Abstract
The Oncology Grand Rounds series is designed to place original reports published in the Journal into clinical context. A case presentation is followed by a description of diagnostic and management challenges, a review of the relevant literature, and a summary of the authors’ suggested management approaches. The goal of this series is to help readers better understand how to apply the results of key studies, including those published in the Journal of Clinical Oncology, to patients seen in their own clinical practice.
CASE PRESENTATION
A 47-year-old premenopausal woman presented with a 1.2 cm palpable mass. Her family history was significant for a paternal aunt diagnosed with postmenopausal estrogen receptor–positive (ER+) breast cancer and her paternal grandmother with ovarian cancer. An ultrasound-guided core needle biopsy identified moderately differentiated infiltrating ductal carcinoma, an ER expression by immunohistochemistry of 99%, a progesterone receptor expression of 55%, and a human epidermal growth factor receptor 2 expression of 1+ (negative). Germline susceptibility testing was performed and identified a pathogenic variant (PV) in ATM, c.538C>T (p.Gln180Ter). She was referred to discuss local management and the appropriateness of breast conservation versus bilateral mastectomy.
CHALLENGES IN DIAGNOSIS AND MANAGEMENT
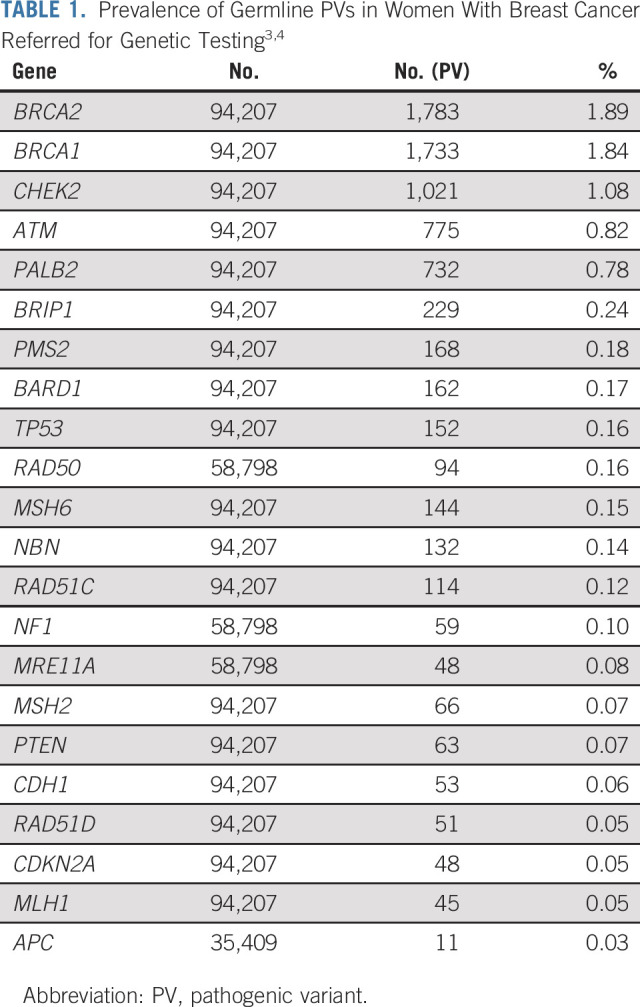
The year 2013 was a momentous one for clinical cancer genetics. In that year, the Supreme Court decision in Association of Molecular Pathology v Myriad Genetics Inc relaxed barriers to competition in germline testing for cancer susceptibility in the United States. The rapid development of massively parallel sequencing for clinical testing reduced the costs of testing while allowing the simultaneous analysis of multiple genes. Multigene panel testing quickly became the standard method of assessing inherited risk,1,2 identifying a number of germline alterations beyond BRCA1/2. Two reports from commercial diagnostic testing laboratories described PVs in approximately 8% of women with breast cancer referred for testing (Table 1).3,4 Although alterations in BRCA1 and BRCA2 were the most common, 2.7% of women were found to carry PV in CHEK2, ATM, and PALB2. Alterations were observed in many other genes. These discoveries raised significant clinical challenges. Women with breast cancer and non-BRCA alterations clearly cannot be managed in the same way as women with BRCA PV because of the different level and spectrum of risk. It has also been challenging to provide recommendations to unaffected female family members who share these moderate penetrance alterations (PV that confers two- to five-fold increased risks, as opposed to high-penetrance alterations in BRCA1, BRCA2, PTEN, and TP53). For some genes (eg, NBN), there has been controversy as to whether there is any association with breast cancer at all. The exact magnitude of risk has been difficult to establish for many genes, and there is limited information about the effectiveness of surveillance interventions in women whose average absolute risk is in the range of that of a woman whose sister was affected by breast cancer (which confers a relative risk of 1.8-2.3).5,6 Guideline groups such as the National Comprehensive Cancer Network in the United States have made empiric surveillance recommendations on the basis of a 20% cumulative lifetime risk threshold, extrapolating from guidance provided for management of high-risk women,7 but the optimal risk threshold and timing of intervention have not been studied. For this reason, the most appropriate management of both affected and unaffected women with moderate penetrance variants is incompletely defined.
TABLE 1.
SUMMARY OF THE RELEVANT LITERATURE
Genes Associated With an Elevated Risk of Breast Cancer
As noted, pathogenic germline variants are quite prevalent in women with breast cancer (Table 1). Huang et al8 analyzed germline data from 1,076 patients with breast cancer enrolled in the Cancer Genome Atlas and identified PV in 9.9%. The pathogenicity of a variant only describes impact on gene function, however, and identifying a functionally significant PV in a gene is not enough to conclude that the gene confers susceptibility. Studies suggest that humans carry between 250 and 300 loss-of-function variants in their genomes, most of which are not associated with disease in the individual.9 It has been challenging to come to agreement on a definitive list of breast cancer predisposition genes. Clinical cancer geneticists came to consensus quickly on high-penetrance genes that confer a > 5-fold relative risk (BRCA1, BRCA2, TP53, PTEN, and CDH1) because PV in these genes causes autosomal dominant cancer syndromes with risks that can be estimated through pedigree analysis. PALB2 is a more recently identified gene for which PVs are associated with four- to eight-fold increases in risk (an average of 53% breast cancer risk by age 80 years).10-12 PALB2 is often therefore considered a high penetrance gene, although it will be discussed here.
At the other end of the risk spectrum are common variants (single nucleotide polymorphisms [SNPs]) identified through large genome-wide association studies that individually confer a minimal increase in risk but that are incontrovertibly associated statistically with breast cancer.13 Although it is not possible to use individual common variants to predict breast cancer risk, there is a strong association with polygenic risk scores (PRSs) derived from hundreds of individual variant genotypes.14 These PRSs are now being incorporated into testing being offered by commercial diagnostic laboratories, although their clinical utility remains unclear.
Between rare high-penetrance genes and common SNPs lies a group of so-called moderate penetrance genes. These are genes in which PVs are found in < 1% of the general population, which confer between two- and five-fold increases in breast cancer risk. Because of the generally modest degrees of increased risk, PV in these genes cannot usually be identified through pedigree analysis as they often do not result in a recognizable Mendelian pattern of disease. They are best thought of as strong risk factors that interact with other factors such as family history, genetic background, mammographic density, and traditional risk factors to generate an individual woman's risk of breast cancer.15 Establishing a definitive list of moderate penetrance breast cancer susceptibility genes has been a challenge. Recent publications from the Breast Cancer Association Consortium and the CARRIERS Consortium describe large population-based case control analyses of more than 80,000 women with breast cancer (Fig 1).10,11 There is a canonical set of genes that are incontrovertibly associated with breast cancer, specifically BRCA1, BRCA2, PALB2, CHEK2, and ATM. Other previously reported associations were not validated, such as BRIP1, FANCC, FANCM, MRE11A, NBN (including the Slavic founder variant 657del5 [c.657_661del, p.Lys219fs]), RAD50, REQL, and RINT1. Risks described for known familial cancer syndrome genes such as CDH1, STK11, and TP53 were lower in these population-based studies than expected, possibly because recognition of these syndromes would remove subjects from both the case and control groups. Despite the size of these studies, the association of several genes with breast cancer remains unclear. This includes BARD1, MSH6, RAD51C, and RAD51D, for which the odds ratio point estimates of the associations are generally < 2.1 and statistically significant in one study but not the other. For BARD1, RAD51C, and RAD51D, associations with ER-negative (ER−) or triple-negative breast cancer are robust despite a lack of clear association with overall risk. The lack of impact on overall risk may result from the lack of association with the more prevalent ER-positive (ER+) breast cancer.
FIG 1.
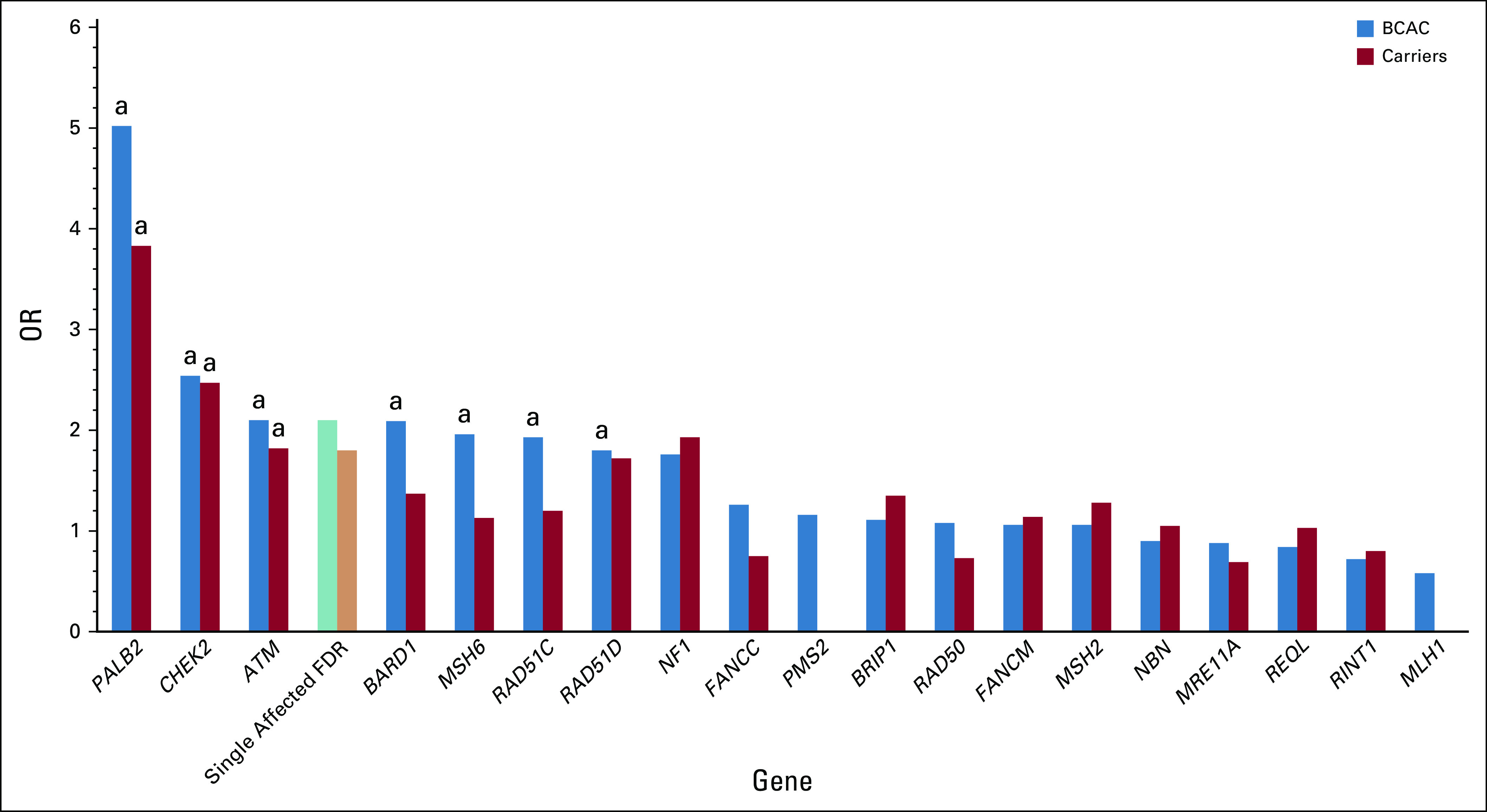
ORs for breast cancer for pathogenic variants in moderate penetrance genes (population-based studies10,11). Relative risks for unselected woman with an affected first-degree relative are indicated by “Single Affected FDR.”5,6 aStatistically significant associations within study. BCAC, Breast Cancer Association Consortium; OR, odds ratio.
MODIFIERS OF BREAST CANCER RISK IN WOMEN WITH MODERATE PENETRANCE PATHOGENIC VARIANTS
CHEK2 was the first moderate penetrance gene to be identified because of a common pathogenic founder variant (c.1100del, p.Thr367fs) found in Northern Europe. In 2004, a consortium calculated that this variant resulted in a 2.34-fold increase in risk (95% CI, 1.72 to 3.20).16 Subsequent studies of this variant and other PVs in CHEK2 illustrated the challenges of incorporating knowledge of moderate penetrance variants into clinical care. For example, the estimated risks associated with CHEK2 varied from study to study not only because of expected statistical variation and differing methods for computing absolute risks but also because of variation in different studies in the distributions of other risk factors. CHEK2 risk is modified by age,17 family history,18 and likely traditional reproductive risk factors and mammographic density.15 There are also genotype-specific variations in risk as some variants, such as CHEK2 c.470T>C (p.Ile157Thr), are associated with average risks that are in the range of a risk SNP rather than a rare variant.19
The degree to which moderate penetrance genes other than CHEK2 are modified by traditional risk factors has not been explored. For several genes (such as BRCA1, BRCA2, PALB2, and CHEK2), the risk relative to the general population appears to decline with age,10 but there is insufficient evidence that this applies for other moderate penetrance genes. It would be anticipated that genes that predispose to ER-positive breast cancer (ATM, CHEK2, and PALB2) would be more subject to modification by reproductive risk factors than genes that predispose to ER-negative cancers (BARD1, RAD51C, and RAD51D), but this hypothesis has not been tested.
There has been an increasing recognition that the risk of moderate penetrance PV is modified by genetic background, as represented by PRSs. This has been well-established for CHEK2 PV,20,21 and recent work has extended the observation to ATM and PALB2.22 The Original Report companion to this article by Gao et al23 further evaluates the impact of PRS on risk in ATM, CHEK2, and PALB2 carriers. In this report, all carriers of PV in PALB2 were calculated to have a lifetime (by age 80 years) risk of > 20% (the US threshold for magnetic resonance imaging [MRI] surveillance), but risks ranged from 21.5% for a carrier with no family history and a PRS in the 10% percentile up to 59.5% for a carrier with a family history and a PRS in the 90% percentile. For ATM and CHEK2 carriers, risks range from 12.8% (ATM) and 15.2% (CHEK2) for women with no family history and a PRS in the 10% percentile up to 40.9% (ATM) and 46.6% (CHEK2) for women with a family history and a PRS in the 90% percentile23 and 31.3% of CHEK2 PV carriers without a family history were calculated to have lifetime breast cancer risks under 20%. For women with a family history, the proportions were 21.2% for ATM and 10.1% for CHEK2.
SECOND CANCER RISKS IN WOMEN WITH MODERATE PENETRANCE PATHOGENIC VARIANTS
Breast Cancer
There are few data regarding second ipsilateral or contralateral breast cancer (CBC) risks associated with moderate penetrance PV. Not surprisingly, studies of CHEK2 c.1100del are the most robust given the prevalence of this variant. The results of the studies are variable, with some reports describing a three- to four-fold increased risk of CBC.24,25 Other studies have not shown such an association.26 PRS may modify CBC risk as it does primary cancer risk, but studies to examine this question have been underpowered.20 ATM variants that are considered pathogenic by clinical diagnostic laboratories are not associated with an increased CBC risk, even after radiation therapy.26 There are no data elucidating CBC risk in women with PALB2 PV. There are no data indicating that women with moderate penetrance PV are at increased risk for ipsilateral tumor recurrence after breast-conserving treatment, although the question has not been studied.
Ovarian Cancer
The most common moderate penetrance breast cancer predisposition genes are ATM, CHEK2, and PALB2. PALB2 has been linked to an increase in ovarian cancer risk in several studies.12,27 This risk is generally similar to that experienced by a woman whose sister has been diagnosed with ovarian cancer.28 In contrast, CHEK2 has not been linked to increased risk. Although ATM has not been definitively associated with an increased risk of ovarian cancer, two studies comparing PV variants in patients with ovarian cancer with public database controls reported 2.25- to 2.85-fold increased risks.29,30 BARD1 is not robustly associated with an increased risk of ovarian cancer.29-31
BRIP1, RAD51C, and RAD51D are clearly moderate penetrance ovarian cancer predisposition genes,29-34 but risk estimates have been labile as PVs are rare. A systematic analysis comparing the prevalence of PV in these genes with public databases generated point estimate odds ratios (ORs) of 4.94 for BRIP1, 5.59 for RAD51C, and 6.94 for RAD51D.35 A large recent collaborative analysis assessing risk in families with RAD51C (relative risk 7.55) and RAD51D (relative risk 7.60) alterations produced estimated absolute ovarian cancer risks by age 80 years of 11% and 13%, respectively. Estimated absolute risks were substantially higher in women with a family history of ovarian cancer.34
Pancreas Cancer
ATM PVs were observed in familial pancreas cancer in 2012 and were present in 1.8% of a prospective ascertainment of patients with the disease.36,37 A study comparing the prevalence of PV in 3,030 patients with pancreas cancer with public database controls calculated an point OR of 5.71, close to the OR for BRCA2 (6.20).38 PALB2 PVs were associated with an OR of 2.37 for pancreas cancer in one study,12 but an elevated risk was not identified in another report.38 Other genes have not been linked.
SUGGESTED APPROACH TO MANAGEMENT
Surgical Treatment
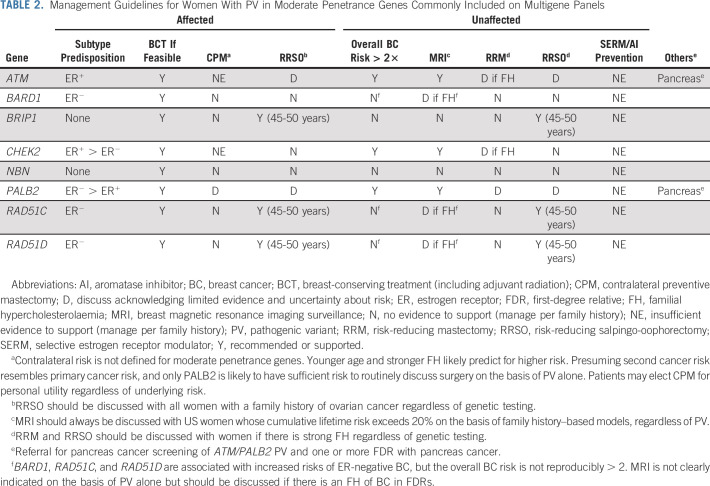
There is no current evidence that women are at increased risk for ipsilateral recurrence or ipsilateral second primary malignancies if they are found to carry a PV in ATM, CHEK2, PALB2, or other genes such as BARD1, RAD51C, or RAD51D (Table 2). Since BRIP1, NBN, and other genes do not play a clear role in breast cancer susceptibility, PV in these genes would also not be expected to affect risk of ipsilateral events. Therefore, women with PV in moderate penetrance genes should be considered candidates for breast-conserving treatment (BCT) if otherwise appropriate. Despite theoretical concerns, women with PV in ATM are not clearly at increased risk of CBC after radiation treatment. Women with ATM PV should not be precluded from considering BCT.
TABLE 2.
Management Guidelines for Women With PV in Moderate Penetrance Genes Commonly Included on Multigene Panels
There is limited evidence that CHEK2 PV may be associated with modest increased risks of CBC, but these risks are likely modified by genetic background (PRS) and may also be modified by endocrine therapy since a large proportion of CHEK2-associated cancers are ER-positive. There are no data to suggest that women with PV in ATM, PALB2, or other genes are at sufficient CBC risk to warrant a general recommendation for contralateral preventive mastectomy. It should be noted that the risks associated with PALB2 are generally like those of BRCA2 and that CBC risk has not been well-studied in women with PALB2 PV. Consideration of contralateral preventive mastectomy may be reasonable in such women, particularly if they are young and with a significant family history of breast cancer.
Systemic Treatment
There have been studies suggesting that women with CHEK2 PV experience worse breast cancer outcomes than women without PV.25,39,40 These studies were performed before the widespread availability of prognostic genomic testing. Decisions about systemic adjuvant therapy should follow established decision pathways without regard to the presence of the CHEK2 PV. There is no evidence that the presence of PV in other genes is associated with worse prognosis or should influence adjuvant therapy decisions.
The products of many of the moderate penetrance susceptibility genes operate in DNA damage repair pathways. PARP inhibitors are known to be effective in women with breast cancer and germline PV in BRCA1 and BRCA2.41,42 A small study, TBCRC048, has suggested that olaparib is highly active in patients with metastatic breast cancer and germline PALB2 alterations but not in women with CHEK2 or ATM PV.43 The activity in women with alterations in other genes has not been studied, although there is reason to be optimistic that PARP inhibitors would be active in patients with PV in RAD51C and RAD51D.
MANAGEMENT OF RISK OF OTHER CANCERS
Although PALB2 and perhaps ATM are associated with modestly increased risks of ovarian cancer, these risks resemble those of a woman with a first-degree relative with ovarian cancer and premenopausal risk-reducing salpingo-oophorectomy (RRSO) is unlikely to be of benefit. Even in women with PV in BRCA2, which confers substantially higher risk, RRSO is often deferred until age 45 years, which supports deferring surgery until after menopause for PALB2 and ATM PV. Women with PV in BRIP1, RAD51C, and RAD51D are at higher risk and should strongly consider RRSO between 45 and 50 years.
The benefit of screening for pancreas cancer is not established. Gastroenterology professional societies recommend consideration of screening for women with PV in ATM or PALB2 who also have one or more first-degree relatives affected with pancreas cancer.44,45
MANAGEMENT OF UNAFFECTED RELATIVES
Unaffected relatives of women with moderate penetrance PV should be referred for genetic counseling and presymptomatic testing. Unaffected women with PV in ATM, CHEK2, or PALB2 have average lifetime risks exceeding 20% and are therefore candidates for enhanced surveillance with MRI in addition to mammography or contrast-enhanced mammography. The current recommendation is to begin such surveillance at age 40 years, although there may be benefits to starting younger, especially if there is a significant family history of breast cancer. PRS-modified absolute risk estimates may be provided by commercial testing laboratories. In the absence of prospective validations, it would be unwise to forego enhanced surveillance on the basis of an adjusted absolute risk under 20%, particularly in the presence of a family history. Women with alterations in other genes, including BARD1, BRIP1, RAD51C, RAD51D, and NBN, should be followed as appropriate for their family history of breast cancer.
In general, the average breast cancer risks conferred by moderate penetrance PV are insufficient to support risk-reducing mastectomy (RRM). Furthermore, the studies of PRS modification indicate that a substantial proportion of women with PV in ATM and CHEK2 have lifetime risks below the 20% threshold used for MRI and it would be inappropriate to risk operating on such women. Conversely, however, caution should be used when employing a PRS-adjusted risk estimate to support RRM until robust validation studies have been published. For women with PV in PALB2, RRM may be considered, particularly if there is a significant family history of breast cancer. In women with PALB2 PV and no family history, the distribution of risk is such that some women may have lifetime risks in the range of 30% or even less. RRM would be an aggressive approach in such women.
Management of ovarian cancer and pancreas cancer risks in unaffected women should follow the earlier suggestions.
SUMMARY
Multigene panel testing identifies PVs in a substantial fraction of women with breast cancer. Cancer risks and management approaches are well-defined for women with PV in high penetrance cancer syndrome genes. For genes that confer more modest degrees of risk, best practices are less clear. In general, treatment of women with breast cancer and PV in moderate penetrance genes should proceed as treatment for women without such alterations. Some genes are associated with sufficient ovarian cancer or pancreas cancer risk to warrant intervention. Unaffected female family members should undergo cancer genetic counseling by a knowledgeable provider to be informed of the most up-to-date risk figures and recommendations for management. In most situations, the breast cancer risks for unaffected women will not warrant serious consideration of RRM although MRI surveillance will often be appropriate and RRSO will need to be considered for some PV.
The patient elected to undergo wide local excision and sentinel node biopsy. Her final stage was pT1cN0M0. Her 21-gene recurrence score was 16, and she elected adjuvant antiestrogen treatment alone. She was referred for adjuvant radiotherapy.
Mark Robson
Consulting or Advisory Role: Change HealthCare
Research Funding: AstraZeneca, Pfizer, Merck
Other Relationship: Research to Practice, Clinical Care Options, Physicans' Education Resource, Invitae, Pfizer
Uncompensated Relationships: Merck, Pfizer, Daiichi Sankyo, Epic Sciences
Open Payments Link: https://openpaymentsdata.cms.gov/physician/612669/summary
No other potential conflicts of interest were reported.
See accompanying article on page 2564
SUPPORT
Supported by Breast Cancer Research Foundation, NIH/NCI Cancer Center Support Grant P30 CQA008748.
AUTHOR'S DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Management of Women With Breast Cancer and Pathogenic Variants in Genes Other Than BRCA1 or BRCA2
The following represents disclosure information provided by the author of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Mark Robson
Consulting or Advisory Role: Change HealthCare
Research Funding: AstraZeneca, Pfizer, Merck
Other Relationship: Research to Practice, Clinical Care Options, Physicans' Education Resource, Invitae, Pfizer
Uncompensated Relationships: Merck, Pfizer, Daiichi Sankyo, Epic Sciences
Open Payments Link: https://openpaymentsdata.cms.gov/physician/612669/summary
No other potential conflicts of interest were reported.
REFERENCES
- 1.Kurian AW, Ward KC, Abrahamse P, et al. Time trends in receipt of germline genetic testing and results for women diagnosed with breast cancer or ovarian cancer, 2012-2019 J Clin Oncol 391631–16402021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurian AW, Ward KC, Hamilton AS, et al. Uptake, results, and outcomes of germline multiple-gene sequencing after diagnosis of breast cancer JAMA Oncol 41066–10722018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buys SS, Sandbach JF, Gammon A, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes Cancer 1231721–17302017 [DOI] [PubMed] [Google Scholar]
- 4.Couch FJ, Shimelis H, Hu C, et al. Associations between cancer predisposition testing panel genes and breast cancer JAMA Oncol 31190–11962017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collaborative Group on Hormonal Factors in Breast Cancer Familial breast cancer: Collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease Lancet 3581389–13992001 [DOI] [PubMed] [Google Scholar]
- 6.Pharoah PD, Day NE, Duffy S, et al. Family history and the risk of breast cancer: A systematic review and meta-analysis Int J Cancer 71800–8091997 [DOI] [PubMed] [Google Scholar]
- 7.Saslow D, Boetes C, Burke W, et al. American Cancer Society guidelines for breast screening with MRI as an adjunct to mammography CA Cancer J Clin 5775–892007 [DOI] [PubMed] [Google Scholar]
- 8.Huang KL, Mashl RJ, Wu Y, et al. Pathogenic germline variants in 10,389 adult cancers Cell 173355–370 e142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing Nature 4671061–10732010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breast Cancer Association Consortium. Dorling L, Carvalho S, et al. Breast cancer risk genes: Association analysis in more than 113,000 women N Engl J Med 384428–4392021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu C, Hart SN, Gnanaolivu R, et al. A population-based study of genes previously implicated in breast cancer N Engl J Med 384440–4512021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Leslie G, Doroszuk A, et al. Cancer risks associated with germline PALB2 pathogenic variants: An international study of 524 families J Clin Oncol 38674–6852020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michailidou K, Lindstrom S, Dennis J, et al. Association analysis identifies 65 new breast cancer risk loci Nature 55192–942017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mavaddat N, Michailidou K, Dennis J, et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes Am J Hum Genet 10421–342019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: A comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors Genet Med 211708–17182019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consortium CBCC-C CHEK2*1100delC and susceptibility to breast cancer: A collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies Am J Hum Genet 741175–11822004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt MK, Hogervorst F, van Hien R, et al. Age- and tumor subtype-specific breast cancer risk estimates for CHEK2*1100delC carriers J Clin Oncol 342750–27602016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cybulski C, Wokolorczyk D, Jakubowska A, et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer J Clin Oncol 293747–37522011 [DOI] [PubMed] [Google Scholar]
- 19.Liu C, Wang Y, Wang QS, et al. The CHEK2 I157T variant and breast cancer susceptibility: A systematic review and meta-analysis Asian Pac J Cancer Prev 131355–13602012 [DOI] [PubMed] [Google Scholar]
- 20.Borde J, Ernst C, Wappenschmidt B, et al. Performance of breast cancer polygenic risk scores in 760 female CHEK2 germline mutation carriers. J Natl Cancer Inst. 2020 doi: 10.1093/jnci/djaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muranen TA, Greco D, Blomqvist C, et al. Genetic modifiers of CHEK2*1100delC-associated breast cancer risk Genet Med 19599–6032017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher S, Hughes E, Wagner S, et al. Association of a polygenic risk score with breast cancer among women carriers of high- and moderate-risk breast cancer genes. JAMA Netw Open. 2020;3:e208501. doi: 10.1001/jamanetworkopen.2020.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao C, Polley EC, Hart SN, et al. Risk of breast cancer among carriers of pathogenic variants in breast cancer predisposition genes varies by polygenic risk score J Clin Oncol 392564–25732021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellemkjaer L, Dahl C, Olsen JH, et al. Risk for contralateral breast cancer among carriers of the CHEK2*1100delC mutation in the WECARE Study Br J Cancer 98728–7332008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weischer M, Nordestgaard BG, Pharoah P, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer J Clin Oncol 304308–43162012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiner AS, Robson ME, Mellemkjaer L, et al. Radiation treatment, ATM, BRCA1/2, and CHEK2*1100delC pathogenic variants and risk of contralateral breast cancer J Natl Cancer Inst 1121275–12792020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song H, Dicks EM, Tyrer J, et al. Population-based targeted sequencing of 54 candidate genes identifies PALB2 as a susceptibility gene for high-grade serous ovarian cancer J Med Genet 58305–3132020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stratton JF, Pharoah P, Smith SK, et al. A systematic review and meta-analysis of family history and risk of ovarian cancer Br J Obstet Gynaecol 105493–4991998 [DOI] [PubMed] [Google Scholar]
- 29.Lilyquist J, LaDuca H, Polley E, et al. Frequency of mutations in a large series of clinically ascertained ovarian cancer cases tested on multi-gene panels compared to reference controls Gynecol Oncol 147375–3802017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu HM, Li S, Black MH, et al. Association of breast and ovarian cancers with predisposition genes identified by large-scale sequencing JAMA Oncol 551–572019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramus SJ, Song H, Dicks E, et al. Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst. 2015;107:djv214. doi: 10.1093/jnci/djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song H, Dicks E, Ramus SJ, et al. Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population J Clin Oncol 332901–29072015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber-Lassalle N, Hauke J, Ramser J, et al. BRIP1 loss-of-function mutations confer high risk for familial ovarian cancer, but not familial breast cancer. Breast Cancer Res. 2018;20:7. doi: 10.1186/s13058-018-0935-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Song H, Leslie G, et al. Ovarian and breast cancer risks associated with pathogenic variants in RAD51C and RAD51D J Natl Cancer Inst 1121242–12502020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suszynska M, Ratajska M, Kozlowski P. BRIP1, RAD51C, and RAD51D mutations are associated with high susceptibility to ovarian cancer: Mutation prevalence and precise risk estimates based on a pooled analysis of ∼30,000 cases. J Ovarian Res. 2020;13:50. doi: 10.1186/s13048-020-00654-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowery MA, Wong W, Jordan EJ, et al. Prospective evaluation of germline alterations in patients with exocrine pancreatic neoplasms J Natl Cancer Inst 1101067–10742018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts NJ, Jiao Y, Yu J, et al. ATM mutations in patients with hereditary pancreatic cancer Cancer Discov 241–462012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu C, Hart SN, Polley EC, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer JAMA 3192401–24092018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greville-Heygate SL, Maishman T, Tapper WJ, et al. Pathogenic variants in CHEK2 are associated with an adverse prognosis in symptomatic early-onset breast cancer JCO Precis Oncol 4472–4852020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt MK, Tollenaar RA, de Kemp SR, et al. Breast cancer survival and tumor characteristics in premenopausal women carrying the CHEK2*1100delC germline mutation J Clin Oncol 2564–692007 [DOI] [PubMed] [Google Scholar]
- 41.Litton JK, Hurvitz SA, Mina LA, et al. Talazoparib versus chemotherapy in patients with germline BRCA1/2-mutated HER2-negative advanced breast cancer: Final overall survival results from the EMBRACA trial Ann Oncol 311526–15352020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robson M, Im SA, Senkus E, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation N Engl J Med 377523–5332017 [DOI] [PubMed] [Google Scholar]
- 43.Tung NM, Robson ME, Ventz S, et al. TBCRC 048: Phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes J Clin Oncol 384274–42822020 [DOI] [PubMed] [Google Scholar]
- 44.Aslanian HR, Lee JH, Canto MI.AGA clinical practice update on pancreas cancer screening in high-risk individuals: Expert review Gastroenterology 159358–3622020 [DOI] [PubMed] [Google Scholar]
- 45.Goggins M, Overbeek KA, Brand R, et al. Management of patients with increased risk for familial pancreatic cancer: Updated recommendations from the International Cancer of the Pancreas Screening (CAPS) consortium Gut 697–172020 [DOI] [PMC free article] [PubMed] [Google Scholar]