Abstract
Chronic kidney disease (CKD) and acute kidney injury (AKI) are common, heterogeneous, and morbid diseases. Mechanistic characterization of CKD and AKI in patients may facilitate a precision medicine approach to prevention, diagnosis, and treatment. The Kidney Precision Medicine Project aims to ethically and safely obtain kidney biopsies from participants with CKD or AKI, create a reference kidney atlas, and characterize disease subgroups to stratify patients based on molecular features of disease, clinical characteristics, and associated outcomes. An additional aim is to identify critical cells, pathways, and targets for novel therapies and preventive strategies. This project is a multicenter prospective cohort study of adults with CKD or AKI who undergo a protocol kidney biopsy for research purposes. This investigation focuses on kidney diseases that are most prevalent and therefore substantially burden the public health, including CKD attributed to diabetes or hypertension and AKI attributed to ischemic and toxic injuries. Reference kidney tissues (for example, living kidney donor biopsies) will also be evaluated. Traditional and digital pathology will be combined with transcriptomic, proteomic, and metabolomics analysis of the kidney tissue as well as deep clinical phenotyping for supervised and unsupervised subgroup analysis and systems biology analysis. Participants will be followed prospectively for ten years to ascertain clinical outcomes. Cell types, locations, and functions will be characterized in health and disease in an open, searchable, online kidney tissue atlas. All data from the Kidney Precision Medicine Project will be made readily available for broad use by scientists, clinicians, and patients.
Keywords: chronic kidney disease, acute kidney injury, precision medicine, diabetes, hypertension
Graphical Abstract
Introduction
Chronic kidney disease (CKD) and acute kidney injury (AKI) are common, morbid diseases that result in significant burdens on patients, their families, and society. Among adults in the United States, the prevalence of CKD is as high as 14% (37 million people), with a prevalence of 25% or more for those with diabetes or hypertension.1,2 AKI affects 10–15% of all hospitalized patients, with particularly high incidence rates among patients with sepsis and other conditions requiring admission to intensive care units.3,4 CKD and AKI can both progress to kidney failure, and people with CKD and AKI experience high rates of cardiovascular events, infections, and mortality.4–7 Globally, as a result, CKD and AKI are associated with tremendous morbidity.6
Remarkable advances have been made in our understanding of CKD and AKI pathophysiology. For example, numerous pathways have been implicated in the development of CKD due to diabetes, APOL1 genotype was identified as a strong determinant of CKD in African Americans, and molecular pathways mediating ischemic kidney damage have been delineated.8–11
Nonetheless, few therapies have been proven to prevent CKD or its progression, and there is currently no proven management approach to prevent or ameliorate the course of AKI.6 Animal models, while critical to evaluating mechanisms of injury, often do not faithfully recapitulate human disease, and interventions effective in these models do not necessarily extend to humans.12,13 In humans, individual disease states and affected pathways are rarely known, in part because kidney tissue is not routinely obtained for analysis in clinical care. Together, these hurdles make it difficult to prioritize targets for drug development, implement effective clinical trials of new therapeutic agents, and translate knowledge to human kidney health.14–17
Rationale for kidney precision medicine
CKD and AKI are diagnosed and classified crudely, based predominantly on estimated glomerular filtration rate (eGFR) and urine protein excretion for CKD and short-term changes in serum creatinine and urine output for AKI.4,18,19 While useful for staging and epidemiology, these clinical evaluations lack granular insight into the underlying molecular mechanisms or structural injury that contributes to heterogeneity of CKD and AKI, including differences that may contribute to known racial and ethnic disparities in kidney disease risk.
The lack of a mechanistic basis in the current kidney disease taxonomy has significant consequences for identification of novel disease mechanisms, development of therapeutic targets, and routine clinical care. For example, important molecular mechanisms of human kidney disease can be difficult to discover when present in a relatively small subset of patients. Moreover, if a therapy effectively targets a relevant molecular mechanism, the benefit of that treatment in a clinical trial or clinical practice may be diluted among patients with different disease pathways not targeted by the intervention.20 This conundrum provides rationale for development of new molecular mechanism-based disease definitions, enabling therapy targeted to the specific molecular processes responsible for kidney injury and its progression in a patient or a subgroup of patients with similar molecular disease phenotype.
Precision medicine refers to prevention and treatment strategies that take individual variability into account.21 When former President Barack Obama announced the Precision Medicine Initiative in 2015, he referred to precision medicine as “delivering the right treatments, at the right time, every time to the right person.22” This approach requires detailed understanding of the molecular basis of health and disease. Advances in technology now offer an unprecedented opportunity to interrogate critical cells and pathways using deep molecular phenotyping, including analysis of DNA, RNA, proteins, and metabolites.
In 2016, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) convened a workshop to explore whether and how precision medicine could be used to improve the diagnosis and management of CKD and AKI. In the workshop, patients, clinicians, academic researchers, bioethicists, and industry representatives discussed opportunities, resources, and barriers for obtaining and interrogating human kidney tissues to advance kidney precision medicine. In the same year, the NIDDK released a series of Requests for Applications for the Kidney Precision Medicine Project (KPMP), which commenced in the fall of 2017.
Overall goals
The KPMP seeks to redefine CKD and AKI by integrating deep molecular phenotypes with clinical characteristics, digital pathology, and clinical outcomes. Specifically, the overall goals of the KPMP are to (1) ethically and safely obtain kidney biopsies from participants with CKD or AKI, (2) create a reference kidney atlas, (3) characterize disease subgroups to stratify patients based on molecular mechanisms of disease and associated outcomes, and (4) identify critical cells, pathways, and targets for novel therapies. Ultimately, this approach may allow diagnosis of key disease pathways on the individual level, identify promising targets for intervention, facilitate evaluation of new interventions through targeted clinical trials, help validate biomarkers for diagnosis and treatment, and promote the application of precision medicine to patients with CKD and AKI.
Achieving the goals of KPMP requires kidney tissue from humans with common forms of CKD and AKI. Patients who have CKD attributed to diabetes or hypertension or AKI attributed to ischemic or toxic injury rarely undergo kidney biopsy, or do so only when their clinical presentation is unusual. As a result, studies of clinically indicated kidney biopsies may over-represent unusual causes of disease or fail to identify the most common disease mechanisms. Effectively evaluating the heterogeneity of CKD and AKI presentations therefore requires enrolling a new cohort of people willing to volunteer to donate kidney tissue who otherwise would likely not undergo kidney biopsies as part of usual clinical care. Accordingly, the first goal of the KPMP is to ethically and safely obtain tissue from such study participants.
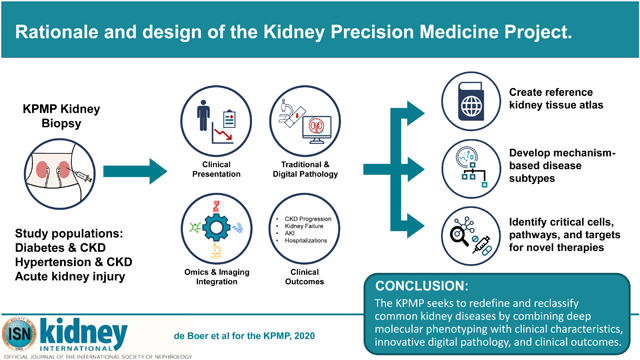
A key deliverable of the KPMP is a kidney atlas that charts cell types and functions in health and disease. The atlas will be an interactive tool that integrates molecular data with clinical presentation, imaging data, and clinical outcomes. A standardized and machine-interpretable kidney nomenclature (ontology) will be developed to support data sharing and integration and construction of new tools for data analysis. Molecular data generated by new assays that are developed, optimized, and implemented by KPMP investigators will be integrated with digital pathology and clinical phenotypes using a systems biology approach to generate new disease subtypes and targets for therapy (Figure 1).23,24
Figure 1. In the Kidney Precision Medicine Project, kidney biopsy tissues will be interrogated with multiple levels of omics technologies, and results will be integrated with digital pathology, clinical characteristics and outcomes to create a reference kidney atlas, develop mechanism-based disease subtypes, and identify critical cells, pathways, and targets for novel therapies.
CKD = chronic kidney disease AKI = acute kidney injury.
Study organization
The KPMP is broadly inclusive of patients, nephrologists, radiologists, pathologists, primary care providers, intensivists, research coordinators, project managers, ethicists, informaticians, research scientists from different disciplines, and others invested in improving health for people with kidney disease. Recruitment sites and tissue interrogation sites are linked and organized by a central hub that contains an administrative core, data coordinating center, and data visualization center (https://kpmp.org/consortium-members/). The study is overseen by three independent bodies, including a Data Safety Monitoring Board, central Institutional Review Board (Washington University, St. Louis), and an NIDDK-convened External Expert Panel.
Patients drive the KPMP
A defining feature of the KPMP has been the establishment of an equitable partnership with patients who have kidney disease. Patients have been integrally involved in the KPMP throughout the full duration and at all levels of the design process.25 Patients with kidney disease were included in the 2016 NIDDK workshop on kidney precision medicine to guide an ethical and feasible approach to research-focused kidney biopsies. Patients from all KPMP sites also were invited to attend the first KPMP Steering Committee meeting, and initial patients subsequently worked with investigators to recruit other representatives to exemplify the diversity of KPMP target populations. As the research protocol was developed, patients informed the study on best practices for engagement and participated in drafting the KPMP informed consent form (Table 1). Protocol development was influenced by patient perspectives on merits and burdens of study procedures. Patients advocated strongly for enrolling a diverse and representative study population, engaging enrolled study participants as research partners, and returning study results in a fully transparent and timely manner. Patients are represented in every KPMP Committee, including the Steering Committee, where they actively participate and lead as peers. These contributions have helped to make the research protocol appropriate for potential participants and ensure that the ultimate products of the KPMP will be optimally oriented toward patient values and preferences.
Table 1.
Examples of patient influence on design of the Kidney Precision Medicine Project
| Design challenge | Patient input |
|---|---|
| Protocol: balance of detail versus participant burden | Willing participants will accept additional procedures |
| Recruitment | Emphasize underrepresented populations, form Community Advisory Boards |
| Communications | Feature frequently asked questions and video testimonials |
| Informed consent process | Include all trusted advisors, including family members and personal physicians |
| Potential complications | Participants must not bear any direct financial costs |
| Return of results | Return all possible results, even if clinical implications are not clear |
| Data and tissue sharing | Make most extensive possible use of all donated tissue and data |
| Organization | Include patients in committees, provide explanations of terms and patient-level summaries |
Study design
The KPMP is a multicenter prospective cohort study of people with CKD or AKI who undergo a protocol kidney biopsy at study entry. The full study protocol and detailed manuals of procedures are freely available at kpmp.org/researcher-resources. The KPMP focuses on kidney diseases that are most prevalent and therefore substantially burden the public health. For CKD, these are presentations attributed clinically to diabetes, hypertension, or both. For AKI, these are presentations attributed to sepsis, ischemic, or toxic injury. Some patients who have been offered a kidney biopsy for clinical indication may elect to participate in the KPMP research protocol instead of a standard clinical biopsy and provide additional tissue for research purposes. However, it is anticipated that most participants who volunteer to participate in the KPMP would not have otherwise undergone a kidney biopsy.
Enrollment strategies include recruiting from clinical care encounters (e.g. clinic, emergency room, or hospital stay), electronic health information resources (e.g. existing registries and electronic health records), and community networks and outreach. Site-specific approaches optimize recruitment strategies according to their local health care settings and target study population(s). The KPMP data coordinating center monitors demographic and disease characteristics of enrolled participants, compares these characteristics to those of the underlying target populations, and feeds these data back to recruitment sites to prioritize recruiting efforts as needed. To this end, community advisory boards at each recruiting site facilitate engagement of local patient populations and advocacy groups.
Informed consent is viewed as a longitudinal process that includes patients, patients’ trusted advisors, clinical providers, and trained KPMP staff and spans days (for AKI) to weeks (for CKD), depending on clinical presentation. The process includes providing access to extensive educational materials in addition to written informed consent and is designed to ensure that participants fully understand the scope of the study and its possible risks.
Inclusion criteria were designed to enroll a population that is broadly representative of these clinical conditions, including early clinical presentations (Table 2). For CKD, participants are limited to those with estimated GFR ≥30 mL/min/1.73m2 to increase the likelihood of identifying abnormalities that initiate and propagate relatively early disease, providing the greatest opportunity for intervention. For AKI, eligibility criteria are designed to select participants with minimal underlying CKD, which could confound identification of mechanisms specific to AKI. Patients undergoing percutaneous kidney biopsy are further required to have elevated serum creatinine that is either sustained or accompanied by evidence of parenchymal injury,26 while patients undergoing laparotomy kidney biopsy (usually in the acute emergency department setting) have a broader set of inclusion criterion designed to include early AKI or high risk of imminent AKI. Special populations are also enrolled. For CKD, people with longstanding type 1 diabetes without evidence of CKD (diabetic kidney disease “resistors”) are evaluated to search for pathways associated with protection from CKD. For AKI, people undergoing exploratory laparotomy who have evidence of early kidney dysfunction or are at high risk of developing AKI postoperatively undergo open biopsy at the time of surgery.
Table 2.
Selected eligibility criteria for the Kidney Precision Medicine Project
| Inclusion criteria | Exclusion criteria | |
|---|---|---|
| CKD | AKI | |
|
One of the following:
|
For percutaneous kidney biopsy:
For open laparotomy kidney biopsy:
|
|
Does not apply to laparotomy biopsies
Exclusion criteria were designed primarily to protect participant safety. These exclusion criteria were based on published literature and clinical experience and focus largely on reducing the risk of bleeding (Table 2). Additional criteria exclude people with kidney diseases already under investigation by other large cohort studies (e.g. people with glomerular disorders and polycystic kidney disease) and those with unusual clinical scenarios for which unique abnormalities may underlie disease but for which an insufficient number of participants would be recruited to draw meaningful conclusions regarding pathogenesis.
Participants are followed for 10 years from the time of kidney biopsy through a combination of annual in-person study visits and interval remote visits to document clinically important events, including loss of kidney function, kidney failure, interval hospitalizations, cardiovascular events, and mortality (Table 3). Participants enrolled for AKI have an additional in-person study visit three months after kidney biopsy to establish a clinical post-AKI baseline. Surveillance of electronic health records and national databases such as the United States Renal Data System and National Death Index will supplement these approaches, especially for longer-term follow-up. An extensive biorepository of serum, plasma, circulating leukocytes preserved for DNA and RNA analysis, timed and spot urine samples, and stool samples is collected at the time of kidney biopsy and at in-person study visits. Biorespository samples are stored for future projects aiming to further understand disease pathogenesis and to develop and validate biomarkers reflecting disease heterogeneity. There are currently no plans to repeat kidney biopsies in the KPMP, but this may be considered in the future.
Table 3.
Outcomes assessed by the Kidney Precision Medicine Project
| Outcome | Metrics |
|---|---|
| Biopsy-related outcomes |
|
| Kidney tissue atlas |
|
| Molecular kidney disease subtypes |
|
| Kidney disease progression |
|
| Outcomes specific to AKI |
|
| Additional clinical outcomes |
|
Kidney biopsy
The centerpiece of the KPMP research protocol is the kidney biopsy, which is used to generate a traditional histology-based diagnosis as well as novel molecular phenotypes. The ethical conduct of kidney biopsies is an absolute requirement of the KPMP and has been a focus of discussion throughout planning and development. In general, during the planning process, physicians and scientists thought that while kidney biopsies in the planned populations may occasionally yield useful information for diagnosis or prognosis, most individual participants were unlikely to directly benefit from their biopsy. Patient representatives expressed a broader concept of potential benefits, valuing knowledge of their disease process even if it does not alter medical therapy as well as the opportunity to contribute altruistically to their communities through new discovery. Regardless of potential benefit, kidney biopsies come with inherent risk, including risk of serious complications such as need for blood transfusions, hospitalization, kidney loss, and death.27,28 The KPMP performed a systematic review of kidney biopsy complications to quantify these risks for potential participants and their providers.29 Moreover, the KPMP aims to minimize these risks and to disclose them through a comprehensive informed consent process that is transparent and respects individual autonomy. In addition, the KPMP will prospectively collect data on both benefits (to participants and clinical providers) and risks in order to refine informed consent over time and better quantify biopsy outcomes for the broader community.
The guiding principle in performing the kidney biopsy is optimizing patient safety. Safety has been emphasized throughout the KPMP by developing a safety culture, specific safety-oriented protocols, and procedures to monitor and adapt the protocol as needed (Table 4). Many elements of the KPMP kidney biopsy protocol are standardized, while some elements are flexible in order to match procedural expertise at individual KPMP recruiting sites. All percutaneous kidney biopsies are performed by experienced, KPMP-certified operators using 16-gauge needles and real-time imaging guidance. KPMP-certified operators must demonstrate that they have completed 35 biopsies over the past two years with a major complication rate less than 10%, and 85% of the biopsies must have been adequate for diagnosis. A maximum of five passes is allowed to attain a goal of three biopsy cores. Operators may be nephrologists or radiologists. For participants undergoing open laparotomy, surgeons will perform biopsies using 16-gauge needles with direct visualization of the kidney. Checklists are used to ensure that all biopsies are only performed under appropriate conditions, including ensuring that vital signs and laboratory parameters are within safety limits. Adverse events are prospectively ascertained and monitored by an internal KPMP Safety and Adjudication Committee as well as an external Data Safety Monitoring Board and single Institutional Review Board.
Table 4.
Safety measures undertaken by the Kidney Precision Medicine Project
| Safety domain | Measures |
|---|---|
| Safety culture |
|
| Protocol development |
|
| Protocol implementation |
|
| Surveillance for adverse effects |
|
| Ongoing review of adverse events to adapt protocol if needed |
|
Kidney tissue processing and analysis
One of the major challenges for KPMP is maximizing the use of limited kidney tissue available for both traditional pathologic diagnosis and multiple molecular evaluations. To this end, pathologists, researchers and nephrologists spanning all KPMP sites undertook a collaborative effort to design a consensus protocol to process kidney biopsy tissue (Figure 2). The protocol was informed by a series of pilot studies and incorporates rigorous quality control at each step. Kidney biopsy tissue is triaged by a pathologist in the biopsy suite, and the most optimal biopsy core is selected for traditional histologic evaluation using algorithms available at kpmp.org/researcher-resources/. This diagnostic core is processed locally at the recruitment site for light microscopy, immunofluorescence, and electron microscopy, and a clinical pathology report based on this core is generated and returned to the participant and clinical care team. Any remaining tissue from this core is then sent to a central biorepository for research use to be made available to tissue interrogation sites for multiplexed imaging studies. Two other cores are processed expeditiously for deep molecular phenotyping and frozen for transportation to a central biorepository at the University of Michigan.
Figure 2. Kidney Precision Medicine Project tissue triage and interrogation schematic.
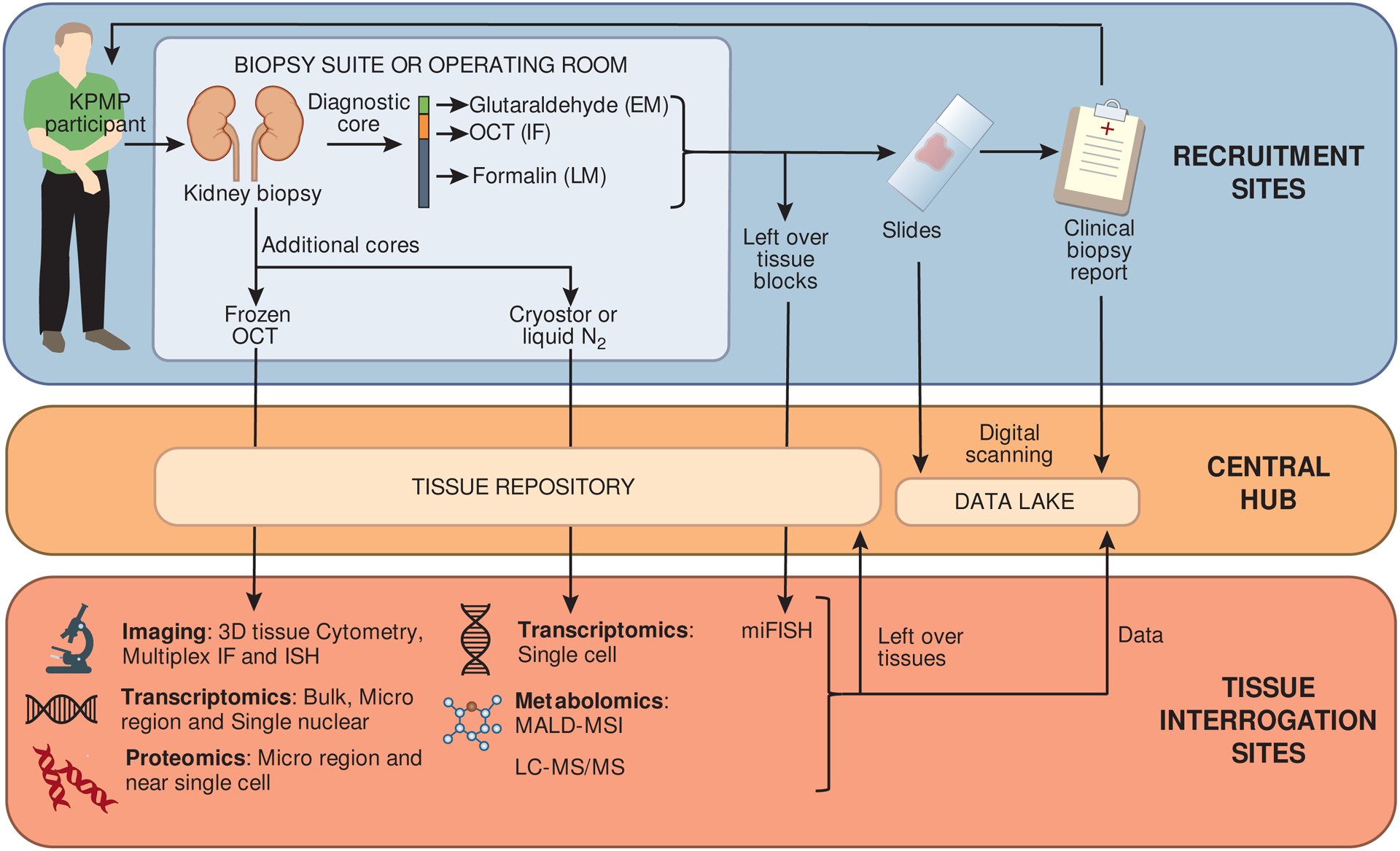
Kidney biopsy tissues will be processed for local clinical pathology review and remote research interrogation according to a detailed, standardized protocol. KPMP = Kidney Precision Medicine Project; EM = electron microscopy; IF = immunofluorescence; LM = light microscopy; OCT = optimal cutting temperature compound; N2 = nitrogen; ISH = in-situ hybridization’ MALD-MSI = matrix-assisted laser desorption ionization mass spectrometry imaging; LC-MS/MS = liquid chromatography-mass spectrometry.
All sites formally demonstrate readiness for kidney biopsy and tissue preservation, including certification to maintain proficiency. All reagents and supplies are shipped as standardized, prelabeled kits from the central biorepository. Slides generated at recruitment sites are scanned centrally for digital viewing and analysis, and tissue earmarked for research studies is sent to the central biorepository and then distributed to tissue interrogation sites.
Technologies applied to kidney tissue span imaging, transcriptomics, proteomics, and spatial metabolomics (Figure 2). These technologies are expected to evolve over time, and additional technologies will be added. Quality control is emphasized throughout processing and analysis, including close attention to preanalytic conditions such as tissue handling and storage, assay reliability, comparisons across technologies, and shift and drift in assays over time.
Reference kidney tissues
Reference kidney tissues interrogated using KPMP technologies is needed to populate a kidney atlas in health and provide context for kidney diseases. Nephrectomy and deceased donor kidney tissues have been used in KPMP pilot studies for optimizing protocol and developing an initial version of a reference atlas.30,31 However, these procurement methods are subject to artifacts including ischemia that may substantially change gene expression and protein and metabolite content in tissues. Therefore, the KPMP is developing partnerships and protocols to obtain better sources of healthy reference kidney tissues from living kidney donors as well as people without clinically evident kidney disease who undergo invasive kidney procedures, for example percutaneous nephrolithotomy for isolated nephrolithiasis. These reference tissues will be processed and interrogated using KPMP protocols and included in the KPMP tissue atlas.
Study outcomes
The KPMP ascertains a range of outcomes consistent with its overarching goals (Table 3). These start with evaluation of the risks and benefits of the kidney biopsy itself because experience with kidney biopsies in the target populations is limited. Kidney biopsy outcomes are ascertained through prospective questionnaires administered to participants, clinicians, and study investigators as well as careful surveillance of adverse events. In addition, development of an open-source kidney tissue atlas and molecular-based disease subtypes are considered explicit deliverables of the KPMP.
The KPMP also follows participants for more traditional health outcomes, including progression of CKD, AKI outcomes (for AKI participants), and other health events, as defined in Table 4. Data for these outcomes are obtained through surveillance of electronic health records, the USRDS registry, and the national death index in addition to in-person and remote study visits. Comprehensive efforts are made to retain participants for clinical follow-up, with KPMP-wide patient representatives and recruitment site community advisory boards playing key roles in developing and implementing comprehensive engagement and retention plans.
Sample size
Currently, there are insufficient data to calculate reliable power estimates or sample sizes needed to achieve the goals of the KPMP. Therefore, the KPMP plans to enroll 200 participants, a number judged to be safe and feasible given the complexity of the clinical protocol, during its first phase (through mid-2022). Biopsies from these participants will be used to evaluate safety, generate initial versions of the reference kidney atlas (along with reference tissues), and identify initial mechanism-based disease subgroups. Moreover, biopsies from these participants will be used to understand variability within kidney cells of similar lineage, between different cell types, across tissue samples from the same individual, between individuals with the same kidney disease subtype, between disease subtypes, and in the associations of disease subtypes with clinical outcomes for the populations studied and technologies employed by the KPMP. These data will guide sample size calculations for future work.
Kidney atlas
A novel kidney tissue atlas characterizing cell types, locations, and functions in health and disease will be a central product of KPMP efforts. The atlas will integrate digital pathology, transcriptomics, proteomics, and metabolomics with clinical data, present a set of curated multiscalar two- and three-dimensional maps of kidney tissue, and offer analysis and visualization tools for users (Figure 3). Ontologies will be developed to semantically integrate these components and link to complementary data resources. The atlas will contain data on clinical disease-free reference tissue to help understand kidney structure and function in new detail as well as datasets containing results from enrolled KPMP participants with CKD or AKI, defined using kidney biopsy and detailed clinical characteristics. State-ofthe-art algorithms will be employed to view digital histology and search structural features in real time. The kidney atlas will be accessible via kpmp.org and will provide access to all processed and raw data (after cleaning) generated by the KPMP. Portals of entry will be established for common anticipated types of users, including clinicians, pathologists, scientists, and patients.
Figure 3. Vision for a kidney tissue atlas.
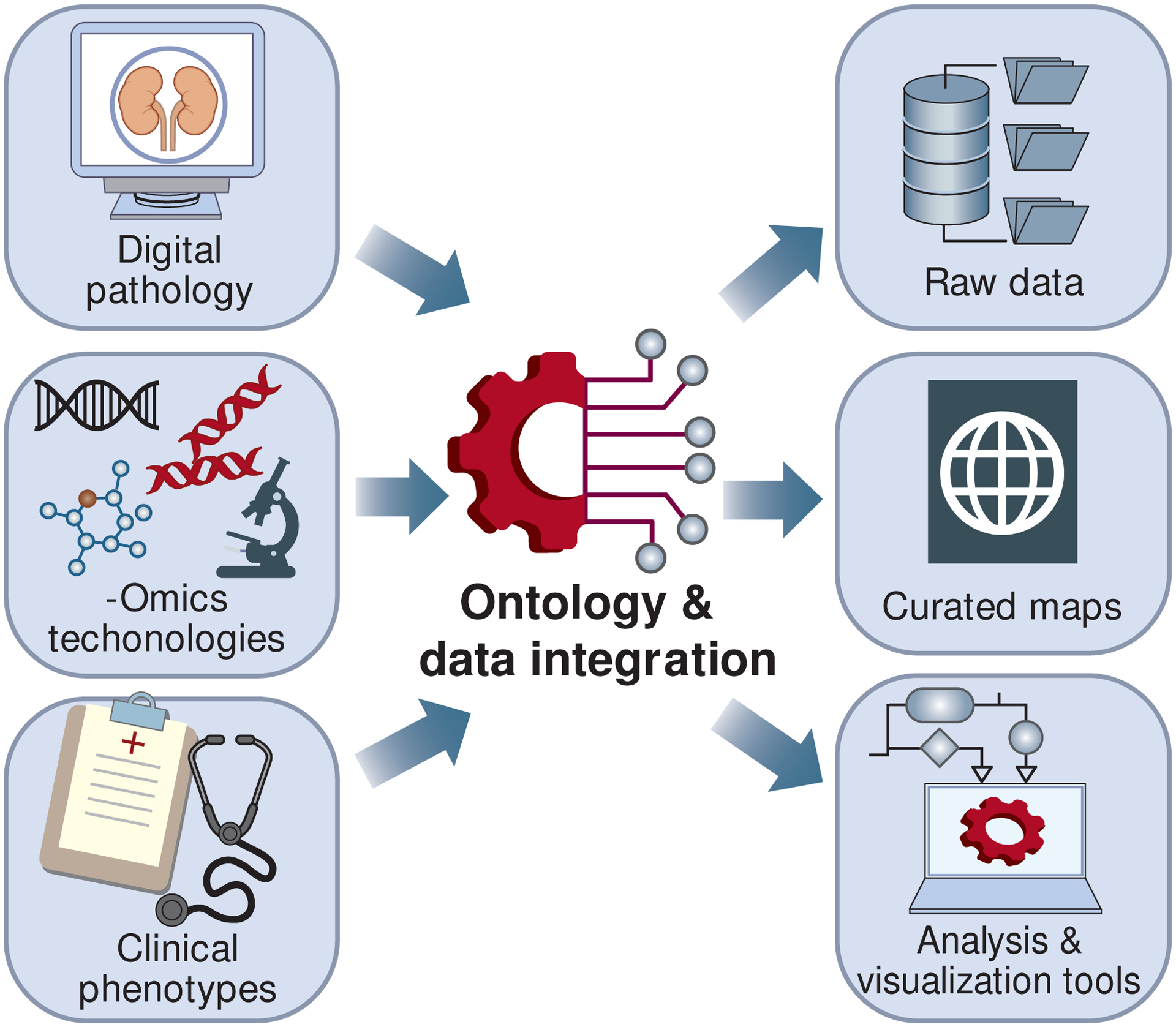
The Kidney Precision Medicine Project will integrate digital pathology, omics data, and clinical data using common ontologies, making raw data, curated maps, and analysis and visualization tools widely available to clinical and research communities.
Open source platform
The KPMP was conceived as an open-source platform to expeditiously share novel data with patients, clinicians, and scientists investigating kidney health and disease or broadly investigating human biology. As such, KPMP investigators are committed to making data publicly available as soon as it passes internal quality controls, beginning in spring, 2020, while protecting participant privacy. To that end, KPMP data are available for download, accessible through the KPMP website, kpmp.org. Data that carry risk of individual identification require a data use agreement for access that stipulates that no attempts will be made to re-identify participants. KPMP data sharing policies, protocols, and manuals of procedures are also freely available from the study website.
To maximize integration, dissemination, and collaboration, the KPMP developed and utilizes ontologies to classify all collected data. Ontologies are systems that classify symptoms, signs, conditions, and their relationships. Using common ontologies can facilitate combining of data across studies.32 The KPMP has taken a leadership role in developing and refining kidney disease ontologies. For example, the KPMP hosted an international kidney-focused ontology meeting in Seattle, WA during the summer of 2018.
Partnerships
The KPMP also encourages new partnerships, including partnerships with academic investigators, healthcare systems, and industry. Such partnerships are actively solicited through request for funding opportunity announcements (https://kpmp.org/opportunity-pool/) and can also be initiated through ancillary study applications to access KPMP participants or samples. Competitive travel grants are offered to present research posters at KPMP Steering Committee meetings, for which young investigators are particularly encouraged to apply. Additionally, KPMP is designed to integrate and synergize with other atlas efforts, such as the HuBMAP (https://commonfund.nih.gov/hubmap), GUDMAP (https://gudmap.org), and Human Cell Atlas (https://www.humancellatlas.org/). KPMP external collaboration and ancillary studies policies are available at kpmp.org.
Challenges
The initial challenges of the KPMP include developing a novel protocol that is safe, ethical, rigorous, feasible, and acceptable to participants. This report highlights basic aspects of this protocol, and further reports will provide additional details regarding patient engagement, tissue processing and pathology, molecular interrogation, and data visualization. Numerous additional challenges remain. Quality control must be maintained throughout the collection and analysis of tissue and data. Finite amounts of kidney tissue must be divided among multiple informative technologies, including promising new technologies that emerge during the study. Sufficient numbers of participants must be enrolled to address all sources of variability, the scope of which is not yet fully defined. Kidney tissue status at a single snapshot in time must be related to diseases that evolve over time and to long-term clinical outcomes. Rich clinical phenotype data on kidney function above and beyond eGFR and albuminuria may be required to create new disease classifications. Data will be made available for public use in a manner that promotes integration with complementary resources, relying on common metadata and ontologies that require further development.
Vision for the future
The KPMP is a new initiative that seeks to redefine and reclassify common kidney diseases by combining deep molecular phenotyping with clinical characteristics, innovative digital pathology, and relevant clinical outcomes. Data generated by the KPMP will improve the knowledge base for a wide range of research within and beyond nephrology, organized in a kidney tissue atlas and disseminated through an open source platform. Near-term goals of this work include developing new mechanism-based subtypes of kidney diseases that are common but heterogenous and to help identify and prioritize targets for intervention. Such advances would support clinical trials, enabling platform trials and other innovative methods of testing new diagnostics and treatments.33
In the process, the KPMP aims to redefine the diagnosis and understanding of both CKD and AKI. It will comprehensively assess the role for kidney biopsy in the diagnosis and treatment of these kidney diseases. New methods of molecular analysis, digital pathology, imaging techniques, and bioinformatics are expected to become standard for diagnosis of kidney disease, and new protocols may be developed to reduce risks of kidney biopsy, facilitate proper selection of patients, and identify those who may benefit the most from the procedure. These contributions should improve the benefit-risk profile of kidney biopsy and increase its use in clinical practice. Patients themselves may be empowered to request the option of a kidney biopsy from their providers. In addition, biomarkers may be developed to accurately identify kidney pathology. Kidney precision medicine offers myriad opportunities to improve science and health, and the KPMP along with similar research initiatives will stimulate interest in nephrology to entice a new generation to innovate upon the KPMP foundation.
Ultimately, the goal of the KPMP is to improve the health of patients with and at-risk for kidney diseases by enabling individual-level diagnosis and treatment. To achieve this goal, KPMP investigators intend to advance kidney precision health by classifying disease subgroups to stratify patients based on molecular mechanisms of disease and identifying critical cells and pathways as targets for novel clinical strategies. Data will be visualized and disseminated through a new kidney tissue atlas and shared openly to catalyze new diagnostic and therapeutic approaches for CKD and AKI.
Acknowledgements
The KPMP is funded by the following grants from the NIDDK: U2C DK114886, UH3DK114861, UH3DK114866, UH3DK114870, UH3DK114908, UH3DK114915, UH3DK114926, UH3DK114907, UH3DK114920, UH3DK114923, UH3DK114933, and UH3DK114937. The KPMP investigators would like to thank members of the Division of Kidney, Urologic, and Hematologic Diseases of the NIDDK who initiated and guided development of the KPMP, the KPMP External Expert Panel, the KPMP Data Safety Monitoring Board, and the Washington University Institutional Review Board. A full list of KPMP investigators is provided in the appendix.
Appendix:
Kidney Precision Medicine Project Investigators and Institutions
American Association of Kidney Patients, Tampa, FL: Richard Knight
Beth Israel Deaconess, Boston, MA: Stewart Lecker, Isaac Stillman
Boston University, Boston, MA: Sushrut Waikar,
Brigham & Women’s Hospital, Boston, MA: Gearoid Mcmahon, Astrid Weins, Samuel Short
Broad Institute, Cambridge, MA: Nir Hacohen, Paul Hoover
Case Western Reserve, Cleveland, OH: Mark Aulisio
Cleveland Clinic, Cleveland, OH: Leslie Cooperman, Leal Herlitz, John O’toole, Emilio Poggio, John Sedor, Stacey Jolly
Columbia University, New York, NY: Paul Appelbaum, Olivia Balderes, Jonathan Barasch, Andrew Bomback, Pietro A. Canetta, Vivette D. D’Agati, Krzysztof Kiryluk, Satoru Kudose, Karla Mehl, Jai Radhakrishnan, Chenhua Weng
Duke University, Durham, NC: Laura Barisoni
European Molecular Biology Laboratory, Heidelberg, Germany: Theodore Alexandrov
Indiana University, Indianapolis, IN: Tarek Ashkar, Daria Barwinska, Pierre Dagher, Kenneth Dunn, Michael Eadon, Michael Ferkowicz, Katherine Kelly, Timothy Sutton, Seth Winfree
John Hopkins University, Baltimore, MD: Steven Menez, Chirag Parikh, Avi Rosenberg, Pam Villalobos, Rubab Malik, Derek Fine, Mohammed Atta, Jose Manuel Monroy Trujillo, Joslin Diabetes Center, Boston, MA: Alison Slack, Sylvia Rosas, Mark Williams
Mount Sinai, New York, NY: Evren Azeloglu, Cijang (John) He, Ravi Iyengar, Jens Hansen
Ohio State University, Columbus, OH: Samir Parikh, Brad Rovin
Pacific Northwest National Laboratories, Richland, WA: Chris Anderton, Ljiljana Pasa-Tolic, Dusan Velickovic, Jessica Lukowski
Parkland Center for Clinical Innovation, Dallas, TX: George (Holt) Oliver
Patient Partners: Joseph Ardayfio, Jack Bebiak, Keith Brown, Taneisha Campbell, Catherine Campbell, Lynda Hayashi, Nichole Jefferson, Robert Koewler, Glenda Roberts, John Saul, Anna Shpigel, Edith Christine Stutzke, Lorenda Wright, Leslie Miegs, Roy Pinkeney
Princeton University, Princeton, NJ: Rachel Sealfon, Olga Troyanskaya
Providence Medical Research Center, Providence Health Care, Spokane, WA: Katherine Tuttle
Stanford University, Palo Alto, CA: Dejan Dobi, Yury Goltsev
University of California San Diego, La Jolla, CA: Blue Lake, Kun Zhang
University of California San Francisco, San Francisco, CA: Maria Joanes, Zoltan Laszik, Andrew Schroeder, Minnie Sarwal, Tara Sigdel
University of Michigan, Ann Arbor, MI: Ulysses Balis, Victoria Blanc, Oliver He, Jeffrey Hodgin, Matthias Kretzler, Laura Mariani, Rajasree Menon, Edgar Otto, Jennifer Schaub, Becky Steck, Chrysta Lienczewski, Sean Eddy
University of Pittsburgh, Pittsburgh, PA: Michele Elder, Daniel Hall, John Kellum, Mary Kruth, Raghav Murugan, Paul Palevsky, Parmjeet Randhawa, Matthew Rosengart, Sunny Sims-Lucas, Mary Stefanick, Stacy Stull, Mitchell Tublin
University of Washington, Seattle, WA: Charles Alpers, Ian de Boer, Ashveena Dighe, Jonathan Himmelfarb, Robyn Mcclelland, Sean Mooney, Stuart Shankland, Kayleen Williams, Kristina Blank, Jonas Carson, Frederick Dowd, Zach Drager, Christopher Park
UT Health San Antonio, Center for Renal Precision Medicine, San Antonio, TX: Kumar Sharma, Guanshi Zhang, Shweta Bansal, Manjeri Venkatachalam,
UT Southwestern Medical Center, Dallas, TX: Asra Kermani, Simon Lee, Christopher Lu, Tyler Miller, Orson Moe, Harold Park, Kamalanathan Sambandam, Francisco Sanchez, Jose Torrealba, Toto Robert, Miguel Vazquez, Nancy Wang
Washington University in St. Louis, St. Louis, MO: Joe Gaut, Sanjay Jain, Anitha Vijayan
Yale University, New Haven, CT: Randy Luciano, Dennis Moledina, Ugwuowo Ugochukwu, Francis Perry Wilson, Sandy Alfano
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
IHdB report consulting for Boehringer-Ingelheim, Cyclerion Therapeutics, George Clinical, Goldfinch Bio, and Ironwood. JK reports consulting for Aethlon; AM Pharma; Astute Medical/bioMérieux; Astellas; Balmes Transplantation; Baxter; Cytosorbents; GE; Hepa Wash; Indalo Therapeutics; Jafron; Lowell Therapeutics; Mallinckrodt; MDSG; Nxstage; Oncogna; Oxford PG; PhotoPhage; Potrero; RenalSense; TES Pharma and equity stake in PhotoPhage. KK report advisory board for Goldfinch Bio, Boston. SR report advisory board for Bayer. JS report relationship with Maze and Goldfinch Bio. KS report equity in SygnaMap. KT report consulting for Eli Lilly, Boehringer-Ingelheim, AstraZeneca, Goldfinch Bio, Gilead, Novo Nordisk, Bayer. SW report relationships with Harvard Clinical Research Institute/Baim, Cerus, Strataca, Venbio, Taekda, CVS, JNJ, Mass Medical International, GSK, Allena, Wolters Kluewer, Oxidien, Kantum, BioMarin. JH (Himmelfarb) report consulting for Pfizer, advisory board for RenalytixAI, Maze Therapeutics, Akebia, Chinook Therapeutics, and founder of AKTIV-X.
Contributor Information
Ian H. de Boer, Department of Medicine, University of Washington, Seattle, WA.
Charles E. Alpers, Department of Pathology, University of Washington, Seattle, WA.
Tarek M. El-Achkar, Department of Medicine, Indiana University, Indianapolis, IN.
Evren Azeloglu, Department of Medicine, Ichan School of Medicine at Mt. Sinai, New York, NY.
Ulysses G. J. Balis, Department of Pathology, University of Michigan, Ann Arbor, MI.
Jonathan M. Barasch, Department of Pathology, Columbia University, New York, NY.
Laura Barisoni, Department of Pathology, Duke University, Durham, NC.
Kristina Blank, Department of Biostatistics, University of Washington, Seattle, WA.
Andrew S. Bomback, Department of Medicine, Columbia University, New York, NY.
Keith Brown, Patient Representative KPMP Steering Committee Member.
Pierre C. Dagher, Department of Medicine, Indiana University, Indianapolis, IN.
Ashveena L. Dighe, Department of Medicine, University of Washington, Seattle, WA.
Michael T. Eadon, Department of Medicine, Indiana University, Indianapolis, IN.
Joseph P. Gaut, Department of Pathology, Washington University St. Louis, St. Louis, MO.
Nir Hacohen, Broad Institute, Cambridge, MA.
Yongqun He, Department of Microbiology and Immunology, University of Michigan, Ann Arbor, MI.
Jeffrey B. Hodgin, Department of Pathology, University of Michigan, Ann Arbor, MI.
Sanjay Jain, Department of Medicine, Washington University School of Medicine.
John A. Kellum, Department of Critical Care Medicine, University of Pittsburgh, Pittsburgh, PA.
Krzysztof Kiryluk, Department of Medicine, Columbia University, New York, NY.
Richard Knight, American Association of Kidney Patients.
Zoltan G. Laszik, University of California, San Francisco, San Francisco, CA.
Robyn L. Mcclelland, Department of Biostatistics, University of Washington, Seattle, WA.
Steven Menez, Department of Medicine Johns Hopkins Medicine, Baltimore, MD.
Dennis Moledina, Yale School of Medicine, New Haven CT.
Sean D. Mooney, Department of Biomedical Informatics and Medical Education, University of Washington, Seattle, WA.
John O’Toole, Department of Nephrology & Hypertension, Cleveland Clinic, Cleveland OH.
Paul M. Palevsky, Renal-Electrolyte Division, Department of Medicine, University of Pittsburgh and Renal Section VA Pittsburgh Healthcare System, Pittsburgh, PA.
Chirag R. Parikh, Department of Medicine Johns Hopkins Medicine, Baltimore, MD.
Emilio Poggio, Department of Nephrology & Hypertension, Cleveland Clinic, Cleveland OH.
Sylvia Rosas, Joslin Diabetes Center, Boston, MA.
Matthew R. Rosengart, Department of Medicine, University of Pittsburgh, Pittsburgh, PA.
Minnie Sarwal, Department of Surgery, University of California, San Francisco.
Jennifer A. Schaub, Department of Internal Medicine, University of Michigan, Ann Arbor, MI.
John R. Sedor, Department of Nephrology & Hypertension, Cleveland Clinic, Cleveland OH.
Kumar Sharma, Department of Medicine, UT Health San Antonio, San Antonio, TX.
Becky Steck, Department of Internal Medicine, University of Michigan, Ann Arbor, MI.
Robert Toto, Department of Internal Medicine, UT Southwestern Medical Center, Dallas, TX.
Olga Troyanskaya, Department of Computer Science, Princeton University, Princeton, NJ.
Katherine Tuttle, Department of Medicine, University of Washington, Seattle, WA.
Miguel Vazquez, Department of Internal Medicine, UT Southwestern Medical Center, Dallas, TX.
Sushrut S. Waikar, Department of Medicine, Boston University Medical Center, Boston, MA.
Kayleen Williams, Department of Biostatistics, University of Washington, Seattle, WA.
Francis Perry Wilson, Yale School of Medicine, New Haven, CT.
Kun Zhang, Institute for Genomic Sciences, University of California, San Diego, San Diego, CA.
Srinivas Ravi Iyengar, Mount Sinai institute for Systems Biomedicine, Mount Sinai, New York, NY.
Matthias Kretzler, Department of Internal Medicine, University of Michigan, Ann Arbor, MI.
Jonathan Himmelfarb, Department of Medicine, University of Washington, Seattle, WA.
References
- 1.Centers for Disease Control and Prevention. Chronic Kidney Disease Surveillance System website. https://nccd.cdc.gov/CKD (accessed August 26, 2019.
- 2.Afkarian M, Zelnick LR, Hall YN, et al. Clinical Manifestations of Kidney Disease Among US Adults With Diabetes, 1988–2014. JAMA 2016; 316(6): 602–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Jaghbeer M, Dealmeida D, Bilderback A, Ambrosino R, Kellum JA. Clinical Decision Support for In-Hospital AKI. J Am Soc Nephrol 2018; 29(2): 654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellum JA, Lameire N, Group KAGW. Diagnosis, evaluation, and management of acute kidney injury: a KDIGO summary (Part 1). Crit Care 2013; 17(1): 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawla LS, Eggers PW, Star RA, Kimmel PL. Acute kidney injury and chronic kidney disease as interconnected syndromes. N Engl J Med 2014; 371(1): 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin A, Tonelli M, Bonventre J, et al. Global kidney health 2017 and beyond: a roadmap for closing gaps in care, research, and policy. Lancet 2017; 390(10105): 1888–917. [DOI] [PubMed] [Google Scholar]
- 7.Fox CS, Matsushita K, Woodward M, et al. Associations of kidney disease measures with mortality and end-stage renal disease in individuals with and without diabetes: a meta-analysis. Lancet 2012; 380(9854): 1662–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breyer MD, Susztak K. The next generation of therapeutics for chronic kidney disease. Nature reviews Drug discovery 2016; 15(8): 568–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genovese G, Friedman DJ, Ross MD, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science (New York, NY 2010; 329(5993): 841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonventre JV, Yang L. Cellular pathophysiology of ischemic acute kidney injury. J Clin Invest 2011; 121(11): 4210–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ralto KM, Rhee EP, Parikh SM. NAD(+) homeostasis in renal health and disease. Nature reviews Nephrology 2020; 16(2): 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alpers CE, Hudkins KL. Mouse models of diabetic nephropathy. Current opinion in nephrology and hypertension 2011; 20(3): 278–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosius FC 3rd, Alpers CE, Bottinger EP, et al. Mouse models of diabetic nephropathy. J Am Soc Nephrol 2009; 20(12): 2503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonventre JV, Basile D, Liu KD, et al. AKI: a path forward. Clinical journal of the American Society of Nephrology : CJASN 2013; 8(9): 1606–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breyer MD, Coffman TM, Flessner MF, et al. Diabetic nephropathy: a national dialogue. Clinical journal of the American Society of Nephrology : CJASN 2013; 8(9): 1603–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaskel F, Batlle D, Beddhu S, et al. Improving CKD therapies and care: a National Dialogue. Clinical journal of the American Society of Nephrology : CJASN 2014; 9(4): 815–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonventre JV, Boulware LE, Dember LM, et al. The kidney research national dialogue: gearing up to move forward. Clinical journal of the American Society of Nephrology : CJASN 2014; 9(10): 1806–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.KDIGO clinical practice guideline for lipid management in chronic kidney disease. Kidney Int Suppl 2013; 3(3): 259–305. [Google Scholar]
- 19.Hall YN, Himmelfarb J. The CKD Classification System in the Precision Medicine Era. Clin J Am Soc Nephrol 2017; 12(2): 346–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambers Heerspink HJ, Oberbauer R, Perco P, et al. Drugs meeting the molecular basis of diabetic kidney disease: bridging from molecular mechanism to personalized medicine. Nephrol Dial Transplant 2015; 30 Suppl 4: iv105–12. [DOI] [PubMed] [Google Scholar]
- 21.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015; 372(9): 793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obama B. Remarks by the President on Precision Medicine. January 30, 2015. Available at: https://obamawhitehouse.archives.gov/the-press-office/2015/01/30/remarks-president-precision-medicine (accessed November, 20 2019).
- 23.Tavassoly I, Goldfarb J, Iyengar R. Systems biology primer: the basic methods and approaches. Essays in biochemistry 2018; 62(4): 487–500. [DOI] [PubMed] [Google Scholar]
- 24.Mulder S, Hamidi H, Kretzler M, Ju W. An integrative systems biology approach for precision medicine in diabetic kidney disease. Diabetes, obesity & metabolism 2018; 20 Suppl 3: 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimmel PL, Jefferson N, Norton JM, Star RA. How Community Engagement Is Enhancing NIDDK Research. Clin J Am Soc Nephrol 2019; 14(5): 768–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perazella MA, Coca SG, Hall IE, Iyanam U, Koraishy M, Parikh CR. Urine microscopy is associated with severity and worsening of acute kidney injury in hospitalized patients. Clin J Am Soc Nephrol 2010; 5(3): 402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corapi KM, Chen JL, Balk EM, Gordon CE. Bleeding complications of native kidney biopsy: a systematic review and meta-analysis. Am J Kidney Dis 2012; 60(1): 62–73. [DOI] [PubMed] [Google Scholar]
- 28.Moledina DG, Luciano RL, Kukova L, et al. Kidney Biopsy-Related Complications in Hospitalized Patients with Acute Kidney Disease. Clinical journal of the American Society of Nephrology : CJASN 2018; 13(11): 1633–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poggio ED, McClelland RL, Blank K, et al. Systematic Review and Meta-analysis of Native Kidney Biopsy Complications. CJASN [in press] 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lake BB, Chen S, Hoshi M, et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun 2019; 10(1): 2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menon R, Otto EA, Hoover P, et al. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI insight 2020; 5(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohler S, Vasilevsky NA, Engelstad M, et al. The Human Phenotype Ontology in 2017. Nucleic acids research 2017; 45(D1): D865–D76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heerspink HJL, Perkovic V. Trial Design Innovations to Accelerate Therapeutic Advances in Chronic Kidney Disease: Moving from Single Trials to an Ongoing Platform. Clin J Am Soc Nephrol 2018; 13(6): 946–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


