Abstract
Epilepsy affects 1 in 150 children under the age of 10 and is the most common chronic pediatric neurological condition; poor seizure control can irreversibly disrupt normal brain development. The present study compared the ability of different machine learning algorithms trained with resting-state functional MRI (rfMRI) latency data to detect epilepsy. Preoperative rfMRI and anatomical MRI scans were obtained for 63 patients with epilepsy and 259 healthy controls. The normal distribution of latency z-scores from the epilepsy and healthy control cohorts were analyzed for overlap in 36 seed regions. In these seed regions, overlap between the study cohorts ranged from 0.44-0.58. Machine learning features were extracted from latency z-score maps using principal component analysis. Extreme Gradient Boosting (XGBoost), Support Vector Machines (SVM), and Random Forest algorithms were trained with these features. Area under the receiver operating characteristics curve (AUC), accuracy, sensitivity, specificity and F1-scores were used to evaluate model performance. The XGBoost model outperformed all other models with a test AUC of 0.79, accuracy of 74%, specificity of 73%, and a sensitivity of 77%. The Random Forest model performed comparably to XGBoost across multiple metrics, but it had a test sensitivity of 31%. The SVM model did not perform >70% in any of the test metrics. The XGBoost model had the highest sensitivity and accuracy for the detection of epilepsy. Development of machine learning algorithms trained with rfMRI latency data could provide an adjunctive method for the diagnosis and evaluation of epilepsy with the goal of enabling timely and appropriate care for patients.
Keywords: machine learning, extreme gradient boosting, pediatric epilepsy, resting-state functional MRI, temporal latency
Introduction
Epilepsy affects 1 in 150 children under the age of 10 and is the most common chronic pediatric neurological condition (1). Unfortunately, antiepileptic drugs alone fail in 1/3 of these children (2,3). Severe developmental delay from alterations in brain network development results from poorly controlled epilepsy (4,5). Early, accurate diagnosis of epilepsy to address these irreversible developmental impacts is a top research priority for two large international forums (6,7). Current paradigms suggest referral to university centers if the epilepsy becomes drug resistant, that is continued seizures despite a trial of two medications (2). Unfortunately, there is a mean 19-20 year time from seizure onset to epilepsy surgery referral for eligible patients to these centers, partly due to a difficulty in accurate identification of epilepsy, especially without an obvious lesion or generalized motor component (7-9). Both international forums proposed referral paradigm changes either at initial diagnosis or after the first anti-epileptic drug fails (6,7). If adopted, this paradigm change could be quite costly as several children will achieve seizure freedom with one medication (10). Therefore, early and accurate diagnosis is absolutely necessary to prevent over or underdiagnosis of epilepsy, particularly in non-lesional or partial seizure cases.
Resting-state functional MRI (rfMRI) has been evaluated as a potential non-invasive method to aid in the diagnosis of epilepsy (11). RfMRI sequences are obtained concurrently with anatomic MRI, as part of the standard epilepsy workup at certain centers. Consequently, there is no increased cost or need for additional procedures. However, concurrent rfMRI acquisition is currently not part of the standard of care (12,13). The blood-oxygen-level dependent (BOLD) signal acquired at rest in rfMRI is associated with resting metabolic neuronal activity (14). As such, resting-state networks (RSNs) can be constructed from the rfMRI signal using temporal latency analysis (15,16). Disturbances in RSNs are thought to be associated with epilepsy (17-19).
Several other techniques can also be used to construct RSNs. Broadly, they can be divided into two categories: Hypothesis-driven and data-driven methods. Hypothesis-driven methods, such as seed-based functional connectivity, correlate the BOLD signal obtained from the region of interest (ROI) with voxels throughout the brain to construct the RSN (14). Alternatively, data-driven methods, such as independent component analysis (ICA), do not require ROIs to construct RSNs (20). Previous studies have shown that ICA-generated RSNs can be used to analyze epilepsy networks (21-23). However, ICA is extremely time consuming, as each component map must be individually inspected and categorized, thus decreasing the potential ease and utility of this method.
As a result, rfMRI latency analysis, a novel data-driven method of RSN construction, has continued to gain popularity in the field. RfMRI latency analysis correlates the timing of the global mean BOLD signal with the BOLD signal in voxels throughout the brain. This allows for the characterization of multiple brain networks by linking areas based on differences in their BOLD signal timing relative to the global mean BOLD signal (15,16). In our previous studies it has been shown that areas with significantly early or late BOLD signal timing compared to the global mean BOLD signal correlate with epilepsy lateralization, and this can be used to accurately lateralize temporal lobe and extratemporal lobe pediatric epilepsy (11,24). However, this method of thresholding rfMRI latency z-scores was unable to explain laterality in every case.
Machine learning has become a popular technique to explore complex relationships in data (25-27). Supervised machine learning algorithms, such as Support Vector Machines (SVM), Random Forest and Extreme Gradient Boosting (XGBoost), use inputs (x) to define the relationship [f(x)] between said input and an output (y) (28,29). Algorithms then apply the learned relationship to predict outputs from new inputs. For example, SVM creates linear hyperplanes, or the most optimal decision boundary, in feature space to separate class categories of interest. SVM utilizes kernels to transform a non-linearly separable input feature space into higher dimensions enabling the linear separation of the input data (22). On the other hand, Random Forest and XGBoost are ensemble algorithms that classify examples by voting on multiple decision trees created by bagging or boosting, respectively (30,31). Boosting is a method where later iterations of the machine learning model improve upon errors made in prior models. Bagging combines predictions from several machine learning models to increase model accuracy. In both cases, weak learners, or decision trees that correctly predict an average of 50% of cases, are combined. The majority decision from the combined decision trees is then used to make the final prediction. Models generated from multiple weak learners as described, create stronger learners that can make more accurate predictions from complex datasets (32).
The aim of the present study was to compare the performance of SVM, Random Forest, and XGBoost machine learning classifier algorithms trained with novel rfMRI latency data to distinguish children with epilepsy from healthy controls.
Materials and methods
Data source and study population
RfMRI and anatomical MRI were obtained following University of Texas at Houston Institutional Review Board (IRB) approval for an anonymized retrospective review of 78 patients with refractory extratemporal epilepsy from a prospectively registered clinical database from the Washington University School of Medicine/St. Louis Children's Hospital (SLCH, n=73) and McGovern Medical School at UTHealth/Children's Memorial Hermann Hospital (CMHH, n=5) between September 2006 and September 2018. RfMRI and anatomical MRI were obtained for 585 healthy control patients with IRB approval from the multi-institutional ADHD 200 dataset of the 1000 Functional Connectomes Project, International Neuroimaging Data-Sharing Initiative hosted by the Neuroimaging Tools and Resources Collaboratory (33).
Informed consent was obtained as required by each database and institution for each subject. Informed consent was provided by the patients' caregivers or by capable patients aged >18 years old for the 78 prospectively registered patients with refractory epilepsy at SLCH and CMHH. Informed consent for healthy control patients were obtained by each study site in the ADHD dataset (34). The imaging parameters for each scanner used to obtain rfMRI data at each of the participating institutions are shown in Table SI.
Patients with refractory epilepsy analyzed in the present study were referred to SLCH or CMHH for evaluation of surgical management. Inclusion criteria required a diagnosis of refractory epilepsy as determined by multidisciplinary epilepsy conference at either SLCH or CMHH and available structural and rfMRI. Patients who did not meet these requirements or whose rfMRI data did not pass quality control were excluded. Quality control consisted of visual inspection of pre-processed rfMRI data. Poorly registered rfMRI to standard atlas volumetric sequences were excluded. In the rfMRI processing section below, further explanations are provided. After application of these criteria, the final study population included 322 subjects. A total of 63 patients with refractory epilepsy were included (age range, 4-25 years; median age, 17 years old; 41 males and 22 females). A total of 259 healthy controls were included in this study (age range, 7-26 years; median age, 10 years old; 25 males and 134 females). Analysis of baseline characteristics of each cohort was performed. A two-tailed Mann-Whitney U test was used to compare the mean age of subjects within each cohort (epilepsy, n=63; healthy controls, n=259; α=0.05). A χ2 test of independence was used to evaluate for an association between sex and epilepsy (epilepsy, n=63; healthy control, n=259; α=0.05).
RfMRI processing
MRI data was processed using the Washington University in St. Louis (Wustl) 4dfpSuite Linux scripts developed by the Washington University School of Medicine Neuroimaging Laboratory, as described previously (11). Briefly, the BOLD signal sequences of the healthy controls and patients with epilepsy were registered to standard atlas volumetric sequences. Images were processed with spatial smoothing using a Gaussian kernel of 6 mm full width half-maximum, temporal low-pass filtering >0.1 Hz, regression of nuisance waveforms and zero-meaning of each voxel time course. Frames with excessive motion were excluded from analysis (16). Latency maps for patients in both the healthy control and epilepsy groups were generated by computing a voxel-wise lagged cross-covariance function. Lag or latency, is the value at which the absolute cross-covariance function displays extremum between the processed rfMRI BOLD signal and the whole-brain mean signal from parabolic interpolation (16). The normal distributions of the epilepsy and healthy control average latency maps were analyzed for overlap in 36 seed regions (35,36).
Feature engineering
Healthy control and epilepsy latency z-score maps were created by voxel-wise z-score calculation using whole-brain healthy control mean and standard deviation latency maps as well as Fslmaths in the FSL suite version 6.0 (11,37). Principal component analysis (PCA) was performed to create features to train the machine learning algorithms. Observations or features in PCA were orthogonally transformed into linearly uncorrelated principal components (PCs) that represented the variability of the training dataset. PCA was performed in order to reduce the dimensions of the original feature space, in an attempt to mitigate ‘the curse of dimensionality’, which states that the predictive power of a classifier initially increases with the use of more dimensions, but at some point, it begins to decrease (38). In the PCA transformations used in the present study, the original 47,950 non-zero voxel high-dimensional feature space was reduced to 118 PCs, which explained 80% of the variance, and 80% explanation of the variance was chosen, with 118 principal components (PCs), to balance adequately reducing the dimensions of the feature space, whilst also maintaining enough features to allow for effective model classification of epilepsy from healthy controls. The high-dimensional voxel feature space for the training dataset containing both epilepsy and healthy controls was transformed with PCA using the parameters stated above. The same transformation was then also applied to the validation and test datasets to generate comparable datasets with 118 PCs for the models to evaluate.
XGBoost training, validation and testing data partitioning
All epilepsy patient disease labels were defined by a consensus at multidisciplinary pediatric epilepsy conferences. Healthy individuals' labels were determined by each participating institution in the ADHD 200 dataset (34). The data was randomized and stratified using a random seed to create a reproducible pseudorandomized dataset with a constant ratio of patients with epilepsy and control individuals. The data was partitioned into train, validation and test datasets at a standard ratio of 60:20:20 with Python 3.6.3 sklearn.model_selection (version 0.20.3, scikit-learn.org/stable/modules/cross_validation.html#cross-validation) train_test_split function. R (version 3.5.0), e1071 (version 1.6-8), randomForest (version 4.6-14) and xgboost (version 0.71.2) libraries were used for SVM, Random Forest, and XGBoost algorithms, respectively (30,31,39-41). The training data features and labels were used to train SVM, Random Forest and XGBoost algorithms.
Hyperparameter optimization
Hyperparameters for SVM, Random Forest and XGBoost were optimized to increase performance accuracy, area under the receiver operating characteristics (ROC) curve (AUC), specificity and F1-score. Grid searching was used to minimize overfitting of the models. Hyperparameters affected training, model creation, model performance and generalizability. Each hyperparameter was explored at a range and step size to assess model performance in a grid-wise manner within a multidimensional hyperparameter space as shown in Table I, Table II, Table III for SVM, Random Forest and XGBoost algorithms, respectively. Optimal hyperparameters were chosen based on overall model performance using validation data. Then, SVM, Random Forest and XGBoost models with optimal hyperparameters were evaluated with unseen test data to assess model generalizability.
Table I.
Description of the Support Vector Machine algorithm hyperparameters and ranges.
| Hyperparameter | Definition | Significance/usefulness | Grid search range |
|---|---|---|---|
| Kernel | Computes the dot product in the feature space using vectors from the original space. | Kernel function allows low-cost operations in the original feature space without computing the coordinates of the data in a higher dimension space. | Linear, radial basis function, polynomial, and sigmoid |
| γ | Distance of influence a single training point has on a kernel. | For high or low γ, training points closer or further from the decision boundary are weighted, respectively. Improved fitting of the decision boundary to training cases increases model generalizability. | 0.1, 1, 10 and 100 |
| Cost | ‘C’-constant of regularization term from the Lagrange formulation (53). | Controls the trade-off between misclassifications and margin width. Simplified decision boundary for low or high cost to improve point classification. | 0.1, 1, 10, 100 and 1,000 |
Table II.
Description of the Random Forest algorithm hyperparameters and ranges.
| Hyperparameter | Definition | Significance/usefulness | Grid search range |
|---|---|---|---|
| Mtry | Number of variables available for splitting at each tree node | The predictor variable/feature importance estimates are affected by mtry. | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, and 15 |
| Ntree | Number of trees to be grown in the model | Number of trees affects model variance. Increasing ntree increases performance and computation cost. | 1, 5, 10, 100, 500, 1,000, 1,500, 2,000 and 2,500 |
Table III.
Description of the Extreme Gradient Boosting algorithm hyperparameters and ranges.
| Hyperparameter | Definition | Significance/usefulness | Grid search range |
|---|---|---|---|
| ETA | ETA is the shrinkage of the learning rate at each step | ETA prevents the model from overfitting by scaling contribution of each tree. | From 0.01 to 1 at a step size of 0.01 |
| Max Depth | Maximum depth of each tree | Max depth affects the complexity of each tree and the overfitting of the model. Deeper trees can overfit the data. | 1, 2, 3, 4 and 5 |
| Minimum Child Weight | The minimum weight required in order to create a new node in each tree | Adjusting the value can prevent overfitting and reduce the model complexity. | 1, 2, 3, 4 and 5 |
| γ | Minimum loss reduction required to create a further partition on a tree's leaf node | Increasing γ causes the model to be more conservative. | 1, 2, 3, 4 and 5 |
| Nround | The number of training rounds within the model | Increasing Nrounds can reduce biases and variance in a model. | Varies depending on model error/loss and 200 Nround early stopping |
ETA, estimated time of arrival (also known as the learning rate in R user documentation for XGBoost).
Performance metrics
In the present study, the positive class represents subjects with epilepsy and the negative class represents the healthy controls. Mean accuracy, AUC, specificity and F1-score were used to evaluate models and each metric carried equal weight. F1-score was the harmonic mean between precision and recall (sensitivity). A model with a high F1-score has both good positive predictive power (precision) and good true positive rate (sensitivity or recall) (42). The F1 score was calculated as follows: F1 score=2x[(precision x recall)/precision + recall)]. The model with the highest mean value was then used to decide the optimal model overall, after evaluation with the validation dataset.
Model selection
Algorithm hyperparameters were tuned by evaluating trained models with validation data. For each algorithm (SVM, Random Forest and XGBoost), the model with the highest accuracy, AUC, specificity and F1-score determined by the mean was then evaluated with unseen test data. Models with a specificity or sensitivity of zero were eliminated. In addition to performance, the generalizability of each model was evaluated with the final unseen test data by taking the absolute value of the difference between test and validation accuracy, AUC, sensitivity, specificity and F1-score. After evaluating models with validation data, the following SVM, Random Forest and XGBoost model hyperparameter settings were selected for evaluation with test data: SVM: γ=1, Cost=0.01, Kernel=Sigmoid; Random Forest: Mtry=12, Ntree=5.0; XGBoost: ETA=0.61, Max Depth=2.0, γ=1.0, Minimum Child Weight=4.0, Nround=11.0.
Results
The final study population (n=322) included 63 patients with epilepsy and 259 healthy controls. Baseline characteristics and cohort analysis are shown in Table IV. Briefly, the epilepsy cohort (n=63; age range, 4-25 years; 41 males and 22 females) consisted of significantly more males than females (P=0.017) with a sex ratio of 1.86 males-to-females. The epilepsy cohort was also significantly older than the healthy controls with a mean age of 14.5 years with a standard deviation of 6.0 (P<0.00001). The healthy control cohort (n=259; age range, 7-26 years; 125 males and 134 females) included slightly more females than males with a sex ratio of 0.93 males-to-females. The healthy control cohort was significantly younger than the epilepsy cohort with a mean age of 10.7 years and a standard deviation of 2.6 (P<0.00001).
Table IV.
Baseline characteristics of the study population and cohort analysis.
| Refractory epilepsy cohort | Healthy control cohort | P-value | |
|---|---|---|---|
| Cohort size, n (%) | 63 (19.6) | 259 (80.4) | |
| Males, n (%) | 41 (65.1) | 125 (48.3) | 0.017a |
| Female, n (%) | 22 (34.9) | 134 (51.7) | |
| Median age (interquartile range) | 17(8) | 10(3) | <0.00001b |
aP≤0.05,
bP<0.0001.
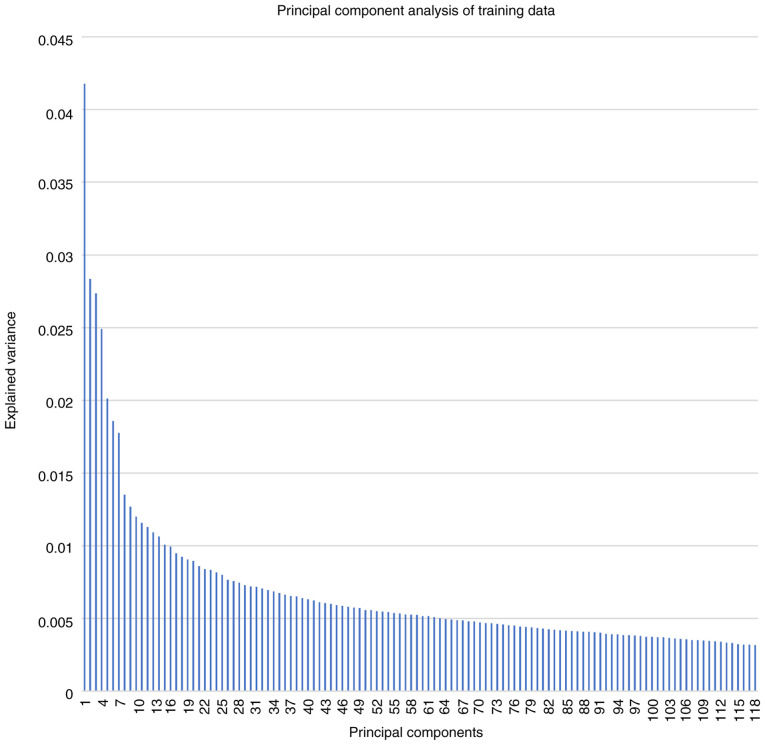
Explained variance for PCs ranged from 4.18% (PC no. 1) to 0.32% (PC no. 118) with a mean and median explained variance of 0.69 and 0.52% as shown in Fig. 1.
Figure 1.
Explained variance of principal component analysis on the training datasets in both patients with epilepsy and healthy controls.
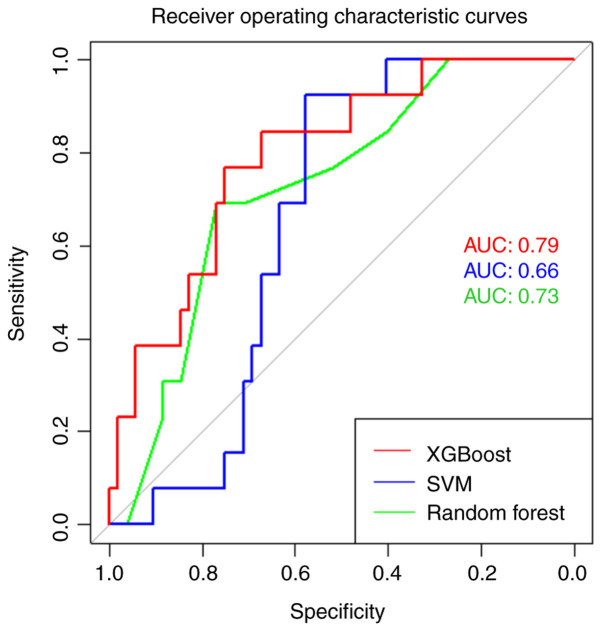
Validation results for each model are presented in Table V. Final test performance and generalizability analyses of the best model from each algorithm are presented in Table VI. The absolute difference between the test-validation results for each model are also presented in Table VI. Lower absolute difference values are desired as they represent less variation between model test and validation results and thus, less overfitting and improved model generalizability. ROC curves were created for each algorithm using test data, and AUC values were calculated as shown in Fig. 2.
Table V.
Validation results from the best SVM, Random Forest and XGBoost models.
| Model | AUC | Accuracy | Sensitivity | Specificity | F1-Score |
|---|---|---|---|---|---|
| SVM | 0.72 | 0.75 | 0.58 | 0.79 | 0.46 |
| Random Forest | 0.86 | 0.84 | 0.33 | 0.96 | 0.44 |
| XGBoost | 0.81 | 0.84 | 0.67 | 0.88 | 0.62 |
SVM, Support Vector Machine; XGBoost, Extreme Gradient Boosting; AUC, area under the receiver operating characteristics curve.
Table VI.
Test results for the best SVM, Random Forest and XGBoost modelsa.
| Model | AUC | Accuracy | Sensitivity | Specificity | F1-Score |
|---|---|---|---|---|---|
| SVM | 0.66 (0.06) | 0.62 (0.13) | 0.38 (0.20) | 0.67 (0.12) | 0.23 (0.18) |
| Random Forest | 0.73 (0.13) | 0.77 (0.07) | 0.31 (0.03) | 0.88 (0.08) | 0.34 (0.10) |
| XGBoost | 0.79 (0.02) | 0.74 (0.11) | 0.77 (0.10) | 0.73 (0.15) | 0.54 (0.07) |
aThe absolute difference of test-validation results for each metric are shown in parentheses. SVM, Support Vector Machine; XGBoost, Extreme Gradient Boosting; AUC, area under the receiver operating characteristics curve.
Figure 2.
Receiver operating characteristic curves for the best SVM, Random Forest and XGBoost models obtained using test data. XGBoost models are shown in red. SVM models are shown in blue. Random Forest models are shown in green. ROC, receiver operating characteristic; AUC, area under the ROC curve SVM, Support Vector Machine; XGBoost, Extreme Gradient Boosting.
XGBoost demonstrated superior performance compared to all the other models tested. XGBoost correctly classified 74% of patients with epilepsy and healthy controls. Importantly, the XGBoost model was sensitive (77%) with a good AUC (0.79), which are both desired characteristics of a screening diagnostic method. XGBoost also outperformed all other tested models in three of the five performance metrics (AUC, sensitivity and F1-score), whereas the Random Forest algorithm performed the best in the remaining two performance metrics (accuracy and specificity). Furthermore, XGBoost-PCA had the greatest generalizability, as demonstrated by the smallest absolute test-validation performance difference for both AUC and F1-score. Random Forest performed well, correctly predicting 77% of patients with a good AUC (0.73). However, the model's performance may be primarily attributed to the correct prediction of control individuals (specificity=88%) as the model poorly predicted patients with epilepsy (sensitivity=31%), which was reflected in a poor mean score of all five metrics. Whilst SVM and Random Forest models were both specific, they were not sensitive, and thus are less effective diagnostic screening tools. The XGBoost model was both sensitive and specific making it the best model for this application.
Discussion
At present, ~1 in 150 children under the age of 10 will be diagnosed with epilepsy (1). One-third of pediatric patients with epilepsy are refractory to treatment with anticonvulsant therapy alone (2). Several of these patients may benefit from additional evaluation at comprehensive epilepsy centers where appropriate treatments can be further explored. Specifically, surgical intervention has demonstrated higher rates of seizure freedom and improved cognitive and developmental outcomes when performed early in appropriate patients (8,43,44). Unfortunately, patients experience a two-decade delay between seizure onset and surgical intervention on average, underlining the need for earlier comprehensive epilepsy evaluation for patients with epilepsy (45,46). Delays in diagnosis and effective treatment of epilepsy place patients at risk of poor development and sudden death from uncontrolled epilepsy (47,48). Whilst several factors may contribute to the referral delay for specialized epilepsy care, the lack of diagnostic methods for early, accurate identification of any kind of epilepsy is a major contributor (6,7). This is especially true in nonconvulsive epilepsy. In the present study, the performance of SVM, Random Forest and XGBoost machine learning algorithms trained with novel rfMRI latency data were evaluated as a classifier to distinguish children with epilepsy from healthy controls.
Previous studies have shown that rfMRI data is correlated with seizure foci laterality and localization (11,21,23,24). Recent machine learning studies demonstrated successful classification of patients with temporal lobe epilepsy from healthy controls, that rfMRI data could be used to improve preoperative planning prior to surgical intervention (21,22). The present study used rfMRI latency analysis, a novel method for RSN construction and analysis, and machine learning to create models that can differentiate children with epilepsy from healthy controls. RfMRI latency analysis has been shown to be correlate with seizure foci laterality, and rfMRI latency analysis has been used to classify epilepsy (11,49).
Recently, an SVM model trained with RSN data obtained from ICA of rfMRI data was able to accurately (97.52%) classify temporal lobe epilepsy with high sensitivity (100%) and specificity (94.4%), but extratemporal epilepsy is far more challenging to classify and treat (11,21). This study also used visual inspection and selection of independent components for RSN construction, which differs from the novel method of constructing RSNs with rfMRI latency analysis used in the present study. The present study demonstrated that RSN trained machine learning algorithms can be used to classify epilepsy in an automated manner using a novel, objective and efficient method of RSN construction that has not yet been demonstrated in the literature to the best of our knowledge.
One aspect of the present study that limited further analysis was the method of feature engineering. PCA transformed the rfMRI latency z-score data into 118 PCs that represent the variance in the training data. However, the use of PC features also prevented analysis of specific pathologies for relevant image features. It was unable to reconstruct the original rfMRI latency z-score spatial data and assign weighting for PC features from the trained XGBoost model. In previous attempts, models trained with the original rfMRI latency voxel z-scores as features did not achieve >50% sensitivity (data not shown). Therefore, PCA was chosen to create a reduced dimension feature space for model training despite these drawbacks. It is hypothesized that the poor performance of the original feature space vs. the PC feature space was due to the relatively small dataset (n=322) compared to the large input features for training the machine learning algorithms (n=47,950).
The primary limitation of the present study was the small amount of training data available at the time of analysis. In future studies, the use of larger training datasets will improve model performance and decrease overfitting, thereby increasing model generalizability (50). The rfMRI data used for analysis was obtained from multiple institutions, using different imaging protocols. However, other publicly accessible healthy rfMRI datasets (ABIDE I and II) could potentially be used to increase the size of the dataset (51,52). Specifics of the imaging parameters used by each scanner to obtain data in the present study are listed in Table SI. The use of different scanner parameters in rfMRI latency data collection could affect model performance, but this also accounts for normal variance present at each institution if this method were to be used to assist in the identification of pediatric epilepsy. Nevertheless, to attempt to mitigate these biases, similar imaging parameters will be used in future studies for all subjects, and the total study population will be increased. In addition, the limitation of the size of the training data affected our ability to age and sex match epilepsy and healthy control cohorts. Additional patients in the healthy control cohort were not eliminated, as it was theorized that it could affect the ability of the models to train when based on a limited dataset. This could affect the features the models use to classify images (such as subtle age and sex brain image differences vs. epilepsy specific image differences). Additionally, the relatively small sample of patients with epilepsy compared with the control individuals could affect how PCA captured the variance in the epilepsy group. However, in the present study PCA was able to capture the variance in the epilepsy cases as both sensitivity and specificity were comparable in the XGBoost model's performance on the test data. However, this was not the case in both Random Forest and SVM, possibly indicating the superiority of XGBoost at detecting more subtle differences in the variance of the PC features between epilepsy and control groups. The potential effects of age, sex and the previously discussed scanner parameters is difficult to assess, and with additional epilepsy and healthy control individuals these potential affects will be mitigated in future studies.
The present study is also limited to binary classification (healthy vs. epilepsy) due to the small sample size and limited representation of rare classification groups in multi-class applications. With additional varied epilepsy rfMRI data, these data distribution effects can be mitigated, and models can be developed to predict epilepsy subgroups (such as surgical outcome and pathological etiology, amongst others), seizure foci lateralization and, ultimately, seizure foci localization.
Refractory epilepsy patient and healthy individuals' data were included in the present study. It was possible to classify these two groups using the Random Forest, SVM and XGBoost models, but it is difficult to assess the ability of these models to classify the diverse spectrum of epilepsy conditions (including non-refractory and temporal epilepsy) from healthy individuals.
In future studies, the use of other machine learning algorithms that will allow for further examination of rfMRI latency, such as semantic segmentation with CNNs will be assessed, which could allow for voxel-wise classification and epileptogenic seizure foci localization. In addition, additional epilepsy data are being collected to expand classification to subgroup analysis and to include non-refractory and temporal epilepsy, with the aim of improving generalizability of the constructed models.
In conclusion, in the present study it was demonstrated that rfMRI latency analysis and machine learning algorithms could be used to identify patients with epilepsy. The XGBoost machine learning model trained with rfMRI latency z-score PC features was better able to distinguish pediatric patients with epilepsy from healthy controls compared with identically trained Random Forest and SVM models. The high sensitivity of the XGBoost test model presented in this study is encouraging. Additional studies with multi-institutional neurological imaging datasets are required to further clarify the value of the machine learning and rfMRI latency in the diagnosis and evaluation of pediatric epilepsy as well as its potential to decrease morbidity and improve prognostication within this patient population.
Supplementary Material
Acknowledgements
We would like to thank Dr Marc E. Raichle (Washington University School of Medicine) and Dr Abraham Z. Snyder (Washington University) for their support in rfMRI analysis.
Funding Statement
Funding: The present study was partly funded by the National Institutes of Health National Institute of Neurological Disorders and Stroke (grant no. R21NS120085).
Availability of data and materials
The code described during the present study is available from the corresponding author on reasonable request. The data will be made available pending approval from the Institutional Review Board at Washington University and McGovern Medical School. Upon approval, the data that support the findings of this study will be made available from the corresponding author upon reasonable request.
Authors' contributions
RDN, MDS and MNS were all involved in study design and co-ordination. RDN wrote the manuscript. RPP, JEL, GVA, MWW, MEF and MNS acquired resting-state functional MRI (rfMRI) data from the Children's Memorial Hermann Hospital. MDS acquired rfMRI data from St. Louis Children's Hospital. LZ and RDN were involved with extreme gradient boosting (XGBoost) model development and algorithm optimization, including tuning of model hyperparameters to achieve optimal performance. LZ, EHK and RDN analyzed the XGBoost model data. LZ assisted with biostatistical analysis for the entire study. LPP, EHK and RDN contributed to the preprocessing of the rfMRI latency data for analysis. AM developed rfMRI latency data analysis software. AM created and revised the protocols used to perform the rfMRI latency data analysis. MDS oversaw rfMRI latency data analysis. SKS performed statistical analysis of the two study cohorts and interpreted these results. RDN and SKS edited the manuscript. MNS coordinated data acquisition from multiple institutions, oversaw rfMRI data latency analysis and manuscript production. All authors contributed to the intellectual concept investigated in this study and have agreed to be accountable for all aspects of their work. All authors have read and approved the final manuscript. RDN and MNS confirm the authenticity of all the raw data.
Ethics approval and consent to participate
IRB approval (approval no. UT Houston IRB #HSC-MS-17-0092) was obtained at both Washington University and McGovern Medical School for images obtained from St. Louis Children's Hospital, Children's Memorial Hermann Hospital, and the multi-institutional ADHD 200 dataset of the 1000 Functional Connectomes Project, International Neuroimaging Data-Sharing Initiative hosted by the Neuroimaging Tools & Resources Collaboratory. Informed consent was obtained from the patient's caregivers or by capable patients aged >18 years old, as required by each database and institution.
Patient consent for publication
All identifiable patient information was removed from the MRI data obtained prior to processing. Then, images were randomly separated into training, validation, and test datasets for comparison. Informed consent was obtained for each study patient from the patient's caregivers or by capable patients aged >18 years old for publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Aaberg KM, Gunnes N, Bakken IJ, Lund Søraas C, Berntsen A, Magnus P, Lossius MI, Stoltenberg C, Chin R, Surén P. Incidence and prevalence of childhood epilepsy: A nationwide cohort study. Pediatrics. 2017;139(e20163908) doi: 10.1542/peds.2016-3908. [DOI] [PubMed] [Google Scholar]
- 2.Kwan P, Brodie MJ. Early identification of refractory epilepsy. N Engl J Med. 2000;342:314–319. doi: 10.1056/NEJM200002033420503. [DOI] [PubMed] [Google Scholar]
- 3.Smith SJ. EEG in the diagnosis, classification, and management of patients with epilepsy. J Neurol Neurosurg Psychiatry. 2005;76 (Suppl 2):ii2–ii7. doi: 10.1136/jnnp.2005.069245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mihara T, Inoue Y, Matsuda K, Tottori T, Otsubo T, Watanabe Y, Hiyoshi T, Kubota Y, Yagi K, Seino M. Recommendation of early surgery from the viewpoint of daily quality of life. Epilepsia. 1996;37 (Suppl 3):S33–S36. doi: 10.1111/j.1528-1157.1996.tb01817.x. [DOI] [PubMed] [Google Scholar]
- 5.Westerveld M, Sass KJ, Chelune GJ, Hermann BP, Barr WB, Loring DW, Strauss E, Trenerry MR, Perrine K, Spencer DD. Temporal lobectomy in children: Cognitive outcome. J Neurosurg. 2000;92:24–30. doi: 10.3171/jns.2000.92.1.0024. [DOI] [PubMed] [Google Scholar]
- 6.Baulac M, de Boer H, Elger C, Glynn M, Kälviäinen R, Little A, Mifsud J, Perucca E, Pitkänen A, Ryvlin P. Epilepsy priorities in Europe: A report of the ILAE-IBE epilepsy advocacy Europe task force. Epilepsia. 2015;56:1687–1695. doi: 10.1111/epi.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg AT, Baca CB, Loddenkemper T, Vickrey BG, Dlugos D. Priorities in pediatric epilepsy research: Improving children's futures today. Neurology. 2013;81:1166–1175. doi: 10.1212/WNL.0b013e3182a55fb9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dlugos DJ. The early identification of candidates for epilepsy surgery. Arch Neurol. 2001;58:1543–1546. doi: 10.1001/archneur.58.10.1543. [DOI] [PubMed] [Google Scholar]
- 9.Gilliam F, Kuzniecky R, Meador K, Martin R, Sawrie S, Viikinsalo M, Morawetz R, Faught E. Patient-oriented outcome assessment after temporal lobectomy for refractory epilepsy. Neurology. 1999;53:687–694. doi: 10.1212/wnl.53.4.687. [DOI] [PubMed] [Google Scholar]
- 10.Guerrini R. Epilepsy in children. Lancet. 2006;367:499–524. doi: 10.1016/S0140-6736(06)68182-8. [DOI] [PubMed] [Google Scholar]
- 11.Shah MN, Nguyen RD, Pao LP, Zhu L, CreveCoeur TS, Mitra A, Smyth MD. Role of resting state MRI temporal latency in refractory pediatric extratemporal epilepsy lateralization. J Magn Reson Imaging. 2019;49:1347–1355. doi: 10.1002/jmri.26320. [DOI] [PubMed] [Google Scholar]
- 12.Guidelines for neuroimaging evaluation of patients with uncontrolled epilepsy considered for surgery. Commission on neuroimaging of the international league against epilepsy. Epilepsia. 1998;39:1375–1376. doi: 10.1111/j.1528-1157.1998.tb01341.x. [DOI] [PubMed] [Google Scholar]
- 13.Recommendations for neuroimaging of patients with epilepsy. Commission on neuroimaging of the international league against epilepsy. Epilepsia. 1997;38:1255–1256. doi: 10.1111/j.1528-1157.1997.tb01226.x. [DOI] [PubMed] [Google Scholar]
- 14.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 15.Mitra A, Snyder AZ, Blazey T, Raichle ME. Lag threads organize the brain's intrinsic activity. Proc Natl Acad Sci USA. 2015;112:E2235–E2244. doi: 10.1073/pnas.1503960112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitra A, Snyder AZ, Hacker CD, Raichle ME. Lag structure in resting-state fMRI. J Neurophysiol. 2014;111:2374–2391. doi: 10.1152/jn.00804.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jirsa VK, Proix T, Perdikis D, Woodman MM, Wang H, Gonzalez-Martinez J, Bernard C, Bénar C, Guye M, Chauvel P, Bartolomei F. The virtual epileptic patient: Individualized whole-brain models of epilepsy spread. Neuroimage. 2017;145:377–388. doi: 10.1016/j.neuroimage.2016.04.049. [DOI] [PubMed] [Google Scholar]
- 18.Pizoli CE, Shah MN, Snyder AZ, Shimony JS, Limbrick DD, Raichle ME, Schlaggar BL, Smyth MD. Resting-state activity in development and maintenance of normal brain function. Proc Natl Acad Sci USA. 2011;108:11638–11643. doi: 10.1073/pnas.1109144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Proix T, Bartolomei F, Guye M, Jirsa VK. Individual brain structure and modelling predict seizure propagation. Brain. 2017;140:641–654. doi: 10.1093/brain/awx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bharath RD, Panda R, Raj J, Bhardwaj S, Sinha S, Chaitanya G, Raghavendra K, Mundlamuri RC, Arimappamagan A, Rao MB, et al. Machine learning identifies ‘rsfMRI epilepsy networks’ in temporal lobe epilepsy. Eur Radiol. 2019;29:3496–3505. doi: 10.1007/s00330-019-5997-2. [DOI] [PubMed] [Google Scholar]
- 22.Boerwinkle VL, Mirea L, Gaillard WD, Sussman BL, Larocque D, Bonnell A, Ronecker JS, Troester MM, Kerrigan JF, Foldes ST, et al. Resting-state functional MRI connectivity impact on epilepsy surgery plan and surgical candidacy: Prospective clinical work. J Neurosurg Pediatr. 2020:1–8. doi: 10.3171/2020.1.PEDS19695. (Online ahead of print) [DOI] [PubMed] [Google Scholar]
- 23.Boerwinkle VL, Mohanty D, Foldes ST, Guffey D, Minard CG, Vedantam A, Raskin JS, Lam S, Bond M, Mirea L, et al. Correlating resting-state functional magnetic resonance imaging connectivity by independent component analysis-based epileptogenic zones with intracranial electroencephalogram localized seizure onset zones and surgical outcomes in prospective pediatric intractable epilepsy study. Brain Connect. 2017;7:424–442. doi: 10.1089/brain.2016.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah MN, Mitra A, Goyal MS, Snyder AZ, Zhang J, Shimony JS, Limbrick DD, Raichle ME, Smyth MD. Resting state signal latency predicts laterality in pediatric medically refractory temporal lobe epilepsy. Childs Nerv Syst. 2018;34:901–910. doi: 10.1007/s00381-018-3770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darcy AM, Louie AK, Roberts LW. Machine learning and the profession of medicine. JAMA. 2016;315:551–552. doi: 10.1001/jama.2015.18421. [DOI] [PubMed] [Google Scholar]
- 26.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 28.Jordan MI, Mitchell TM. Machine learning: Trends, perspectives, and prospects. Science. 2015;349:255–260. doi: 10.1126/science.aaa8415. [DOI] [PubMed] [Google Scholar]
- 29.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 30.Chen T, Guestrin C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, pp785-794, 2016. [Google Scholar]
- 31.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 32.Fern A, Givan R. Online ensemble learning: An empirical study. Mach Learn. 2003;53:71–109. [Google Scholar]
- 33.Mennes M, Biswal BB, Castellanos FX, Milham MP. Making data sharing work: The FCP/INDI experience. Neuroimage. 2013;82:683–691. doi: 10.1016/j.neuroimage.2012.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellec P, Chu C, Chouinard-Decorte F, Benhajali Y, Margulies DS, Craddock RC. The neuro bureau ADHD-200 preprocessed repository. Neuroimage. 2017;144:275–286. doi: 10.1016/j.neuroimage.2016.06.034. [DOI] [PubMed] [Google Scholar]
- 35.Brier MR, Thomas JB, Snyder AZ, Benzinger TL, Zhang D, Raichle ME, Holtzman DM, Morris JC, Ances BM. Loss of intranetwork and internetwork resting state functional connections with Alzheimer's disease progression. J Neurosci. 2012;32:8890–8899. doi: 10.1523/JNEUROSCI.5698-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowerko D. https://www.mathworks.com/matlabcentral/fileexchange/49823-calc_overlap_twonormal(s1,s2,mu1,mu2,xstart,xend,xinterval). MATLAB Central File Exchange. Retrieved July 19, 2021. [Google Scholar]
- 37.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 38.Verleysen M, François D. The Curse of Dimensionality in Data Mining and Time Series Prediction. Vol 3521. IWANN 2005: Computational Intelligence and Bioinspired Systems, pp758-770, 2005. [Google Scholar]
- 39.Meyer D, Dimitriadou E, Hornik K, Weingessel A, Leisch F. e1071: Misc functions of the department of statistics, probability theory group (Formerly: E1071), TU Wien, 2017. [Google Scholar]
- 40. R Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2019. Available from: http://www.r-project.org/index.html. [Google Scholar]
- 41. RStudio Team: RStudio: Integrated Development for R. RStudio, Inc., Boston MA (Computer Software v0.98.1074), 2015. [Google Scholar]
- 42.Sasaki Y. The truth of the F-measure. Teach Tutor Mater, 2007. [Google Scholar]
- 43.Baca CB, Vickrey BG, Vassar S, Hauptman JS, Dadour A, Oh T, Salamon N, Vinters HV, Sankar R, Mathern GW. Time to pediatric epilepsy surgery is related to disease severity and nonclinical factors. Neurology. 2013;80:1231–1239. doi: 10.1212/WNL.0b013e3182897082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loddenkemper T, Holland KD, Stanford LD, Kotagal P, Bingaman W, Wyllie E. Developmental outcome after epilepsy surgery in infancy. Pediatrics. 2007;119:930–935. doi: 10.1542/peds.2006-2530. [DOI] [PubMed] [Google Scholar]
- 45.Berg AT, Vickrey BG, Testa FM, Levy SR, Shinnar S, DiMario F, Smith S. How long does it take for epilepsy to become intractable? A prospective investigation. Ann Neurol. 2006;60:73–79. doi: 10.1002/ana.20852. [DOI] [PubMed] [Google Scholar]
- 46.Trevathan E, Gilliam F. Lost years: Delayed referral for surgically treatable epilepsy. Neurology. 2003;61:432–433. doi: 10.1212/wnl.61.4.432. [DOI] [PubMed] [Google Scholar]
- 47.Elger CE, Helmstaedter C, Kurthen M. Chronic epilepsy and cognition. Lancet Neurol. 2004;3:663–672. doi: 10.1016/S1474-4422(04)00906-8. [DOI] [PubMed] [Google Scholar]
- 48.Sperling MR. Sudden unexplained death in epilepsy. Epilepsy Curr. 2001;1:21–23. doi: 10.1046/j.1535-7597.2001.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen RD, Kennady EH, Smyth MD, Zhu L, Pao LP, Swisher SK, Rosas A, Mitra A, Patel RP, Lankford J, et al. Convolutional neural networks for pediatric refractory epilepsy classification using resting-state functional magnetic resonance imaging. World Neurosurg. 2021;149:e1112–e1122. doi: 10.1016/j.wneu.2020.12.131. [DOI] [PubMed] [Google Scholar]
- 50.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference and Prediction. Springer, New York, NY, 2016. [Google Scholar]
- 51.Di Martino A, O'Connor D, Chen B, Alaerts K, Anderson JS, Assaf M, Balsters JH, Baxter L, Beggiato A, Bernaerts S, et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci Data. 2017;4(170010) doi: 10.1038/sdata.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M, et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 2014;19:659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schölkopf B, Smola AJ. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond. Adaptive Computation and Machine Learning Series, 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code described during the present study is available from the corresponding author on reasonable request. The data will be made available pending approval from the Institutional Review Board at Washington University and McGovern Medical School. Upon approval, the data that support the findings of this study will be made available from the corresponding author upon reasonable request.