ABSTRACT
Background: This study investigated the potential molecular mechanism of circular RNA HIPK3 (circHIPK3) in breast cancer (BCa). Methods: BCa cells were transfected with miR-326 mimic, miR-326 inhibitor, circHIPK3, sicircHIPK3. The expressions of circHIPK3 and miR-326 in BCa tissues and BCa cell lines were determined by RT-qPCR. Cell viability, colony formation, migration, invasion, and apoptosis of the cells were detected by CCK-8 and colony formation, wound-healing, transwell and flow cytometric assays, respectively. The relationship between circHIPK3 and miR-326 was analyzed and confirmed by circInteractome, dual-luciferase reporter, RT-qPCR, Pearson’s correlation assays. Western blot and RT-qPCR were performed to determine the expressions of apoptosis-related molecules (Bcl-2, Bax, and cleaved Caspase-3) and EMT-related molecules (E-cadherin, N-cadherin, and Vimentin) in the BCa cells and tumor tissues. The tumor growth in mice was examined in a xenograft tumor model in which Ki-67 expression was determined by immunohistochemistry (IHC). Results: In BCa, the expression of circHIPK3 was up-regulated and that of miR-326 was down-regulated. CircHIPK3 knockdown inhibited the cell proliferation, invasion, and migration. MiR-326 was the direct target of circHIPK3, and was inversely correlated with circHIPK3 expression. CircHIPK3 overexpression promoted proliferation, migration, invasion, apoptosis resistance, and tumor growth and up-regulated Ki-67 expression, at the same time, the expressions of Bcl-2, N-cadherin, Vimentin were up-regulated, and those of Bax, cleaved Caspase-3 and E-cadherin were inhibited. These above expressions were partially reversed by miR-326 overexpression. Conclusion: CircHIPK3 sponges miR-326 to promote BCa growth and metastasis. The current findings provide a novel therapeutic target for treating BCa.
KEYWORDS: Circular RNA circHIPK3, miR-326, growth, metastasis, breast cancer
Introduction
Breast cancer (BCa) is a frequently diagnosed cancer and a major cause of cancer-related mortality among women in the world [1]. The operation, chemotherapy, and molecular targeting treatment for BCa management have been improved in recent years, but overall prognosis of BCa patients is still unfavorable [2]. Distant metastasis is one of the primary reasons of therapeutic failure [3], and more than 5% of the BCa patients will develop incurable metastasis [4]. To explore novel therapeutic targets and strategies for BCa treatment, it is particularly important to identify effective BCa markers and understand the underlying molecular mechanism of the growth and metastasis of BCa.
Circular RNAs (circRNAs), which are class of conserve and stable endogenous non-coding RNAs, are formed through alternative splicing from a precursor mRNA and are widely expressed in mammals [5,6]. Compared with traditional linear RNAs such as lncRNAs, circRNAs have a covalently closed loop without 5′ to 3′ polarity or polyadenylated tail [7]. CircRNAs have been found to be involved in a variety of physiological and pathological processes [8], showing the potential to serve as molecular markers for the diagnosis and therapy of human diseases, including in cancers [9]. CircHIPK3 originating from HIPK3 is involved in a variety of biological or pathological processes [10]. In tumors, circHIPK3 exerts its effect via sponging miRNAs to modulate cancer development [11,12,13]. So far, the biological role and potential mechanism of circHIPK3 in the carcinogenesis of BCa are less reported.
MicroRNAs (miRNA) are small non-coding RNAs that regulate gene expressions at post-transcriptional level [14]. MiRNAs play important roles in different biological processes [15]. Aberrant miRNAs are closely involved in the pathogenesis of most human cancers, and dysregulated miRNAs have been found to function as oncogenes or tumor suppressors [16]. MiR-326 is a tumor suppressor to represses the initiation and development of some malignant tumors, such as prostatic carcinoma [17], glioma [18], and gastric cancer [19]. Moreover, study indicated that miR-326 also plays an important role in BCa by participating in chemotherapy resistance of BCa cells through regulating expression of multidrug resistance-associated protein 1 [20]. Moreover, Ghaemi et al. [21] found that miR-326 suppresses BCa via targeting ErbB/PI3K signaling pathway. MiR-326 is also a promising therapeutic target for BCa [22]. CircRNAs acts as miRNA sponges to modulate gene expression [23,24].
The current study explored the relationships between circHIPK3 and miR-326, and their interaction in the development of BCa, hoping to provide a novel theoretical basis for the early diagnosis and treatment of BCa.
Materials and method
Ethics statement
The current research was approved by the Ethics Committee of the Xiangya Hospital, Central South University (XH20170422011). All the patients with BCa signed the informed consents before the surgical treatment. All animal experiments obtained the approval from the Committee of Experimental Animals of Xiangya Hospital, Central South University (XH20190605023) and operated in accordance with national guidelines for the care and use of laboratory animals.
Clinical specimens
BCa tissues and adjacent non-cancerous tissues (ANT) from a total of 48 patients confirmed as having BCa by histopathology in Xiangya Hospital, Central South University were collected between May 2017 and May 2019. The patients did not receive preoperative chemoradiotherapy prior to the surgery.
Cell culture
MEGM Bullet Kit (CC-3150) was purchased from Lonza/Clonetics Corporation). McCoy’s 5a Medium Modified (30–2007), and Eagle’s Minimum Essential Medium (30–2003) and Leibovitz’s L-15 Medium were purchased from American type culture collection (ATCC). RPMI Medium 1640 (10,491), DMEM (11,965), FBS (11,011–8611) and Penicillin-Streptomycin Liquid (P1400) were ordered from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). All the cell lines, including human normal mammary epithelial cell line MCF-10A and BCa cell lines (MCF7, SK-BR-3, BT549, BT20, MDA-MB-231, MDA-MB-453), were ordered from ATCC. MCF-10A, SK-BR-3, BT549, MCF7, and BT20 cells were cultured in Mammary Epithelial Cell Growth Medium (MEGM) Bullet Kit, McCoy’s 5a Medium Modified, RPMI Medium 1640, Eagle’s Minimum Essential Medium, and DMEM, respectively. MDA-MB-231 and MDA-MB-453 cells were cultured in Leibovitz’s L-15 Medium. All the mediums were added with 10% FBS and 1% Penicillin-Streptomycin Liquid. The cells were incubated at 37°C in a 5% CO2 atmosphere.
Cell transfection
To up-regulate the expression of circHIPK3 in the BCa cells, human circHIPK3 cDNA was amplified and inserted into a pLVX-cir vector by Genomeditech (Shanghai, China). An empty plasmid served as a negative control (NC) for circHIPK3 over-expression plasmids. Small-interfering RNAs (si-circHIPK3; siB180306094916-1-5) and its corresponding siRNA negative control (siNC; siN0000002-1-5) were obtained from RiboBio Co., Ltd. (Guangzhou, China). MiR-326 mimic (M), miR-326 inhibitor (I), and their corresponding negative control (mimic control (MC), and inhibitor control (IC)) were synthesized by GenePharma (Shanghai, China). For in vitro experiments, cell transfection with siRNA and mimic/inhibitor were transient transfection. MCF7 and BT20 cells were transfected for 48 h using Lipofectamine 3000 (Invitrogen). For in vivo experiments, lentiviral-based circHIPK3 overexpression (circHIPK3) and empty vector control (NC) were purchased from Biomics (Jiangsu, China).
Dual-luciferase reporter assay
The binding sites between circHIPK3 and miR-326 were predicted by CircInteractome. Wild-type (wt) circHIPK3 fragments containing the predicted miR-326–binding site and mutant (mut) circHIPK3 fragments (with the mutated miR-326–binding site) were designed and synthesized by RiboBio Co., Ltd. (Guangzhou, China). The fragments were inserted into the pGL3 vectors plasmid (E1761, Promega), and the product luciferase reporter plasmids were named as circHIPK3-WT and circHIPK3-MUT. MCF7 and BT20 cells were co-transfected with circHIPK3-WT, or circHIPK3-MUT plasmids and miR-326 inhibitor, or inhibitor control. After transfection for 48 h, the luciferase activities were detected by the dual-luciferase reporter Assay system (E1910, Promega).
RNA preparation and RT–qPCR
RT-qPCR assay was performed as described previously [25]. Briefly, total RNAs from whole-cell lysates, the nuclear and cytoplasmic fractions, and tissues were isolated using PARIS™ Kit (AM1921, Thermo Fisher). For RNase R treatment, 2 mg RNA was incubated with or without Ribonuclease R (RNase R; R0301, Geneseed, Guangzhou, China). The cDNAs were synthesized using the PrimeScript RT Master Mix (Takara, Dalian, China) to detect the amount of mRNAs and circRNAs. Real-time PCR were performed using SYBR Premix Ex Taq II (Takara) in a LightCycler/LightCycler 480 System (Roche Diagnostics). miRNA Extraction Kit (B1802, HaiGene), miRNA cDNA Synthesis Kit (B1802, HaiGene) and miRNA qPCR Kit (TAP01724, HaiGene) were used to determine the expression of miRNA, following the manufacture’s protocol. The primer sequences (shown in Tables 1 and Tables 2) were purchased from Sangon Biotech (Shanghai, China). The expressions of genes were calculated by 2−ΔΔCt method [26] and normalized to that of GAPDH and U6. GAPDH and U6 were considered as an internal standard for mRNA/circRNA and miRNA, respectively.
Table 1.
Primer sequences used for quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) in human
| Genes | Primer sequences(5 ´-3 ´) | |
|---|---|---|
| circHIPK3 | forward | TATGTTGGTGGATCCTGTTCGGCA |
| reverse | TGGTGGGTAGACCAAGACTTGTGA | |
| HIPK3 | forward | TCACAAGTCTTGGTCTACCCA |
| reverse | CACATAGGTCCGTGGATAGTTTC | |
| miR-362 | forward | GCCGAAACACACCTATTCAAG |
| reverse | TATGGTTTTGACGACTGTGTGAT | |
| Bax | forward | CCCGAGAGGTCTTTTTCCGAG |
| reverse | CCAGCCCATGATGGTTCTGAT | |
| Bcl-2 | forward | GGTGGGGTCATGTGTGTGG |
| reverse | CGGTTCAGGTACTCAGTCATCC | |
| E-Cadherin | forward | CGAGAGCTACACGTTCACGG |
| reverse | GGGTGTCGAGGGAAAAATAGG | |
| N-Cadherin | forward | TCAGGCGTCTGTAGAGGCTT |
| reverse | ATGCACATCCTTCGATAAGACTG | |
| Vimentin | forward | GACGCCATCAACACCGAGTT |
| reverse | CTTTGTCGTTGGTTAGCTGGT | |
| U6 | forward | CTCGCTTCGGCAGCACA |
| reverse | ACGCTTCACGAATTTGCGT | |
| GAPDH | forward | GGAGCGAGATCCCTCCAAAAT |
| reverse | GGCTGTTGTCATACTTCTCATGG | |
Table 2.
Primer sequences used for quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) in mice
| Genes | Primer sequences(5 ´-3 ´) | |
|---|---|---|
| circHIPK3 | forward | GGATCGGCCAGTCATGTATC |
| reverse | ACCGCTTGGCTCTACTTTGA | |
| miR-362 | forward | GCCGAAACACACCTATTCAAG |
| reverse | TATGGTTTTGACGACTGTGTGAT | |
| Bax | forward | AGACAGGGGCCTTTTTGCTAC |
| reverse | AATTCGCCGGAGACACTCG | |
| Bcl-2 | forward | GCTACCGTCGTGACTTCGC |
| reverse | CCCCACCGAACTCAAAGAAGG | |
| U6 | forward | CTCGCTTCGGCAGCACA |
| reverse | ACGCTTCACGAATTTGCGT | |
| GAPDH | forward | AGGTCGGTGTGAACGGATTTG |
| reverse | GGGGTCGTTGATGGCAACA | |
Western blot
Total proteins were extracted using RIPA buffer (P0013, Beyotime, China) with PMSF (ST506, Beyotime, China), and the protein concentration was determined applying BCA kit (P0009, Beyotime, China). Then, the proteins were separated on 10% SDS-PAGE gel. The membranes were then treated by the primary antibody (listed in Table 3) at 4°C overnight and then by the corresponding secondary antibody (ab205718 or ab205719) at room temperature for 1 h. The protein bands were developed by chemiluminescence through exposing to ECL solution (P0018FS, Shanghai, Beyotime), and visualized by Image J software (Image J 1.8.0, National Institute of Health). The protein expression was normalized to that of GAPDH.
Table 3.
List of primary antibodies used for western blots
| Protein | Antibody | Catalog Number | Company | Antibody Dilution |
|---|---|---|---|---|
| cleaved Caspase-3 | Rabbit Anti-Cleaved Caspase-3 antibody | ab2302 | Abcam | 1:500 |
| Bcl-2 | Rabbit Anti-Bcl-2 antibody | ab59348 | Abcam | 1:1000 |
| Bax | Rabbit Anti-Bax antibody | ab32503 | Abcam | 1:500 |
| E-Cadherin | Rabbit Anti-E Cadherin antibody | ab40772 | Abcam | 1:1000 |
| N-Cadherin | Rabbit Anti-N Cadherin antibody | ab18203 | Abcam | 1:1000 |
| Vimentin | Rabbit Anti-Vimentin antibody | ab92547 | Abcam | 1:1000 |
| GAPDH | Mouse Anti-GAPDH antibody | ab8245 | Abcam | 1:1000 |
Cell proliferation assay
Cell proliferation was measured by CCK-8 kit (C0039, Beyotime). 1000 transfected MCF7 and BT20 cells were incubated in 96-well plates with completed medium (at 100 μL/well) for different times. Then, the cells were incubated with 10 μL of CCK-8 solution for another 4 h in the dark. Finally, the absorbance at 450 nm was read using a microplate reader (SpectraMax iD5, Molecular Devices, US) to determine cell proliferation. Cell clone formation was evaluated by performing colony formation assay. MCF7 and BT20 cells were cultured at 1 × 103 cells/well in 60-mm dishes. After 17 days, the cells were fixed with methanol for 30 min and dyed by 0.5% crystal violet solution. Colony number with more than 50 cells was counted under an inverted microscope (Ts2r-FL, Nikon, Japan).
Wound-healing assay
The migration capability of MCF7 and BT20 cells was determined by wound healing assay. MCF7 and BT20 cells (1×105) were seeded into a 6-well plate and cultured. After reaching 80–90% confluence, the cells were scratched by a 10 μL tip, washed by serum-free medium, and incubated for 48 h. The migration rate was measured under an inverted microscope (100 ×, Ts2r-FL, Nikon, Japan).
Transwell assay
Invasion of MCF7 and BT20 cells were evaluated by performing Transwell (8-μm pores, Corning Inc., Corning, USA). Briefly, 1 × 105 cells were cultured in the matrigel (BD Biosciences)-coated upper chamber containing 200 μL serum-free medium, whereas 600 μL medium with 10% FBS was added to the lower chambers. After incubation for 48 h, non-invaded cells were removed by cotton swab, whereas invaded cells were fixed in 4% methanol for 30 min, dyed by 0.1% crystal violet (C0121, Beyotime) for 20 min, and counted under a 250 × microscope (Ts2r-FL).
Flow cytometric assay
The apoptosis of BCa cells was detected using an apoptosis detection kit (C1062L, Beyotime). Briefly, the transfected MCF7 and BT20 cells (5 × 104) were centrifuged at 1000 g for 5 min. After discarding the supernatant, Annexin V-FITC and 10 μL propidium iodide (PI) were added into the cells according to the protocols provided by the manufacturer for further culture at 20–25°C for 20 min in the dark. The apoptosis was determined by a flow cytometer Accuri™ C6 (BD Biosciences).
Animal experiments in vivo
Animals purchased from the Beijing HFK Bioscience Co., Ltd. were maintained at pathogen-free environment. For xenograft growth of orthotopic animal model assay, MCF7 and BT20 cells (1 × 107 cells in 100 μL) were co-transfected with circHIPK3, and subcutaneously injected into the left flanks of 25 male BALB/c athymic nude mice (6-week-old; male). After inoculation for 10 days, miR-326 mimic or mimic control (100 μM) mixed with mixed with Lipofectamine 2000 were diluted in 100 μL PBS and subcutaneously injected once every other day until the 34th day (totally 12 times). There were 6 mice in each experimental group, and the tumor size of the mice was weekly measured. On the 34th day, the mice were sacrificed and the tumors were weighted. The tumor tissues were used for immunohistochemistry of Ki-67 (proliferation marker) for the detection of its expression, and RT-qPCR was used for determining the expressions of circHIPK3 and miR-326.
Immunohistochemistry (IHC)
The tissues were paraffin-embedded and sectioned for IHC staining to determine the expression of Ki-67, according to the study of Wang F et al. [27]. After pre-treatment with heat mediated antigen retrieval for 20 min in a microwave oven, the endogenous peroxidase activity of the sections (4-μm thick) were blocked by incubating the tissues in 3% H2O2 for 30 min. The sections were treated with 1 µg/mL primary antibody (Rabbit Anti-Ki67 antibody (ab15580, Abcam)) for 15 min at room temperature and subsequently treated with corresponding secondary antibody (ab205718, Abcam)) at room temperature for 30 min. Next, the sections were developing using Diaminobenzidine (DAB) solution (P0203, Beyotime), counterstained by Hematoxylin Staining Solution (C0107, Beyotime), then mounted with DPX, and observed under a microscope (IX71; Olympus, Tokyo, Japan).
Statistical analyses
All the results are shown as means ± standard deviation (SD). Two-tailed Student’s t test and one-way ANOVA were used to compare the data of two and multiple groups. Pearson’s correlation analysis was used to determine the correlations. P < 0.05 was considered as statistically significant.
Results
Expression of circHIPK3 was significantly up-regulated in BCa
The data from RT-qPCR revealed that circHIPK3 was high-expressed in BCa tissues compared with ANT (Figure 1(a) , p < 0.001), and also high-expressed in BCa cell lines (MCF7, SK-BR-3, BT549, BT20, MDA-MB-231, MDA-MB-453), particularly in MCF7 and BT20 cell lines (Figure 1(b), p < 0.05 or p < 0.01 or p < 0.001) when compared with MCF-10A cell line. Thus, MCF7 and BT20 cell lines were used in following experiments.
Figure 1.
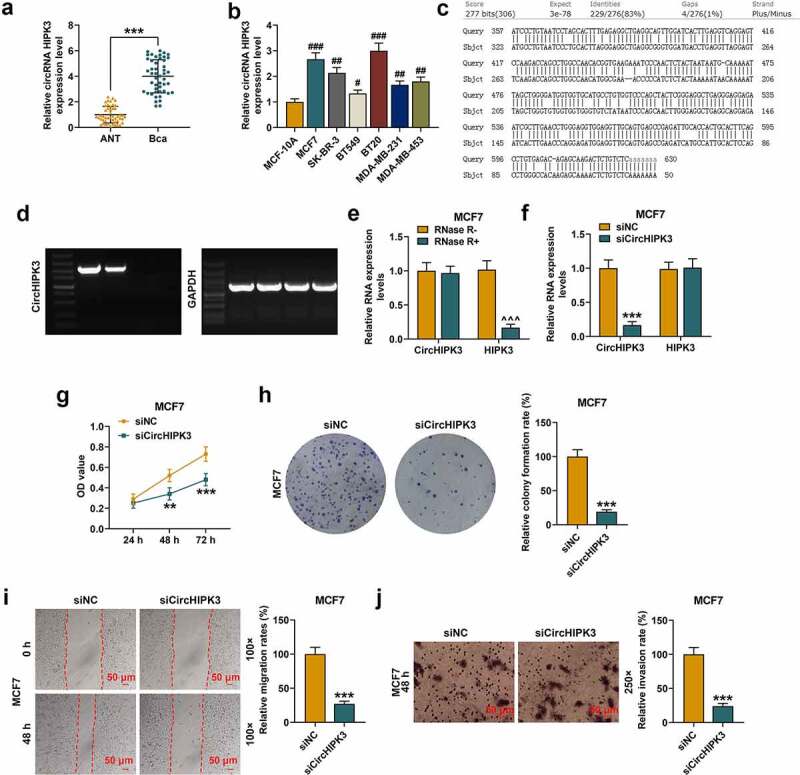
CircHIPK3 knockdown suppressed the malignant biological behaviors of MCF7 cells. (a) The expression of circHIPK3 in BCa tissues and adjacent non-cancerous tissues (ANT) was determined by RT-qPCR. (b) The expression of circHIPK3 in human normal mammary epithelial cell line MCF-10A, and BCa cell lines (MCF7, SK-BR-3, BT549, BT20, MDA-MB-231, MDA-MB-453) was determined by RT-qPCR. (c) CircHIPK3 (has_circ_000284) was located at the second exon of HIPK3 (chr11:33307958–33309057). The highly reverse complementary sequences (83% identity over 200 nucleotides) was found by aligning the sequence of intron 1 to that of intron 2 of the HIPK3 gene. (d) PCR was applied to detect the primers designed for circHIPK3 amplification. (e) The expressions of circHIPK3 and HIPK3 mRNA in MCF7 cells treated with or without RNase R were detected by RT-qPCR. (f) The expression of circHIPK3 in MCF7 cells after transfection with siCircHIPK3 or siNC was determined by RT-qPCR. (g) CCK-8 assay detected the viabilities MCF7 cells with siCircHIPK3 or siNC transfection. (h) Colony formation assay showed the colony formation of MCF7 cells with siCircHIPK3 or siNC transfection. (i) Wound-healing assay detected the inhibited migration of MCF7 cells with circHIPK3 knockdown. (j) Transwell assay detected that the invasion of MCF7 cells with circHIPK3 knockdown was inhibited. **P < 0.01 or ***P < 0.001 vs. siNC; ***P < 0.001 vs. ANT; #P < 0.05 or ##P < 0.01 or ###P < 0.001 vs. MCF-10A. ^^^P < 0.001 vs. RNase R- (without RNase R). GAPDH was used as an internal control. Data are shown as mean±SD, n = 3. BCa: RT-qPCR: reverse transcription-quantitative polymerase chain reaction; NC, negative control; MC, mimic control; M, miR-326 mimic; siCircHIPK3: small interfering circular RNA HIPK3; siNC, small interfering negative control; CCK-8, cell counting kit-8
Furthermore, it is well-known that the formation of circRNAs is commonly attributed to the sequences which can be capable of pairing to form RNA duplexes and significantly enhancing backsplicing [28]. Therefore, to confirm circHIPK3 is a circular RNA, we analyze the formation of circHIPK3. CircHIPK3 (has_circ_000284) was located at the second exon of HIPK3 (chr11:33307958–33309057). We investigated the characterization of circHIPK3 was by aligning the sequence of intron 1 to that of intron 2 of the HIPK3 gene, we found highly reverse complementary sequences (83% identity over 200 nucleotides) (Figure 1(c)). Then, we design primers for circHIPK3 amplification. First, PCR primers were designed to be divergent on linear cDNA thereby rendering amplification impossible. However, primers designed to be divergent on cDNA derived from circRNA these primers are convergent, leading to a circRNA-specific amplicon. The PCR results were showed inFigure 1(d). Besides, our study has demonstrated that the treatment of RNase R has no significant effects on the expression levels of circHIPK3. All these results indicated that circHIPK3 is circular. To verify the gene detected was circular gene (CircHIPK3) rather than linear gene (HIPK3), the gene expression after RNase R Treatment was determined. The results showed that circHIPK3 was resistant to RNase R, and that the mRNA expression of HIPK3 was obviously reduced in MCF7 (Figure 1(e), p < 0.001) and BT20 cells (Figure S1A, p < 0.001) after the RNase R treatment.
Down-regulated circHIPK3 suppressed the proliferation, migration and invasion of BCa cells
SicircHIPK3 inhibited the expression of circHIPK3 in MCF7 (Figure 1(f), p < 0.001) and BT20 (Figure S1B, p < 0.001) cells. Then the potential role of circHIPK3 in regulating the cellular behaviors of BCa cells was explored. According to CCK-8 assay results, circHIPK3 silencing significantly inhibited the viabilities of MCF7 cells (Figure 1(g), p < 0.01 or 0.001) and BT20 cells (Figure S1C, p < 0.01 or 0.001) at 48 and 72 h. Meanwhile, colony formation showed that circHIPK3 silencing significantly inhibited colony formation of MCF7 cells (Figure 1(h), p < 0.001) and BT20 cells (Figure S1D, p < 0.001) as well as the migration and invasion of MCF7 cells (Figure 1(i,j), p < 0.001) and BT20 cells (Figure S1E and F, p < 0.001).
MiR-326 was the target of circHIPK3 in BCa cells
The possible molecular mechanism of circHIPK3 in BCa was explored. The RT-qPCR assay was performed to detect the subcellular localization of circHIPK3 in BCa cells, and it was found that circHIPK3 and circular form of circHIPK3 were widely distributed in the cytoplasm of both MCF7 (Figure 2(a), p < 0.001) and BT20 cells (Figure S2A, p < 0.001). From the prediction of CircInteractome, circHIPK3 had potential binding sites to miR-326 (Figure 2(b)), and the prediction was further confirmed by a dual-luciferase assay using miR-326 inhibitor. The results demonstrated that the luciferase activities of MCF7 (Figure 2(c), p < 0.05) and BT20 cells (Figure S2B, p < 0.01) co-transfected with wild-type circHIPK3 and miR-326 inhibitor were increased compared with the control. Meanwhile, the luciferase activities of MCF7 and BT20 cells co-transfected with mutant circHIPK3 and miR-326 inhibitor did not change significantly. Moreover, silencing circHIPK3 significantly promoted the expression of miR-326 of MCF7 (Figure 2(d), p < 0.01) and BT20 cells (Figure S2C, p < 0.01). Additionally, miR-326 was noticeably down-regulated in BCa tissues compared that in ANT (Figure 2(e), p < 0.001). We also found that circHIPK3 expression was inversely correlated with miR-326 expression in ANT (Figure 2(f), p < 0.001) and BCa (Figure 2(g), p < 0.001). These data indicated that circHIPK3 may directly target miR-326 to down-regulate its expression in BCa.
Figure 2.
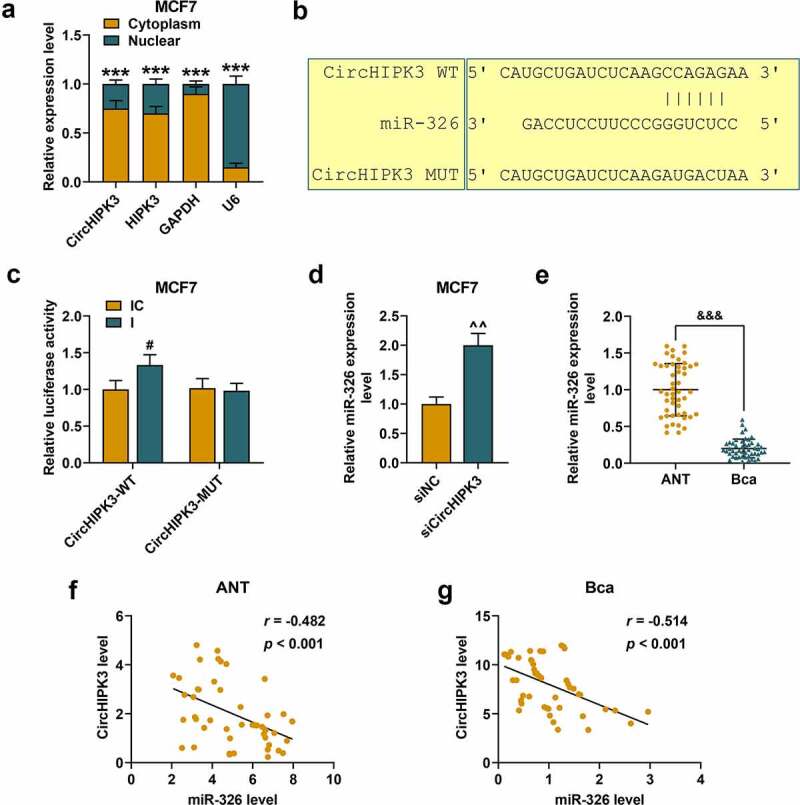
MiR-326 was targeted by circHIPK3 in MCF7 cells. (a) Cytoplasmic and nuclear RNA fractions were isolated from MCF7 cells. Relative expressions of circHIPK3 in the cell cytoplasm or nuclei were examined by RT-qPCR. GAPDH was used as the cytoplasmic control, and U6 was used as a nuclear control. (b) The binding sites for miR-326 in circHIPK3 were predicted by circInteractome. (c) Luciferase reporter assay showed that miR-326 inhibitor promoted the luciferase activity of circHIPK3 in MCF7 cells. (d) Relative expression of miR-326 was detected after transfection with sicircHIPK3 in MCF7 cells by RT-qPCR. U6 was used as an internal control. (e) Relative miR-326 expression was detected in the BCa tissues and adjacent non-cancerous tissues (ANT) was determined by RT-qPCR. (f and g) The correlation between circHIPK3 and miR-326 in BCa tissues and ANT was determined by Pearson’s correlation analysis. ***P < 0.001 vs. Cytoplasm; #P < 0.05 vs. IC; ^^P < 0.01 vs. siNC; &&& P < 0.001 vs. ANT. Data are shown as mean±SD, n = 3. Mut, mutant; WT, wild-type; siCircHIPK3, small interfering circular RNA HIPK3; siNC, small interfering negative control; IC, inhibitor control. RT-qPCR, reverse transcription-quantitative polymerase chain reaction
MiR-326 overexpression partially reversed the effect of CircHIPK3 on BCa cells
CircHIPK3 overexpression significantly inhibited the expressions of miR-326 in MCF7 (Figure 3(a), p < 0.01) and BT20 cells (Figure S3A, p < 0.01), which were partially reversed by miR-326 mimic (P < 0.001). MiR-326 mimic obviously promoted the expressions of miR-326 in MCF7 (Figure 3(a), p < 0.001) and BT20 cells (Figure S3A, p < 0.01), which, however, were partially reversed by CircHIPK3 overexpression (P < 0.01). The viabilities of MCF7 (Figure 3(b), p < 0.05) and BT20 cells (Figure S3B, p < 0.05) were noticeably increased by circHIPK3 overexpression, but the effects were partially reversed by miR-326 overexpression (Figure 3(b) and S3B, p < 0.01). MiR-326 overexpression in MCF7 and BT20 cells obviously decreased the cell viabilities, which were partially reversed by circHIPK3 overexpression (Figure 3(b) and S3B, p < 0.05). Similarly, the results of colony formation assay demonstrated that circHIPK3 overexpression in MCF7 (Figure 3(c), p < 0.01) and BT20 cells (Figure S3C, p < 0.01) significantly increased colony formation rates, however, miR-326 overexpression partly reduced the colony formation (Figure 3c and S3C, p < 0.01).
Figure 3.
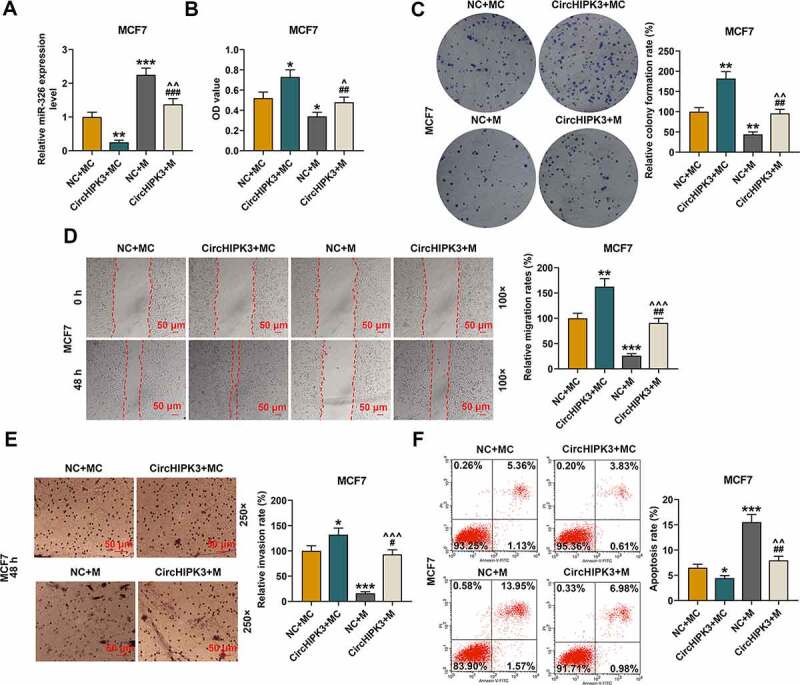
MiR-326 overexpression partially reversed the effect of CircHIPK3 on MCF7 cells. (a) RT-qPCR determined the expression of miR-326 in MCF7 cells transfected with miR-326 mimic and /or circHIPK3. U6 was used as an internal control. (b) CCK-8 assay indicated the viabilities of MCF7 cells transfected with miR-326 mimic and /or circHIPK3. (c) Colony formation assay indicated the colony formation of MCF7 cells transfected with miR-326 and /or circHIPK3. (d) Wound-healing assay showed that the migration of MCF7 cells transfected with miR-326 and /or circHIPK3 was inhibited. (e) Transwell assay showed that the invasion of MCF7 cells transfected with miR-326 and /or circHIPK3 was inhibited. (f) Flow cytometry assay determined the apoptosis of MCF7 cells transfected with miR-326 and /or circHIPK3. *P < 0.05 or **P < 0.01 or ***P < 0.001 vs. NC+MC; ##P < 0.01 or ###P < 0.001 vs. CircHIPK3+ MC; ^P < 0.05 ^^P < 0.01 ^^^P < 0.001 vs. NC+M. Data are shown as mean±SD, n = 3. NC, negative control; MC, mimic control; M, miR-326 mimic; RT-qPCR, reverse transcription-quantitative polymerase chain reaction
According to wound-healing and transwell assays, it was found that the cell migration and invasion of MCF7 (Figure 3(d,e), p < 0.05 or 0.01) and BT20 cells (Figure S3D and S3E, p < 0.05 or 0.01) promoted by circHIPK3 overexpression were partially reduced by miR-326 overexpression, but then increased by circHIPK3 overexpression to some extent (for MCF7: Figure 3(d,e), p < 0.001; for BT20: Figure S3D and S3E, p < 0.01 or 0.001). According to flow cytometric assays, circHIPK3 overexpression greatly decreased the apoptosis of MCF7 (Figure 3(f), p < 0.05) and BT20 cells (Figure S3F, p < 0.01), but miR-326 overexpression partly increased the apoptosis (Figures 3(f) and S3F, p < 0.01). Noticeably, miR-326 overexpression significantly increased apoptosis of MCF7 (Figure 3(f), p < 0.001) and BT20 cells (Figure S3F, p < 0.001), which were partially reversed by CircHIPK3 overexpression (Figure 3(f) and Figure S3F, p < 0.01).
The results of Western blot and RT-qPCR showed that for MCF7 cells with circHIPK3 overexpression, the expressions of cleaved Caspase-3 (Figure 4(a), p < 0.001) and Bax were significantly decreased (Figure 4(a,b), p < 0.05 or 0.01), and that of Bcl-2 was also increased (Figure 4(a,b), p < 0.05 or 0.01), which were all partially reversed by miR-326 overexpression (Figure 4(a,b), p < 0.05 or 0.01 or 0.001). Similarly, circHIPK3 overexpression in BT20 cells significantly decreased the expressions of cleaved Caspase-3 (Figure S4A, p < 0.01) and Bax (Figure S4A and B, p < 0.001) but increased that of Bcl-2 (Figure S4A and B, p < 0.05), which were all partially reversed by miR-326 overexpression (Figure S4A and B, p < 0.01 or 0.001). Additionally, circHIPK3 overexpression decreased the expression of E-cadherin, but increased the expressions of N-cadherin and Vimentin of MCF7 (Figure S4C and D, p < 0.05 or 0.01 or 0.001) and BT20 cells (Figure S4C and D, p < 0.01 or 0.001), which were partially reversed by miR-326 overexpression (P < 0.05 or 0.001).
Figure 4.
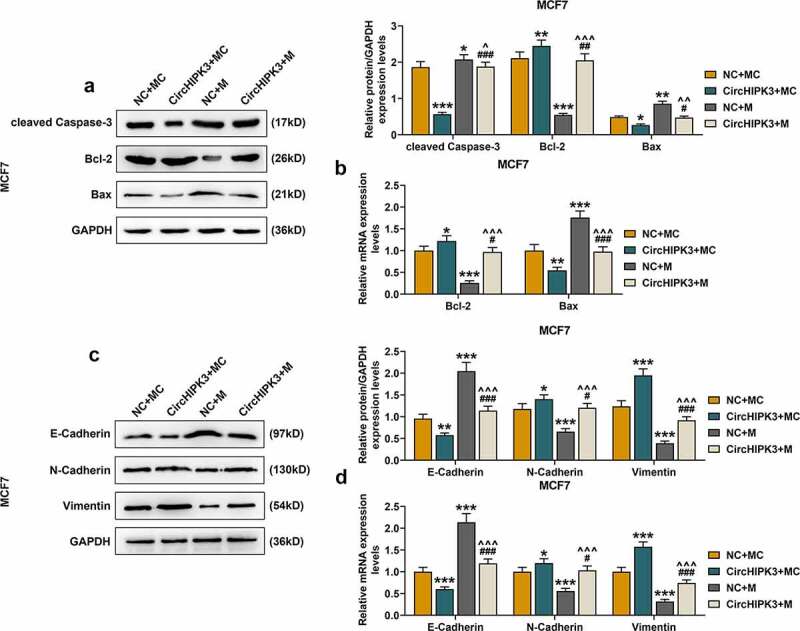
MiR-326 overexpression partially reversed the effect of CircHIPK3 on the expression of apoptosis- and metastasis-related proteins in MCF7 cells. (a) Western blot detected the expressions of Bax, Bcl-2 and cleaved Caspase-3 in MCF7 cells transfected with miR-326 and /or circHIPK3. (b) RT-qPCR detected the expressions of Bax and Bcl-2 in MCF7 cells transfected with miR-326 and /or circHIPK3. (c) Western blot detected the expressions of E-Cadherin, N-Cadherin and Vimentin in MCF7 cells transfected with miR-326 and /or circHIPK3. (d) RT-qPCR indicated the expressions of E-Cadherin, N-Cadherin and Vimentin in MCF7 cells transfected with miR-326 and /or circHIPK3. GAPDH was used as an internal control. *P < 0.05 or **P < 0.01 or ***P < 0.001 vs. NC+MC; #P < 0.05 or ##P < 0.01 or ###P < 0.01 vs. CircHIPK3+ MC; ^P < 0.05 ^^P < 0.01 ^^^P < 0.001 vs. NC+M. Data are shown as mean±SD, n = 3. NC, negative control; MC, mimic control; M, miR-326 mimic; RT-qPCR, reverse transcription-quantitative polymerase chain reaction
MiR-326 overexpression partially reversed the effect of circHIPK3 on promoting BCa tumorigenicity in vivo
The data revealed that circHIPK3 overexpression obviously increased BCa tumor diameters (Figures 5(a) and S5A) and weights of the mice (Figure 5(b) and S5B, p < 0.001) compared with those of control group, however, these effects were partially reversed by miR-326 overexpression (Figures 5(a,b) and S5A and B, p < 0.001). MiR-326 overexpression obviously reduced the diameters (Figures 5(a) and S5A) and weights (Figures 5(b) and S5B, p < 0.001) of BCa tumor compared with those of control group, however, these effects were partially reversed by circHIPK3 overexpression (Figures 5(a,b), and S5A and B, p < 0.001).
Figure 5.
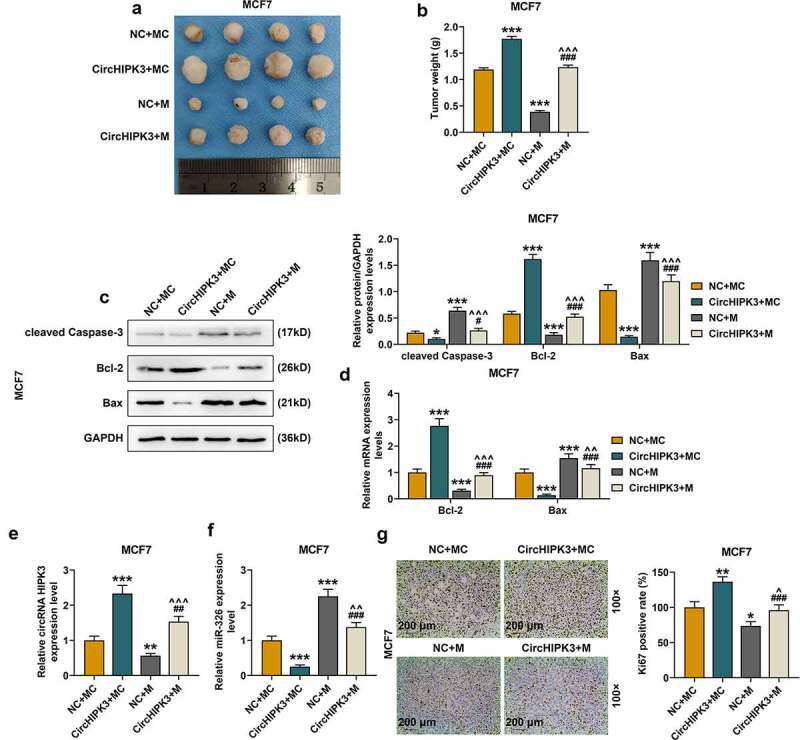
MiR-326 overexpression partially reversed the effect of CircHIPK3 on promoting BCa tumorigenicity in vivo. (a) Representative tumors from nude mice after injection of controls cells or MCF7 cells transfected with miR-326 and /or circHIPK3. (b) Tumor weights were measured. (c) Western blot detected the expressions of Bax, Bcl-2 and cleaved Caspase-3 in tumor from nude mice after the injection of controls cells or MCF7 cells transfected with miR-326 and /or circHIPK3. (d) RT qPCR determined the expressions of Bax, Bcl-2 in tumor from nude mice after the injection of controls cells or MCF7 cells transfected with miR-326 and /or circHIPK3. (e) RT-qPCR detected the expression of circHIPK3 in tumor from nude mice after the injection of controls cells or MCF7 cells transfected with miR-326 and /or circHIPK3. (f) RT-qPCR detected the expression of miR-326 in tumor from nude mice after the injection of controls cells or MCF7 cells transfected with miR-326 and /or circHIPK3. (g) Ki67 expression were determined by immunohistochemistry in each group. GAPDH was used as an internal control. *P < 0.05 or **P < 0.01 or ***P < 0.001 vs. NC+MC; #P < 0.05 or ###P < 0.01 vs. CircHIPK3+ MC; ^^P < 0.01 or ^^^P < 0.001 vs. NC+M. Data are shown as mean±SD, n = 3. NC, negative control; MC, mimic control; M, miR-326 mimic; RT-qPCR, reverse transcription-quantitative polymerase chain reaction
CircHIPK3 overexpression reduced the expressions of cleaved Caspase-3 and Bax but increased the expression of Bcl-2, which were partially reversed by miR-326 overexpression (Figure 5(c,d)). Furthermore, in tumor tissues of the circHIPK3 group at the end of the experiment, the expression of circHIPK3 was markedly increased and partially reduced by miR-326 overexpression (Figures 5(e) and S5E, p < 0.01). The expression of miR-326 was markedly decreased in the tumor tissues of the miR-326 mimic group (Figures 5(e) and S5E, p < 0.01), but was partially increased by circHIPK3 overexpression (Figures 5(e) and S5E, p < 0.001). Moreover, the expression of miR-326 in tumor tissues was markedly decreased by circHIPK3 overexpression (Figures 5(f) and S5F, p < 0.001), but the effect of which was partially reversed by miR-326 overexpression (Figure 5(f) and S5F, p < 0.001). In addition, immunohistochemistry results showed that compared with control group, the tumor tissues collected from the circHIPK3 group showed more Ki-67 positive (Figures 5(g) and S5G), which was partially reversed by miR-326 overexpression (Figures 5(g) and S5G).
Discussion
Studies reported that CircHIPK3 is dysregulated in some cancers. CircHIPK3 is high-expressed in colorectal cancer [11], gallbladder cancer [29], lung cancer [30], epithelial ovarian cancer [31], prostate cancer [10], and nasopharyngeal carcinoma [32]. However, the circHIPK3 expression also shows specificity to different cancers. CircHIPK3 is down-regulated in bladder cancer [12] and gastric cancer [33], but its expression in BCa is hardly reported, and to the best of our knowledge, the current study was the first to directly demonstrate that circHIPK3 expression was increased in BCa tissues and cell lines. Our data indicated that circHIPK3 was a potential therapeutic gene for BCa and tumor oncogene for the disease progression.
Circular RNAs are highly stable in vivo, and are mainly distributed in the cytoplasm [34]. Consistently, the present study showed that circHIPK3 was stable after treatment with RNase R, and that circHIPK3 and HIPK3 mainly located in the cytoplasm of BCa cells. These results suggested that circHIPK3 was a circRNA generated from HIPK3 mRNA. Evidence supported that some dysregulated circRNAs are closely involved in BCa, and play significant role in the cancer progression. For example, circKIF4A and circEPSTI1 serves as prognostic factors and mediators to modulate the development of triple-negative BCa [35,36]; circular RNA hsa_circ_001783 regulates breast cancer development through sponging miR-200c-3p [37]. Similarly, in the present study, reducing circHIPK3 expression suppressed cell proliferation, migration and invasion of BCa cells. CircRNAs are not simply the junk-products of pre-mRNA splicing [5]. From circularRNA profiling, a study indicated that circHIPK3 modulates cell growth through sponging multiple miRNAs [25]. Therefore, we speculated that circHIPK3 serves as a ceRNA to modulate target gene miRNA, thus influencing BCa development.
The results from circInteractome database showed that circHIPK3 had binding sites to miR-326. By performing dual-luciferase assay, the results confirmed that miR-326 inhibitor increased the luciferase intensity by the wild-type 3′-UTR of circHIPK3 in BCa cells, but the luciferase intensity of mutant 3'-UTR of circHIPK3 was not changed by miR-326 inhibitor. Furthermore, the expression of miR-326 was found up-regulated after silencing circHIPK3 in BCa cells, and was inversely correlated with the expression level of circHIPK3 in BCa tissues and adjacent non-cancerous tissues. Moreover, miR-326 expression was down-regulated in BCa cells. Therefore, these results demonstrated that miR-326 was a direct target gene for circHIPK3.
In the present study, we found that miR-326 expression was inhibited by circHIPK3 overexpression in BCa cells, but was reversed by miR-326 overexpression. CircHIPK3 overexpression promoted the viability, colony formation, migration, invasion of BCa cells, but inhibited the cell apoptosis. However, the effects of circHIPK3 overexpression on the BCa cells were partially reversed by miR-326 overexpression. Although we have explored the role of circHIPK3 in cancer phenotype at cellular level, molecular expression involved in BCa progression was not fully understood. We therefore determined the expressions of pro-apoptotic gene (Bax), anti-apoptotic gene (Bcl-2), cleaved Caspase-3, and epithelial-to-mesenchymal transition (EMT)-related markers (E-Cadherin, N-Cadherin and Vimentin). In the present study, circHIPK3 decreased the expressions of Bax, cleaved Caspase-3, E-Cadherin, but increased the expressions of Bcl-2, N-Cadherin and Vimentin in BCa cells. As expected, these effects were reversed by miR-326 overexpression.
Increasing evidence demonstrated that cellular apoptosis is a programmed cell-death dysregulated in various malignant tumors [38]. Cell apoptosis is regulated by multiple factors, especially by Bcl-2 and Bax and the activation of apoptotic cascade through caspase-3, and cleaved caspase-3 protein is an executioner of apoptotic pathway and is also required for induction of apoptosis [39]. EMT is involved in the migration and invasion features of tumors and plays a significant role in cancer metastasis [40,41]. EMT is characterized by decreased E-Cadherin expression and increased N-Cadherin expression [42]. Vimentin is widely found in normal mesenchymal cells and maintains cellular integrity [43]. In this study, circHIPK3 sponging miR-326 inhibited apoptosis and promoted metastasis through regulating apoptotic molecules (Bcl-2, Bax and cleaved Caspase-3) and (EMT)-related markers (E-Cadherin, N-Cadherin and Vimentin).
Moreover, the tumor growth in vivo was promoted by circHIPK3 overexpression, but was reversed by miR-326 overexpression. The effects of circHIPK3 and miR-326 on apoptotic molecules (Bcl-2, Bax and cleaved Caspase-3) in vivo were consistent with those in vitro. The expression of ki-67 was promoted by circHIPK3 overexpression, but was reversed by miR-326 overexpression. These data suggested the BCa tumorigenicity was promoted by circHIPK3 overexpression, which was reversed by miR-326 overexpression.
However, this study still has some limitations. Studies have demonstrated the interaction between circRNAs and protein coding genes. However, our study only focused on the mechanisms of the circRNA functioning as a sponge for miRNA, but whether circHIPK3 was associated with a protein coding gene was not investigated.
Conclusion
In conclusion, the present study found that circHIPK3 overexpression sponging miR-326 promoted the proliferation and metastasis, inhibited apoptosis of BCa cells, and promoted BCa tumorigenicity in vivo.
Supplementary Material
Funding Statement
No funding was received.
Data availability statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
COI-statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Mathews MJ, Mead RN, Galizio M.. Effects of N-Methyl-D-aspartate (NMDA) antagonists ketamine, methoxetamine, and phencyclidine on the odor span test of working memory in rats. Exp Clin Psychopharmacol. 2018. Feb;26(1):6–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li W, Jia G, Qu Y, et al. Long non-coding RNA (LncRNA) HOXA11-AS promotes breast cancer invasion and metastasis by regulating epithelial-mesenchymal transition. Med Sci Monit. 2017. Jul 13;23:3393–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lemler DJ, Lynch ML, Tesfay L, et al. DCYTB is a predictor of outcome in breast cancer that functions via iron-independent mechanisms. BCR. 2017. Mar 7;19(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015. Mar;34(1):145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016. Apr;17(4):205–211. [DOI] [PubMed] [Google Scholar]
- [6].Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017. Aug 10;36(32):4551–4561. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [7].Liu X, Zhong Y, Li J, et al. Circular RNA circ-NT5C2 acts as an oncogene in osteosarcoma proliferation and metastasis through targeting miR-448. Oncotarget. 2017. Dec 29;8(70):114829–114838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kristensen LS, Hansen TB, Veno MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018. Feb 1;37(5):555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017. May 23;16(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cai C, Zhi Y, Wang K, et al. CircHIPK3 overexpression accelerates the proliferation and invasion of prostate cancer cells through regulating miRNA-338-3p. Onco Targets Ther. 2019;12:3363–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018. Apr 1;9(4):417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [12].Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017. Sep;18(9):1646–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lu H, Han X, Ren J, et al. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol Ther. 2020;21(2):113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].He L, Tang M, Xiao T, et al. Obesity-associated miR-199a/214 cluster inhibits adipose browning via PRDM16-PGC-1alpha transcriptional network. Diabetes. 2018. Dec;67(12):2585–2600. [DOI] [PubMed] [Google Scholar]
- [15].Yang BF, Lu YJ, Wang ZG. MicroRNAs and apoptosis: implications in the molecular therapy of human disease. Clin Exp Pharmacol Physiol. 2009. Oct;36(10):951–960. [DOI] [PubMed] [Google Scholar]
- [16].Dong Y, Liu Y, Jiang A, et al. MicroRNA-335 suppresses the proliferation, migration, and invasion of breast cancer cells by targeting EphA4. Mol Cell Biochem. 2018. Feb;439(1–2):95–104. [DOI] [PubMed] [Google Scholar]
- [17].Liang X, Li Z, Men Q, Liang X, Li Z, Men Q, et al . miR-326 functions as a tumor suppressor in human prostatic carcinoma by targeting Mucin1. Biomed Pharmacothe. 2018. Dec;108:574–583. [DOI] [PubMed] [Google Scholar]
- [18].Zhou J, Xu T, Yan Y, et al. MicroRNA-326 functions as a tumor suppressor in glioma by targeting the Nin one binding protein (NOB1). PLoS One. 2013;8(7):e68469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ji S, Zhang B, Kong Y, et al. miR-326 inhibits gastric cancer cell growth through downregulating NOB1. Oncol Res. 2017. Jul 5;25(6):853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liang Z, Wu H, Xia J, et al. Involvement of miR-326 in chemotherapy resistance of breast cancer through modulating expression of multidrug resistance-associated protein 1. Biochem Pharmacol. 2010. Mar 15;79(6):817–824. [DOI] [PubMed] [Google Scholar]
- [21].Ghaemi Z, Soltani BM, Mowla SJ. MicroRNA-326 functions as a tumor suppressor in breast cancer by targeting ErbB/PI3K signaling pathway. Front Oncol. 2019;9:653. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [22].Du Y, Shen L, Zhang W, et al. Functional analyses of microRNA-326 in breast cancer development. Biosci Rep. 2019. Jul 31;39(7). DOI: 10.1042/BSR20190787 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [23].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013. Mar 21;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- [24].Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016. Apr 6;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001. Dec;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- [27].Wang F, Ying HQ, He BS, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015. Apr 10;6(10):7899–7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang XO, Wang HB, Zhang Y, et al. Complementary sequence-mediated exon circularization. Cell. 2014. Sep 25;159(1):134–147. [DOI] [PubMed] [Google Scholar]
- [29].Kai D, Yannian L, Yitian C, et al. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem Biophys Res Commun. 2018. Sep 5;503(2):863–869. [DOI] [PubMed] [Google Scholar]
- [30].Yu H, Chen Y, Circular JP. RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018. Nov 30;506(3):455–462. [DOI] [PubMed] [Google Scholar]
- [31].Liu N, Zhang J, Zhang LY, et al. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018. Jun;22(12):3713–3718. [DOI] [PubMed] [Google Scholar]
- [32].Shuai M, Hong J, Huang D, et al. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018. Nov;16(5):6495–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ghasemi S, Emadi-Baygi M, Nikpour P. Down-regulation of circular RNA ITCH and circHIPK3 in gastric cancer tissues. Turk J Med Sci. 2019. Apr 18;49(2):687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015. Aug;25(8):981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tang H, Huang X, Wang J, et al. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer. 2019. Feb 11;18(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen B, Wei W, Huang X, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8(14):4003–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu Z, Zhou Y, Liang G, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019. Jan 22;10(2):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Feng X, Zhang C, Yang Y, et al. Role of miR-181a in the process of apoptosis of multiple malignant tumors: a literature review. Adv Clin Exp Med. 2018. Feb;27(2):263–270. [DOI] [PubMed] [Google Scholar]
- [39].Dolka I, Krol M, Sapierzynski R. Evaluation of apoptosis-associated protein (Bcl-2, Bax, cleaved caspase-3 and p53) expression in canine mammary tumors: an immunohistochemical and prognostic study. Res Vet Sci. 2016. Apr;105:124–133. [DOI] [PubMed] [Google Scholar]
- [40].Jakobsen KR, Demuth C, Sorensen BS, et al. The role of epithelial to mesenchymal transition in resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Transl Lung Cancer Res. 2016. Apr;5(2):172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Xiao Z, Chen M, Yang J, et al. [MTBP regulates migration and invasion of prostate cancer cells in vitro]. Nan Fang Yi Ke Da Xue Xue Bao. 2019. Jan 30;39(1):6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ramamurthy VP, Ramalingam S, Gediya LK, et al. The retinamide VNLG-152 inhibits f-AR/AR-V7 and MNK-eIF4E signaling pathways to suppress EMT and castration-resistant prostate cancer xenograft growth. FEBS J. 2018. Mar;285(6):1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011. Sep;68(18):3033–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.


