Abstract
Background
Localized stress and cell death in chronic inflammatory diseases may release tissue-specific lipids into the circulation causing the blood plasma lipidome to reflect the type of inflammation. However, deep lipid profiles of major chronic inflammatory diseases have not been compared.
Methods
Plasma lipidomes of patients suffering from two etiologically distinct chronic inflammatory diseases, atherosclerosis-related vascular disease, including cardiovascular (CVD) and ischemic stroke (IS), and systemic lupus erythematosus (SLE), were screened by a top-down shotgun mass spectrometry-based analysis without liquid chromatographic separation and compared to each other and to age-matched controls. Lipid profiling of 596 lipids was performed on a cohort of 427 individuals. Machine learning classifiers based on the plasma lipidomes were used to distinguish the two chronic inflammatory diseases from each other and from the controls.
Findings
Analysis of the lipidomes enabled separation of the studied chronic inflammatory diseases from controls based on independent validation test set classification performance (CVD vs control - Sensitivity: 0.94, Specificity: 0.88; IS vs control - Sensitivity: 1.0, Specificity: 1.0; SLE vs control – Sensitivity: 1, Specificity: 0.93) and from each other (SLE vs CVD ‒ Sensitivity: 0.91, Specificity: 1; IS vs SLE - Sensitivity: 1, Specificity: 0.82). Preliminary linear discriminant analysis plots using all data clearly separated the clinical groups from each other and from the controls, and partially separated CVD severities, as classified into five clinical groups. Dysregulated lipids are partially but not fully counterbalanced by statin treatment.
Interpretation
Dysregulation of the plasma lipidome is characteristic of chronic inflammatory diseases. Lipid profiling accurately identifies the diseases and in the case of CVD also identifies sub-classes.
Funding
Full list of funding sources at the end of the manuscript.
Keywords: Vascular diseases, Systemic lupus erythematosus, Dyslipidemia, Lipid profiling, Lipid biomarker
1. Introduction
Lipids in the plasma reflect diet and metabolic characteristics of an individual [1] but are also known to regulate inflammatory responses both positively and negatively [2], [3], [4]. In the case of inflammatory processes cell stress and death also release internal cellular lipids into the blood. It might, therefore, be expected that chronic inflammatory diseases alter the plasma lipidome in a manner that is characteristic of the chemistry of the cells/tissues primarily involved in the disease. With this reasoning we examined the lipidomes of patients suffering from cardiovascular diseases (CVD), ischemic stroke (IS), and systemic lupus erythematosus (SLE) and compared them with the lipidome of age-matched controls who were not known to suffer, or to have suffered, from any of these diseases.
The role of inflammation and lipids in atherosclerosis is well established [5], [6], [7]. There is a reasonably large amount of literature [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29] on the profiling of lipids in plasma and atherosclerotic plaques in CVD cohorts. The emphasis of previous studies has been on the association of plasma lipid species with CVD-risk stratification and CVD-related mortality [8,10,11,[13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]], improved classification of stable and unstable CVD states [25], correlation with established diagnostic tools for CVD [22,26], genetic risk factors for CVD [10,22,27], association of CVD with co-morbidities [8,18,28,29], and association of CVD with changes in lipid biochemistry in the plasma [30]. In general, the different lipidomic profiling studies differ in the methods and definition of clinical outcome subjected to lipidome-based classification. This means that the clinical outcome and lipids profiled differ across studies. Consequently, consensus targets from multiple studies are still unobtainable. Nevertheless, common to all studies are the highly sensitive changes in lipid profiles depending on the clinical outcome evaluated. Table S1 summarizes individual lipids associated with CVD or CVD risk factors from past studies.
SLE is a chronic inflammatory disease. Its etiology displays multifactorial characteristics and the molecular mechanisms of this disorder are largely unknown [31], [32], [33]. A notable aspect of this disease is the excessive production of reactive oxygen species that oxidize cellular lipids producing derivatives that cause dyslipidemia and dyslipoproteinemia [34,35]. The dyslipoproteinemia signature has been identified in SLE patients with markedly increased age-specific incidence of cardiovascular disease [35]. Lipidomics of the plasma of SLE patients has been previously reported [36,37].
In this work we enquired whether it would be possible to characterize the lipidomic profile of different manifestations of essentially the same clinical disease (different degrees of CVD and IS) and whether the lipidomic profile of two distinct diseases that only had inflammation as a common characteristic could be distinguished from each other and used as a diagnostic identifier. Furthermore, the accuracy as a diagnostic identifier was evaluated on an independent test data set.
Despite the preliminary efforts and promising results based on Liquid Chromatography-Mass Spectrometry (LC-MS) lipid profiling, identifying individuals at risk for stroke and cardiovascular events from a healthy control population remains a challenge. Time invested per sample constitutes a barrier for large scale clinical validation and implementation of lipid profiles (e.g. the Liquid Chromatography (LC) dimension of previous studies is time intensive (see [38] and references therein)). In this study we evaluated classification performance of lipid profiling based on shotgun Mass Spectrometry (MS) without LC lipid separation prior to MS. A total of 596 lipids were profiled in a total of 427 individuals.
2. Methods
2.1. Patient samples
Plasma samples were obtained from a total of 427 individuals. Baseline characteristics are outlined in Table 1. Control (n = 85) were taken from the population of the Coimbra and Lisbon, Portugal, regions. They were healthy volunteers who satisfied the criterion that they had never had any CVD- or SLE-related health complaints. The CVD patients (n = 217) were divided into 6 groups. CVD1 (n = 67) contains individuals who went to the hospital with chest pain but had no indicators for stable angina pectoris, unstable angina pectoris or myocardial infarction. CVD2 (n = 76) are patients with stable angina pectoris (SAP). CVD1 and CVD2 are defined according to the ACCF/AHA/ACP/AATS/PCNA/SCAI/STS guidelines [39]. CVD3 (n = 14) contains patients with unstable angina pectoris, CVD4 (n = 39) are patients who suffered an acute myocardial infarction with no ST-elevation in ECG, and CVD5 (n = 21) are patients who suffered acute myocardial infarction with ST-elevation in ECG [40,41]. CVD3, CVD4, and CVD5, together, may be classified as patients with an acute coronary syndrome (ACS). CVD1 through CVD5 groups were all obtained from Hospital Santa Cruz, Carnaxide, Portugal. Acute ischemic stroke (IS) (n = 21) were patients admitted at the emergency room of the Centro Hospitalar de Lisboa Ocidental, Lisbon, Portugal, who suffered from acute ischemic stroke. The SLE cohort (n = 104) were patients from Hospital Dr. Fernando Fonseca, Amadora, Portugal. The inclusion criteria were all patients diagnosed with the pathology and above 18 years old. The exclusion criteria were the existence of serious renal and hepatic pathologies, cancer or existence of infectious diseases. All the lupus patients had active disease but were all stable on medication. None was flaring at the time of the study (no A or B in BILAG) [42] nor were any patients in clinical remission (D and E only in BILAG). The study design is summarized in Fig. 1.
Table 1.
Baseline characteristics of the analyzed lipid cohort. NA indicates Not Available.
| Control (N=85) | CVD1 (N=67) | CVD2 (N=76) | CVD3 (N=14) | CVD4 (N=39) | CVD5 (N=21) | IS (N=21) | SLE (N=104) | Total (N=427) | |
|---|---|---|---|---|---|---|---|---|---|
| Sex | |||||||||
| Missing | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| F | 52 (61%) | 36 (54%) | 22 (29%) | 3 (21%) | 11 (28%) | 3 (14%) | 14 (67%) | 95 (91%) | 236 (55.2%) |
| M | 33 (39%) | 30 (45%) | 54 (71%) | 11 (79%) | 28 (72%) | 18 (86%) | 7 (33%) | 9 (9%) | 190 (44.5%) |
| Age (Years) | |||||||||
| Missing | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 9 | 10 |
| Mean (SD) | 45 (17) | 69 (12) | 67 (12) | 67 (10) | 67 (14) | 63 (12) | 74 (10) | 47 (16) | 59 (17) |
| Range | 22 - 82 | 36 – 90 | 33-87 | 50-79 | 36-92 | 37-87 | 49-94 | 20-82 | 20-94 |
| Weight (kg) | |||||||||
| Mean (SD) | 68 (14) | 73 (14) | 77 (13) | 80 (14) | 75 (14) | 80 (12) | NA | NA | 74 (14) |
| valid (missing) | 83 (2) | 66 (1) | 73 (3) | 13 (1) | 37 (2) | 21 (0) | 0 (22) | NA | 293 (31) |
| Height (cm) | |||||||||
| Mean (SD) | 167 (9) | 164 (9) | 167 (9) | 170 (7) | 165 (9) | 171 (9) | NA | NA | 166 (8.8) |
| valid (missing) | 80 (5) | 66 (1) | 73 (3) | 13 (1) | 37 (2) | 21 (0) | 0 (22) | NA | 290 (34) |
| BMI (kg/m²) | 104 | ||||||||
| Mean (SD) | 25 (4) | 27 (4) | 27 (4) | 28 (4) | 28 (5) | 28 (4) | NA | NA | 26.7 (4.3) |
| valid (missing) | 80 (5) | 66 (1) | 73 (3) | 13 (1) | 37 (2) | 21 (0) | 0 (22) | NA | 290 (34) |
| P value vs control | 0.03 | 0.004 | 0.04 | 0.02 | 0.02 | NA | NA | ||
| Hypertension | |||||||||
| No | 70 (82%) | 16 (24%) | 11 (14%) | 2 (14%) | 7 (18%) | 6 (29%) | 4 (19%) | 13 (12%) | 129 |
| Yes | 15 (18%) | 51 (76%) | 64 (84%) | 12 (86%) | 30 (77%) | 15 (71%) | 17 (81) | 0 (0%) | 204 |
| Missing | 0 (0%) | 0 (0%) | 1 (1%) | 0 (0%) | 2 (5%) | 0 (0%) | 0 (0%) | 91 (88%) | 94 |
| Dyslipidemia | |||||||||
| No | 85 (100%) | 24 (36%) | 17 (22%) | 3 (21%) | 16 (41%) | 14 (67%) | 10 (48%) | 13 (12%) | 182 |
| Yes | 0 (0%) | 43 (64%) | 58 (76%) | 11 (79%) | 21 (54%) | 7 (33%) | 11 (52%) | 0 (0%) | 151 |
| Missing | 0 (0%) | 0 (0%) | 1 (1%) | 0 (0%) | 2 (5%) | 0 (0%) | 0 (0%) | 91 (88%) | 94 |
| Statin Use | |||||||||
| No | 79 (93%) | 27 (40%) | 18 (24%) | 2 (14%) | 15 (38%) | 13 (62%) | 15 (71%) | 75 (72%) | 273 |
| Yes | 6 (7.1%) | 37 (55%) | 49 (64%) | 11 (79%) | 18 (46%) | 5 (24%) | 6 (29%) | 24 (23%) | 132 |
| Missing | 0 (0%) | 3 (5%) | 9 (12%) | 1 (7%) | 6 (15%) | 3 (14%) | 0 (0%) | 5 (4.8%) | 22 |
| Anti-Hypertensives | |||||||||
| No | 70 (82%) | 33 (49%) | 14 (18%) | 1 (7%) | 0 (0%) | 0 (0%) | 8 (38%) | 104 (100%) | 230 |
| Yes | 15 (18%) | 31 (46%) | 42 (55%) | 2 (14%) | 2 (5%) | 0 (0%) | 13 (62%) | 105 | |
| Missing | 0 (0%) | 3 (5%) | 20 (26%) | 11 (79%) | 37 (95%) | 21 (100%) | 0 (0%) | 92 | |
| Anti-coagulant | |||||||||
| No | 85 (100%) | 40 (60%) | 10 (13%) | 0 (0%) | 1 (3%) | 0 (0%) | 14 (67%) | 104 (100%) | |
| Yes | 0 (0%) | 23 (34%) | 47 (62%) | 3 (21%) | 1 (3%) | 0 (0%) | 7 (33%) | ||
| Missing | 0 (0%) | 4 (6%) | 19 (25%) | 11 (79%) | 37 (95%) | 21 (100%) | 0 (0%) | ||
| Immuno-supressors | |||||||||
| No | 85 (100%) | 67 (100%) | 76 (100%) | 14 (100%) | 39 (100%) | 21 (100%) | 21 (100%) | 42 (40%) | 365 |
| Yes | 56 (54%) | 56 | |||||||
| Missing | 6 (6%) | 6 | |||||||
| Anti-diabetic | |||||||||
| No | 84 (99%) | 52 (78%) | 46 (60%) | 9 (64%) | 27 (69%) | 18 (86%) | 15 (71%) | 98 (94%) | 349 |
| Yes | 1 (1 %) | 14 (21%) | 30 (39%) | 4 (29%) | 12 (31%) | 3 (14%) | 6 (29%) | 1 (1%) | 71 |
| Missing | 1 (1%) | 0 (0%) | 1 (7%) | 0.00% | 0 (0%) | 5 (4.8%) | 7 | ||
| Steroids | |||||||||
| No | 85 (100%) | 67 (100%) | 76 (100%) | 14 (100%) | 39 (100%) | 21 (100%) | 21 (100%) | 30 (29%) | 353 |
| Yes | 69 (66%) | 69 | |||||||
| Missing | 5 (5%) | 5 |
Fig. 1.
Scheme of the study design. Eighty-five healthy control subjects, two-hundred seventeen cardiovascular disease (CVD) subjects (including acute myocardial infarction with and without ST-elevation in ECG and stable and unstable angina pectoris), twenty one acute ischemic stroke (IS) subjects and one hundred four systemic lupus erythematosus (SLE) patients were included in this study. Individual data [age, height, weight, body mass index (BMI), clinical history and medication] were collected from all cohorts at baseline. Blood samples from all subjects were obtained and processed for plasma isolation. Then, plasma lipid extraction was performed for mass spectrometry lipidomics (shotgun lipidomics) to obtain the lipid profile of all individuals involved in this study. The obtained data were analysed by PCA, LDA and PLS models were tested for classification performance on independent data set. The R package limma was used for statistical analysis of differential regulated lipids.
2.2. Ethics
A written informed-consent was obtained from all the participants in this study or from their legal representatives. The study was approved by the Ethical Review Board of the Faculty of Medicine of the New University of Lisbon (n°06/2015/CEFCM) and the Ethics Committee for Health of the Centro Hospitalar de Lisboa Ocidental, that includes the Hospital Santa Cruz, the Hospital Egas Moniz and Hospital São Francisco Xavier, and the Ethics Committee for Health of the Hospital Fernando Fonseca. All experiments were performed in accordance with the guidelines and regulations including, the Universal Declaration on Bioethics and Human Rights of UNESCO, 2005; The Charter of Fundamental rights of the EU, 2012; Ethical principles for medical research involving human subjects - Declaration of Helsinki, 2013; EU Regulation 2016/679 and Good Clinical Practice guidelines (Directive 2001/20/EC) and EU Clinical Trials Directive (2005/28/EC). Moreover, they complied with national legislations for the scientific use of human biological samples (Law N° 12/2005 and N° 131/2014).
2.3. Blood samples collection and processing
Blood samples were drawn into tubes containing an anti-coagulant (heparin or EDTA) immediately after admission into the hospital and signing of the informed consent. The samples were kept at 4 °C and processed within 24 h from collection. Plasma was obtained by centrifugation of the blood at 500 g for 10 min at 4 °C, frozen at −80 °C and stored at this temperature until they were used for the lipidomic analysis.
2.4. Definition of age matched controls
The total control cohort consisted of individuals with ages between 22 and 82 years. The whole control cohort (n = 85) was used for comparison with SLE patients.
We applied the Kolmogorov–Smirnov test in R statistical programming language to optimize the similarity of the age distribution in the control group with each of the disease groups [43,44]. Linear scanning of age thresholds from low to high identifies a age threshold with a minimum –log10(p value). In the neighborhood of the minimum, we have insufficient evidence to claim that the two distributions are different. The scanning was performed with steps of one year (Fig. S1). The minimum is where the two distributions should have the highest similarity. However, we are also interested in optimizing the number of controls to obtain better statistical sampling. We therefore apply the threshold that gives the largest number of controls and with a two-sided Kolmogorov–Smirnov test statistic below 0.01 for CVD (Fig. S1a) and IS (Fig. S1b). This resulted in the age threshold ≥50 years for CVD and ≥55 for IS. The minimum –log10 P value for SLE was obtained when using all controls (Fig. S1c). These age matched control groups were used together with data from diseased groups for constructing the partial least square (PLS) models and for the principal component analysis (PCA) analysis to ensure that the observed separation was not age related. For linear discriminant analysis (LDA) and linear regression analysis all controls were used. For linear regression the models were adjusted for age, gender and statin use. Statin use was defined based on clinical records.
2.5. Lipid extraction for mass spectrometry lipidomics
Mass spectrometry-based lipid analysis was performed at Lipotype GmbH (Dresden, Germany) as previously described [38]. Briefly, plasma was diluted 1:50 v/v in water (LC-MS grade, Sigma) and 50 µL of this diluted plasma (equivalent to 1 μL of undiluted plasma) was mixed with 130 μL of 150 mM ammonium bicarbonate solution and 810 μL of methyl tert-butyl ether/methanol (7:2, v/v) was added. 21 µL of an internal standard mixture was pre-mixed with the mixture of organic solvents. As internal standards cholesterol (Chol) D6, cholesteryl ester (CE) 20:0, ceramide (Cer) 18:1;2/17:0, diacylglycerol (DAG) 17:0/17:0, phosphatidylcholine (PC) 17:0/17:0, phosphatidylethanolamine (PE) 17:0/17:0, lysophosphatidylcholine (LPC) 12:0, lysophosphatidylethanolamine (LPE) 17:1, triacylglycerol (TAG) 17:0/17:0/17:0, and sphingomyelin (SM) 18:1;2/12:0 were used. The plate was then sealed with a teflon-coated lid, shaken at 4 °C for 15 min, and spun down (3000 g, 5 min) to facilitate separation of the liquid phases. One hundred microliters of the organic phase was transferred to an infusion plate and dried in a speed vacuum concentrator. Dried lipids were re-suspended in 40 μL of 7.5 mM ammonium acetate in chloroform/methanol/propanol (1:2:4, v/v/v) and the wells were sealed with an aluminum foil to avoid evaporation and contamination during infusion. All liquid handling steps were performed using a Hamilton STARlet robotic platform with the Anti Droplet-Control feature for pipetting of organic solvents.
2.6. MS data acquisition
Details of MS data acquisition have been described by Surma et al [38]. Samples were analyzed by direct infusion in a QExactive mass spectrometer (Thermo Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences). 5 µL were infused with gas pressure and voltage set to 1.25 psi and 0.95 kV, respectively. Samples were analyzed in both positive and negative ion modes with a resolution of Rm/z=200 = 280000 for MS and Rm/z=200 = 17,500 for Tandem mass spectrometry (MSMS) experiments, in a single acquisition. MSMS was triggered by an inclusion list encompassing corresponding MS mass ranges scanned in 1 Da increments. Both MS and MSMS data were combined to monitor CE, DAG and TAG ions as ammonium adducts; PC, ether phosphatidylcholine (PC O-), as acetate adducts; and PE, ether phosphatidylethanolamine (PE O-) and phosphatidylinositol (PI) as deprotonated anions. MS only was used to monitor LPE as deprotonated anion; Cer, SM and LPC as acetate adducts and cholesterol as an ammonium adduct.
2.7. Lipid nomenclature
The following annotations were used: Lipid class-<sum of carbon atoms>:<sum of double bonds>;<sum of hydroxyl groups>, i.e. SM-34:1;2 means an SM lipid with 34 carbon atoms, 1 double bond and 2 hydroxyl groups in the ceramide backbone. Lipid molecular subspecies annotation [45] contains additional information on the exact identity of their fatty acids. For example PC 18:1;0_16:0;0 denotes a phosphatidylcholine with one acyl chain having 18 carbon atoms, 1 double bond, 0 hydroxylation, and a second acyl chain with 16 carbon atoms, 0 double bonds, 0 hydroxylation. The exact position of the fatty acids in relation to the glycerol backbone (sn-1 or sn-2) cannot be discriminated. CE 18:1;0 denotes a cholesteryl ester with an 18:1;0 fatty acid. Lipid identifiers of the SwissLipids database [45] and LIPID MAPS Shorthand Notation [46] are provided in the supplemental dataset (Table S2).
2.8. Post-processing
Data were analyzed with in-house developed lipid identification software based on LipidXplorer [47,48]. All information concerning the characteristic fragments used to identify lipid classes is shown in Table S3. The mass errors for precursor ion was 1ppm, for fragment ions around 5 ppm. Data post-processing and normalization were performed using an in-house developed data management system. This management system is programed in PHP, C++ and Javasript and contains in-house developed deconvolution and normalization algorithms. Only lipid identifications with a signal-to-noise ratio >5, and a signal intensity 5-fold higher than in corresponding blank samples were considered for further data analysis. Using 8 reference samples per 96-well plate batch, lipid amounts were corrected for batch variations. Amounts were also corrected for analytical drift, if the p-value of the slope was below 0.05 with an R2 greater than 0.6 and the relative drift was above 5%. The full data set contained quantitative information from 623 lipids. Lipids with a concentration less than 0.5 μM were considered “not analyzable” (NA) and, for the purposes of the present work were considered to be at half of the minimum detectable value and zero variance lipids were filtered out providing a data set of 596 lipids which served as the input to subsequent multivariate analyses.
Statistical analysis was performed in R statistical programming language. The R function seed was applied to enable producible random number generations before each analysis. Shapiro–Wilk normality test was applied to decide whether a Student's t-test or Wilcoxon test should be applied.
Principal component analysis was performed by single value decomposition in R statistical programming language. All the lipid expression values were quantile normalized with the “normalize quantiles” function from the library preprocessCore. The quantitative values were added one, log2 transformed and subtracted by row means. The base R function “svd” were used to calculate principal components. The two main components were plotted in a scatter plot using R.
Significant regulated lipids were defined by the R package “limma version 3.42.0” [49]. The data expression values obtained by MS were added one and log2 transformed. Next the quantitative values were normalized across samples using robust quantile normalization [50]. Both the raw and the quantile normalized values were analyzed by the R package “limma version 3.42.0” to determine significant regulated lipids between control and patient cohorts. Linear regression models included terms to correct for gender and statin treatment and only age matched controls were included for statistical regulation analysis involving IS and CVD cases. All controls were included for comparison with the SLE group. The SLE group was sub divided into two groups according to the Systemic Lupus International Collaborating Clinics (SLICC) group criteria (SLICC ≥ 4 versus SLICC < 4) [51]. Statistical differential regulated lipids were determined using the R package “limma version 3.42.0” and p values were adjusted for multiple testing by the method of Benjamini and Hochberg [52]. Principal component analysis was performed using R base functions on centered quantile normalized quantification values. Overlap between significant regulated lipids was performed with the R package “VennDiagram”.
2.9. Supervised classification
LDA was performed using the R package “caret version 6.0.84” [53] as interface to “MASS version 7.3.51.4”. LDA was performed using either all 596 zero variance filtered lipids or 206 lipids selected with more than 50% non-NAs per clinical group. Additionally, the parameters for training and testing of LDA were set to remove lipids with total intensity across all samples below 30. The value 30 was obtained by stepwise optimization to establish best possible separations. The correlation cut off was set to 1 which means that correlated variables were not eliminated (caret parameter pair-wise absolute correlation cutoff). Setting the correlation cut off slightly lower, e.g. 0.9-1 had no effect on the separation in the LDA plots. The results from the two approaches resulted in similar classification performance and for simplicity only the results from the 596 zero variance filtered lipids is presented.
PLS analysis was performed using the R package “caret version 6.0.84” [53] as interface to “pls version 2.7.1“. The full data set 427 individuals were split into disease groups with age matched controls as described in section “Definition of age matched controls”. Training and test sets were prepared by first selecting the appropriate controls for the classification task involving CVD, IS and SLE. For each of these pairwise comparisons the smallest group sizes were determined. The size of the training data was two times 75% of the smallest group size. The function “strata” from the library “sampling” were used to ensure a random and balanced training set containing an equal number of cases of controls and diseased. For CVD, 26 controls with age ≥50 years and 26 CVD cases were selected as a training set. The remaining data set were used as a validation set (8 controls and 191 CVD). This analysis was also performed using the full control cohort or an age threshold above 37 years to have more controls in the validation set. These additional analyses are not presented here but the classification performance was similar. The training set consisted of 16 controls ≥55 and 16 IS cases for the classification of IS and control. The validation set for IS versus control contained 9 controls and 5 IS cases. For classification of SLE versus control, 64 controls and 64 SLE cases were used as training data. The remaining 21 controls and 40 SLE cases were used as validation set. The data were preprocessed before PLS model training. The “caret” function “nearZeroVar” identified features with near zero variance which were filtered out. Correlation between features were calculated with base R function “cor” and features with a correlation above 75% identified with the “caret” function “findCorrelation” were filtered out.
The caret preprocessing parameters specified were: "center – subtracts the mean from predictor values”, and "scale – divide predictor values with the standard deviation”. The model was optimized by 10-fold cross validation repeated 10 times using accuracy as the metric for optimization. ROC curves and area under the curve were estimated using the R package “pROC” [54] for model performance evaluation on the left out validation set.
2.10. Funding
Funding sources had no role in study design, data collection, data analysis, interpretation, decision to publish or preparation of the manuscript.
3. Results
Baseline characteristics: Table 1 summarizes the baseline characteristics. The CVD cohort analyzed in this study consisted of 217 participants with mean age of 67 years (range 33–92 years). A total of 21 IS patients were included with a similar age range as the CVD patients. 104 SLE patients were also included in this study. Statistical comparisons were in all cases performed with age-matched controls.
Lipidomic analysis: Based on a single shot MS analysis of 1 µL of plasma, 623 lipids spanning 15 lipid classes were identified and quantified. The analysis presented in the current study used only 596 of these lipids and demonstrated, as discussed below, that accurate classification of SLE, IS, CVD and CVD sub-groups from controls was achievable in spite of the fact that there were several shared dysregulated lipids among the different cohorts.
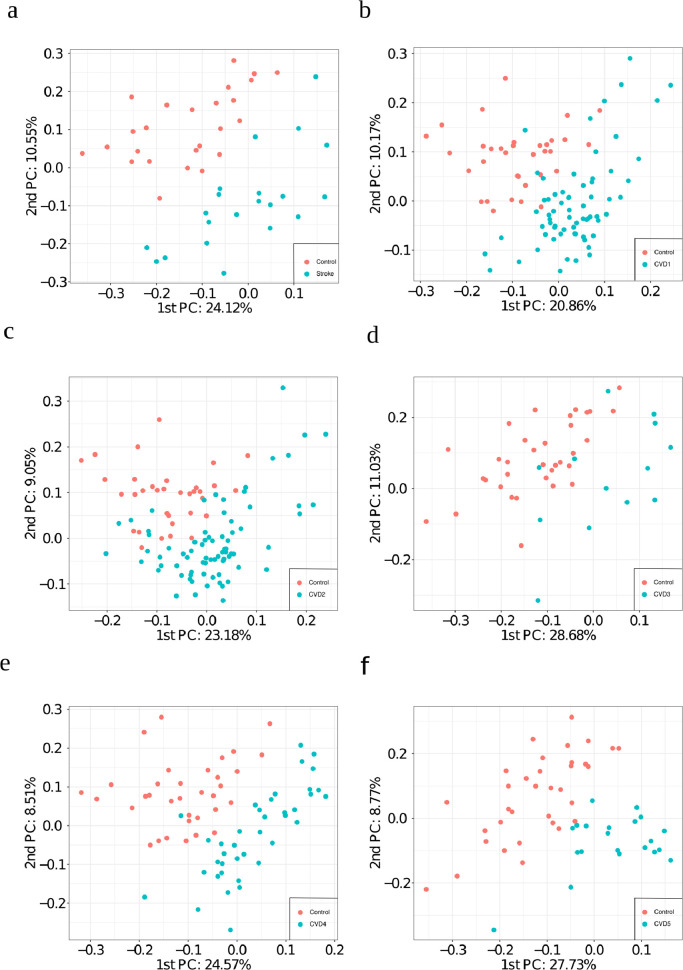
Unsupervised analysis: PCA based on all 596 selected lipids provided a reasonable separation between aged-matched controls versus IS, CVD1, CVD2, CVD3, CVD4 and CVD5 (Fig. 2a–f). CVD4 and CVD5 are almost fully separable from controls based on the first two principal components. CVD3 versus control resulted in the poorest separation by the first two principal components. Depending on the condition either PC1 or PC2 provided the most separation. This strongly suggests that the largest or second largest variance component in lipid abundance correlate with these disease conditions. Notably, this PCA analysis was performed using quantitative values from all lipids without any biased pre-selection of lipids known to associate to CVD and IS.
Fig. 2.
Principal component analysis using all 596 selected lipids as input. Plot of the first and second principal component for a) IS vs control (n=46), b) CVD1 vs control_50 (n=101), c) CVD2 vs control_50 (n=110), d) CVD3 vs control_50 (n=48), e) CVD4 vs control_50 (n=73), and f) CVD5 vs control_50 (n=55).
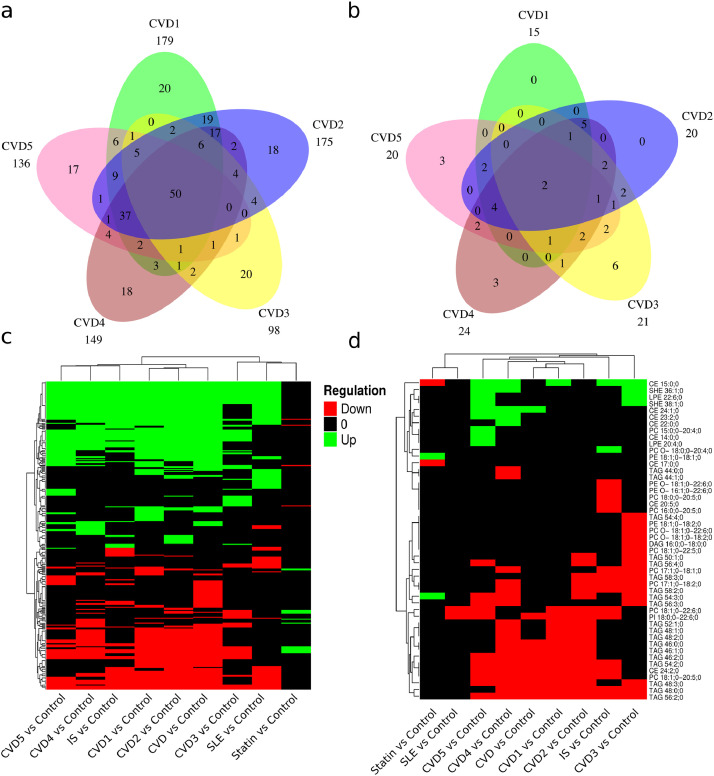
Association of lipids with clinical diagnosis. Comparison of SLE sub-groups SLE_SLICC criteria ≥ 4 vs < 4 resulted in no significant lipids after correction of multiple testing (Table S4). Pairwise comparison between age matched controls and the patient groups adjusted for gender and statin use revealed the following number of significant dysregulated lipids after correction of multiple testing (First value: padjusted <0.05 / Second value: padjusted <0.05 and at least two-fold regulated, Table S4): 179/15 CVD1, 175/20 CVD2, 98/21 CVD3, 149/24 CVD4, 136/20 CVD5, 128/18 IS and 113/2 SLE. Among controls the number of significant regulated lipids after correction of multiple testing (padjusted <0.05/padjusted <0.05 and at least two-fold regulated) were 19/4 when comparing controls with and without statin treatment (Table S4). Only about 2–20% of the significant (dys)regulated lipids displayed an effect size bigger or equal to two-fold, prompting the question if the lipids with small effect size are diagnostically relevant.
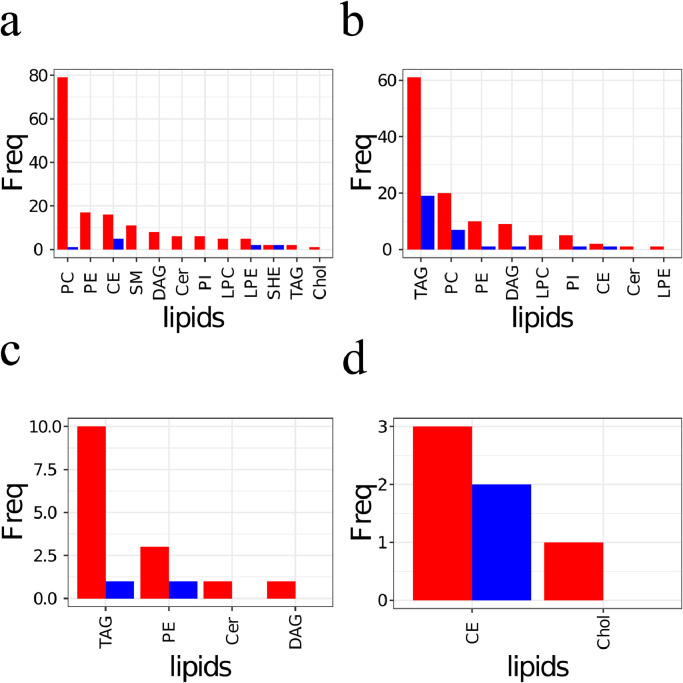
Therefore, Venn diagrams were used to compare the overlap of significant dysregulated lipids (Fig. 3a) and significant dysregulated lipids with an effect size more than two-fold (Fig. 3b). We observed that a considerable number of significant dysregulated lipids with effect size less than two-fold were shared across pathological groups (Fig. 3ab). Heatmaps were constructed to address the direction of the shared dysregulated lipids (Fig. 3cd). These heatmaps only depict lipids that are regulated in more than one pairwise comparison. The direction of dysregulation of lipids displays large similarities for the CVD, IS and SLE groups. In contrast, lipids regulated between controls with and without statin treatment displayed a reverse pattern of regulation compared to the pathological groups (Fig. 3cd). Next, the frequency of lipid classes of the significant regulated lipids for pathological groups versus statin regulation were compared (Fig. 4abcd). Predominantly the lipid classes PC, PE, CE, SM including Chol were up-regulated in the pathological conditions (Fig. 4a) which to some extent match the statin down-regulation of CE and Chol (Fig. 4d). Similarly, the down regulation of lipid classes TAG, PC, PE and DAG in pathological conditions match, to some degree, the observed up-regulation of TAG, PE, DAG and Cer for statin treated control subjects.
Fig. 3.
Significant regulated lipids across comparisons. Venn comparison of significant regulated lipids after correction of multiple testing (a) and with two fold regulation (b) for CVD groups (n=217). c) Heatmap depicting the direction of regulation for significant regulated lipids across comparisons (n=427). d) Heatmap depicting the direction of regulation for significant regulated lipids with more than two fold regulation across comparisons.
Fig. 4.
Frequency of lipid classes among significant regulated lipids. Significant up (a) and down (b) regulated lipid classes for CVD versus control (n=217). Significant up (c) and down (d) regulated lipid classes for statin treated versus not-treated controls (n=85). Red bars indicate significant regulated lipid class frequencies after correction of multiple testing. Blue bars indicate significant regulated lipid class frequencies after correction of multiple testing and filtering for at least two-fold regulation Panel (a) includes sterol hemi-esters (SHE), the end product of oxidation of polyunsaturated fatty acid esters of cholesterol.
In general, pairwise comparison for disease groups or disease sub-groups versus control resulted in statistical differences in lipid expression (Table S4). This was also the case when different disease groups were compared pairwise against each other. However, sub-comparisons of CVD (e.g. CVD1 versus CVD5) or SLE (e.g. SLICC ≥ 4 versus SLICC < 4) resulted in fewer significant regulated lipids after correcting for multiple testing. Especially, SLICC ≥ 4 versus SLICC < 4 comparison resulted in no significant regulated lipids after correction for multiple testing.
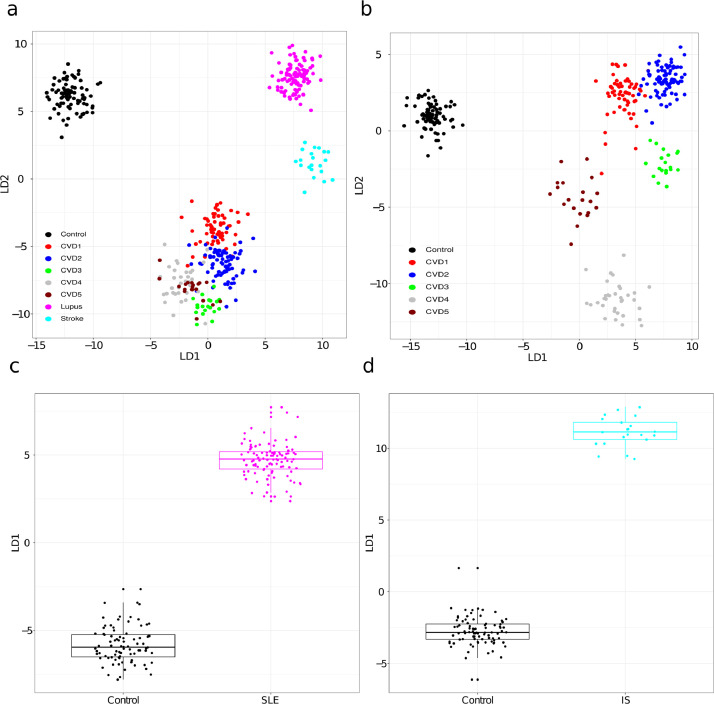
Supervised analysis. Preliminary LDA based on the 596 selected lipids and all data displayed strong potential for building classifiers for separating the pathological groups and control (Fig. 5). Even the individual CVD sub-groups were fairly effectively separated in the LDA plots (Fig. 5ab). Analyzing the CVD groups and controls separately further supported the potential of lipids to stratify CVD severity groups (Fig. 5b). LDA separates SLE from controls with a pvalue < 2.2e-16 according to student T test (Fig. 5c). Fig. 5d depicts the LDA plot for controls versus IS. The first LDA component significantly separates IS patients from controls (p value < 1.5e-12, Wilcoxon test). The PCA (Fig. 2) and LDA (Fig. 5) plots encouraged us to build and test PLS models. As a proof-of-concept three PLS classifiers with high classification performance when tested on an independent data set were established (Fig. 6).
Fig. 5.
Separation of the disease groups by LDA based on selected lipids. a) LDA plots demonstrating separation of CVD, SLE, IS and control. b) LDA base separation of CVD groups and control samples. c) First LDA component versus SLE and control. d) First LDA component versus IS and control. Shapiro–Wilk normality test implied that the data in d was not normal distributed for controls and therefore a Wilcoxon test was applied for this specific case.
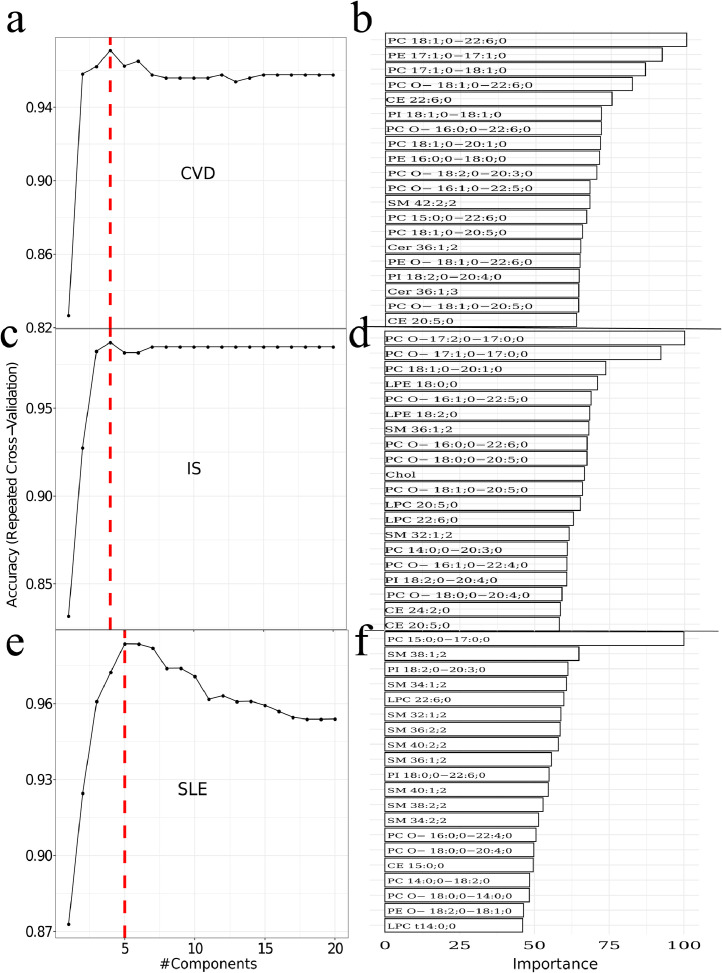
Fig. 6.
Training of PLS models. Left panel: Optimal number of PLS components for model based on a) CVD versus control, c) IS versus control, and e) SLE versus control. Right panel (b,d,f): Indicate the 20 most important lipids in the PLS models. Red labels indicate overall higher abundance in disease whereas green labels indicate overall higher abundance in controls.
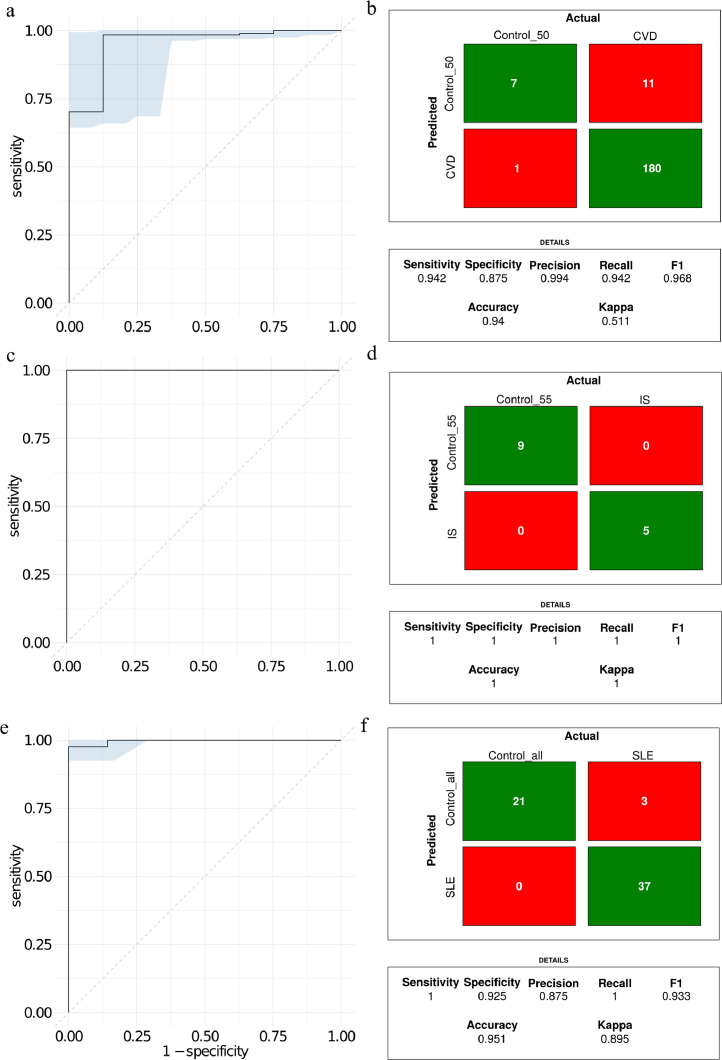
The optimal number of PLS component for separation of CVD and controls was four (Fig. 6a). The right panel (Fig. 6b) displays the 20 most important lipids contributing to the PLS components. The width of each bar is indicative of the lipid´s importance in the model. The optimal PLS component for IS versus control were four (Fig. 6c) and the 20 highest ranked lipids in terms of importance are depicted in the right panel (Fig. 6d). For SLE versus controls five PLS components were optimal (Fig. 6e). The 20 most important lipids for SLE (Fig. 6f) displayed some similarity to the most relevant lipids for IS versus control (Fig. 6bdf). For example, the lipids SM 32:1;2, LPC 22:6;0, SM 36:1;2, and PC O− 18:0;0−20:4;0 were including in the models for IS and SLE. The three PLS models for separating IS, CVD and SLE from age matched controls was validated by using a validation set left out from the training and optimization of the PLS models (Fig. 7). The ROC curve for classification performance of CVD versus controls is depicted in Fig. 7a. The area under the curve was 0.95. The confusion matrix depicted in Fig. 7b indicates that 187 out of 199 of the left-out data set were correctly classified. The model for CVD1 versus controls displayed a slightly worse classification performance with five out of 52 misclassified (Accuracy ≈ 90%, area under the curve was 0.97). Although, LDA based on all data displayed partial to complete separation for CVD1 versus CVD2 (Fig. 5ab) the PLS model was not much better than random when tested on the independent data set (area under the ROC curve=0.53).
Fig. 7.
Classification performance illustrated with ROC curves and confusion matrix for a) CVD versus control, b) IS versus control, and c) SLE versus control.
The ROC curve for the PLS model for classification of IS versus controls when tested on the independent validation data set is depicted in Fig. 7cd. The IS model misclassified zero patients in the independent validation out of a total of 14 samples (Accuracy=100%). The ROC curve for the PLS model for classification of SLE versus controls displayed an area under the curve of 1 and no misclassified subjects. The PLS model for SLE versus control demonstrated slightly improved classification performance compared to CVD versus controls with a total of 3 misclassified cases out of 61 (Fig. 7ef).
In conclusion, we present the accuracy of separation on independent lipidomics test data, of the cohorts studied – namely, control, SLE, IS, and CVD1 through CVD5. The accuracy of separating SLE and CVD cases was above 0.91. The accuracy of separating IS versus SLE and IS versus CVD was 0.78. Fig. 8 summarizes pairwise classification accuracies for all CVD subgroups versus controls when evaluated on an independent test data set. All CVD cases were separated from the controls with accuracies above 0.80. The high accuracies obtained in our study may result from the higher number of lipids profiled. Pairwise classifications of CVD1 versus CVD2 and CVD4 versus CVD5 resulted in an accuracy which was only slightly better than random. These pairwise classification accuracies are concurrent with the number of significantly identified lipids in the pairwise comparisons obtained from the linear regression models (Table S4). Overall, these findings suggest that plasma lipidomics profiles have the potential to accurately distinguish chronic inflammatory diseases from controls and that the lipidomic profiles are characteristics of the pathophysiological states.
Fig. 8.
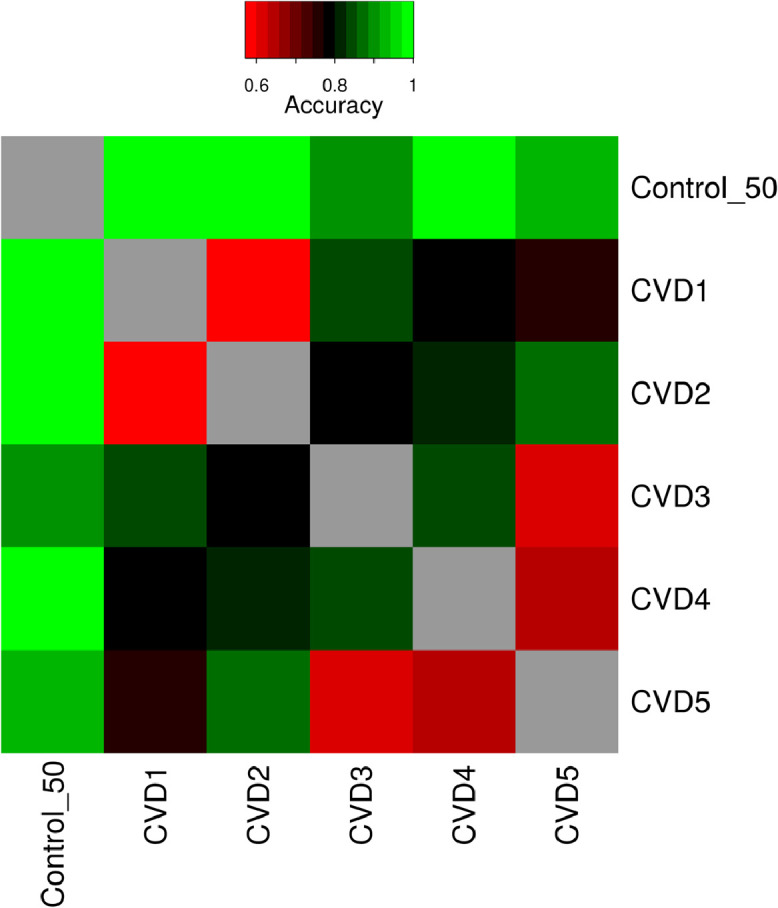
Summary of classification performance for all pairwise comparison of control and CVD1 to CVD5. The heatmap depicts the classification accuracy obtained on an independent test set for each of the pairwise comparisons.
4. Discussion
Blood plasma is the medium through which the physiological steady state of lipid distribution in the body is maintained. These lipids may be of dietary or auto-synthetic origin but are also the result of degradative chemical processes (oxidation, lysis, modification in the plasma, etc.) and cell death due to acute or chronic processes such as inflammation. The exact steady state concentration of each lipid in the plasma is therefore a combination of dietary, physiological-chemical, genetic, and patho-physiological states. Many lipidic products are known to positively or negatively influence inflammatory processes [3,4]. Lipidomics may therefore hold a promise as a valuable tool for identifying and distinguishing between different patho-physiological states. There are perhaps well over a thousand different lipids and their derivative products in the plasma. Therefore, the more lipids are identified and quantified, the better will be our capacity to diagnose the physiological state. This view seems to be reinforced by recent developments [55].
We have, therefore, used lipidomic data on three separate cohorts – a cohort consisting of people who suffer from SLE, a cohort of people who suffer from atherosclerosis-related vascular disease (CVD and IS), and an age-matched Control cohort characterized by the fact that they never sought medical help for any of the disease states of the other two cohorts. The CVD cohort consisted of five patient groups, CVD-1 through CVD-5, as described earlier. The patients of CVD, IS, and SLE have a common characteristic – chronic inflammation – but distinct primary etiologies. Inflammation plays a major role in the genesis of atherosclerotic vascular disease, which underlies the vast majority of clinical manifestations of coronary and cerebral arterial disease represented in the studied population. Inflammatory cells and cytokines are present from the early stages of atherosclerosis, contributing decisively to its development [56,57]. The infiltration of atherogenic low density lipoprotein-cholesterol (LDL-C) particles from the endothelium modified by mechanical and oxidative stress associated with several vascular risk factors triggers the inflammatory response. Monocytes bind to the endothelium expressing adhesion molecules and chemokines direct their migration into the intima followed by differentiation into macrophages and phagocytosis of LDL-C particles with formation of foam cells. Once the process of atheroma formation begins, inflammation promotes progression and instability of the atheroma leading to rupture and thrombus formation triggering the vascular event. SLE is an autoimmune disease characterized by aberrant B and T cell responses [58,59] with increased production of autoantibodies [60]. Although inflammation is also involved and an interferon signature is associated with SLE [61] a dysfunctional adaptive immune system is mainly responsible for the pathogenesis of this disease. Overall, immune dysfunction contributes to accelerated atherosclerosis in patients with SLE [62].
The comparison of these two patho-physiological states therefore provides an efficient way to evaluate the ability of lipidomics to distinguish between them and possibly serve eventually as a diagnostic tool. A total of 596 lipids divided into 15 lipid classes were analyzed using shotgun lipidomics and adequate statistical methods were used to analyze the results. We note that our aim has not been to identify the lipids characteristic of one pathological state or another, although some conclusions in this regard may also be drawn, but rather to use the data to create distinct diagnostic groups that correspond with the distinct pathological states.
The improvement in lipid identification and quantification from plasma samples presented in this study led to improved classification performance of SLE, CVD and CVD sub-groups, and IS from controls. This reinforces the potential of plasma lipidomic profiles as biomarkers for cardiovascular risk stratification proposed in previous studies [24]. We observed clear commonalities between dysregulated lipids in CVD and in IS and to a lesser extent in SLE (Fig. 3cd). This observation supports the idea of a common etiology for CVD and IS which involve atherosclerotic lesions in blood vessels in both cases. It also may partially explain why CVD is overrepresented in patients with SLE and SLE patients demonstrate accelerated atherosclerosis [63], [64], [65], [66]. The reverse regulation observed for lipids in statin treated versus CVD and IS suggests that statins to some extent stabilize the lipid profiles towards lipid profiles of disease-free controls. Nevertheless, the most frequent up-regulated lipid classes in CVD were only moderately affected by statin (Fig. 4a versus 4d). Chol and CE were observed lower in controls treated with statin which is in concordance with the known effect on statin as an inhibitor of HMG-CoA reductase [67]. PE, an anti-inflammatory lipid, was higher in statin treated and in line with statins anti-inflammatory properties. TAG and DAG were observed higher in controls treated statin than without. Higher dose of statin is reported to lower TAG and DAG [67]. The opposite pattern observed for TAG and DAG in our study might be caused by the cofounding factor introduced by the tendency to treat patients with high TAG and DAG with statins. Ideally this experiment should be performed as a randomized control study. We observed a high frequency of the lipid class PC up regulated in CVD patients. PCs are described as anti-inflammatory which may appear counterintuitive. However, a recent study found a causal link between obesity-associated increase in de novo PC synthesis and PC turnover and pro-inflammatory activation of adipose tissue macrophages [68]. The large number of dysregulated PCs does not appear affected by statin treatment and may constitute attractive pharmaceutical targets for future treatment modalities. CVD1 constitutes patients who complain about chest pain but no pathological diagnostic indicators for CVD were identified. The trained classifiers on lipid abundance were, however, able to distinguish between control and CVD1. However, it must be noted that the classification between CVD1 and CVD2 were basically close to random. This suggests that lipid profiling is the first clinical indicator that can diagnose CVD1 individuals. This finding may have both therapeutic and diagnostic impact and must be further explored in future studies.
In conclusion, we present the accuracy of separation on independent lipidomics test data, of the cohorts studied – namely, control, SLE, IS, and CVD1 through CVD5. The accuracy of separating SLE and CVD cases was above 0.91. The accuracy of separating IS versus SLE and IS versus CVD was 0.78. Fig. 8 summarizes pairwise classification accuracies for all CVD subgroups versus controls when evaluated on an independent test data set. All CVD cases were separated from the controls with accuracies above 0.80. The high accuracies obtained in our study may result from the higher number of lipids profiled. However, the cohorts used were not ideal in terms of sex distribution across CVD conditions and controls. Furthermore, the age matched controls had a slightly lower average age than the CVD and stroke groups. No statistical differences between the Control groups and CVD groups for body mass index (BMI) was identified (Table 1). The samples for each individual disease condition were also not obtained from multiple centers which may introduce bias.
Pairwise classifications of CVD1 versus CVD2 and CVD4 versus CVD5 resulted in an accuracy which was only slightly better than random. These pairwise classification accuracies are concurrent with the number of significantly identified lipids in the pairwise comparisons obtained from the linear regression models (Table S4). Overall, these findings suggest that plasma lipidomics profiles have the potential to accurately distinguish chronic inflammatory diseases from controls and that the lipidomic profiles are characteristics of the pathophysiological states. The necessary criterion is that the lipidomic data contain as many lipids as possible. We suggest that, given the ease of shotgun lipidomic quantification of a very large number of lipids in plasma and the high accuracy of the separation and identification of chronic inflammatory pathologies upon analysis of the lipidomic data, the methods described in this work could be a valuable tool in early diagnostic methodology.
Study limitations and caveats. This study is limited by the number of samples per group especially when subdividing into cardiovascular subgroups. Therefore, the list of lipids in Fig. 6 should be considered as indicative and not definitive. Gender balance is unbalanced across stroke and cardiovascular disease (see Table 1). For linear regression models statin use, age and gender were corrected for in the linear regression model. The age distributions of controls were optimized for similarity to the age distribution of the disease groups for the analyses involving PCA and PLS models. Females were more prevalent in controls (61.2%), IS (67%) and SLE (91%) and males more prevalent in CVD (65% males). Therefore, gender might constitute a confounding factor in the separation of the disease groups from controls in the PCA and PLS analysis. This might especially be a problem for the SLE disease group. However, linear regression models with or without the gender variable did not suggest gender to have a major effect on the number of significant regulated lipids. For example, the number of significant regulated lipids after correction of multiple testing were 201 and 203 with and without correction for gender. In the case of SLE versus control the number of significant regulated lipids after correction of multiple testing were 113 and 140 with and without gender correction. The samples for each individual disease condition were not obtained from multiple centers which may introduce bias.
The reverse regulation of lipids when comparing statin treated controls with CVD and IS is an explorative result and the observed regulation might originate due to confounding factors. This observation should preferably be validated in a randomized control study.
We understand the present study as an exploratory preliminary study and clearly more inflammatory diseases need to be compared. Recommendations with regard to diagnostics will require much larger cohorts with better matching of age, sex, statin use, BMI, etc. This will be a process that will require many more years and preferably collaborative work between many laboratories.
Contributors
R. Matthiesen: Data analysis and interpretation of data for the work; Data verification and Drafting, writing and revising the work
C. Lauber: Data acquisition; Data analysis and Data verification
J. L. Sampaio: Data acquisition
N. Domingues: Data acquisition
L. Alves: Data acquisition
M. J. Gerl: Data analysis and interpretation of data for the work; Revising the work and Data verification
M.S. Almeida: Conception and design of the work
G. Rodrigues: Data acquisition
P.A. Gonçalves: Conception and design of the work
J. Ferreira: Conception and design of the work
C. Borbinha: Data acquisition
J.P. Marto: Data acquisition
M. Neves: Data acquisition
F. Batista: Data acquisition
M.Viana-Baptista: Conception and design of the work
J. Alves: Conception and design of the work
K. Simons: Conception and design of the work
W.L. C. Vaz: Conception and design of the work; Data analysis and interpretation of data for the work; Drafting, writing and revising the work
O.V. Vieira: Conception and design of the work; Data acquisition; Interpretation of data for the work; Drafting, writing and revising the work
R. Matthiesen, C. Lauber and M. J. Gerl have verified the underlying data.
All authors have approved the final version of the manuscript and agreed with all aspects concerning accuracy and integrity of the work.
Declaration of Competing Interest
KS is CEO and shareholder of Lipotype GmbH. CL and MG are employees of Lipotype GmbH. All other authors declare that they do not have any competing interests.
Acknowledgments
Acknowledgements
This work was supported by PTDC/MED-PAT/29395/2017 financially supported by Fundação para a Ciência e a Tecnologia (FCT), through national funds and co-funded by FEDER under the PT2020 Partnership. ND was a holder of PhD fellowship from the FCT (Ref. No.: SFRH/BD/51877/2012), attributed by the Inter-University Doctoral Programme in Ageing and Chronic Disease (PhDOC). LA was a holder of a FCT PhD fellowship (PD/BD/114254/2016), attributed by the ProRegem Doctoral Programme in 2016.
Data Sharing Statement
Anonymized data described in the manuscript are available in supplementary materials.
Appendix. Supplementary materials
All data and R analysis is provided in the link: https://github.com/ruma1974/lipidProfillingRM/tree/master. It includes an R package with additional plotting functionality.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103504.
Contributor Information
Rune Matthiesen, Email: rune.matthiesen@nms.unl.pt.
Otilia V. Vieira, Email: otilia.vieira@nms.unl.pt.
Appendix. Supplementary materials
References
- 1.Frahnow T, Osterhoff MA, Hornemann S, Kruse M, Surma MA, Klose C. Heritability and responses to high fat diet of plasma lipidomics in a twin study. Sci Rep. 2017;7(1):3750. doi: 10.1038/s41598-017-03965-6. Epub 2017/06/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokomizo T. Introduction: special issue-lipids in inflammation. Int Immunol. 2019;31(9):555–557. [Google Scholar]
- 3.Zhang C, Wang K, Yang L, Liu R, Chu Y, Qin X. Lipid metabolism in inflammation-related diseases. Analyst. 2018;143(19):4526–4536. doi: 10.1039/c8an01046c. Epub 2018/08/22. [DOI] [PubMed] [Google Scholar]
- 4.Dasilva G, Medina I. Lipidomic methodologies for biomarkers of chronic inflammation in nutritional research: omega-3 and omega-6 lipid mediators. Free Radic Biol Med. 2019;144:90–109. doi: 10.1016/j.freeradbiomed.2019.03.017. Epub 2019/03/25. [DOI] [PubMed] [Google Scholar]
- 5.Hansson GK, Robertson AK. Soderberg-Naucler C. Inflammation and atherosclerosis. Ann Rev Pathol. 2006;1:297–329. doi: 10.1146/annurev.pathol.1.110304.100100. Epub 2007/11/28. [DOI] [PubMed] [Google Scholar]
- 6.Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med. 2011;17(11):1410–1422. doi: 10.1038/nm.2538. Epub 2011/11/09. [DOI] [PubMed] [Google Scholar]
- 7.Stegemann C, Drozdov I, Shalhoub J, Humphries J, Ladroue C, Didangelos A. Comparative lipidomics profiling of human atherosclerotic plaques. Circ Cardiovasc Genet. 2011;4(3):232–242. doi: 10.1161/CIRCGENETICS.110.959098. Epub 2011/04/23. [DOI] [PubMed] [Google Scholar]
- 8.Mundra PA, Barlow CK, Nestel PJ, Barnes EH, Kirby A, Thompson P. Large-scale plasma lipidomic profiling identifies lipids that predict cardiovascular events in secondary prevention. JCI insight. 2018;3(17) doi: 10.1172/jci.insight.121326. Epub 2018/09/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding M, Rexrode KM. A review of lipidomics of cardiovascular disease highlights the importance of isolating lipoproteins. Metabolites. 2020;10(4) doi: 10.3390/metabo10040163. Epub 2020/04/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson JC, Jiang XC, Tabas I, Tall A, Shea S. Plasma sphingomyelin and subclinical atherosclerosis: findings from the multi-ethnic study of atherosclerosis. Am J Epidemiol. 2006;163(10):903–912. doi: 10.1093/aje/kwj140. Epub 2006/04/14. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez C, Sandin M, Sampaio JL, Almgren P, Narkiewicz K, Hoffmann M. Plasma lipid composition and risk of developing cardiovascular disease. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0071846. e71846Epub 2013/08/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz SO, Sanchez-Quesada JL, de Freitas V, Leite-Moreira A, Barros AS, Reis A. Exploratory analysis of large-scale lipidome in large cohorts: are we any closer of finding lipid-based markers suitable for CVD risk stratification and management? Anal Chim Acta. 2021;1142:189–200. doi: 10.1016/j.aca.2020.10.037. Epub 2020/12/08. [DOI] [PubMed] [Google Scholar]
- 13.Sigruener A, Kleber ME, Heimerl S, Liebisch G, Schmitz G, Maerz W. Glycerophospholipid and sphingolipid species and mortality: the Ludwigshafen Risk and Cardiovascular Health (LURIC) study. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085724. e85724. Epub 2014/01/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation. 2014;129(18):1821–1831. doi: 10.1161/CIRCULATIONAHA.113.002500. Epub 2014/03/14. [DOI] [PubMed] [Google Scholar]
- 15.Tarasov K, Ekroos K, Suoniemi M, Kauhanen D, Sylvanne T, Hurme R. Molecular lipids identify cardiovascular risk and are efficiently lowered by simvastatin and PCSK9 deficiency. J Clin Endocrinol Metab. 2014;99(1):E45–E52. doi: 10.1210/jc.2013-2559. Epub 2013/11/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng JM, Suoniemi M, Kardys I, Vihervaara T, de Boer SP, Akkerhuis KM. Plasma concentrations of molecular lipid species in relation to coronary plaque characteristics and cardiovascular outcome: Results of the ATHEROREMO-IVUS study. Atherosclerosis. 2015;243(2):560–566. doi: 10.1016/j.atherosclerosis.2015.10.022. Epub 2015/11/03. [DOI] [PubMed] [Google Scholar]
- 17.Laaksonen R, Ekroos K, Sysi-Aho M, Hilvo M, Vihervaara T, Kauhanen D. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J. 2016;37(25):1967–1976. doi: 10.1093/eurheartj/ehw148. Epub 2016/04/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alshehry ZH, Mundra PA, Barlow CK, Mellett NA, Wong G, McConville MJ. Plasma lipidomic profiles improve on traditional risk factors for the prediction of cardiovascular events in Type 2 diabetes mellitus. Circulation. 2016;134(21):1637–1650. doi: 10.1161/CIRCULATIONAHA.116.023233. Epub 2016/10/21. [DOI] [PubMed] [Google Scholar]
- 19.Havulinna AS, Sysi-Aho M, Hilvo M, Kauhanen D, Hurme R, Ekroos K. Circulating ceramides predict cardiovascular outcomes in the population-based FINRISK 2002 Cohort. Arterioscler Thromb Vasc Biol. 2016;36(12):2424–2430. doi: 10.1161/ATVBAHA.116.307497. Epub 2016/10/22. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Long W, Li D, Wang D, Zhong Y, Mu D. Plasma 7-ketocholesterol levels and the risk of incident cardiovascular events. Heart. 2017;103(22):1788–1794. doi: 10.1136/heartjnl-2016-310914. Epub 2017/05/12. [DOI] [PubMed] [Google Scholar]
- 21.Razquin C, Liang L, Toledo E, Clish CB, Ruiz-Canela M, Zheng Y. Plasma lipidome patterns associated with cardiovascular risk in the PREDIMED trial: A case-cohort study. Int J Cardiol. 2018;253:126–132. doi: 10.1016/j.ijcard.2017.10.026. Epub 2018/01/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harshfield EL, Koulman A, Ziemek D, Marney L, Fauman EB, Paul DS. An unbiased lipid phenotyping approach to study the genetic determinants of lipids and their association with coronary heart disease risk factors. J Proteome Res. 2019;18(6):2397–2410. doi: 10.1021/acs.jproteome.8b00786. Epub 2019/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anroedh S, Hilvo M, Akkerhuis KM, Kauhanen D, Koistinen K, Oemrawsingh R. Plasma concentrations of molecular lipid species predict long-term clinical outcome in coronary artery disease patients. J Lipid Res. 2018;59(9):1729–1737. doi: 10.1194/jlr.P081281. Epub 2018/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang DD, Toledo E, Hruby A, Rosner BA, Willett WC, Sun Q. Plasma ceramides, mediterranean diet, and incident cardiovascular disease in the predimed trial (Prevencion con Dieta Mediterranea) Circulation. 2017;135(21):2028–2040. doi: 10.1161/CIRCULATIONAHA.116.024261. Epub 2017/03/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meikle PJ, Wong G, Tsorotes D, Barlow CK, Weir JM, Christopher MJ. Plasma lipidomic analysis of stable and unstable coronary artery disease. Arterioscler Thromb Vasc Biol. 2011;31(11):2723–2732. doi: 10.1161/ATVBAHA.111.234096. Epub 2011/09/10. [DOI] [PubMed] [Google Scholar]
- 26.Ellims AH, Wong G, Weir JM, Lew P, Meikle PJ, Taylor AJ. Plasma lipidomic analysis predicts non-calcified coronary artery plaque in asymptomatic patients at intermediate risk of coronary artery disease. Eur Heart J Cardiovas Imaging. 2014;15(8):908–916. doi: 10.1093/ehjci/jeu033. Epub 2014/03/13. [DOI] [PubMed] [Google Scholar]
- 27.Bellis C, Kulkarni H, Mamtani M, Kent JW,Jr, Wong G, Weir JM. Human plasma lipidome is pleiotropically associated with cardiovascular risk factors and death. Circ Cardiovasc Genet. 2014;7(6):854–863. doi: 10.1161/CIRCGENETICS.114.000600. Epub 2014/11/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramo JT, Ripatti P, Tabassum R, Soderlund S, Matikainen N, Gerl MJ. Coronary artery disease risk and lipidomic profiles are similar in hyperlipidemias with family history and population-ascertained hyperlipidemias. J Am Heart Assoc. 2019;8(13) doi: 10.1161/JAHA.119.012415. e012415. Epub 2019/07/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin X, Willinger CM, Keefe J, Liu J, Fernandez-Ortiz A, Ibanez B. Lipidomic profiling identifies signatures of metabolic risk. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.10.046. Epub 2019/12/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerl MJ, Vaz WLC, Domingues N, Klose C, Surma MA, Sampaio JL. Cholesterol is inefficiently converted to cholesteryl esters in the blood of cardiovascular disease patients. Sci Rep. 2018;8(1):14764. doi: 10.1038/s41598-018-33116-4. Epub 2018/10/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T, Mohan C. Caution in studying and interpreting the lupus metabolome. Arthr Res Ther. 2020;22(1):172. doi: 10.1186/s13075-020-02264-2. Epub 2020/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira HB, Pereira AM, Melo T, Paiva A, Domingues MR. Lipidomics in autoimmune diseases with main focus on systemic lupus erythematosus. J Pharm Biomed Anal. 2019;174:386–395. doi: 10.1016/j.jpba.2019.06.005. Epub 2019/06/18. [DOI] [PubMed] [Google Scholar]
- 33.Shah D, Mahajan N, Sah S, Nath SK, Paudyal B. Oxidative stress and its biomarkers in systemic lupus erythematosus. J Biomed Sci. 2014;21:23. doi: 10.1186/1423-0127-21-23. Epub 2014/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reichlin M, Fesmire J, Quintero-Del-Rio AI, Wolfson-Reichlin M. Autoantibodies to lipoprotein lipase and dyslipidemia in systemic lupus erythematosus. Arthritis Rheum. 2002;46(11):2957–2963. doi: 10.1002/art.10624. Epub 2002/11/13. [DOI] [PubMed] [Google Scholar]
- 35.Borba EF, Bonfa E, Vinagre CG, Ramires JA, Maranhao RC. Chylomicron metabolism is markedly altered in systemic lupus erythematosus. Arthritis Rheum. 2000;43(5):1033–1040. doi: 10.1002/1529-0131(200005)43:5<1033::AID-ANR11>3.0.CO;2-B. Epub 2000/05/19. [DOI] [PubMed] [Google Scholar]
- 36.Hu C, Zhou J, Yang S, Li H, Wang C, Fang X. Oxidative stress leads to reduction of plasmalogen serving as a novel biomarker for systemic lupus erythematosus. Free Radic Biol Med. 2016;101:475–481. doi: 10.1016/j.freeradbiomed.2016.11.006. Epub 2016/11/12. [DOI] [PubMed] [Google Scholar]
- 37.Lu L, Hu C, Zhao Y, He L, Zhou J, Li H. Shotgun lipidomics revealed altered profiles of serum lipids in systemic lupus erythematosus closely associated with disease activity. Biomolecules. 2018;8(4) doi: 10.3390/biom8040105. Epub 2018/10/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Surma MA, Herzog R, Vasilj A, Klose C, Christinat N, Morin-Rivron D. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur J Lipid Sci Technol: EJLST. 2015;117(10):1540–1549. doi: 10.1002/ejlt.201500145. Epub 2015/10/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fihn SD, Gardin JM, Abrams J, Berra K, Blankenship JC, Dallas AP. ACCF/AHA/ACP/AATS/PCNA/SCAI/STS guideline for the diagnosis and management of patients with stable ischemic heart disease: executive summary: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, and the American College of Physicians, American Association for Thoracic Surgery, Preventive Cardiovascular Nurses Association, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol 2012. 2012;60(24):2564–2603. doi: 10.1016/j.jacc.2012.07.012. Epub 2012/11/28. [DOI] [PubMed] [Google Scholar]
- 40.Thygesen K, Alpert JS, Jaffe AS, Chaitman BR, Bax JJ, Morrow DA. Fourth universal definition of myocardial infarction (2018) Circulation. 2018;138(20):e618–ee51. doi: 10.1161/CIR.0000000000000617. Epub 2018/12/21. [DOI] [PubMed] [Google Scholar]
- 41.Amsterdam EA, Wenger NK, Brindis RG, Casey DE,Jr, Ganiats TG, Holmes DR,Jr. 2014 AHA/ACC guideline for the management of patients with non-ST-elevation acute coronary syndromes: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;130(25):e344–e426. doi: 10.1161/CIR.0000000000000134. Epub 2014/09/25. [DOI] [PubMed] [Google Scholar]
- 42.Hay EM, Bacon PA, Gordon C, Isenberg DA, Maddison P, Snaith ML. The BILAG index: a reliable and valid instrument for measuring clinical disease activity in systemic lupus erythematosus. Q J Med. 1993;86(7):447–458. Epub 1993/07/01. [PubMed] [Google Scholar]
- 43.Conover JW. John Wiley & Sons; New York: 1971. Practical nonparametric statistics; pp. 295–301. [Google Scholar]
- 44.Conover WJ. John Wiley & Sons; New York: 1971. Nonparametric statistics; pp. 309–314. [Google Scholar]
- 45.Aimo L, Liechti R, Hyka-Nouspikel N, Niknejad A, Gleizes A, Gotz L. The SwissLipids knowledgebase for lipid biology. Bioinformatics. 2015;31(17):2860–2866. doi: 10.1093/bioinformatics/btv285. Epub 2015/05/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liebisch G, Fahy E, Aoki J, Dennis EA, Durand T, Ejsing CS. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J Lipid Res. 2020;61(12):1539–1555. doi: 10.1194/jlr.S120001025. Epub 2020/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herzog R, Schuhmann K, Schwudke D, Sampaio JL, Bornstein SR, Schroeder M. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS One. 2012;7(1):e29851. doi: 10.1371/journal.pone.0029851. Epub 2012/01/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12(1):R8. doi: 10.1186/gb-2011-12-1-r8. Epub 2011/01/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stati Appl Genet Mol Biol. 2004;3(Article3) doi: 10.2202/1544-6115.1027. Epub 2006/05/02. [DOI] [PubMed] [Google Scholar]
- 50.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. Epub 2003/01/23. [DOI] [PubMed] [Google Scholar]
- 51.Petri M, Orbai AM, Alarcon GS, Gordon C, Merrill JT, Fortin PR. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 2012;64(8):2677–2686. doi: 10.1002/art.34473. Epub 2012/05/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B. 1995;57(1):289–300. [Google Scholar]
- 53.Kuhn M. Building predictive models in R using the caret Package. J Stat Softw. 2008;28(5):1–26. [Google Scholar]
- 54.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC. pROC: an open-source package for R and S plus to analyze and compare ROC curves. BMC Bioinformatics. 2011:12. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burnett JR, Hooper AJ, Hegele RA. Remnant cholesterol and atherosclerotic cardiovascular disease risk. J Am Coll Cardiol. 2020;76(23):2736–2739. doi: 10.1016/j.jacc.2020.10.029. Epub 2020/12/05. [DOI] [PubMed] [Google Scholar]
- 56.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105(9):1135–1143. doi: 10.1161/hc0902.104353. Epub 2002/03/06. [DOI] [PubMed] [Google Scholar]
- 57.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–1695. doi: 10.1056/NEJMra043430. Epub 2005/04/22. [DOI] [PubMed] [Google Scholar]
- 58.Yap DYH, Chan TM. B cell abnormalities in systemic lupus erythematosus and lupus nephritis-role in pathogenesis and effect of immunosuppressive treatments. Int J Mol Sci. 2019;20(24) doi: 10.3390/ijms20246231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katsuyama T, Tsokos GC, Moulton VR. Aberrant T Cell signaling and subsets in systemic lupus erythematosus. Front Immunol. 2018;9 doi: 10.3389/fimmu.2018.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yaniv G, Twig G, Shor DB, Furer A, Sherer Y, Mozes O. A volcanic explosion of autoantibodies in systemic lupus erythematosus: a diversity of 180 different antibodies found in SLE patients. Autoimmun Rev. 2015;14(1):75–79. doi: 10.1016/j.autrev.2014.10.003. Epub 2014/12/03. [DOI] [PubMed] [Google Scholar]
- 61.Bezalel S, Guri KM, Elbirt D, Asher I, Sthoeger ZM. Type I interferon signature in systemic lupus erythematosus. Isr Med Assoc J. 2014;16(4):246–249. Epub 2014/05/20. [PubMed] [Google Scholar]
- 62.Skaggs BJ, Hahn BH, McMahon M. Accelerated atherosclerosis in patients with SLE–mechanisms and management. Nat Rev Rheumatol. 2012;8(4):214–223. doi: 10.1038/nrrheum.2012.14. Epub 2012/02/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petri MA, Barr E, Magder LS. Development of a systemic lupus erythematosus cardiovascular risk equation. Lupus Sci Med. 2019;6(1) doi: 10.1136/lupus-2019-000346. Epub 2019/11/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McMahon M, Hahn BH, Skaggs BJ. Systemic lupus erythematosus and cardiovascular disease: prediction and potential for therapeutic intervention. Expert Rev ClinImmunol. 2011;7(2):227–241. doi: 10.1586/eci.10.98. Epub 2011/03/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hak AE, Karlson EW, Feskanich D, Stampfer MJ, Costenbader KH. Systemic lupus erythematosus and the risk of cardiovascular disease: results from the nurses' health study. Arthritis Rheum. 2009;61(10):1396–1402. doi: 10.1002/art.24537. Epub 2009/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeller CB, Appenzeller S. Cardiovascular disease in systemic lupus erythematosus: the role of traditional and lupus related risk factors. Curr Cardiol Rev. 2008;4(2):116–122. doi: 10.2174/157340308784245775. Epub 2008/05/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu A, Wu Q, Guo J, Ares I, Rodriguez JL, Martinez-Larranaga MR. Statins: adverse reactions, oxidative stress and metabolic interactions. Pharmacol Ther. 2019;195:54–84. doi: 10.1016/j.pharmthera.2018.10.004. Epub 2018/10/16. [DOI] [PubMed] [Google Scholar]
- 68.Petkevicius K, Virtue S, Bidault G, Jenkins B, Cubuk C, Morgantini C. Accelerated phosphatidylcholine turnover in macrophages promotes adipose tissue inflammation in obesity. eLife. 2019;8 doi: 10.7554/eLife.47990. Epub 2019/08/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.