Abstract
Nasopharyngeal swabs are considered the preferential collection method for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnostics. Less invasive and simpler alternative sampling procedures, such as saliva collection, are desirable. We compared saliva specimens and nasopharyngeal (NP) swabs with respect to sensitivity in detecting SARS-CoV-2. A nasopharyngeal and two saliva specimens (collected by spitting or oral swabbing) were obtained from >2500 individuals. All samples were tested by RT-qPCR, detecting RNA of SARS-CoV-2. The test sensitivity was compared on the two saliva collections with the nasopharyngeal specimen for all subjects and stratified by symptom status and viral load. Of the 2850 patients for whom all three samples were available, 105 were positive on NP swab, whereas 32 and 23 were also positive on saliva spitting and saliva swabbing samples, respectively. The sensitivity of the RT-qPCR to detect SARS-CoV-2 among NP-positive patients was 30.5% (95% CI, 1.9%–40.2%) for saliva spitting and 21.9% (95% CI, 14.4%–31.0%) for saliva swabbing. However, when focusing on subjects with medium to high viral load, sensitivity on saliva increased substantially: 93.9% (95% CI, 79.8%–99.3%) and 76.9% (95% CI, 56.4%–91.0%) for spitting and swabbing, respectively, regardless of symptomatic status. Our results suggest that saliva cannot readily replace nasopharyngeal sampling for SARS-CoV-2 diagnostics but may enable identification of the most contagious cases with medium to high viral loads.
Massive RT-qPCR–based testing for the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA is a key element in the strategy to control the current coronavirus disease 2019 (COVID-19) pandemic. Currently, collecting samples from the upper respiratory tract is recommended for diagnostic testing by the World Health Organization and (American and European) Centers for Disease Control and Prevention, with nasopharyngeal (NP) swabs being considered the standard collection procedure1 , 2 (https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html, last accessed April 8, 2021). Although extremely sensitive, this sampling procedure is relatively invasive, causing discomfort and anxiety in individuals undergoing the procedure, and relies on trained health care workers wearing full personal protective equipment to obtain samples.
Before the outbreak of SARS-CoV-2, several studies have reported on the utility of saliva as a diagnostic specimen for testing respiratory viruses.3, 4, 5, 6 In addition, studies related to SARS-CoV-2 have shown that the virus binds to angiotensin-converting enzyme 2 receptors that are present in epithelial cells of the oral mucosa, suggesting the use of saliva as a potential sample for SARS-CoV-2 detection. The noninvasive nature of saliva collection in a simple container makes this specimen a valuable biomaterial. Besides, saliva sampling could be a solution in resource-limiting settings with respect to health care personnel, and could reduce the amount of contact required between a health care provider and the patient, lowering the risk of transmission and personal protective equipment use. As saliva sampling is patient friendly, it can also be of value when testing in children during a SARS-CoV-2 outbreak in schools.
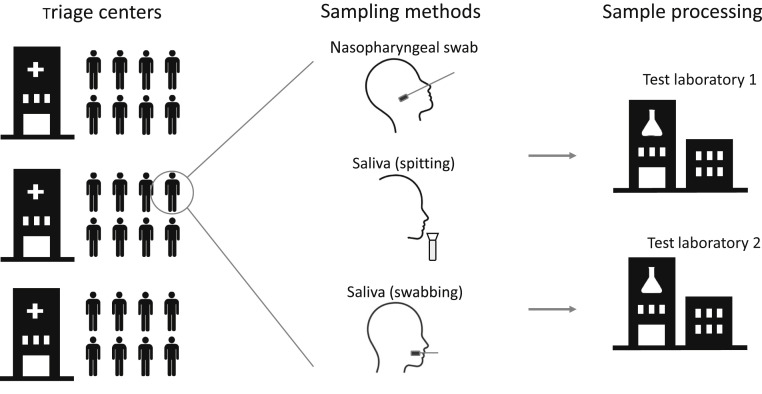
Although several recent studies have documented the potential utility of saliva for diagnostic testing of SARS-CoV-2,7, 8, 9, 10, 11, 12, 13 these studies experience one or more limitations (ie, nonpaired study design, small cohorts, and testing in biased populations, such as previously confirmed positive cases and/or hospitalized patients). Herein, we set out to prospectively evaluate the potential use of saliva samples for diagnostic testing of SARS-CoV-2 using a large population of >2500 individuals in triage centers in Belgium. Individuals were sampled using two saliva collection devices and a matching nasopharyngeal swab, and samples were analyzed by two test laboratories to independently verify conclusions (Figure 1 ).
Figure 1.
Overview of study design. Study participants were sampled at triage centers using a nasopharyngeal swab and two different saliva collection devices. Samples were processed at two different test laboratories using independent sample processing workflows.
Materials and Methods
Study Design
This study has been reported using the Standards for Reporting of Diagnostic Accuracy Studies guidelines 2015.14 As part of the Belgian national testing platform, asymptomatic and symptomatic individuals suggestive of COVID-19 at centralized triage centers in Belgium were prospectively enrolled. More than 2500 individuals were tested in these triage centers from June 2020 to July 2020, at the end of the first infection wave. All individuals aged ≥18 years who presented at triage centers were considered eligible.
Sample and Data Collection
Study samples were collected by trained mobile teams. Individuals were sampled using three different procedures: i) a nasopharyngeal swab sample representing the standard comparator for SARS-CoV-2 diagnostics, ii) a saliva sample collected through self-sampling with a commercial saliva spitting device (Saliva RNA Collection and Preservation Device; Norgen Biotek, Thorold, ON, Canada), and iii) a saliva sample collected through self-sampling with a commercial oral swabbing device (Oracollect RNA; DNA Genotek, Kanata, Ontario, Canada). For the saliva spitting device (Norgen Biotek), the collected volume of saliva was 2 mL. For the saliva swabbing device (DNA Genotek), the collected volume of saliva was approximately 300 μL. NP sample collection was performed using iClean NP swabs (Chenyang Global, Shenzhen, China). All samples were collected in a transport buffer that inactivates the virus and stabilizes the RNA [Norgen Biotek or DNA Genotek buffer for the respective saliva samples and 2 mL of DNA/RNA shield buffer (Zymo Research, Irvine, CA), custom prefilled in Vacuette tubes (Greinder Bio-One, Vilvoorde, Belgium) for the nasopharyngeal samples]. Participants in the study were asked not to eat, drink, smoke, or use chewing gum 30 minutes preceding saliva sampling. Saliva samples were collected according to the manufacturer's instructions. These instructions were available as an instruction sheet with each saliva collection device. Instructions were communicated to each participant by a health care professional before sampling. Participants were not instructed to produce deep throat saliva or gargle before saliva collection. For the swabbing device, participants were instructed to place the swab between the right cheek and gum, swab 10 times, and repeat for the left cheek. After sample collection, a short survey was completed and data were collected on age group, ease of use of the saliva devices, comfort of saliva sampling versus nasopharyngeal sampling, and symptomatic status. To enquire about symptomatic status, the case definition of Sciensano, the Belgian Institute for Public Health, was used. The case definition stated that a possible case of COVID-19 had at least one of the following main symptoms that occurred acutely without other plausible cause: cough, dyspnea, thoracic pain, anosmia, or dysgeusia; or at least two of the following symptoms that occurred without other plausible cause: fever, muscle strain, fatigue, rhinitis, sore throat, headache, anorexia, watery diarrhea, acute confusion, or sudden fall; or worsening of chronic respiratory symptoms (chronic obstructive pulmonary disease, asthma, or chronic cough) without other plausible cause. This study was approved by the ethical review committee of the University Hospital of Leuven on May 29, 2020, as S64125., Vilvoorde, Belgium.
Test Methods
SARS-CoV-2 testing was performed by two independent test laboratories, applying different RNA extraction and RT-qPCR workflows (see below). Note that, because of logistics reasons, not all samples were analyzed by both laboratories. After sample collection, samples were shipped to one of the laboratories, where the required volume of sample for RNA extraction was removed from the sample collection tube. Sample collection tubes were subsequently shipped to the other test laboratory for analysis. Before sample transfer, samples were vortex mixed and centrifuged. For highly viscous samples, aspiration was performed at low speed.
Nucleic Acid Extraction and RT-qPCR in Laboratory 1
RNA was extracted using the Total RNA Purification Kit (Norgen Biotek; number 24300), according to the manufacturer's instructions, using 200 μL viral transport medium [for the nasopharyngeal swab, DNA/RNA Shield (number R1100-250; Zymo Research) or 200 μL saliva collected with the spitting or swabbing device (ie, saliva mixed with transport buffer)] as input in the 96-well filter plate. Samples were supplemented with 200 μL lysis buffer, 200 μL ethanol, 4 μL of a proprietary 700-nucleotides spike-in control RNA (5000 copies, produced through in vitro transcription), and carrier RNA [200 ng of yeast tRNA (Roche, Vilvoorde, Belgium; number 10109517001)]. Filter plates were further processed with a centrifuge (5810R with rotor A-4-81; both from Eppendorf [Aarschot, Belgium]). RNA was eluted from the filter plates using 50 μL elution buffer (nuclease-free water), resulting in approximately 45 μL eluate. RNA extractions were simultaneously performed for 94 patient samples and 2 negative controls (nuclease-free water). To the eluate of one of the negative control wells, 7500 (digital PCR value assigned) RNA copies of positive control RNA (Synthetic SARS-CoV-2 RNA Control 2; Twist Biosciences, San Francisco, CA; number 102024) were added to serve as positive PCR control.
RNA eluate (6 μL) was used as input for a 20-μL duplex RT-qPCR in a CFX384 real-time quantitative PCR instrument using 10 μL iTaq one-step RT-qPCR mastermix (Bio-Rad, Temse, Belgium; number 1725141), according to the manufacturer's instructions. Reactions were set up using 400 nmol/L final concentration of primers and 250 nmol/L of a hydrolysis probe. Primers and probes were synthesized by Integrated DNA Technologies (Coralville, IA) using clean-room GMP production. For detection of the SARS-CoV-2 virus, the Charité E gene assay was used (FAM)15; for the internal control, a proprietary hydrolysis probe assay (HEX) was used. Quantification cycle (Cq) values were generated using the FastFinder software version 3.300.5 (UgenTec, Hasselt, Belgium). The FastFinder software was also used to call a sample positive or negative for SARS-CoV-2. Only batches with a clean negative control and a positive control in the expected range were approved. Proper RNA extraction and RT-qPCR was confirmed by observing spike-in RNA signal in each sample well in the expected range.
Nucleic Acid Extraction and RT-qPCR in Laboratory 2
RNA extraction was performed using a magnetic bead–based RNA extraction method developed by University of Liège (CoRNA kit) and according to the recommended protocol. For nasopharyngeal swab samples, 200 μL of sample was transferred to 11 μL of a proteinase K solution (20 mL/mL). For both saliva devices, 100 μL of saliva was transferred to 175 μL of a lysis buffer mix [11:164 (vol/vol) of proteinase K solution (20 mg/mL) + lysis buffer]. All samples were spiked with the MS2 phage as internal control [10 μL, concentration proprietary information from supplier Thermo Fisher Scientific (Waltham, MA) kit A47814], and in presence of carrier RNA (10 μL; 20 ng/μL; Merck/Roche, Vilvoorde, Belgium; number 10109517001) to increase RNA extraction efficiency. The multiplex RT-qPCR was performed on 5 μL of RNA eluate using TaqPath COVID-19 Combo Kit (comprising ORF1ab, N gene, S gene, and MS2 as internal control; number A47814; Thermo Fisher Scientific), TaqPath positive control kit (containing a stock of 104 copies SARS-CoV-2/μL; number A47816; Thermo Fisher Scientific), and TaqPath 1-Step Multiplex Master Mix (no ROX; number A28523; Thermo Fisher Scientific), following the manufacturer's instructions. Positive control was diluted to 25 copies/μL in control dilution buffer, and 2 μL (50 copies) was further diluted in 3 μL nuclease-free water, which was added to the well of the RT-PCR plate. Cq values were generated using the FastFinder software version 3.300.5. The FastFinder software was also used to call a sample positive or negative for SARS-CoV-2. Results were approved when a clean negative control and a positive control in the expected range were obtained. Correct RNA extraction and RT-qPCR setup was also confirmed by controlling MS2 amplification in each sample well (applying an MS2 Cq cutoff of 33).
Digital PCR Quantification of Positive Control RNA Samples
Digital PCR was performed on a QX200 instrument (Bio-Rad) using the One-Step RT-ddPCR Advanced Kit for Probes (Bio-Rad; number 1864022), according to the manufacturer's instructions. Briefly, 22 μL prereactions were prepared, consisting of 5 μL 4× supermix, 2 μL reverse transcriptase, 6 μL positive control RNA (see further), 15 mmol/L dithiothreitol, 900 nmol/L of each forward and reverse primer, and 250 nmol/L E-gene hydrolysis probe (FAM).15 A total of 20 μL of the prereaction was used for droplet generation using the QX200 Droplet Generator (Bio-Rad), followed by careful transfer to a 96-well PCR plate for thermocycling: 60 minutes at 46 °C for reverse transcription, 10 minutes at 95 °C for enzyme activation, 40 cycles of 30 seconds of denaturation at 95 °C and 1 minute of annealing/extension at 59 °C, and finally 10 minutes at 98 °C for enzyme deactivation. Droplets were analyzed by the QX200 Droplet Reader (Bio-Rad) and QuantaSoft software (Bio-Rad).
With an input of 4000 RNA copies per reaction (Armored RNA Quant SARS-CoV-2 Panel; Asuragen, Austin, TX; number 52036), the digital PCR result was 875 cDNA copies (or 21.88% of the expected number). Of note, RNA was not extracted on the Armored RNA material; instead, a short heat release of RNA was done per the manufacturer's instructions. With an RNA input of 750 copies per reaction (Synthetic SARS-CoV-2 RNA Control 2; Twist Biosciences; number 102024), the digital PCR result was 150 cDNA copies (or 20% of the expected number).
Viral Load Cuffoff Concentration Determination
The Cq value cutoff for viral load classification was determined on the basis of the Cq correlation between NP and saliva samples. The cutoff represents the NP viral load above which saliva samples show highest sensitivity for SARS-CoV-2 detection in NP positive samples, and below which sensitivity for SARS-CoV-2 detection in saliva decreases to almost 0%. This analysis resulted in an E-gene Cq cutoff of 24.5 and an N-gene Cq cutoff of 25.5 for laboratory 1 and 2, respectively.
To convert the laboratory 1 Cq value cutoff to SARS-CoV-2 RNA copies/mL of viral transport medium, a six-point 10-fold serial dilution of Armored RNA was generated in triplicate (from 2.19 × 108 to 2.19 × 103 digital PCR value assigned copies/mL), followed by RNA extraction and RT-qPCR (using the laboratory 1 method). On the basis of the slope of −3.381, the y intercept of 45.387, and the r 2 value of 0.995, the laboratory 1 Cq value cutoff corresponds to 1.51 × 106 copies/mL viral transport medium. This viral copy number corresponds to a viral load that is typically associated with infectious individuals in literature. For instance, van Kampen et al16 demonstrated that the probability of isolating infectious SARS-CoV-2 was <5% when the viral load was <6.63 log10 RNA copies/mL. Therefore, this was referred as high viral load.
Sample Size Calculation
The sample size was computed for assessment of a hypothesis on noninferior SARS-CoV-2 positivity on saliva compared with on nasopharyngeal specimen in paired testing, as proposed by Tang et al,17 using target values from a systematic review.18 A confidence of 95%, a power of 80%, a sensitivity of the test in NP samples of 95%, a proportion of saliva-negative/NP-positive samples of 5%, and 0.90 were accepted as benchmark for the relative positivity rate (saliva/NP), which yielded 84 SARS-CoV-2–positive subjects needed. These could be found in a study population of 841 to 8410 subjects, assuming a prevalence of 1% to 10%. Given the substantially larger contrast in test positivity between saliva and NP specimens, study enrollment was stopped after reaching 2850 inclusions.
Data Analysis
All results presented in the article are based on data generated by laboratory 1, unless stated otherwise. Only patients with available results for the three specimens were included. Patient paired data were used to construct 2 × 2 contingency tables. The sensitivity of SARS-CoV-2 testing was defined by the proportion of saliva-positive patients (index positive) among those who were positive on NP swab (reference positive). The test positivity ratio was also computed as the proportion with a positive index test/the proportion with a positive reference or comparator test. The 95% CIs for binomial data were computed as well as for ratios of paired proportions. Three separate analyses were performed: one comparing spit samples with NP samples, a second comparing swab samples with NP samples, and a third comparing spit and swab samples. NP samples were considered the standard comparator or reference. In addition, the estimations were stratified by viral load (categorized as high and low) and symptoms (categorized as symptomatic and asymptomatic).
All statistical analyses were performed using Stata statistical software version 14.2 (Stata, College Station, TX). Statistical significance was defined at P < 0.05.
Data Availability
RT-qPCR data from test laboratory 1 and test laboratory 2 as well as the survey data are provided in Supplemental Tables S1, S2, and S3.
Results
Patient Characteristics
In total, 2954 individuals were recruited between June 2020 and July 2020. Data were excluded from 104 (3.5%) participants because of missing NP results. A total of 2268 individuals were sampled with a nasopharyngeal swab and both saliva collection methods, whereas 2469 and 2649 matched samples were available for spit and swab samples, respectively. The median age group of participants was between 31 and 40 years old, and symptomatic status data were available for 2071 individuals.
Comfort and Ease of Saliva Self-Sampling Devices
Although saliva sampling was generally perceived as more comfortable than nasopharyngeal sampling (Supplemental Figure S1A), study participants scored the ease of use of the saliva swabbing device significantly higher than that of the saliva spitting device (P < 0.0001, U-test) (Supplemental Figure S1B).
Saliva Sampling Identifies Individuals with Medium to High Viral Load
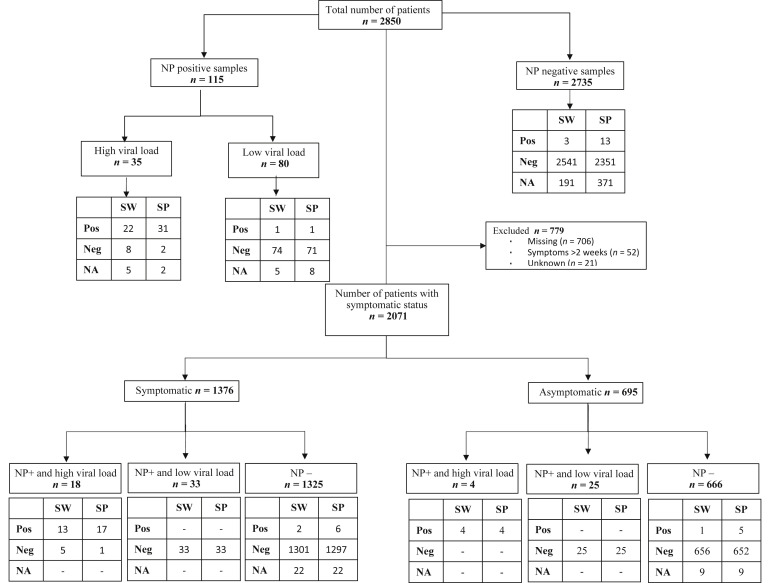
Of 2850 nasopharyngeal swab samples analyzed by laboratory 1, 115 (4.0%) were SARS-CoV-2 positive (Figure 2 ). In positive NP samples, 30.4% (35/115) showed high viral load. There were 105 of 115 nasopharyngeal-positive samples for which a matching saliva spitting sample was available, and 105 of 115 nasopharyngeal-positive samples for which a matching saliva swabbing sample was available (Figure 2). We observed 32 of 105 (sensitivity, 30.5%; 95% CI, 21.9%–40.2%) in the spitting sample and 23 of 105 (sensitivity, 21.9%; 95% CI, 14.4%–31.0%) in the swabbing samples that were SARS-CoV-2 positive, indicating reduced overall sensitivity in saliva for SARS-CoV-2 detection (Figures 2 and 3 and Table 1 ).
Figure 2.
Study flow chart and results of RT-qPCR of patients enrolled in the saliva SARS-CoV-2 diagnostic test accuracy study by specimen type, viral load level, and symptomatic status (laboratory 1). NA, not available; Neg, negative; NP, nasopharyngeal samples; Pos, positive; SP, spit sample; SW, swab samples.
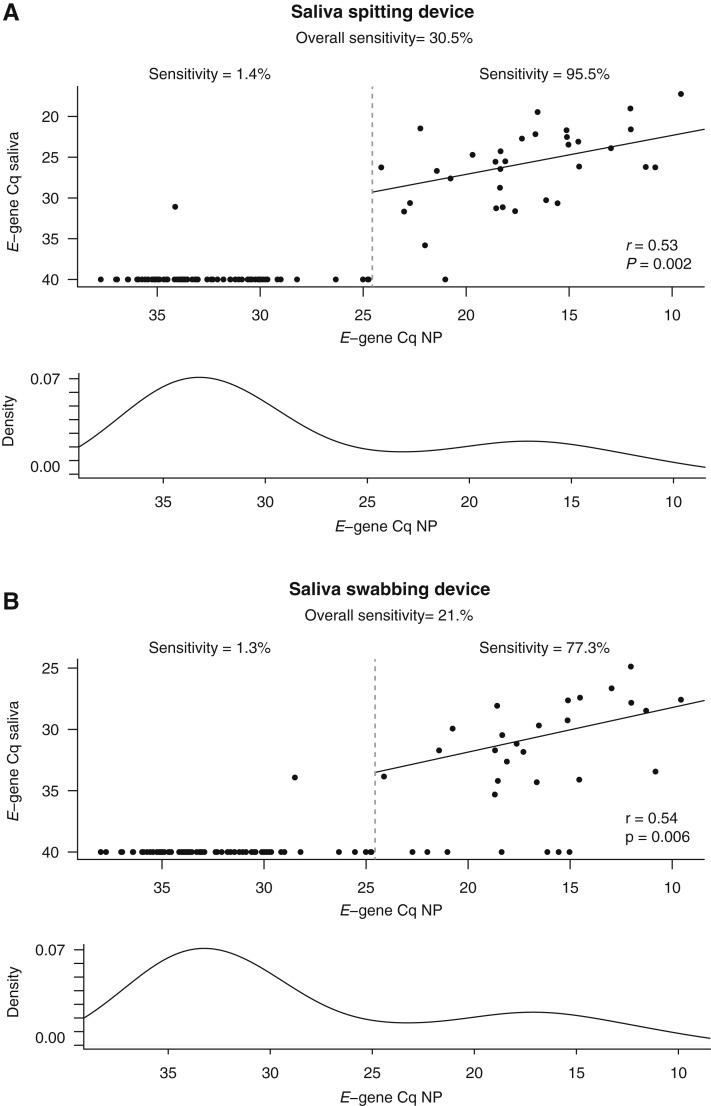
Figure 3.
E-gene quantification cycle values from laboratory 1 in SARS-CoV-2–positive nasopharyngeal samples (NP) and matching saliva samples. A: Results obtained with the saliva spitting device. B: Results obtained with the saliva swabbing device. Plots indicate the correlation coefficient (r) value and P value (Pearson correlation test) calculated using only those samples with a nasopharyngeal E-gene Cq value of <24.5 (threshold marked with a dashed line). Bottom graphs in each panel represent the E-gene Cq-value distribution in the NP-positive samples (y axis represents kernel density estimate).
Table 1.
Sensitivity and Test Positivity Ratios of SARS-CoV-2 Testing on NP Specimens versus Saliva (Collected by Swabs or Spitting) and on Swab Saliva Samples versus Spitting Saliva Samples by Viral Load Level and Symptom Status (Laboratory 1)
| Comparisons |
Viral load categories | Symptom status | Reference positive |
Reference negative |
Sensitivity, % |
Test positivity ratio |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | Reference | Index positive | Index negative | Index positive | Index negative | TP/(TP + FN) | 95% CI | (TP and FP)/(TP + FN) | 95% CI | ||
| Swab | NP specimen | All | All | 23 | 82 | 3 | 2541 | 21.9 | 14.4–31.0 | 0.25 | 0.15–0.31 |
| Spit | NP specimen | All | All | 32 | 73 | 13 | 2351 | 30.5 | 21.9–40.2 | 0.43 | 0.23–0.41 |
| Swab | Spit | All | All | 23 | 14 | 0 | 2231 | 62.2 | 44.8–77.5 | 0.62 | 0.48–0.80 |
| Swab | NP specimen | All | Yes | 13 | 38 | 2 | 1301 | 25.5 | 14.3–39.6 | 0.29 | 0.16–0.41 |
| Spit | NP specimen | All | Yes | 17 | 34 | 6 | 1297 | 33.3 | 20.8–47.9 | 0.45 | 0.23–0.49 |
| Swab | Spit | All | Yes | 15 | 8 | 0 | 1309 | 65.2 | 42.7–83.6 | 0.65 | 0.48–0.88 |
| Swab | NP specimen | All | No | 4 | 25 | 1 | 656 | 13.8 | 3.9–31.7 | 0.17 | 0.06–0.34 |
| Spit | NP specimen | All | No | 4 | 25 | 5 | 652 | 13.8 | 3.9–31.7 | 0.31 | 0.06–0.34 |
| Swab | Spit | All | No | 5 | 4 | 0 | 668 | 55.6 | 21.2–83.6 | 0.56 | 0.31–1.00 |
| Swab | NP specimen | High | All | 22 | 8 | 0 | 0 | 73.3 | 54.1–87.7 | 0.73 | 0.59–0.91 |
| Spit | NP specimen | High | All | 31 | 2 | 0 | 0 | 93.9 | 79.8–99.3 | 0.94 | 0.86–1.02 |
| Swab | Spit | High | All | 20 | 6 | 0 | 2 | 76.9 | 56.4–91.0 | 0.77 | 0.62–0.95 |
| Swab | NP specimen | High | Yes | 13 | 5 | 0 | 0 | 72.2 | 46.5–90.3 | 0.72 | 0.54–0.96 |
| Spit | NP specimen | High | Yes | 17 | 1 | 0 | 0 | 94.4 | 72.7–99.9 | 0.94 | 0.81–1.06 |
| Swab | Spit | High | Yes | 13 | 4 | 1 | 0 | 76.5 | 50.1–93.2 | 0.82 | 0.59–1.00 |
| Swab | NP specimen | High | No | 4 | 0 | 0 | 0 | 100.0 | 39.8–100.0 | 1.00 | 1.00–1.00 |
| Spit | NP specimen | High | No | 4 | 0 | 0 | 0 | 100.0 | 39.8–100.0 | 1.00 | 1.00–1.00 |
| Swab | Spit | High | No | 4 | 0 | 0 | 0 | 100.0 | 39.8–100.0 | 1.00 | 1.00–1.00 |
FN, false negative; FP, false positive; NP, nasopharyngeal; TP, true positive.
However, a significantly higher nasopharyngeal viral load was observed in patients with a true-positive saliva sample compared with patients with a false-negative saliva sample (spitting device: log2 fold change = 14.89, P = 3.79 × 10−15; swabbing device: log2 fold change = 14.7, P = 3.67 × 10−12; U-test) (Supplemental Figure S2).
Individuals with an E-gene Cq >24.5 in the nasopharyngeal sample (corresponding to 1.51 × 106 copies/mL viral transport medium, as determined by digital PCR and further referred to as low viral load) almost always presented with a negative saliva sample [sensitivity, 1.4% (95% CI, 0.07%–7.5%); and sensitivity, 1.3% (95% CI, 0.07%–7.2%) in the saliva spitting and saliva swabbing sample, respectively] (Figure 3). In contrast, for individuals with a high viral load (E-gene Cq < 24.5 in the nasopharyngeal sample), concordance between the nasopharyngeal and matching saliva sample improved dramatically, especially for the saliva spitting device, resulting in high sensitivity in this subgroup [sensitivity, 95.5% (95% CI, 77.2%–99.9%); and sensitivity, 77.3% (95% CI, 54.6%–92.2%) for the saliva obtained by spitting and swabbing, respectively] (Figure 3 and Table 1). In addition, a significant positive correlation was observed between E-gene Cq values in the nasopharyngeal and saliva samples for those individuals with high viral load (spitting device: r = 0.53, P = 0.002; swabbing device: r = 0.54, P = 0.006; Pearson correlation). Notably, similar findings were obtained on the basis of test results generated by laboratory 2 (Supplemental Figure S3 and Supplemental Table S4). In the medium to high viral load subgroup, saliva spitting resulted in a sensitivity of 96.9% (95% CI, 83.8%–99.9%), whereas saliva swabbing resulted in a sensitivity of 60.7% (95% CI, 40.6%–78.5%).
To assess whether the poor sensitivity in saliva for SARS-CoV-2 detection could be due to sampling issues during saliva collection, the human gene RPS18 was quantified using RT-qPCR on a representative set of RNA samples used for SARS-CoV-2 detection. No differences in RPS18 levels were observed between saliva-positive and saliva-negative samples for any of the saliva sampling devices, suggesting that sampling issues do not explain the false-negative saliva samples (Supplemental Figure S4).
SARS-CoV-2 Detection in Saliva, according to Symptom Status
Presence or absence of symptoms of COVID-19 in 2123 study participants was registered. From these participants, 1376 (64.8%) were symptomatic, 695 (32.7%) were asymptomatic, and 52 (2.5%) indicated they experienced symptoms 2 weeks before the test. The latter group was excluded from further analyses because of the limited number of individuals.
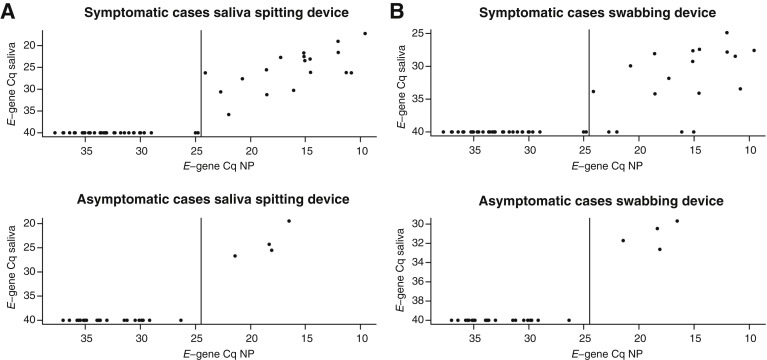
The proportions of individuals who were SARS-CoV-2 positive in the nasopharyngeal sample were similar in the symptomatic and asymptomatic groups (3.7% and 4.2%, respectively); however, the symptomatic group was enriched with high viral load samples (E-gene Cq < 24.5; P = 0.042; Fisher exact test). As a result, sensitivity in saliva for SARS-CoV-2 detection was higher among symptomatic cases [sensitivity, 33.3% (95% CI, 20.8%–47.9%); and sensitivity, 25.5% (95% CI, 14.3%–39.6%) for spitting and swabbing saliva device, respectively] compared with asymptomatic cases [sensitivity, 13.8% (95% CI, 3.9%–31.7%) for both the spitting and swabbing saliva device] (Figure 4 ).
Figure 4.
E-gene quantification cycle values from laboratory 1 in nasopharyngeal-positive samples and matching saliva samples from symptomatic and asymptomatic cases. A: Results obtained with the saliva spitting device. B: Results obtained with the saliva swabbing device. NP, nasopharyngeal samples.
Among individuals with high viral load in NP samples, the sensitivity in the saliva samples was high, irrespective of symptomatic status. Sensitivity was 94.4% (95% CI, 77.2%–99.9%) and 100.0% (95% CI, 39.8%–100%) in symptomatic and asymptomatic individuals in the spitting sample, whereas the sensitivity from the swabbing saliva sample was 72.2% (95% CI, 46.5%–90.3%) in symptomatic subjects and 100.0% (95% CI, 39.8%–100%) in asymptomatic subjects. The sensitivity and test positive ratio are summarized in Table 1 (and in Supplemental Table S4 for laboratory 2).
Discussion
The literature on the use of saliva for SARS-CoV-2 detection is rapidly evolving and expanding. Saliva sampling for COVID-19 diagnostics has been put forward as an alternative for nasopharyngeal sampling in several independent studies. A rapid review of the literature estimated a pooled sensitivity of SARS-CoV-2 testing on saliva versus nasopharyngeal samples as high as 97%.18 More recently, a meta-analysis of 16 unique studies, representing 5922 patients, reported a pooled sensitivity for SARS-CoV-2 detection in saliva of 83.2%.19 Herein, we compared the sensitivity of two different saliva collection devices with the nasopharyngeal swab in >2500 individuals who were sampled at different triage centers in Belgium.
In contrast to the current literature, a substantially lower SARS-CoV-2 test positivity rate was observed in saliva than in nasopharyngeal samples. However, when focusing on individuals with a high viral load (>1.51 × 106 copies/mL viral transport medium), sensitivity improved dramatically, especially in saliva samples produced through spitting. There are several potential reasons for the discrepancy between results presented herein and current literature reports. First, the study population may be different. Most of the studies reported in literature predominantly include symptomatic patients (whether or not hospitalized) who are more likely to have a high viral load. Our study included individuals visiting triage centers to obtain diagnostic testing for SARS-CoV-2, because either they presented with COVID-19 symptoms or they had been in close contact with an infected individual. These individuals were not critically ill and often did not have symptoms. Second, although our study compared two different devices for saliva collection, the possibility that these devices are less suitable for saliva collection compared with what has been used in other studies cannot be excluded. Of note, differential sensitivity was observed between both devices and it was concluded that saliva sampling through spitting is more sensitive than swabbing. Whether the order of saliva collection (participants were asked to first swab, then spit) could impact these results remains to be investigated. A study conducted in British Columbia collected samples from outpatient testing centers and evaluated saline mouth rinse/gargle (alias, swish and gargle approach) to collect saliva samples compared with neat saliva collection and found that the swish/gargle method had a higher sensitivity than neat saliva, 97.5% (95% CI, 86.9%–99.9%) versus 78.8% (95% CI, 61%–91%). It is unclear why the authors observed a significant difference in the sensitivity, but the saliva collection method could influence sensitivity of the test result. Note that, in this study, saliva samples were collected without gargling or throat clearing, actions that may further improve the sensitivity of SARS-CoV-2 detection in saliva. Finally, other covariates, including time of the day of sampling, stage of infection at sampling, and timing relative to an epidemiologic wave (ie, varying reproduction number), may also impact results. With respect to the latter, patients were sampled at the end of the first (spring) wave in Belgium, a period of low prevalence and low individual viral load [compared with the beginning of the second (autumn) wave20]. More studies are required to further investigate the impact of these factors. Notably, testing different but biologically related samples from each patient (saliva and oral swab) provides an internal validation of our study results, as both sample types lead to highly similar conclusions.
Our study also had some limitations. First, more detailed clinical and demographic data would have been helpful to evaluate if other factors could explain the difference in sensitivity between saliva and the nasopharyngeal swab. In addition, the inclusion of hospitalized patients may have allowed a more in-depth analysis on the relation between disease severity and detection sensitivity in saliva. Finally, longitudinal saliva collections in positive individuals could shed light on the dynamics of SARS-CoV-2 detection rates in function of disease progression.
In summary, this study suggests that saliva sampling cannot replace the standard nasopharyngeal swab for the diagnosis of SARS-CoV-2 in the population studied herein. Nevertheless, because of its ease of use and compatibility with self-sampling, saliva sampling could play a role in systematic screening campaigns that aim to identify asymptomatic cases with medium to high viral loads. However, on the basis of results presented herein, such screening campaigns would fail to identify low positives. Whether, and to what extent, these low positives are capable of spreading the virus requires further investigation.
Acknowledgments
We thank the physicians, the nurses, and the paramedics of the Belgian Armed Forces and other teams involved in the massive sampling campaigns in the different triage centers; Anke Maertens for logistics support; Pierre Wattiau (GSK Biologicals) and Olivier Vandeputte (GSK Biologicals) for constructive discussions on study design and results; and Els Dequeker (KU Leuven) for supervising the ethical approval and processing of the survey forms.
Footnotes
Supported by the VALCOR (Validation of SARS-CoV-2 Assays) project, funded by Sciensano, Brussels, Belgium (M.A. and S.K.D.).
P.M., M.G., S.K.D, M.A., J.S., and J.V. contributed equally to this work.
Disclosures: P.M. performs consultancy for Biogazelle and owns Biogazelle stock. J.V. is cofounder and chief scientific officer of Biogazelle and owns Biogazelle stock. J.H. is cofounder and chief technology officer of Biogazelle and owns Biogazelle stock.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2021.07.017.
Author Contributions
P.M., M.G., E.A., J.-P.P., J.P., V.H., H.M., J.S., and J.V. conceptualized and supervised the study. P.M., M.G., J.H., C.V., M.B., V.D.W., J.S., and J.V. generated and processed primary data. P.M., M.G., S.K.D., M.A., J.S., and J.V. analyzed the data. M.A. performed the sample size computation. S.K.D. and M.A. conducted the statistical analysis of the accuracy data. M.G. and P.M. verified the underlying data. J.-P.P., J.P., P.S., and S.D. organized and supervised sample collection at triage centers. All authors contributed to discussions of results and manuscript writing, and approved the final version of the manuscript.
Supplemental Data
A: Saliva versus nasopharyngeal (NP) sampling comfort, scored by study participants on a scale from 1 (more comfortable) to 5 (less comfortable) for both saliva sampling devices. B: Ease of use of both saliva sampling devices, scored by study participants on a scale of 1 (easy to use) to 5 (difficult to use).
A: Laboratory 1 E-gene quantification cycle value in nasopharyngeal samples for which the matching saliva spitting sample is a true positive (TP) or a false negative (FN). B: Laboratory 1 E-gene quantification cycle value in nasopharyngeal samples for which the matching saliva swabbing sample is a TP or an FN.
N-gene quantification cycle values quantified by laboratory 2 in nasopharyngeal (NP) positive samples and matching saliva samples. A: Results obtained with the saliva spitting device. B: Results obtained with the saliva swabbing device. Plots indicate the correlation coefficient (r) value and P value (Pearson correlation test) calculated using only those samples with a nasopharyngeal E-gene Cq value of <25.5 (threshold marked with a dashed line). Bottom graphs in each panel represent the N-gene quantification cycle-value distribution in the NP-positive samples (y axis represents kernel density estimate).
Abundance levels of RPS18 in SARS-CoV-2 positive and negative saliva samples (A and B) and nasopharyngeal swab samples (C). SARS-CoV-2 positivity is determined on the basis of results from laboratory 1.
References
- 1.European Centre for Disease Prevention and Control Considerations for the use of saliva as sample material for COVID-19 testing, n.d. https://www.ecdc.europa.eu/en/publications-data/considerations-use-saliva-sample-material-covid-19-testing Available at: (accessed June 15, 2021)
- 2.World Health Organization . World Health Organization; Geneva, Switzerland: 2020. Laboratory Testing for Coronavirus Disease (COVID-19) in Suspected Human Cases: Interim Guidance, 19 March 2020. [Google Scholar]
- 3.Kim Y.G., Yun S.G., Kim M.Y., Park K., Cho C.H., Yoon S.Y., Nam M.H., Lee C.K., Cho Y.J., Lim C.S. Comparison between saliva and nasopharyngeal swab specimens for detection of respiratory viruses by multiplex reverse transcription-PCR. J Clin Microbiol. 2016;55:226–233. doi: 10.1128/JCM.01704-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L., Chen Q.Y., Li Y.Y., Wang Y.F., Yang Z.F., Zhong N.S. Comparison among nasopharyngeal swab, nasal wash, and oropharyngeal swab for respiratory virus detection in adults with acute pharyngitis. BMC Infect Dis. 2013;13:281. doi: 10.1186/1471-2334-13-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim C., Ahmed J.A., Eidex R.B., Nyoka R., Waiboci L.W., Erdman D., Tepo A., Mahamud A.S., Kabura W., Nguhi M., Muthoka P., Burton W., Breiman R.F., Njenga M.K., Katz M.A. Comparison of nasopharyngeal and oropharyngeal swabs for the diagnosis of eight respiratory viruses by real-time reverse transcription-PCR assays. PLoS One. 2011;6:e21610. doi: 10.1371/journal.pone.0021610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson J.L., Lee B.E., Kothapalli S., Craig W.R., Fox J.D. Use of throat swab or saliva specimens for detection of respiratory viruses in children. Clin Infect Dis. 2008;46:e61–e64. doi: 10.1086/529386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., Fasano M., Sessa F., Tettamanti L., Carinci F., Maurino V., Rossi A., Tagliabue A., Baj A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Sungkanuparph S., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2020;27:285.e1–285.e4. doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landry M.L., Criscuolo J., Peaper D.R. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. 2020;130:104567. doi: 10.1016/j.jcv.2020.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a noninvasive specimen for detection of sars-cov-2. J Clin Microbiol. 2020;58:e00776-20. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao M., Rashid F.A., Sabri F.S.A.H., Jamil N.N., Zain R., Hashim R., Amran F., Kok H.T., Samad M.A.A., Ahmad N. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS-CoV-2. Clin Infect Dis. 2020;72:e352–e356. doi: 10.1093/cid/ciaa1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T., Sato K., Oguri S., Taki K., Senjo H., Sugita J., Hayasaka K., Konno S., Nishida M., Teshima T. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020;81:e145–e147. doi: 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P., et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen J.F., Korevaar D.A., Altman D.G., Bruns D.E., Gatsonis C.A., Hooft L., Irwig L., Levine D., Reitsma J.B., de Vet H.C.W., Bossuyt P.M.M. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6:e012799. doi: 10.1136/bmjopen-2016-012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G.J.C., Haagmans B.L., Van Der Veer B., Van Den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P.G., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Kampen J.J.A., van de Vijver D.A.M.C., Fraaij P.L.A., Haagmans B.L., Lamers M.M., Okba N., van den Akker J.P.C., Endeman H., Gommers D.A.M.P.J., Cornelissen J.J., Hoek R.A.S., van der Eerden M.M., Hesselink D.A., Metselaar H.J., Verbon A., de Steenwinkel J.E.M., Aron G.I., van Gorp E.C.M., van Boheemen S., Voermans J.C., Boucher C.A.B., Molenkamp R., Koopmans M.P.G., Geurtsvankessel C., van der Eijk A.A. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang N.S., Tang M.L., Chan I.S.F. On tests of equivalence via non-unity relative risk for matched-pair design. Stat Med. 2003;22:1217–1233. doi: 10.1002/sim.1213. [DOI] [PubMed] [Google Scholar]
- 18.Peeters E, Kaur Dhillon Ajit Singh S, Vandesompele J, Mestdagh P, Hutse V, Arbyn M: Rapid systematic review of the sensitivity of SARS-CoV-2 molecular testing on saliva compared to nasopharyngeal swabs. medRxiv, [Epub] 2020. doi: 10.1101/2020.08.05.20168716 [DOI]
- 19.Butler-Laporte G., Lawandi A., Schiller I., Yao M., Dendukuri N., McDonald E.G., Lee T.C. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS-CoV-2: a systematic review and meta-analysis. JAMA Intern Med. 2021;181:353–360. doi: 10.1001/jamainternmed.2020.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verwilt J, Hellemans J, Sante T, Mestdagh P, Vandesompele J: Evaluation of efficiency and sensitivity of 1D and 2D sample pooling strategies for SARS-CoV-2 RT-qPCR screening purposes. medRxiv, 2021 [Epub]. doi: 10.1101/2020.07.17.20152702 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: Saliva versus nasopharyngeal (NP) sampling comfort, scored by study participants on a scale from 1 (more comfortable) to 5 (less comfortable) for both saliva sampling devices. B: Ease of use of both saliva sampling devices, scored by study participants on a scale of 1 (easy to use) to 5 (difficult to use).
A: Laboratory 1 E-gene quantification cycle value in nasopharyngeal samples for which the matching saliva spitting sample is a true positive (TP) or a false negative (FN). B: Laboratory 1 E-gene quantification cycle value in nasopharyngeal samples for which the matching saliva swabbing sample is a TP or an FN.
N-gene quantification cycle values quantified by laboratory 2 in nasopharyngeal (NP) positive samples and matching saliva samples. A: Results obtained with the saliva spitting device. B: Results obtained with the saliva swabbing device. Plots indicate the correlation coefficient (r) value and P value (Pearson correlation test) calculated using only those samples with a nasopharyngeal E-gene Cq value of <25.5 (threshold marked with a dashed line). Bottom graphs in each panel represent the N-gene quantification cycle-value distribution in the NP-positive samples (y axis represents kernel density estimate).
Abundance levels of RPS18 in SARS-CoV-2 positive and negative saliva samples (A and B) and nasopharyngeal swab samples (C). SARS-CoV-2 positivity is determined on the basis of results from laboratory 1.
Data Availability Statement
RT-qPCR data from test laboratory 1 and test laboratory 2 as well as the survey data are provided in Supplemental Tables S1, S2, and S3.