Abstract
Reversible protein phosphorylation is a widespread post-translational modification fundamental for signaling across all domains of life. Tyrosine (Tyr) phosphorylation has recently emerged as being important for plant receptor kinase (RK)-mediated signaling, particularly during plant immunity. How Tyr phosphorylation regulates RK function is however largely unknown. Notably, the expansion of protein Tyr phosphatase and SH2 domain-containing protein families, which are the core of regulatory phospho-Tyr (pTyr) networks in choanozoans, did not occur in plants. Here, we summarize the current understanding of plant RK Tyr phosphorylation focusing on the critical role of a pTyr site (‘VIa-Tyr’) conserved in several plant RKs. Furthermore, we discuss the possibility of metazoan-like pTyr signaling modules in plants based on atypical components with convergent biochemical functions.
Keywords: phosphorylation/dephosphorylation, receptor tyrosine kinases, receptors
Tyrosine phosphorylation is important to all organisms
All organisms must perceive external cues for adaptation of their physiology to changing environmental conditions. Either cell-surface or cytosolic receptors perceive external cues. Mechanisms for external stimulus perception and signal transduction (transmembrane signaling) are diverse, and the molecular components involved display distinct patterns of evolutionary radiation across kingdoms. Animals employ receptor tyrosine kinases (RTKs), Toll-like receptors (TLRs), ion channel-linked receptors, and G-protein coupled receptors for transmembrane signaling whereas plants rely mainly on receptor kinases (RKs) and receptor proteins (RPs) [1, 2]. A key molecular event in RK and RTK-mediated transmembrane signaling is the conversion of external ligand binding into the activation of cytosolic protein kinase domains that trigger signal transduction via a phospho-relay mechanism. Phosphorylation is a critical post-translational modification (PTM) that controls diverse cellular processes such as enzyme activities and protein–protein interactions, protein localization and trafficking, and protein degradation [3–5]. Protein phosphorylation at either serine (Ser, S), threonine (Thr, T) or tyrosine (Tyr, Y) residues is catalyzed by members of the eukaryotic protein kinase (EPK) family, to which the cytoplasmic domain of RKs and RTKs belong. The EPK family is subdivided into two classes of catalytic domains having specificity for Ser/Thr or Tyr as the substrate phospho-acceptor [6, 7]. Owing to the action of dual-specificity kinases (i.e. Ser/Thr kinases that can also phosphorylate Tyr), Tyr phosphorylation is prevalent in the proteomes of organisms that lack canonical Tyr kinase (TK) domains [8–11]. Surprisingly, the relative abundance of phosphotyrosine (pTyr) is similar in species with and without TKs [8, 10, 12–15]. In the last years, Tyr phosphorylation on plant RKs was shown to be critical, but its mechanistic importance for signal transduction remains unclear [16–19].
In the present review, we compare, and contrast RK-based signaling mechanisms in plants and metazoans, with focus on the role of Tyr phosphorylation in plants and familiarize the reader with plant RK-mediated growth and immune signaling. The recent discovery of a conserved pTyr site in plant RKs and of non-canonical protein tyrosine phosphatases in plants represent exciting new developments in the field of plant signal transduction [16–21]. These observations highlight a potential conserved, pTyr-based regulatory mechanism across the Arabidopsis thaliana (hereafter, Arabidopsis) RK family, and suggest that plants may have independently evolved a choanozoan-like pTyr-based signaling system wherein evolutionarily distinct proteins have convergent biochemical properties. Nevertheless, it is still unclear whether plant pTyr-based signaling functions in a manner analogous to that in choanozoans, particularly concerning the existence of pTyr-binding proteins in plants. Future investigation of plant Tyr phosphorylation requires a multi-faceted quantitative approach to inform on this poorly understood PTM in plants.
Plant RK signaling shares analogy to metazoan TLR and RTK signaling
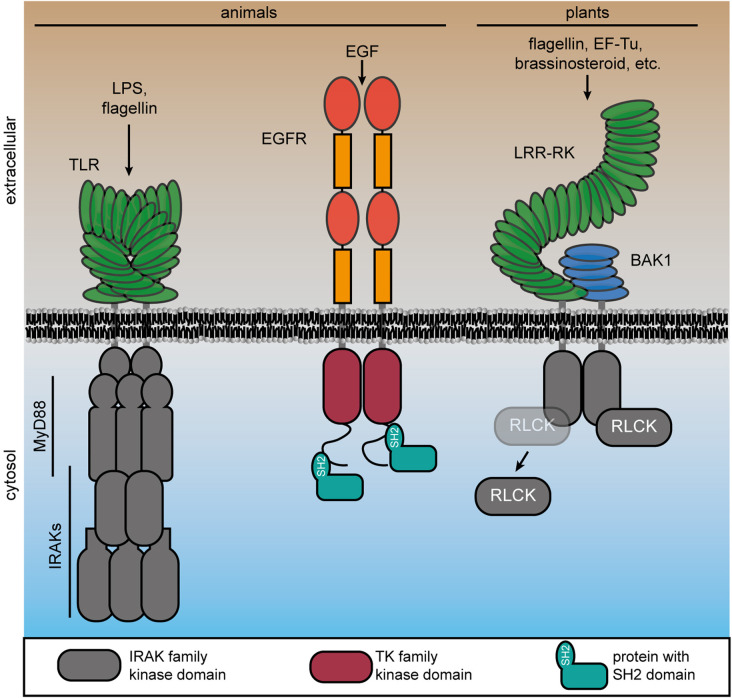
RTKs represent the major class of transmembrane protein kinases consisting of an extracellular ligand-binding domain, a single pass transmembrane helix, and an intracellular TK domain. They underwent evolutionary diversification in the choanozoans, and the rapid expansion of RTK families correlates with the evolution of multicellularity [11, 22, 23]. The emergence of TK domains, and RTKs respectively, is the beginning of the evolution of a sophisticated pTyr-based signaling mechanism in choanozoans, requiring three functional modules: (1) the tyrosine phosphorylating TK or RTK respectively, (2) an effector protein, containing a pTyr-binding domain, e.g. Src homology 2 (SH2) or pTyr-binding (PTB) domain, and (3) a phosphotyrosine phosphatase (PTP), that dephosphorylates pTyr. These three functional modules can be conceptualized as ‘writer’, ‘reader’ and ‘eraser’ modules, respectively [11, 22].
Following ligand perception, RTKs dimerize and the TK kinase domains auto(trans)phosphorylate on Tyr residues in their C-terminal cytoplasmic domain [24]. Specific SH2 and PTB domain-containing proteins bind to pTyr in the cytoplasmic domain of the RTK, depending on pTyr-site adjacent motifs [25] (Figure 1). Binding of the readers to the writers establishes the functional signaling complex and propagates the perceived signal to divergent downstream pathways via MAPK and cytosolic TK cascades, and second messenger systems [25]. Recruitment of specific readers to different pTyr sites is a critical step in establishing quantitative outputs of RTK complexes, and mis-recruitment of signaling components to pTyr sites due to a mutation in the specificity-determining adjacent motif underlies some types of cancer [26]. Eventually, PTPs act generally as negative regulators of RTK signaling activity by dephosphorylating pTyr. Thereby, the binding of the pTyr-binding proteins to the TK domain, and thus the constitution of the functional signaling complex, is inhibited [22].
Figure 1. Transmembrane signaling mechanisms in choanozoans and plants involving protein kinases.
Upon recognition of pathogen-associated molecular patterns, animal TLRs dimerize and recruit cytosolic adaptor proteins (e.g. MyD88) and subsequently IRAKs. In RTKs, the kinase domain is fused to the extracellular domain by a single pass transmembrane helix. RTKs form a complex upon ligand perception, here shown for epidermal growth factor (EGF) receptor (EGFR). Subsequent activation of the intracellular TK domain, Tyr-autophosphorylation in their C-terminal tails leads to recruitment of cytoplasmic signaling components with pTyr-binding SH2 domain. Plant RKs also have an extracellular receptor domain that is directly coupled by a single pass transmembrane helix to the intracellular kinase domain that is phylogenetically related to IRAKs. Ligand perception induces complex formation of the RK (here a LRR-RK) and its co-receptor (here BAK1, as an example). In response to receptor complex formation, receptor-like cytoplasmic kinases (RLCKs), which belong also to the IRAK family, dynamically associate with the complex.
Plant RKs share similar domain architectures to RTKs and consist of diverse extracellular ligand-binding domains, a single-pass transmembrane helix, and a cytosolic protein kinase or pseudokinase domain (Figure 1) [24, 27, 28]. RPs consist of an extracellular ligand-binding domain anchored in the plasma membrane but lack a cytosolic domain of any kind. Instead, RPs associate with adaptor RKs to form a bipartite functional RK unit [29–31]. Rather than possessing a cytosolic TK domain, plant RK protein kinase domains are phylogenetically related to the metazoan Pelle Ser/Thr-kinase family that includes INTERLEUKIN-1 (IL-1) RECEPTOR ASSOCIATED KINASES (IRAKs) [32–35]. In animals, IRAKs associate with the intracellular domains of TLR immune receptors through interaction with adaptor proteins such as MyD88 or TIR-domain-containing adapter-inducing interferon-β (TRIF), and function to activate signaling downstream of TLR-mediated pathogen-associated molecular pattern (PAMP) recognition (Figure 1) [36, 37]. Plant RKs — at least those with leucine-rich repeat (LRR)-type ectodomains — can thus be conceptualized as an ‘all-in-one’ Myddosome complex wherein a TLR ligand perceiving module is directly tethered to an intracellular IRAK signaling module [29]. Despite the distinct architecture of RTKs, TLRs, and plant RKs, the recruitment of additional proteins during receptor complex activation is a common theme in transmembrane signaling (Figure 1).
Interestingly, even though plants lack canonical TKs, Tyr phosphorylation is widespread in many plant species [12, 13, 38]. In the last two decades, specific pTyr residues were identified by shot-gun phospho-proteomics or by targeted analysis of individual proteins, including several plant RKs [12, 16–19, 39–43]. pTyr sites are distributed throughout plant RK cytosolic domains, i.e. juxtamembrane domain, kinase subdomains and the C-terminal tail (Table 1), highlighting the potential for multiple regulatory functions of pTyr in RK-mediated signaling. Analysis of RK cytosolic domains in vitro indicates that RK Tyr phosphorylation is mostly derived from autophosphorylation activities [19, 39]. Furthermore, structural analysis of the brassinosteroid (BR) receptor BR INSENSITIVE 1 (BRI1) kinase domain revealed structural features in the activation loop reminiscent of Ser/Thr-kinase as well as TKs. In particular, the BRI1 activation loop conformation is similar to Ser/Thr-kinases, whereas the phosphate-binding pocket for the BRI1 activation loop-residue pS1044 rather resembles TKs, highlighting the dual-specificity nature of plant RKs [44]. Thus, phosphorylation of Tyr residues in plant RKs is reminiscent of choanozoan RTK signaling, but the molecular details of how plant pTyr-based signaling is executed and regulated remain undiscovered.
Table 1. Reported phospho-tyrosine residues on plant receptor kinases.
| Protein | Residue | Kinase subdomain | Auto/trans-phosphorylation | LC–MS/MS | Sequence and phosphorylation specific antibody | Site-directed mutagenesis | Impaired kinase activity (Autorad) | Reference |
|---|---|---|---|---|---|---|---|---|
| BRI1 | Y831 | JM | Auto | - | Yes | Yes3 | Yes4 | Oh et al. [39] |
| Y956 | V | Auto | - | Yes | Yes3 | Yes | Oh et al. [39] | |
| Y1072 | IX | Auto | - | Yes3 | Yes3 | Yes | Oh et al. [39] | |
| BAK1 | Y4432 | VIII | - | Yes | - | - | - | Mergner et al. [9] |
| Y403 | Tyr-VIa | Auto | Yes3 | Yes3 | - | No | Perraki et al. [19] | |
| EFR | Y836 | Tyr-VIa | -5 | Yes1 | Yes | Yes3 | No | Macho et al. [16] |
| LORE1 | Y600 | Tyr-VIa | Auto5 | Yes3 | Yes | Yes3 | Yes | Luo et al. [17] |
| CERK1 | Y428 | Tyr-VIa | Auto5 | Yes3 | Yes | Yes3 | No | Liu et al. [18], Suzuki et al. [42] |
| Y557 | XI | Auto5 | Yes3 | Yes | Yes3 | - | ||
| PSKR1 | Y888 | VIII | - | Yes | - | No | Yes4 | Muleya et al. [45] |
| HAESA | Y724 | III | Auto | - | - | Yes3 | No | Taylor et al. [43] |
| AT1G51805 | Y544 | JM | - | Yes | - | - | - | Mergner et al. [9] |
| AT5G62710 | Y266 | JM | - | Yes | - | - | - | Mergner et al. [9] |
| AT5G56890 | Y1047 | C-tail | - | Yes | - | - | - | Mergner et al. [9] |
| AT2G30940 | Y181 | I | - | Yes | - | - | - | Sugiyama et al. [46] |
| AT1G60630 | Y343 | - | Yes6 | - | - | - | Mithoe et al. [47] | |
| AT1G66980 | Y991 | IX | - | Yes6 | - | - | - | Mithoe et al. [47] |
Using selective reaction monitoring;
BAK1, SERK1, SERK2: ambiguous origin;
In vitro identification onl;y
Peptide as kinase substrate. BRI1: SP11, PSKR: Sox;
Ligand-dependent changes in phosphorylation;
Enrichment with pY antibody;
Abbreviations: JM: Juxtamembrane, Ecto: Ectodomain, BRI1: BRASSINOSTEROID INSENSITIVE 1, BAK1: BRI1-ASSOCIATED KINASE, EFR: EF-TU RECEPTOR, LORE: LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION, CERK1: CHITIN ELICITOR RECEPTOR KINASE 1, PSKR1: PHYTOSULFOKIN RECEPTOR 1.
Plant RK Tyr phosphorylation regulates development and immunity
In recent years, several studies reported the importance of Tyr phosphorylation on RKs for plant development and immunity [16–19, 39, 40, 42]. Using a combination of mass spectrometry and sequence-specific pTyr antibodies, one of the first plant RKs for which Tyr phosphorylation was reported is BRI1 [39].
Importance of Tyr phosphorylation for BR signaling
Upon perception of BR, BRI1 forms a complex with its cognate co-receptors from the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) family, e.g. BRI1 ASSOCIATED KINASE 1 (BAK1)/SERK3 and BAK1-LIKE 1 (BKK1)/SERK4 [48, 49]. Following complex formation and activation of the intracellular kinase domains, receptor-like cytoplasmic kinases (RLCKs) CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1) and BR SIGNALING KINASE 1 (BSK1) are activated through transphosphorylation. CDG1 and BSK1 control the activity of the phosphatase BRI1 SUPPRESSOR 1 (BSU1), which negatively regulates the glycogen synthase kinase 3/Shaggy-related kinase BR INSENSITIVE 2 (BIN2) [20, 50, 51]. In turn, BIN2 controls the localization and stability of the transcription factors BRASSINAZOLE RESISTANT 1 (BZR1) and BZR2 through phosphorylation, thereby impairing the expression of BR-responsive genes [52]. Deactivation of BIN2 by BSU1 leads to stabilization and re-localization of BZR1/BZR2 to the nucleus, and eventually to induction of BR-responsive genes [53, 54]. (Figure 1A)
Impairing BR signaling through compromising BR binding by the BRI1 ectodomain, decreasing kinase activity or protein stability or impairing correct localization of BRI1 results in stunted growth and round leaf shape of the plant, e.g. in the bri1-5 mutant [55]. Mutation of identified Tyr phosphorylation sites to phospho-ablative phenylalanine impaired BRI1 kinase activity in most cases, demonstrating their importance for catalytic activity [39]. Phosphorylation of BRI1-Y831 in the juxtamembrane region is not essential for kinase activity, but it is required for proper BR signaling in vivo [39]. Complementation of the weak bri1-5 mutant with the phospho-ablative mutant BRI1Y831F results in rescue of the dwarfism phenotype but only partially restores the aberrant leaf shape phenotype and induces early flowering, whereas phospho-mimic substitutions behave like wild-type BRI1. How BRI1Y831F affects individual outcomes of BR signaling is uncertain. Whether the mutation BRI1Y831F impairs localization or interaction with other proteins has not been investigated. Mis-localization or aberrant interaction of BRI1Y831F seem unlikely given that BRI1Y831F mutant plants do not exhibit the strong dwarf phenotype typical of BR insensitive mutants.
Additional Tyr residues of BRI1 — namely Y956 and Y1072 — are also phosphorylated and essential for BR signaling as Y-to-F mutations at these sites compromise kinase activity [39]. Nevertheless, whether the phosphorylation of these residues or the residue itself is essential for kinase activity is unknown.
Tyr phosphorylation in plant immunity
Dual-specificity kinase activity was reported for additional RKs, including the BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), which acts as co-receptor for multiple LRR-RKs, inclusive of BRI1 and several pattern recognition receptors (PRRs) involved in plant immunity [39]. The significance of Tyr kinase activity in plant immunity was ambiguous until recently, when Tyr phosphorylation was shown critical for signal transduction [16–19].
In plant immunity, several RKs with divergent ectodomains function as PRRs, recognizing different PAMPs and damage-associated molecular patterns (DAMPs). Bacterial flagellin and elongation factor thermo unstable (EF-Tu) are perceived by the LRR-RKs FLAGELLIN SENSING 2 (FLS2) and EF-TU RECEPTOR (EFR), respectively, both of which recruit BAK1 as a co-receptor [56–58] (Figure 1). A subset of RKs with lysin motif (LysM) ectodomains recognize the microbial cell wall components chitin and peptidoglycan. In Arabidopsis, LysM-CONTAINING RECEPTOR-LIKE KINASE 5 (LYK5) and LYK4, or LysM DOMAIN-CONTAINING GPI-ANCHORED PROTEIN 1 (LYM1) form a complex with CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) for the perception of chitin or peptidoglycan, respectively [59–62]. A lectin-type ectodomain RK, LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION (LORE), perceives free fatty acids [63]. Ligand perception by PRRs leads to activation of the intracellular kinase domains that propagate signaling to RLCKs, resulting in activation of diverse molecular events, including Ca2+-influx, an apoplastic oxidative burst, MAPK activation and transcriptional reprogramming [64–70], leading to pattern-triggered immunity (PTI; Figure 2). EFR, FLS2, and BAK1 activate several PTI outputs through the RLCKs BOTRYTIS INDUCED KINASE 1 (BIK1) and PBS-LIKE KINASE 1 (PBL1) [27, 64], whereas chitin signaling occurs through both BIK1 and PBL27 [64, 71], and free fatty acid-induced signaling occurs through PBL34-36 [17].
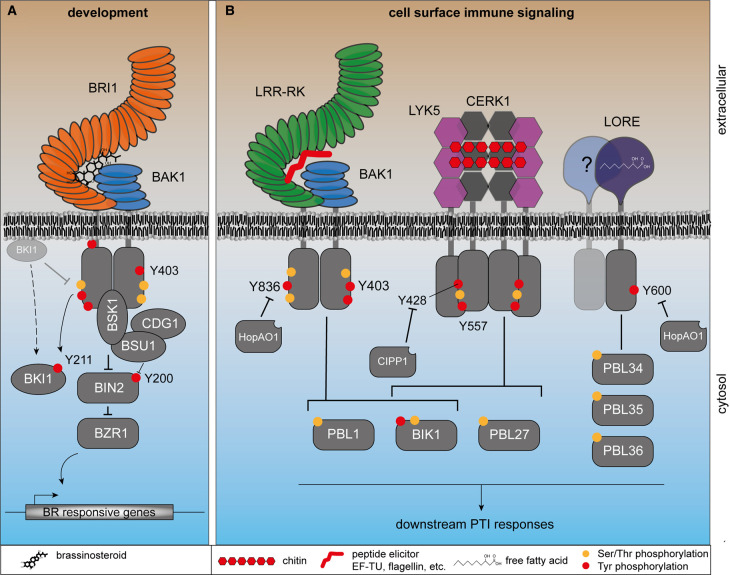
Figure 2. Phosphorylation of plant RKs involved in immunity, growth and development.
(A) Brassinosteroid signaling is activated by tyrosine autophosphorylation and transphosphorylation on BKI1, which triggers its dissociation from BRI1 (dashed arrow) to allow full activation of BRI1 kinase activity. This activates BSU1, dephosphorylating BIN2 pY200 in the activation loop and eventually promotes nuclear localization of the transcription factor BZR1 for transcriptional reprogramming. (B) In cell surface immunity, receptor kinases recognize their respective ligand, which leads to dimerization or oligomerization, activation of the cytosolic kinase domains, and phosphorylation on a key conserved Tyr residue (BAK1-Y403, EFR-Y836, CERK1- Y428 and LORE-Y600). RKs relay the signal to receptor-like cytoplasmic kinases that activate a battery of cellular responses including MAPK phosphorylation, Ca2+ influx and reactive oxygen species production. The signaling cascades are regulated by phosphorylation on Ser, Thr (orange dots) and Tyr (red dots). Receptor activation is regulated by plant (CIPP1) or microbial (HopAO1) phosphatases.
For several PRRs, namely CERK1, LORE, EFR, and BAK1, Tyr phosphorylation on the cytoplasmic domain is essential for signaling. Interestingly, Tyr phosphorylation of these PRR-RKs occurs on a conserved residue (BAK1-Y403, EFR-Y836, CERK1-Y428, LORE-Y600) in subdomain VIa, which we refer to as VIa-Tyr (Figure 3A), at the C-terminal end of the αE-helix in the C-lobe of the kinase domain [16–19] (Figure 3B). Y-to-F substitution of the VIa-Tyr compromises receptor function and consequently immunity. Furthermore, pathogenic bacteria use a type III-secretion system to inject effector proteins into plant cells to interfere with PTI signaling and their recognition by the plant. The bacterial type III-secreted effector protein HopAO1 dephosphorylates EFR pY836 and LORE pY600 to suppress immunity substantiating the importance of this phospho-residue for immune signaling [16, 17].
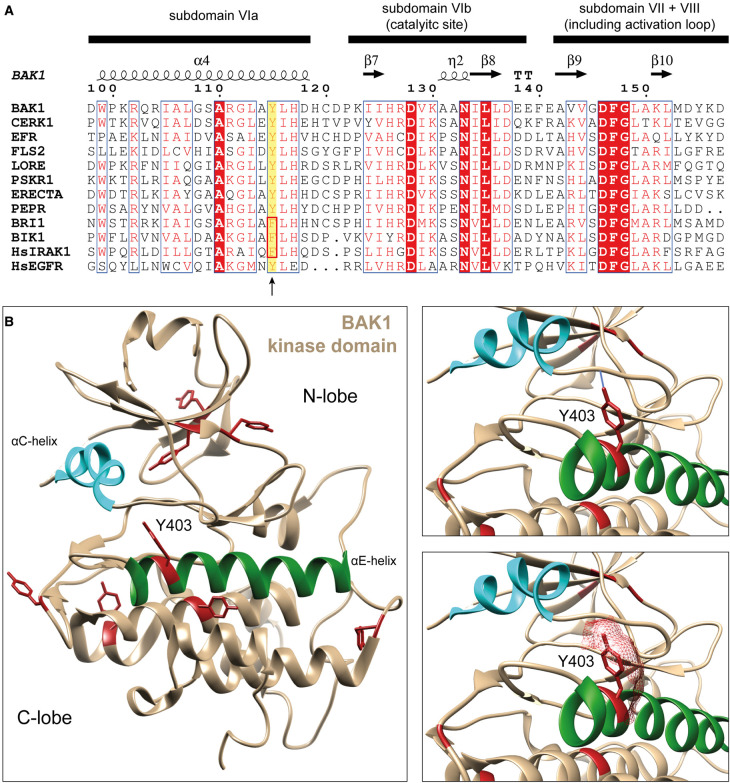
Figure 3. BAK1-Y403 conservation across plant RKs and its structural presentation.
(A) Multiple sequence alignment of plant RK kinase domains, showing the conservation of BAK1-Y403 across plant RKs (highlighted with yellow background). This residue is not conserved in the BAK1Y403F insensitive brassinosteroid signaling pathway. Alignment was visualized using ESPript3.0 [72]. (B) Crystal structure of BAK1 kinase domain (PDB: 3UIM) [73]. All tyrosine residues are shown in red. The αC-helix and αE-helix are highlighted in cyan and dark green, respectively. On the right, the upper panel shows H-bond analysis of the BAK1 Y403 hydroxyl group, predicting an H-bond with C-terminal base of the αC-helix. In the lower panel, the surface contribution of BAK1-Y403 is depicted, showing its solvent exposure. Structures were visualized using Chimera [74].
Apart from the phosphorylation site conservation, CERK1-Y428 phospho-kinetics differ compared with EFR-Y836 and LORE-Y600. EFR-Y836 and LORE-Y600 become phosphorylated only following ligand perception. Conversely, CERK1-Y428 is phosphorylated in the absence of chitin [18]. Chitin triggers CERK1-pY428 dephosphorylation by CERK1-INTERACTING PROTEIN PHOSPHATASE 1 (CIPP1) for deactivation of signaling. CERK1-Y428 is re-phosphorylated afterward to return CERK1 to the stand-by state [18]. The dynamics of BAK1-Y403 phosphorylation are unknown, although it is anticipated to occur following ligand perception like Y836 on EFR. For CERK1 and LORE, VIa-Tyr phosphorylation may directly regulate kinase activity, explaining compromised immunity in plants expressing Y-to-F mutants of these receptors [17, 18]. By comparison, Y-to-F substitution in EFR and BAK1 does not or only slightly reduces kinase activity, and thus the molecular basis of BAK1-Y403 and EFR-Y836 phosphorylation in regulating the receptor complex is unclear [16, 19].
Besides phosphorylation on the conserved VIa-Tyr, phosphorylation of CERK1-Y557 is essential for a subset of chitin-triggered immune responses, i.e. the oxidative burst but not MAPK activation. CERK1Y557F has wild-type kinase activity, consistent with partially functional immune signaling. In contrast with CERK1-Y428, its phosphorylation is ligand dependent, rather resembling EFR-Y836 and LORE-Y600 phosphorylation. Phosphorylation of CERK1-Y557 controls the dissociation of BIK1 from CERK1 following chitin perception, but does not impair PBL27 recruitment [18]. An impaired dissociation of BIK1 from CERK1Y557F suggests that CERK1-pY557 induces either conformational changes of the kinase domain or electrostatic repulsion, promoting BIK1 release.
Functional dichotomy of BAK1 Y403
Notably, all tested LRR-RK-mediated signaling pathways that use BAK1 as a co-receptor are sensitive to BAK1Y403Fand bak1-5 if the ligand-binding RK also possesses the VIa-Tyr, which extends to LRR-RKs involved in plant development, e.g. ERECTA, HAESA, HAESA-LIKE 2, and PHYTOSULFOKINE RECEPTORs [19, 75, 76]. BAK1-5 is a hypoactive BAK1 variant harboring a C-to-Y mutation at position 408 within the protein kinase domain [19, 77]. Molecular modeling suggests that the Y408 side chain of BAK1-5 might interfere with BAK1-Y403 phosphorylation through steric hinderance, which is supported by reduced BAK1-Y403 autophosphorylation in the BAK1-5 variant [19]. In contrast, BR signaling is insensitive to bak1-5 and the BAK1Y403F variant. The brassinosteroid receptor BRI1 harbors a Phe at the VIa-Tyr position (Figure 2), suggesting a VIa-Tyr-dependent functional dichotomy of the common co-receptor BAK1 [19]. It is yet unknown how BAK1Y403F and BAK1-5 contribute to the functional dichotomy of BAK1. One possibility is that full BAK1 kinase activity is required for signaling by VIa-Tyr-type LRR-RKs but is dispensable for BRI1 signaling. In fact, BAK1-5 and BAK1Y403F exhibit slightly reduced kinase activity, and phosphorylation of BAK1 Y403 might contribute to full kinase activation. Despite the fact that BAK1-5's reduced Y403 phosphorylation might be a consequence of reduced kinase activity, the phospho-ablative BAK1Y403F variant also reduces kinase activity slightly [19], indeed hinting at a contribution of BAK1-pY403 to full activation of BAK1 kinase activity. Even though full BAK1 kinase activity is dispensable, at least weak BAK1 kinase activity is required for BR signaling as kinase-inactive BAK1 variants cannot complement the seedling lethality of bak1-4/bkk1 and have a dominant negative effect on the hypoactive bri1-5 mutant [78]. This requirement of BAK1 kinase activity for BR signaling further substantiates that BAK1Y403F is an active kinase in vivo. It is mechanistically unclear though, how BAK1 pY403 could contribute to its full kinase activation. Interestingly, the active-state conformation of the BAK1 kinase domain may be regulated by folding transitions of the αC-helix [79], and it would be interesting to determine whether Tyr phosphorylation contributes to this mechanism since the VIa-Tyr (BAK1-Y403) side-chain hydrogen bonds with the peptide backbone of the αC-β4-loop (Figure 3B). Detailed biochemical and structural analysis of the Tyr-phosphorylated and -unphosphorylated BAK1 kinase domain may thus shed light on a common VIa-Tyr-based mechanism for regulating plant RK activation.
Tyr phosphorylation beyond the cell-surface receptor complex
Downstream of BRI1, components of the BR signaling pathway are also Tyr phosphorylated (Figure 2A). BRI1 KINASE INHIBITOR 1 (BKI1) — a membrane-associated, BRI1-interacting protein that inhibits BRI1 activation — is phosphorylated in a BR-dependent manner on Y211 within its membrane-targeting motif. Tyr-phosphorylated BKI1 dissociates from the membrane, releasing BRI1 inhibition to permit BR pathway activation [40]. BIN2 phosphorylates BR responsive transcription factors, promoting their retention in the cytosol. Dephosphorylation of pY200 in the BIN2 activation loop by the phosphatase BSU1 inactivates BIN2 leading to nuclear accumulation of BR responsive transcription factors, and BR-dependent transcriptional responses [20].
Similar to BR signaling, Tyr phosphorylation also extends to downstream components of immune signaling. BAK1 transphosphorylates BIK1 on Y243 and Y250 in vitro, and BIK1 also autophosphorylates on Y250 [80, 81]. Even though BIK1-Y243 and -Y250 are located in the activation loop, their mutations only slightly impair kinase activity but compromises BIK1 function in immunity [80].
MAPKs act downstream of RKs and are involved in signaling in response to diverse biotic and abiotic stimuli, e.g. pathogen infection, UV radiation, heat stress, etc. [82]. A certain stimulus is activating different MAPK cascades in which a MAPK KINASE KINASE (MAPKKK) is phosphorylating a MAPK KINASE (MAPKK) on either Ser or Thr in the MAPKK activation loop. Subsequently, the MAPKK, a dual-specificity kinase, dual-phosphorylates MAPKs on Thr and Tyr of the conserved T[ED]Y motif in the activation loop. MAPK activity results in the regulation of transcription factor activity, metabolic enzymes and other kinases, and eventually contributes to plant immunity, stress tolerance and development [83]. Deactivation of MAPKs occurs through the action of PTPs [84], dual specificity phosphatases (DSP) [84, 85] or protein Ser/Thr phosphatases (PSTP) [86], which themselves are often regulated through their substrate MAPK [87]. Knock-out alleles of PTPs and DSPs regulating MAPK activity result in defects of growth, development and responses to biotic and abiotic stress [87]. For example, MAPK PHOSPHATASE 1 (MKP1) is involved in RK-mediated PTI signaling. The mkp1 knock-out mutant exhibits elevated MAPK activation and ROS production in response to PAMP treatment and increases resistance against the bacterial phytopathogen Pseudomonas syringae [88–91]. Thus, Tyr phosphorylation plays also a significant regulatory role downstream of RKs during signaling.
A pTyr signaling set for plants?
Before TK emergence in choanozoans, Tyr phosphorylation occurred via the unusual activity of dual-specificity Ser/Thr kinases towards Tyr. This sporadic Tyr phosphorylation likely drove the evolution of PTPs to alleviate detrimental allosteric effects caused by pTyr [22, 92]. After PTP expansion, diversification of proteins containing pTyr-binding SH2 domains facilitated the pTyr-dependent assembly of protein complexes — a key step in the evolution of pTyr signaling. In many early eukaryotes, such as the slime mold Dictyostelium discoideum, a limited number of PTPs and SH2-containing proteins, and dual-specificity protein kinases comprise the pTyr signaling tool kit [22]. With the emergence, expansion and diversification of TKs and RTKs in choanozoans, the reader and eraser modules rapidly expanded and diversified as well. The modules combined with each other and other functional domains, building complex multidomain proteins driving a sophisticated signaling mechanism [11, 22]. Perhaps surprisingly, and despite the presence of dual-specificity protein kinases, neither the PTP nor SH2/PTB families expanded in plant genomes raising the question of how Tyr phosphorylation mediates signaling and how it is regulated in plants. One intriguing possibility is that evolutionarily distinct proteins with convergent biochemical properties fulfill the roles of PTP and pTyr-binding proteins.
Regulation of pTyr by protein phosphatases in Arabidopsis
In the well-established choanozoan pTyr-based signaling mechanism, PTPs act as eraser, negatively regulating signaling activity of the RTKs and downstream TKs in general [22]. PTPs regulating pTyr-based signaling are the founding member of the diverse PTP superfamily, which comprises four different families (Class I–III Cys-based and Asp-based PTPs). Class I–III Cys-based PTPs have the catalytic signature motif (H)CX5R in common. The canonical PTPs involved in the choanozoan pTyr-based signaling mechanism form a subgroup in Class I Cys-based PTPs neighboring the subgroup of DSPs, which dephosphorylate various proteinaceous and non-proteinaceous substrates [93]. Also, the single member of Class II Cys-based PTPs low molecular weight PTP (LMWPTP), is involved in the regulation of choanozoan pTyr-based signaling. Additionally, some DSP members can also dephosphorylate pTyr, even though it is not their main substrate [93].
Arabidopsis harbors several Cys-based PTP genes in its genome, majorly belonging to Class I Cys-based PTPs [94] (Figure 4). Homologs of canonical PTPs and LMWPTP are represented in Arabidopsis by only two proteins, one canonical PTP (AtPTP1) and one chloroplastic LMWPTP [94, 95] (Figure 4). AtPTP1 has pTyr phosphatase activity [96], but this appears to be physiologically non-essential because the loss of AtPTP1 function is not detrimental to plant growth or responses to environmental stimuli, suggesting the presence of other non-canonical PTPs [84], or a role in yet untested specific conditions.
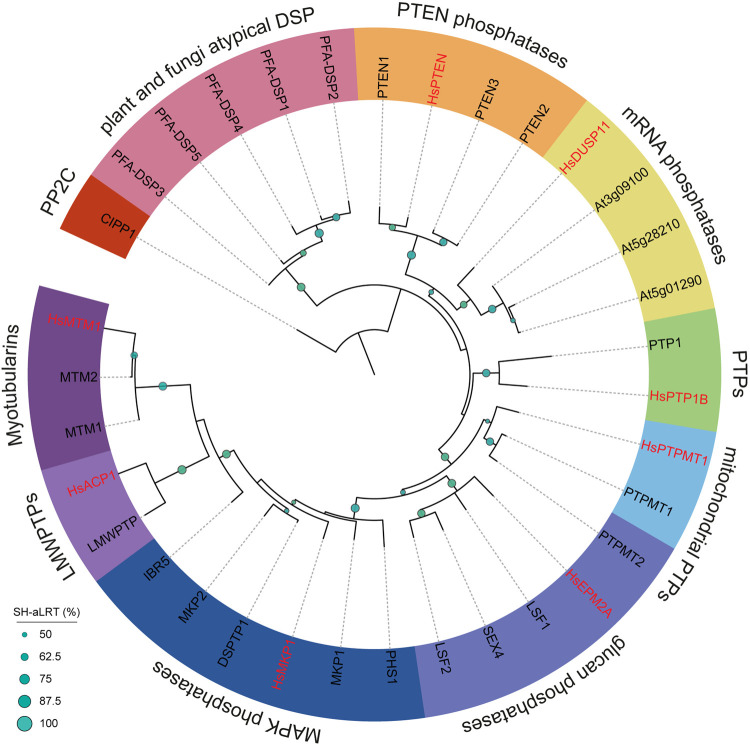
Figure 4. Phylogeny of Cys-based phosphatases in Arabidopsis thaliana.
Arabidopsis possesses homologs of H. sapiens Class I and II Cys-based phosphatases but only one for each pY-specific PTP, PTP1B and LMWPTP. The other phosphatases are dual-specificity phosphatases dephosphorylating Ser, Thr and Tyr, as well as mRNA, lipids and glucans with different efficiencies. Plant and fungal atypical dual specificity phosphatases (PFA-DSPs) are related to H. sapiens Cys-based phosphatases but have no direct homolog. Multiple sequence alignment of PTPs (as annotated on UniProt) were constructed using MUSCLE. Phylogeny was inferred using IQtree [106] and the tree visualized by iTOL [107]. Branch support: Shimodaira–Hasegawa approximate log-likelihood ratio test (SH-aLRT) in %. CIPP1 (AT1G34750, outgroup), HsDUSP11 (HGNC:3066), PTEN1 (AT5G39400), PTEN2 (AT3G19420), PTEN3 (AT3G50110), PFA-DSP1 (AT1G05000), PFA-DSP2 (AT2G32960), PFA-DSP3 (AT3G02800), PFA-DSP4 (AT4G03960), PFA-DSP5 (AT5G16480), PTP1 (AT1G71860), MTM1 (AT3G10550), MTM2 (AT4G27940), HLP (AT3G62010), PHS1 (AT5G23720), MKP1 (AT3G55270), IBR5 (AT2G04450), DSPTP1 (AT3G23610), MKP2 (AT3G06110), LSF1 (AT3G01510), LSF2 (AT3G10940), SEX4 (AT3G52180), LMWPTP (AT3G44620), PTPMT1 (AT2G35680), PTPMT2 (AT5G56610), HsDUSP11 (HGNC:3066), HsPTEN (HGNC:9588), HsPTPMT1 (HGNC:26965), HsPTP1B (HGNC:9642), HsMTM1 (HGNC:7448), HsMKP1 (HGNC:3064), HsACP1 (HGNC:122), PsHopAO1 (UniProt: H1ZWZ1).
The DSPs of Class I Cys-based PTPs are further expanded than Class I canonical PTPs in Arabidopsis [94] (Figure 4). DSPs dephosphorylate proteinaceous Ser/Thr/Tyr and non-proteinaceous substrates, e.g. phosphoinositol phosphates (PIPs), mRNA, and glucans. Indeed, the Arabidopsis DUAL SPECIFICTY PROTEIN PHOSPHATASE 1 (AtDSPTP1), and members of PLANT AND FUNGI ATYPICAL DUAL SPECIFICITY PHOSPHATASES (PFA-DSP) subfamily show pTyr dephosphorylation activity [85, 97, 98]. AtPFA-DSPs also dephosphorylate phosphatidylinositols (PIPs), which is the primary substrate of PHOSPHATASE AND TENSIN HOMOLOGS (PTENs) in animals [97–100]. Their activity toward pTyr may complement the function of AtPTP1 to support a bona fide pTyr-based signaling mechanism in Arabidopsis.
Interestingly, Ser/Thr phosphatases of the protein phosphatase type-2C (PP2C) and phosphoprotein phosphatase (PPP) family, such as CIPP1, BSU1, RHIZOBIALES-LIKE PHOSPHATASE HOMOLOG 2 (RLPH2), and Shewanella-like protein phosphatase 1 (SLP1), respectively, have PTP activity in vitro [18, 20, 21]. CIPP1 acts on CERK1-pY428, and is thus a good candidate to dephosphorylate other RK VIa-Tyr sites such as EFR-pY836, BAK1-pY403 and LORE-pY600. Nevertheless, thorough enzymatic characterization of BSU1 and CIPP1 PTP activity is lacking to fully substantiate their function as bona fide PTPs. Biochemical and structural characterization of RLPH2 supports its role as a pTyr-specific PPP-type phosphatase [21, 101], although the physiological substrates of RLPH2 are unknown.
Do plants have pTyr-binding proteins?
The regulation of pTyr through PTPs may have promoted the emergence of pTyr-binding domains, i.e. SH2 and PTB domains [22]. The choanozoan SH2 domain characteristically folds into an αβββ(β)α architecture. The β-strands form an antiparallel β-sheet, which separates the α-helices on either site. In the canonical SH2-binding mode, pTyr is coordinated by residues in the conserved Gx[YF]xxR motif of βB and bound between the αA-helix and the β-sheets [25, 102]. On the opposite side of the β-sheet, the SH2 domains harbors a specificity determining region that engages with amino acids carboxy-terminal of the substrates' pTyr [25, 103]. Specificity can be further increased by in-tandem arranged SH2 domains that bind two spaced pTyrs on the substrate [104].
The PTB domain consists of two antiparallel β-sheets forming a β-sandwich and has a C-terminal α-helix [105]. They bind pTyr peptides - similar to SH2 domains — based on motif selection. In contrast with SH2 domains, the motifs lie N-terminal of the pTyr and consist of a core NPX[YpY] motif [25, 105]. This motif is coordinated between the C-terminal α-helix and the β5-sheet and establishes hydrogen bonds to the β5-sheet [25, 105]. In addition, PTB domains also bind to phospholipids through an alternative binding surface and other, non-pTyr peptides [25, 108, 109].
In plants, a few protein domains with predicted SH2 homology share the characteristic secondary αβββα-structure [14] and no homologous PTB domains were found [25]. These domains are found in SUPPRESSOR OF TY 6 LIKE (SPT6L), for which the βD sheet is poorly conserved, and SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION LIKE (STATL) proteins [110]. Besides the proto-SH2 domains of SPT6L and STATL, plants do not possess any other canonical SH2-like domain [110]. SPT6L proto-SH2 domains are likely ancestral and may bind pTyr, though this capability is under debate [111]. In any case, given the conservation of SPT6 in organisms lacking RKs and their function in RNA polymerase II-dependent transcription [112, 113], it is unlikely that SPT6L proteins are involved in transmembrane signaling in plants.
Metazoan STAT proteins function in the Janus kinase (JAK)-STAT pathway that is, for example, activated downstream of cytokine perception [114]. Stimulated cytokine receptors activate JAK leading to receptor phosphorylation, STAT recruitment, and finally STAT Tyr-phosphorylation [114]. Tyr-phosphorylated STATs then dimerize and relocalize to the nucleus where they modulate transcription [114]. The DNA-binding domain of plant STAT-like proteins is poorly conserved, as are the Tyr phosphorylation sites, which are important for STAT dimerization, suggesting that the plant isoforms lack pTyr-dependent function. Although AtSTATLs can pull down Tyr-phosphorylated proteins in vitro [110], it is not clear whether this depends on pTyr of the prey protein or on the SH2 domain of AtSTATLs [115]. Thus, inferring the function of plant STATL proteins from metazoan STATs is difficult and requires further biochemical investigation.
Notably, SH2 and phosphotyrosine-binding domain (PTB) are not the only functional domains that can bind pTyr in animals. Besides typical pTyr-binding domains, some C2 domains, as well as Hakai pTyr-binding (HYB) domain and catalytically inactive PTP domains bind pTyr [116]. Furthermore, pyruvate kinase M2 (PKM2) and glycerol-3-phosphate dehydrogenase can bind pTyr through non-canonical mechanisms [117, 118]. PKM2 binds pTyr-peptides via its allosteric fructose-1,6-bisphosphate (FBP)-binding pocket, promoting release of the allosteric activator FBP [118]. Commonly, these non-canonical pTyr-binding proteins bind pTyr through positively charged pockets that are deep enough to accommodate the bulky Tyr side chain. Even though C2 and HYB domains exist in plants, their pTyr-binding capability has not been tested. It remains an open question whether plant pTyr-based signaling employs pTyr-binding proteins in a manner analogous to RTK mediated signaling, but the occurrence of pTyr-binding in proteins lacking SH2 or PTB domains raises the possibility that non-canonical pTyr-binding proteins are present in plants as well. Mass spectrometry-based proteomics will be a critical tool in uncovering the identity of plant pTyr-binding proteins. Indeed, methods using immobilized pTyr-peptide libraries for untargeted identification of novel pTyr-binding proteins [117] and for targeted analysis of pTyr sites of interest [119] have already been developed but have not yet been applied in plants. Furthermore, these approaches could also be used to directly test whether plant proto-SH2 domain-containing proteins interact with pTyr.
Concluding remarks
Although Tyr phosphorylation was reported in plants more than 30 years ago [120] and despite its abundance in plant phosphoproteomes, very little is known about how this post-translational modification contributes to signal transduction in plants. The emergence of PTPs and SH2 domains in D. discoideum indicates that the activity of dual-specificity protein kinases is sufficient to drive the evolution of pTyr-based signaling. It is therefore unexpected that these two protein domains are mostly absent from plants given the extent of Tyr phosphorylation. Thus, the requirement of Tyr phosphorylation for RK-mediated signaling poses two critical questions for plant biologists: First, how do pTyr residues function to relay a signal to downstream effector proteins? Second, how is pTyr-based signal transduction attenuated? In our view, there are two possible scenarios (Figure 5) for how pTyr could facilitate plant signaling: (1) pTyr exerts mainly conformational effects that control the behavior of pTyr proteins concerning either catalytic activity, protein–protein interactions, or subcellular localization; (2) the roles of core pTyr signaling proteins — writers, readers, and erasers — are fulfilled by non-canonical components and the molecular identity of proteins with ‘reader’ function in particular, has not been discovered. In both scenarios, the writer and eraser functions are fulfilled by dual-specificity protein kinases and phosphatases, respectively.
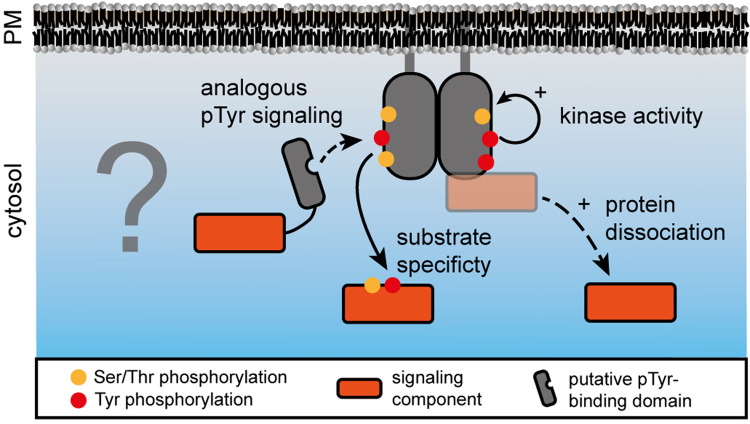
Figure 5. Potential roles of pTyr in plant transmembrane signaling.
Tyrosine phosphorylation in plants may support a pTyr-based signaling mechanism analogous to that in animals but the pTyr-binding proteins and PTPs remain unknown. In contrast with pTyr-specific recruitment of signaling components, tyrosine phosphorylation may induce the dissociation of signaling components that are associated with the receptor or co-receptor in a resting state. Alternatively, tyrosine phosphorylation could contribute to kinase activity or its substrate specificity by affecting structural properties of the kinase domain.
Elucidating the function of this important PTM has been challenging partly owing to the difficulty of in vivo identification of pTyr sites. Advanced mass spectrometer technology and optimized experimental pipelines will aid our understanding of the dynamics of Tyr phosphorylation in plants, particularly during RK-mediated signaling [9]. To complement the continued identification of novel regulatory pTyr sites on RKs involved in diverse aspects of plant physiology, phosphoproteomics must be coupled to experimental approaches aimed at revealing the biochemical action of pTyr in plant signal transduction. We envision a multi-faceted quantitative approach to evaluate the effect of Tyr phosphorylation on protein–protein interactions in vivo and in vitro, on protein subcellular localization, and on the catalytic activity of Tyr-phosphorylated proteins where applicable. Indeed, our understanding of Tyr phosphorylation in plants is only in its infancy and helping this important PTM come of age represents an exciting challenge for the plant science community.
Acknowledgements
All members of the Zipfel laboratory are thanked for inputs and discussion.
Abbreviations
- BAK1
BRI1 ASSOCIATED KINASE 1
- BIK1
BOTRYTIS INDUCED KINASE 1
- BIN2
BR INSENSITIVE 2
- BR
brassinosteroid
- BSU1
BRI1 SUPPRESSOR 1
- BZR1
BRASSINAZOLE RESISTANT 1
- CERK1
CHITIN ELICITOR RECEPTOR KINASE 1
- CIPP1
CERK1-INTERACTING PROTEIN PHOSPHATASE 1
- DSP
dual specificity phosphatases
- EFR
EF-TU RECEPTOR
- EPK
eukaryotic protein kinase
- IRAKs
INTERLEUKIN-1 (IL-1) RECEPTOR ASSOCIATED KINASES
- JAK
Janus kinase
- LMWPTP
low molecular weight PTP
- LORE
LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION
- LRR
leucine-rich repeat
- MKP1
MAPK PHOSPHATASE 1
- PAMP
pathogen-associated molecular pattern
- PIPs
phosphoinositol phosphates
- PKM2
pyruvate kinase M2
- PPP
phosphoprotein phosphatase
- PRRs
pattern recognition receptors
- RK
receptor kinase
- TK
Tyr kinase
- TLRs
Toll-like receptors
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
Research related to this review was funded by the Gatsby Charitable Foundation, the University of Zurich, the European Research Council under the grant agreement no. 309858 (grant ‘PHOSPHinnATE’), the Swiss National Science Foundation (grant agreement no. 31003A_182625), and a joint European Research Area Network for Coordinating Action in Plant Sciences (ERA-CAPS) grant (‘SICOPID’) from UK Research and Innovation (BB/S004734/1).
References
- 1.Shiu, S.H. and Bleecker, A.B. (2001) Plant receptor-like kinase gene family: diversity, function, and signaling. Sci. STKE 2001, re22 10.1126/stke.2001.113.re22 [DOI] [PubMed] [Google Scholar]
- 2.Trusov, Y. and Botella, J.R. (2016) Plant G-proteins come of age: breaking the bond with animal models. Front. Chem. 4, 24 10.3389/fchem.2016.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishi, H., Shaytan, A. and Panchenko, A.R. (2014) Physicochemical mechanisms of protein regulation by phosphorylation. Front. Genet. 5, 270 10.3389/fgene.2014.00270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Offringa, R. and Huang, F. (2013) Phosphorylation-dependent trafficking of plasma membrane proteins in animal and plant cells. J. Integr. Plant Biol. 55, 789–808 10.1111/jipb.12096 [DOI] [PubMed] [Google Scholar]
- 5.Humphrey, S.J., James, D.E. and Mann, M. (2015) Protein phosphorylation: a major switch mechanism for metabolic regulation. Trends Endocrinol. Metab. 26, 676–687 10.1016/j.tem.2015.09.013 [DOI] [PubMed] [Google Scholar]
- 6.Hanks, S.K. and Hunter, T. (1995) The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 10.1096/fasebj.9.8.7768349 [DOI] [PubMed] [Google Scholar]
- 7.Taylor, S.S., Radzio-Andzelm, E. and Hunter, T. (1995) How do protein kinases discriminate between serine/threonin and tyrosine? Structural insights from the insulin receptor protein-tyrosine kinase. FASEB J. 9, 1255–1266 10.1096/fasebj.9.13.7557015 [DOI] [PubMed] [Google Scholar]
- 8.Sugden, C., Urbaniak, M.D., Araki, T. and Williams, J.G. (2015) The dictyostelium prestalk inducer differentiation-inducing factor-1 (DIF-1) triggers unexpectedly complex global phosphorylation changes. Mol. Biol. Cell 26, 805–820 10.1091/mbc.E14-08-1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mergner, J., Frejno, M., List, M., Papacek, M., Chen, X., Chaudhary, A., et al. (2020) Mass-spectrometry-based draft of the arabidopsis proteome. Nature 579, 409–414 10.1038/s41586-020-2094-2 [DOI] [PubMed] [Google Scholar]
- 10.Yachie, N., Saito, R., Sugiyama, N., Tomita, M. and Ishihama, Y. (2011) Integrative features of the yeast phosphoproteome and protein-protein interaction map. PLoS Comput. Biol. 7, e1001064 10.1371/journal.pcbi.1001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pincus, D., Letunic, I., Bork, P. and Lim, W.A. (2008) Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proc. Natl Acad. Sci. U.S.A. 105, 9680–9684 10.1073/pnas.0803161105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Wijk, K.J., Friso, G., Walther, D. and Schulze, W.X. (2014) Meta-analysis of arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 26, 2367–2389 10.1105/tpc.114.125815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahsan, N., Wilson, R.S., Rao, R.S.P., Salvato, F., Sabila, M., Ullah, H.et al. (2020) Mass spectrometry-based identification of phospho-Tyr in plant proteomics. J. Proteome Res. 19, 561–571 10.1021/acs.jproteome.9b00550 [DOI] [PubMed] [Google Scholar]
- 14.de la Fuente van Bentem, S. and Hirt, H. (2009) Protein tyrosine phosphorylation in plants: more abundant than expected? Trends Plant Sci. 14, 71–76 10.1016/j.tplants.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 15.Olsen, J.V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P.et al. (2006) Global, In vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 10.1016/j.cell.2006.09.026 [DOI] [PubMed] [Google Scholar]
- 16.Macho, A.P., Schwessinger, B., Ntoukakis, V., Brutus, A., Segonzac, C., Roy, S., et al. (2014) A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343, 1509–1512 10.1126/science.1248849 [DOI] [PubMed] [Google Scholar]
- 17.Luo, X., Wu, W., Liang, Y., Xu, N., Wang, Z., Zou, H.et al. (2020) Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J. 39, e102856 10.15252/embj.2019102856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, J., Liu, B., Chen, S., Gong, B.Q., Chen, L., Zhou, Q., et al. (2018) A tyrosine phosphorylation cycle regulates fungal activation of a plant receptor Ser/Thr kinase. Cell Host Microbe 23, 241–253.e6 10.1016/j.chom.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 19.Perraki, A., DeFalco, T.A., Derbyshire, P., Avila, J., Séré, D., Sklenar, J., et al. (2018) Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 561, 248–252 10.1038/s41586-018-0471-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, T.W., Guan, S., Sun, Y., Deng, Z., Tang, W., Shang, J.X.et al. (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–1260 10.1038/ncb1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhrig, R.G., Labandera, A.M., Muhammad, J., Samuel, M. and Moorhead, G.B. (2016) Rhizobiales-like phosphatase 2 from arabidopsis thaliana is a novel phospho-tyrosine-specific phospho-protein phosphatase (PPP) family protein phosphatase. J. Biol. Chem. 291, 5926–5934 10.1074/jbc.M115.683656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim, W.A. and Pawson, T. (2010) Phosphotyrosine signaling: evolving a New cellular communication system. Cell 142, 661–667 10.1016/j.cell.2010.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning, G., Plowman, G.D., Hunter, T. and Sudarsanam, S. (2002) Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514–520 10.1016/S0968-0004(02)02179-5 [DOI] [PubMed] [Google Scholar]
- 24.Lemmon, M.A. and Schlessinger, J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaffe, M.B. (2002) Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 3, 177–186 10.1038/nrm759 [DOI] [PubMed] [Google Scholar]
- 26.Lundby, A., Franciosa, G., Emdal, K.B., Refsgaard, J.C., Gnosa, S.P., Bekker-Jensen, D.B., et al. (2019) Oncogenic mutations rewire signaling pathways by switching protein recruitment to phosphotyrosine sites. Cell 179, 543–560.e26 10.1016/j.cell.2019.09.008 [DOI] [PubMed] [Google Scholar]
- 27.Liang, X. and Zhou, J.-M. (2018) Receptor-Like cytoplasmic kinases: central players in plant receptor kinase–mediated signaling. Annu. Rev. Plant Biol. 69, 267–299 10.1146/annurev-arplant-042817-040540 [DOI] [PubMed] [Google Scholar]
- 28.Hohmann, U., Lau, K. and Hothorn, M. (2017) The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68, 109–137 10.1146/annurev-arplant-042916-040957 [DOI] [PubMed] [Google Scholar]
- 29.Couto, D. and Zipfel, C. (2016) Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 10.1038/nri.2016.77 [DOI] [PubMed] [Google Scholar]
- 30.He, Y., Zhou, J., Shan, L. and Meng, X. (2018) Plant cell surface receptor-mediated signaling - A common theme amid diversity. J. Cell Sci. 131, jcs209353 10.1242/jcs.209353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, J.D.G. and Dangl, J.L. (2006) The plant innate immune system. Nature 444, 323–329 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 32.Shiu, S., Karlowski, W., Pan, R., Tzeng, Y.-H., Mayer, K.F.X. and Li, W.-H. (2004) Comparative analysis of the receptor-like kinase family in arabidopsis and rice. Plant Cell 16, 1220–1234 10.1105/tpc.020834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gish, L.A. and Clark, S.E. (2011) The RLK/Pelle family of kinases. Physiol. Behav. 66, 117–127 10.1111/j.1365-313X.2011.04518.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiu, S.H. and Bleecker, A.B. (2001) Receptor-like kinases from arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl Acad. Sci. U.S.A. 98, 10763–10768 10.1073/pnas.181141598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiu, S.H. and Bleecker, A.B. (2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in arabidopsis. Plant Physiol. 132, 530–543 10.1104/pp.103.021964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai, T. and Akira, S. (2010) The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 11, 373–384 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 37.Kawasaki, T. and Kawai, T. (2014) Toll-like receptor signaling pathways. Front. Immunol. 5, 1–8 10.3389/fimmu.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mithoe, S.C. and Menke, F.L.H. (2011) Phosphoproteomics perspective on plant signal transduction and tyrosine phosphorylation. Phytochemistry 72, 997–1006 10.1016/j.phytochem.2010.12.009 [DOI] [PubMed] [Google Scholar]
- 39.Oh, M.H., Wang, X., Kota, U., Goshe, M.B., Clouse, S.D. and Huber, S.C. (2009) Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in arabidopsis. Proc. Natl Acad. Sci. U.S.A. 106, 658–663 10.1073/pnas.0810249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaillais, Y., Hothorn, M., Belkhadir, Y., Dabi, T., Nimchuk, Z.L., Meyerowitz, E.M.et al. (2011) Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25, 232–237 10.1101/gad.2001911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macho, A.P., Lozano-Durán, R. and Zipfel, C. (2015) Importance of tyrosine phosphorylation in receptor kinase complexes. Trends Plant Sci. 20, 269–272 10.1016/j.tplants.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 42.Suzuki, M., Watanabe, T., Yoshida, I., Kaku, H. and Shibuya, N. (2018) Autophosphorylation site Y428 is essential for the in vivo activation of CERK1. Plant Signal. Behav. 13, e1435228 10.1080/15592324.2018.1435228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, I., Wang, Y., Seitz, K., Baer, J., Bennewitz, S., Mooney, B.P.et al. (2016) Analysis of phosphorylation of the receptor-like protein kinase HAESA during arabidopsis floral abscission. PLoS ONE 11, e0147203 10.1371/journal.pone.0147203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bojar, D., Martinez, J., Santiago, J., Rybin, V., Bayliss, R. and Hothorn, M. (2014) Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 78, 31–43 10.1111/tpj.12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muleya, V., Marondedze, C., Wheeler, J.I., Thomas, L., Mok, Y.F., Griffin, M.D.W. et al. (2016) Phosphorylation of the dimeric cytoplasmic domain of the phytosulfokine receptor, PSKR1. Biochem. J. 473, 3081–3098 [DOI] [PubMed] [Google Scholar]
- 46.Sugiyama, N., Nakagami, H., Mochida, K., Daudi, A., Tomita, M., Shirasu, K. and Ishihama, Y. (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mithoe, S.C., Boersema, P.J., Berke, L., Snel, B., Heck, A.J.R. and Menke, F.L.H. (2012) Targeted quantitative phosphoproteomics approach for the detection of phospho-tyrosine signaling in plants. J. Proteome Res. 11, 438–448 [DOI] [PubMed] [Google Scholar]
- 48.Ma, X., Xu, G., He, P. and Shan, L. (2016) SERKing coreceptors for receptors. Trends Plant Sci. 21, 1017–1033 10.1016/j.tplants.2016.08.014 [DOI] [PubMed] [Google Scholar]
- 49.He, K., Gou, X., Yuan, T., Lin, H., Asami, T., Yoshida, S.et al. (2007) BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-Death pathways. Curr. Biol. 17, 1109–1115 10.1016/j.cub.2007.05.036 [DOI] [PubMed] [Google Scholar]
- 50.Tang, W., Kim, T.-W., Oses-Prieto, J.A., Sun, Y., Deng, Z., Zhu, S.et al. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in arabidopsis. Science 321, 557–560 10.1126/science.1156973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim, T.W., Guan, S., Burlingame, A.L. and Wang, Z.Y. (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43, 561–571 10.1016/j.molcel.2011.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryu, H., Kim, K., Cho, H., Park, J., Choe, S. and Hwang, I. (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in arabidopsis brassinosteroid signaling. Plant Cell 19, 2749–2762 10.1105/tpc.107.053728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolf, S. (2020) Deviating from the beaten track: new twists in brassinosteroid receptor function. Int. J. Mol. Sci. 21, 1561 10.3390/ijms21051561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, Z.Y., Zhu, J.Y. and Sae-Seaw, J. (2013) Brassinosteroid signaling. Development 140, 1615–1620 10.1242/dev.060590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H.et al. (1999) Brassinosteroid-insensitive dwarf mutants of arabidopsis accumulate brassinosteroids. Plant Physiol. 121, 743–752 10.1104/pp.121.3.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D.G., Felix, G.et al. (2004) Bacterial disease resistance in arabidopsis through flagellin perception. Nature 428, 764–767 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
- 57.Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J.D.G., Boller, T.et al. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts agrobacterium-Mediated transformation. Cell 125, 749–760 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- 58.Gómez-Gómez, L. and Boller, T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in arabidopsis. Mol. Cell 5, 1003–1011 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- 59.Cao, Y., Liang, Y., Tanaka, K., Nguyen, C.T., Jedrzejczak, R.P., Joachimiak, A.et al. (2014) The kinase LYK5 is a major chitin receptor in arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766 10.7554/eLife.03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012) Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164 10.1126/science.1218867 [DOI] [PubMed] [Google Scholar]
- 61.Willmann, R., Lajunen, H.M., Erbs, G., Newman, M.A., Kolb, D., Tsuda, K., et al. (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl Acad. Sci. U.S.A. 108, 19824–19829 10.1073/pnas.1112862108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wan, J., Tanaka, K., Zhang, X.C., Son, G.H., Brechenmacher, L., Nguyen, T.H.N.et al. (2012) LYK4, a lysin motif receptor-like kinase, is important for chitin signaling and plant innate immunity in arabidopsis. Plant Physiol. 160, 396–406 10.1104/pp.112.201699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kutschera, A., Dawid, C., Gisch, N., Schmid, C., Raasch, L., Gerster, T., et al. (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in arabidopsis plants. Science 364, 178–181 10.1126/science.aau1279 [DOI] [PubMed] [Google Scholar]
- 64.Zhang, J., Li, W., Xiang, T., Liu, Z., Laluk, K., Ding, X., et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a pseudomonas syringae effector. Cell Host Microbe 7, 290–301 10.1016/j.chom.2010.03.007 [DOI] [PubMed] [Google Scholar]
- 65.Ranf, S., Eschen-Lippold, L., Frhlich, K., Westphal, L., Scheel, D. and Lee, J. (2014) Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. Ann. Bot. 14, 374 10.1186/s12870-014-0374-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henty-Ridilla, J.L., Li, J., Day, B. and Staiger, C.J. (2014) ACTIN DEPOLYMERIZING FACTOR4 regulates actin dynamics during innate immune signaling in arabidopsis. Plant Cell 26, 340–352 10.1105/tpc.113.122499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014) The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase rbohD to control plant immunity. Cell Host Microbe 15, 329–338 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 68.Liu, Z., Wu, Y., Yang, F., Zhang, Y., Chen, S., Xie, Q.et al. (2013) BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl Acad. Sci. U.S.A. 110, 6205–6210 10.1073/pnas.1215543110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu, D., Wu, S., Gao, X., Zhang, Y., Shan, L. and He, P. (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl Acad. Sci. U.S.A. 107, 496–501 10.1073/pnas.0909705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kadota, Y., Sklenar, J., Derbyshire, P., Stransfeld, L., Asai, S., Ntoukakis, V., et al. (2014) Direct regulation of the NADPH oxidase RBOHD by the PRR-Associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55 10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- 71.Shinya, T., Yamaguchi, K., Desaki, Y., Yamada, K., Narisawa, T., Kobayashi, Y., et al. (2014) Selective regulation of the chitin-induced defense response by the arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79, 56–66 10.1111/tpj.12535 [DOI] [PubMed] [Google Scholar]
- 72.Robert, X. and Gouet, P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yan, L., Ma, Y., Liu, D. et al. (2012) Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1. Cell Res. 22, 1304–1308 10.1038/cr.2012.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C. et al. (2004) UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 75.Meng, X., Chen, X., Mang, H., Liu, C., Yu, X., Gao, X.et al. (2015) Differential function of arabidopsis SERK family receptor-like kinases in stomatal patterning. Curr. Biol. 25, 2361–2372 10.1016/j.cub.2015.07.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Meng, X., Zhou, J., Tang, J., Li, B., de Oliveira, M.V.V., Chai, J.et al. (2016) Ligand-Induced receptor-like kinase complex regulates floral organ abscission in arabidopsis. Cell Rep. 14, 1330–1338 10.1016/j.celrep.2016.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwessinger, B., Roux, M., Kadota, Y., Ntoukakis, V., Sklenar, J., Jones, A.et al. (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7, e1002046 10.1371/journal.pgen.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, X., Kota, U., He, K., Blackburn, K., Li, J., Goshe, M.B.et al. (2008) Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15, 220–235 10.1016/j.devcel.2008.06.011 [DOI] [PubMed] [Google Scholar]
- 79.Moffett, A.S., Bender, K.W., Huber, S.C. and Shukla, D. (2017) Molecular dynamics simulations reveal the conformational dynamics of arabidopsis thaliana BRI1 and BAK1 receptor-like kinases. J. Biol. Chem. 292, 12643–12652 10.1074/jbc.M117.792762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin, W., Li, B., Lu, D., Chen, S., Zhu, N., He, P.et al. (2014) Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates arabidopsis innate immunity. Proc. Natl Acad. Sci. U.S.A. 111, 3632–3637 10.1073/pnas.1318817111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu, J., Wei, X., Yan, L., Liu, D., Ma, Y., Guo, Y., et al. (2013) Identification and functional analysis of phosphorylation residues of the arabidopsis BOTRYTIS-INDUCED KINASE1. Protein Cell 4, 771–781 10.1007/s13238-013-3053-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krysan, P.J. and Colcombet, J. (2018) Cellular complexity in MAPK signaling in plants: questions and emerging tools to answer them. Front. Plant Sci. 871, 1674 10.3389/fpls.2018.01674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Colcombet, J. and Hirt, H. (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413, 217–226 10.1042/BJ20080625 [DOI] [PubMed] [Google Scholar]
- 84.Bartels, S., Anderson, J.C., Gonzalez Besteiro, M.A., Carreri, A., Hirt, H., Buchala, A.et al. (2009) MAP KINASE PHOSPHATASE1 and PROTEIN TYROSINE PHOSPHATASE1 are repressors of salicylic acid synthesis and SNC1-mediated responses in arabidopsis. Plant Cell 21, 2884–2897 10.1105/tpc.109.067678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gupta, R., Huang, Y., Kieber, J. and Luan, S. (1998) Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from arabidopsis. Plant J. 16, 581–589 10.1046/j.1365-313x.1998.00327.x [DOI] [PubMed] [Google Scholar]
- 86.Schweighofer, A., Kazanaviciute, V., Scheikl, E., Teige, M., Doczi, R., Hirt, H., et al. (2007) The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in arabidopsis. Plant Cell 19, 2213–2224 10.1105/tpc.106.049585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bartels, S., Besteiro, M.A.G., Lang, D. and Ulm, R. (2010) Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci. 15, 322–329 10.1016/j.tplants.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 88.Jiang, L., Anderson, J.C., González Besteiro, M.A. and Peck, S.C. (2017) Phosphorylation of arabidopsis MAP kinase phosphatase 1 (MKP1) is required for PAMP responses and resistance against bacteria. Plant Physiol. 175, 1839–1852 10.1104/pp.17.01152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anderson, J.C., Bartels, S., Besteiro, M.A.G., Shahollari, B., Ulm, R. and Peck, S.C. (2011) Arabidopsis MAP kinase phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J. 67, 258–268 10.1111/j.1365-313X.2011.04588.x [DOI] [PubMed] [Google Scholar]
- 90.Anderson, J.C., Wan, Y., Kim, Y.M., Pasa-Tolic, L., Metz, T.O. and Peck, S.C. (2014) Decreased abundance of type III secretion systeminducing signals in arabidopsis mkp1 enhances resistance against pseudomonas syringae. Proc. Natl Acad. Sci. U.S.A. 111, 6846–6851 10.1073/pnas.1403248111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Escudero, V., Torres, M.Á, Delgado, M., Sopeña-Torres, S., Swami, S., Morales, J., et al. (2019) Mitogen-activated protein kinase phosphatase 1 (MKP1) negatively regulates the production of reactive oxygen species during arabidopsis immune responses. Mol. Plant Microbe Interact. 32, 464–478 10.1094/MPMI-08-18-0217-FI [DOI] [PubMed] [Google Scholar]
- 92.Hunter, T. (2014) The genesis of tyrosine phosphorylation. Cold Spring Harb. Perspect. Biol. 6, a020644 10.1101/cshperspect.a020644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alonso, A., Sasin, J., Bottini, N., Friedberg, I., Friedberg, I., Osterman, A.et al. (2004) Protein tyrosine phosphatases in the human genome. Cell 117, 699–711 10.1016/j.cell.2004.05.018 [DOI] [PubMed] [Google Scholar]
- 94.Kerk, D., Templeton, G. and Moorhead, G.B.G. (2008) Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, Green algae, and higher plants. Plant Physiol. 146, 351–367 10.1104/pp.107.111393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Helm, S., Dobritzsch, D., Rödiger, A., Agne, B. and Baginsky, S. (2014) Protein identification and quantification by data-independent acquisition and multi-parallel collision-induced dissociation mass spectrometry (MSE) in the chloroplast stroma proteome. J. Proteomics 98, 79–89 10.1016/j.jprot.2013.12.007 [DOI] [PubMed] [Google Scholar]
- 96.Xu, Q., Fu, H.H., Gupta, R. and Luan, S. (1998) Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in arabidopsis. Plant Cell 10, 849–857 10.1105/tpc.10.5.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aceti, D.J., Bitto, E., Yakunin, A.F., Proudfoot, M., Bingman, C.A., Frederick, R.O., et al. (2008) Structural and functional characterization of a novel phosphatase from the arabidopsis thaliana gene locus At1g05000. Proteins Struct. Funct. Genet. 73, 241–253 10.1002/prot.22041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Romá-Mateo, C., Sacristán-Reviriego, A., Beresford, N.J., Caparrós-Martín, J.A., Culiáñez-Macià, F.A., Martín, H.et al. (2011) Phylogenetic and genetic linkage between novel atypical dual-specificity phosphatases from non-metazoan organisms. Mol. Genet. Genomics 285, 341–354 10.1007/s00438-011-0611-6 [DOI] [PubMed] [Google Scholar]
- 99.Pribat, A., Sormani, R., Rousseau-Gueutin, M., Julkowska, M.M., Testerink, C., Joubès, J., et al. (2012) A novel class of PTEN protein in arabidopsis displays unusual phosphoinositide phosphatase activity and efficiently binds phosphatidic acid. Biochem. J. 441, 161–171 10.1042/BJ20110776 [DOI] [PubMed] [Google Scholar]
- 100.Gupta, R., Ting, J.T.L., Sokolov, L.N., Johnson, S.A. and Luan, S. (2002) A tumor suppressor homolog, AtPTEN1, is essential for pollen development in arabidopsis. Plant Cell 14, 2495–2507 10.1105/tpc.005702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Labandera, A.M., Glen Uhrig, R., Colville, K., Moorhead, G.B. and Ng, K.K.S. (2018) Structural basis for the preference of the arabidopsis thaliana phosphatase RLPH2 for tyrosine-phosphorylated substrates. Sci. Signal. 11, eaan8804 10.1126/scisignal.aan8804 [DOI] [PubMed] [Google Scholar]
- 102.Liu, B.A. and Nash, P.D. (2012) Evolution of SH2 domains and phosphotyrosine signalling networks. Philos. Trans. R. Soc. B Biol. Sci. 367, 2556–2573 10.1098/rstb.2012.0107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bradshaw, J.M. and Waksman, G. (1999) Calorimetric examination of high-affinity Src SH2 domain-tyrosyl phosphopeptide binding: dissection of the phosphopeptide sequence specificity and coupling energetics. Biochemistry 38, 5147–5154 10.1021/bi982974y [DOI] [PubMed] [Google Scholar]
- 104.Ottinger, E.A., Botfield, M.C. and Shoelson, S.E. (1998) Tandem SH2 domains confer high specificity in tyrosine kinase signaling. J. Biol. Chem. 273, 729–735 10.1074/jbc.273.2.729 [DOI] [PubMed] [Google Scholar]
- 105.Eck, M.J., Dhe-Paganon, S., Trüb, T., Nolle, R.T. and Shoelson, S.E. (1996) Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell 85, 695–705 10.1016/S0092-8674(00)81236-2 [DOI] [PubMed] [Google Scholar]
- 106.Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A., Minh, B.Q. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44, W232–W235 10.1093/nar/gkw256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Letunic, I. and Bork, P. (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Research 47, W256–W259 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ravichandran, K.S., Zhou, M.M., Pratt, J.C., Harlan, J.E., Walk, S.F., Fesik, S.W.et al. (1997) Evidence for a requirement for both phospholipid and phosphotyrosine binding via the Shc phosphotyrosine-binding domain in vivo. Mol. Cell. Biol. 17, 5540–5549 10.1128/MCB.17.9.5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dho, S.E., Jacob, S., Wolting, C.D., French, M.B., Rohrschneider, L.R. and McGlade, C.J. (1998) The mammalian numb phosphotyrosine-binding domain. characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J. Biol. Chem. 273, 9179–9187 10.1074/jbc.273.15.9179 [DOI] [PubMed] [Google Scholar]
- 110.Gao, Q., Hua, J., Kimura, R., Headd, J.J., Fu, X.Y. and Chin, E.Y. (2004) Identification of the linker-SH2 domain of STAT as the origin of the SH2 domain using two-dimensional structural alignment. Mol. Cell. Proteomics 3, 704–714 10.1074/mcp.M300131-MCP200 [DOI] [PubMed] [Google Scholar]
- 111.Sdano, M.A., Fulcher, J.M., Palani, S., Chandrasekharan, M.B., Parnell, T.J., Whitby, F.G.et al. (2017) A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription. Elife 6, e28723 10.7554/eLife.28723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen, C., Shu, J., Li, C., Thapa, R.K., Nguyen, V., Yu, K., et al. (2019) RNA polymerase II-independent recruitment of SPT6L at transcription start sites in arabidopsis. Nucleic Acids Res. 47, 6714–6725 10.1093/nar/gkz465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoh, S.M., Cho, H., Pickle, L., Evans, R.M. and Jones, K.A. (2007) The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 21, 160–174 10.1101/gad.1503107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Salas, A., Hernandez-Rocha, C., Duijvestein, M., Faubion, W., McGovern, D., Vermeire, S.et al. (2020) JAK–STAT pathway targeting for the treatment of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 323–337 10.1038/s41575-020-0273-0 [DOI] [PubMed] [Google Scholar]
- 115.William, J.G. and Zvelebil, M. (2004) SH2 domains in plants imply new signalling secanrios. Trends Plant Sci. 9, 161–163 10.1016/j.tplants.2004.02.001 [DOI] [PubMed] [Google Scholar]
- 116.Kaneko, T., Joshi, R., Feller, S.M. and Li, S.S.C. (2012) Phosphotyrosine recognition domains: the typical, the atypical and the versatile. Cell Commun. Signal. 10, 32 10.1186/1478-811X-10-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Christofk, H.R., Wu, N., Cantley, L.C. and Asara, J.M. (2011) Proteomic screening method for phosphopeptide motif binding proteins using peptide libraries. J. Proteome Res. 10, 4158–4164 10.1021/pr200578n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Christofk, H.R., Vander Heiden, M.G., Wu, N., Asara, J.M. and Cantley, L.C. (2008) Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature 452, 181–186 10.1038/nature06667 [DOI] [PubMed] [Google Scholar]
- 119.Schulze, W.X. and Mann, M. (2004) A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 279, 10756–10764 10.1074/jbc.M309909200 [DOI] [PubMed] [Google Scholar]
- 120.Elliot, D.C. and Geytenbeek, M. (1985) Identification of products of protein phosphorylation in T37-transformed cells and comparison with normal cells. Biochim. Biophys. Acta 845, 317–323 10.1016/0167-4889(85)90194-6 [DOI] [Google Scholar]