Abstract
Background
Intratumoral injection of cyclic dinucleotide (CDN) agonists of the stimulator of interferon genes (STING) pathway engages innate immune activation and priming of adaptive immune effectors to foster local and distal tumor clearance. Despite proven therapeutic efficacy in preclinical models, a thorough understanding of how CDNs reprogram suppressive myeloid stroma in mouse and man is lacking.
Methods
Here, we perform deep transcript-level and protein-level profiling of myeloid-derived suppressor cells and M2 macrophages following stimulation with CDNs of ascending potency. Additionally, we leverage orthotopic Kras+/G12DTP53+/R172HPdx1-Cre (KPC) derived models of pancreatic adenocarcinoma (PDAC) to determine the capacity for locally administered CDNs to sensitize PDAC to immune checkpoint blockade. We use bioluminescent in vivo imaging and 30-parameter flow cytometry to profile growth kinetics and remodeling of the tumor stroma post-therapy.
Results
Highly potent synthetic STING agonists repolarize suppressive myeloid populations of human and murine origin in part through inhibition of Myc signaling, metabolic modulation, and antagonism of cell cycle. Surprisingly, high-potency synthetic agonists engage qualitatively unique pathways as compared with natural CDNs. Consistent with our mechanistic observations, we find that intratumoral injection of the highest activity STING agonist, IACS-8803, into orthotopic pancreatic adenocarcinoma lesions unmasks sensitivity to checkpoint blockade immunotherapy. Dimensionality reduction analyses of high parameter flow cytometry data reveals substantial contributions of both myeloid repolarization and T cell activation underlying the in vivo therapeutic benefit of this approach.
Conclusions
This study defines the molecular basis of STING-mediated myeloid reprogramming, revealing previously unappreciated and qualitatively unique pathways engaged by CDNs of ascending potency during functional repolarization. Furthermore, we demonstrate the potential for high potency CDNs to overcome immunotherapy resistance in an orthotopic, multifocal model of PDAC.
Keywords: immunotherapy, immunity, innate, tumor microenvironment, alarmins, myeloid-derived suppressor cells
Background
Successful antitumor immunity requires proinflammatory activation of the tumor microenvironment (TME) in order to overcome innate and adaptive immune tolerance. For ‘hot’ tumors possessing a sufficiently dense but functionally suppressed T cell infiltrate, blockade of immune checkpoint receptors including CTLA-4 or PD-1 (ICB) can be efficacious. In ‘cold’ tumors like pancreatic ductal adenocarcinoma (PDAC), however, effector T cells are excluded or disabled by an immunosuppressive myeloid and fibroblast stroma, leading to ICB failure.1 2 While vaccination can amplify peripheral antitumor T cell frequencies,3 4 local desmoplasia remains a major barrier to T cell entry, longevity, and effector function at the malignant pancreatic niche.5 6 We hypothesize that both proinflammatory reprogramming of the tumor stroma and enhanced immune priming via vaccination are required to unmask sensitivity to checkpoint blockade in PDAC.
Direct intratumoral delivery of innate immune adjuvants, in situ vaccination, can theoretically achieve both adaptive immune priming and stromal remodeling.7 8 In the context of cancer, this requires two critical events: (1) tumor antigen cross-presentation by Batf3+ dendritic cells (DCs) to prime CD8 T cells and (2) inflammatory polarization of tumor-associated macrophages (TAM) and myeloid-derived suppressor cells (MDSC) to phenotypes that facilitate, rather than suppress, T cell mediated immunity. Type I interferons are essential to induce priming of antitumor T cells,9 10 and in cancer, the stimulator of interferon genes (STING) pathway is considered essential for type I IFN elaboration.11 12 Predictably, intratumoral administration of STING agonists complements ICB in many established preclinical tumor models.13–17 However, whether and how STING agonists affect function of suppressive tumor myeloid cells remains enigmatic. Several reports have associated in situ STING activation with inflammatory modulation of tumor myeloid phenotypes, generally measured as conversion of ‘M2’ to ‘M1’ markers on TAMs.14 18 The molecular mechanisms driving repolarization remain undescribed, and little is known regarding effects of STING activation on MDSC populations. Thus, a deeper understanding of how STING agonists modulate myeloid function is needed.
In this study, we sought to dissect the underlying molecular mechanisms by which STING agonists induce proinflammatory conversion of the immunosuppressive PDAC myeloid stroma. We evaluated four CDNs: cyclic di-GMP (CDG), 2′3′-cGAMP (cGAMP), ML-RR-S2-CDA (ADU-S100; abbreviated ML-RR), and a newly described CDN with high relative potency IACS-8803 (8803), for efficacy and myeloid polarization efficiency in a novel murine model of PDAC.19 Also, we performed extensive multiomic analysis of ex vivo generated MDSCs and M2-like macrophages of murine and human origin on stimulation with each agonist. Integration of these data reveal unappreciated roles for STING agonists in counteracting Myc signaling, altering cell cycle dynamics, and modulating metabolic programming. Finally, we used our novel agonist 8803 to demonstrate that CDN-mediated in situ vaccination can potentiate dual checkpoint blockade in a multifocal, orthotopic model of pancreatic cancer. By using 30-color flow cytometry and dimensionality-reduction algorithms, we elucidate how each therapy remodels the tumor stroma and impacts anti-PDAC adaptive immunity. These data provide a deeper mechanistic understanding of the molecular pathways engaged downstream of STING activation to functionally repolarize tumor myeloid cells and demonstrate how CDNs of the highest potency can cooperate with T cell checkpoint blockade to enable effective immunotherapy of PDAC.
Materials and methods
Mice
Male C57BL/6 and B6(Cg)-Tyrc-2J /J albino mice were purchased from The Jackson Laboratory, and STING Gt/Gt mice were a kind gift from Dr Kimberly Schluns. Mice were used between 5 and 8 weeks of age.
Cell lines
The mT4-2D cell line was a kind gift from Dr David Tuveson20 and was maintained in complete Dulbecco′s Modified Eagle′s Medium (DMEM) containing 10% fetal bovine serum (Gibco), 100 U/mL penicillin, 100 mg/mL streptomycin sulfate, and 2 mM L-glutamine. THP-1 Dual and J774-Dual reporter cells were obtained from InvivoGen and cultured according to manufacturer’s instructions. HEK-Blue reporter cells were a kind gift from Dr Simon Yu at the University of Texas MD Anderson Institute for Applied Cancer Science and cultured in cDMEM supplemented with zeocin (100 µg/mL) and blasticidin (30 µg/mL) for antibiotic selection.
Cyclic dinucleotides (CDNs)
CDG and 2′3′ cGAMP were purchased from InvivoGen and reconstituted in water according to manufacturer’s instructions. ML-RR-CDA was synthesized by Wuxi AppTec and reconstituted in water. IACS-8803 was designed in collaboration with the University of Texas MD Anderson Cancer Center and synthesized by Wuxi AppTec19 and was reconstituted in water or PBS for in vitro or in vivo use.
Antibodies and chemotherapy
Antibodies used in in vivo studies were purchased from BioXCell or Leinco Technologies and were administered via intraperitoneal (IP) injection at the following concentrations: αCTLA-4 clone 9H10 (100 µg/mouse) and αPD-1 clone RMP1-14 (250 µg/mouse). For lymphocyte depletion, 2.43 (CD8), GK1.5 (CD4), and PK136 (NK1.1) antibodies were given at 250 µg/mouse every 3 days for the duration of the study. Chemotherapy agents gemcitabine (Gemzar; Eli Lily) and nab-paclitaxel (Abraxane; Celgene) were obtained from the University of Texas MD Anderson pharmacy and were reconstituted according to manufacturer’s instructions. Both agents were administered IP at 120 mg/kg. Antibodies used for flow cytometry were obtained from BD Biosciences, BioLegend, or Roche.
STING reporter cell assays
All STING reporter cell assays were conducted according to manufacturer’s protocols. In short, CDN or control agonists were added at desired concentrations in 20 µL PBS to desired wells of flat-bottom 96-well plates. HEK-Blue, J774-Dual, THP-1 Dual, or THP-1 parental cell lines were harvested, washed, counted, and resuspended for addition of 5×104 – 1×105 cells per well in 180 µL recommended media. Cells were incubated with CDN for 20–24 hours, when supernatants were harvested for Luciferase or SEAP assays using QuantiLuc or QuantiBlue solutions according to manufacturer’s instructions. For ELISA, THP-1 parental cells were incubated with CDN for 72 hours, then supernatants were harvested, and IFNβ levels were quantified using the PBL Human IFNβ ELISA kit according to manufacturer’s protocols.
cMyc reporter cell assays
The HCT116 cMyc reporter cell assays (BPS Bioscience) were conducted according to manufacturer’s protocols. In short, 25 000 HCT116 cells were plated into flat bottom 96-well plates the night before the assay. Assays were performed with a threefold serial dilution of either the provided b-catenin inhibitor (ICG-001), STING agonists IACS-8803 or cGAMP (Invivogen), or FSL-1 (Invivogen). After an overnight incubation, cMyc activity was assessed by Luciferase expression follow addition of One-Step Luciferase substrate (Promega).
MDSC generation, proliferation assays, and T cell suppression assays
Bone marrow was isolated from naïve male C57BL/6 mice through aspiration of the femur and/or tibia. Red blood cells were lysed using RBC lysis buffer (Sigma), and 4×106 cells were plated in 10 mL in non-TC treated 10 cm petri dishes (Falcon) in RPMI containing 10% Fetal Bovine Serum (FBS), 100 U/mL penicillin, 100 mg/mL streptomycin sulfate, recombinant mouse GM-CSF (40 ng/mL; BioLegend) and IL-6 (40 ng/mL; BioLegend), and 2-Mercaptoethanol (β-ME; 55 µM) to induce MDSC differentiation. After 4 days in culture, bone marrow derived MDSC (BM-MDSC) suspension cells were harvested, washed with cRPMI to remove cytokine, and replated in 10 mL cRPMI with no cytokine at 4×105 cells/mL in 10 cm petri dishes. CDN were added at 1–10 µg/mL as indicated for 24-hour repolarization.
MDSC proliferation assays and calculation of proliferation index
Bone marrow MDSCs were generated as above (GM-CSF and IL-6 for 4 days) and treated with STING agonists or vehicle control for 48 hours following CFSE labeling prior to analysis on a flow cytometer. Proliferation index is calculated by the average number of cell divisions that a cell in the original population has undergone. It was obtained by the following formula that measures the ratio of the number of cells that underwent division to the total number of cells in the system. , where i=number of cell division peak as determined by CFSE dilution, Ni=number of cells in that division peak.
For suppression assays, naïve splenocytes were obtained from male C57BL/6 mice, and CD8 T cells were isolated by negative selection with the Miltenyi Mouse CD8 T Cell Isolation Kit. CD8 T cells were labeled with CellTrace CFSE (ThermoFisher) as described. The 1×105 CFSE-labeled CD8 T cells were plated at indicated ratios with unstimulated or repolarized BM-MDSC in 200 µL cDMEM containing 10% FBS, 50 U/mL IL-2, 27.5 µM β-ME, and 2 µg/mL αCD28 (clone 37.51; BioLegend) in round bottom 96-well plates coated with αCD3 (BioXCell clone 145–2 C11; coated overnight at 10 µg/mL in PBS at 4°C). After 72–96 hours, CD8 T cells were analyzed for CFSE dilution by flow cytometry.
Human macrophage generation
Healthy donor buffy coats were obtained from the University of Texas MD Anderson Cancer Center Blood Bank. Peripheral blood mononuclear cells were isolated by ficoll gradient centrifugation (Histopaque 1077; Sigma), and CD14+ monocytes were isolated by negative selection using the Miltenyi Classical Monocyte Isolation Kit. Monocytes were differentiated into M2c macrophages as described.21 In short, monocytes were plated in T25 flasks (1×106 cells/mL) in RPMI containing 20% FBS and 100 ng/mL recombinant human M-CSF for 6 days, with a media refresh on day 4. On day 6, media was replaced, with the addition of recombinant human TGF-β (10 ng/mL) and IL-10 (10 ng/mL). On day 8, adherent cells were washed with PBS and cultured with cRPMI + 100 ng/mL M-CSF + 10 µg/mL indicated CDN for repolarization over 72 hours. Adherent macrophages were harvested with Detachin cell detachment solution (Genlantis) for downstream applications. All recombinant human cytokines were purchased from PeproTech and reconstituted according to manufacturer’s instructions.
Luminex, RNA microarray, and RPPA sample preparation
Repolarized human M2c macrophages or murine BM-MDSC were generated as described. Supernatants from repolarized cultures were isolated, spun to remove contaminating cells, then frozen at −80oC. All harvested supernatants were thawed on ice and were analyzed with the Cytokine & Chemokine 36-Plex Mouse ProcartaPlex or Cytokine/Chemokine/Growth Factor 45-Plex Human ProcartaPlex Panels (Invitrogen/ThermoFisher) using a Luminex MAGPIX machine according to manufacturer’s instructions. For RNA, 4×105 – 2×106 repolarized cells were harvested, lysed, and homogenized for RNA isolation with the QIAGEN RNeasy Mini Kit according to manufacturer’s protocols. RNA quality and concentration was determined by Bioanalyzer (Agilent), and RNAseq was performed through Illumina. For RPPA, a minimum of 1×106 cells were washed twice with cold PBS, and dry pellets were stored at −80oC. Cell lysis and analysis was conducted at the University of Texas MD Anderson Functional Proteomics RPPA Core Facility.
Orthotopic pancreatic tumor implantation and IVIS imaging
Subconfluent cultures of mT4-2D, mT4-LA, or mT4-LS were harvested with 0.05% trypsin, washed with PBS, and resuspended at desired concentration in ice cold PBS containing 30% Matrigel (Corning) by volume for implantation. Each mouse was anesthetized through isoflurane inhalation and was administered 3 µg buprenorphine hydrochloride analgesic (Sigma) by IP injection. Following sterilization of the surgical field with 70% isopropyl alcohol, a small (~1–2 cm) incision was made on the left flank skin as well as the underlying peritoneal lining. Blunt-end forceps were used to access the spleen and accompanying pancreas. Fifty microliters of cell suspension or CDN solution was administered into the head of the pancreas using a U-40 insulin syringe with 29.5-gage needle. The peritoneal wound and surrounding skin were cleaned with betadine antiseptic swabs, and the wound was closed using 4–0 absorbable sutures for the peritoneal incision and the Autoclip Wound Closing System for the skin (Braintree Scientific). Mice were allowed to recover under a heat lamp until displaying normal ambulance and were monitored daily for signs of pain or discomfort. Wound clips were removed 10–14 days following application.
Mice were imaged weekly to monitor tumor growth using an IVIS Spectrum in vivo imaging system (PerkinElmer) and randomized based on equivalent distribution of larger versus smaller tumors into groups prior to initiating therapy. Mice were anesthetized with isoflurane and received 200 µL D-Luciferin (15 mg/mL in PBS; GoldBio) by IP injection. Bioluminescent images were acquired 11–12 min following administration of D-Luciferin. All imaging was conducted in the University of Texas MD Anderson Small Animal Imaging Facility.
Flow cytometry analysis of tumor immune infiltrates
mT4-2D or mT4-LA cells were harvested with 0.05% trypsin, washed with PBS, counted, and resuspended in ice cold PBS containing 30% Matrigel for implantation of 2.5×105 cells in 100 µL subcutaneously or 3.5×104 cells in 50 µL orthotopically as described. Mice were treated with intratumoral CDN), checkpoint blockade antibodies, or chemotherapy according to treatment schedules included in associated figures. Following euthanasia, primary tumors were harvested, massed, and finely diced into 70 µm filters within 6 cm petri dishes. Diced tumors were enzymatically digested in a 37oC incubator for 30 min in digestion media consisting of X-Vivo15 media (Lonza) supplemented with collagenase H (1 mg/mL; Sigma) and DNAse (160 µg/mL; Roche), then tumors were physically mashed through 70 µm filters to create single cell suspensions. Total cells were counted, then live immune cells were purified by ficoll gradient centrifugation (Histopaque 1119; Sigma). Samples were fixed overnight and permeabilized using the FoxP3/Transcription Factor Staining Buffer Set (eBioscience/ThermoFisher) for staining with fluorescently labeled antibodies in 96-well U-bottom plates. Stained samples were analyzed using a BD LSRII or X-30 prototype flow cytometer.
Statistics
Unless otherwise indicated, all statistical analyses comparing two independent groups were performed using Student’s t-test. For Kaplan-Meier survival curves, Log-Rank Mantel Cox test was used to test for differences between treatment groups. For high dimensional flow cytometry analyses comparing cluster frequencies between multiple treatment groups, two-way analysis of variance with Tukey’s correction for multiple comparisons was used.
Results
Evaluating CDNs of ascending potency in a transplantable model of PDAC
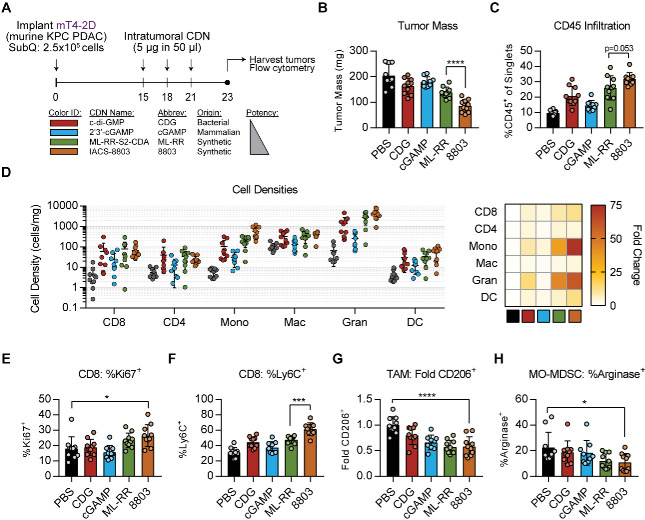
The novel CDN STING agonist IACS-8803 exhibits enhanced potency relative to the clinical ML-RR compound in established in vitro and in vivo settings.19 We sought to leverage the diversity of STING-activating CDNs available to probe whether higher potency STING activation impacted the PDAC tumor stroma in unique ways. We first performed intratumoral injection of CDNs into mT4-2D PDAC tumors—a cell line derived from Kras+/G12DTP53+/R172HPdx1-Cre (KPC) organoid cultures.20 We focused on four CDNs: natural agonists 3′-3′-cyclic di-GMP (CDG) and 2′3′-cGAMP (cGAMP), and synthetic agonists ML-RR and 8803 (figure 1A). Tumors were harvested following three injections with 5 µg of indicated CDN for mass measurement and analysis by multiparameter flow cytometry (figure 1A). We found that, relative to vehicle injection, ML-RR and 8803 induce regression of mT4-2D tumors, with 8803 demonstrating superior efficacy (figure 1B). Local delivery of 8803 triggers expansion of the CD45+ immune infiltrate (figure 1C), which is dominated by CD11b+Ly6G+Ly6CmidF4/80− granulocytes (figure 1D). This granulomatous response occurs at similar or reduced levels in tumors exposed to ML-RR or CDG but is significantly less with cGAMP treatment. CD8 T cells increase in number after 8803 exposure and express more Ki67 and Ly6C (figure 1D–F). Expansion of CD11b+Ly6C+Ly6G-F4/80mid/− monocytes with reduced arginase expression also occurs after 8803 (figure 1D, H). TAM (CD11b+Ly6C-Ly6G-F4/80+) do not expand, but the expression of the M2 marker CD206 is significantly reduced in the post-treatment TAM compartment (figure 1G). Together, these data indicate 8803 is the most potent therapeutic CDN among those tested in ectopic KPC-derived tumors and that in situ vaccination with 8803 precipitates inflammatory modulation of the PDAC TME including numeric and phenotypic enhancement of both lymphoid and myeloid populations. These data also suggest that therapeutic benefit in ‘cold’ TMEs may scale proportional to the potency of the applied STING agonist. Of note, we compared both IACS-8802 and 8803 to cGAMP to determine to what extent higher concentrations of the weaker agonist might generate functional equivalence and found that between a 10X and 50X excess of cGAMP could achieve high level IRF3 activation (online supplemental figure 1). However, even a 100X excess of cGAMP could not equal 8802/8803 level NFκB activation in short term assays, although equivalence could be obtained over longer incubations.
Figure 1.
In vivo characterization of 8803 in subcutaneous KPC-derived PDAC. (A) Mice received subcutaneous injection of 2.5×105 mT4-2D cells in 30% matrigel, then were injected intratumorally with 5 µg CDN on days 15, 18, and 21 before tumor harvest on day 23. Tumors were massed, counted, and processed for flow cytometry analysis as described in Methods. Data shown represent (B) tumor mass, (C) CD45+ infiltration, (D) overall composition of analyzed CD45+ cells, (E) cellular densities, (F) Ki67 and (G) Ly6C expression on tumor CD8 T cells, (H) normalized CD206 expression on macrophages, and (I) arginase expression in MO-MDSC. Data are cumulative of two independent experiments with five mice per group. Statistical significance was calculated using Student’s t-test. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001. CDN, cyclic dinucleotide; MDSC, myeloid-derived suppressor cells; ns, not significant; PDAC, pancreatic adenocarcinoma.
jitc-2021-003246supp001.pdf (5.4MB, pdf)
High potency CDNs diminish proliferation and cMyc signaling in MDSC
Given these observations, we sought a deeper understanding of the mechanisms by which synthetic CDNs alter the function, phenotype, and molecular programming of the myeloid lineage. For this, we studied effects of CDNs on in vitro differentiated suppressive myeloid populations. First, we generated bone marrow derived MDSC (BM-MDSC) using the prevalent protocol (GM-CSF and IL-6) that yields a ~2:1 ratio of granulocytic to monocytic MDSC, approximating their murine in vivo prevalence (online supplemental figure 2A).22 These BM-MDSC cultures suppress T cell proliferation in vitro, which we validated in a CFSE-based T cell suppression assay (online supplemental figure 2B). To determine if STING activation modulates MDSC suppression, we stimulated BM-MDSC with CDNs for 24 hours, removed CDN from the culture, then evaluated BM-MDSC T cell suppression capacity. We find 8803-treated BM-MDSC lose suppressive function in a dose-dependent manner (online supplemental figure 2B). Comparing different CDNs, we find the relative effects mirror the therapeutic potencies observed in our in vivo studies (figure 2A and online supplemental figure 2C). These data indicate CDNs are capable of reversing the T cell suppressive activities of BM-MDSC.
Figure 2.
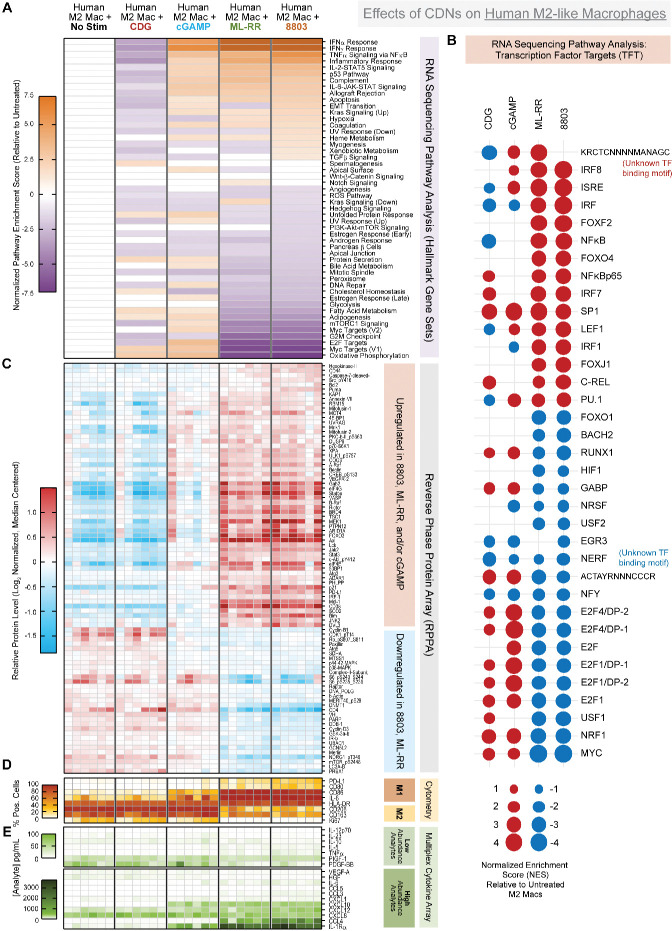
Profiling effects of CDNs on murine bone marrow derived MDSCs. (A) Summary of T cell suppression assay using BM-MDSCs stimulated with the indicated CDN for 24 hours at 2.5 µg/mL. CFSE dilution at the 1:1 MDSC:T cell ratio is shown, and per cent of live CD8 T cells proliferating at each indicated ratio is shown by heat map. For B–E, BM-MDSC were stimulated by indicated CDN at 2.5 µg/mL for 24 hours, followed by RNA, whole cell pellet, or supernatant harvesting for downstream multiomic analysis. (B) Gene set enrichment analysis results of RNA sequencing data represented as log2 normalized pathway enrichment score for each condition as compared with untreated BM-MDSC. Focus on hallmark gene sets. (C) Same as in figure part B), focused on transcription factor targets gene sets. (D) Relative protein levels as measured by reverse phase protein array, following log2 normalization and median centering of the data. (E) Secreted analyte concentrations in culture supernatants measured using Luminex multiplex cytokine/chemokine analysis. Data were manually segregated to display relevant low abundance and high abundance analytes. Data are cumulative of MDSCs from 4 to 8 murine donors in two independent batches. CDN, cyclic dinucleotide; MDSC, bone marrow derived MDSC.
We next sought to dissect the molecular mechanisms underlying CDN-mediated MDSC refunctionalization. Our approach involved generation of orthogonal datasets detailing effects of individual CDNs on MDSC transcription, protein expression, and cytokine/chemokine secretion. To this end, we activated BM-MDSC with each CDN for 24 hours then harvested bulk RNA for RNA sequencing (RNAseq), whole cell pellets for reverse phase protein array (RPPA), and culture supernatants for multiplex cytokine analysis. These data are represented in figure 2 as an integrated map of BM-MDSC activity at each cellular dimension; transcription (figure 2B, C), translation (figure 2D), and function (figure 2A, E) in response to each CDN.
We first performed gene set enrichment analysis of the RNAseq data, focusing on the Hallmark and Transcription Factor Target gene sets (figure 2B–C), using unstimulated BM-MDSC as our reference. We report the top pathways activated by CDN exposure in BM-MDSC are associated with IFNα, IFNγ, and NFκB signaling (figure 2B). In addition, we report the most negatively enriched pathways to be E2F targets, G2M checkpoint, and mitotic spindle gene sets, suggesting STING engages cell cycle inhibition. Interestingly, this antiproliferative signature is specifically observed on exposure to synthetic CDNs ML-RR and 8803 (figure 2B, C). These data are validated at the protein level in our RPPA dataset, where BM-MDSC exposed to ML-RR or 8803 exhibit significantly reduced levels of CDC42, CDC25C, CDC6, and phosphorylated CDK family members (figure 2D). The capacity of IACS-8803 but not cGAMP to significantly suppress proliferation of murine MDSC was confirmed in vitro (online supplemental figure 3). Interestingly, we find the antiproliferative signature correlates more closely with downregulation of the Myc targets (v1) Hallmark gene set (figure 2B). While c-Myc has been identified as a regulator of MDSC cell cycle,23 this is to our knowledge the first association between STING activation, Myc inhibition, and reduced proliferation in MDSCs.
We additionally find CDNs promote secretion of myeloid-homing chemokines by MDSCs. We report two ‘tiers’ of elicited chemokines based on relative abundance following stimulation by ML-RR or 8803. The high abundance chemokines are CCL2, CCL5, CXCL10, CCL4, and CCL7, which increase from 1000 to 2000-fold on stimulation (figure 2E). These chemokines share redundant and non-overlapping roles in recruitment of monocytes, macrophages, and select T cell subsets. The lower abundance chemokines CXCL2, CXCL1, and CCL3 also increase 10–100 fold on stimulation (figure 2E). In addition to chemokines, synthetic CDNs activate production and release of diverse inflammatory factors, indicative of a shift away from an immunosuppressive phenotype. A total of 8803 elicits robust production of IL-6 and Tumour Necrosis Factor alpha (TNFα), while 8803 and ML-RR both incite release of IL-18. To a lesser extent, synthetic CDNs also induce IL-12 family members IL-12p70, IL-23, and IL-27 (figure 2E).
These data also reveal a number of unexpected and unappreciated effects of CDNs on BM-MDSC. First, we report that cGAMP, ML-RR, and 8803 activate a transcriptional signature associated with oxidative phosphorylation (OXPHOS), which correlates with a reduction in the fatty acid oxidation (FAO) (figure 2B). Transcription factor targets analysis reveals modulation of unexpected transcription factors downstream of STING including enrichment of the CREB family member ATF3 and the PAX family of transcription factors, and negative enrichment for SRF, NRF1 and NRF2, and NF-Y. Intriguingly, this analysis also reveals 8803 induces enrichment of genes with an unknown transcription factor binding motif upstream (KRCTCNNNNMANAGC). Whether this motif is an unappreciated binding sequence of canonical downstream STING targets in the Interferon Regulatory Factor (IRF) or Nuclear Factor kappa-light-chain-enhancer of activated B cells (NFκB) families is an area of active investigation.
Finally, these integrated data suggest qualitative differences in signaling programs modulated by CDNs of ascending potency. While highly potent synthetic CDNs ML-RR and 8803 are largely overlapping in Hallmark gene sets, we find cGAMP elicits a unique—rather than quantitatively intermediate—signature. This is clear when comparing E2F target, G2M checkpoint, mitotic spindle, and Myc target gene sets, which are strongly downregulated by ML-RR and 8803 yet moderately upregulated by cGAMP (figure 2B). Additionally, in our RPPA dataset, cGAMP stimulation promotes a strong positive feedback loop to increase STING protein levels that greatly exceeds the level of upregulation achieved by more potent agonists ML-RR or 8803, as well as the less potent CDG (figure 2C). These data suggest transcriptional signatures engaged by CDNs of ascending potency can diverge in a qualitative—not simply quantitative—manner.
High potency CDNs decrease proliferation and cMyc signaling in human M2c-polarized macrophages
To investigate how CDNs polarize human macrophages, we performed an analogous multiomic analysis of human M2-like macrophages exposed to different CDNs. For this, we generated M2-like macrophages from human CD14+ monocytes cultured in the presence of M-CSF, IL-10, and TGF-β, yielding macrophages with high expression of CD68, CD163, CD206, CD204, and IRF4 (online supplemental figure 4A, B).21 We stimulated these M2c-like macrophages with CDNs for 3 days before analysis. We found unstimulated M2c macrophages to exhibit classical M2-like morphology characterized by firmly adherent, elongated cellular architecture. In contrast, addition of CDNs results in a shift in morphology toward a less adherent, ‘fried egg’ M1-like shape.24 25 The degree to which this morphological shift occurred—as measured approximately by side scatter characteristics on analysis by flow cytometry—was correlated with CDN potency. (online supplemental figure 4C, D).
As previously mentioned, we quantified effects of CDNs on human M2c-like macrophage transcription by RNAseq (figure 3A–B), translation by RPPA (figure 3C), and function by flow cytometry and multiplex cytokine analysis (figure 3D–E). Consistent with previous findings, synthetic CDNs ML-RR and 8803 and the natural CDN cGAMP activate type I interferon and NFκB signaling in human M2-like macrophages (figure 3A). In agreement with our murine BM-MDSC data, synthetic CDNs uniquely inhibit proliferation in human M2-like macrophages as evidenced by negative enrichment of E2F targets, G2M checkpoints, and mitotic spindle Hallmark gene sets (figure 3A), protein-level downregulation of cyclin B1, cyclin D3, and phospho-CDK1 (pT14) (figure 3C), and downregulation of Ki67 by flow cytometry (figure 3D). Again, this antiproliferative state correlates with strong negative enrichment of Myc targets Hallmark gene sets. Myc has been implicated in supporting the proliferative nature and phenotypic polarization of M2 macrophages26 27; thus, we again discover a novel association between potent CDN stimulation, c-Myc inhibition, and reduced proliferation in cells of the myeloid lineage.
Figure 3.
Profiling effects of CDNs on human M2c-polarized macrophages. Human M2c macrophages were differentiated from PBMC monocytes as described in Methods, then stimulated by indicated CDN at 10 µg/mL in the presence of supportive recombinant M-CSF for 72 hours. RNA, whole cell pellets, and/or supernatants were harvested for downstream multiomic analysis. (A) Gene set enrichment analysis results of RNA sequencing data represented as log2 normalized pathway enrichment score for each condition as compared with untreated M2c macrophages. Focus on hallmark gene sets. (B) Same as in figure part A, focused on transcription factor targets gene sets. (C) Relative protein levels as measured by reverse phase protein array, following log2 normalization and median centering. (D) Validation of phenotypic and functional marker expression at the protein level by flow cytometry, with heatmap coloring representing percent cells expressing the indicated marker, relative to isotype control gating. (E) Secreted analyte concentrations in culture supernatants measured using Luminex multiplex cytokine/chemokine analysis. Data were manually segregated to display relevant low abundance and high abundance analytes. Data are cumulative of macrophages from six unique donors in two independent batches. CDNs, cyclic dinucleotides.
By flow cytometry profiling, we report induction of M1 markers CD80, CD86, and HLA-DR, concomitant with downregulation of M2 markers CD163 and CD206 following exposure to 8803, ML-RR, and cGAMP by flow cytometry (figure 3D). In supernatants from these cultures, we find synthetic CDNs achieve downregulation of known tumor-supportive or anti-inflammatory factors including CXCL8/IL-8, IL-10, VEGF-A, and PDGF-BB (figure 3D). In contrast, 8803 and ML-RR induce moderate production of proinflammatory cytokines TNFα and IL-6, together with substantial release of CCL4, CXCL12, and CXCL10 chemokines. Interestingly, of the 45 analytes detected in this assay, IL-1 decoy receptor IL-1Rα is the most abundantly released factor in human M2-like macrophages exposed to synthetic CDNs (figure 3D). In total, these observations confirm CDNs are capable of reprogramming the human M2-like macrophage phenotype and secretome.
We additionally report CDNs alter human M2-like macrophage metabolic programs at the transcriptional level. We find OXPHOS to be the most downregulated gene set on ML-RR and 8803 stimulation in M2 macrophages (figure 3A). Interestingly, we find exposure to ML-RR or 8803 induces a coordinate downregulation of mTORC1, glycolysis, and FAO Hallmark gene sets (figure 3A), indicative of a transcriptionally ‘hypometabolic’ state. We did find protein-level upregulation of proautophagic factors Beclin-1, phospho-ULK1 (pS757), ATG3, and FOXO3 by RPPA (figure 3C), and positive enrichment of the KEGG regulation of autophagy gene set on stimulation by 8803 or ML-RR (NES=2.39 and 2.63). Our data indicate this is restricted to synthetic CDNs, as cGAMP stimulation did not antagonize OXPHOS, mTORC1, or FAO at the transcriptional level (figure 3A).
A number of unexpected targets exist downstream of STING activation in human macrophages. The top positively enriched hit on ML-RR or 8803 stimulation in Transcription Factor Target analysis is KRCTCNNNNMANAGC, the same unknown binding motif previously detected for murine BM-MDSC (figures 2C and 3B). We found multiple Forkhead box transcription factor (FOX) family gene sets enriched on stimulation with ML-RR or 8803 including FOXF2, FOXJ1, FOXO4, and FOXO1 (figure 3B). Moreover, we report synthetic CDNs induce enrichment of genes regulated by BACH2 and RUNX1, coincident with downregulation of the RUNX1 repressor NERF/ELF-2 (figure 3B).28 Synthesizing these data, we report that CDN-induced macrophage repolarization involves elements of the canonical M2/M1 transition yet extends beyond to include: (1) inhibition of Myc signaling, (2) metabolic rewiring toward a hypometabolic, proautophagic state, (3) enhanced secretion of proinflammatory factors and (4) activation of a diverse set of transcriptional enhancers of macrophage maturation and inflammatory function that include—but are not limited to—NFκB and IRF family members.
Synthetic STING agonists induce cMyc downregulation
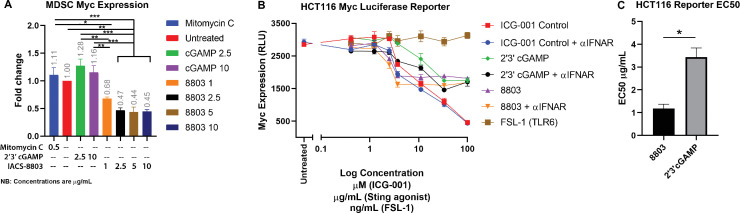
Our multiomic data indicating cMyc suppression by potent STING agonists reveal a novel mechanism of action for these drugs with both proinflammatory immunoregulatory potential at the level of myeloid stroma and, in cases where STING remains intact, direct antitumor potential. To validate these findings, we generated MDSC from murine bone marrow, treated them with either cGAMP or 8803, and then measured cMyc transcript levels by RT-PCR. Consistent with the -omic data, we find that IACS-8803 significantly suppressed MDSC expression of cMyc, while cGAMP did not (figure 4A). HCT116 is a human colorectal cancer line that retains sensitivity to STING agonists and is available with an integrated cMyc Luciferase reporter. Using this reporter system, we found that both 8803 and cGAMP could induce cMyc reporter suppression, although not to the levels of the β-catenin inhibitor control, while the TLR6 agonist FSL-1 had no activity (TLR6 is highly expressed by HCT116) (figure 4B). Blockade of interferon-α/β receptor had no impact on Myc downregulation demonstrating that this was not a secondary effect of IFN release. While myeloid cells appeared insensitive to cGAMP in terms of cMyc levels, these tumor cells did respond but with less efficiency than to 8803 (figure 4C).
Figure 4.
Synthetic STING agonists induce cMyc downregulation. (A) Bone marrow derived MDSC were incubated with the indicated drug at the concentration shown (μg/mL) for 48 hours, and then cMyc expression levels were measured using TaqMan (Invitrogen) and are shown relative to the untreated control. (B) The HCT116 Myc reporter cell line (BPS Bioscience) was treated overnight with the indicated drug at the concentration shown. Luciferase reporter expression was measured using the Promega one-step system. The polyclonal PA1-24777 antibody (Invitrogen) was added at 10 µg/mL to block interferon-α/β receptor engagement where indicated. (C) EC50s are shown for the triplicate repeats of IACS-8803 and 2’3’-cGAMP from the experiment in (B). Statistical significance was calculated using analysis of variance (A) or Student’s t-test (C). *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ns, not significant; STING, stimulator of interferon genes.
Combining local 8803 with checkpoint blockade cures multifocal mT4-LS PDAC
Immunosuppressive macrophages and MDSC have been associated with poor prognosis and resistance to checkpoint blockade in PDAC.29 30 Our data suggest that STING agonists could provide therapeutic benefit in PDAC through proinflammatory remodeling the suppressive myeloid stroma to actively support CD8 T cell recruitment, activation, effector function and persistence. Furthermore, by leveraging the known capacity for STING activation to expand CD8 T cells via enhancement of DC antigen presentation, we hypothesized that in situ CDN vaccination at a primary PDAC lesion would promote systemic CD8 T cell responses that can be protected by checkpoint inhibition for ‘abscopal’ control of distal, uninjected PDAC metastases, as we previously demonstrated in a bilateral model of prostate cancer.15 To evaluate these hypotheses, we again used mT4-2D, a PDAC cell line derived from KPC tumor organoid cultures.20 As is common of KRAS-driven spontaneous murine tumor models, we found mT4-2D to possess an extremely low mutational burden with similarly few predicted neoantigenic epitopes (online supplemental figure 5A, B). Retroviral transduction of an optimized firefly luciferase gene facilitated longitudinal monitoring of orthotopically implanted mT4-2D-luciferase (termed mT4-LA) using IVIS bioluminescent imaging. We found low doses of <5×104 mT4-LA cells capable of aggressive local invasion and metastatic spread to the liver, peritoneum, and mesentery, with mice succumbing to disease within 3 weeks. Additionally, we isolated a single cell clone from the mT4-LA population with reduced in vivo growth kinetics compared with mT4-LA following both subcutaneous and orthotopic implantation (mT4-LS for ‘slow’ growth; online supplemental figure 5C). We leveraged the larger therapeutic window afforded by the mT4-LS clone to screen potential combination approaches, then performed validation of effective therapies in the highly aggressive orthotopic mT4-LA model.
To evaluate the efficacy of in situ vaccination with 8803 in combination with checkpoint blockade against multifocal PDAC, we simultaneously implanted mice with both orthotopic and subcutaneous mT4-LS tumors to model a primary and metastatic lesion. Mice received localized 8803 or vehicle within the orthotopic tumor by survival surgery on days 10 and 22 postimplantation and received checkpoint antibody blockade of CTLA-4 or PD-1 by IP injection on days 10, 14, and 18 (figure 5A). We found monotherapy with αCTLA-4, αPD-1, or intrapancreatic 8803 capable of curing 50% of mice of both lesions in this model, with combination αCTLA-4/αPD-1 not demonstrating significant benefit over monotherapies. However, the combination of intratumoral 8803 with concurrent checkpoint blockade cured all mice tested of both lesions, regardless of the checkpoint being targeted (figure 5B, C). IVIS imaging reveals a rapid kinetic of tumor regression following combination therapy, with tumors falling below the level of detection within 6 days of treatment. These data demonstrate that local STING activation by 8803 within orthotopically PDAC can potentiate checkpoint blockade and induce robust curative immunity against both injected and distal uninjected mT4-LS tumors.
Figure 5.
Combining local 8803 with checkpoint blockade cures multifocal mT4-LS PDAC. (A) Implantation and treatment schedule. Intrapancreatic delivery of 8803 involves survival surgery to inject 8803 directly into primary mT4-LS lesions. (B) Representative longitudinal IVIS imaging of mice treated as in figure part A. (C) Survival data indicating therapeutic additivity between 8803 and CTLA-4 or PD-1 (D) or the combination of CTLA-4 and PD-1 (E). Data are cumulative of two independent experiments with 5–10 mice per group. Statistical significance was calculated using the log-rank Mantel-Cox test. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ns, not significant.
8803 potentiates checkpoint blockade against orthotopic mT4-LA PDAC independent of chemotherapy
Given the cooperativity observed between intratumoral 8803 and systemic checkpoint blockade in the responsive mT4-LS model, we tested whether synergy would be observed in the aggressive, immune refractory mT4-LA model. We implanted mice simultaneously with orthotopic and subcutaneous mT4-LA to model primary and metastatic lesions and intratumorally injected 8803 or vehicle on days 10 and 22 postimplantation. Mice received αCTLA-4 and/or αPD-1 IP on days 10, 14, 18, 22, and 26 (figure 6A). In this model, local 8803 induced transient regressions that moderately extend survival compared with PBS-treated mice; however, no mice were cured. Monotherapy with αCTLA-4 or αPD-1 does not deliver significant survival benefit, and the combination of either αCTLA-4 or αPD-1 with 8803 does not significantly extend survival compared with monotherapies (figure 6B, C and online supplemental figure 6A). Therefore, in contrast to the mT4-LS model, 8803 does not significantly synergize with individual checkpoint blockade against aggressive mT4-LA tumors. Dual blockade of CTLA-4 and PD-1, however, extends survival compared with PBS treatment, and the intratumoral 8803 and αCTLA-4/αPD-1 combination shows significant survival benefit over 8803 or αCTLA-4/αPD-1 alone (figure 5D and online supplemental figure 6A). Additionally, this combination shows an abscopal immune trend evidenced by reduced growth kinetics at the uninjected subcutaneous tumor (online supplemental figure 6B). These data demonstrate that 8803 can be delivered into aggressive metastatic tumors and synergize with dual checkpoint blockade to mediate local and potentially also systemic control of mT4-LA PDAC tumors.
Figure 6.
Intrapancreatic 8803 synergizes with dual checkpoint blockade to prolong survival in mT4-LA independent of chemotherapy. (A) Implantation and treatment schedule. (B–D) Survival of mice bearing orthotopic and subcutaneous mT4-LA treated as described in figure part A, with n=5–10 mice per group. (E) Mice bearing only orthotopic mT4-LA (0.35×105 cells) were treated as described in figure part A with additional standard of care chemotherapy consisting of gemcitabine (120 mg/kg) and nab-paclitaxel (120 mg/kg) (Gem/nP) administered on days 10 and 22 in indicated groups. Longitudinal tumor growth by IVIS imaging and overall survival are shown. (F) Survival data associated with images presented in figure part E. Data are cumulative of two independent experiments each with 5–10 mice per group. (G) A single mouse cured by 8803 + αCTLA-4/αPD-1 in figure part E was rechallenged with 0.35×106 mT4-LA cells as previously described, in parallel with five naïve B6 albino mice. Tumor regression is shown via IVIS imaging. Statistical significance was calculated using the Log-rank Mantel-Cox test. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ns, not significant.
We next hypothesized that concurrent administration of first-line chemotherapy agents gemcitabine (Gem) and Abraxane (nab-paclitaxel; nP) could further potentiate tumor control. We implanted mice orthotopically with mT4-LA and evaluated whether Gem/nP administered IP (120 mg/kg) on days 10 and 22 could enhance the efficacy of intratumoral 8803 and αCTLA-4/αPD-1 dual checkpoint blockade. In this study, subcutaneous ‘pseudo-metastatic’ lesions were not implanted due to the highly metastatic nature of orthotopic mT4-LA and the tendency of sub-cutaneous tumors to ulcerate early in progression requiring premature euthanasia of long-term responders (online supplemental figure 6C). Chemotherapy with Gem/nP alone delivers no observable benefit by IVIS imaging or survival and does not significantly boost the efficacy of 8803 or αCTLA-4/αPD-1 (online supplemental figure 6D). When combined with both 8803 and αCTLA-4/αPD-1, Gem/nP promotes early survival, with no deaths until day 41 postimplantation. However, this early benefit does not translate to a long-term survival advantage over mice treated with 8803 plus αCTLA-4/αPD-1 alone (figure 6E, F). Pertinently, we observe signs of increased treatment-related toxicity in mice exposed to Gem/nP, with 31.5% of mice failing to recover from the final therapeutic surgery (online supplemental figure 6E). A smaller proportion of mice receiving Gem/nP with 8803 or αCTLA-4/αPD-1 exhibit similar morbidity, while all mice receiving combination 8803 and αCTLA-4/αPD-1 recover without incident. Therefore, we conclude that standard of care chemotherapy is not required to achieve maximum therapeutic benefit when combining local 8803 with systemic dual checkpoint blockade with αCTLA-4 and αPD-1 against orthotopic mT4-LA PDAC tumors. Of note, in this experiment, a single mouse was cured following treatment with 8803 and αCTLA-4/αPD-1. To determine whether this therapeutic approach engendered immunological memory, this mouse was rechallenged with 0.35×105 mT4-LA cells again into the orthotopic site, and tumor growth was monitored (figure 6G). Interestingly, this mouse spontaneously rejected its tumor, whereas equivalent tumors grew rapidly in naïve mice. This case suggests robust antitumor immunity elicited by in situ vaccination with 8803 in combination with dual checkpoint inhibition is capable of leading to fully protective immune memory.
Dimensionality reduction of high-parameter flow cytometry data reveals inflammatory remodeling of the PDAC stroma by 8803 and checkpoint blockade
To deeply probe the immune microenvironment of PDAC tumors at baseline and in response to 8803-mediated in situ vaccination and checkpoint blockade, we developed a 30-parameter immunophenotyping antibody panel for use with a BD FACSymphony A3. Orthotopic mT4-LA tumors were harvested 20 days postimplantation, a therapeutic response inflection point in our survival studies, in mice receiving therapies outlined in figure 6E–F. At this timepoint, we observe significant reductions in overall mass and increased CD45+ immune infiltration in tumors isolated from mice treated with 8803 and αCTLA-4/αPD-1 relative to checkpoint treated or untreated mice, but only trending differences between mice receiving 8803 and αCTLA-4/αPD-1 versus those receiving concomitant Gem/nP chemotherapy (online supplemental figure 7A, B). Of note, antibody depletion of lymphocyte subsets suggests a critical dependence on CD8 T cells for therapeutic benefit with lesser contributions of CD4 T cells and NK cells (online supplemental figure 7C).
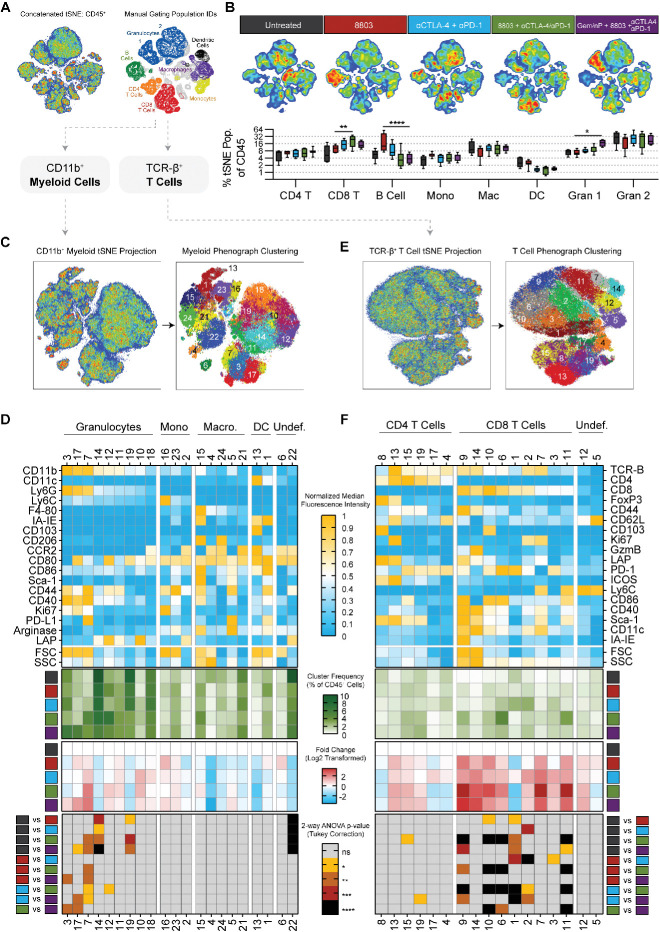
To visualize global changes in these data, we performed t stochastic neighbor embedding (tSNE) analysis of 5×104 CD45+ infiltrating immune cells concatenated from each sample (figure 7A). Through retrospective marker visualization and manual gating validation, we identified metaclusters in the master tSNE plot representing known immune cell populations including CD8 and CD4 T cell subsets, B cells, monocytes, macrophages, granulocytes, and DCs (figure 7A and online supplemental figure 7D). We report the combination of intratumoral 8803 with systemic αCTLA-4/αPD-1 supports the greatest expansion of tumor-infiltrating CD8 T cells, threefold and twofold as a fraction of immune infiltrate relative to untreated and 8803 tumors, respectively (figure 7B). Unexpectedly, intratumoral delivery of 8803 elicits a roughly fourfold expansion of B220+MHC-II+CD11b−CD11c− B cells in the tumor. We find this is unique to 8803 monotherapy, as relative B cell frequencies remain unchanged when 8803 is delivered in combination with systemic checkpoint blockade (figure 7B). Furthermore, Gem/nP induces a significant expansion of a CD11b+Ly6G+ population that clusters distinctly from other granulocytes in tSNE space (figure 7A, B). These data together reveal unique effects of each therapeutic component on the PDAC TME and suggest the combination of in situ 8803 and systemic αCTLA-4/αPD-1 is most effective at expanding the CD8 T cell compartment in aggressive orthotopic mT4-LA tumors.
Figure 7.
Dimensionality reduction of high-parameter flow cytometry reveals inflammatory remodeling of the PDAC stroma following 8803 and checkpoint blockade. (A) Visual summary of tSNE analysis including validation of metaclusters via manual population identification. (B) Visualized deconvolution of tSNE map by treatment group and quantified metacluster frequencies as a percent of infiltrating CD45+ cells in each treatment group. (C) Phenograph clustering of all CD11b+ tumor myeloid populations. (D) Summary of all myeloid cluster data including a phenotyping heat map of each cluster using median fluorescence intensity of each marker normalized to the minimum and maximum expression levels of that marker, average cluster frequencies as a percent of infiltrating CD45+ cells in each treatment group, log2 normalized fold change in reference to untreated group frequencies, and a heat map summary of statistical significance calculations comparing each cluster frequency (% of CD45) between each treatment group. (E) Phenograph clustering of all TCR-β+ tumor T cell populations. (F) Summary of all T cell data as described for D. Data represent 5–10 mice in each treatment group and is representative of two individual experiments. Statistical significance was calculated using a two-way analysis of variance with Tukey’s correction for multiple comparisons. *P<0.05, **p<0.01, ***p<0.001, ****p<0.0001. ns, not significant; PDAC, pancreatic ductal adenocarcinoma; tSNE, t stochastic neighbor embedding.
We next harnessed the high-dimensional nature of these data to more precisely define changes in specific immune subpopulations in response to 8803, checkpoint blockade, and/or Gem/nP. For this, we manually gated and concatenated T cell (CD45+TCR-B+) and myeloid cell (CD45+TCR-B−CD11b+) compartments from each sample, computed tSNE projections of both compartments in parallel, and used the Phenograph algorithm to define cellular subpopulations in an unsupervised fashion (figure 7C, E).
In the myeloid stroma, we find substantial changes occurring across the granulocyte compartment, as well as in a single putative macrophage cluster. We observe two granulocyte metaclusters that are largely divergent in expression of LAP/TGF-β, CD40, and Ki67. The smaller metacluster (composed of clusters 3, 7, and 17) encompasses putative proinflammatory neutrophils, owing to low LAP/ TGF-β expression, higher proliferative capacity, and increased expression of CD40. These clusters are each expanded in mice receiving Gem/nP, and cluster 7 is also significantly expanded relative to controls in mice receiving 8803 and checkpoint blockade. Cluster 7 differs from clusters 3 and 17 by its increased expression of PD-L1, possibly due to increased inflammation in the context of combination therapy. In contrast, clusters 19 and 14 represent an ‘N2-like’ or PMN-MDSC phenotype characterized by LAP/ TGF-β expression and reduced CD40 and constitute roughly 20% of the immune infiltrate at baseline. These cells are reduced to 10% by 8803 C/P and Gem/nP-8803-C/P. Similarly, a single macrophage population (cluster 22) exhibiting LAP/ TGF-β exhibits a trending reduction in treated animals relative to baseline, suggestive of possible in situ polarization. Taken in the context of our in vitro studies, these data support that STING targeting in vivo can remodel the PDAC myeloid compartment and that effects of 8803 can be further augmented when combined with checkpoint blockade and/or chemotherapy.
Next, focusing on the T cell compartment, we observe few changes in CD4 T cells but substantial effects on the frequencies and phenotypes of tumor infiltrating CD8 T cells by our combination approach. After multiple testing correction, no statistically significant changes are observed in the frequencies of two Treg clusters 8 and 13, while effector CD4 T cell cluster 15 is moderately expanded in mice treated with the combination of 8803 C/P. In contrast, of nine distinct CD8 T cell clusters identified by Phenograph, six exhibit statistically significant changes in frequency relative to baseline in multiple treatment groups. The majority of CD8 clusters increase in frequency, with four of five clusters most highly expanded in mice receiving 8803 and αCTLA-4/αPD-1. These are: (1) cluster 9: a highly activated blasting CD8 T cell with production of granzyme B, high expression of CD44, PD-1, and non-canonical activation markers CD86, CD11c, and MHC-II; (2) cluster 10: representing a CD103+ tissue resident CD8 T cell; (3) cluster 6: defined by high expression of PD-1 and CD86; and (4) cluster 11: defined by low PD-1 expression and increased levels of Ly6C, which can indicate central memory-like CD8 T cells. A similar phenotype was observed in CD8s from 8803-treated subcutaneous PDAC tumors earlier in this study (figure 1).31 These data demonstrate the capacity for 8803-mediated in situ vaccination to cooperate with checkpoint blockade to mobilize a phenotypically diverse, numerically rich effector CD8 T cell response in a highly aggressive, poorly antigenic orthotopic model of pancreatic cancer. This enhanced CD8 response is accompanied by—and likely related to—a proinflammatory remodeling of the suppressive PDAC myeloid stroma. In this fashion, potent STING agonists have utility in rendering PDAC sensitive to checkpoint blockade immunotherapy.
Discussion
CDN agonists of the STING pathway are progressing through the clinical sphere for use as in situ vaccine agents across many cancers; however, the molecular mechanisms underlying their well-documented therapeutic potential are not fully understood. Here, we sought to comprehensively describe how STING agonists of distinct potencies functionally repolarize immunosuppressive myeloid lineages, as these cells play a critical role in establishment of tumor immune privilege. We integrated orthogonal transcriptional, translational, and functional datasets to map the myeloid response to CDNs at a heretofore unprecedented depth and in so doing identified key signaling programs engaged during inflammatory polarization of murine MDSCs and human M2 macrophages. Our analysis of these data has generated at least four novel insights that merit further investigation.
First, we report a novel association between synthetic CDN stimulation and inhibition of Myc signaling, both in human M2 macrophages and murine MDSC. In macrophages, cMyc is induced by M2-polarizing factors including IL-4, IL-13, IL-10, and TGF-β, can be highly expressed in TAM and regulates transcription of genes associated with alternative macrophage activation including CD206, PPARγ, STAT6, TGF-β, VEGFα, and Hif1α.26 Additionally, interactome mapping of RNAseq data from murine tumor infiltrating MDSC implicates cMyc in the control of MDSC cell cycle dynamics.23 Our data suggest that STING agonist driven proinflammatory repolarization of myeloid stroma is, at least in part, linked to inhibition of cMyc. Beyond this novel association, we demonstrate that STING agonists are capable of inhibiting tumor cell-intrinsic cMyc activity and that may contribute to the therapeutic effect of intratumorally delivered CDNs in a manner that could be more effectively exploited through precision medicine approaches. That −Myc is amplified in 28% of all TCGA samples,32 yet is infamously ‘undruggable’,33 warrants further investigation into the potential for CDNs to modulate tumor cMyc activity clinically. Future work will be required to further understand the contributions of cMyc suppression versus more direct immune activation to synthetic STING agonist potency against cancers sensitive to both modalities.
Second, we report a novel association between CDN stimulation and metabolic reprogramming within suppressive myeloid cells. Interestingly, transcriptional signatures for metabolic pathways enriched in murine BM-MDSC by each CDN were distinct from those observed in human M2 macrophages. This was clearly observed in the positive versus negative enrichment of an OXPHOS gene signature in MDSC and macrophages, respectively. It is likely that this incongruence is a cell type-specific phenomenon, as OXPHOS is a key bioenergetic pathway for alternatively activated macrophages,34 35 while MDSCs are thought to rely primarily on FAO.36 While MDSC appear to turn to OXPHOS in response to synthetic CDNs, macrophages transcriptionally downregulate multiple metabolic pathways: OXPHOS, glycolysis, FAO, and mTORC1 signaling. A recent study demonstrated that cMyc activation can promote FAO, an observation that may link the cMyc and FAO downregulation we observed in these myeloid compartments.37 We hypothesize autophagy becomes a central bioenergetic source for CDN-stimulated macrophages. Evolutionarily, STING is linked to autophagy. The proautophagic activity of STING predates induction of IRF or NFκB signaling.38 Together, these observations open a novel avenue of study to understand how CDN-mediated metabolic changes control myeloid cell function and phenotype in the TME.
Third, we learned that transcriptional, protein, and functional signatures of STING activation in myeloid cells via CDNs of ascending potency are not linearly correlated with known in vitro potency. Specifically, the cGAMP activation pattern diverged strongly from an intermediate state between weaker CDG and stronger ML-RR or 8803. This divergence was observed at key nodes: in enrichment in Myc and E2F target genes, in protein levels of STING in BM-MDSC, in cellular morphology of macrophages poststimulation, and in the induced frequencies of granulocytes and monocytes post-intra-tumoral injection into mT4 tumors. We do not currently understand the mechanistic basis for this behavior, thus further investigation is warranted, as an ability to fine-tune the functional effects of STING activation by modifying CDN structure could be theoretically leveraged to optimize therapeutic benefit in patients.
In pancreatic cancer, deficient antitumor immunity is a result of both numerically deficient CD8 T cell responses and a dense immunosuppressive myeloid stroma. We hypothesized that local delivery of STING agonists could help address both deficiencies. Other groups have attempted to target STING in PDAC for this reason, and we believe our results largely complement and build on these previous observations. Baird et al 14 demonstrated intratumoral RR-CDG synergizes with tumor-targeted radiation therapy to regress PancO2 tumors and observed CDN exposure modulating expression of select M1/M2 cytokines in ex vivo cultured macrophages. Smith et al 39 used a biopolymer to deliver CDG together with CAR T cells and αCD3/αCD28/αCD137 beads locally onto orthotopically implanted KPC tumors, demonstrating the requirement for sustained intratumoral dosing of CDNs for optimal effect in visceral PDAC lesions. Most recently, Jing et al 40 studied the effects of systemically administered DMXAA on orthotopically implanted KPC tumor growth and found this non-CDN STING agonist to be therapeutically effective, inducing proinflammatory activation of the PDAC stroma, including macrophages. The multiplex cytokine analyses we report here are largely concordant with those of this prior study; however, our expanded panel and comprehensive multiomic analyses more deeply define the precise phenotype of pro-inflammatory myeloid repolarization resulting from CDN-mediated STING activation. Our therapeutic studies also provide a translatable alternative to their systemic delivery approach, demonstrating synthetic CDNs can be locally administered to visceral PDAC lesions with beneficial effect in the context of checkpoint blockade immunotherapy.
In total, our studies enhance our understanding of mechanisms underlying the therapeutic activity of CDNs. We present a comparative analysis of transcriptional, translational, and functional consequences of CDN stimulation on two critical immunosuppressive myeloid populations: MDSCs and M2 macrophages. These data reveal novel associations between CDN stimulation and both Myc signaling and cellular metabolism and identify numerous signaling components previously unknown to play a role in STING responses. Additionally, we report that synthetic CDNs can engage qualitatively different signaling pathways to those engaged by natural CDNs in these cells, which cannot be explained by mere differences in functional potency. The clinical potential of recently described non-CDN systemic STING relative to these CDN STING agonists may be informed by future comparative studies of their potential to impact the novel myeloid cMyc, proliferative and metabolic pathways described here. Focusing on a novel, highly potent CDN, we find in situ myeloid repolarization through STING activation likely nucleates CD8 T cell expansion thus promoting tumor control in the context of TG cell checkpoint blockade. These studies elucidate critical pathways controlling the suppressive to proinflammatory myeloid polarization axis and implicate STING as a potential therapeutic target for patients with immune ‘cold’ tumors like PDAC.
Acknowledgments
We would like to thank Dr David Tuveson for sharing the mT4-2D pancreatic cancer cell line and Dr James Allison for providing DNA sequencing of that line.
Footnotes
Twitter: @caseyager
Contributors: Conceptualization: CRA and MAC; methodology: CRA; formal analysis: KR, CC, and CRA; investigation, CRA, AB, CRD, PJ, RBS, BM, RS, and MAD; resources: MEDF and PJ; writing – original draft: CRA; writing – review and editing: MAC and CRA; visualization: CRA; supervision: MAC.
Funding: These studies were supported by a Pancreatic Cancer Action Network Translational Research Grant (18-65-CURR). Partial support was also provided by an MD Anderson Sheikh Ahmed Bin Zayed Al Nahyan Grant and by the Department of Defense Peer Reviewed Cancer Research Program Career Development Award CA140792. CRA was supported by NIH TL1 fellowships (TL1TR000369 and TL1TR000371).
Competing interests: MAC reports personal fees from ImmunoGenesis, Inc, personal fees from Alligator Bioscience, Inc, personal fees from ImmunOS, Inc, grants and personal fees from ImmunoMet, Inc, personal fees from Oncoresponse, Inc, personal fees from Pieris, Inc, personal fees from Nurix, Inc, personal fees from Aptevo, Inc, personal fees from Servier, Inc, personal fees from Kineta, Inc, personal fees from Salarius, Inc, personal fees from Xencor, Inc, personal fees from Agenus, Inc, personal fees from Mereo, outside the submitted work; in addition, MAC has a patent 'Methods and Composition for Localized Secretion of Anti-CTLA-4 Antibodies' licensed to multiple licensees, and a patent 'Dual specificity antibodies which bind both PD-L1 and PD-L2 and prevent their binding to PD-1' licensed to ImmunoGenesis, Inc. MAC, MEDF and PJ have a patent 'Cyclic Dinucleotides as Agonists of Stimulator of Interferon Gene Dependent Signaling' licensed to ImmunoGenesis, Inc.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
All mice were housed in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care and NIH standards. All experiments were conducted according to protocols approved by the University of Texas MD Anderson Cancer Center Institutional Animal Care and Use Committee.
References
- 1. Royal RE, Levy C, Turner K, et al. Phase 2 trial of single agent ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother 2010;33:828–33. 10.1097/CJI.0b013e3181eec14c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brahmer JR, Tykodi SS, Chow LQM, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455–65. 10.1056/NEJMoa1200694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laheru D, Lutz E, Burke J, et al. Allogeneic granulocyte macrophage colony-stimulating factor-secreting tumor immunotherapy alone or in sequence with cyclophosphamide for metastatic pancreatic cancer: a pilot study of safety, feasibility, and immune activation. Clin Cancer Res 2008;14:1455–63. 10.1158/1078-0432.CCR-07-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le DT, Wang-Gillam A, Picozzi V, et al. Safety and survival with GVAX pancreas prime and Listeria Monocytogenes-expressing mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J Clin Oncol 2015;33:1325–33. 10.1200/JCO.2014.57.4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009;324:1457–61. 10.1126/science.1171362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ene-Obong A, Clear AJ, Watt J, et al. Activated pancreatic stellate cells sequester CD8+ T cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology 2013;145:1121–32. 10.1053/j.gastro.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marabelle A, Tselikas L, de Baere T, et al. Intratumoral immunotherapy: using the tumor as the remedy. Ann Oncol 2017;28:xii33–43. 10.1093/annonc/mdx683 [DOI] [PubMed] [Google Scholar]
- 8. Hammerich L, Binder A, Brody JD. In situ vaccination: cancer immunotherapy both personalized and off-the-shelf. Mol Oncol 2015;9:1966–81. 10.1016/j.molonc.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunn GP, Bruce AT, Sheehan KCF, et al. A critical function for type I interferons in cancer immunoediting. Nat Immunol 2005;6:722–9. 10.1038/ni1213 [DOI] [PubMed] [Google Scholar]
- 10. Diamond MS, Kinder M, Matsushita H, et al. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med 2011;208:1989–2003. 10.1084/jem.20101158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woo S-R, Fuertes MB, Corrales L, et al. Sting-Dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 2014;41:830–42. 10.1016/j.immuni.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ahn J, Xia T, Rabasa Capote A, et al. Extrinsic Phagocyte-Dependent sting signaling dictates the immunogenicity of dying cells. Cancer Cell 2018;33:862–73. 10.1016/j.ccell.2018.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corrales L, Glickman LH, McWhirter SM, et al. Direct activation of sting in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep 2015;11:1018–30. 10.1016/j.celrep.2015.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baird JR, Friedman D, Cottam B, et al. Radiotherapy combined with novel STING-Targeting oligonucleotides results in regression of established tumors. Cancer Res 2016;76:50–61. 10.1158/0008-5472.CAN-14-3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ager CR, Reilley MJ, Nicholas C, et al. Intratumoral sting activation with T-cell checkpoint modulation generates systemic antitumor immunity. Cancer Immunol Res 2017;5:676–84. 10.1158/2326-6066.CIR-17-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Francica BJ, Ghasemzadeh A, Desbien AL, et al. Tnfα and radioresistant stromal cells are essential for therapeutic efficacy of cyclic dinucleotide sting agonists in Nonimmunogenic tumors. Cancer Immunol Res 2018;6:422–33. 10.1158/2326-6066.CIR-17-0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sivick KE, Desbien AL, Glickman LH, et al. Magnitude of Therapeutic STING Activation Determines CD8+ T Cell-Mediated Anti-tumor Immunity. Cell Rep 2018;25:3074–85. 10.1016/j.celrep.2018.11.047 [DOI] [PubMed] [Google Scholar]
- 18. Chandra D, Quispe-Tintaya W, Jahangir A, et al. Sting ligand c-di-GMP improves cancer vaccination against metastatic breast cancer. Cancer Immunol Res 2014;2:901–10. 10.1158/2326-6066.CIR-13-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ager CR, Zhang H, Wei Z, et al. Discovery of IACS-8803 and IACS-8779, potent agonists of stimulator of interferon genes (STING) with robust systemic antitumor efficacy. Bioorg Med Chem Lett 2019;29:126640. 10.1016/j.bmcl.2019.126640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boj SF, Hwang C-I, Baker LA, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015;160:324–38. 10.1016/j.cell.2014.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gabrusiewicz K, Rodriguez B, Wei J, et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 2016;1. 10.1172/jci.insight.85841. [Epub ahead of print: 25 02 2016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marigo I, Bosio E, Solito S, et al. Tumor-Induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity 2010;32:790–802. 10.1016/j.immuni.2010.05.010 [DOI] [PubMed] [Google Scholar]
- 23. Aliper AM, Frieden-Korovkina VP, Buzdin A, et al. Interactome analysis of myeloid-derived suppressor cells in murine models of colon and breast cancer. Oncotarget 2014;5:11345–53. 10.18632/oncotarget.2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Georgouli M, Herraiz C, Crosas-Molist E, et al. Regional activation of myosin II in cancer cells drives tumor progression via a secretory cross-talk with the immune microenvironment. Cell 2019;176:757–74. 10.1016/j.cell.2018.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McWhorter FY, Wang T, Nguyen P, et al. Modulation of macrophage phenotype by cell shape. Proc Natl Acad Sci U S A 2013;110:17253–8. 10.1073/pnas.1308887110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pello OM, De Pizzol M, Mirolo M, et al. Role of c-myc in alternative activation of human macrophages and tumor-associated macrophage biology. Blood 2012;119:411–21. 10.1182/blood-2011-02-339911 [DOI] [PubMed] [Google Scholar]
- 27. Pello OM, Chèvre R, Laoui D, et al. In vivo inhibition of c-myc in myeloid cells impairs tumor-associated macrophage maturation and pro-tumoral activities. PLoS One 2012;7:e45399. 10.1371/journal.pone.0045399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cho J-Y, Akbarali Y, Zerbini LF, et al. Isoforms of the Ets transcription factor NERF/ELF-2 physically interact with AML1 and mediate opposing effects on AML1-mediated transcription of the B cell-specific blk gene. J Biol Chem 2004;279:19512–22. 10.1074/jbc.M309074200 [DOI] [PubMed] [Google Scholar]
- 29. Zhu Y, Herndon JM, Sojka DK, et al. Tissue-Resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity 2017;47:597. 10.1016/j.immuni.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y, Velez-Delgado A, Mathew E, et al. Myeloid cells are required for PD-1/PD-L1 checkpoint activation and the establishment of an immunosuppressive environment in pancreatic cancer. Gut 2017;66:124–36. 10.1136/gutjnl-2016-312078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hänninen A, Maksimow M, Alam C, et al. Ly6C supports preferential homing of central memory CD8+ T cells into lymph nodes. Eur J Immunol 2011;41:634–44. 10.1002/eji.201040760 [DOI] [PubMed] [Google Scholar]
- 32. Schaub FX, Dhankani V, Berger AC, et al. Pan-Cancer alterations of the Myc oncogene and its proximal network across the cancer genome atlas. Cell Syst 2018;6:282–300. 10.1016/j.cels.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dang CV, Reddy EP, Shokat KM, et al. Drugging the 'undruggable' cancer targets. Nat Rev Cancer 2017;17:502–8. 10.1038/nrc.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang F, Zhang S, Vuckovic I, et al. Glycolytic stimulation is not a requirement for M2 macrophage differentiation. Cell Metab 2018;28:463–75. 10.1016/j.cmet.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jha AK, Huang SC-C, Sergushichev A, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015;42:419–30. 10.1016/j.immuni.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 36. Hossain F, Al-Khami AA, Wyczechowska D, et al. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunol Res 2015;3:1236–47. 10.1158/2326-6066.CIR-15-0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Casciano JC, Perry C, Cohen-Nowak AJ, et al. Myc regulates fatty acid metabolism through a multigenic program in claudin-low triple negative breast cancer. Br J Cancer 2020;122:868–84. 10.1038/s41416-019-0711-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gui X, Yang H, Li T, et al. Autophagy induction via sting trafficking is a primordial function of the cGAS pathway. Nature 2019;567:262–6. 10.1038/s41586-019-1006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith TT, Moffett HF, Stephan SB, et al. Biopolymers codelivering engineered T cells and sting agonists can eliminate heterogeneous tumors. J Clin Invest 2017;127:2176–91. 10.1172/JCI87624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jing W, McAllister D, Vonderhaar EP, et al. Sting agonist inflames the pancreatic cancer immune microenvironment and reduces tumor burden in mouse models. J Immunother Cancer 2019;7:115. 10.1186/s40425-019-0573-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003246supp001.pdf (5.4MB, pdf)
Data Availability Statement
Data are available in a public, open access repository.