Abstract
In addition to mutations or aberrant expression in the protein coding genes, mutations and misregulation of non-coding RNAs, in particular long noncoding RNAs (lncRNA), appear to play major roles in cancer. Genome-wide association studies (GWAS) of tumor samples have identified a large number of lncRNAs associated with various types of cancer. Alterations in lncRNA expression and their mutations promote tumorigenesis and metastasis. LncRNAs may exhibit tumor suppressive and promoting (oncogenic) functions. Due to their genome-wide expression patterns in a variety of tissues and their tissue-specific expression characteristics, lncRNAs hold strong promise as novel biomarkers and therapeutic targets for cancer. In this article, we have reviewed the emerging functions and association of lncRNAs in different types of cancer and discussed their potential implications in cancer diagnosis and therapy.
Keywords: Non-coding RNA, Long noncoding RNA, lncRNA, Cancer, Epigenetics, Biomarker, Therapeutics
2. Introduction
Cancer is a complex disease associated with a variety of genetic mutations, epigenetic alterations, chromosomal translocations, deletions, and amplification (1). Non-coding RNAs (ncRNAs) are an emerging class of transcripts which are coded by the genome but are mostly not translated into proteins (2). Though not translated, ncRNAs are crucial players in a variety of cellular and physiological functions (3). In particular, long non-coding RNAs (ncRNAs that are >200 nt long) play key roles in regulating chromatin dynamics, gene expression, growth, differentiation, and development (4). It is now well-recognized that more than 75% of the human genome is functional and encodes large numbers of ncRNAs (5). Based on the ENCODE project, it is estimated that the human genome encodes more than 28,000 distinct long noncoding RNAs (lncRNAs) many of which are still being discovered and are yet to be annotated (6). While understanding the functions of so many lncRNAs and their detailed characterization are challenging tasks, analysis of transcriptome profiles using next-generation sequencing in the last few years has revealed that thousands of lncRNAs are aberrantly expressed or mutated in various cancers (7).
Although lncRNAs are emerging as a major class of noncoding transcripts, the discovery of tremendously large numbers of lncRNAs and their diverse functions and complexity pose a major challenge to effectively classify them in different categories. At this point, lncRNAs are broadly classified based on their genomic localization, modes of action, and function. Intronic lncRNAs originate from the introns of protein coding genes; intergenic lncRNAs (lincRNAs) originate from the region between two protein-coding genes; enhancer lncRNAs (elncRNAs) originate from the promoter enhancer regions; bidirectional lncRNAs are localized within the vicinity of a coding transcript of the opposite strand; sense-overlapping lncRNAs overlap with one or more introns and exons of different protein coding genes in the sense strand of the DNA; antisense-transcripts originate from the antisense-strands of the DNA, and they may or may not be complementary to protein coding sequences in the sense-strand (7,8). Functionally, lncRNAs are classified as signaling, decoy, guide, and scaffold lncRNAs (9). Signaling lncRNAs are associated with specific signaling pathways and their expression indicates an active signaling event, irrespective of their roles (direct/indirect) in the signaling process (9). For example, the expression of XIST signals X-inactivation in females (10). Decoy lncRNAs act like molecular sinks for transcription factors and repressors. They interact with and titrate away transcription factors from binding to the target gene promoters facilitating gene activation or silencing (9). Examples of decoy lncRNAs include GAS5 (growth arrest specific 5), TERRA (telomeric repeat-containing RNAs), and others. (9). Guide lncRNAs bind to the regulatory or enzymatically active protein complexes and direct them to specific target gene promoters or genomic loci regulating downstream signaling events and gene expressions. Examples of guide lncRNAs include AIR, CCND1 (cyclin D promoter associated lncRNA), lincRNA-p21, and others (8,9). Scaffold lncRNAs act as a central platform to which various protein complexes tether and get directed to specific genomic location or target gene promoter regulating gene expression and chromosomal dynamics. Examples of scaffold lncRNAs are HOTAIR, TERC, and others.
Beyond traditional ncRNAs, circular RNAs (circRNAs) are also emerging as a novel class of endogenous noncoding RNAs that form covalently-closed continuous loops instead of traditional linear forms. CircRNAs are conserved across species and are found to be associated with a variety of important biological processes and human diseases including cancer. CircRNAs appear to function as microRNA (miRNA) sponges and are involved in the regulation of mRNA splicing, transcription, and gene expression (11,12). Generally, circRNAs are classified as exonic, intronic, and retained-intronic circRNAs. They may be derived from exons, introns, untranslated regions, antisense transcripts, and intergenic regions. CircRNA biogenesis has been explained by various models, incorporating a range of spliceosomes and RNA binding proteins. The most accepted model suggests that circRNA biogenesis involves joining of a 5’ splice site and a 3’ splice site as the result of back splicing (13,14). Because of their unique structure, circRNAs are resistant to nucleases and are stable with a relatively long half-life. They may exist in tissues, serum, and urine, indicating their potential as novel biomarkers for human cancer. CircRNAs are implicated in a variety of cancers including laryngeal cancer, gastric cancer, hepatocellular cancer, bladder cancer, and esophageal cancer, among others (11,15,16). For example, circular RNA ciRS-7, which acts as a sponge for miR-7, is involved in promoting colorectal cancer through inhibiting the repression of oncogenes such as YY1 by tumor suppressor miR-7 (15). CiRS-7 is an endogenous circular RNA highly expressed in the brain and transcribed antisense to the CDR1 (cerebellum degeneration-related antigen 1) gene (12). CircRNAs such as circ-ITCH, hsa_circ_002059 and hsa_circ_0001649 are downregulated in colorectal cancer, gastric cancer, and hepatocellular cancer, whereas circ-VCAN, circTCF25, and circ-KLDGC10 are upregulated in glioma, bladder cancer, and hepatocellular cancer (11,12,16–18). CircRNAs such as ci-ankrd52 and circular -ANRIL are examples of circular lncRNAs (19,20). Similar to lncRNAs, many circRNAs display aberrant expression in various cancers and possess strong promise toward development of novel biomarkers and therapeutics.
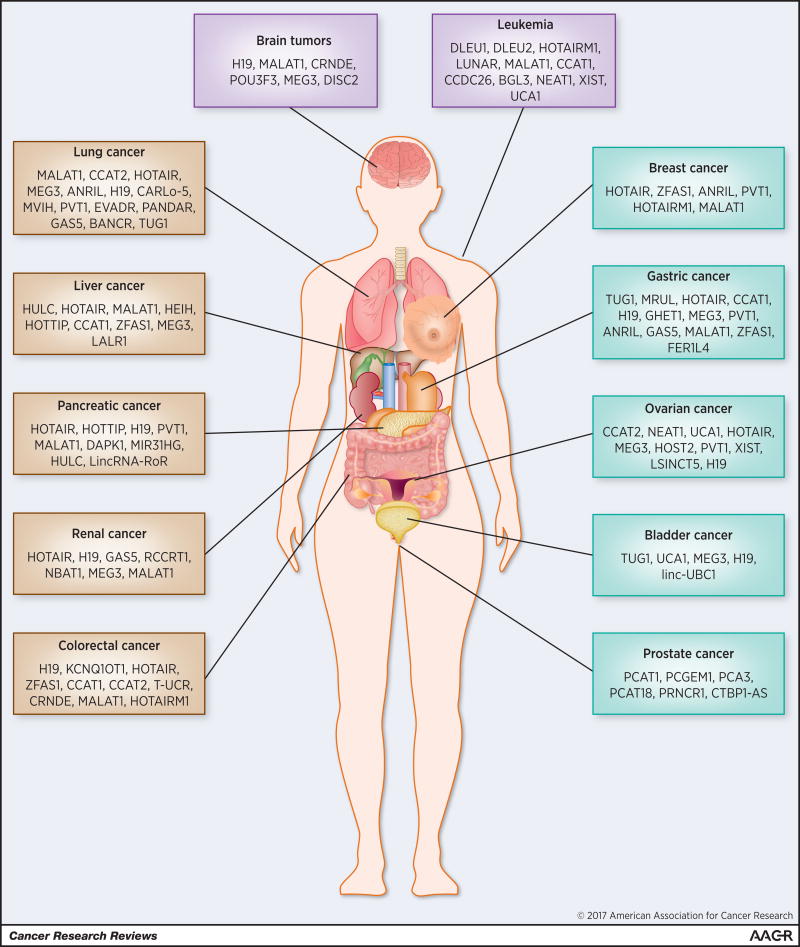
Thus, in addition to protein coding genes, ncRNAs, in particular lncRNAs, are rapidly emerging as a novel class of transcripts associated with a variety of cellular and biological processes including gene regulation and chromatin dynamics. They are abundantly expressed and widely associated with a variety of cancers, and the aberrant expression and mutations are closely linked to tumorigenesis, metastasis, and tumor stage (21–23). Moreover, they are specifically expressed in certain types of cancer and detected in circulating blood and/or urine (24–26). LncRNAs are a novel class of potential biomarkers and therapeutic targets for the treatment of cancer. In this article, we have reviewed the functions of various lncRNAs in different types of cancer and discussed their potential implications in diagnosis and therapy (Figure 1).
Figure 1.
LncRNAs associated with various types of cancer
3. LncRNAs in prostate cancer
Prostate cancer is the most common cancer and the second leading cause of cancer deaths in American men. The American Cancer Society estimates about 181,000 new cases of prostate cancer and 26,000 deaths from prostate cancer in the US in 2016. There is an urgent need to develop novel diagnostic biomarkers and effective therapies for prostate cancer. Genome-wide RNA-Seq analyses identified many lncRNAs that are up- or down-regulated in prostate cancer (27). Several lncRNAs such as PCA3, PCGEM1, and PCAT-1 are highly specific to prostate cancer (28) (Figure 1, Table 1).
Table 1.
LncRNAs: their mechanism of action and significance in cancer
| LncRNA | Cancer type | Mechanism of action and function | Refs |
|---|---|---|---|
| PCA3 (a.k.a. DD3) | Prostate | Steroid receptor-regulated lncRNA; Induces RNA editing via interaction with PRUNE2-pre-mRNA to form a double-stranded RNA duplex and ADAR proteins; Knockdown results in reduced cell growth and survival and induction of apoptotic cells; (↑) | (29,31,33,34,251) |
| PCGEM1 | Prostate | Promotes colony formation, cell proliferation; promotes chemo-resistance via inhibition of PARP cleavage and delaying the induction of tumor suppressors p53 and p21; Regulates AR target genes expression, in conjunction with lncRNA PRNCR1, AR, histone methylase DOT1L; and Pygopus family PHD finger 2 (PYGO2); Knockdown results in reduced proliferation and increased apoptosis; (↑) | (35–37) |
| PCAT-1 | Prostate | Promotes cell proliferation, downregulates genes and tumor suppressor genes; sensitizes prostate cancer cells towards PARP 1 inhibitors; post-transcriptionally upregulates c-Myc; (↑) | (27,39) |
| HOTAIR | Breast, hepatocellular, colorectal, pancreatic, lung, ovarian, liver | Scaffolding lncRNA, silences genes via interaction with PRC2 and LSD1, aids in protein degradation via interaction with E3 ubiquitin ligases; Knockdown reduces tumor invasiveness, disrupts of EMT; (↑) | (45–53,57,58,255) |
| ANRIL | Breast, gastric, lung, liver | Controls cell proliferation and senescence via regulating tumor suppressors CDKN2A/B; Represses the INK4A locus via interaction with CBX7 and PRC2; Knockdown lowers multidrug resistance, reduces proliferation, and invasiveness; (↑) | (66–78) |
| MALAT1 (a.k.a. NEAT2) | Lung, prostate, breast, colorectal, liver, gastric, leukemia, brain, renal | Undergoes processing to produce a short and long RNA transcript; localized into nuclear speckles; influences SR-protein phosphorylation and modulates alternative splicing; regulates of EMT gene expression; associates with SUZ12 and regulates N-cadherin and E-cadherin expression. Knockdown reduces cell growth, invasion, and migration, and differentiation into cystic tumors; (↑) | (83–90) |
| NEAT1 | Leukemia, ovarian | Regulates ADARB2 expression via protein sequestration into paraspeckles; Knockdown results in inhibition of cell growth; (↑) | (95,96) |
| H19 | Bladder, brain, gastric, renal, lung, ovarian, colorectal, pancreatic | Pivotal in embryonic development and tumorigenesis; maternally expressed and paternally imprinted; precursor of miRNAs (miR-675), P53 represses the H19 gene and the H19-derived miR-675 inhibits p53; Interacts with EZH2, MBD1 and induces gene repression; Knockdown reduces tumor size and metastasis; (↑) | (1,101–117) |
| KCNQ1OT1 | Colorectal, hepatocellular, pediatric adrenocortical; Beckwith-Wiedemann syndrome. | Paternally imprinted; interacts with PRC1, PRC2, and G9a and silences KCNQ1 via induction in histone and DNA methylation. Imprinting disruption of the CDKN1C/KCNQ1OT1 domain is involved in the development of both BWS and cancer. Knockdown results in loss of imprinting in the 5’-domain of KCNQ1OT1; (↑) | (120–124) |
| T-UCRs | Colorectal, Barrett's adenocarcinoma, bladder, liver | CpG-island hypermethylation induced T-UCR silencing is common in many tumors; Inhibits miR-596 via interaction with YY1, inhibits miR-193b; Overexpression inhibits migration and invasion; (↑) | (129–131) |
| CCAT1 | Colorectal, leukemia, gastric, Lung, esophageal squamous cell carcinoma | Acts as a sponge for let-7 and miR-155, regulates c-Myc, HOXB13, SPRY4; Knockdown reduces cell proliferation and migration; (↑) | (136,138) |
| HULC | Hepatocellular, pancreatic | Acts as a microRNA sponge and sequesters miR-372; potential biomarker for HCC; Knockdown inhibits cell proliferation and increases chemosensitivity; (↑) | (143,144) |
| HEIH | Hepatocellular | Linked with hepatitis-B-virus associated HCC recurrence; Regulates cell cycle regulatory genes p15, p16, p21 via interaction with EZH2; Knockdown reduces cell proliferation and suppresses tumor growth (↑) | (141,147,148) |
| HOTTIP | Prostate, liver, pancreatic | Controls the HOXA locus via interaction with WDR5/MLL; Knockdown suppresses chemoresistance, and mesenchymal characteristics; (↑) | (152–154) |
| UCA1 | Bladder, leukemia, ovarian, breast | Potential urine biomarker; promotes chemoresistance; Recruits SWI/SNF to the TCF7 promoter, induces Wnt/β-catenin signaling and ER redistribution; Knockdown increases chemo-sensitivity, reduces cell migration and tumor size; (↑) | (157,158) |
| DLEU1, DLEU2 | Leukemia | Deleted in lymphocytic Leukemia; regulate NF-kB activity, acts as a precursor for miR-15a and miR-16-1 in leukemia; (↓) | (163,164) |
| LUNAR1 | Leukemia, B-cell lymphoma | Promotes T-ALL growth by inducing IGF1R expression, regulates IGF1R via interaction with mediator complex; Knockdown reduces cell proliferation and viability; (↑) | (165,166) |
| BGL3 | Leukemia | Regulates Bcr-Abl through sponging miRNAs (miR-17, miR-93, miR-20a, miR-20b, miR-106a, and miR-106b) and via c-Myc-dependent DNA methylation; (↓) | (167) |
| HOTAIRM1 | Breast, leukemia, colorectal | Controls myeloid autophagy and maturation via interaction with PRC2 and UTX/MLL; Knockdown results in retardation of myeloid cell differentiation; (↑) | (169–171) |
| XIST | Ovarian, leukemia | Inactivates X chromosome via coating and interaction with PRC1/2, YY1, CTCF, etc.; Knockdown results in enhanced sensitivity to Taxol; (↑) | (173,174) |
| FER1L4 | Gastric, endometrial | Regulates PTEN and the PI3K-AKT pathway by behaving as a ceRNA for miR-106a-5p; Overexpression reduces cell growth and colony formation; (↓) | (168,188) |
| NBAT1 | Renal, neuroblastoma | Silences neuronal-specific NRSF/REST through association with PRC2; Overexpression results in differentiation of neuronal precursors; (↓) | (189,190) |
| GAS5 | Breast, renal, prostate, endometrial | Acts as decoy for glucocorticoid receptor (GR), inhibits transcriptional induction by GR, causes growth arrest and apoptosis, induces PTEN via inhibiting miR-103; (↓) | (191–193) |
| TERRA | Pancreatic, cervical, gastric, breast | Facilitates heterochromatin formation via interaction with TRF1 and TRF2, aids in telomerase function by providing a RNA template; (↓) | (198–200) |
| ZFAS1 | Breast, colorectal, gastric, liver | Interacts with CDK1/cyclin B, EZH2, LSD1/CoREST, acts as a sponge for miR-150, promotes cell proliferation; Knockdown results in inhibition of cell proliferation, migration, and colony formation; (↑) | (202–204) |
| PVT1 | Breast, pancreatic, ovarian, gastric, lung | Promote proliferation via interaction with NOP2 with the aid of TGFβ1, enhances c-Myc stability via inhibiting its phosphorylation; Knockdown results in reduced cell proliferation and chemoresistance; (↑) | (206–208) |
| MEG3 | Renal, gastric, ovarian, liver, lung, brain, bladder | Represses MDM2, aids in p53 accumulation, represses genomic loci of genes associated with transforming growth factor-β (TGF-β) pathway via cooperating with PRC2; Overexpression results in apoptosis and inhibition of proliferation; (↓) | (211–214) |
| TUG1 | Bladder, gastric, lung | Silences cell cycle associated genes via interaction with PRC2; Knockdown results in inhibition of cell proliferation, invasion, and colony formation; (↑) | (2,3,268–270) |
| Linc-RoR | Breast, ,pancreatic, hepatocellular, endometrial, nasopharyngeal | Induces epithelial-mesenchymal transition, drug resistance and invasiveness of cancer cells; promotes invasion, metastasis and tumor growth through activating ZEB1 pathway; (↑) | (178) |
(↑) Upregulated in cancer (oncogenic); (↓) Downregulated in cancer (tumor suppressor)
PCA3
PCA3 (Prostate Cancer Antigen 3) (a.k.a. DD3), a steroid receptor-regulated lncRNA transcribed from 9q21.22, is overexpressed in 95% of prostate cancer cases and is detected with high specificity in the urine of patients with malignant and benign prostate cancer (29) (30,31)(Figure 1, Tables 1 and 2). PCA3 and Hedgehog receptor PTCH (also implicated in prostate cancer) are highly upregulated in the circulating prostate cancer cells of androgen refractory patients (32–34). Prune2 (a tumor suppressor and a target of PCA3) and PCA3 expressions are inversely correlated in prostate cancer (34). PCA3 binds to PRUNE2-pre-mRNA to form a double-stranded RNA duplex that recruits adenosine deaminase (ADA), inducing RNA editing through acting on RNA (ADAR) proteins (34).
Table 2.
LncRNAs as cancer biomarkers
| Cancer | LncRNA Biomarker | Potential Implications | Site of Detection | Refs |
|---|---|---|---|---|
| Prostate cancer | PCA3 | Detection; Prognosis | Urine; Tumor | (30,218) |
| LincRNA-p21 | Detection; Stratification | Urine | (192) | |
| PCAT-18 | Metastasis | Plasma | (28) | |
| MALAT1 | Risk of tumorigenesis; Detection | Urine; Plasma | (233,234) | |
| PVT1 | Aggressiveness | Tumor | (219) | |
| TRPM2 | Early identification; aggressiveness | (220) | ||
| Breast cancer | ZFAS1 | Detection | Tumor | (205) |
| HOTAIR | Detection | Serum | (221) | |
| RP11-445H22.4 | Detection | (235) | ||
| HIF1A-AS2; AK124454 | Recurrence | Tumor | (222,223) | |
| Lung cancer | MALAT1 | Early detection; Risk of metastasis | Whole blood; Tumor | (87,224,225) |
| SPRY4-IT1; ANRIL; NEAT1 | Early detection | Plasma | (92) | |
| UCA1 | Detection | Plasma; Tumor | (226) | |
| Colorectal cancer | HOTAIR | Risk of tumorigenesis | Tumor | (227) |
| HOTAIR; CCAT1; CCAT2 | Detection | Serum | (228) | |
| FER1L4 | Recurrence; Metastasis | Plasma | (229) | |
| XLOC_006844; LOC152578; XLOC_000303 | Risk of tumorigenesis | (230) | ||
| Hepatocellular cancer | HOTAIR | Recurrence after transplant | Tumor | (51) |
| uc001ncr; AX800134 | Detection (especially early-stage) | Serum | (145) | |
| HULC; Linc00152 | Detection; Metastasis | Plasma | (231) | |
| RP11-160H22.5; XLOC014172; LOC149086, HEIH | Risk of tumorigenesis, prognostic factor for recurrence and survival | (232) | ||
| XLOC014172; LOC149086 | Risk of metastasis | (232) | ||
| Bladder cancer | UCA1 | Detection | Urine | (236) |
| H19 | Early recurrence | Tumor | (101) | |
| HOTAIR | Overall survival | (237) | ||
| Leukemia | CRNDE | Identification of subtypes of AML (acute myeloid leukemia) (M2 or M3) | Bone marrow, Lymph nodes | (238) |
| Ovarian cancer | NEAT1 | Invasiveness; Prognosis | Tumor | (239) |
| Renal cancer | LET; PVT1; PANDAR; PTENP1; LINC00963 | Early detection | Serum | (240) |
| Cervical cancer | HOTAIR | Prognosis; Recurrence | Serum | (241) |
| Esophageal cancer | POU3F3; HNF1A-AS1; SPRY4-IT1 | Early screening | Plasma | (242) |
| Gastric cancer | H19 | Early screening | Plasma | (243) |
| LINC00152 | Detection; Invasion | Gastric juice; Tumor | (244) | |
| UCA1 | Early detection; Prognosis prediction | (245) | ||
| CUDR; LSINCT-5; PTENP1 | Detection | Serum | (246) | |
| AA174084 | Early diagnosis | Tumor; Plasma; Gastric juice | (247) |
PCGEM1
PCGEM1 (Prostate Cancer Gene Expression Marker 1) is a 1.6 kb long lncRNA from the 2q32 locus. It is a highly prostate tissue-specific and androgen-regulated lncRNA that is overexpressed in prostate cancer and promotes cell proliferation and colony formation (35–37) (Figure 1 and Table 1). PCGEM1 expression inhibits doxorubicin-induced apoptosis and promotes chemo-resistance via inhibition of PARP (poly-ADP-ribose polymerase) cleavage and delaying the induction of tumor suppressors p53 and p21 (36). Another lncRNA PRNCR1 (prostate cancer noncoding RNA1), in conjunction with PCGEM1, regulates gene expression by promoting epigenetic modifications (36). PRNCR1 binds to acetylated androgen receptor (AR) at the enhancer, and recruits histone H3K79 methyltransferase DOT1L (disruptor of telomeric silencing 1-like) which methylates AR that aids in the recruitment of PCGEM1 to the AR N-terminal and modulates target gene expression (35). Similarly, PCGEM1 recruits the Pygopus family PHD finger 2 (PYGO2) to the enhancer-promoter regions of AR gene and regulates AR-induced gene expression (38).
PCAT-1
PCAT-1 (Prostate Cancer-Associated ncRNA Transcript 1) is a 7.8 kb long intergenic lncRNA (originating from 8q24 locus) that is overexpressed in and highly specific to high-grade localized and metastatic prostate cancer (28,38,39) (Figure 1, Tables 1 and 2). It is independent of chromosome 8q24 amplification that is often observed in other cancers. There is a converse correlation between the expression of PCAT-1 and EZH2 (a histone H3K27-specific methyltransferase and interacting component of polycomb repressive complex 2 (PRC2)) (27). EZH2 (enhancer of zeste homolog 2) knockdown upregulates PCAT-1 (27). PRC2 binds the PCAT-1 promoter and suppresses PCAT-1 expression (27). PCAT-1 induces cell proliferation and downregulates the expression of genes including tumor suppressor gene BRCA2. PCAT-1 sensitizes prostate cancer cells towards PARP 1 inhibitors. PCAT-1 post-transcriptionally upregulates c-Myc that promotes prostate cancer cell proliferation (28,38).
Various other lncRNAs including MALAT1, GAS5, PCAT-18, CTBP1-AS, ANRIL, PVT1, and SCHLAP1 are also linked to prostate cancer (28,38) (Table 1). PCAT-18 is a highly prostate-specific transcript upregulated in prostate cancer and regulated by AR (28). CTBP1-AS is an androgen-responsive lncRNA and an antisense transcript of the CTBP1 gene (40). Overexpression of CTBP1-AS inhibits the expression of cell cycle regulators such as p53 and Smad3 in prostate cancer cells, resulting in cell proliferation (41,42).
4. Breast cancer
Breast cancer is the most common and the second deadliest cancer among women. It is estimated that 246,660 new cases and 40,450 deaths occurred from breast cancer in the US in 2016. LncRNAs implicated in breast cancer include HOTAIR, ANRIL, ZFAS1, HOTAIRM1, PVT1, MALAT1, and LNP1, among others (43,44) (Figure 1, Tables 1 and 2).
HOTAIR
HOTAIR (HOX Transcript Antisense Intergenic RNA) is one of the most well-studied lncRNAs that is overexpressed in a variety of cancers including breast, colorectal, hepatocellular, gastrointestinal, and non-small cell lung carcinomas (4,45–51) (Table 1). HOTAIR, a 2.2 kb antisense lncRNA, interacts with two major gene silencing factors: PRC2 and LSD1 (lysine specific demethylase 1). PRC2 is a multiprotein complex comprised of EZH2 (H3K27-methylase), SUZ12, EED, and RbAp46/48 (52–54). LSD1 interacts with co-repressors REST and CoREST (54,55). H3K27-methylation by EZH2 and H3K4-demethylation by LSD1 are both critical to gene silencing (54). HOTAIR recruits PRC2 and LSD1 at the target gene, inducing gene silencing via H3K27-methylation and H3K4-demethylation (54,56). BRCA1, a critical player in DNA damage response and breast cancer, also interacts with EZH2 which in turn interacts with HOTAIR (54,57,58). Thus, BRCA1 and HOTAIR are both interacting partners of EZH2 and may have competitive roles in gene expression and DNA damage response (59). HOTAIR is also implicated in assembling E3-ubiquitin ligases during protein degradation (4,7,53). HOTAIR, EZH2, and LSD1 are all highly expressed in breast and other cancers. HOTAIR represses tumor suppressors such as PGR (Progesterone Receptor), PCDH10 (Protocadherin10), PCDHB5 (Protocadherin Beta 5), and JAM2 (Junctional Adhesion Molecule 2) (52). Post-translational functions of the HOTAIR have also been identified. HOTAIR induces ubiquitin-mediated proteolysis via interaction with E3 ubiquitin ligases Dzip3 and Mex3b, along with their respective ubiquitination substrates Ataxin-1 and Snurportin-1 (60). This leads to the degradation of Ataxin-1 and Snurportin-1 (60). Being an oncogenic lncRNA, its expression is correlated to tumor invasiveness and metastasis (53). HOTAIR serves as a diagnostic and prognostic marker for multiple cancers. HOTAIR also regulates the expression of miRNAs such as miR-130a (in gallbladder cancer cells) and others (4). Studies from our lab show that HOTAIR is required for the viability of breast cancer cells and its expression is transcriptionally regulated by estradiol via coordination of estrogen receptors (ERs) and ER-coregulators such as the MLL (mixed lineage leukemia)-family of histone methyltransferases and CBP/p300 (45,61–65). HOTAIR is also a target of endocrine disruption by estrogenic endocrine disruptors such as bisphenol-A (BPA) and diethylstilbestrol (DES) that may contribute to cancer (45,61,62).
ANRIL
ANRIL (Antisense Noncoding RNA) (a.k.a. CDKN2B-AS) is encoded in the chromosome 9p21 region at the INK4 locus (Tables 1 and 2) (66–78). Polymorphisms in the INK4 locus serve as a hotspot for a variety of diseases including cardiovascular disease, cancer, and diabetes. ANRIL is an antisense transcript of the CDKN2B gene (cyclin-dependent kinase inhibitor 2B) and controls cell proliferation and senescence via regulating its neighboring tumor suppressors CDKN2A/B by epigenetic mechanisms. This occurs through interacting with CBX7 (a PRC1 component) and SUZ12 (a PRC2 component) to induce gene silencing at the INK4b-ARF-INK4a locus (66). It also represses tumor suppressor p15. ANRIL is overexpressed in a variety of cancers including leukemia, breast cancer, and prostate cancer where CDKN2A/B show opposite patterns of expression (79).
5. Lung cancer
Lung cancer is the leading cause of cancer deaths and the second most common cancer in both men and women. Deaths caused by lung cancer exceed those of prostate, breast, and colon cancer combined. LncRNAs implicated in lung cancer include MALAT1, CCAT2, HOTAIR, AK126698, MEG3, SOX2-OT, HNF1A-AS1, ANRIL, H19, CARLo-5, MVIH, PVT1, EVADR, SPRY4-IT1, PANDAR, GAS5, BANCR, TUG1, and others (Figure 1 and Table 1) (80–82).
MALAT1
MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript) (a.k.a. NEAT2 Nuclear Enriched Abundant Transcript 2), a 7.5 kb long lncRNA, was originally found to be overexpressed in primary non-small cell lung cancers (83–91). MALAT1 is expressed in many tissues and is evolutionarily conserved among mammals. MALAT1 undergoes post-transcriptional processing to produce a short RNA (cytoplasmic mascRNA, MALAT1-associated small cytoplasmic RNA) and a long MALAT1 transcript that are localized to nuclear speckles and influence the level of phosphorylated splicing-associated SR (Serine Arginine) proteins. MALAT1 is also overexpressed in other cancers including bladder carcinoma, breast cancer, prostate cancer, and ovarian cancer, and is a potential biomarker and therapeutic target (85,91). Genome-wide analyses identified multiple mutations in the SRSF1-binding sites of MALAT1 in breast cancer, suggesting an alternation in the splicing pattern in these cancers (91).
Similar to NEAT2, NEAT1 transcripts are also associated with nuclear paraspeckles and are involved in transcriptional and post-transcriptional regulation of the expression of genes such as ADARB2 (Adenosine Deaminase, RNA Specific B2) (92–96). NEAT1 has two isoforms: a 3.7 kb (NEAT-1-1) and a 23 kb (NEAT-1-2) long isoform that are widely expressed in several tissues and overexpressed in breast cancer and acute myeloid leukemia. NEAT1 knockdown affects the viability and morphology of Burkitts lymphoma cells (97).
6. Colorectal cancer
Colorectal cancer (CRC) is currently the third most common malignancy worldwide. LncRNAs associated with colorectal cancer include H19, KCNQ1OT1, HOTAIR, MALAT1, ZFAS1, CCAT1, CCAT2, OCC-1, CCAT1-L, CRNDE, T-UCR, and others (98–100) (Figure 1 and Table 1).
H19
H19 (2.7 kb) is one of the first lncRNAs discovered and a pivotal player in embryonic development and tumorigenesis (1,101–117). It is a maternally expressed and paternally imprinted gene located near the telomeric region of chromosome 11p15.5 adjacent to IGF2 (Insulin like Growth Factor 2) gene. H19 is conserved between rodents and humans. MiR-675, a highly conserved miRNA that regulates a variety of transcripts, resides within exon-1 of the H19 gene (104). H19 acts as a decoy for miRNAs, modulating their availability and activity. It interacts with transcription-repressors such as EZH2 and MBD1 (methyl-CpG–binding domain protein 1) and induces repression by recruiting them to target genes (including H19’s reciprocally imprinted gene IGF2) (104). H19 is an oncogenic RNA associated with tumorigenesis starting from the early stages to metastasis (101,102,107,115, 118, 119). Tumor suppressor p53 and H19 are mutually counter-regulated (59). P53 represses the H19 gene and the H19-derived miR-675 inhibits p53 and p53-dependent protein expression (116). The p53-H19 interplay appears to play major roles in tumorigenesis and metastasis (102,103). H19 expression is induced by hypoxic stress and linked with epithelial to mesenchymal transition (EMT), and its overexpression leads to the activation of genes involved in angiogenesis, cell survival, and proliferation, triggering malignancies such as liver, breast, colorectal, esophageal, lung, pancreatic, gastric, bladder, and cervical carcinomas (101,102,108).
KCNQ1OT1
KCNQ1OT1 (KCNQ1 Overlapping Transcript 1) is a 91 kb nuclear antisense lncRNA that is imprinted from the paternal allele and originates from intron 11 of the KCNQ1 gene (potassium voltage-gated channel subfamily Q member 1) (120–125). The KCNQ1OT1 domain is regulated by a functionally independent imprinting control region (ICR) located in an intron of KCNQ1 (125). The promoter of the KCNQ1OT1 gene, located within the ICR locus, undergoes methylation on the maternally-inherited chromosome and demethylation on the paternally-inherited chromosome. Therefore, it preferentially allows the KCNQ1OT1 gene expression from the paternal allele (123,125). It interacts with chromatin modifying enzymes like PRC1, PRC2, and G9a and regulates the silencing of KCNQ1 via induction of histone and DNA methylation (123,125). The aberration in KCNQ1OT1 is associated with Beckwith-Wiedemann syndrome, colorectal, hepatocellular, and pediatric adrenocortical tumors (125,126).
T-UCRs
T-UCR lncRNAs are about 200 to 779 nt in length and are generated from ultraconserved regions (UCRs) and show tissue-specific expression patterns (127,128). T-UCR lncRNAs are altered in a variety of cancers including colorectal carcinoma, chronic lymphocytic leukemia, neuroblastomas, hepatocellular carcinoma, and prostate cancer (128). They play a key role in the suppression of miRNAs such as miR-596 and miR-193b involved in carcinogenesis and apoptosis, respectively (129–132). Modulation of T-UCR expression promotes colorectal carcinoma progression (4,7,133). Notably, the CpG island hypermethylation-induced epigenetic silencing of tumor suppressor miRNAs appears to be closely associated with a variety of cancers. Recent studies also demonstrate that in addition to miRNAs, various lncRNAs such as T-UCRs are silenced via CpG island hypermethylation, which is a common feature of many tumor types (133,134). Further, the CpG island methylation-induced silencing of protein coding and noncoding sequences in the sense strand as well as antisense-transcripts (many antisense lncRNA) are closely associated with human tumors. For example antisense lncRNA VIM-AS1 (vimentin antisense 1), which is regulated via R-loop (three-stranded RNA-DNA hybrid) formation, is silenced in colorectal cancer through CpG island hypermethylation (135).
CCAT1
CCAT1 (Colon Cancer-Associated Transcript-1) (a.k.a. CARLo-5) is an oncogenic lncRNA located at 8q24.21. CCAT1 expression is induced by c-Myc that binds to its promoter. CCAT1 epigenetically downregulates c-Myc by acting as a competing endogenous RNA (ceRNA) for miR-155 that represses c-Myc expression. It is also involved in the regulation of HOXB13 and SPRY4 (136–138). CCAT1 has been implicated in acute myeloid leukemia (AML), colorectal, esophageal, lung, and other cancers (139).
7. Liver cancer
Hepatocellular carcinoma (HCC) is a leading cause of cancer deaths with an incidence that has tripled since 1980. Although many lncRNAs are implicated in HCC, the most studied are MALAT1, HULC, HEIH, and HOTAIR that are known to be upregulated in HCC (140,141). Other lncRNAs implicated in liver cancer are lncTCF7, CCAT1, MEG3, CUDR, LALR1, and others (142) (Figure 1 and Table 1).
HULC
HULC (Highly Up-regulated in Liver Cancer), a 1.6kb oncogenic lncRNA, is overexpressed in HCC (100) (143,144). Augmented levels of the HULC transcript are observed in metastatic liver nodules from colon cancer. HULC is upregulated in both tumors and plasma of HCC patients, and is a potential biomarker for HCC. The SNP (single nucleotide polymorphism) in HULC is associated with HCC susceptibility in HBV (Hepatitis B Virus) carriers (145). HULC might function to downregulate the activity of miR-372 by acting as an endogenous sponge (146). Suppression of miR-372 by HULC represses the translational inhibition of miR-372 target genes. HULC promoter possesses a binding site for transcription factor cAMP response element binding (CREB) and its expression is potentially regulated by CREB phosphorylation (146).
HEIH
HEIH (High Expression in HCC), a 1.6 kb SP1-regulated long lncRNA located in the 5q34.3 locus, is differentially expressed in HCC, closely associated with HCC recurrence, and a prognostic factor for HCC (141,147,148). HEIH interacts with EZH2 and regulates EZH2 target genes including cell-cycle regulatory genes p15, p16, p21, and p57 (147). Knockdown of HEIH reduces cell proliferation and suppresses tumor growth (147).
Other lncRNAs implicated in liver cancer are DILC, H19, TCF7, HOTTIP, and ZFAS1 (141,149). DILC (downregulated in liver cancer) is a tumor suppressor whose expression is inversely related to those of EpCAM (epithelial cell adhesion molecule), CD24, and CD90 in hepatoma spheroids (150). HOTTIP (HOXA Transcript at the distal Tip) upregulation is associated with liver cancer metastasis (151,152). HOTTIP, in conjunction with the WDR5/MLL complex, mediates the trimethylation of H3K4 and HOXA gene expression (141,153,154).
8. Bladder cancer
Bladder cancer is the tenth most common malignancy in women and the fourth most common in men. LncRNAs implicated in bladder cancer are TUG1, UCA1, MALAT1, MEG3, H19, linc-UBC1, and others (155) (Figure 1 and Table 1).
UCA1
UCA1 (Urothelial Cancer Associated-1), transcribed from 19p13.12, was originally cloned from the human bladder cell line, and is overexpressed in embryonic tissues, bladder cancers, and other cancers (156) (157,158). It promotes chemoresistance through promoting the expression of wingless-type MMTV integration site family member 6 (Wnt6) (157). It also plays a role in β-catenin translocation into the nucleus and TCF7 regulation via interaction with SWI/SNF (switch/sucrose non-fermentable) in other types of cancer (159). UCA1 is a potential urine biomarker for noninvasive diagnosis of bladder cancer. MALAT1 associates with SUZ12 and regulates N-cadherin and E-cadherin expression, promotes tumor growth and metastasis, and forms a fusion gene in renal carcinoma (155).
9. Leukemia
Defects in hematopoietic stem cell differentiation and proliferation cause leukemia. A variety of lncRNAs are implicated in leukemia that include DLEU1, DLEU2, LUNAR1, HOTAIRM1, MALAT1, CCAT1, CCDC26, BGL3, NEAT1, NALT, UCA1, and others (160) (Figure 1 and Table 1). LncRNA mutations such as internal tandem duplications in the FLT3 (FMS-like tyrosine kinase 3) gene (FLT3-ITD) and mutations in the NPM1, CEBPA, IDH2, ASXL1, and RUNX1 genes are also linked to recurrent leukemia (161,162).
DLEU1 and DLEU2
LncRNAs DLEU1 and DLEU2 (Deleted in lymphocytic Leukemia 1 and 2), originating from the 13q14.3 region, are often deleted in solid tumors and hematopoietic malignancies like chronic lymphocytic leukemia (CLL) and lymphomas (163). DLEU1 and DLEU2 regulate NF-kB activity by regulating genes that affect NF-kB activity. The promoter regions of DLEU1 and DLEU2 exhibit demethylation or activation marks in CLL (163). DLEU2 acts as a precursor for various miRNAs such as miR-15a and miR-16-1 that are involved in CLL (164).
LUNAR1
LUNAR1 (Leukemia-induced Non-coding Activator RNA-1), derived from 15q26.3, is a NOTCH-regulated oncogenic lncRNA in T-cell acute lymphoblastic leukemia (T-ALL), and it promotes T-ALL cell growth by enhancing IGF1R expression and IGF1 signaling. LUNAR1 recruits the mediator complex on the IGF1R promoter and regulates its transcription. Abnormal NOTCH1 signaling is closely associated with human T-ALL (165,166).
BGL3
BGL3 (Beta Globin Locus transcript 3) is a 3.6 kb lncRNA derived from chromosome 11p15.4. BGL3 expression in leukemic cells is negatively regulated by Bcr-Abl through c-Myc-mediated DNA methylation (167). Conversely, BGL3 regulates Bcr-Abl through sequestering miR-17, miR-93, miR-20a, miR-20b, miR-106a, and miR-106b (167). These miRNAs are known to repress the expression of phosphatase and tensin homolog (PTEN) (168).
HOTAIRM1
HOTAIRM1 (HOTAIR Myeloid-specific 1), a 483 bp lncRNA transcribed from the HOXA cluster, is expressed in the myeloid lineage. Inhibition of HOTAIRM1 downregulates numerous HOXA genes critical for hematopoiesis (169–171). HOTAIRM1 has a similar expression pattern as that of HOXA1 and HOXA2 in thymus, muscle, colon, lung, kidney, spleen, etc. (172). HOTAIRM1 is induced by all trans retinoic acid (RA) and is involved in RA-induced myeloid differentiation. HOTAIRM1 regulates myeloid differentiation genes CD11b and CD18, and also interacts with chromatin modifying enzymes including PRC1, PRC2, and CBX1 (171).
XIST
XIST (X-Inactive Specific Transcript) induces X-inactivation and is aberrantly expressed in leukemia (161). Homozygous and heterozygous deletion of XIST in hematopoietic stem cells leads to the development of blood cancers, suggesting that aberrant X inactivation promotes carcinogenesis (161). It regulates genes in various other cancers via interaction with PRC1, PRC2, YY1, and CTCF, among others (129,149) (173,174). UCA1 knockdown negatively affects the proliferation of acute myeloid leukemia (AML) cells in vitro (149,175).
10. Other cancers
A large number of lncRNAs are identified in various other types of cancers, however, their detailed functions and specificity remain elusive (7) (Figure 1, Tables 1 and 2). For examples, pancreatic cancer, which accounts for 7% of cancer deaths worldwide, is associated with lncRNAs HOTAIR, HOTTIP, H19, PVT1, MALAT1, linc-RoR and others (176–178). Ovarian cancer, being the fifth deadliest cancer in women, is associated with abnormal expression of lncRNAs such as CCAT2, NEAT1, UCA1, HOTAIR, MEG3, HOST2, PVT1, and others (179,180). The lncRNAs implicated in renal cancer include HOTAIR, H19, GAS5, CADM-AS1, RCCRT1, NBAT1, MEG3, SPRY-IT1, MALAT1, and others (181,182). The lncRNAs implicated in gastric cancer include TUG1, MRUL, HOTAIR, MALAT1, CCAT1, H19, MEG3, HULC, PVT1, ANRIL, GAS5, and others (183–185). The expression of lncRNAs H19, MALAT1, CRNDE, and POU3F3 is positively correlated with malignant glioma (186). MEG3 is a tumor suppressor lncRNA that is highly expressed in normal brain tissue and downregulated in gliomas (187). FER1L4 (Fer-1-Like Protein 4) is a tumor suppressor lncRNA involved in the regulation of PTEN and inhibition of Akt phosphorylation in endometrial cancer (188). NBAT1 (Neuroblastoma-Associated Transcript 1) represses the expression of neuronal-specific transcription factor NRSF/REST through association with PRC2 (189,190).
GAS5
(Growth Arrest Specific 5) and SRA (Steroid receptor RNA Activator) are two lncRNAs implicated in hormone signaling (191–194). GAS5 produces two splice variant lncRNAs, and its introns also give rise to several snoRNAs (small nucleolar RNA) involved in the biosynthesis of ribosomal RNA from its introns. GAS5 interacts with glucocorticoid receptor (GR) and suppresses the expression of GR-regulated genes (195). It causes growth arrest and apoptosis and induces PTEN via inhibiting miR-103 (191). GAS5 acts as a tumor suppressor and its misregulation and genetic aberrations are associated with breast cancer, prostate cancer, leukemia, gastric cancer, and others (196). The lncRNA SRA interacts with various steroid hormone receptors and stimulates transcriptional activation, and is associated with breast, uterine, ovarian, and prostate cancers (197).
TERRA
TERRA (Telomeric Repeat-containing RNAs) is a set of lncRNAs (ranging in size from 100 bp–9 kb) transcribed from telomeres. LncRNAs containing UUAGGG repeats are generally called TERRA (198–201). TERRA interacts with telomere-associated TRF1 and TRF2 (telomere repeat factors 1 and 2), subunits of the origin recognition complex (ORC), heterochromatin protein 1 (HP1), H3K9-methylated histone, and facilitates heterochromatin formation at telomeres. TERRA is known to negatively regulate telomerase and act as a tumor suppressor (200,201).
ZFAS1
ZFAS1 (ZNFX1 Antisense RNA 1) is a spliced and polyadenylated lncRNA transcribed from the 5’ end of ZNFX1. It is derived from chromosome 20q13.13, and is implicated in different types of cancer including gastric cancer, colorectal cancer, and hepatocellular cancer, among others. It interacts with CDK1 and cyclin B to control p53-dependent cell cycle regulation (202). In addition, it promotes cell proliferation by recruiting EZH2 and LSD1/CoREST to the promoters of genes including KLF2 (Kruppel like factor 2) and NKD2 (naked cuticle 2) to regulate their expression (203). It also acts as a sponge for tumor suppressor miR-150 (204). Knockdown of ZFAS1 results in the repression of cell proliferation, migration and colony formation (203,205).
PVT1
PVT1 (Plasmacytoma Variant Translocation 1) is an oncogenic, intergenic lncRNA derived from 8q24.21 with multiple splice isoforms (206–208). It is upregulated in different types of cancer such as ovarian cancer, cervical cancer, and pancreatic cancer, among others. It suppresses the phosphorylation of Myc, thereby enhancing its stability (209). Further, it promotes proliferation via interaction with NOP2 (nucleolar protein 2 homolog) with the help of TGFβ (transforming growth factor β) (206). PVT1 promotes cell proliferation and invasion in gastric cancer by recruiting EZH2 to repress the expression of tumor suppressor genes p15 and p16 (207). It associates with a multifunctional DNA and RNA binding protein called nucleolin involved in oncogene expression and ribosomal biogenesis, among other activities (210).
MEG3
MEG3 (Maternally Expressed 3) is an imprinted, tumor suppressive lncRNA transcribed from chromosome 14q32.2 (211–214). It is a polyadenylated lncRNA overexpressed in human pituitary, but downregulated in cancer cells (212). Overexpression of MEG3 in bladder cancer cells has been shown to induce autophagy and increase cell proliferation (215). MEG3 is involved in the accumulation of tumor suppressor p53 and regulation of TGF-β pathway genes involved in cell invasion, immune regulation, etc. It also interacts with PRC2 to repress MDM2 (murine double minute 2) which contributes to p53 accumulation (214,216).
11. LncRNAs as biomarkers and in gene therapy
Numerous lncRNAs are aberrantly expressed in various tumors and some appear to be cancer-specific. Many lncRNAs (or their processed fragments) are stable in body fluids and detectable in the plasma and urine of cancer patients (24,217). Their levels are indicative of the severity of the disease. All these factors render lncRNAs an attractive choice for their applications as non-invasive biomarkers and therapeutic targets for the treatment of cancer (Table 2) (28,30,92,145,205,218–247). LncRNAs differ from protein-coding genes in many respects. First, due to their greater abundance than protein coding genes, a modulation in larger number of lncRNA expression may be observed in a given subtype of cancer, which provides a larger window for the detection of subtype-specific lncRNA-based biomarker. Subtype/tissue-specific lncRNA expressions are crucial for developing novel diagnostic biomarker and personalized therapy (43,238). LncRNAs, being large in size, may fold into complex secondary/tertiary structures and scaffolds through which they may interact with various proteins, transcriptional regulators, mRNA (complementary), and DNA sequences, which may aid in cancer initiation and progression. The presence of a large number of regulatory interaction sites in lncRNAs provides a wider platform for developing novel structure-based cancer drugs. Furthermore, given their participation in diverse cell signaling pathways and tissue-specific expression, lncRNAs can be utilized to formulate novel strategies for specific cancer subtype diagnosis and targeting (248,249).
Few lncRNAs are already implicated as biomarkers and some of them are in clinical trials (223,250) (Table 2). For example, lncRNA PCA3 which is highly upregulated and specific to prostate cancer is detectable in urine with levels that correspond to the severity of prostate cancer (30,31,218). Since it can be detected in urine, PCA3 has advantages over the widely-used serum-based prostate cancer biomarker PSA (prostate-specific antigen) for noninvasive diagnosis of prostate cancer (251). Additionally, PCAT-1, PRNCR1, PCGEM, PlncRNA1, and PCAT-18 are highly expressed in prostate tumors and are potential diagnostic markers (Table 2) (44,252). Circulating HOTAIR may also be used to diagnose breast cancer (221). ZFAS1, HIF1a-AS2, and others are also implicated as biomarkers for breast cancer (Table 2). Similarly, MALAT1, UCA1, ANRIL, and NEAT1 can be used to predict early stage as well as metastatic lung cancers (Table 2) (85). The expression of HOTAIR, CCAT1, FER1L4, and others is linked to colorectal cancer (CRC) (Table 2). CpG-island methylation of T-UCR promoter is also linked to CRC diagnosis. LncRNAs H19, HULC, HEIH, linc00152, and MVIH are highly upregulated in hepatocellular cancer (HCC) and are valuable HCC biomarkers (Table 2) (253). HULC expression correlates with histological grade and oncoprotein hepatitis B Virus X (HBx) (254). Hepatitis B virus (HBV)-positive hepatocellular cancer can be detected using lncRNAs uc001ncr and AX800134. Uc001ncr and AX800134 have a 100% detection rate in HCC patients (145). HOTAIR overexpression may be used to predict the recurrence of HCC and is highly expressed in 65.7% of recurrence HCC patients (140,255). UCA1, H19 and HOTAIR expression may be used as a biomarker to detect bladder cancer (Table 2) (175). CRNDE is expressed in the APL (acute promyelocytic leukemia) subtype of AML ten times more than the other subtypes. This makes CRNDE a suitable biomarker to detect the APL subtype of AML (238). LET, PVT1, PANDAR, and PTENP1 expression is linked to renal cancer (Table 2). Thus, lncRNAs appear to be promising novel diagnostic and prognostic markers for a variety of cancers (Table 2), however, there are still many challenges and validations required for their clinical applications.
As lncRNA expressions are differentially modulated in different types of cancer and their expression levels correlate with tumorigenesis, tumor aggressiveness, and stages, they are potential targets for cancer therapy. There are several ways by which lncRNAs may be targeted to modulate their expression: a) LncRNA transcript degradation/destabilization by using lncRNA-specific siRNAs, antisense oligonucleotide (ASO), gapmers, and ribozymes; b) Modulating lncRNA transcription by altering the lncRNA-coded promoter activity (e.g., via inhibition of transcription factors binding to respective promoters); c) Blocking interactions between lncRNAs and regulatory factors - small synthetic molecules/peptides can be developed that are designed to block the binding of lncRNAs with protein, DNA, RNA, or other interacting complexes by associating with specific binding pockets; and d) Functional disruption of lncRNAs using aptamers that can be selected to bind at specific structural regions to target lncRNAs and antagonize their association with their binding partners (256,257). For example, siRNA-mediated downregulation of HOTAIR expression leads to reduced tumor cell viability and invasiveness and induction of apoptosis in breast tumors (221). CCAT2 is upregulated in colorectal cancer and has been targeted by specific miRNAs to suppress colorectal cancer growth (258–260). Antisense-mediated silencing of MALAT1 prevents in vivo lung cancer metastasis (85). Breast cancer progression can be hindered through systemic knockdown of MALAT1 using antisense oligonucleotide (85,91,194). Antisense-mediated lncRNA targeting has shown to be promising in the treatment of other disorders like Angelman’s syndrome through silencing lncRNA UBE3A-AS (261,262). Oncogenic lncRNA H19 is overexpressed in a variety of cancers such as pancreatic tumors. The H19 promoter has been used to express diphtheria toxin (DTA) in pancreatic cancer cells (118,119,263). Administration of pancreatic tumors with a H19-DTA plasmid construct resulted in a significant decrease in tumor size and metastasis. The H19 (and IGF2) regulatory sequences can be used to inhibit the growth and metastasis of CRC. Overall, lncRNA-based targeted cancer therapies are promising, however, at present, they are at their infancy and require further development of experimental strategies, siRNA/antisense delivery strategies, screening novel small molecules libraries, and many clinical trials prior to their success in targeted, lncRNA-based gene therapy.
Apart from evaluating the direct significance of lncRNAs in cancer diagnosis and therapy, they can also be considered for improving therapeutic efficacy and development of combination therapy. Therapeutic resistance (such as chemo- or radio-resistance) is a major challenge in cancer treatment; however, this could be improved by increasing the therapeutic sensitivity of tumors by modulating a critical cell signaling pathway that confers resistance. As lncRNAs are closely associated with many cell signaling processes, the modulation of their expression could be done to improve the therapeutic sensitivity of tumors. One approach is to resensitize chemoresistant cells by modulating factors associated with DNA damage response pathways. For example, knockdown of HOTAIR enhances the sensitivity of cancer cells to chemotherapeutic agents like cisplatin and doxorubicin (264–266). Cisplatin-mediated upregulation of HOTAIR in human lung adenocarcinoma cells suppressed p21 (WAF1/CIP1) signaling pathway and caused a G0/G1 arrest by modulating the p53 expression and HOXA1 methylation (157,267). LncRNA TUG1 (Taurine Upregulated Gene 1, (2,3,268–270)) overexpression is responsible for the chemoresistance of lung cancer cells. TUG1 regulates the expression of LIM-kinase 2b and other cell cycled-associated genes through recruiting EZH2 to its promoter. TUG1 knockdown has been shown to enhance chemosensitivity in lung cancer (271). Silencing CRNDE results in the suppression of cell proliferation and chemoresistance in colorectal cancer. CRNDE inhibits the expression of miR-181a-5p, which in turn silences Wnt/β-catenin signaling (272). Similarly, HOTTIP promotes chemoresistance via activation of Wnt/β-catenin signaling (273). GAS5 modulates chemoresistance in gastric cancer by acting as a sponge for miR-23a that inhibits the expression of metallothionein 2A (MT2A) (274). In a similar role, CCAT1 sponges let-7c-mediated release of Bcl-xL. This involves EMT and resistance to docetaxel (137). MALAT1 knockdown causes re-sensitization of glioblastoma multiforme cells to temozolomide. The MALAT1-mediated chemoresistance in glioblastoma multiforme cells is made possible via inhibition of miR-203, thereby activating the expression of thymidylate synthase (275). Other lncRNAs that may be targeted to increase the chemosensitivity of tumors include HULC (gastric cancer), H19 (breast cancer), ODRUL (osteosarcoma), OMRUL (lung cancer), and PVT1 (pancreatic cancer) (209,276–278). Thus, it is evident that the modulation of lncRNA expression can be exploited to improve the therapeutic sensitivity of tumors and may also be used for combination therapy.
12. Conclusions
LncRNAs are emerging stars in cancer, diagnosis, and therapy (279). The discovery of huge numbers of lncRNA, their wide-range of expression patterns in various types of cancer, their tumor-specificity, and their stability in circulating body fluids (plasma and urine) provide a new foundation for developing diagnosis and therapies for cancer. LncRNA expression may also be used to predict the cancer prognosis and patients outcome. LncRNAs are major regulators of chromatin dynamics and gene regulation, associated with a variety of cell signaling pathways, and their expressions are influenced by a variety of factors including hormones, nutrients, age, and sex (161,280–283). Aberrant expression, mutations and SNPs of lncRNAs are associated with tumorigenesis and metastasis. Some lncRNAs act as oncogenes, whereas others act as tumor suppressors (284). Oncogenic lncRNAs include PCA3, PCGEM1, PCAT1, PCAT18, CTBP-AS, SCHLAP1, HOTAIR, ANRIL, MALAT1, NEAT1, H19, KCNQ1OT1, lncTCF-7, HOTTIP, HULC, HEIH, TUG1, UCA1, PVT1, and LSINCT5 (279). Tumor suppressor lncRNAs include GAS5, MEG3, DILC, NBAT-1, DLEU1, DLEU2, TERRA, BGL3, and others. Novel lncRNAs are still being discovered (285). Thus, lncRNAs holds strong promise towards the discovery of novel diagnostics and therapeutics for cancer. However, there are still many challenges. First, given the large number of lncRNAs and their up- or down-regulation in various cancers, it is crucial to identify the most important lncRNAs associated with a specific types/subtype of cancer. Second, the field of lncRNAs is at its infancy at this point; the structural and functional information on most lncRNAs remain uncharacterized. Without detailed understanding on the structure and functions of lncRNAs, developing lncRNA-based therapies is like “shooting in the dark”. Additionally, unlike protein-coding genes, lncRNAs are poorly conserved across different species; therefore, the structural and functional information as well as the promising therapeutic strategies developed using in vitro and animal models may not be easily extended to immediate human application and may need detailed clinical studies. To fully explore the potential of lncRNAs in cancer diagnosis and targeted therapy, it is important to characterize each lncRNA in detail, identify their cellular functions, roles in diseases, and SNPs. The cause-effect relationships of each lncRNA need to be established for determining their tissue-specificity and linking them to tumor stage. The future studies on the use of lncRNAs as biomarkers and therapeutics should focus not only on their identification and functional characterization, but also on optimizing isolation procedures, characterizing variations by internal and external factors using large numbers of statistically significant patient cohorts, and development of proper animal models for testing and validations, prior to clinical trials. Development of technologies for efficient detection of lncRNAs and their tissue-specific delivery methods are critical to the success of the diagnostics and therapeutics. Recent advancements in CRISPR/Cas9 technologies for gene knockout, knock-in, and point mutations may facilitate understanding the biological roles of lncRNAs and aid in the development of lncRNA-based targeted cancer therapy. Nevertheless, discovering novel lncRNAs, identifying their function and association with various cancer subtypes, developing novel lncRNA-based strategies for diagnosis and targeted therapies appear very promising, bring a new paradigm in cancer research, and may emerge as a major therapeutic strategy for the treatment of cancer in the near future.
Acknowledgments
Funding: Mandal research is supported by National Institute of Health (1R15 ES019129-01).
We thank Mandal lab members for helpful discussions. We sincerely apologize to all the colleagues whose contributions are not cited here due to the limitation in the scope and length of the article.
References
- 1.Glassman ML, de Groot N, Hochberg A. Relaxation of imprinting in carcinogenesis. Cancer Genet Cytogenet. 1996;89:69–73. doi: 10.1016/0165-4608(95)00364-9. [DOI] [PubMed] [Google Scholar]
- 2.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhan A, Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856:151–64. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanfilippo PG, Hewitt AW. Translating the ENCyclopedia Of DNA Elements Project findings to the clinic: ENCODE's implications for eye disease. Clin Exp Ophthalmol. 2014;42:78–83. doi: 10.1111/ceo.12150. [DOI] [PubMed] [Google Scholar]
- 6.Tragante V, Moore JH, Asselbergs FW. The ENCODE project and perspectives on pathways. Genet Epidemiol. 2014;38:275–80. doi: 10.1002/gepi.21802. [DOI] [PubMed] [Google Scholar]
- 7.Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. 2014;9:1932–56. doi: 10.1002/cmdc.201300534. [DOI] [PubMed] [Google Scholar]
- 8.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–33. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Kevin C, Chang Howard Y. Molecular Mechanisms of Long Noncoding RNAs. Molecular cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. 2011;130:223–36. doi: 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Y, He D, Peng Z, Peng W, Shi W, Wang J, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Vicens Q, Westhof E. Biogenesis of Circular RNAs. Cell. 2014;159:13–4. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–87. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–12. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016;32:309–16. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Vitiello M, Tuccoli A, Poliseno L. Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol (Dordr) 2015;38:17–28. doi: 10.1007/s13402-014-0180-x. [DOI] [PubMed] [Google Scholar]
- 22.Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornfeld JW, Bruning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi T, Gao G, Cao Y. Long Noncoding RNAs as Novel Biomarkers Have a Promising Future in Cancer Diagnostics. Dis Markers. 2016;2016:9085195. doi: 10.1155/2016/9085195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015;28:529–40. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crea F, Watahiki A, Quagliata L, Xue H, Pikor L, Parolia A, et al. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 2014;5:764–74. doi: 10.18632/oncotarget.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warrick JI, Tomlins SA, Carskadon SL, Young AM, Siddiqui J, Wei JT, et al. Evaluation of tissue PCA3 expression in prostate cancer by RNA in situ hybridization--a correlative study with urine PCA3 and TMPRSS2-ERG. Mod Pathol. 2014;27:609–20. doi: 10.1038/modpathol.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chevli KK, Duff M, Walter P, Yu C, Capuder B, Elshafei A, et al. Urinary PCA3 as a predictor of prostate cancer in a cohort of 3,073 men undergoing initial prostate biopsy. J Urol. 2014;191:1743–8. doi: 10.1016/j.juro.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Vedder MM, de Bekker-Grob EW, Lilja HG, Vickers AJ, van Leenders GJ, Steyerberg EW, et al. The added value of percentage of free to total prostate-specific antigen, PCA3, and a kallikrein panel to the ERSPC risk calculator for prostate cancer in prescreened men. Eur Urol. 2014;66:1109–15. doi: 10.1016/j.eururo.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albino G, Capoluongo E, Rocchetti S, Palumbo S, Zuppi C, Cirillo-Marucco E. Evaluation of the diagnostic and predictive power of PCA3 in the prostate cancer. A different best cut-off in each different scenario. Preliminary results. Arch Ital Urol Androl. 2014;86:306–10. doi: 10.4081/aiua.2014.4.306. [DOI] [PubMed] [Google Scholar]
- 33.Dijkstra S, Leyten GH, Jannink SA, de Jong H, Mulders PF, van Oort IM, et al. KLK3, PCA3, and TMPRSS2-ERG expression in the peripheral blood mononuclear cell fraction from castration-resistant prostate cancer patients and response to docetaxel treatment. Prostate. 2014;74:1222–30. doi: 10.1002/pros.22839. [DOI] [PubMed] [Google Scholar]
- 34.Salameh A, Lee AK, Cardo-Vila M, Nunes DN, Efstathiou E, Staquicini FI, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S A. 2015;112:8403–8. doi: 10.1073/pnas.1507882112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25:135–41. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 37.Hung CL, Wang LY, Yu YL, Chen HW, Srivastava S, Petrovics G, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci U S A. 2014;111:18697–702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh AL, Tuzova AV, Bolton EM, Lynch TH, Perry AS. Long noncoding RNAs and prostate carcinogenesis: the missing 'linc'? Trends Mol Med. 2014;20:428–36. doi: 10.1016/j.molmed.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–8. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang R, Asangani IA, Chakravarthi BV, Ateeq B, Lonigro RJ, Cao Q, et al. Role of transcriptional corepressor CtBP1 in prostate cancer progression. Neoplasia. 2012;14:905–14. doi: 10.1593/neo.121192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weichenhan D, Plass C. The evolving epigenome. Hum Mol Genet. 2013;22:R1–6. doi: 10.1093/hmg/ddt348. [DOI] [PubMed] [Google Scholar]
- 42.Moiola CP, De Luca P, Zalazar F, Cotignola J, Rodriguez-Segui SA, Gardner K, et al. Prostate tumor growth is impaired by CtBP1 depletion in high-fat diet-fed mice. Clin Cancer Res. 2014;20:4086–95. doi: 10.1158/1078-0432.CCR-14-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5:9864–76. doi: 10.18632/oncotarget.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Wang X. Role of long noncoding RNAs in malignant disease (Review) Mol Med Rep. 2016;13:1463–9. doi: 10.3892/mmr.2015.4711. [DOI] [PubMed] [Google Scholar]
- 45.Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR is Transcriptionally Induced by Estradiol. J Mol Biol. 2013;425:3707–22. doi: 10.1016/j.jmb.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui L, Xie XY, Wang H, Chen XL, Liu SL, Hu LN. [Expression of long non-coding RNA HOTAIR mRNA in ovarian cancer] Sichuan Da Xue Xue Bao Yi Xue Ban. 2013;44:57–9. [PubMed] [Google Scholar]
- 47.Geng Y, Xie S, Li Q, Ma J, Wang G. Large Intervening Non-Coding RNA HOTAIR is Associated with Hepatocellular Carcinoma Progression. J Int Med Res. 2011;39:2119–28. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 48.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–25. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, et al. Long Intergenic Noncoding RNA HOTAIR Is Overexpressed and Regulates PTEN Methylation in Laryngeal Squamous Cell Carcinoma. The American journal of pathology. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 50.Nakagawa T, Endo H, Yokoyama M, Abe J, Tamai K, Tanaka N, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436:319–24. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 51.Yang Z, Zhou L, Wu L-M, Lai M-C, Xie H-Y, Zhang F, et al. Overexpression of Long Non-coding RNA HOTAIR Predicts Tumor Recurrence in Hepatocellular Carcinoma Patients Following Liver Transplantation. Ann Surg Oncol. 2011;18:1243–50. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 52.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xhemalce B. From histones to RNA: role of methylation in cancer. Brief Funct Genomics. 2013;12:244–53. doi: 10.1093/bfgp/els064. [DOI] [PubMed] [Google Scholar]
- 57.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long Noncoding RNA HOTAIR Regulates Polycomb-Dependent Chromatin Modification and Is Associated with Poor Prognosis in Colorectal Cancers. Cancer Res. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 58.Milhem MM, Knutson T, Yang S, Zhu D, Wang X, Leslie KK, et al. Correlation of MTDH/AEG-1 and HOTAIR Expression with Metastasis and Response to Treatment in Sarcoma Patients. J Cancer Sci Ther. 2011:S5. [PMC free article] [PubMed] [Google Scholar]
- 59.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol. 2014;141:160–70. doi: 10.1016/j.jsbmb.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhan A, Mandal SS. Estradiol-Induced Transcriptional Regulation of Long Non-Coding RNA, HOTAIR. Methods Mol Biol. 2016;1366:395–412. doi: 10.1007/978-1-4939-3127-9_31. [DOI] [PubMed] [Google Scholar]
- 63.Ansari KI, Hussain I, Das HK, Mandal SS. Overexpression of human histone methylase MLL1 upon exposure to a food contaminant mycotoxin, deoxynivalenol. FEBS J. 2009;276:3299–307. doi: 10.1111/j.1742-4658.2009.07055.x. [DOI] [PubMed] [Google Scholar]
- 64.Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is overexpressed in breast cancer and transcriptionally regulated by estrogen via involvement of histone methylases MLL3 and MLL4. J Mol Endocrinol. 2012;48:61–75. doi: 10.1530/JME-11-0078. [DOI] [PubMed] [Google Scholar]
- 65.Bhan A, Hussain I, Ansari KI, Bobzean SA, Perrotti LI, Mandal SS. Histone methyltransferase EZH2 is transcriptionally induced by estradiol as well as estrogenic endocrine disruptors bisphenol-A and diethylstilbestrol. J Mol Biol. 2014;426:3426–41. doi: 10.1016/j.jmb.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 66.Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71:5365–9. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iacobucci I, Sazzini M, Garagnani P, Ferrari A, Boattini A, Lonetti A, et al. A polymorphism in the chromosome 9p21 ANRIL locus is associated to Philadelphia positive acute lymphoblastic leukemia. Leuk Res. 2011;35:1052–9. doi: 10.1016/j.leukres.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 68.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Healy J, Bélanger H, Beaulieu P, Larivière M, Labuda D, Sinnett D. Promoter SNPs in G1/S checkpoint regulators and their impact on the susceptibility to childhood leukemia. Blood. 2007;109:683–92. doi: 10.1182/blood-2006-02-003236. [DOI] [PubMed] [Google Scholar]
- 72.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 SNPs Associated with Multiple Disease Phenotypes Correlate with ANRIL Expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Holdt LM, Beutner F, Scholz M, Gielen S, Gäbel G, Bergert H, et al. ANRIL Expression Is Associated With Atherosclerosis Risk at Chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–7. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 74.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25:444–8. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 75.Schaefer AS, Richter GM, Groessner-Schreiber B, Noack B, Nothnagel M, El Mokhtari NE, et al. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009;5:e1000378. doi: 10.1371/journal.pgen.1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lan WG, Xu DH, Xu C, Ding CL, Ning FL, Zhou YL, et al. Silencing of long non-coding RNA ANRIL inhibits the development of multidrug resistance in gastric cancer cells. Oncol Rep. 2016;36:263–70. doi: 10.3892/or.2016.4771. [DOI] [PubMed] [Google Scholar]
- 77.Sun Y, Zheng ZP, Li H, Zhang HQ, Ma FQ. ANRIL is associated with the survival rate of patients with colorectal cancer, and affects cell migration and invasion in vitro. Mol Med Rep. 2016;14:1714–20. doi: 10.3892/mmr.2016.5409. [DOI] [PubMed] [Google Scholar]
- 78.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–9. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 79.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 80.Peng Z, Zhang C, Duan C. Functions and mechanisms of long noncoding RNAs in lung cancer. Onco Targets Ther. 2016;9:4411–24. doi: 10.2147/OTT.S109549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu CH, Hsu CL, Lu PC, Lin WC, Juan HF, Huang HC. Identification of lncRNA functions in lung cancer based on associated protein-protein interaction modules. Sci Rep. 2016;6:35939. doi: 10.1038/srep35939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei MM, Zhou GB. Long Non-coding RNAs and Their Roles in Non-small-cell Lung Cancer. Genomics Proteomics Bioinformatics. 2016;14:280–8. doi: 10.1016/j.gpb.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–93. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guo F, Li Y, Liu Y, Wang J, Li G. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin (Shanghai) 2010;42:224–9. doi: 10.1093/abbs/gmq008. [DOI] [PubMed] [Google Scholar]
- 85.Gutschner T, Hämmerle M, Eißmann M, Hsu J, Kim Y, Hung G, et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han Y, Liu Y, Nie L, Gui Y, Cai Z. Inducing Cell Proliferation Inhibition, Apoptosis, and Motility Reduction by Silencing Long Noncoding Ribonucleic Acid Metastasis-associated Lung Adenocarcinoma Transcript 1 in Urothelial Carcinoma of the Bladder. Urology. 2013;81 doi: 10.1016/j.urology.2012.08.044. 209.e1-.e7. [DOI] [PubMed] [Google Scholar]
- 87.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 88.Lai M-c, Yang Z, Zhou L, Zhu Q-q, Xie H-y, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–6. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 89.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: Progress and prospects. The International Journal of Biochemistry & Cell Biology. 2013;45:1895–910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 90.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–97. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hu X, Bao J, Wang Z, Zhang Z, Gu P, Tao F, et al. The plasma lncRNA acting as fingerprint in non-small-cell lung cancer. Tumour Biol. 2016;37:3497–504. doi: 10.1007/s13277-015-4023-9. [DOI] [PubMed] [Google Scholar]
- 93.Jiang P, Wu X, Wang X, Huang W, Feng Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget. 2016 doi: 10.18632/oncotarget.9712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016 doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–83. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q, et al. NEAT1 is Required for Survival of Breast Cancer Cells Through FUS and miR-548. Gene Regul Syst Bio. 2016;10:11–7. doi: 10.4137/GRSB.S29414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Blume CJ, Hotz-Wagenblatt A, Hullein J, Sellner L, Jethwa A, Stolz T, et al. p53-dependent non-coding RNA networks in chronic lymphocytic leukemia. Leukemia. 2015;29:2015–23. doi: 10.1038/leu.2015.119. [DOI] [PubMed] [Google Scholar]
- 98.Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong H, et al. Long non-coding RNAs in colorectal cancer. Oncotarget. 2016;7:5226–39. doi: 10.18632/oncotarget.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu MD, Qi P, Du X. Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Mod Pathol. 2014;27:1310–20. doi: 10.1038/modpathol.2014.33. [DOI] [PubMed] [Google Scholar]
- 100.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 101.Ariel I, Sughayer M, Fellig Y, Pizov G, Ayesh S, Podeh D, et al. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol. 2000;53:320–3. doi: 10.1136/mp.53.6.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc Oncogene Directly Induces the H19 Noncoding RNA by Allele-Specific Binding to Potentiate Tumorigenesis. Cancer Res. 2006;66:5330–7. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 103.Berteaux N, Lottin S, Monté D, Pinte S, Quatannens B, Coll J, et al. H19 mRNA-like Noncoding RNA Promotes Breast Cancer Cell Proliferation through Positive Control by E2F1. J Biol Chem. 2005;280:29625–36. doi: 10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 104.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–6. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castle JC, Armour CD, Lower M, Haynor D, Biery M, Bouzek H, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hashimoto K, Azuma C, Tokugawa Y, Nobunaga T, Aki TA, Matsui Y, et al. Loss of H19 imprinting and up-regulation of H19 and SNRPN in a case with malignant mixed Mullerian tumor of the uterus. Hum Pathol. 1997;28:862–5. doi: 10.1016/s0046-8177(97)90162-3. [DOI] [PubMed] [Google Scholar]
- 107.Hibi K, Nakamura H, Hirai A, Fujikake Y, Kasai Y, Akiyama S, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480–2. [PubMed] [Google Scholar]
- 108.Kondo M, Suzuki H, Ueda R, Osada H, Takagi K, Takahashi T. Frequent loss of imprinting of the H19 gene is often associated with its overexpression in human lung cancers. Oncogene. 1995;10:1193–8. [PubMed] [Google Scholar]
- 109.Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, et al. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis. 2002;23:1885–95. doi: 10.1093/carcin/23.11.1885. [DOI] [PubMed] [Google Scholar]
- 110.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-lail R, Fellig Y, et al. The oncofetal H19 RNA connection: Hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–51. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 112.Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proceedings of the National Academy of Sciences. 1984;81:5523–7. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pachnis V, Brannan CI, Tilghman SM. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J. 1988;7:673–81. doi: 10.1002/j.1460-2075.1988.tb02862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Poirier F, Chan CT, Timmons PM, Robertson EJ, Evans MJ, Rigby PW. The murine H19 gene is activated during embryonic stem cell differentiation in vitro and at the time of implantation in the developing embryo. Development. 1991;113:1105–14. doi: 10.1242/dev.113.4.1105. [DOI] [PubMed] [Google Scholar]
- 115.Takeuchi S, Hofmann W-K, Tsukasaki K, Takeuchi N, Ikezoe T, Matsushita M, et al. Loss of H19 imprinting in adult T-cell leukaemia/lymphoma. Br J Haematol. 2007;137:380–1. doi: 10.1111/j.1365-2141.2007.06581.x. [DOI] [PubMed] [Google Scholar]
- 116.Yoshimizu T, Miroglio A, Ripoche M-A, Gabory A, Vernucci M, Riccio A, et al. The H19 locus acts in vivo as a tumor suppressor. Proceedings of the National Academy of Sciences. 2008;105:12417–22. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Amit D, Hochberg A. Development of targeted therapy for a broad spectrum of cancers (pancreatic cancer, ovarian cancer, glioblastoma and HCC) mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. Int J Clin Exp Med. 2012;5:296–305. [PMC free article] [PubMed] [Google Scholar]
- 119.Sorin V, Ohana P, Gallula J, Birman T, Matouk I, Hubert A, et al. H19-promoter-targeted therapy combined with gemcitabine in the treatment of pancreatic cancer. ISRN Oncol. 2012;2012:351750. doi: 10.5402/2012/351750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, et al. LIT1, an Imprinted Antisense RNA in the Human KvLQT1 Locus Identified by Screening for Differentially Expressed Transcripts Using Monochromosomal Hybrids. Hum Mol Genet. 1999;8:1209–17. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 121.Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T, et al. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer Sci. 2006;97:1147–54. doi: 10.1111/j.1349-7006.2006.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pandey RR, Ceribelli M, Singh PB, Ericsson J, Mantovani R, Kanduri C. NF-Y Regulates the Antisense Promoter, Bidirectional Silencing, and Differential Epigenetic Marks of the Kcnq1 Imprinting Control Region. J Biol Chem. 2004;279:52685–93. doi: 10.1074/jbc.M408084200. [DOI] [PubMed] [Google Scholar]
- 123.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 Antisense Noncoding RNA Mediates Lineage-Specific Transcriptional Silencing through Chromatin-Level Regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 124.Zhang H, Zeitz MJ, Wang H, Niu B, Ge S, Li W, et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J Cell Biol. 2014;204:61–75. doi: 10.1083/jcb.201304152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Robbins KM, Chen Z, Wells KD, Rivera RM. Expression of KCNQ1OT1, CDKN1C, H19, and PLAGL1 and the methylation patterns at the KvDMR1 and H19/IGF2 imprinting control regions is conserved between human and bovine. J Biomed Sci. 2012;19:95. doi: 10.1186/1423-0127-19-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.DeBaun MR, Niemitz EL, McNeil DE, Brandenburg SA, Lee MP, Feinberg AP. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am J Hum Genet. 2002;70:604–11. doi: 10.1086/338934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 128.Peng JC, Shen J, Ran ZH. Transcribed ultraconserved region in human cancers. RNA Biol. 2013;10:1771–7. doi: 10.4161/rna.26995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Terreri S, Durso M, Colonna V, Romanelli A, Terracciano D, Ferro M, et al. New Cross-Talk Layer between Ultraconserved Non-Coding RNAs, MicroRNAs and Polycomb Protein YY1 in Bladder Cancer. Genes (Basel) 2016;7 doi: 10.3390/genes7120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang C, Wang Z, Zhou J, Liu S, Wu C, Huang C, et al. TUC.338 promotes invasion and metastasis in colorectal cancer. Int J Cancer. 2017;140:1457–64. doi: 10.1002/ijc.30542. [DOI] [PubMed] [Google Scholar]
- 131.Jiang BC, Yang T, He LN, Tao YX, Gao YJ. Altered T-UCRs expression profile in the spinal cord of mice with neuropathic pain. Transl Perioper Pain Med. 2016;1:1–10. [PMC free article] [PubMed] [Google Scholar]
- 132.Braconi C, Valeri N, Kogure T, Gasparini P, Huang N, Nuovo GJ, et al. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108:786–91. doi: 10.1073/pnas.1011098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liz J, Portela A, Soler M, Gomez A, Ling H, Michlewski G, et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell. 2014;55:138–47. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 135.Boque-Sastre R, Soler M, Oliveira-Mateos C, Portela A, Moutinho C, Sayols S, et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci U S A. 2015;112:5785–90. doi: 10.1073/pnas.1421197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang E, Han L, Yin D, He X, Hong L, Si X, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen J, Zhang K, Song H, Wang R, Chu X, Chen L. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget. 2016;7:62474–89. doi: 10.18632/oncotarget.11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Guo X, Hua Y. CCAT1: an oncogenic long noncoding RNA in human cancers. J Cancer Res Clin Oncol. 2017;143:555–62. doi: 10.1007/s00432-016-2268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–28. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 141.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–7. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 142.Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78. doi: 10.1016/j.pharmthera.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Panzitt K, Tschernatsch MMO, Guelly C, Moustafa T, Stradner M, Strohmaier HM, et al. Characterization of HULC, a Novel Gene With Striking Up-Regulation in Hepatocellular Carcinoma, as Noncoding RNA. Gastroenterology. 2007;132:330–42. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 144.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–11. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang K, Guo WX, Li N, Gao CF, Shi J, Tang YF, et al. Serum LncRNAs Profiles Serve as Novel Potential Biomarkers for the Diagnosis of HBV-Positive Hepatocellular Carcinoma. PLoS One. 2015;10:e0144934. doi: 10.1371/journal.pone.0144934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–83. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–89. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 148.Zhang Y, Li Z, Zhang Y, Zhong Q, Chen Q, Zhang L. Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. Int J Clin Exp Med. 2015;8:12956–62. [PMC free article] [PubMed] [Google Scholar]
- 149.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–66. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 150.Wang X, Sun W, Shen W, Xia M, Chen C, Xiang D, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. 2016;64:1283–94. doi: 10.1016/j.jhep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 151.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: from function to translation. Trends Cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang SR, Yang JK, Xie JK, Zhao LC. Long noncoding RNA HOTTIP contributes to the progression of prostate cancer by regulating HOXA13. Cell Mol Biol (Noisy-le-grand) 2016;62:84–8. [PubMed] [Google Scholar]
- 153.Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–23. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016 doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen P, Wan D, Zheng D, Zheng Q, Wu F, Zhi Q. Long non-coding RNA UCA1 promotes the tumorigenesis in pancreatic cancer. Biomed Pharmacother. 2016;83:1220–6. doi: 10.1016/j.biopha.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 157.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, et al. Long non-coding RNA UCA1 increases chemoresistance of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281:1750–8. doi: 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- 158.Liu H, Wang G, Yang L, Qu J, Yang Z, Zhou X. Knockdown of Long Non-Coding RNA UCA1 Increases the Tamoxifen Sensitivity of Breast Cancer Cells through Inhibition of Wnt/beta-Catenin Pathway. PLoS One. 2016;11:e0168406. doi: 10.1371/journal.pone.0168406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–25. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 160.Hu S, Shan G. LncRNAs in Stem Cells. Stem Cells Int. 2016;2016:2681925. doi: 10.1155/2016/2681925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Alvarez-Dominguez JR, Hu W, Gromatzky AA, Lodish HF. Long noncoding RNAs during normal and malignant hematopoiesis. Int J Hematol. 2014;99:531–41. doi: 10.1007/s12185-014-1552-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–8. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mertens D, Philippen A, Ruppel M, Allegra D, Bhattacharya N, Tschuch C, et al. Chronic lymphocytic leukemia and 13q14: miRs and more. Leuk Lymphoma. 2009;50:502–5. doi: 10.1080/10428190902763509. [DOI] [PubMed] [Google Scholar]
- 164.Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013;9:e1003373. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606. doi: 10.1016/j.cell.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Peng W, Feng J. Long noncoding RNA LUNAR1 associates with cell proliferation and predicts a poor prognosis in diffuse large B-cell lymphoma. Biomed Pharmacother. 2016;77:65–71. doi: 10.1016/j.biopha.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 167.Guo G, Kang Q, Zhu X, Chen Q, Wang X, Chen Y, et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34:1768–79. doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
- 168.Xia T, Chen S, Jiang Z, Shao Y, Jiang X, Li P, et al. Long noncoding RNA FER1L4 suppresses cancer cell growth by acting as a competing endogenous RNA and regulating PTEN expression. Sci Rep. 2015;5:13445. doi: 10.1038/srep13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M, et al. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016;20:2036–44. doi: 10.1111/jcmm.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang XQ, Dostie J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014;11:777–87. doi: 10.4161/rna.28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wei S, Zhao M, Wang X, Li Y, Wang K. PU.1 controls the expression of long noncoding RNA HOTAIRM1 during granulocytic differentiation. J Hematol Oncol. 2016;9:44. doi: 10.1186/s13045-016-0274-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cerase A, Pintacuda G, Tattermusch A, Avner P. Xist localization and function: new insights from multiple levels. Genome Biol. 2015;16:166. doi: 10.1186/s13059-015-0733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Huang KC, Rao PH, Lau CC, Heard E, Ng SK, Brown C, et al. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol Cancer Ther. 2002;1:769–76. [PubMed] [Google Scholar]
- 175.Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–27. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 176.Kladi-Skandali A, Michaelidou K, Scorilas A, Mavridis K. Long Noncoding RNAs in Digestive System Malignancies: A Novel Class of Cancer Biomarkers and Therapeutic Targets? Gastroenterol Res Pract. 2015;2015:319861. doi: 10.1155/2015/319861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, et al. Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer. 2011;10:141. doi: 10.1186/1476-4598-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ren C, Li X, Wang T, Wang G, Zhao C, Liang T, et al. Functions and Mechanisms of Long Noncoding RNAs in Ovarian Cancer. Int J Gynecol Cancer. 2015;25:566–9. doi: 10.1097/IGC.0000000000000413. [DOI] [PubMed] [Google Scholar]
- 180.Zhong Y, Gao D, He S, Shuai C, Peng S. Dysregulated Expression of Long Noncoding RNAs in Ovarian Cancer. Int J Gynecol Cancer. 2016;26:1564–70. doi: 10.1097/IGC.0000000000000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Blondeau JJ, Deng M, Syring I, Schrodter S, Schmidt D, Perner S, et al. Identification of novel long non-coding RNAs in clear cell renal cell carcinoma. Clin Epigenetics. 2015;7:10. doi: 10.1186/s13148-015-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Seles M, Hutterer GC, Kiesslich T, Pummer K, Berindan-Neagoe I, Perakis S, et al. Current Insights into Long Non-Coding RNAs in Renal Cell Carcinoma. Int J Mol Sci. 2016;17:573. doi: 10.3390/ijms17040573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Gu W, Gao T, Sun Y, Zheng X, Wang J, Ma J, et al. LncRNA expression profile reveals the potential role of lncRNAs in gastric carcinogenesis. Cancer Biomark. 2015;15:249–58. doi: 10.3233/CBM-150460. [DOI] [PubMed] [Google Scholar]
- 184.Wan X, Ding X, Chen S, Song H, Jiang H, Fang Y, et al. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumour Biol. 2015;36:521–32. doi: 10.1007/s13277-015-3136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang E, He X, Yin D, Han L, Qiu M, Xu T, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. doi: 10.1038/cddis.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kraus TF, Greiner A, Guibourt V, Lisec K, Kretzschmar HA. Identification of Stably Expressed lncRNAs as Valid Endogenous Controls for Profiling of Human Glioma. J Cancer. 2015;6:111–9. doi: 10.7150/jca.10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113:1868–74. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 188.Qiao Q, Li H. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem Biophys Res Commun. 2016;478:507–12. doi: 10.1016/j.bbrc.2016.06.160. [DOI] [PubMed] [Google Scholar]
- 189.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–37. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 190.Xue S, Li QW, Che JP, Guo Y, Yang FQ, Zheng JH. Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2015;8:3765–74. [PMC free article] [PubMed] [Google Scholar]
- 191.Guo C, Song WQ, Sun P, Jin L, Dai HY. LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J Biomed Sci. 2015;22:100. doi: 10.1186/s12929-015-0213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Isin M, Uysaler E, Ozgur E, Koseoglu H, Sanli O, Yucel OB, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6:168. doi: 10.3389/fgene.2015.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 194.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–5. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li W, Zhai L, Wang H, Liu C, Zhang J, Chen W, et al. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7:27778–86. doi: 10.18632/oncotarget.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pickard MR, Williams GT. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes (Basel) 2015;6:484–99. doi: 10.3390/genes6030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Yan R, Wang K, Peng R, Wang S, Cao J, Wang P, et al. Genetic variants in lncRNA SRA and risk of breast cancer. Oncotarget. 2016;7:22486–96. doi: 10.18632/oncotarget.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat–Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 199.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA Binding to TRF2 Facilitates Heterochromatin Formation and ORC Recruitment at Telomeres. Mol Cell. 2009;35:403–13. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Maicher A, Kastner L, Luke B. Telomeres and disease: enter TERRA. RNA Biol. 2012;9:843–9. doi: 10.4161/rna.20330. [DOI] [PubMed] [Google Scholar]
- 201.Arora R, Brun CM, Azzalin CM. TERRA: Long Noncoding RNA at Eukaryotic Telomeres. Prog Mol Subcell Biol. 2011;51:65–94. doi: 10.1007/978-3-642-16502-3_4. [DOI] [PubMed] [Google Scholar]
- 202.Thorenoor N, Faltejskova-Vychytilova P, Hombach S, Mlcochova J, Kretz M, Svoboda M, et al. Long non-coding RNA ZFAS1 interacts with CDK1 and is involved in p53-dependent cell cycle control and apoptosis in colorectal cancer. Oncotarget. 2016;7:622–37. doi: 10.18632/oncotarget.5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Nie F, Yu X, Huang M, Wang Y, Xie M, Ma H, et al. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2016 doi: 10.18632/oncotarget.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181–91. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 205.Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–91. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Colombo T, Farina L, Macino G, Paci P. PVT1: a rising star among oncogenic long noncoding RNAs. Biomed Res Int. 2015;2015:304208. doi: 10.1155/2015/304208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu E, Liu Z, Zhou Y, Mi R, Wang D. Overexpression of long non-coding RNA PVT1 in ovarian cancer cells promotes cisplatin resistance by regulating apoptotic pathways. Int J Clin Exp Med. 2015;8:20565–72. [PMC free article] [PubMed] [Google Scholar]
- 209.You L, Chang D, Du HZ, Zhao YP. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem Biophys Res Commun. 2011;407:1–6. doi: 10.1016/j.bbrc.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 210.Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 Contributes to the Cervical Cancer Phenotype and Associates with Poor Patient Prognosis. PLoS One. 2016;11:e0156274. doi: 10.1371/journal.pone.0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gejman R, Batista DL, Zhong Y, Zhou Y, Zhang X, Swearingen B, et al. Selective Loss of MEG3 Expression and Intergenic Differentially Methylated Region Hypermethylation in the MEG3/DLK1 Locus in Human Clinically Nonfunctioning Pituitary Adenomas. J Clin Endocrinol Metab. 2008;93:4119–25. doi: 10.1210/jc.2007-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, et al. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–20. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- 213.Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, et al. Maternally Expressed Gene 3, an Imprinted Noncoding RNA Gene, Is Associated with Meningioma Pathogenesis and Progression. Cancer Res. 2010;70:2350–8. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 Non-coding RNA. J Biol Chem. 2007;282:24731–42. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 215.Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, et al. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol Biosyst. 2013;9:407–11. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 216.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014;455:43–57. doi: 10.1016/j.bbrc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, et al. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–8. [PubMed] [Google Scholar]
- 219.Ilboudo A, Chouhan J, McNeil BK, Osborne JR, Ogunwobi OO. PVT1 Exon 9: A Potential Biomarker of Aggressive Prostate Cancer? Int J Environ Res Public Health. 2015;13 doi: 10.3390/ijerph13010012. ijerph13010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Orfanelli U, Jachetti E, Chiacchiera F, Grioni M, Brambilla P, Briganti A, et al. Antisense transcription at the TRPM2 locus as a novel prognostic marker and therapeutic target in prostate cancer. Oncogene. 2015;34:2094–102. doi: 10.1038/onc.2014.144. [DOI] [PubMed] [Google Scholar]
- 221.Zhang L, Song X, Wang X, Xie Y, Wang Z, Xu Y, et al. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res Treat. 2015;152:199–208. doi: 10.1007/s10549-015-3431-2. [DOI] [PubMed] [Google Scholar]
- 222.Jiang YZ, Liu YR, Xu XE, Jin X, Hu X, Yu KD, et al. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Cancer Res. 2016;76:2105–14. doi: 10.1158/0008-5472.CAN-15-3284. [DOI] [PubMed] [Google Scholar]
- 223.TA(E)C-GP Versus A(E)C-T for the High Risk TNBC Patients and Validation of the mRNA-lncRNA Signature. https://ClinicalTrials.gov/show/NCT02641847.
- 224.Weber DG, Johnen G, Casjens S, Bryk O, Pesch B, Jockel KH, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Guo F, Yu F, Wang J, Li Y, Li Y, Li Z, et al. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed Rep. 2015;3:309–12. doi: 10.3892/br.2015.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int J Clin Exp Med. 2015;8:11824–30. [PMC free article] [PubMed] [Google Scholar]
- 227.Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H, et al. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30:303–10. doi: 10.1093/mutage/geu076. [DOI] [PubMed] [Google Scholar]
- 228.Zhao W, Song M, Zhang J, Kuerban M, Wang H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int J Clin Exp Pathol. 2015;8:14131–40. [PMC free article] [PubMed] [Google Scholar]
- 229.Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F, et al. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer Sci. 2015;106:1323–32. doi: 10.1111/cas.12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shi J, Li X, Zhang F, Zhang C, Guan Q, Cao X, et al. Circulating lncRNAs associated with occurrence of colorectal cancer progression. Am J Cancer Res. 2015;5:2258–65. [PMC free article] [PubMed] [Google Scholar]
- 231.Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687–96. doi: 10.1159/000430387. [DOI] [PubMed] [Google Scholar]
- 232.Tang J, Jiang R, Deng L, Zhang X, Wang K, Sun B. Circulation long non-coding RNAs act as biomarkers for predicting tumorigenesis and metastasis in hepatocellular carcinoma. Oncotarget. 2015;6:4505–15. doi: 10.18632/oncotarget.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5:11091–102. doi: 10.18632/oncotarget.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949–59. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 235.Xu N, Chen F, Wang F, Lu X, Wang X, Lv M, et al. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: a Chinese population-based study. Tumour Biol. 2015;36:7659–65. doi: 10.1007/s13277-015-3469-0. [DOI] [PubMed] [Google Scholar]
- 236.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–8. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 237.Shang C, Guo Y, Zhang H, Xue YX. Long noncoding RNA HOTAIR is a prognostic biomarker and inhibits chemosensitivity to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother Pharmacol. 2016;77:507–13. doi: 10.1007/s00280-016-2964-3. [DOI] [PubMed] [Google Scholar]
- 238.Melo CP, Campos CB, Rodrigues Jde O, Aguirre-Neto JC, Atalla A, Pianovski MA, et al. Long non-coding RNAs: biomarkers for acute leukaemia subtypes. Br J Haematol. 2016;173:318–20. doi: 10.1111/bjh.13588. [DOI] [PubMed] [Google Scholar]
- 239.Chen ZJ, Zhang Z, Xie BB, Zhang HY. Clinical significance of up-regulated lncRNA NEAT1 in prognosis of ovarian cancer. Eur Rev Med Pharmacol Sci. 2016;20:3373–7. [PubMed] [Google Scholar]
- 240.Wu Y, Wang YQ, Weng WW, Zhang QY, Yang XQ, Gan HL, et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis. 2016;5:e192. doi: 10.1038/oncsis.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Li J, Wang Y, Yu J, Dong R, Qiu H. A high level of circulating HOTAIR is associated with progression and poor prognosis of cervical cancer. Tumour Biol. 2015;36:1661–5. doi: 10.1007/s13277-014-2765-4. [DOI] [PubMed] [Google Scholar]
- 242.Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, et al. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441–7. doi: 10.1007/s13277-014-1709-3. [DOI] [PubMed] [Google Scholar]
- 245.Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C, Ye H, et al. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Oncol. 2015;17:640–6. doi: 10.1007/s12094-015-1290-2. [DOI] [PubMed] [Google Scholar]
- 246.Dong L, Qi P, Xu MD, Ni SJ, Huang D, Xu QH, et al. Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int J Cancer. 2015;137:1128–35. doi: 10.1002/ijc.29484. [DOI] [PubMed] [Google Scholar]
- 247.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–8. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 248.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 250.A Study on the Gastrointestinal Disease and Helicobacter Pylori Controlled Long Non-coding RNA. https://ClinicalTrials.gov/show/NCT03057171.
- 251.Bourdoumis A, Papatsoris AG, Chrisofos M, Efstathiou E, Skolarikos A, Deliveliotis C. The novel prostate cancer antigen 3 (PCA3) biomarker. Int Braz J Urol. 2010;36:665–8. doi: 10.1590/s1677-55382010000600003. discussion 9. [DOI] [PubMed] [Google Scholar]
- 252.Li C, Yang L, Lin C. Long noncoding RNAs in prostate cancer: mechanisms and applications. Mol Cell Oncol. 2014;1:e963469. doi: 10.4161/23723548.2014.963469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Shi L, Peng F, Tao Y, Fan X, Li N. Roles of long noncoding RNAs in hepatocellular carcinoma. Virus Res. 2016;223:131–9. doi: 10.1016/j.virusres.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 254.Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2012;7:e35145. doi: 10.1371/journal.pone.0035145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–50. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 256.Lavorgna G, Vago R, Sarmini M, Montorsi F, Salonia A, Bellone M. Long non-coding RNAs as novel therapeutic targets in cancer. Pharmacol Res. 2016;110:131–8. doi: 10.1016/j.phrs.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 257.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–79. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Cai Y, He J, Zhang D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. Onco Targets Ther. 2015;8:2657–64. doi: 10.2147/OTT.S90485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Redis RS, Vela LE, Lu W, Ferreira de Oliveira J, Ivan C, Rodriguez-Aguayo C, et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol Cell. 2016;61:520–34. doi: 10.1016/j.molcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zheng J, Zhao S, He X, Zheng Z, Bai W, Duan Y, et al. The up-regulation of long non-coding RNA CCAT2 indicates a poor prognosis for prostate cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2016 doi: 10.1016/j.bbrc.2016.08.120. [DOI] [PubMed] [Google Scholar]
- 261.Johnstone KA, DuBose AJ, Futtner CR, Elmore MD, Brannan CI, Resnick JL. A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum Mol Genet. 2006;15:393–404. doi: 10.1093/hmg/ddi456. [DOI] [PubMed] [Google Scholar]
- 262.Tan WH, Bird LM. Pharmacological therapies for Angelman syndrome. Wien Med Wochenschr. 2016 doi: 10.1007/s10354-015-0408-z. [DOI] [PubMed] [Google Scholar]
- 263.Scaiewicz V, Sorin V, Fellig Y, Birman T, Mizrahi A, Galula J, et al. Use of H19 Gene Regulatory Sequences in DNA-Based Therapy for Pancreatic Cancer. J Oncol. 2010;2010:178174. doi: 10.1155/2010/178174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Chen H, Xin Y, Zhou L, Huang JM, Tao L, Cheng L, et al. Cisplatin and paclitaxel target significant long noncoding RNAs in laryngeal squamous cell carcinoma. Med Oncol. 2014;31:246. doi: 10.1007/s12032-014-0246-7. [DOI] [PubMed] [Google Scholar]
- 265.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–55. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Zhao W, Dong S, Duan B, Chen P, Shi L, Gao H, et al. HOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. Am J Transl Res. 2015;7:1295–302. [PMC free article] [PubMed] [Google Scholar]
- 267.Fang S, Gao H, Tong Y, Yang J, Tang R, Niu Y, et al. Long noncoding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab Invest. 2016;96:60–8. doi: 10.1038/labinvest.2015.123. [DOI] [PubMed] [Google Scholar]
- 268.Han Y, Liu Y, Gui Y, Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:555–9. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 269.Young TL, Matsuda T, Cepko CL. The Noncoding RNA Taurine Upregulated Gene 1 Is Required for Differentiation of the Murine Retina. Curr Biol. 2005;15:501–12. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 270.Li Z, Shen J, Chan MT, Wu WK. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49:471–5. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Niu Y, Ma F, Huang W, Fang S, Li M, Wei T, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer. 2017;16:5. doi: 10.1186/s12943-016-0575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Mol Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Li Z, Zhao L, Wang Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/beta-catenin pathway. Am J Transl Res. 2016;8:2385–93. [PMC free article] [PubMed] [Google Scholar]
- 274.Liu X, Jiao T, Wang Y, Su W, Tang Z, Han C. Long non-coding RNA GAS5 acts as a molecular sponge to regulate miR-23a in gastric cancer. Minerva Med. 2016 [PubMed] [Google Scholar]
- 275.Chen W, Xu XK, Li JL, Kong KK, Li H, Chen C, et al. MALAT1 is a prognostic factor in glioblastoma multiforme and induces chemoresistance to temozolomide through suppressing miR-203 and promoting thymidylate synthase expression. Oncotarget. 2017 doi: 10.18632/oncotarget.15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zhu KP, Zhang CL, Shen GQ, Zhu ZS. Long noncoding RNA expression profiles of the doxorubicin-resistant human osteosarcoma cell line MG63/DXR and its parental cell line MG63 as ascertained by microarray analysis. Int J Clin Exp Pathol. 2015;8:8754–73. [PMC free article] [PubMed] [Google Scholar]
- 277.Zhang CL, Zhu KP, Shen GQ, Zhu ZS. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumour Biol. 2016;37:2737–48. doi: 10.1007/s13277-015-4130-7. [DOI] [PubMed] [Google Scholar]
- 278.Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang E, et al. LncRNA H19 confers chemoresistance in ERalpha-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 2016;7:81452–62. doi: 10.18632/oncotarget.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–46. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 280.Alvarez-Dominguez JR, Bai Z, Xu D, Yuan B, Lo KA, Yoon MJ, et al. De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Long Non-coding RNA Regulators of Brown Adipocyte Development. Cell Metab. 2015;21:764–76. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015;11:151–60. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110:3387–92. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Alvarez-Dominguez JR, Hu W, Yuan B, Shi J, Park SS, Gromatzky AA, et al. Global discovery of erythroid long noncoding RNAs reveals novel regulators of red cell maturation. Blood. 2014;123:570–81. doi: 10.1182/blood-2013-10-530683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Baldassarre A, Masotti A. Long non-coding RNAs and p53 regulation. Int J Mol Sci. 2012;13:16708–17. doi: 10.3390/ijms131216708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]