aldrich) for the duration of hypoxia exposure. For necroptosis inhibition, BMDMs were treated with necrostatin-1 (50 µM) or vehicle for the duration of hypoxia exposure. BMDMs cultured in a temperature- and humidity-controlled tissue culture incubator supplied with room air (21% O2) and 5% CO2 were used as normoxic controls.
In vitro efferocytosis assay
To measure BMDM efferocytosis of apoptotic Jurkat T cells, BMDMs were labeled with MitoTracker Red (100 nM) in serum-free DMEM for 5 min and Jurkat T cells were labeled with Calcein AM (5 nM) in PBS for 15 min. For induction of apoptosis, Jurkat T cells were UV-treated (254 nM) with an EL Series Ultraviolet Hand Lamp (UVP) for 7 min and then incubated for 2 h. Adherent BMDMs were overlaid with ACs at a ratio of 5 ACs: 1 BMDM. Nonengulfed ACs were removed from adherent phagocytes 3 h after cocultivation by sequential washes. Engulfment was confirmed by microscopic analysis and the percent efferocytosis was calculated as the number of Calcein AM positive macrophages divided by the total number macrophages. Levels of IL-10 in cell culture supernatant was determined by ELISA (BD Biosciences) 3–12 h after efferocytosis. In some experiments, 20 µM TOFA or vehicle was added to BMDMs after washing off nonengulfed ACs to block postengulfment ACC function.
Gene knockdown
One day before transfection, BMDMs (2.5 × 105 cells/well) were seeded in a 24-well tissue culture plate and allowed to adhere overnight. Using HiPerFect transfection reagent (Qiagen) in Opti-MEM serum-free medium, BMDMs were treated with scrambled control or 10 nM of four preselected siRNAs (Qiagen) specific for Cpt1a or Hilpda for 24–48 h.
BODIPY–fatty acid trafficking
BMDMs (106 cells/dish) were seeded to 35-mm glass-bottom dishes (Cellvis) and allowed to adhere overnight. The next day, Jurkat T cells were labeled with 5 µM BODIPY FL C16 (4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-hexadecanoic acid) in PBS for 30 min at 37°C in a tissue culture incubator. Cells were centrifuged and washed three times with serum containing RPMI media to remove unbound BODIPY, counted, and irradiated to induce apoptosis. BODIPY-labeled, apoptotic Jurkat T cells were added to BMDMs at a ratio of five ACs to one BMDM. After 3 h of co-culture, nonengulfed ACs were removed by sequential washes and BMDMs were cultured for an additional 1 h. BMDMs were fixed and mounted with VECTASHIELD antifade mounting media with DAPI. Images were collected using a Plan Fluor 40x Oil objective (NA 1.3) on a Nikon Ti2 inverted Widefield fluorescence microscope. Each sample was imaged at least four times at randomly selected focal points. A four-step (5 micron per step) Z-series was acquired using identical settings for all samples to preserve relative differences. Image processing was performed using FIJI (ImageJ). A Z-projection for each series was generated using the default ImageJ Max Intensity function. Brightness and contrast were equally adjusted for each image and each channel. The number of fluorescent AC particles in each cell were quantified using the maximum intensity projections for image. The fluorescence signal in the BODIPY (green) channel was subjected to a threshold to remove background fluorescent and segment individual particles. Only signal within the BMDM cell body (identified using background auto fluorescence of the cell) was scored. Total cell number for each analyzed field was determined using the DAPI-stained nuclei. Data represent the analysis of at least four fields per sample (more than 100 cells per sample per experiment).
In vitro respiratory analyses
To measure OCR of efferocytic macrophages, 106 BMDMs were seeded to a tissue culture–treated 6-well plate (Falcon) and allowed to adhere overnight. The next day, apoptotic Jurkat T cells were added to BMDMs at a ratio of five ACs to one BMDM. Cells were co-cultured for 3 h, and nonengulfed ACs were removed by sequential washes with PBS. BMDMs were removed using Cellstripper, and viable cells were enumerated by counting cells on a Luna II automated cell counter (Logos Biosystems) with trypan blue exclusion. BMDMs (105 cells/well) were seeded to a Cell-Tak–coated, XFe96 cell culture microplate, centrifuged at 200 ×g for 1 min, and allowed to adhere in a 37°C, non-CO2 incubator for an additional 1 h. Experiments were conducted in prewarmed DMEM without phenol red, glucose, and sodium pyruvate and containing 2 mM L-glutamine, pH 7.4, using a Seahorse XFe96 analyzer (Agilent). After baseline measurements in triplicate, the following chemicals (all from Sigma-Aldrich) were injected in this order: (1) glucose (25 mM), (2) oligomycin (1.5 mM), (3) sodium pyruvate (1 mM) and carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (1.5 mM), and (4) rotenone (100 nM) and antimycin A (1 mM). To measure extracellular acidification rate (ECAR), BMDMs (105 cells/well) were seeded to a tissue culture–treated XFe96 cell culture microplate and allowed to adhere overnight. The next day, BMDMs were treated with LPS (10 ng/ml) for 3 h. Experiments were conducted in prewarmed DMEM medium without phenol red, glucose, and sodium pyruvate and containing 2 mM L-glutamine, pH 7.4, using a Seahorse XFe96 analyzer (Agilent). After baseline measurements in triplicate, the following chemicals (all from Sigma-Aldrich) were injected in this order: (1) glucose (25 mM), (2) oligomycin (1.5 mM), and (3) 2-DG (50 mM) to inhibit glycolysis. Measurements were taken in triplicate after the addition of each chemical. OCR and ECAR analyses were generated by Wave Desktop software (Agilent).
MerTK cleavage assay
For serum levels of solMER (Becker et al., 2021), mice were anesthetized with avertin (0.1 mg/kg i.p.), and whole blood was collected into 1.5-ml microfuge tubes by retroorbital bleeding with a heparinized capillary tube (Fisherbrand). Blood was allowed to clot at room temperature for 15 min, and the clot was removed by centrifuging at 5,000 rpm for 15 min at 4°C. Serum was stored at −80°C until analyses. For levels of solMER from BMDM cultures, BMDMs were plated at a density of 5 × 105 cells/well in a 24-well plate in 500 µl DMEM with 10% FBS and 1% penicillin/streptomycin. To induce MerTK cleavage, BMDMs were treated with LPS (100 ng/ml) for 1 h. Culture media was collected to assay for solMER by ELISA, and cells were harvested for MerTK cell surface expression analysis by flow cytometry. To block glycolysis, 2-DG (25 mM) was added or culture media was changed to DMEM without glucose 30 min before addition of LPS. Levels of solMER were analyzed in diluted serum (1:100) or undiluted culture supernatant using the Mouse Mer DuoSet ELISA (R&D Systems) according to the manufacturer’s instructions. Absorbances were read at 450 nm on an iMark Microplate Reader (Bio-Rad).
Quantitative real-time PCR
RNA was extracted from cardiac extracts or primary macrophages using Trizol (Invitrogen) according to the manufacturer’s instructions. In brief, 1 ml Trizol (Invitrogen) was added to LV myocardium, and tissue was homogenized on ice using a Tissue-Tearor homogenizer (Biospec) set at 35,000 rpm for 15 s. For BMDMs, 500 µl Trizol was added to 5 × 106 cells/well in a 24-well plate and gently pipetted to lyse the cells. RNA was measured using a NanoDrop spectrophotometer (ThermoFisher), and 1 µg of RNA was transcribed to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative PCR was performed using SYBR Green Master Mix (Bio-Rad) on a QuantStudio 3 Real-Time PCR System (Applied Biosystems). Results are expressed as ΔΔCt values normalized to β2m. Primers used for semiquantitative/real-time PCR were supplied by Integrated DNA Technologies and are as previously described (Xie et al., 2017, Lian et al., 2019, Teitz et al., 2013, van Dierendonck et al., 2020, Honarpisheh et al., 2016, Rankin et al., 2009, Thomas et al., 2016, Stanfield et al., 2021) and listed in Table S2.
Statistics
Statistical analyses were performed with GraphPad Prism 9 software (GraphPad Software). Comparisons between two groups were performed using a two-tailed, unpaired t test with a 95% confidence interval. For comparisons of more than two variables, a one-way or two-way ANOVA was used with a 95% confidence interval; when necessary, a Tukey test was used to correct for multiple comparisons. The statistical difference in animal survival after MI was determined by a log-rank (Mantel–Cox) test. For in vivo experiments, experimental sample size is indicated in figure and figure legends and represent pooled data from two or more independent experiments. For in vitro experiments, experimental sample size is indicated in figure and figure legends and are representative data from two or more independent experiments. All data are presented as mean ± SEM. Criteria for significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) are located in the figure legends. Analyses labeled “ns” are not statistically significant.
Online supplemental material
Fig. S1 shows flow cytometry of HIF-1α and HIF-2α expression in cardiac macrophages and single-cell RNA sequencing of infarct scar–associated cells during human ischemic cardiomyopathy. Fig. S2 shows LysM-Cre activity in cardiac cell populations, infarct sizes in control animals, and HIF-1α and HIF-2α expression in cardiac macrophages and BMDMs exposed to normoxia or hypoxia. Fig. S3 shows neutrophil apoptosis and macrophage proliferation after MI. Fig. S4 shows the working model of the role of HIF-1α and HIF-2α in macrophages after MI. Table S1 shows the top 20 differentially expressed genes in each cluster from the single-cell RNA sequencing of human ischemic cardiomyopathy. Table S2 shows the resources and reagents used in this study.
Supplementary Material
shows differential gene expression used to identify unique cell clusters during human ischemic cardiomyopathy.
lists the reagents and resources used in this study.
Acknowledgments
We thank Anna Huskin, RN, BSN, CCRC, Program Development Manager in the Clinical Trials Unit of the Bluhm Cardiovascular Institute, and Patrick McCarthy, MD.
This work was supported by the American Heart Association (grant CDA24110032 to M. DeBerge) and the National Institutes of Health (grant F32HL127958 to M. DeBerge and grant R01HL122309 to E.B. Thorp). These studies used sequencing services provided by NUSeq Core at Northwestern University. Imaging work was performed at the Northwestern University Center for Advanced Microscopy, generously supported by National Cancer Institute cancer center support grant P30CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. Publication of this research was supported by the Sidney and Bess Eisenberg Memorial Fund.
Author contributions: M. DeBerge and E.B. Thorp designed the research studies. M. DeBerge, S. Dehn, and D.P. Sullivan conducted murine experiments and acquired data. C. Lantz performed bioinformatic analyses of human single-cell RNA-sequencing data. A.M. van der Laan and H.W.M. Niessen obtained and prepared human acute MI specimens. M.E. Flanagan stained and analyzed human acute MI specimens. L.D. Wilsbacher obtained and processed the human ischemic cardiomyopathy specimens. All authors contributed to data analyses. M. DeBerge and E.B. Thorp wrote the manuscript, and all authors contributed to manuscript revision.
References
- A-Gonzalez, N., Quintana J.A., García-Silva S., Mazariegos M., González de la Aleja A., Nicolás-Ávila J.A., Walter W., Adrover J.M., Crainiciuc G., Kuchroo V.K., et al. 2017. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214:1281–1296. 10.1084/jem.20161375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarup, A., Pedersen T.X., Junker N., Christoffersen C., Bartels E.D., Madsen M., Nielsen C.H., and Nielsen L.B.. 2016. Hypoxia-Inducible Factor-1α Expression in Macrophages Promotes Development of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 36:1782–1790. 10.1161/ATVBAHA.116.307830 [DOI] [PubMed] [Google Scholar]
- Becker, A.C., Lantz C.W., Forbess J.M., Epting C.L., and Thorp E.B.. 2021. Cardiopulmonary Bypass-Induced Inflammation and Myocardial Ischemia and Reperfusion Injury Stimulates Accumulation of Soluble MER. Pediatr. Crit. Care Med. 10.1097/PCC.0000000000002725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel, V.D., Guillaume J.-L., Lambiotte R., and Lefebvre E.. 2008. Fast unfolding of communities in large networks. J. Stat. Mech. P10008:P10008. 10.1088/1742-5468/2008/10/P10008 [DOI] [Google Scholar]
- Bøtker, H.E., Kharbanda R., Schmidt M.R., Bøttcher M., Kaltoft A.K., Terkelsen C.J., Munk K., Andersen N.H., Hansen T.M., Trautner S., et al. 2010. Remote ischaemic conditioning before hospital admission, as a complement to angioplasty, and effect on myocardial salvage in patients with acute myocardial infarction: a randomised trial. Lancet. 375:727–734. 10.1016/S0140-6736(09)62001-8 [DOI] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, B., Thorp E.B., Doran A.C., Subramanian M., Sansbury B.E., Lin C.-S., Spite M., Fredman G., and Tabas I.. 2016. MerTK cleavage limits proresolving mediator biosynthesis and exacerbates tissue inflammation. Proc. Natl. Acad. Sci. USA. 113:6526–6531. 10.1073/pnas.1524292113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavadas, M.A.S., Mesnieres M., Crifo B., Manresa M.C., Selfridge A.C., Keogh C.E., Fabian Z., Scholz C.C., Nolan K.A., Rocha L.M.A., et al. 2016. REST is a hypoxia-responsive transcriptional repressor. Sci. Rep. 6:31355. 10.1038/srep31355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarov, S., Lim H.Y., Tan L., Lim S.Y., See P., Lum J., Zhang X.-M., Foo S., Nakamizo S., Duan K., et al. 2019. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science. 363:eaau0964. 10.1126/science.aau0964 [DOI] [PubMed] [Google Scholar]
- Chandel, N.S., Maltepe E., Goldwasser E., Mathieu C.E., Simon M.C., and Schumacker P.T.. 1998. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. USA. 95:11715–11720. 10.1073/pnas.95.20.11715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.-C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H.A., Rao N.A., Aghajanirefah A., et al. 2014. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 345:1250684. 10.1126/science.1250684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney, K.D., Infante J.R., Lam E.T., Figlin R.A., Rini B.I., Brugarolas J., Zojwalla N.J., Lowe A.M., Wang K., Wallace E.M., et al. 2018. Phase I Dose-Escalation Trial of PT2385, a First-in-Class Hypoxia-Inducible Factor-2α Antagonist in Patients With Previously Treated Advanced Clear Cell Renal Cell Carcinoma. J. Clin. Oncol. 36:867–874. 10.1200/JCO.2017.74.2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer, T., Yamanishi Y., Clausen B.E., Förster I., Pawlinski R., Mackman N., Haase V.H., Jaenisch R., Corr M., Nizet V., et al. 2003. HIF-1α is essential for myeloid cell-mediated inflammation. Cell. 112:645–657. 10.1016/S0092-8674(03)00154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerge, M., Yeap X.Y., Dehn S., Zhang S., Grigoryeva L., Misener S., Procissi D., Zhou X., Lee D.C., Muller W.A., et al. 2017. MerTK Cleavage on Resident Cardiac Macrophages Compromises Repair After Myocardial Ischemia Reperfusion Injury. Circ. Res. 121:930–940. 10.1161/CIRCRESAHA.117.311327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerge, M., Yu S., Dehn S., Ifergan I., Yeap X.Y., Filipp M., Becker A., Luo X., Miller S., and Thorp E.B.. 2020. Monocytes prime autoreactive T cells after myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 318:H116–H123. 10.1152/ajpheart.00595.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehn, S., DeBerge M., Yeap X.-Y., Yvan-Charvet L., Fang D., Eltzschig H.K., Miller S.D., and Thorp E.B.. 2016. HIF-2α in Resting Macrophages Tempers Mitochondrial Reactive Oxygen Species To Selectively Repress MARCO-Dependent Phagocytosis. J. Immunol. 197:3639–3649. 10.4049/jimmunol.1600402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick, S.A., and Epelman S.. 2016. Chronic Heart Failure and Inflammation: What Do We Really Know? Circ. Res. 119:159–176. 10.1161/CIRCRESAHA.116.308030 [DOI] [PubMed] [Google Scholar]
- Dick, S.A., Macklin J.A., Nejat S., Momen A., Clemente-Casares X., Althagafi M.G., Chen J., Kantores C., Hosseinzadeh S., Aronoff L., et al. 2019. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat. Immunol. 20:29–39. 10.1038/s41590-018-0272-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., Khalil M., Kiedrowski M., O’Connor C., Petrovic E., Zhou X., and Penn M.S.. 2010. Critical role for leukocyte hypoxia inducible factor-1alpha expression in post-myocardial infarction left ventricular remodeling. Circ. Res. 106:601–610. 10.1161/CIRCRESAHA.109.208967 [DOI] [PubMed] [Google Scholar]
- Du, W., Zhang L., Brett-Morris A., Aguila B., Kerner J., Hoppel C.L., Puchowicz M., Serra D., Herrero L., Rini B.I., et al. 2017. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat. Commun. 8:1769. 10.1038/s41467-017-01965-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitel, I., Stiermaier T., Rommel K.P., Fuernau G., Sandri M., Mangner N., Linke A., Erbs S., Lurz P., Boudriot E., et al. 2015. Cardioprotection by combined intrahospital remote ischaemic perconditioning and postconditioning in ST-elevation myocardial infarction: the randomized LIPSIA CONDITIONING trial. Eur. Heart J. 36:3049–3057. 10.1093/eurheartj/ehv463 [DOI] [PubMed] [Google Scholar]
- Fadok, V.A., Bratton D.L., Konowal A., Freed P.W., Westcott J.Y., and Henson P.M.. 1998. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J. Clin. Invest. 101:890–898. 10.1172/JCI1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, P.J., Liyanage S.E., Cristante E., Sampson R.D., Dick A.D., Ali R.R., and Bainbridge J.W.. 2017. Hypoxia inducible factors are dispensable for myeloid cell migration into the inflamed mouse eye. Sci. Rep. 7:40830. 10.1038/srep40830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister, C., and Satija R.. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20:296. 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf, I., Gerhardt L.M.S., Tan T.C., Winter C., Holderried T.A.W., Chousterman B.G., Iwamoto Y., Liao R., Zirlik A., Scherer-Crosbie M., et al. 2014. Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ. Res. 114:1611–1622. 10.1161/CIRCRESAHA.114.303204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honarpisheh, M., Desai J., Marschner J.A., Weidenbusch M., Lech M., Vielhauer V., Anders H.-J., and Mulay S.R.. 2016. Regulated necrosis-related molecule mRNA expression in humans and mice and in murine acute tissue injury and systemic autoimmunity leading to progressive organ damage, and progressive fibrosis. Biosci. Rep. 36:e00425. 10.1042/BSR20160336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howangyin, K.-Y., Zlatanova I., Pinto C., Ngkelo A., Cochain C., Rouanet M., Vilar J., Lemitre M., Stockmann C., Fleischmann B.K., et al. 2016. Myeloid-Epithelial-Reproductive Receptor Tyrosine Kinase and Milk Fat Globule Epidermal Growth Factor 8 Coordinately Improve Remodeling After Myocardial Infarction via Local Delivery of Vascular Endothelial Growth Factor. Circulation. 133:826–839. 10.1161/CIRCULATIONAHA.115.020857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiyaz, H.Z., Williams E.P., Hickey M.M., Patel S.A., Durham A.C., Yuan L.-J., Hammond R., Gimotty P.A., Keith B., and Simon M.C.. 2010. Hypoxia-inducible factor 2α regulates macrophage function in mouse models of acute and tumor inflammation. J. Clin. Invest. 120:2699–2714. 10.1172/JCI39506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan, M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J.M., Lane W.S., and Kaelin W.G. Jr. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 292:464–468. 10.1126/science.1059817 [DOI] [PubMed] [Google Scholar]
- Kido, M., Du L., Sullivan C.C., Li X., Deutsch R., Jamieson S.W., and Thistlethwaite P.A.. 2005. Hypoxia-inducible factor 1-alpha reduces infarction and attenuates progression of cardiac dysfunction after myocardial infarction in the mouse. J. Am. Coll. Cardiol. 46:2116–2124. 10.1016/j.jacc.2005.08.045 [DOI] [PubMed] [Google Scholar]
- Kim, J.W., Tchernyshyov I., Semenza G.L., and Dang C.V.. 2006a. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3:177–185. 10.1016/j.cmet.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Kim, W.Y., Safran M., Buckley M.R., Ebert B.L., Glickman J., Bosenberg M., Regan M., and Kaelin W.G. Jr. 2006b. Failure to prolyl hydroxylate hypoxia-inducible factor α phenocopies VHL inactivation in vivo. EMBO J. 25:4650–4662. 10.1038/sj.emboj.7601300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen, M., Lee J.W., Seo S.-W., Brodsky K.S., Kreth S., Yang I.V., Buttrick P.M., Eckle T., and Eltzschig H.K.. 2018. Hypoxia-inducible factor 2-alpha-dependent induction of amphiregulin dampens myocardial ischemia-reperfusion injury. Nat. Commun. 9:816. 10.1038/s41467-018-03105-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen, P., Hirsilä M., Günzler V., Kivirikko K.I., and Myllyharju J.. 2004. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J. Biol. Chem. 279:9899–9904. 10.1074/jbc.M312254200 [DOI] [PubMed] [Google Scholar]
- Koudstaal, S., Oerlemans M.I.F.J., Van der Spoel T.I.G., Janssen A.W.F., Hoefer I.E., Doevendans P.A., Sluijter J.P.G., and Chamuleau S.A.J.. 2015. Necrostatin-1 alleviates reperfusion injury following acute myocardial infarction in pigs. Eur. J. Clin. Invest. 45:150–159. 10.1111/eci.12391 [DOI] [PubMed] [Google Scholar]
- Lantz, C., Radmanesh B., Liu E., Thorp E.B., and Lin J.. 2020. Single-cell RNA sequencing uncovers heterogenous transcriptional signatures in macrophages during efferocytosis. Sci. Rep. 10:14333. 10.1038/s41598-020-70353-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., Uttarwar L., Gao B., Charbonneau M., Shi Y., Chan J.S.D., Dubois C.M., and Krepinsky J.C.. 2015. High Glucose Up-regulates ADAM17 through HIF-1α in Mesangial Cells. J. Biol. Chem. 290:21603–21614. 10.1074/jbc.M115.651604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, G., Li X., Zhang L., Zhang Y., Sun L., Zhang X., Liu H., Pang Y., Kong W., Zhang T., et al. 2019. Macrophage metabolic reprogramming aggravates aortic dissection through the HIF1α-ADAM17 pathway. EBioMedicine. 49:291–304. 10.1016/j.ebiom.2019.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde, M., Lutz M., Carter N., Sosna J., Jacoby C., Vucur M., Gautheron J., Roderburg C., Borg N., Reisinger F., et al. 2014. RIP3, a kinase promoting necroptotic cell death, mediates adverse remodelling after myocardial infarction. Cardiovasc. Res. 103:206–216. 10.1093/cvr/cvu146 [DOI] [PubMed] [Google Scholar]
- Maier, A., Wu H., Cordasic N., Oefner P., Dietel B., Thiele C., Weidemann A., Eckardt K.-U., and Warnecke C.. 2017. Hypoxia-inducible protein 2 Hig2/Hilpda mediates neutral lipid accumulation in macrophages and contributes to atherosclerosis in apolipoprotein E-deficient mice. FASEB J. 31:4971–4984. 10.1096/fj.201700235R [DOI] [PubMed] [Google Scholar]
- Maxwell, P.H., Wiesener M.S., Chang G.-W., Clifford S.C., Vaux E.C., Cockman M.E., Wykoff C.C., Pugh C.W., Maher E.R., and Ratcliffe P.J.. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 399:271–275. 10.1038/20459 [DOI] [PubMed] [Google Scholar]
- Nailwal, H., and Chan F.K.-M.. 2019. Necroptosis in anti-viral inflammation. Cell Death Differ. 26:4–13. 10.1038/s41418-018-0172-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazon, A., Goldrath A.W., Nizet V., and Johnson R.S.. 2014. HIF transcription factors, inflammation, and immunity. Immunity. 41:518–528. 10.1016/j.immuni.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin, E.B., Rha J., Selak M.A., Unger T.L., Keith B., Liu Q., and Haase V.H.. 2009. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol. Cell. Biol. 29:4527–4538. 10.1128/MCB.00200-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand, J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., and Vilo J.. 2016. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44(W1):W83-9. 10.1093/nar/gkw199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann, M., Delgobo M., Gaal C., Büchner L., Steinau P., Reshef D., Gil-Cruz C., Horst E.N.T., Kircher M., Reiter T., et al. 2019. Myocardial infarction triggers cardioprotective antigen-specific T helper cell responses. J. Clin. Invest. 129:4922–4936. 10.1172/JCI123859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffelmacher, T., Clarke A., Richter F.C., Stranks A., Pandey S., Danielli S., Hublitz P., Yu Z., Johnson E., Schwerd T., et al. 2017. Autophagy-Dependent Generation of Free Fatty Acids Is Critical for Normal Neutrophil Differentiation. Immunity. 47:466–480.e5. 10.1016/j.immuni.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, D., Schön C., Boerries M., Didrihsone I., Ritterhoff J., Kubatzky K.F., Völkers M., Herzog N., Mähler M., Tsoporis J.N., et al. 2014. S100A1 is released from ischemic cardiomyocytes and signals myocardial damage via Toll-like receptor 4. EMBO Mol. Med. 6:778–794. 10.15252/emmm.201303498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin, C.V., and Lemke G.. 2010. TAM receptor signaling and autoimmune disease. Curr. Opin. Immunol. 22:740–746. 10.1016/j.coi.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiku, P., Willson J.A., Dickinson R.S., Murphy F., Harris A.J., Lewis A., Sammut D., Mirchandani A.S., Ryan E., Watts E.R., et al. 2017. Prolyl hydroxylase 2 inactivation enhances glycogen storage and promotes excessive neutrophilic responses. J. Clin. Invest. 127:3407–3420. 10.1172/JCI90848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Zas, I., Lemarié J., Zlatanova I., Cachanado M., Seghezzi J.-C., Benamer H., Goube P., Vandestienne M., Cohen R., Ezzo M., et al. 2021. Cytotoxic CD8+ T cells promote granzyme B-dependent adverse post-ischemic cardiac remodeling. Nat. Commun. 12:1483. 10.1038/s41467-021-21737-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, B., and Fulda S.. 2015. Reactive oxygen species regulate Smac mimetic/TNFα-induced necroptotic signaling and cell death. Oncogene. 34:5796–5806. 10.1038/onc.2015.35 [DOI] [PubMed] [Google Scholar]
- Semenza, G.L.2012. Hypoxia-inducible factors in physiology and medicine. Cell. 148:399–408. 10.1016/j.cell.2012.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.C.T., Davidson S.M., Lim S.Y., Simpkin J.C., Hothersall J.S., and Yellon D.M.. 2007. Necrostatin: a potentially novel cardioprotective agent? Cardiovasc. Drugs Ther. 21:227–233. 10.1007/s10557-007-6035-1 [DOI] [PubMed] [Google Scholar]
- Sousa, G.R., Kosiborod M., Bluemke D.A., and Lipes M.A.. 2020. Cardiac Autoimmunity Is Associated With Subclinical Myocardial Dysfunction in Patients With Type 1 Diabetes Mellitus. Circulation. 141:1107–1109. 10.1161/CIRCULATIONAHA.119.044539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield, B.A., Purves T., Palmer S., Sullenger B., Welty-Wolf K., Haines K., Agarwal S., and Kasotakis G.. 2021. IL-10 and class 1 histone deacetylases act synergistically and independently on the secretion of proinflammatory mediators in alveolar macrophages. PLoS One. 16:e0245169. 10.1371/journal.pone.0245169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szobi, A., Gonçalvesová E., Varga Z.V., Leszek P., Kuśmierczyk M., Hulman M., Kyselovič J., Ferdinandy P., and Adameová A.. 2017. Analysis of necroptotic proteins in failing human hearts. J. Transl. Med. 15:86. 10.1186/s12967-017-1189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill, G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H., et al. 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 496:238–242. 10.1038/nature11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitz, T., Inoue M., Valentine M.B., Zhu K., Rehg J.E., Zhao W., Finkelstein D., Wang Y.-D., Johnson M.D., Calabrese C., et al. 2013. Th-MYCN mice with caspase-8 deficiency develop advanced neuroblastoma with bone marrow metastasis. Cancer Res. 73:4086–4097. 10.1158/0008-5472.CAN-12-2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, A., Belaidi E., Aron-Wisnewsky J., van der Zon G.C., Levy P., Clement K., Pepin J.-L., Godin-Ribuot D., and Guigas B.. 2016. Hypoxia-inducible factor prolyl hydroxylase 1 (PHD1) deficiency promotes hepatic steatosis and liver-specific insulin resistance in mice. Sci. Rep. 6:24618. 10.1038/srep24618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, A.A.R., Elks P.M., Marriott H.M., Eamsamarng S., Higgins K.R., Lewis A., Williams L., Parmar S., Shaw G., McGrath E.E., et al. 2014. Hypoxia-inducible factor 2α regulates key neutrophil functions in humans, mice, and zebrafish. Blood. 123:366–376. 10.1182/blood-2013-05-500207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp, E.B.2021. Macrophage Metabolic Signaling during Ischemic Injury and Cardiac Repair. Immunometabolism. 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp, E., Vaisar T., Subramanian M., Mautner L., Blobel C., and Tabas I.. 2011. Shedding of the Mer tyrosine kinase receptor is mediated by ADAM17 protein through a pathway involving reactive oxygen species, protein kinase Cδ, and p38 mitogen-activated protein kinase (MAPK). J. Biol. Chem. 286:33335–33344. 10.1074/jbc.M111.263020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan, A.M., Ter Horst E.N., Delewi R., Begieneman M.P.V., Krijnen P.A.J., Hirsch A., Lavaei M., Nahrendorf M., Horrevoets A.J., Niessen H.W.M., and Piek J.J.. 2014. Monocyte subset accumulation in the human heart following acute myocardial infarction and the role of the spleen as monocyte reservoir. Eur. Heart J. 35:376–385. 10.1093/eurheartj/eht331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dierendonck, X.A.M.H., de la Rosa Rodriguez M.A., Georgiadi A., Mattijssen F., Dijk W., van Weeghel M., Singh R., Borst J.W., Stienstra R., and Kersten S.. 2020. HILPDA Uncouples Lipid Droplet Accumulation in Adipose Tissue Macrophages from Inflammation and Metabolic Dysregulation. Cell Rep. 30:1811–1822.e6. 10.1016/j.celrep.2020.01.046 [DOI] [PubMed] [Google Scholar]
- Walmsley, S.R., Chilvers E.R., Thompson A.A., Vaughan K., Marriott H.M., Parker L.C., Shaw G., Parmar S., Schneider M., Sabroe I., et al. 2011. Prolyl hydroxylase 3 (PHD3) is essential for hypoxic regulation of neutrophilic inflammation in humans and mice. J. Clin. Invest. 121:1053–1063. 10.1172/JCI43273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, E., Yeap X.Y., Dehn S., Terry R., Novak M., Zhang S., Iwata S., Han X., Homma S., Drosatos K., et al. 2013. Enhanced efferocytosis of apoptotic cardiomyocytes through myeloid-epithelial-reproductive tyrosine kinase links acute inflammation resolution to cardiac repair after infarction. Circ. Res. 113:1004–1012. 10.1161/CIRCRESAHA.113.301198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, L., Xue X., Taylor M., Ramakrishnan S.K., Nagaoka K., Hao C., Gonzalez F.J., and Shah Y.M.. 2014. Hypoxia-inducible factor/MAZ-dependent induction of caveolin-1 regulates colon permeability through suppression of occludin, leading to hypoxia-induced inflammation. Mol. Cell. Biol. 34:3013–3023. 10.1128/MCB.00324-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, C., Yagai T., Luo Y., Liang X., Chen T., Wang Q., Sun D., Zhao J., Ramakrishnan S.K., Sun L., et al. 2017. Activation of intestinal hypoxia-inducible factor 2α during obesity contributes to hepatic steatosis. Nat. Med. 23:1298–1308. 10.1038/nm.4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Weinberg S., DeBerge M., Gainullina A., Schipma M., Kinchen J.M., Ben-Sahra I., Gius D.R., Yvan-Charvet L., Chandel N.S., et al. 2019. Efferocytosis Fuels Requirements of Fatty Acid Oxidation and the Electron Transport Chain to Polarize Macrophages for Tissue Repair. Cell Metab. 29:443–456.e5. 10.1016/j.cmet.2018.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows differential gene expression used to identify unique cell clusters during human ischemic cardiomyopathy.
lists the reagents and resources used in this study.