ABSTRACT
Passive transfer of broadly neutralizing antibodies is showing promise in the treatment and prevention of HIV-1. One class of antibodies, the VRC01 class, appears especially promising. To improve VRC01-class antibodies, we combined structure-based design with a matrix-based approach to generate VRC01-class variants that filled an interfacial cavity, used diverse third-complementarity-determining regions, reduced potential steric clashes, or exploited extended contacts to a neighboring protomer within the envelope trimer. On a 208-strain panel, variant VRC01.23LS neutralized 90% of the panel at a geometric mean IC80 less than 1 μg/ml, and in transgenic mice with human neonatal-Fc receptor, the serum half-life of VRC01.23LS was indistinguishable from that of the parent VRC01LS, which has a half-life of 71 d in humans. A cryo-electron microscopy structure of VRC01.23 Fab in complex with BG505 DS-SOSIP.664 Env trimer determined at 3.4-Å resolution confirmed the structural basis for its ~10-fold improved potency relative to VRC01. Another variant, VRC07-523-F54-LS.v3, neutralized 95% of the 208-isolated panel at a geometric mean IC80 of less than 1 μg/ml, with a half-life comparable to that of the parental VRC07-523LS. Our matrix-based structural approach thus enables the engineering of VRC01 variants for HIV-1 therapy and prevention with improved potency, breadth, and pharmacokinetics.
KEYWORDS: Antibody VRC01, broadly neutralizing antibody, HIV-1 envelope trimer, matrix-based design, polyreactivity, prophylaxis, treatment
Introduction
Advances in B-cell sequencing and high-throughput neutralization have enabled the isolation of potent and broadly neutralizing HIV-1 antibodies, with multiple animal models demonstrating their efficacy in preventing infection and with clinical studies beginning to demonstrate their ability to suppress virus in infected donors.1–5 Among this new generation of neutralizing antibodies are PG9, PGDM1400, and VRC26.25, which target the V1V2 apex of the HIV-1 Env;6–8 PGT121,10–1074, and BG18, that target the glycan-V3 epitope;9–11 VRC01, N6, N49P7, 3BNC117, and 1–18 that target the CD4-binding site;12–16 4E10, 10E8, DH511, and VRC42, which target the membrane-proximal external region17–20; 35O22, PGT151 and 8ANC195 that target the gp120 and gp41 interface region;21–23 and SF12, which recognizes the silent face of HIV-1.24
Passive transfer of these antibodies has shown varying degrees of protection against SHIV challenges in nonhuman primates (NHPs),25–31 and suppression of viremia in HIV-1-infected donors.32–35 In NHP studies, the effective serum concentration of the antibodies required for protection in vivo was estimated to be ~200-fold higher than the geometric mean IC50 measured in the TZM-bl neutralization assay.1 Recently, results of two randomized trials, HVTN 704/HPTN 085 and HVTN 703/HPTN 081, that evaluated antibody-mediated prevention against HIV-1 exposure in humans were reported. In these studies, 10 or 30 mg/kg of VRC01 was administered to 2,699 men and 1,924 women every 8 weeks for ten infusions in total, and the results indicated that VRC01 conferred protection effectively against strains with an IC80 less than 1 µ/ml.36 Altogether, accumulating data show a clear need for antibodies with high neutralization potency and breadth for antibody-mediated prevention and treatment.
Multiple approaches have been used to improve the potency and breadth of isolated antibodies, including structure-based design,37–39 surface-matrix screening,40 and library-based optimization.41 In this study, we sought to further increase the potency and breadth of VRC01-class antibodies, as they exhibit some of the best potencies and breadths against currently circulating isolates. Moreover, VRC01-class antibodies such as 3BNC117, VRC01, VRC01LS, and VRC07-523LS have shown favorable safety profiles in clinical trials32,42–46 with the extended half-life variant VRC01“LS” reaching 71 ± 18 d.47–49 We applied a structure-based matrix approach to generate variants of VRC01, VRC07-523LS, VRC08, N6, 3BNC117 and N49P7, and assessed neutralization potency and breadth, polyreactivity, manufacturability, and pharmacokinetics in human neonatal Fc receptor (FcRn) transgenic mice. On a diverse cross-clade panel of 208 HIV-1 strains,50 the best of the obtained variants showed more than 10-fold higher potency than VRC01, with favorable pharmacokinetic properties.
Results
A matrix of structure-based designs focuses on hotspots dominating VRC01-class antibody recognition
To improve the potency and breadth of VRC01-class antibodies, we chose to start with antibody VRC01, as this antibody, especially the extended half-life version VRC01LS, has already demonstrated superior physical and pharmacokinetic properties47 (Figure 1). Studies to improve the function of VRC01-class antibodies by structure-based rational design have been reported previously. Four hotspots dominating recognition have been investigated: (1) Gly54 in the heavy chain; Diskin et al. showed filling a hydrophobic pocket at the interface between gp120 and antibody NIH45-46 by substituting Gly54 with Trp (G54W) to improve its affinities to gp120s and neutralization potency by up to 4-fold and 10-fold, respectively37 (Figure 1a); (2) CDR H3; the CDR H3 of NIH45-46 and VRC07, four amino acids longer than the CDR H3 of VRC01, allows NIH45-46 and VRC07 to engage gp120 with greater contact surface than VRC0137,38 (Figure 1b); (3) light-chain N-terminus; truncation of VRC01 light-chain Glu1 and Ile2 with Val3 to Ser mutation (V3S) improve the potency of VRC07-523 variants38 (Figure 1c); and (4) VRC03-framework region 3 (03FR3) loop; addition of this region increases potency while reducing polyreactivity. Truncation of the 03FR3 loop in VRC03 resulted in loss of its binding to BG505 SOSIP.664 and reduced binding to JRFL SOSIP.66439 (Figure 1d).
Figure 1.
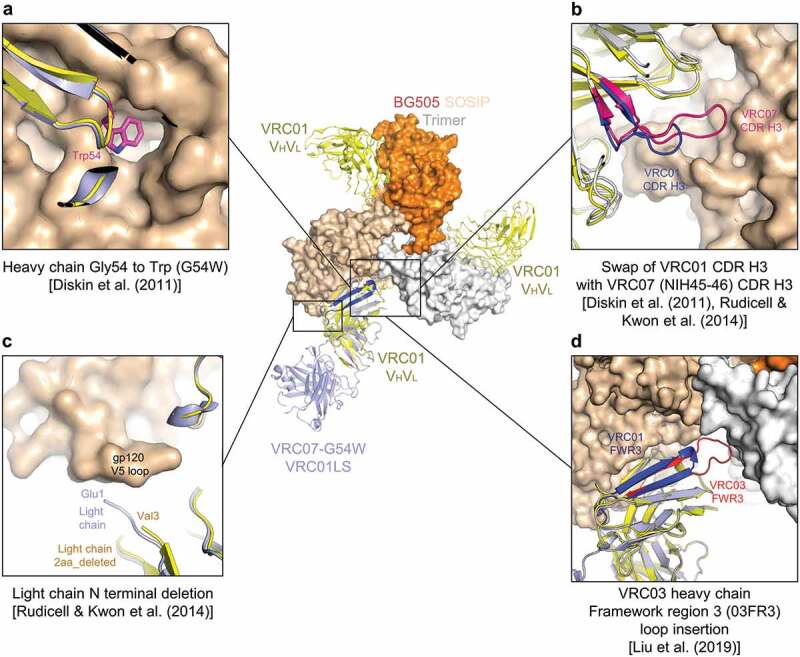
Structure-based approach focuses on recognition hotspots for VRC01-class antibodies. (a) Heavy-chain Gly54 to Trp mutation to mimic Phe43CD4 occupying a hydrophobic pocket on gp120. (b) Swap of VRC01 CDR H3 with VRC07 CDR H3 to increase binding surface between gp120 and the CDR H3. (c) Deletion of three residues of N-terminal light chain to better accommodate various lengths and conformations of the V5 region of gp120. (d) Replacement of framework region 3 with VRC03 framework region 3 (03FR3) to extend its interaction to the neighboring protomer. The structure of core gp120 in complex with VRC07-G54W (PDB ID: 4OLZ) is shown, superimposed on the structure of BG505 DS-SOSIP in complex with VRC01 Fab (PDB ID: 6NNF)
While each of these hotspots has been tested separately, antibodies containing mutations on these hotspots in combination have not been assessed for their potency, breadth, and pharmacokinetics. We therefore generated a 4 × 3 matrix of VRC01 variants displaying four combinations of G54W, 03FR3, or VRC07-CDR H3 mutation in the heavy chain with the light chain as (1) wild-type, (2) incorporating Glu1 and Ile2 deleted and V3S mutation (2aa_del_V3S), or (3) incorporating a deletion of the first three residues (3aa_del) (Figure 2a and Table S1), and assessed their neutralization potency and breadth on a representative 12-virus panel (Figure 2b). We found VRC01.23LS containing G54W and 03FR3 mutation in the heavy chain and 3aa_del in the light chain to be the most potent among the 12 variants tested, with its geometric mean IC50 improved more than 50-fold compared to VRC01LS. Overall, the heavy-chain G54W mutation improved the potency about fivefold, and the breadth from 58% to 83% compared to the parental VRC01LS, and 03FR3 insertion improved the potency about twofold, and the breadth from 83% to 91%. However, swapping the CDR H3 with that of NIH45-46 did not increase the potency and breadth. In addition, we observed a clear trend in rank-order for potency of the light-chain variants, with 3aa_del the most potent followed by 2aa_del_V3S and by wild-type.
Figure 2.
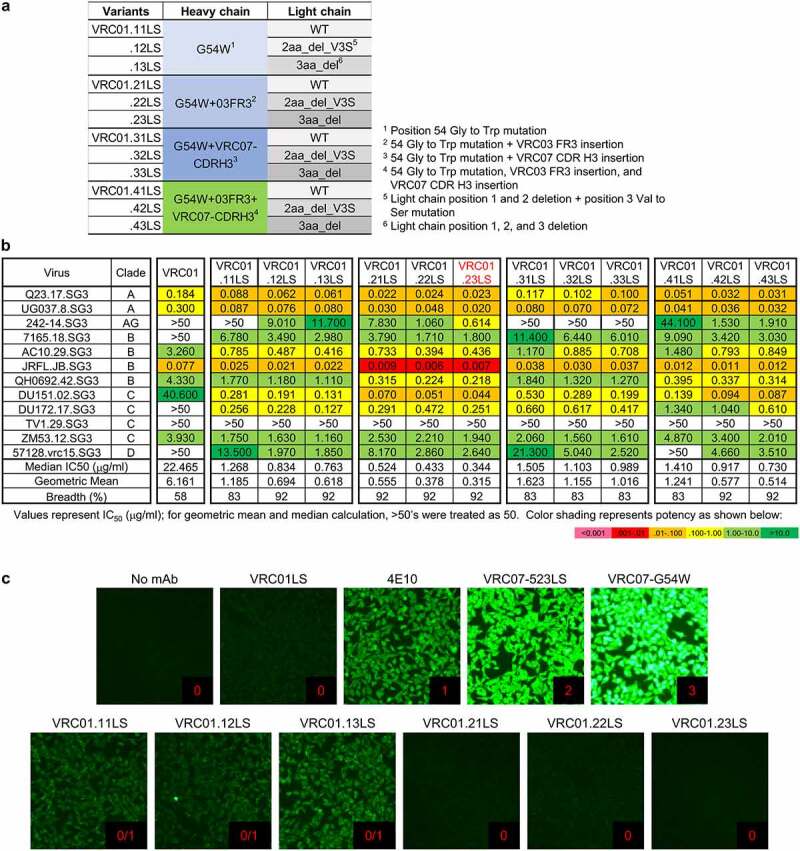
A matrix of structure-based designs identifies a variant, VRC01.23LS, with improved potency and low HEp-2 reactivity. (a) Four different VRC01LS heavy chain variants were paired with three different VRC01LS light chains to generate 12 VRC01LS variants. (b) Neutralization IC50 values of 12 VRC01LS variants assessed on 12 diverse strains of HIV-1. Variant VRC01.23LS neutralized 11 out of 12 viruses with median IC50 of 0.34 μg/ml. (c) HEp-2 cell staining assay against VRC01LS variants was performed at a concentration of 25 μg/ml along with control antibodies, VRC01LS, 4E10, VRC07-523LS, and VRC07-G54W. Control antibodies were assigned a score between 0 and 3. Antibodies scored greater than 1 at 25 µg/ml were considered polyreactive
Next, we assessed the polyreactivity of the variants using HEp-2 cells and a cardiolipin enzyme-linked immunosorbent assay (ELISA). Contrary to what has been observed for antibodies NIH45-46 and VRC07, whose polyreactive was substantially increased by G54W mutation,38 G54W in antibody VRC01 did not induce much polyreactivity (see variants VRC01.11LS, VRC01.12LS and VRC01.13LS in Figure 2c). Notably, when both 03FR3 and G54W mutations were introduced, no detectable polyreactivity was observed in HEp-2 cell staining and cardiolipin-binding assay (Figure 2c and Table S2). Thus, a matrix of structure-based designs yielded VRC01.23LS, a variant with ~70-fold increased potency, as assessed on a 12-isolate panel versus the parent VRC01LS, along with low polyreactivity.
Additional VRC01-class variants with improved potency and breadth
In light of the substantial improvement in potency of VRC01.23LS versus VRC01LS, we sought to apply the matrix-based approach to improve other VRC01-class antibodies such as 3BNC117, VRC07-523LS, N6, N49P713 and VRC08,51 some of which showed favorable clinical properties.42,44 Since we found that heavy-chain G54W and 03FR3 mutation yielded the most potent VRC01LS variant, we introduced both G54W and 03FR3 mutations to the heavy chains of these antibodies and paired them with light-chain wild-type, 2aa_del, or 3aa_del (Figure 3a and Table S1).
Figure 3.
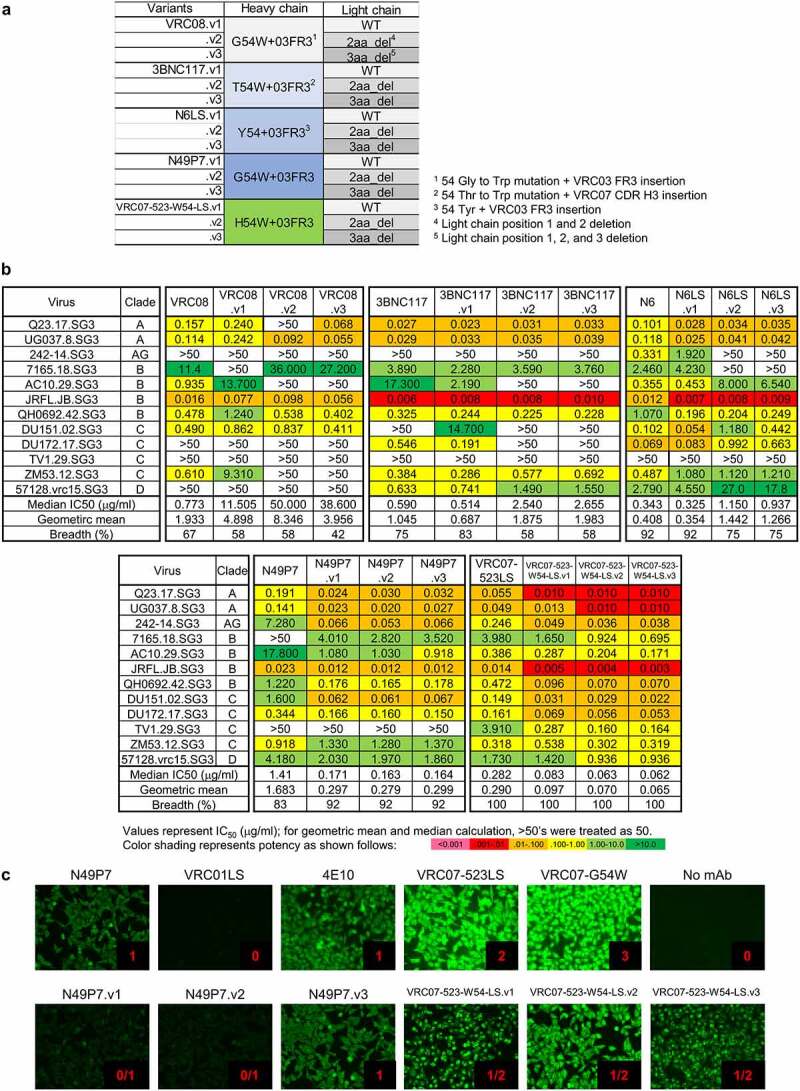
N49P7 and VRC07-523LS variants with improved potency and some HEp-2 reactivity. (a) Matrix of VRC01-class antibody variants. (b) Neutralization IC50 values of N49P7 variants and VRC07-523LS variants against a 12-virus panel. (c) HEp-2 cell staining assay against N49P7 and VRC07 variants was performed in the concentration of 25 μg/ml along with control antibodies, VRC01LS, 4E10, VRC07-523LS, and VRC07-G54W. Control antibodies were assigned a score between 0 and 3. Test antibodies scored greater than 1 at 25 µg/ml were considered polyreactive
As some VRC01-class antibodies have residues other than the germline-encoded Gly at position 54 in the heavy chain, mimicking Phe43CD4-gp120 interactions, (3BNC117, N6, and VRC07-523LS have Thr, Tyr, and His at position 54, respectively), we reasoned that a Trp mutation at residue 54 in these antibodies might not achieve a level of improvement similar to what has been observed with VRC01. Thus, we retained a tyrosine at position 54 in N6 (Figure 3a), but changed Thr and His to Trp in both 3BNC117 and VRC07-523LS. When tested, all variants with heavy-chain Trp54 and 03FR3 mutation showed improvements in potency, ranging from 1.4- to 6-fold, except for VRC08 (Figure 3b). Improvements in potency as a result of light-chain mutations were observed for N49P7 and VRC07-523LS variants, with light-chain mutations in 3BNC117, N6 and VRC08 decreasing potency and breadth. Overall, N49P7.v2 and VRC07-523-W54-LS.v3 showed ~6- and ~4.5-fold improvement in potency, respectively (Figure 3b). These variants also showed a decrease in polyreactivity; specifically, while N49P7 showed a moderate degree of polyreactivity, N49P7.v1 and N49P7.v2 showed reduced HEp-2 cell binding in the presence of G54W mutation (Figure 3c). Likewise, the polyreactivities of VRC07-523-W54-LS variants were not elevated to the same level as shown in VRC07-G54W in the presence of the H54W mutation, indicating that the 03FR3 insertion contributed to the reduced polyreactivity as reported previously.39 Overall, the structure-based matrix approach to optimize 3BNC117, N6, N49P7, VRC08, and VRC07-523LS yielded variants with about ~5-fold improvement (as low as median IC50 on the 12 virus-panel of 0.062 μg/ml for VRC07-523-W54-LS.v3 versus 0.344 μg/ml for VRC01.23LS), though the more potent variants did show some polyreactivity.
Truncation of Disordered Light-Chain N-termini Increases Potency, While Truncation of Ordered Light Chain N-termini Decreases Potency
To understand the mechanism underpinning the improved potency by light-chain truncation with only select antibodies, we analyzed the crystal structures of VRC01-class antibodies in complex with gp120. Notably, we observed the N-termini of the light chains of VRC01 (PDB ID: 3NGB), VRC07 (PDB ID: 4OLU), and N49P7 (PDB ID: 6BCK), whose potencies were improved by N-terminal truncation, to be disordered, whereas we observed the N-termini of the light chains of VRC08 (PDB ID: 4XMP), 3BNC117 (PDB ID: 4JPV), and N6 (PDB ID: 5TE6), whose potencies were impaired by N-terminal truncation, to be well ordered (Figure 4). Thus, potency seemed to be improved only for VRC01 class antibodies for which the light chain was disordered, suggesting that ordered light chains could accommodate interactions with the V5 loop or that disordered light chains sterically clashed with the V5 loop. However, this proposition comes with two caveats: (1) the crystal structures of these VRC01-class antibodies in complex with clade A/E 93TH057 gp120, except VRC08 which is in complex with a clade A Q842.d12 gp120, do not represent all N-terminal states of the antibodies engaging different gp120s, and (2) lack of density at the N-termini of VRC01, VRC07, and N49P7 could arise because of the structures determined at relatively lower resolutions. Therefore, we further explored if stability of antibodies is associated with changes in potency, as it has been reported that improved antibody stability by optimizing contacts between variable heavy and light chains increased affinity and expression.52 We measured the thermal stability of the antibody variants using differential scanning fluorimetry (DSF) and found the melting temperature (Tm) of VRC08.v3 (VRC08-G54W+03FR3/3aa_del light chain), whose potency was impaired by light-chain truncation, to be about 10°C lower than that of VRC08.v1 (VRC08-G54W+03FR3/wild-type (WT) light chain). However, the Tms of the rest of antibodies did not show any significant differences between 3aa_del-light chain variants and their wild-type counterparts, including 3BNC117.v3 and N6LS.v3, which showed reduced potency with the N-terminal truncation (Table S3). Thus, changes in potency due to truncation of light chain could not be fully explained by changes in thermal stability, suggesting either accommodability by light chain of V5 loops of diverse strains or loss of direct contact with gp120 to likely be the main attributor to changes in potency by light-chain truncation.
Figure 4.
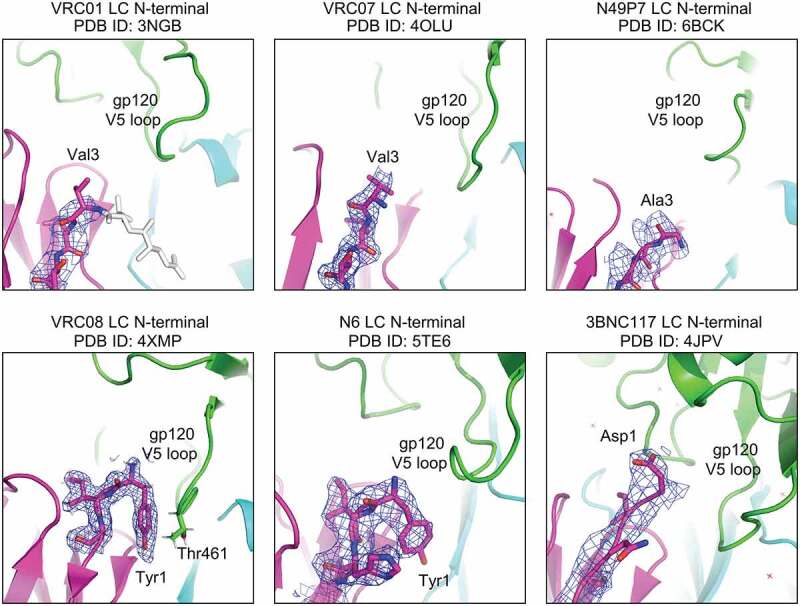
Truncation of disordered N-termini of VRC01-class light chains increased potency, while truncation of ordered light chains impaired potency. The N-termini of VRC01-class antibody light chains were shown in purple with 2fo-fc electron density maps in blue mesh contoured at 1 σ
Large panel neutralization and serum half-life in human-FcRn transgenic mice
To confirm the improved neutralization potency and breadth observed with small panel and to better assess the suitability of their use in clinical trials, we tested the potency and breadth of VRC01.23LS, N49P7.v2, and VRC07-523-W54-LS.v3 on a cross-clade panel of 208-HIV-1 strains.50 VRC01.23LS neutralized 96% of the 208 viruses at a geometric mean IC50 of 0.042 μg/ml, which amounted to a 6% improvement in breadth and 7.5-fold improvement in potency over VRC01 (Figure 5a). More importantly, VRC01.23LS neutralized 94% of the virus tested with a geometric mean IC50 of <1 μg/ml, while VRC01 neutralized 74% of the viruses tested with a geometric mean IC50 of <1 μg/ml. Meanwhile, N49P7.v2 neutralized 97% of the viruses tested with a geometric mean IC50 of 0.082 μg/ml and 93% of the breadth with a geometric mean IC50 of <1 μg/ml, while the parental N49P7 neutralized 93% of the viruses with a geometric mean IC50 of 0.27 μg/ml (Figure 5a, 5b, Table S4).
Figure 5.

VRC01-class antibodies: neutralization potency and breadth against multiclade panel of viruses and serum half-life in human FcRn transgenic mice. (a) Summary of neutralization potency and breadth of VRC01-class antibodies against a panel of 208 pseudoviruses (b) Scattered plots of neutralization potency and breadth with black bars representing median IC50s. A horizontal dotted line extending from the median IC50 of VRC07-523-W54-LS.v3 was drawn as a reference for comparison. (c) Pharmacokinetics of VRC01-class antibodies in human FcRn transgenic mice. Error bars represent SD
VRC07-523-W54-LS.v3 showed even greater improvement in potency and breadth. VRC07-523-W54-LS.v3 neutralized 97% of the viruses tested with the geometric mean IC50 of 0.024 μg/ml; remarkably, 96% of the viruses could be neutralized by VRC07-523-W54-LS.v3 with a geometric mean IC50 of <1 μg/ml. We also assessed a variant of VRC07-523-W54-LS.v3, VRC07-523-F54-LS.v3 (Table S1), in which the Trp at position 54 was replaced with Phe, as VRC07-523-F54-LS.v3 was less polyreactive than VRC07-523-W54-LS.v3 (Figure S1). We observed VRC07-523-F54-LS.v3 to be almost as potent as VRC07-523-W54-LS.v3 (median IC50 of 0.027 μg/ml versus 0.023 μg/ml) (Figure 5a).
In terms of serum half-life, which we assessed in transgenic mice with human FcRn, VRC01.23LS showed the serum half-life to be comparable to VRC01LS. VRC07-523-W54-LS.v3, however, exhibited a shorter serum half-life than VRC01LS and VRC07-523LS, which was not surprising as it was more polyreactive than VRC01LS (Figure 5c). But it is very encouraging to find the serum half-life of VRC07-523-F54-LS.v3 to be comparable to its parental VRC07-523LS (Figure 5c), which in humans shows a half-life of 38 d after intravenous infusion.42
Cryo-EM structure revealed functional relevance of VRC01.23LS design components
To visualize the recognition of each design component in VRC01.23LS, we determined the cryo-EM structure of the VRC01.23LS antigen-binding fragment (Fab) in complex with BG505 DS-SOSIP.664 at 3.4-Å resolution (Figure 6 and Table S5). Three VRC01.23 Fab molecules bound to three CD4-binding sites in the BG505 DS-SOSIP Env trimer (Figure S2), which contained the prefusion-stabilizing mutations C201 and C433 (DS).53 This mode of recognition resembled the crystal structure of BG505 DS-SOSIP in complex with VRC01 scFv and 35O22 scFv and 3H/109L Fab complexes (PDB ID: 6NNF), except for heavy-chain position 54 where Gly was replaced with Trp (Figure 6a) and the light chain N-terminal where 3aa of position 1 to 3 were deleted (Figure 6b). The cryo-EM structure clearly showed Trp54 with its side chain filling the entrance to the Phe43 cavity54 on gp120 and the first N-terminal light-chain residue, Leu4, to be 9.5-Å (Cα-Cα distance) from the closest residue on gp120, Thr461 (Figure 6b). The former could allow VRC01.23LS to bind the Env trimer with increased affinity and the latter may enable VRC01.23LS to better accommodate divergent V5 loops.
Figure 6.

Cryo-EM structure of VRC01.23 Fab in complex with BG505 DS-SOSIP trimer at 3.4-Å resolution. (a) Heavy-chain Gly54 to Trp in its electron density map (b) The N-terminus of light chain VRC01.23LS starting at Leu4 in its electron density map. Cα-Cα distance of 9.5 Å between Leu4 and Thr461 in gp120 was shown in a dotted line. (c) The protruding loop region of the 03FR3 was shown in stick representation. (d) The hydrogen bonding networks between glycan at position 276 of gp120 and Arg66 of VRC01.23 light chain were shown in red dotted lines
The extension of VRC03 framework 3 loop (03FR3) insertion was evident in the reconstruction density, as it protruded to reach the neighboring gp120 protomer (Figure 6c). The inserted loop provided an additional contact surface of 106-Å2 on gp120. However, no specific hydrogen bonds or charge–charge interactions between the insert and the gp120 were observed. In addition to the specific design components, the cryo-EM structure revealed an interesting interaction between Arg66 of the light chain of VRC01.23 and the N-linked glycan core emanating from Asn276 of the BG505 Env; an extensive hydrogen bonding network was observed that differed from what has been observed previously55 (Figure S3), suggesting Arg66, in addition to the other identified recognition hotspots, to be a key site for Env-trimer recognition (Figure 6d).
Manufacturing and biophysical risk assessment
Lastly, as the improved VRC01-class antibodies described in this study may need to be manufactured, we assessed the biophysical properties for two of the most promising variants, VRC01.23LS and VRC07-523-F54-LS.v3 (Table S6). Some properties of VRC01.23LS, particularly, its melting temperature, as determined by differential scanning calorimetry, and its colloidal stability were compromised compared to VRC01LS, as these properties were assessed with VRC01.23LS in phosphate-buffered saline (PBS), at pH 7.4, which is not an ideal buffer and pH. The antibody in optimized buffer, comprising 10 mM acetate-phosphate at pH 5.5, 50 mM arginine, and 50 mM glutamic acid, allowed us to concentrate the antibody up to 120 mg/ml with no visible particles. Furthermore, we were able to develop a stable Chinese hamster ovary cell line that yielded 4.5–5.5 g/L of a VRC01.23LS variant. As such, the variants demonstrated acceptable characteristics to advance for product development.
Discussion
An ideal HIV-1 antibody for passive immunization should possess high neutralization potency and breadth, long serum half-life, and good manufacturability. In this study, we sought to improve the potency and breadth of VRC01-class antibodies by screening a matrix of variants with mutations identified by structure-based rational design.
We first chose to improve the potency and breadth of antibody VRC01LS because it has shown optimal biophysical properties for manufacturing, polyreactivity, and pharmacokinetics among the VRC01-class antibodies we have developed. Diskin et al. first showed that G54W mutation, a mimic of the Phe43CD4-gp120 interaction, enhanced the potency of NIH45-46 ~10-fold.37 However, increased potency by G54W was accompanied with increased polyreactivity. Likewise, the G54W mutation in antibody VRC07 improved the potency about fourfold but with substantially increased polyreactivity, indicating that Trp at position 54 is associated with increased polyreactivity, and therefore should be avoided. Nevertheless, we explored G54W mutation to improve VRC01-like antibodies because it improved the potency of NIH45-46 substantially, and we also found polyreactivity induced by G54W mutation to be antibody specific – some VRC01-class antibodies with Trp or residues other than Gly at position 54 are not polyreactive. Furthermore, Liu et al. demonstrated that 03FR3 insertion reduces polyreactivity.39 Indeed, G54W at VRC01LS showed substantially lower polyreactivity against HEp-2 cells than VRC07-G54W (Figure 2c), with a potency improved more than ~8-fold against a 208-virus panel. Polyreactivity was further reduced with 03FR3 insertion. However, replacing the CDR H3 of VRC01 with that of VRC07 did not improve the potency. For light-chain variants, the 3aa-del consistently increased the potency more than the 2aa-del or light chain wild-type, regardless of the heavy chains to which it was paired. Overall, the VRC01.23LS variant, incorporating G54W mutation and 03FR3 insertion in heavy chain with 3aa_del at the light-chain N-terminus displayed potency about tenfold increased relative to VRC01. More importantly, VRC01.23LS did not show any detectable polyreactivity with nearly the same serum half-life as that of VRC01LS.
The same mutations, G54W and the 03FR3 insertion, incorporated into the VRC07-523LS heavy chain along with 3aa-del mutation in light chain, yielded the even more potent VRC01-class antibody, VRC07-523-W54-LS.v3, which neutralized 98% of 208 viruses tested at a geometric mean IC50 of 0.024 μg/ml. As has been observed in VRC01.23LS, the 03FR3 insertion reduced the polyreactivity of VRC07-523-W54-LS.v3 to a level similar to that of VRC07-523LS (Figure 3c and Table S2). However, polyreactivity of VRC07-523-W54-LS.v3 was noticeably higher than that of VRC01LS and VRC01.23LS; therefore, we replaced its Trp 54 with Phe in an attempt to reduce polyreactivity. Indeed, VRC07-523-F54-LS.v3 showed reduced polyreactivity (Figure S1) with nearly the same potency and breadth of VRC07-523-W54-LS.v3 (Figure 5a). Furthermore, VRC07-523-F54-LS.v3 showed a markedly enhanced serum half-life over VRC07-523-W54-LS.v3 (Figure 5c), as expected from the correlation between reduced polyreactivity and increased serum half-life.54
Antibody N49P7 with G54W and the 03FR3 insertion also displayed about a fivefold increase in potency against a 12-virus panel. The 2aa- or 3aa-del mutations in light chain further increased the potency. Overall, N49P7.v2 showed about threefold improvement in potency with a geometric mean IC50 of <1 μg/ml against 93% of 208 viruses tested, while the parental N49P7 neutralized 79% of the panel with a geometric mean IC50 of <1 ug/ml (Figure 5a). VRC08, 3BNC117, and N6, however, did not show as much improvement in potency as VRC01.23LS, VRC07-523-W54-LS.v3, and N49P7 did, which was not surprising given that 3BNC117 and N6 have the naturally occurring Thr and Tyr at position 54, respectively. Contrary to what was observed in VRC01.23LS, VRC07-523-W54-LS.v3, and N49P7, light-chain deletion mutations in VRC08, 3BNC117, and N6 caused impaired potency and breadth. Interestingly, we observed that improvement in potency by light-chain truncation is dependent on the degree of order of the light-chain N-termini. The crystal structures of the antibodies in complex with gp120 showed the N-termini of antibodies with improved potency by light-chain truncation were disordered, while the antibodies that decreased potency by light-chain truncation had ordered N-termini. We also observed that the first three residues of VRC08 and N6 light-chain contact neighboring residues more substantially compared to VRC01, VRC07, and N49P7 (Figure S4), whose potencies were improved by light-chain truncation. However, determining the exact mechanism underpinning the reduced potency of 3BNC117 and N6 variants by light-chain truncation has been complicated by the presence of Trp54 or 03FR3 mutation, as Liu et al. found the 03FR3 insertion to 3BNC117 reduced the potency.
In summary, we have engineered VRC01-class antibodies, VRC01.23LS and VRC07-523-F54-LS.v3, with improved potency and breadth without compromising either physical properties or pharmacokinetics by designing and testing a matrix of antibody variants generated by structure-based design. Such an approach may be applicable to the optimization of broadly neutralizing antibodies of different classes, such as against the V2-apex or glycan-V3 supersites of vulnerability on gp120, or the membrane-proximal or fusion peptide supersites of vulnerability on gp41, by exploiting the critical sites that each of these antibody classes uses to neutralize HIV-1. With VRC01-class antibodies, we also demonstrated that the serum half-life of an antibody could be extended by reducing polyreactivity. The highly favorable potency, breadth, and half-life of these newly obtained matrix-optimized VRC01-class antibodies suggest that they may have increased prophylactic efficacy for HIV-1 prevention versus prior generations of VRC01-class antibodies.
Materials and methods
Construct design, protein expression, and purification
Plasmids encoding heavy-chain or light-chain variant genes were generated by site-directed mutagenesis or overlapping PCR using pVRC8400 vectors containing WT heavy-chain or light-chain genes as templates. The variants were expressed by transient transfection in EXPI293 cells (Thermo Fisher) using Turbo293 transfection reagent (SPEED BioSystems) according to the manufacturer’s recommendation. The plasmid encoding heavy-chain or light-chain variant genes were mixed with the transfection reagents, added to 100 ml of cells at 2.5 × 106/ml, and incubated in a shaker incubator at 120 rpm, 37°C, 9% CO2 overnight. Cell cultures were incubated at 120 rpm, 37°C, 9% CO2 for additional 4 d. At 5 d post-transfection, cell culture supernatant was harvested and purified with a Protein A (GE Healthcare) column. The eluted variants were brought to neutral pH with 1 M Tris-HCl, pH 8.0 and were dialyzed against PBS overnight, and were confirmed by SDS-PAGE before use.
Neutralization
Single-round-of-replication Env pseudoviruses were prepared, titers were determined, and the pseudoviruses were used to infect TZM-bl target cells as described previously.56 Neutralization of monoclonal antibodies was determined using a multiclade panel of 12 HIV-1 Env-pseudoviruses including clade A (2), clade AG (1), clade B (4), clade C (4), and clade D (1), and using a 208-isolate panel.50 Each antibody was assayed at fivefold dilutions starting at 50 μg/ml. The neutralization titers were calculated as a reduction in luminescence units compared with control wells, and reported as 50% or 80% inhibitory concentration (IC50 or IC80) in micrograms per milliliter.
Polyreactivity
Polyreactivity was determined by ANA HEp-2 Staining Analysis (ZEUS Scientific Cat. No: FA2400) and anticardiolipin ELISA (Inova Diagnostics Cat. No.: 708625). For the HEp-2 assay, all antibodies were tested at 25 and 50 μg/ml as per manufacturer’s protocol and imaged on a Nikon Ts2R microscope for 500 ms. Scores from 0 to 3 were defined with four control antibodies VRC01-LS, 4E10, VRC07-523LS, and VRC07-G54W. Test antibodies were scored by visual estimation of staining intensity in comparison to the control antibodies. Scores equal to or greater than 1 at 25 μg/ml were classified as polyreactive, and between 0 and 1 as mildly polyreactive. In the cardiolipin ELISA, antibodies were tested at a starting concentration of 100 μg/ml, followed by threefold dilutions. IgG phospholipid (GPL) units were calculated from the standard curve. GPL score < 20 was considered as not reactive, 20–80 as low positive and >80 as high positive.
Serum half-life in human FcRn transgenic mouse model
Human FcRn transgenic mice (C57BL/6, B6.mFcRn−/− hFcRn Tg32 line from The Jackson Laboratory) were used to assess the pharmacokinetics of VRC01 and VRC07 variant antibodies. Each animal was infused intravenously with 5 mg antibody/kg of body weight. Whole blood samples were collected at d 1, 2, 5, 7, 9, 14, 21, 28, and 35. Serum was separated by centrifugation. Serum antibody levels were measured by ELISA as described previously.38 All mice were bred and maintained under pathogen-free conditions at an American Association for Assessment and Accreditation of Laboratory Animal Care International-accredited animal facility at the National Institute of Allergy and Infectious Diseases and housed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals. All mice were between 6 and 13 weeks of age. The study protocol was evaluated and approved by the National Institutes of Health Animal Care and Use Committee (ASP VRC-18-747).
Cryo-EM data collection and processing
BG505 DS-SOSIP.664 was prepared as previously described53 and incubated with molar excess of antibody Fabs and 2.3 μl of the complex at 1 mg/ml concentration was deposited on a C-flat grid (protochip.com). The grid was vitrified using an FEI Vitrobot Mark IV with a wait time of 30 s, blot time of 3 s and blot force of 1. Automated data collection on a Titan Krios electron microscope was performed with Leginon57 with a Gatan K2 Summit direct detection device. Exposures were collected in movie mode for a 10 s with the total dose of 70.41 e–/Å2 fractionated over 50 raw frames. Images were preprocessed using Appion58,59; individual frames were aligned and dose-weighted using MotionCor2.60 CTFFind461,62 was used to estimate the CTF and DoG Picker58,59 was used for particle picking. RELION63 was used for particle extraction. CryoSPARC 2.1264 was used for 2D classifications, ab initio 3D reconstruction, homogeneous refinement, and nonuniform 3D refinement. Initial 3D reconstruction was performed using C1 symmetry, confirming 3 Fab molecules per trimer, whereupon C3 symmetry was applied for the final reconstruction and refinement. Coordinates from PDB ID 6NNF39 was used for initial fit to the reconstructed map. This was followed by simulated annealing and real space refinement in Phenix65 and then iteratively processed with manual fitting of the coordinates in Coot.66 Geometry and map fitting were evaluated throughout the process using Molprobity67 and EMRinger.68 PyMOL (www.pymol.org) was used to generate figures.
Manufacturability assessment
Manufacturability for VRC01.23LS and VRC07-523-F54-LS.v3 was assessed by visual inspection, dynamic light scattering, thermal transitions by dynamic light scattering, differential scanning calorimetry, circular dichroism, and isothermal chemical denaturation as previously described.40
Nano differential scanning fluorimetry
The thermal stability of antibody variants was determined by using Prometheus NT.48 instrument (NanoTemper Technologies). Variants were diluted to the concentration of 0.5 mg/ml in PBS, pH 7.4, loaded into capillaries, and placed on the sample holder. The temperature gradient was set from 30°C to 90°C with an increment of 1°C/min, and the intrinsic fluorescence intensity at wavelengths of 330 and 350 nm was measured. Data were analyzed using NanoTemper software.
Statistical analysis and the antibody numbering scheme
The statistical analyses were performed using GraphPad Prism. The Kabat numbering scheme was used for the numbering of amino acid residues in antibodies.
Supplementary Material
Acknowledgments
We thank J. Stuckey for assistance with figures, members of the Electron Microscopy Group at the New York Structural Biology Center for assistance with data collection, and members of the Virology Laboratory and Vector Core, Vaccine Research Center, for discussions and comments on the manuscript. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 envelope plasmids used in the 208-strain panel. We thank K. McKee, C. Moore, S. O’Dell, G. Padilla, S.D. Schmidt, C. Whittaker, and A.B. McDermott for assistance with neutralization assessments on the 208-strain panel. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID). Some of this work was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310) with additional support from the Agouron Institute (F00316) and NIH S10 OD019994-01.
Funding Statement
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (US) [ZIA AI005022-19].
Abbreviations
| ANA | Antinuclear antibody |
| CDRs | Complementarity-determining regions |
| Cryo-EM | Cryo-electron microscopy |
| CTF | Contrast transfer function |
| DoG | Difference of Gaussian |
| ELISA | Enzyme-linked immunosorbent assay |
| Fab | Antigen-binding fragment |
| Fc | Fragment crystallizable |
| FcRn | Neonatal Fc receptor |
| FR | Framework region |
| GPL | IgG phospholipid |
| HEp-2 | Human epithelial type-2 |
| HIV | Human immunodeficiency virus |
| HPTN | HIV prevention trials network |
| HVTN | HIV vaccine trials network |
| IC50 | 50% inhibitory concentration |
| IC80 | 80% inhibitory concentration |
| NHPs | Nonhuman primates |
| PBS | Phosphate-buffered saline |
| PCR | Polymerase chain reaction |
| PDB | Protein data bank |
| scFv | Single-chain variable fragment |
| SHIV | Simian-human immunodeficiency virus |
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data and code availability
Cryo-EM maps and fitted coordinates of BG505 DS-SOSIP-VRC01.23 Fab complex have been deposited with database codes EMDB-21208 and PDB ID: 6VI0, respectively.
Consortia VRC Production Program
Christine Anderson1, Sashikanth Banappagari1, Niutish Bastani1, Michael Bender1, Amy L. Chamberlain1, Ria Caringal1, Rajoshi Chaudhuri1, Jonathan W. Cooper1, Marianna Fleischman1, Raju Gottumukkala1, Hsing-Ho (Vasha) Hsu1, Elihu Ihms1, Tina Khin1, Lisa Kueltzo1, Alaina Lapanse1, Q. Paula Lei1, Yile Li1, Slobodanka Manceva1, Gabriel Moxey1, Lan Nguyen1, Aakash Patel1, Rahul Ragunathan1, Andrew Shaddeau1, William Shadrick1, Sudesh Upadhyay1.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
References
- 1.Sok D, Burton DR.. Recent progress in broadly neutralizing antibodies to HIV. Nat Immunol. 2018;19:1179–12. doi: 10.1038/s41590-018-0235-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pegu A, Hessell AJ, Mascola JR, Haigwood NL. Use of broadly neutralizing antibodies for HIV-1 prevention. Immunol Rev. 2017;275(1):296–312. doi: 10.1111/imr.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Julg B, Barouch DH. Neutralizing antibodies for HIV-1 prevention. Curr Opin HIV AIDS. 2019;14(4):318–24. doi: 10.1097/COH.0000000000000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruell H, Klein F. Antibody-mediated prevention and treatment of HIV-1 infection. Retrovirology. 2018;15:73. doi: 10.1186/s12977-018-0455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaworski JP, Cahn P. Preventive and therapeutic features of broadly neutralising monoclonal antibodies against HIV-1. Lancet HIV. 2018;5(12):e723–e31. doi: 10.1016/S2352-3018(18)30174-7. [DOI] [PubMed] [Google Scholar]
- 6.Walker LM, Phogat SK, Chan-Hui P-Y, WagneR D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326(5950):285–89. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang G-Y, Pancera M, Cale EM, Ernandes MJ, Louder MK, et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol. 2017;2:76–91. doi: 10.1126/sciimmunol.aal2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sok D, Pauthner M, Briney B, Lee JH, Saye-Francisco KL, Hsueh J, Ramos A, Le KM, Jones M, Jardine JG, et al. A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity. 2016;45(1):31–45. doi: 10.1016/j.immuni.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freund NT, Wang H, Scharf L, Nogueira L, Horwitz JA, Bar-On Y, Golijanin J, Sievers SA, Sok D, Cai H, et al. Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller. Sci Transl Med. 2017;9(373):9. doi: 10.1126/scitranslmed.aal2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Kang BH, Ishida E, Zhou T, Griesman T, Sheng Z, Wu F, Doria-Rose NA, Zhang B, McKee K, et al. Identification of a CD4-binding-site antibody to HIV that evolved near-pan neutralization breadth. Immunity. 2016;45:1108–21. doi: 10.1016/j.immuni.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sajadi MM, Dashti A, Rikhtegaran Tehrani Z, Tolbert WD, Seaman MS, Ouyang X, Gohain N, Pazgier M, Kim D, Cavet G, et al. Identification of near-pan-neutralizing antibodies against HIV-1 by deconvolution of plasma humoral responses. Cell. 2018;173:1783–95 e14. doi: 10.1016/j.cell.2018.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–37. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schommers P, Gruell H, Abernathy ME, Tran MK, Dingens AS, Gristick HB, Barnes CO, Schoofs T, Schlotz M, Vanshylla K, et al. Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody. Cell. 2020;180:471–89 e22. doi: 10.1016/j.cell.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10:359–69. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- 18.Williams LD, Ofek G, Schatzle S, McDaniel JR, Lu X, Nicely NI, Wu L, Lougheed CS, Bradley T, Louder MK, et al. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci Immunol. 2017;2(7):2. doi: 10.1126/sciimmunol.aal2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–12. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krebs SJ, Kwon YD, Schramm CA, Law WH, Donofrio G, Zhou KH, Gift S, Dussupt V, Georgiev IS, Schatzle S, et al. Longitudinal analysis reveals early development of three MPER-directed neutralizing antibody lineages from an HIV-1-infected individual. Immunity. 2019;50:677–91 e13. doi: 10.1016/j.immuni.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science. 2016;352(6287):828–33. doi: 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang C-H, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40(5):657–68. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang G-Y, Druz A, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41–gp120 interface. Nature. 2014;515(7525):138–42. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoofs T, Barnes CO, Suh-Toma N, Golijanin J, Schommers P, Gruell H, West AP Jr., Bach F, Lee YE, Nogueira L, et al. Broad and potent neutralizing antibodies recognize the silent face of the HIV envelope. Immunity. 2019;50(6):1513–29 e9. doi: 10.1016/j.immuni.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15(8):951–54. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR, Farzan M. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 2009;5(5):e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6(2):207–10. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 28.Shingai M, Donau OK, Plishka RJ, Buckler-White A, Mascola JR, Nabel GJ, Nason MC, Montefiori D, Moldt B, Poignard P, et al. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211(10):2061–74. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pegu A, Yang Z-Y, Boyington JC, Wu L, Ko S-Y, Schmidt SD, McKee K, Kong W-P, Shi W, Chen X, et al. Neutralizing antibodies to HIV-1 envelope protect more effectively in vivo than those to the CD4 receptor. Sci Transl Med. 2014;6(243):243ra88. doi: 10.1126/scitranslmed.3008992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, et al. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian–human immunodeficiency virus infection. Nat Med. 2000;6(2):200–06. doi: 10.1038/72309. [DOI] [PubMed] [Google Scholar]
- 31.Moldt B, Rakasz EG, Schultz N, Chan-Hui P-Y, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012;109(46):18921–25. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendoza P, Gruell H, Nogueira L, Pai JA, Butler AL, Millard K, Lehmann C, Suarez I, Oliveira TY, Lorenzi JCC, et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature. 2018;561:479–84. doi: 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP Jr., Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–91. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med. 2015;7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med. 2017;23:185–91. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corey L, Gilbert PB, Juraska M, Montefiori DC, Morris L, Karuna ST, Edupuganti S, Mgodi NM, deCamp AC, Rudnicki E, et al. Two Randomized Trials of Neutralizing Antibodies to Prevent HIV-1 Acquisition. N Engl J Med. 2021;384:1003–14. doi: 10.1056/NEJMoa2031738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diskin R, Scheid JF, Marcovecchio PM, West AP Jr., Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–93. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudicell RS, Kwon YD, Ko SY, Pegu A, Louder MK, Georgiev IS, Wu X, Zhu J, Boyington JC, Chen X, et al. Enhanced potency of a broadly neutralizing HIV-1 antibody in vitro improves protection against lentiviral infection in vivo. J Virol. 2014;88:12669–82. doi: 10.1128/JVI.02213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, Lai YT, Zhang P, Louder MK, Pegu A, Rawi R, Asokan M, Chen X, Shen CH, Chuang GY, et al. Improvement of antibody functionality by structure-guided paratope engraftment. Nat Commun. 2019;10:721. doi: 10.1038/s41467-019-08658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon YD, Chuang GY, Zhang B, Bailer RT, Doria-Rose NA, Gindin TS, Lin B, Louder MK, McKee K, O’Dell S, et al. Surface-matrix screening identifies semi-specific interactions that improve potency of a near pan-reactive HIV-1-neutralizing antibody. Cell Rep. 2018;22:1798–809. doi: 10.1016/j.celrep.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherf GM, Cochran JR. Applications of yeast surface display for protein engineering. Methods Mol Biol. 2015;1319:155–75. doi: 10.1007/978-1-4939-2748-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaudinski MR, Houser KV, Doria-Rose NA, Chen GL, Rothwell RSS, Berkowitz N, Costner P, Holman LA, Gordon IJ, Hendel CS, et al. Safety and pharmacokinetics of broadly neutralising human monoclonal antibody VRC07-523LS in healthy adults: a phase 1 dose-escalation clinical trial. Lancet HIV. 2019;6:e667–e79. doi: 10.1016/S2352-3018(19)30181-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ledgerwood JE, Coates EE, Yamshchikov G, Saunders JG, Holman L, Enama ME, DeZure A, Lynch RM, Gordon I, Plummer S, et al. Safety, pharmacokinetics and neutralization of the broadly neutralizing HIV-1 human monoclonal antibody VRC01 in healthy adults. Clin Exp Immunol. 2015;182:289–301. doi: 10.1111/cei.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bar-On Y, Gruell H, Schoofs T, Pai JA, Nogueira L, Butler AL, Millard K, Lehmann C, Suarez I, Oliveira TY, et al. Safety and antiviral activity of combination HIV-1 broadly neutralizing antibodies in viremic individuals. Nat Med. 2018;24:1701–07. doi: 10.1038/s41591-018-0186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen YZ, Lorenzi JCC, Krassnig L, Barton JP, Burke L, Pai J, Lu C-L, Mendoza P, Oliveira TY, Sleckman C, et al. Relationship between latent and rebound viruses in a clinical trial of anti–HIV-1 antibody 3BNC117. J Exp Med. 2011;334(9):2311–24. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayer KH, Seaton KE, Huang Y, Grunenberg N, Isaacs A, Allen M, Ledgerwood JE, Frank I, Sobieszczyk ME, Baden LR, et al. Safety, pharmacokinetics, and immunological activities of multiple intravenous or subcutaneous doses of an anti-HIV monoclonal antibody, VRC01, administered to HIV-uninfected adults: results of a phase 1 randomized trial. PLoS Med. 2015;182(11):e1002435. doi: 10.1111/cei.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaudinski MR, Coates EE, Houser KV, Chen GL, Yamshchikov G, Saunders JG, Holman LA, Gordon I, Plummer S, Hendel CS, et al. Safety and pharmacokinetics of the Fc-modified HIV-1 human monoclonal antibody VRC01LS: a Phase 1 open-label clinical trial in healthy adults. PLoS Med. 2011;333(1):e1002493. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko S-Y, Pegu A, Rudicell RS, Yang Z-Y, Joyce MG, Chen X, Wang K, Bao S, Kraemer TD, Rath T, et al. Enhanced neonatal Fc receptor function improves protection against primate SHIV infection. Nature. 2014;514(7524):642–45. doi: 10.1038/nature13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zalevsky J, Chamberlain AK, Horton HM, Karki S, Leung IW, Sproule TJ, Lazar GA, Roopenian DC, Desjarlais JR. Enhanced antibody half-life improves in vivo activity. Nat Biotechnol. 2011;333(2):1633–37. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doria-Rose NA, Louder MK, Yang Z, O’Dell S, Nason M, Schmidt SD, McKee K, Seaman MS, Bailer RT, Mascola JRHIV-1. HIV-1 neutralization coverage is improved by combining monoclonal antibodies that target independent epitopes. J Virol. 2018;24(6):1701–07. doi: 10.1038/s41591-018-0186-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, et al. Maturation and diversity of the VRC01-antibody lineage over 15 years of chronic HIV-1 infection. Cell. 2015;161(3):470–85. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warszawski S, Borenstein Katz A, Lipsh R, Khmelnitsky L, Ben Nissan G, Javitt G, Dym O, Unger T, Knop O, Albeck S, et al. Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces. PLoS Comput Biol. 2019;15(8):e1007207. doi: 10.1371/journal.pcbi.1007207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat Struct Mol Biol. 2015;22(7):522–31. doi: 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393(6686):648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165:813–26. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131–46. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 58.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166(2):205–13. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14(4):331–32. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang ZK. Gctf: real-time CTF determination and correction. J Struct Biol. 2016;193(1):1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2018;215(2):216–21. doi: 10.1084/jem.20180936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2018;561(3):519–30. doi: 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14(3):290–96. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 65.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2016;165(1):813–26. doi: 10.1016/j.cell.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 66.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(12):2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 67.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32(Web Server):W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, Fraser JS. EMRinger: side chain–directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2020;180(10):471–489.e22. doi: 10.1016/j.cell.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps and fitted coordinates of BG505 DS-SOSIP-VRC01.23 Fab complex have been deposited with database codes EMDB-21208 and PDB ID: 6VI0, respectively.


