Summary
The RNA polymerase-binding protein DksA, together with the alarmone nucleotides (p)ppGpp, mediates the stringent response to nutrient starvation in Borrelia burgdorferi. To date, the contribution of DksA to B. burgdorferi infection remains unknown. We report here that DksA is essential for B. burgdorferi to infect a mammalian host. dksA expression was highly induced during infection. Moreover, a dksA-deficient mutant was incapable of infecting mice. The mutant displayed growth defects when cultured in vitro, and resistance to osmotic pressure was markedly reduced. These phenotypes were fully restored to those of the wild type when dksA mutation was complemented. We further showed that DksA controlled expression of virulence-associated lipoprotein OspC, likely via the central alternative sigma factor RpoS. Synthesis of RpoS was abolished in the dksA mutant, but rpoS transcription remained unaffected. Additionally, we found that expression of clpX, clpA, clpP, and clpP2 was significantly increased in the mutant, suggesting that DksA may post-transcriptionally regulate rpoS expression via its effect on ClpXP and/or ClpAP proteases. These combined data demonstrate that DksA regulates B. burgdorferi virulence at least partially through its influence on RpoS and OspC. This study thus elucidates that, in addition to function as a stringent response regulator, DksA promotes the transcription and/or translation of genes contributing to B. burgdorferi infectivity.
Keywords: Lyme disease, Borrelia burgdorferi, Pathogenesis, Virulence, Gene regulation
Graphical Abstract
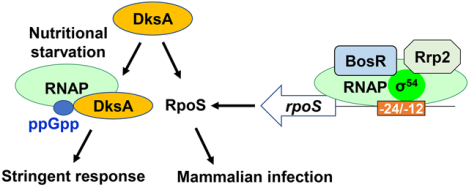
Borrelia burgdorferi utilizes numerous strategies to survive in ticks and mammalian hosts. In this study, we report an essential role of DksA in the infection of B. burgdorferi. The loss of DksA not only impairs bacterial growth and resistance to osmotic pressure, but also renders the bacterium incompetent to infect mice. We also found that DksA regulated synthesis of RpoS, which provides new insights into another regulatory layer controlling RpoS in B. burgdorferi.
Introduction
Borrelia burgdorferi, the etiological agent for Lyme disease, survives in nature through a complex enzootic life cycle involving Ixodes ticks and many mammalian hosts (Burgdorfer et al., 1982, De Silva & Fikrig, 1995, Schwan, 1996, Steere, 1993, Steere et al., 2004). After B. burgdorferi is acquired by newly hatched larvae, the bacterium resides in the tick midgut through the molt. When infected nymphs take a blood meal from a naive mammalian host, spirochetes are transmitted to the host to initiate an infection. During its transit between tick and mammals, B. burgdorferi encounters a myriad of environmental challenges such as changes in temperature, pH, osmolarity, and nutrient resources [see Reviews (Radolf et al., 2012, Rosa et al., 2005, Samuels, 2011)]. To survive these hostile conditions, B. burgdorferi has evolved dedicated mechanisms to respond to environmental changes and fine-tune its cellular processes.
Stringent response is a physiological mechanism utilized by bacteria to adapt to nutrient deprivation and many other stresses (Irving & Corrigan, 2018, Gourse et al., 2018). Upon nutrient depletion, bacteria must change their metabolism and adjust their growth in order to promote a balance between cell survival and proliferation. This is usually achieved by reprogramming global gene expression in response to nutrient starvation. Specifically, when sensing nutrient depletion, bacteria down-regulate genes involved in the synthesis of translational machinery (such as rRNA and tRNA) and up-regulate genes for amino acid biosynthesis (Gaca et al., 2015, Gottesman, 2019, Gourse et al., 2018, Irving & Corrigan, 2018, Zhu et al., 2019). Global gene expression changes during stringent response is mediated by the alarmone nucleotides guanosine pentaphosphate (pppGpp) and guanosine tetraphosphate (ppGpp), collectively termed (p)ppGpp (Gaca et al., 2015, Zhu et al., 2019, Irving & Corrigan, 2018, Gourse et al., 2018). In bacterial species such as E. coli, synthesis of (p)ppGpp is dependent on the monofunctional synthetase RelA and the bifunctional synthetase/hydrolase SpoT. In B. burgdorferi and many others, a single enzyme with both synthase and hydrolase activities called Rel or RSH (RelA/SpoT homolog) catalyzes the production of (p)ppGpp (Atkinson et al., 2011, Concepcion & Nelson, 2003, Bugrysheva et al., 2005). Upon production, (p)ppGpp binds to RNA polymerase (RNAP) and redirects global gene transcription in response to nutrient starvation. More specifically, (p)ppGpp binds to RNAP via two binding sites (Ross et al., 2016, Gourse et al., 2018). Binding Site 1 is located between the ω and β’ subunits; binding of (p)ppGpp to Site 1 may induce RNAP conformational changes to influence the lifetime of the open complex. Binding Site 2 is located in the secondary channel of RNAP; occupation of this site by (p)ppGpp, together with a small protein called DksA, affects the stability of the open complex and influences the initiation of gene transcription.
The DnaK suppressor protein DksA is an ~18 kDa protein found in a wide variety of microorganisms (Perederina et al., 2004, Gourse et al., 2018). This small cytosolic protein was originally identified as a suppressor for the temperature-sensitive growth and filamentation of an E. coli dnaK deletion mutant (Kang & Craig, 1990). Subsequently, DksA has been implicated in many cellular processes such as amino acid biosynthesis, translation, cell division, stress responses, and pathogenesis. Structural analyses indicate that DksA contains two domains including a long coiled-coil domain with two conserved aspartic acidic residues and a globular domain with a zinc finger motif (Parshin et al., 2015, Perederina et al., 2004). It is believed that DksA does not regulate gene transcription through DNA binding. Rather, it exerts its function on gene expression by directly binding to the (p)ppGpp-binding Site 2 on RNAP (Parshin et al., 2015, Perederina et al., 2004, Ross et al., 2016, Gourse et al., 2018). Binding of both DksA and (p)ppGpp to RNAP determines the synergistic effects of DksA and (p)ppGpp on global stringent response. In addition to their coordinated involvement in stringent response, DksA and (p)ppGpp have been reported to possess diverse roles and regulate gene expression independent of each other.
Notably, bb0168 in B. burgdorferi encodes a potential DksA homolog (Boyle et al., 2019, Fraser et al., 1997). This protein, together with (p)ppGpp, are believed to mediate stringent response in the Lyme disease spirochete (Drecktrah et al., 2018, Bugrysheva et al., 2011, Caimano et al., 2016, Drecktrah et al., 2015, Bugrysheva et al., 2002, Bugrysheva et al., 2003, Boyle et al., 2019). When the (p)ppGpp synthetase/hydrolase RelBbu was inactivated, the mutant showed survival defects in ticks during the transition from unfed to fed nymphs (Drecktrah et al., 2015). Global transcriptome analyses revealed that expression of glycerol and chitobiose utilization pathways, as well as many other genes involved in survival and persistence of B. burgdorferi in ticks, were altered in the RelBbu mutant (Bugrysheva et al., 2015, Drecktrah et al., 2018, Drecktrah et al., 2015). Recently, Boyle et al. created a dksA-deficient mutant in B. burgdorferi and found that the level of (p)ppGpp was elevated in the mutant (Boyle et al., 2019). Accordingly, microarray analyses revealed that, in addition to modulating expression of genes independent of (p)ppGpp, DksA also influences expression of an array of (p)ppGpp-dependent genes, suggesting that this protein regulates the stringent response in B. burgdorferi (Boyle et al., 2019). Despite these important findings, the role of DksA in B. burgdorferi infection and pathogenesis remains unknown, primarily due to the hitherto unavailability of a gene mutant containing all essential plasmids. In this study, we report the successful generation of a dksA deletion mutant retaining all endogenous plasmids of B. burgdorferi. Characterization of the mutant and its complemented counterparts highlights an essential role of this protein in the infection of B. burgdorferi. Our data also revealed a role of DksA in modulating the expression of several important virulence determinants.
Results
Expression of dksA is influenced by environmental factors.
To investigate the environmental influence on dksA expression, B. burgdorferi was cultivated under different conditions and gene expression was analyzed using qRT-PCR. First, we profiled the expression kinetics of dksA throughout the growth phases. As shown in Fig. 1A, when compared with gene expression at early-logarithmic phase, dksA expression was not significantly changed at stationary phase, but was down-regulated ~3.3 fold at mid-logarithmic growth. We also assessed the impact of temperature change on gene expression. Relative to gene expression in spirochetes cultivated at 23 °C, dksA transcription was only slightly changed (~1.5 fold) when B. burgdorferi was grown at 37 °C (Fig. 1B). When gene expression was examined in spirochetes cultivated under pH 7.6 or pH 6.8, comparable levels of dksA transcription were observed (Fig. 1C). As DksA has been implicated in starvation response in B. burgdorferi, we further examined whether nutrient starvation affected dksA expression. B. burgdorferi was first cultivated in BSK-II medium. When growth reached stationary phase, spirochetes were collected (referred to as stationary-phase spirochetes) by centrifugation and then starved in RPMI medium (referred to as starved spirochetes). Compared with gene expression in stationary-phase spirochetes, expression of dksA was upregulated ~3.7 fold in starved spirochetes; but this difference is not statistically significant (Fig. 1D).
Fig. 1. Analyses of dksA expression through qRT-PCR analyses.
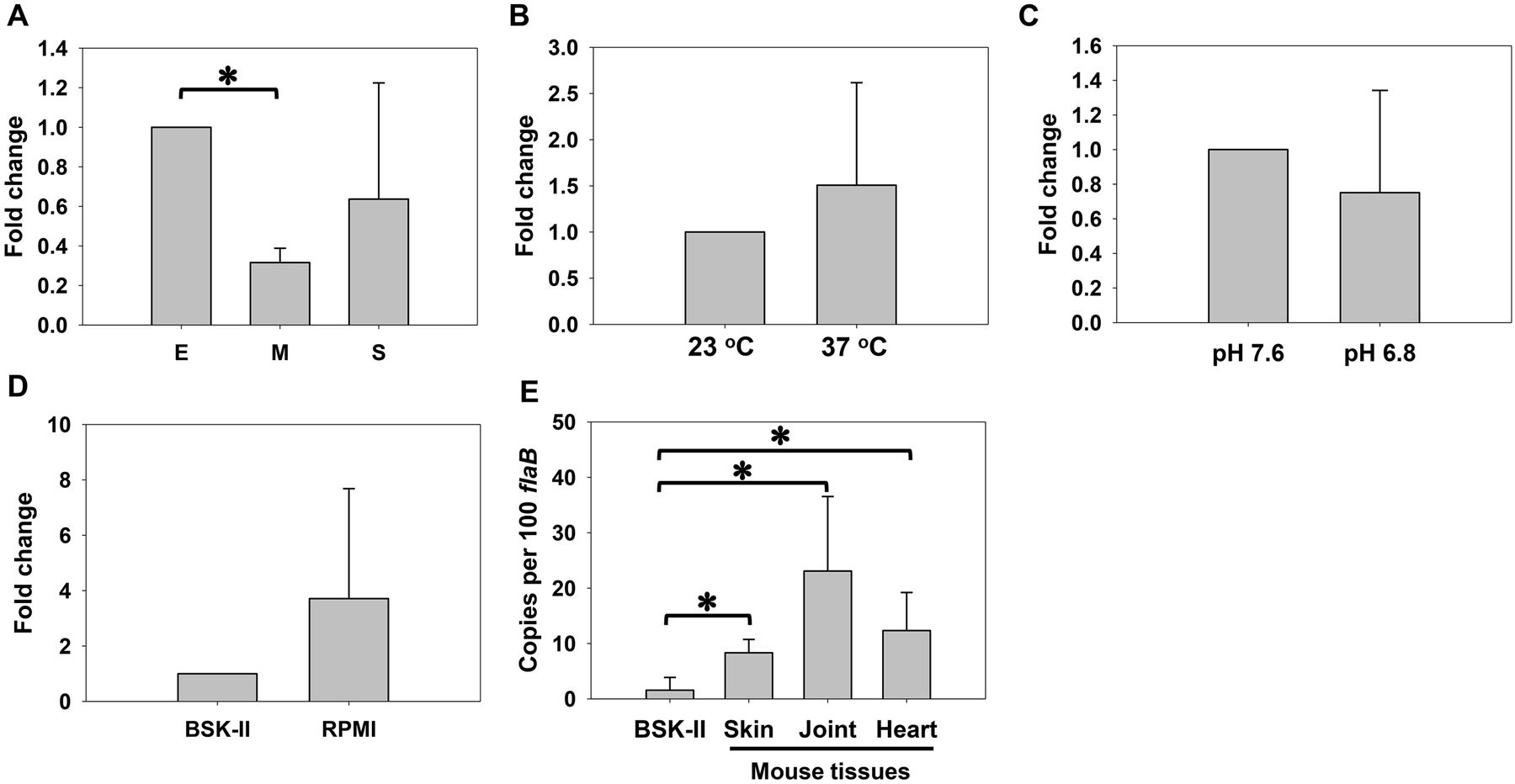
Relative quantification of dksA expression (normalized to 16S rRNA) in B. burgdorferi cultivated at different growth phases (E, early logarithmic phase; M, mid-logarithmic phase; S, stationary phase) (A), different temperatures (B), different pH (C), or starved in RPMI for 6 h (D). (E) dksA transcripts were analyzed in mouse samples and stationary-phase spirochetes (grown in BSK-II at 37 °C/pH 7.6) by qRT-PCR via absolute quantification. Skin, heart, and joints were isolated from mice infected with B. burgdorferi at 3 weeks post-infection. The values represent the average copy number of dksA transcripts normalized per 100 copies of flaB transcripts. All data were collected from three independent experiments, and the bars represent the mean values ± standard deviation. The asterisk indicates statistical significance using one-way ANOVA (P < 0.05).
dksA is highly expressed in infected animals.
To unravel the role of DksA in B. burgdorferi infection, expression of dksA was examined in infected animals. Copy numbers of B. burgdorferi flaB and dksA were determined in mouse skin, joints, and heart using absolute quantification qRT-PCR. As shown in Fig. 1E, relative to gene expression in spirochetes cultivated in BSK-II, dksA transcription was highly induced in infected animals. Approximately 3.7-, 16.4-, and 9.2-copies of dksA transcripts per 100 flaB transcripts were detected in skin, joints, and heart, respectively, whereas only ~0.3 dksA transcripts per 100 flaB transcripts were detected in spirochetes cultivated in vitro (Fig. 1E).
Generation of dksA deletion mutant and complemented strains.
In order to study the role of dksA in the biology of B. burgdorferi, we created a dksA deletion mutant by introducing the suicide plasmid pOY598 into strain CE162 (Fig. 2A). Through allelic exchange, a 246-bp internal fragment of the 378-bp open reading frame (ORF) of dksA was replaced with the PflgB-Kan cassette, yielding the kanamycin-resistant strain OY413. To complement the dksA mutant, the suicide vector pOY662 was created by linking dksA to the PflgB-aadA cassette (Fig. 2A). After electroporation of pOY662 into OY413, the corresponding streptomycin-resistant complemented strain OY463 was created, in which a wild type (WT) copy of dksA was restored at its native chromosomal locus. Another complemented strain OY479 was created by inserting the PdksA-dksA-PflgB-aadA cassette into the dispensable bbb20-bbb21 locus (Fig. 2A). Plasmid profiling analysis revealed that OY413, OY463, and OY479 retained all plasmids contained in WT CE162. The inactivation and complementation of dksA in these strains were confirmed by using PCR. PCR employing dksA specific primers amplified a fragment in WT and both complemented strains, but not in the mutant (Fig. 2B). The kanamycin resistance gene was amplified in the mutant and the trans-complemented strain OY479, whereas the streptomycin resistance gene was amplified only in the complemented strains OY463 and OY479 (Fig. 2B). RT-PCR employing dksA specific primers was performed to detect gene expression in these strains. As expected, dksA transcripts were detected in both WT and the complemented strains, but not in the mutant (Fig. 2C).
Fig. 2. Inactivation and complementation of dksA in B. burgdorferi.
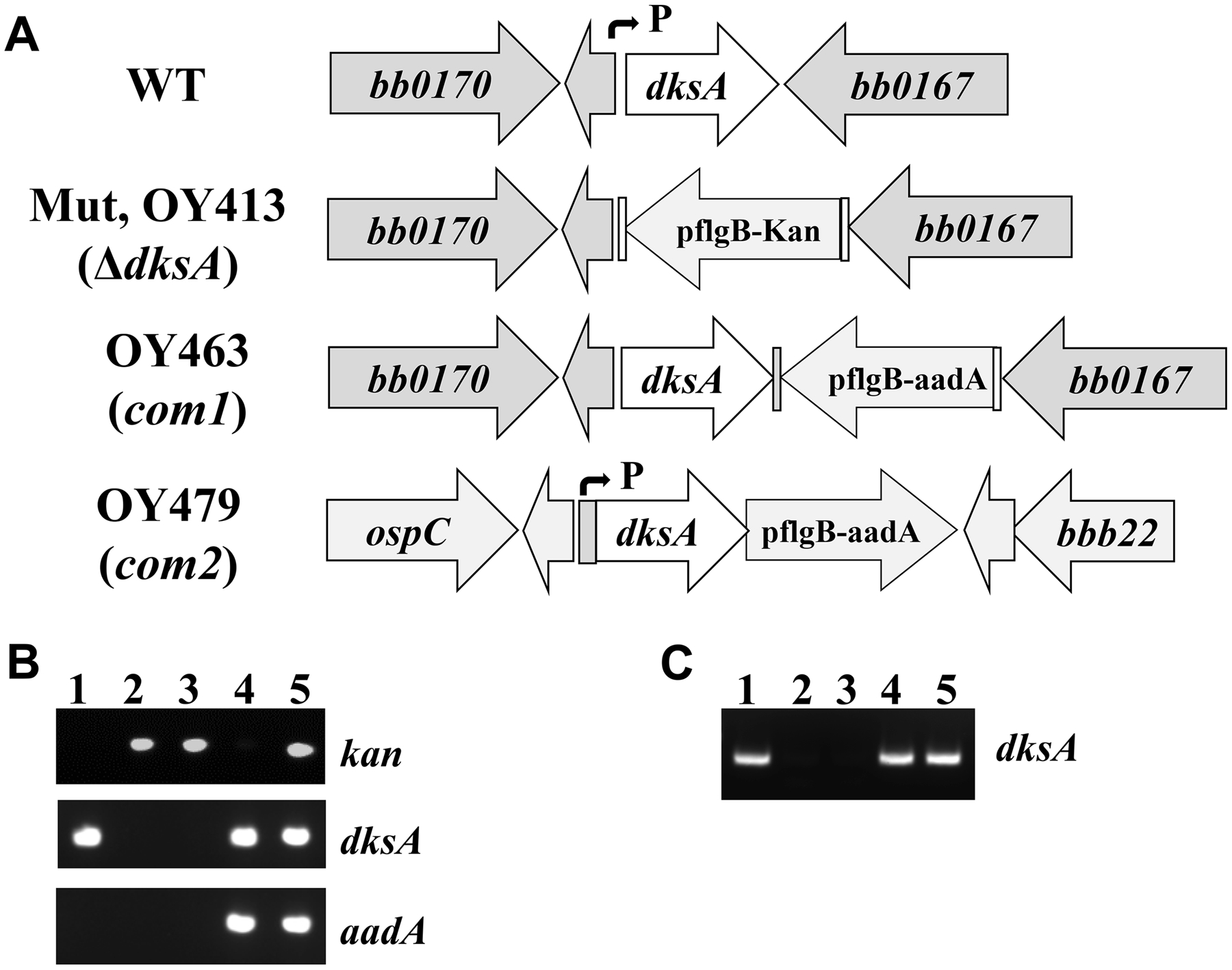
(A) Schematic representation of the dksA locus in WT strain CE162, the mutant OY413, and the complemented strains OY463 and OY479. Genes are shown as thick arrows circumscribing the respective gene numbers. The small arrows indicate the approximate positions of the dksA promoter. (B, C) PCR (B) and RT-PCR (C) analyses of WT CE162, the dksA mutant, and the complemented strains. The specific target gene names are indicated on the right. Lane 1, WT CE162; lanes 2 and 3, two representative clones M1 and M2 of the mutant; lane 4, OY463; lane 5, OY479.
DksA is essential for B. burgdorferi infection in mice.
To assess the importance of DksA in mammalian infection, C3H mice were challenged via needle inoculation with 10,000 spirochetes per mouse of WT CE162, the dksA mutant, or the complemented strains. After three weeks, mice were sacrificed, and skin, heart, and joint samples were harvested. Spirochete burdens in mouse tissues were assessed by qPCR. Compared with the number of flaB in samples collected from mice infected with WT CE162, significantly lower flaB copies were detected in skin, heart, and joints from mice inoculated with the mutant strain (Mut) (Fig. 3). Spirochete burdens in mice inoculated with the complemented strain OY463 (Com1) or OY479 (Com2) were significantly higher than those in mice inoculated with the mutant, even though they were not comparable to the bacterial numbers in mice infected with WT CE162. Infection in animals was also determined by culturing tissues specimens in BSK-II medium. Whereas motile spirochetes were recovered from all animals inoculated with either WT or the complemented strains, bacterial growth was not observed in cultures from mice infected with the dksA mutant (Table 1). In addition, mice were also injected with the dksA mutant at a dose of 107 spirochetes per animal. When infection was determined using the cultivation method, no spirochete was recovered from these animals (Table 1).
Fig. 3. DksA is required by B. burgdorferi to infect mice.
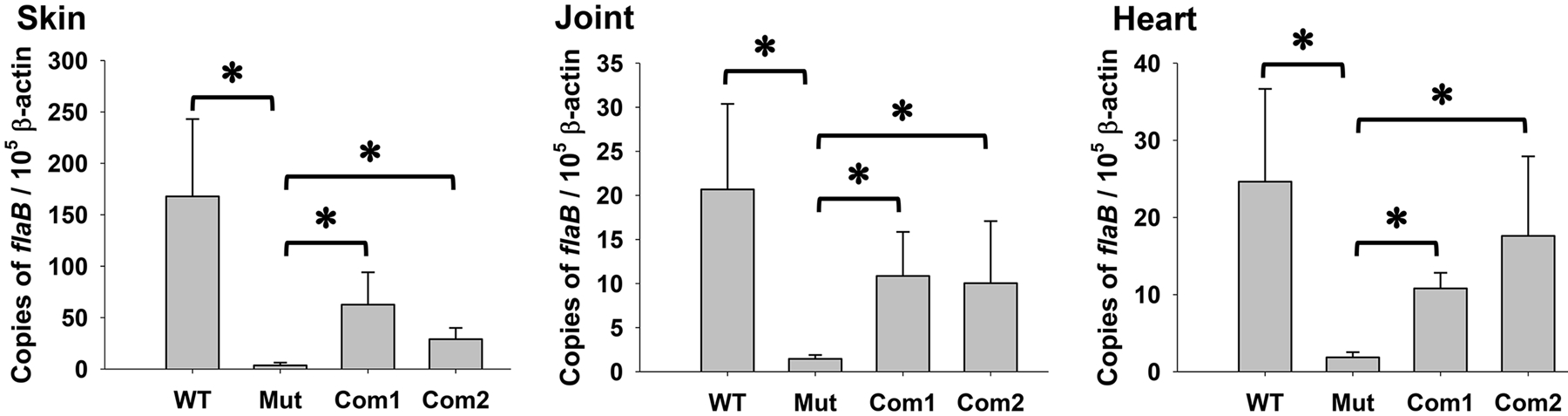
Mice were injected with WT CE162, the dksA mutant (Mut), OY463 (Com1), or OY479 (Com2) at 10,000 spirochetes per animal. Mice were sacrificed at 3 weeks post-inoculation and samples were collected as indicated in Table 1. Spirochete burdens were quantified using qPCR via absolute quantification. The values represent the average copy number of B. burgdorferi flaB normalized per 105 mouse β-actin gene copies. Data are presented as the mean values ± standard deviation. The asterisk indicates statistical significance using one-way ANOVA (P < 0.05).
Table 1.
Infectivity of B. burgdorferi in mice.a
| Strain | Description | Dose | No. of cultures positive / total No. of specimens examined | No. of mice infected / total No. of mice | |||
|---|---|---|---|---|---|---|---|
| Skin | Joint | Heart | All sites | ||||
| CE162 | WT B. burgdorferi | 104 | 3/3 | 3/3 | 3/3 | 9/9 | 3/3 |
| OY413 | ΔdksA | 104 | 0/12 | 0/12 | 0/12 | 0/36 | 0/12 |
| OY413 | ΔdksA | 107 | 0/6 | 0/6 | 0/6 | 0/18 | 0/6 |
| OY463 | dksA-com1 | 104 | 3/3 | 3/3 | 2/3 | 8/9 | 3/3 |
| OY479 | dksA-com2 | 104 | 3/3 | 2/2b | 2/2b | 7/7 | 3/3 |
Data were collected from three independent experiments. WT: wild type; ΔdksA: dksA deletion mutant; dksA-com1, dksA-com2: complemented strains.
Culture data from several samples were not determined due to contamination.
DksA is required for the optimal growth of B. burgdorferi in BSK-II medium.
The morphology of spirochetes cultured in BSK-II was examined using dark-field microscopy, and cell length was measured using the Olympus cellSens imaging software. Whereas all strains exhibited similar spirochetal morphology under our tested conditions, the dksA mutant spirochetes were significantly longer than WT and the complemented strains (Fig. 4A and B). To assess the effect of DksA deficiency on bacterial growth, B. burgdorferi was cultured in BSK-II at 35 °C and 37 °C and growth curve was determined. Compared with WT and the complemented strains, the dksA mutant displayed a growth phenotype at both temperatures (Fig. 4C and D). Both WT and the complemented strains attained a density of ~3.0 × 108 spirochetes per ml at day 7 post-inoculation. However, the mutants displayed a longer lag phase and attained a cell density of ~1.0 × 108 spirochetes per ml at day 7 post-inoculation. These results are consistent with the previous report by Boyle et al (Boyle et al., 2019).
Fig. 4. DksA is required for optimal growth of B. burgdorferi in vitro.
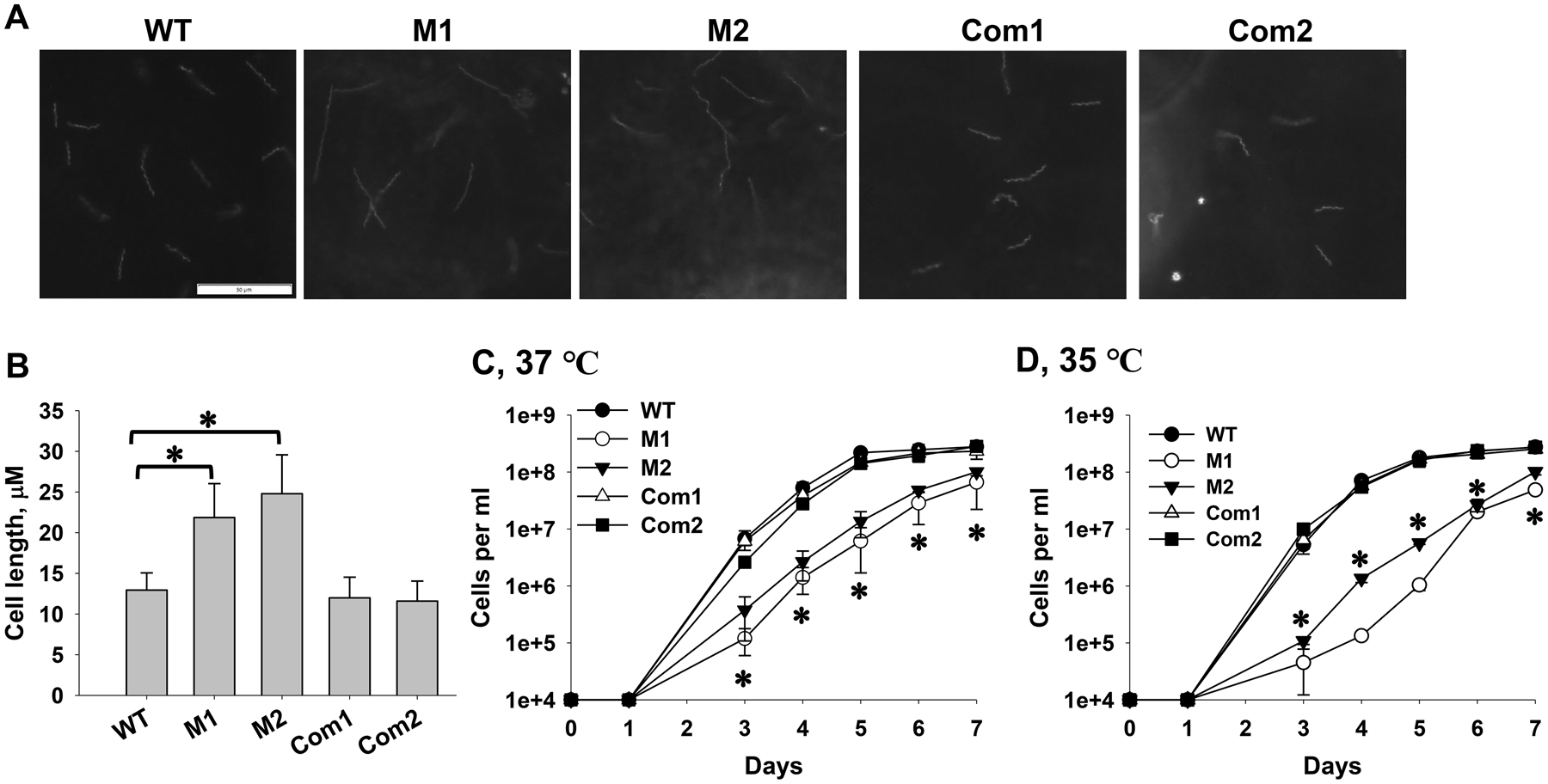
WT, two dksA mutant clones M1 and M2, the complemented strains OY463 (Com1) and OY479 (Com2) were grown in BSK-II at 37 °C (A-C) or 35 °C (D). Spirochetes were enumerated daily using dark-field microscopy. Images were taken for stationary-phase spirochetes by using an Olympus DP22 microscope digital camera (A), and cell length was measured using the Olympus cellSens Imaging Software (B). In (B), data are presented as the mean values ± standard deviation from fifty spirochetes for each strain. The asterisk indicates that the difference is statistically significant (P < 0.05). (C, D) Growth curve analyses of B. burgdorferi. Data are presented as the mean values ± standard deviation from three biological replicates. The asterisk indicates statistical significance (P < 0.05) between the mutants (M1 and M2) and WT CE162 or the complemented strains.
DksA is required for the tolerance of B. burgdorferi to osmotic stress.
To understand how DksA contributes to mammalian infection, we examined the tolerance of B. burgdorferi to osmotic stress. Specifically, WT, two independent clones (M1 and M2) of the dksA mutant, and the complemented strains OY463 and OY479 were cultured in modified BSK-II medium supplemented with various concentrations of NaCl, and bacterial growth curves were determined. Relative to those of WT and the complemented strain, growth of the mutants in medium containing 150-mM NaCl were significantly inhibited (Fig. 5A). WT and the complemented strains, but not the mutants, were capable of growing in medium containing 180- or 200-mM NaCl (Fig. 5B and C).
Fig. 5. DksA is involved in the tolerance of B. burgdorferi to osmotic stress.
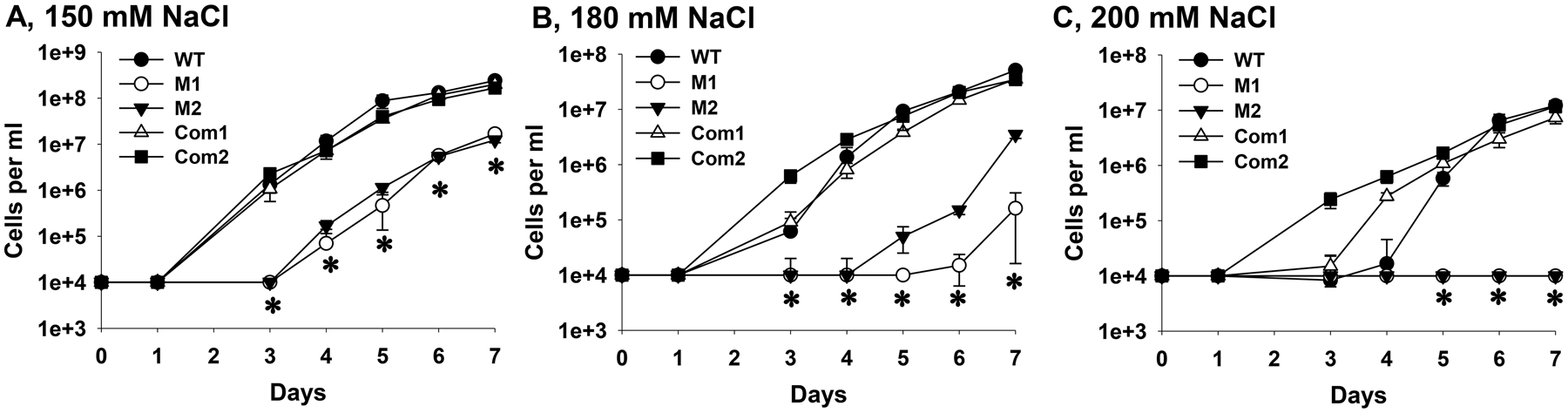
B. burgdorferi was inoculated at 10,000 spirochetes per ml into modified BSK-II with 150 mM (A), 180 mM (B), or 200 mM (C) of NaCl. Spirochetes were enumerated daily by dark-field microscopy. Data are presented as the mean values ± standard deviation from three biological replicates. The asterisk indicates statistical significance (P < 0.05) between the mutants (M1 and M2) and WT CE162 or the complemented strains. WT, CE162; M1 and M2, two representative clones of the mutant; Com1, OY463; Com2, OY479.
DksA regulates the expression of rpoS and ospC in B. burgdorferi.
To understand the regulatory effect of DksA on virulence gene expression in B. burgdorferi, we analyzed the expression of ospC using qRT-PCR. In many bacteria such as E. coli, rRNA levels are regulated by (p)ppGpp and DksA-mediated stringent response. However, Boyle et al. reported that the levels of 16S rRNA transcripts were not affected by the mutation of dksA in B. burgdorferi (Boyle et al., 2019). Specifically, by comparing gene transcription between WT and the dksA mutant using qRT-PCR, the authors found that 16S rRNA was more stable than other commonly used reference genes such as flaB and rpoD. We also performed qRT-PCR and compared the Ct values of 16S rRNA, flaB, and eno among WT CE162, the dksA mutant and complemented strains. Consistent with the findings by Boyle et al. (Boyle et al., 2019), our results showed that the Ct values of 16S rRNA were less responsive to dksA mutation than flaB and eno (Fig. 6A). Therefore, we also used 16S rRNA, as in the previous research (Boyle et al., 2019), to normalize the qRT-PCR data.
Fig. 6. DksA regulates rpoS and ospC expression.
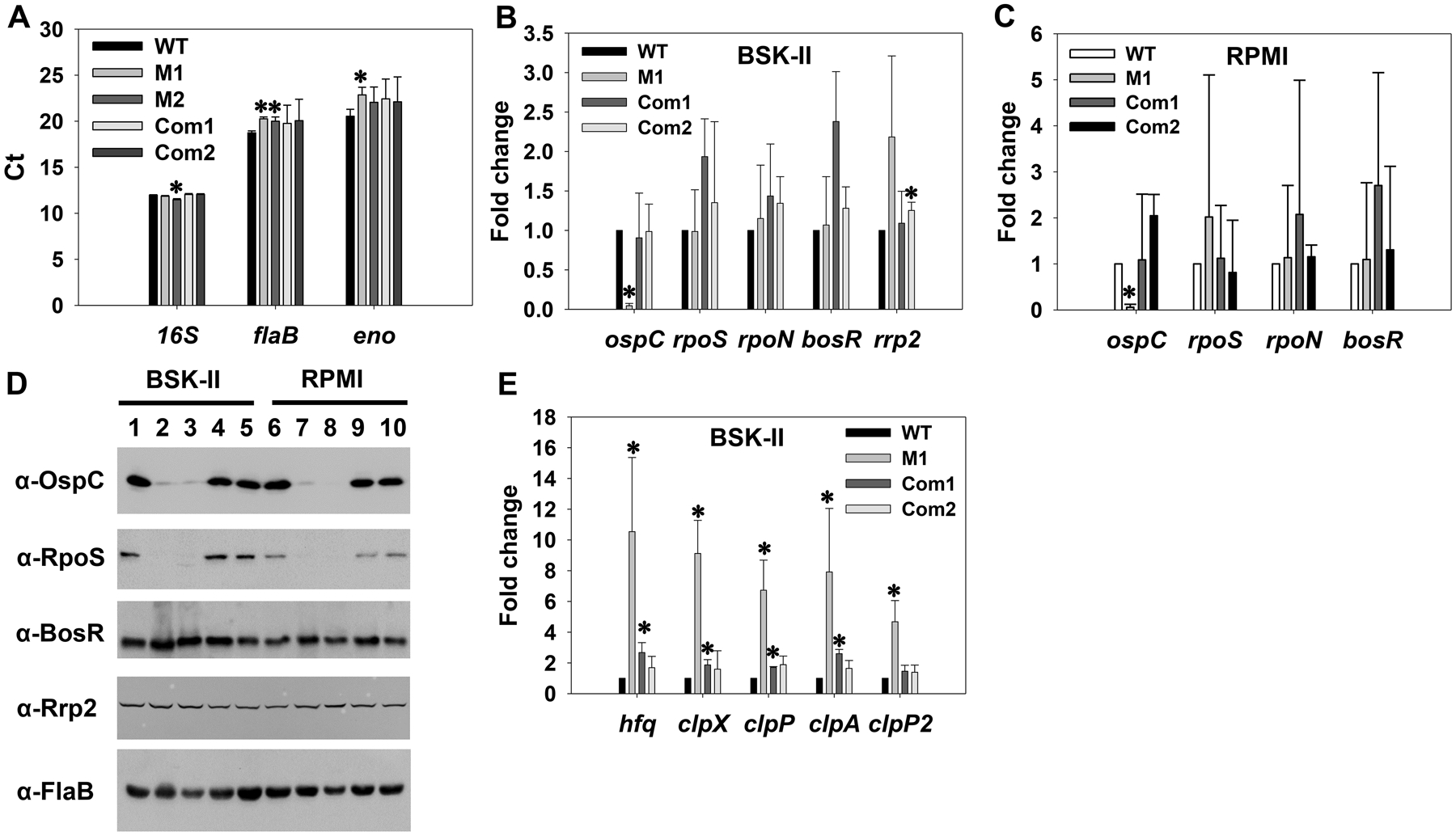
B. burgdorferi was cultivated in BSK-II and spirochetes were collected at stationary phase. In addition, spirochetes were starved in RPMI for 6 h. Gene expression was assessed via qRT-PCR (A, B, C, E) or immunoblot (D). (A) comparison of Ct values (threshold cycle) for reference genes. Gene expression was analyzed using qRT-PCR. Data were obtained from three biological replicates and presented as the mean measurements ± standard deviations. The asterisk indicates statistical significance (P < 0.05) from WT CE162. In (B, C, E), data were normalized using 16S rRNA as an internal control. Bars represent the mean measurements ± standard deviations from three biological replicates. The asterisk indicates statistical significance (P < 0.05) from the WT strain. WT, CE162; M1 and M2, two representative clones of the mutant; Com1, the complemented strain OY463; Com2, the complemented strain OY479. In (D), approximately 4 × 107 spirochetes were loaded onto each lane of a 12.5% SDS-polyacrylamide gel. After proteins were resolved by SDS-PAGE, they were transferred to nitrocellulose membranes and analyzed by immunoblot. Specific antibodies, denoted as α, are indicated on the left. Lanes 1 and 6, WT CE162; lanes 2 and 7, M1 clone of the mutant OY413; lanes 3 and 8, M2 clone of the mutant; lanes 4 and 9, the complemented strain OY463; lanes 5 and 10, the complemented strain OY479.
As shown in Fig. 6B and 6C, in both stationary-phase spirochetes cultivated in BSK-II and starved spirochetes, transcription of ospC was dramatically decreased in the dksA mutant. We also measured protein levels through immunoblot. Compared with that in WT, the level of OspC was substantially reduced in the mutant deficient in dksA; only a trace amount of OspC was detected in the mutant (Fig. 6D). To verify the decrease of ospC expression in the dksA mutant was due solely to the inactivation of dksA, gene expression was analyzed in the complemented strains. As shown in Fig. 6B–D, both mRNA and protein levels of OspC were fully restored when the dksA mutation was complemented.
In B. burgdorferi, expression of ospC is dependent on the alternative sigma factor RpoS; and rpoS transcription requires the other alternative sigma factor RpoN (i.e., σ54) along with the enhancer binding protein Rrp2 and the Fur/BosR homolog BosR [see Reviews (Stevenson & Seshu, 2018, Samuels, 2011, Radolf et al., 2012, Troxell & Yang, 2013)]. We thus analyzed the expression of these regulators in the dksA mutant via qRT-PCR and immunoblot analyses. As shown in Fig. 6B, in stationary-phase spirochetes cultivated in BSK-II, transcription of rpoS remained unchanged in the dksA mutant. Surprisingly, RpoS protein was essentially undetected in the dksA mutant, whereas genetic complementation readily restored RpoS to a WT-level (Fig. 6D). Moreover, the transcript levels of rpoN, bosR and rrp2 in the dksA mutant were comparable to those in WT and the complemented strains (Fig. 6B). In addition, similar levels of Rrp2 or BosR were detected among WT, the mutant and the complemented strains (Fig. 6D). We also analyzed gene expression in starved spirochetes, and similar results were obtained (Fig. 6C and 6D).
To understand how DksA regulates RpoS, we measured expression of hfq, clpX, clpP, clpA, and clpP2 in the dksA mutant; these factors have been reported to affect rpoS expression at the post-transcriptional level in B. burgdorferi or other bacteria. To this end, B. burgdorferi was cultivated in BSK-II and harvested at the stationary phase. Gene expression was analyzed using qRT-PCR. As shown in Fig. 6E, transcription of hfq was significantly increased ~11 fold in the mutant. Moreover, expression of clpX, clpA, clpP, and clpP2 was significantly upregulated ~9.1-, 7.9-, 6.7-, or 4.7-fold, respectively, in the mutant. When mutation was complemented, gene expression was restored to near WT-levels.
Discussion
In this study, we provide evidence that the stringent response regulator DksA is essential for B. burgdorferi to infect animals. The first line of evidence supporting our conclusion emanates from analyzing the expression of dksA. When dksA expression was analyzed in tissue samples isolated from animals infected with B. burgdorferi, high levels of dksA transcripts were detected in these samples, suggesting that this gene may be important for animal infection. To directly determine the role of DksA during animal infection, we successfully created a mutant lacking DksA in low-passage, virulent strain CE162. This mutant retains all essential plasmids contained in its parental strain. When the infectivity of the dksA mutant was analyzed using a murine model, our data revealed that inactivation of dksA rendered B. burgdorferi completely noninfectious in mice. Motile spirochetes were readily recovered from all mice injected with 10,000 WT spirochetes. In contrast, when mice were injected with the mutant at a dose of 10,000- or even as high as 107 spirochetes per mouse, no animal was infected. To ensure that the phenotypes observed in the dksA mutant were due solely to the inactivation of dksA, gene mutation was complemented using two different approaches. First, an WT-copy of dksA was restored to its native chromosomal locus. Second, the PdksA-dksA cassette was inserted into the dispensable bbb20-bbb21 locus of cp26. Transcription of dksA in both complemented strains were under the control of its native promoter, thereby facilitating a similar WT-level of dksA expression in the complemented strains. When the infectivity of these two complemented strains were assessed, our data showed that the infectivity phenotype was fully restored. These combined data strongly support that DksA is essential for the infection of B. burgdorferi in a mammalian host.
To survive in nature and to maintain its tick-mammal life cycle, B. burgdorferi must contend with multiple environmental stresses such as osmotic pressure (Brisson et al., 2012, Pal & Fikrig, 2003, Stewart & Rosa, 2018, Estrada-Pena et al., 2018, Bernard et al., 2019, Samuels, 2011, Radolf et al., 2012). To understand why the dksA mutant lost its ability to infect mammalian hosts, we examined the effect of DksA deficiency on the tolerance of B. burgdorferi to osmotic stress. Our results showed that, compared with WT and the complemented strain, the dksA mutant was much more susceptible to osmotic stress. These results support that DksA is critical for the fitness and survival of B. burgdorferi under stressful conditions. Our data also showed that growth of B. burgdorferi in BSK-II at the mammal-specific temperature (i.e., 37 °C) was affected by dksA inactivation. Compared with WT spirochetes, the mutant displayed a prolonged lag phase. Under the microscope, the dksA mutant is significantly longer than WT and the complemented strains. Despite that the precise reasons for these phenotypes remain currently unknown, they are probably resulted from the defect of the mutant in uptake and utilizing nutrients to support cell division and replication, which suggests that DksA may contribute to the replication and/or growth kinetics of B. burgdorferi.
In a wide variety of bacteria, the alternative sigma factor RpoS is a master regulator for general stress response at stationary phase or under nutritional starvation conditions (Gottesman, 2019, Hengge, 2009, Battesti et al., 2011). However, in B. burgdorferi, RpoS is not involved in general stress response or adaptation to stationary phase (Caimano et al., 2004, Caimano et al., 2019). Rather, this protein plays a central role in mouse infection and controls expression of many virulence-associated lipoproteins such as OspC [see Reviews (Stevenson & Seshu, 2018, Samuels, 2011, Radolf et al., 2012, Troxell & Yang, 2013)]. To discern the mechanistic details underlying the essential attributes of DksA, we analyzed expression of rpoS and RpoS-dependent ospC in the dksA mutant. Consistent with the previous report (Boyle et al., 2019), our results showed that transcription of ospC, but not rpoS, was dramatically reduced in the dksA mutant. Moreover, our immunoblot analyses revealed that synthesis of OspC was substantially diminished in the mutant. More surprisingly, in both stationary-phase spirochetes and starved spirochetes, RpoS was abolished when dksA was inactivated, and genetic complementation restored protein expression to a WT level. These data suggest that DksA regulates rpoS expression at the post-transcriptional level.
Usually, cellular protein levels are determined by two processes including protein synthesis and protein degradation. RpoS synthesis in B. burgdorferi has been reported to be post-transcriptionally regulated by the small non-coding RNA DsrABb and the chaperone Hfq (Lybecker et al., 2010, Lybecker & Samuels, 2007). Specifically, the level of RpoS was found to be reduced in a hfq mutant at stationary phase, which is similar to our findings for the dksA mutant. In contrast, RpoS levels were reduced in a dsrABb mutant only at log-phase, but not at stationary phase. DksA may activate the expression of Hfq, thereby improving RpoS translational efficiency. However, our results revealed that expression of hfq was elevated, rather than reduced, in the dksA mutant, suggesting that DksA may regulate RpoS independent of Hfq. We also tested the hypothesis that DksA might affect the degradation of RpoS in B. burgdorferi. In an array of bacteria, RpoS degradation is mediated by the AAA+ ATP-dependent protease ClpXP [see Reviews (Battesti et al., 2011, Hengge, 2009)]. Moreover, ClpAP, another Clp protease complex, was reported to degrade RpoS in the soil bacterium Azotobacter vinelandii (Muriel-Millan et al., 2017). Homologs of all these Clp proteases have been identified in B. burgdorferi, despite that the contributions of these protease to RpoS degradation need to be confirmed. Among these proteases, BB0611 (ClpP) and BB0612 (ClpX) are encoded by a gene operon and may form the ClpXP complex, whereas BB0369 (ClpA) and BB0757 (ClpP2) may form the ClpAP complex (Fraser et al., 1997, Mason et al., 2020). We thus examined the expression of these factors in the dksA mutant. Our data showed that expression of clpX, clpP, clpA, and clpP2 were highly upregulated in the dksA mutant. Therefore, the reduction of RpoS levels in the dksA mutant is probably due to increased levels of Clp proteases and consequently, increased RpoS degradation. It is currently impossible to assess the contribution of DksA to RpoS degradation using protein turnover assays, as RpoS is undetected in the dksA mutant. Nevertheless, our results suggest that DksA may impact RpoS levels via its effects on the ClpXP and/or ClpAP protease complexes. Future work, such as analyzing gene expression in a dksA and clpXP or clpAP double or triple mutant, may help elucidate how DksA plays a Clp proteases-dependent role in RpoS regulation.
Previous global transcriptome analyses have revealed that DksA and (p)ppGpp coordinately regulate expression of numerous B. burgdorferi genes involved in stringent response (Bugrysheva et al., 2015, Drecktrah et al., 2015, Boyle et al., 2019, Drecktrah et al., 2018). In addition, DksA and (p)ppGpp regulate transcription of different sets of genes, suggesting that these two factors have independent roles in B. burgdorferi. This notion is further buttressed by our present study. Specifically, whereas RelBbu is dispensable for murine infection by needle inoculation (Drecktrah et al., 2015), DksA is required by B. burgdorferi to infect mice, supporting that DksA likely plays multifaceted roles in the pathogenesis of B. burgdorferi. We propose that three different mechanisms may account for the attenuated infectivity phenotype of the dksA mutant. First, DksA post-transcriptionally regulates RpoS which, in turn, influences the production of OspC and other RpoS-dependent virulence determinants. Moreover, DksA may regulate genes involved in cell replication and growth. In addition, genes regulated by DksA may be needed by B. burgdorferi to cope with environmental and metabolic stress challenges. Previous microarray analyses (Boyle et al., 2019) have revealed numerous genes regulated by DksA only, but not by (p)ppGpp. Future characterization of these genes exclusively regulated by DksA may uncover novel virulence factors for this important human pathogen. Continued efforts are also warranted to address the contribution of DksA and the DksA regulon to the tick phase infection of B. burgdorferi. This study thus not only has elucidated a key factor governing B. burgdorferi virulence, but also provides new insights into another regulatory layer controlling the central RpoS and the RpoS regulon in B. burgdorferi.
Materials and Methods
Bacterial strains and culture conditions.
The infectious clonal isolate CE162 (Caimano et al., 2004) was used as the WT strain in this study. B. burgdorferi was routinely cultivated at 35 °C and 5% CO2 in BSK-II medium (Pollack et al., 1993) supplemented with 6% rabbit serum (Pel-Freeze, Rogers, AR). To examine the impact of environmental factors on gene expression, B. burgdorferi was cultured at 23 °C or 37 °C, and the pH value of BSK-II was adjusted to pH 6.8. When applicable, antibiotics were added to the media at the following concentrations: kanamycin, 160 μg/ml; streptomycin, 100 μg/ml. Escherichia coli was grown in Lysogeny broth (LB) medium with appropriate antibiotics: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; or spectinomycin, 100 μg/ml. E. coli strain TOP 10 (Thermo Fisher Scientific, Grand Island, NY) was utilized as the cloning host for routine molecular cloning and plasmid propagation. All plasmid constructs utilized in this study were confirmed by using PCR amplification, restriction digestion, and sequence analysis.
Construction of the dksA mutant and complemented strains.
The dksA deletion mutant OY413 was created through homologous recombination. In brief, the pMB1 origin of replication was amplified from pUC19 (Thermo Fisher Scientific). The PflgB-Kan cassette was amplified from our bmtA mutant (Ouyang et al., 2009a). Furthermore, a ~1024-bp 5’ arm and a ~1177-bp 3’ arm flanking dksA were PCR amplified from CE162. Following purification, these four DNA fragments were assembled using the GeneArt seamless cloning and assembly kit (Thermo Fisher Scientific). In the resulting construct pOY598, the PflgB-Kan cassette was oriented opposing dksA transcription. Purified pOY598 was then transformed into B. burgdorferi strain CE162 through electroporation (Samuels et al., 2018, Samuels, 1995). Transformants were selected using kanamycin and confirmed by PCR and RT-PCR analyses. The plasmid content of B. burgdorferi was determined by multiplex PCR as previously described (Bunikis et al., 2011, Xiang et al., 2017).
Two strategies were employed to complement the dksA mutation. First, a suicide plasmid pOY662 was created to complement dksA mutation at its native chromosome locus. To this end, a ~1526-bp 5’ arm encompassing ~986-bp sequences upstream of dksA and the entire dksA ORF, and a ~1177-bp 3’ arm downstream of dksA were PCR amplified from CE162. Moreover, the PflgB-aadA cassette was amplified from our bmtA complemented strain (Ouyang et al., 2009a). These three fragments were then purified and assembled with the pMB1 origin of replication using the GeneArt seamless cloning and assembly kit. In the resultant pOY662, pflgB-aadA was oriented in the opposite direction of dksA. Purified pOY662 was transformed into the dksA mutant OY413, generating the streptomycin-resistant complemented strain OY463.
The dksA mutant was also complemented by inserting dksA with its native promoter (PdksA) (Boyle et al., 2019) into the dispensable bbb20-bbb21 intergenic region of the endogenous plasmid cp26. First, a ~1200-bp upstream region and a ~1170-bp downstream region flanking the bbb20-bbb21 intergenic region (Dunham-Ems et al., 2009) were amplified from CE162. Purified DNA was then assembled into linearized pUC19L (Thermo Fisher Scientific), creating pOY625. Next, one fragment encompassing the promoter of dksA and the dksA ORF (i.e., PdksA-dksA) was amplified from CE162. PdksA-dksA was digested with XbaI and BglII, and then cloned into pJD54 digested with same restriction enzymes, yielding pOY683. The PdksA-dksA-PflaB-aadA cassette was excised from pOY683 using AscI, and ligated into pOY625, creating the final suicide plasmid construct pOY685. Purified pOY685 was electroporated into the dksA mutant OY413, generating the streptomycin-resistant complemented strain OY479.
Growth curve analyses.
Bacterial growth was analyzed as previously described (Ouyang et al., 2009b, Ouyang et al., 2008, Ouyang et al., 2009a). In brief, B. burgdorferi was initially cultivated in BSK-II media. When growth reached late logarithmic phase (~ 5 × 107 to 1 × 108 cells per ml), spirochetes were inoculated into fresh BSK-II medium at 1 × 104 cells per ml. Cells were counted every 24 hours using dark-field microscopy.
Osmotic stress tolerance assay.
This assay was performed as previously described (Curtis et al., 2018, Mason et al., 2020). In brief, B. burgdorferi was grown in BSK-II until the late logarithmic phase. Spirochetes were inoculated into modified BSK-II containing varying concentrations of NaCl. Growth was enumerated by dark-field microscopy.
Serum starvation experiments.
Starvation of B. burgdorferi was performed as previously described (Caimano et al., 2004, Drecktrah et al., 2015, Boyle et al., 2019). Briefly, when B. burgdorferi grown in BSK-II medium reached stationary phase, spirochetes were collected by centrifugation and resuspended in RPMI 1640 medium without L-glutamine (Sigma-Aldrich, St. Louis, MO). Spirochetes were then incubated at 37 °C for 6 h and harvested by centrifugation for gene expression analyses.
Mouse infection studies.
The infectivity of dksA mutant was assessed by using the murine needle-challenge model of Lyme borreliosis (Barthold et al., 1991, Barthold et al., 1993, Barthold et al., 1990). All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of South Florida in compliance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. C3H/HeN mice (Charles River Laboratories) were intradermally injected with B. burgdorferi. At 3 weeks post-inoculation, mice were euthanized, and samples of skin, heart, and joints were collected. Infection of B. burgdorferi was determined by culturing tissue specimens in BSK-II supplemented with 1× Borrelia antibiotic mixture (BAM, Sigma). The resulting growth was monitored by using dark-field microscopy.
Bacterial load in mouse tissue samples was determined by using quantitative PCR (qPCR). Briefly, mouse specimens were homogenized using the Bead Mill 4 Homogenizer (Thermo Fisher Scientific). DNA was extracted using the Thermo GeneJET Genomic DNA Purification Kit (Thermo Fisher Scientific). qPCR was performed using the PowerUP SYBR Green Master Mix (Thermo Fisher Scientific). As previously described (Pal et al., 2004, Pal et al., 2008, Ouyang et al., 2012, Mason et al., 2020), the absolute quantification method was employed to calculate the copy number of murine β-actin gene and B. burgdorferi flaB gene in each sample.
Analysis of dksA expression in infected animals through qRT-PCR.
As aforementioned, mice were injected with B. burgdorferi CE162 at a dose of 10,000 spirochetes per animal. Ear punch biopsy samples were taken at 2 weeks post-infection and cultured in BSK-II with 1 × BAM to confirm bacterial infection. Mice were sacrificed at 3 weeks post-infection, and skin, heart, and joints were collected and homogenized using the Bead Mill 4 Homogenizer. Total RNA was isolated by using Trizol (Thermo Fisher Scientific) and purified using the RNeasy Mini Kit (Qiagen, Valencia, CA). After genomic DNA was digested by using Turbo DNase (Thermo Fisher Scientific), RNA was purified further using GeneJET RNA Cleanup and Concentration Micro Kit (Thermo Fisher Scientific). cDNA was synthesized by using the SuperScript IV reverse transcriptase (Thermo Fisher Scientific) according to the instructions. qPCR was performed using the PowerUP SYBR Green Master Mix. Transcript copies of dksA and flaB present in mouse specimens were determined by using qPCR via the absolute quantification method. Standard curves for flaB and dksA were created using 10-fold serial dilutions of pOY16 (pGEM-Teasy-flaB) or pOY685, respectively, as the template in qPCR, and transcript copy number was calculated by using the Absolute Quantification Analysis program.
Analysis of gene expression in B. burgdorferi grown in BSK-II medium through qRT-PCR.
B. burgdorferi grown under various conditions were harvested by centrifugation. Total RNA isolation, removal of genomic DNA, cDNA synthesis and qPCR were carried out essentially as described above. Relative quantification (ΔΔCT) was employed to compare gene expression in in vitro-grown spirochetes. As in the previous study by Boyle et al. (Boyle et al., 2019), we used B. burgdorferi 16S rRNA as the endogenous control for data normalization.
SDS-PAGE and immunoblot analysis
SDS-PAGE and immunoblot analysis were carried out as previously described (Ouyang et al., 2009b, Ouyang et al., 2008). Briefly, B. burgdorferi grown in BSK-II medium was harvested by centrifugation when bacterial growth reached the stationary phase (referred as stationary-phase spirochetes). In addition, starved spirochetes were also collected. Cells were washed thrice with PBS and resuspended in SDS sample buffer. A volume of whole cell lysate equivalent to 4 × 107 bacteria was loaded per lane on a 12.5% acrylamide gel. Resolved proteins were transferred to nitrocellulose membrane for immunoblot analysis. FlaB, Rrp2, RpoS, OspC, and BosR were detected using anti-FlaB antibody, anti-Rrp2 monoclonal antibody, anti-RpoS monoclonal antibody, anti-OspC monoclonal antibody, or a polyclonal antibody against BosR, respectively. Immunoblots were developed by chemiluminescence using ECL Plus Western Blotting Detection system (Amersham Biosciences).
Statistical analysis.
Data from different experimental groups were analyzed by using ANOVA where statistical significance was established when P < 0.05.
Acknowledgments
We thank Shidoya Parrilla and Candace Sukie for technical help. This work was supported by funding from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (AI146909 and AI119437 to Z.O.).
Footnotes
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- Atkinson GC, Tenson T & Hauryliuk V, (2011) The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6: e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, Beck DS, Hansen GM, Terwilliger GA & Moody KD, (1990) Lyme borreliosis in selected strains and ages of laboratory mice. J Infect Dis 162: 133–138. [DOI] [PubMed] [Google Scholar]
- Barthold SW, de Souza MS, Janotka JL, Smith AL & Persing DH, (1993) Chronic Lyme borreliosis in the laboratory mouse. Am J Pathol 143: 959–971. [PMC free article] [PubMed] [Google Scholar]
- Barthold SW, Persing DH, Armstrong AL & Peeples RA, (1991) Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am J Pathol 139: 263–273. [PMC free article] [PubMed] [Google Scholar]
- Battesti A, Majdalani N & Gottesman S, (2011) The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65: 189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard Q, Thakur M, Smith AA, Kitsou C, Yang X & Pal U, (2019) Borrelia burgdorferi protein interactions critical for microbial persistence in mammals. Cell Microbiol 21: e12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WK, Groshong AM, Drecktrah D, Boylan JA, Gherardini FC, Blevins JS, Samuels DS & Bourret TJ, (2019) DksA Controls the Response of the Lyme Disease Spirochete Borrelia burgdorferi to Starvation. J Bacteriol 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson D, Drecktrah D, Eggers CH & Samuels DS, (2012) Genetics of Borrelia burgdorferi. Annual review of genetics 46: 515–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva J, Dobrikova EY, Godfrey HP, Sartakova ML & Cabello FC, (2002) Modulation of Borrelia burgdorferi stringent response and gene expression during extracellular growth with tick cells. Infect Immun 70: 3061–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva J, Dobrikova EY, Sartakova ML, Caimano MJ, Daniels TJ, Radolf JD, Godfrey HP & Cabello FC, (2003) Characterization of the stringent response and relBbu expression in Borrelia burgdorferi. J Bacteriol 185: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Bryksin AV, Godfrey HP & Cabello FC, (2005) Borrelia burgdorferi rel is responsible for generation of guanosine-3’-diphosphate-5’-triphosphate and growth control. Infect Immun 73: 4972–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Godfrey HP, Schwartz I & Cabello FC, (2011) Patterns and regulation of ribosomal RNA transcription in Borrelia burgdorferi. BMC Microbiol 11: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugrysheva JV, Pappas CJ, Terekhova DA, Iyer R, Godfrey HP, Schwartz I & Cabello FC, (2015) Characterization of the RelBbu Regulon in Borrelia burgdorferi Reveals Modulation of Glycerol Metabolism by (p)ppGpp. PLoS One 10: e0118063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis I, Kutschan-Bunikis S, Bonde M & Bergstrom S, (2011) Multiplex PCR as a tool for validating plasmid content of Borrelia burgdorferi. J Microbiol Methods 86: 243–247. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E & Davis JP, (1982) Lyme disease-a tick-borne spirochetosis? Science 216: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Caimano MJ, Drecktrah D, Kung F & Samuels DS, (2016) Interaction of the Lyme disease spirochete with its tick vector. Cell Microbiol 18: 919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Eggers CH, Hazlett KR & Radolf JD, (2004) RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect Immun 72: 6433–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimano MJ, Groshong AM, Belperron A, Mao J, Hawley KL, Luthra A, Graham DE, Earnhart CG, Marconi RT, Bockenstedt LK, Blevins JS & Radolf JD, (2019) The RpoS Gatekeeper in Borrelia burgdorferi: An Invariant Regulatory Scheme That Promotes Spirochete Persistence in Reservoir Hosts and Niche Diversity. Front Microbiol 10: 1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concepcion MB & Nelson DR, (2003) Expression of spoT in Borrelia burgdorferi during serum starvation. J Bacteriol 185: 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MW, Hahn BL, Zhang K, Li C, Robinson RT & Coburn J, (2018) Characterization of Stress and Innate Immunity Resistance of Wild-Type and Δp66 Borrelia burgdorferi. Infect Immun 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva AM & Fikrig E, (1995) Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am J Trop Med Hyg 53: 397–404. [DOI] [PubMed] [Google Scholar]
- Drecktrah D, Hall LS, Rescheneder P, Lybecker M & Samuels DS, (2018) The Stringent Response-Regulated sRNA Transcriptome of Borrelia burgdorferi. Front Cell Infect Microbiol 8: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Lybecker M, Popitsch N, Rescheneder P, Hall LS & Samuels DS, (2015) The Borrelia burgdorferi RelA/SpoT Homolog and Stringent Response Regulate Survival in the Tick Vector and Global Gene Expression during Starvation. PLoS Pathog 11: e1005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH, Balic A & Radolf JD, (2009) Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J Clin Invest 119: 3652–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Pena A, Alvarez-Jarreta J & Cabezas-Cruz A, (2018) Reservoir and vector evolutionary pressures shaped the adaptation of Borrelia. Infect Genet Evol 66: 308–318. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO & Venter JC, (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390: 580–586. [DOI] [PubMed] [Google Scholar]
- Gaca AO, Colomer-Winter C & Lemos JA, (2015) Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 197: 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, (2019) Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J Biol Chem 294: 11685–11700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Chen AY, Gopalkrishnan S, Sanchez-Vazquez P, Myers A & Ross W, (2018) Transcriptional Responses to ppGpp and DksA. Annu Rev Microbiol 72: 163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R, (2009) Proteolysis of σS (RpoS) and the general stress response in Escherichia coli. Res Microbiol 160: 667–676. [DOI] [PubMed] [Google Scholar]
- Irving SE & Corrigan RM, (2018) Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology 164: 268–276. [DOI] [PubMed] [Google Scholar]
- Kang PJ & Craig EA, (1990) Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol 172: 2055–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC, Abel CA, Feig AL & Samuels DS, (2010) Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 78: 622–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lybecker MC & Samuels DS, (2007) Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol Microbiol 64: 1075–1089. [DOI] [PubMed] [Google Scholar]
- Mason C, Thompson C & Ouyang Z, (2020) The Lon-2 protease of Borrelia burgdorferi is critical for infection in the mammalian host. Mol Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel-Millan LF, Moreno S, Gallegos-Monterrosa R & Espin G, (2017) Unphosphorylated EIIANtr induces ClpAP-mediated degradation of RpoS in Azotobacter vinelandii. Mol Microbiol 104: 197–211. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Blevins JS & Norgard MV, (2008) Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology 154: 2641–2658. [DOI] [PubMed] [Google Scholar]
- Ouyang Z, He M, Oman T, Yang XF & Norgard MV, (2009a) A manganese transporter, BB0219 (BmtA), is required for virulence by the Lyme disease spirochete, Borrelia burgdorferi. Proc Natl Acad Sci U S A 106: 3449–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U & Norgard MV, (2009b) BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol Microbiol 74: 1331–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Narasimhan S, Neelakanta G, Kumar M, Pal U, Fikrig E & Norgard MV, (2012) Activation of the RpoN-RpoS regulatory pathway during the enzootic life cycle of Borrelia burgdorferi. BMC Microbiol 12: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal U, Dai J, Li X, Neelakanta G, Luo P, Kumar M, Wang P, Yang X, Anderson JF & Fikrig E, (2008) A differential role for BB0365 in the persistence of Borrelia burgdorferi in mice and ticks. J Infect Dis 197: 148–155. [DOI] [PubMed] [Google Scholar]
- Pal U & Fikrig E, (2003) Adaptation of Borrelia burgdorferi in the vector and vertebrate host. Microbes Infect 5: 659–666. [DOI] [PubMed] [Google Scholar]
- Pal U, Li X, Wang T, Montgomery RR, Ramamoorthi N, Desilva AM, Bao F, Yang X, Pypaert M, Pradhan D, Kantor FS, Telford S, Anderson JF & Fikrig E, (2004) TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119: 457–468. [DOI] [PubMed] [Google Scholar]
- Parshin A, Shiver AL, Lee J, Ozerova M, Schneidman-Duhovny D, Gross CA & Borukhov S, (2015) DksA regulates RNA polymerase in Escherichia coli through a network of interactions in the secondary channel that includes Sequence Insertion 1. Proc Natl Acad Sci U S A 112: E6862–6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I & Vassylyev DG, (2004) Regulation through the secondary channel--structural framework for ppGpp-DksA synergism during transcription. Cell 118: 297–309. [DOI] [PubMed] [Google Scholar]
- Pollack RJ, Telford SR 3rd & Spielman A, (1993) Standardization of medium for culturing Lyme disease spirochetes. J Clin Microbiol 31: 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B & Hu LT, (2012) Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa PA, Tilly K & Stewart PE, (2005) The burgeoning molecular genetics of the Lyme disease spirochaete. Nat Rev Microbiol 3: 129–143. [DOI] [PubMed] [Google Scholar]
- Ross W, Sanchez-Vazquez P, Chen AY, Lee JH, Burgos HL & Gourse RL, (2016) ppGpp Binding to a Site at the RNAP-DksA Interface Accounts for Its Dramatic Effects on Transcription Initiation during the Stringent Response. Mol Cell 62: 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS, (1995) Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol 47: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS, (2011) Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65: 479–499. [DOI] [PubMed] [Google Scholar]
- Samuels DS, Drecktrah D & Hall LS, (2018) Genetic Transformation and Complementation. Methods Mol Biol 1690: 183–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan TG, (1996) Ticks and Borrelia: model systems for investigating pathogen-arthropod interactions. Infect Agents Dis 5: 167–181. [PubMed] [Google Scholar]
- Steere AC, (1993) Current understanding of Lyme disease. Hosp Pract (Off Ed) 28: 37–44. [DOI] [PubMed] [Google Scholar]
- Steere AC, Coburn J & Glickstein L, (2004) The emergence of Lyme disease. J Clin Invest 113: 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson B & Seshu J, (2018) Regulation of Gene and Protein Expression in the Lyme Disease Spirochete. Curr Top Microbiol Immunol 415: 83–112. [DOI] [PubMed] [Google Scholar]
- Stewart PE & Rosa PA, (2018) Physiologic and Genetic Factors Influencing the Zoonotic Cycle of Borrelia burgdorferi. Curr Top Microbiol Immunol 415: 63–82. [DOI] [PubMed] [Google Scholar]
- Troxell B & Yang XF, (2013) Metal-dependent gene regulation in the causative agent of Lyme disease. Front Cell Infect Microbiol 3: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Yang Y, Du J, Lin T, Chen T, Yang XF & Lou Y, (2017) Investigation of ospC Expression Variation among Borrelia burgdorferi Strains. Front Cell Infect Microbiol 7: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Pan Y & Dai X, (2019) (p)ppGpp: the magic governor of bacterial growth economy. Curr Genet 65: 1121–1125. [DOI] [PubMed] [Google Scholar]


