A NAC transcription factor modulates the trade-offs between growth, immunity, and thermotolerance via inducible expression and differential interactions in pepper.
Abstract
Plant responses to pathogen attacks and high-temperature stress (HTS) are distinct in nature but generally share several signaling components. How plants produce specific responses through these common signaling intermediates remains elusive. With the help of reverse-genetics approaches, we describe here the mechanism underlying trade-offs in pepper (Capsicum annuum) between growth, immunity, and thermotolerance. The NAC-type transcription factor CaNAC2c was induced by HTS and Ralstonia solanacearum infection (RSI). CaNAC2c-inhibited pepper growth, promoted immunity against RSI by activating jasmonate-mediated immunity and H2O2 accumulation, and promoted HTS responses by activating Heat shock factor A5 (CaHSFA5) transcription and blocking H2O2 accumulation. We show that CaNAC2c physically interacts with CaHSP70 and CaNAC029 in a context-specific manner. Upon HTS, CaNAC2c–CaHSP70 interaction in the nucleus protected CaNAC2c from degradation and resulted in the activation of thermotolerance by increasing CaNAC2c binding and transcriptional activation of its target promoters. CaNAC2c did not induce immunity-related genes under HTS, likely due to the degradation of CaNAC029 by the 26S proteasome. Upon RSI, CaNAC2c interacted with CaNAC029 in the nucleus and activated jasmonate-mediated immunity but was prevented from activating thermotolerance-related genes. In non-stressed plants, CaNAC2c was tethered outside the nucleus by interaction with CaHSP70, and thus was unable to activate either immunity or thermotolerance. Our results indicate that pepper growth, immunity, and thermotolerance are coordinately and tightly regulated by CaNAC2c via its inducible expression and differential interaction with CaHSP70 and CaNAC029.
Introduction
In their natural habitats, plants are continuously exposed to biotic and abiotic stresses, either individually or in combination. Plants, therefore, need to appropriately cope with these challenges to maximize fitness by prioritizing the allocation of limited recourses between growth and response to stress. The resulting trade-off is thought to be regulated by crosstalk between signaling pathways (Fujita et al., 2006; Sharma et al., 2013; Nejat and Mantri, 2017) and is likely to be modulated according to dynamic changes in the severity of different stresses (Lozano-Duran et al., 2013). However, the nature of the involved factors and their possible modes of action remain elusive.
High-temperature stress (HTS) and pathogen attack are frequently encountered by plants growing in tropical or subtropical climates and lead to severe retardation in growth and development, sometimes even death. Under the constant selective pressure of these stresses, plants have evolved sophisticated defense systems. Upon pathogen infection, conserved, and ubiquitous receptors—generally termed pattern recognition receptors (PRRs)—at the plant plasma membrane and fast-evolving intracellular R proteins perceive pathogen-derived pathogen-associated molecular patterns (PAMPs) and effectors, thereby activating PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI), respectively (Jones and Dangl, 2006). Under these conditions, plant cells undergo a global reprogramming of their metabolism and adopt a defense mode rather than a growth mode. Plant immune responses generally include the production of reactive oxygen intermediates (ROIs) such as superoxide anion (), nitric oxide (NO), and hydrogen peroxide (H2O2; Schreck and Baeuerle, 1991; Chandra et al., 1996; McDowell and Dangl, 2000) leading to a hypersensitive response (HR; Dangl, 1998; Delledonne et al., 2001). In addition, infected plants produce antimicrobial compounds (Jabs et al., 1997) and pathogenesis-related proteins (Fobert and Després, 2005; Breen et al., 2017) to fight off the infection.
Unlike immune responses initiated by pathogen infection mainly at the cell surface, plants perceive heat via sensors in various cellular locations such as plasma membrane-localized channels, a histone variant sensor in the nucleus, unfolded protein sensors in the endoplasmic reticulum (ER), and the cytosol, the red light photoreceptor phytochrome B (PhyB) and ER membrane-associated basic leucine zipper transcription factors (Mittler et al., 2012; Srivastava et al., 2014; Song et al., 2017). These thermosensors activate thermotolerance by inducing the production of Heat Shock Proteins (HSPs) that act as chaperones to help resolubilize protein aggregates after heat stress, as well as the biosynthesis of antioxidants or reactive oxygen species (ROS) scavengers that offer protection from oxidative damage (Kotak et al., 2007; Chen et al., 2016; Yu et al., 2019).
Although the two pathways involve distinct sensors, plant responses to pathogens and HTS share signaling components such as Ca2+ signaling, ROS, phytohormones such as jasmonic acid (JA) and salicylic acid (SA; Li et al., 2011; Liu et al., 2015a, 2015b) and Nucleotide Binding Site (NBS)-Leucine-Rich Repeat (LRR) proteins (Kim et al., 2015). This overlap hints at the potential for extensive trade-off between plant responses to pathogens and HTS. Indeed, these two processes have been suggested to be closely related (Lee et al., 2012): for example, plant immunity is generally dampened by HTS (Janda et al., 2019). Specific defense responses will thus require the selective activation of the appropriate shared components. However, how these crucial regulators achieve their specific regulation and maintain a balance between plant responses to HTS and pathogen attack remains largely unknown.
Given that plant defense responses against pathogen attacks or HTS generally include massive transcriptional reprogramming, transcription factors (TFs) are likely to act as crucial players in these processes (Hua, 2009; Moore et al., 2011; Fragkostefanakis et al., 2015; Liu et al., 2015a; Xue et al., 2015; Du et al., 2017; Birkenbihl et al., 2018). However, the precise roles of TFs in balancing plant responses to pathogen attacks and HTS and the underlying molecular details remain to be elucidated. The No Apical Meristem (NAM)/Arabidopsis Transcription Activation Factor (ATAF)/Cup-Shaped Cotyledon (CUC) NAC family of TFs constitute one of the largest plant TF families. They are characterized by a conserved N-terminal NAC domain and a diversified C-terminal transcription regulatory region and have been classified into eight subfamilies (Puranik et al., 2012). NAC TFs are implicated in the regulation of plant responses to stress conditions via binding a specific recognition site [CGT (G/A)] in the promoters of their target genes (Fang et al., 2016; Khedia et al., 2018). NAC TFs involved in stress responses belong to one subgroup (Fang et al., 2008; Nakashima et al., 2012; Negi et al., 2018). Although NAC proteins clearly participate in plant responses to heat stress or thermotolerance (Guan et al., 2014; Fang et al., 2015; Dong et al., 2020; Liu et al., 2020) and plant immunity (Perochon et al., 2019; Chang et al., 2020), to date, no individual NAC TF has been demonstrated to influence the balance between plant immunity and heat stress response.
A typical example of balance (or trade-off) between plant immunity and heat stress responses comes from pepper, a member of the Solanaceae family and a vegetable of great agricultural importance worldwide. Pepper originated from the tropical and subtropical regions of Central and South America and is widely grown during the warm seasons or in greenhouses, where it frequently suffers from bacterial wilt caused by Ralstonia solanacearum infections, a soil-borne pathogen that invades plants exclusively through their roots (Mansfield et al., 2012; Jiang et al., 2017). Pepper plants are also routinely exposed to HTS, causing severe growth retardation (Usman et al., 2014). We have shown previously that pepper responses to infection by R. solanacearum and HTS share a number of components, including calcium-dependent protein kinase 15 (CaCDPK15; Shen et al., 2016), the WRKY TFs CaWRKY6 (Cai et al., 2015), and CaWRKY40 (Dang et al., 2013), as well as the TF basic region/leucine zipper motif 63 (CabZIP63; Shen et al., 2016).
In the present study, we demonstrate that CaNAC2c, a member of the NAC family in pepper, not only participates in the regulation of pepper responses to RSI and HTS but also in the regulation of the trade-off between growth and responses to HTS or R. solanacearum infection (RSI). In addition, CaNAC2c acts as a crucial regulator coordinating growth, immunity, and thermotolerance at both the transcriptional and post-transcriptional levels by differential and context-specific interactions with CaNAC029 and CaHSP70.
Results
Expression profiling of pepper NAC genes during HTS and RSI
It was previously shown that NAC TFs that play roles during plant response to heat stress or pathogen attack can be identified based on their transcriptional signature in response to these stresses (Nakashima et al., 2012). To identify candidate pepper NAC genes involved in coordinating responses to HTS and RSI, we explored the expression profiles of 90 NAC genes in pepper plants exposed to HTS and to RSI based on publicly available data from Pepper Hub (http://www.hnivr.org/; Liu et al., 2017) and our unpublished transcriptome deep-sequencing (RNA-seq) data. One NAC gene (Capana06g001739) was upregulated by both HTS and RSI (Supplemental Figure S1, A). We validated the observed upregulation of this gene by RT-qPCR analysis on root samples collected from pepper plants challenged with RSI or HTS (Supplemental Figure S1, B). This NAC and three closely related NAC genes encoded proteins with a conserved N-terminal NAC domain and a diversified C-terminal transcription regulatory (TR) region (Supplemental Figure S2, A). They shared the highest sequence identity with Arabidopsis (Arabidopsis thaliana) NAC2 (Supplemental Figure S2, B and Supplemental Table S2). Accordingly, we named these genes CaNAC2a to CaNAC2d, CaNAC2c being induced by both HTS and RSI. CaNAC2c and CaNAC2d showed a sequence identity of 73%, the highest among these four NAC genes (Supplemental Figure S2, C and D). We explored the response of these four NAC2-like genes to HTS and RSI in our RNA-seq dataset of pepper plants challenged with RSI: CaNAC2c was strongly up-regulated by both HTS and RSI. Notably, CaNAC2d was only induced by RSI in roots (Supplemental Figure S2, E). NAC TFs can exhibit different subcellular localizations (Mathew et al., 2016). To determine where CaNAC2c resides in the cell, we overexpressed 35Spro:Yellow Fluorescent Protein (YFP) or 35Spro:CaNAC2c-YFP in Nicotiana benthamiana leaf epidermal cells. We detected YFP fluorescence exclusively in the nucleus for CaNAC2c-YFP, whereas free YFP was observed throughout the cell, including the cytosol and the nucleus, indicating that CaNAC2c is a nucleus-localized protein (Supplemental Figure S3).
CaNAC2c is a positive regulator of thermotolerance and R. solanacearum resistance but a negative regulator of plant growth
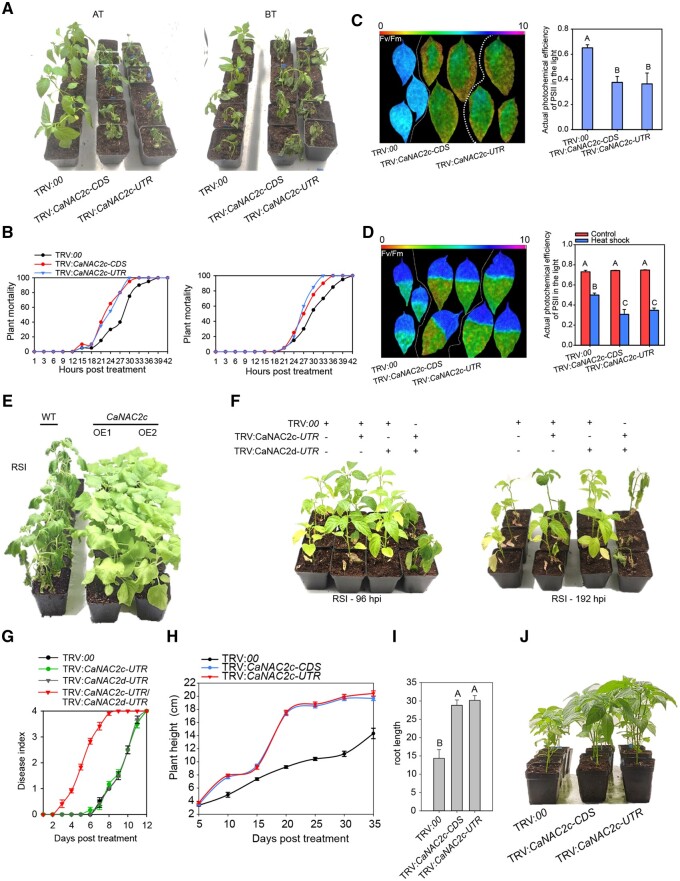
To determine the role of CaNAC2c in response to HTS or RSI, we first performed a loss-of-function assay by gene silencing using virus-induced gene silencing(VIGS), to avoid the possible off-targeting, we selected two specific fragments in the 3′-UTR and the open reading frame(ORF) of CaNAC2c for vector construction, the result showed that CaNAC2c transcript levels were greatly reduced by two distinct vectors targeting different portions of the CaNAC2c mRNA (Supplemental Figure S4, A and B). To detect the specificity of the CaNAC2c silencing, we tested the transcript levels of CaNAC07, CaNAC08, and CaNAC59 that belong to different NAC subfamily in pepper genome, and found that the silencing of CaNAC2c did not reduce the transcript levels of the tested genes compared to the mock treatment(Supplemental Figure S5), indicating the specificity of CaNAC2c silencing. Upon exposure to HTS, CaNAC2c-silenced plants exhibited decreased basal and acquired thermotolerance, as evidenced by a pronounced wilting phenotype and high mortality rates (Figure 1, A and B). CaNAC2c-silenced plants also displayed increased ion leakage in response to HTS, as measured by conductivity, relative to control plants transformed with the empty pTRV vector (TRV:00, Supplemental Figure S4, C and D). In addition, DAB and NBT staining revealed much higher levels of H2O2 and ROS accumulation, respectively, in the leaves and stems of CaNAC2c-silenced plants compared to the control plants (Supplemental Figure S4, E and F). In agreement with these physiological responses, CaNAC2c-silenced plants were more affected by HTS than control plants, as seen by lower Fv/Fm and photosystem II (PSII) photochemical efficiency in the light (фPSII), indicator of thermotolerance and thermostability of the photosynthetic apparatus, respectively (Yan et al., 2008; Wang et al., 2014; Guan et al., 2018) immediately after HTS (Figure 1, C and D). In a complementary approach, we generated N. benthamiana lines overexpressing CaNAC2c (CaNAC2c-OE). We generated transgenic N. benthamiana plants of 35S:CaNAC2c, a total of eight T0 plants were acquired, by strict self-pollination, their corresponding T1 and T2 lines were acquired. Two T2 lines, CaNAC2c-OE1# and CaNAC2c-OE2#, with high levels of CaNAC2c transcripts, were selected for further assay. In contrast to pepper plants silenced for CaNAC2c, these N. benthamiana CaNAC2c-OE plants were more tolerant to HTS compared to wild type (Supplemental Figure S6 and S7), indicating that CaNAC2c acts as a positive regulator in basal and acquired thermotolerance. To evaluate the role of CaNAC2c in the regulation of thermotolerance in different pepper germplasms, we silenced CaNAC2c comparatively in GZN13 (a heat-sensitive inbred line), HN42, and 79 (an inbred line with high level of thermotolerance), we found that these silencings all significantly reduced thermotolerance of the three pepper lines (Supplemental Figure S8).
Figure 1.
Phenotypes associated with CaNAC2c silencing in pepper plants and overexpression in N. benthamiana plants on thermotolerance and resistance to RSI (R. solanacearum infection). A, CaNAC2c-silenced pepper plants display reduced basal (BT) and acquired (AT) thermotolerance (pretreated with 37°C for 12 h and recovery for 12 h). B, Plant mortality (24 plants were calculated) exposed to HTS from 1 to 42 h post-treatment (hpt). C, Fv/Fm and △F/Fmʹ in leaves of CaNAC2c-silenced pepper plants challenged with HTS. D, Fv/Fm and △F/Fmʹ in leaves of CaNAC2c-silenced pepper plants challenged with HTS by dipping half of the leaf in water set to 42°C. E, Increased resistance of N. benthamiana plants overexpressing CaNAC2c to RSI relative to control plants. F and G, Simultaneous silencing of CaNAC2c and CaNAC2d produces an additive decrease in the resistance of pepper plants to RSI (24 plants were calculated for disease index). H, Height of CaNAC2c-silenced and control pepper plants. I, Root length of CaNAC2c-silenced and control pepper plants. J, Overall morphology of CaNAC2c-silenced and control pepper plants 35 d after sowing. In E, F, and J, the images were digitally extracted. In (C, D, G, H, and I), data presented are means ± standard error (SE) of four replicates, different capital letters indicate significant differences among means (P < 0.01), as calculated with Fisher’s protected LSD test.
To investigate the function of CaNAC2c in response to RSI, we inoculated TRV control plants and CaNAC2c-silenced pepper plants with R. solanacearum, but observed no differences (Data has been exposed to CNGB: https://db.cngb.org/search/?q=CNP0001104). By contrast, we noticed clear wilting symptoms in N. benthamiana plants challenged with RSI at 7 dpi (days post-inoculation), whereas N. benthamiana CaNAC2c-OE plants exhibited only slight wilting symptoms, as shown by the lower disease index values and lower R. solanacearum titer (Figure 1, E and Supplemental Figure S9). The silencing of CaNAC2c did not result in obvious symptoms upon RSI, while its overexpression in N. benthamiana plants did promote immunity against RSI, suggesting that CaNAC2c may function redundantly with other factors in response to RSI. As CaNAC2c was highly related to CaNAC2d and both genes were upregulation by RSI (Supplemental Figure S2), we hypothesized that they may act redundantly. To test this hypothesis, we silenced both CaNAC2c and CaNAC2d in pepper and measured the response of these plants to RSI and HTS. Although the silencing of CaNAC2d alone did not significantly affect thermotolerance or immunity against RSI, the combined silencing of CaNAC2c and CaNAC2d produced clear wilting symptoms as early as 3 dpi and reaching the highest disease index value at 8 dpi, while plants silenced for either gene individually behaved as control plants (Figure 1, F and G and Supplemental Figure S10). Collectively, these results indicate that CaNAC2c and CaNAC2d function redundantly in pepper immunity against RSI.
Notably, CaNAC2c-silenced plants grew larger than the control plants: CaNAC2c-silenced plants were taller, had longer roots, stems, and leaves, produced more leaves and flowers (Figure 1, H–J and Supplemental Figure S11). While lowering CaNAC2c expression improved plant fitness, raising CaNAC2c expression had the opposite effect, as seen in N. benthamiana CaNAC2c-OE plants and their smaller leaves, shorter roots and stems, fewer flowers, and leaves (Supplemental Figure S12), supporting the role of CaNAC2c as a negative regulator of plant growth.
CaNAC2c-mediated resistance to RSI is repressed upon HTS by ABA signaling
To better understand how CaNAC2c enhances thermotolerance and improves resistance to RSI, we measured relative transcript levels for a number of marker genes in CaNAC2c-silenced pepper plants, N. benthamiana CaNAC2c-OE plants upon HTS, and in the leaves of pepper plants transiently overexpressing CaNAC2c (CaNAC2c-TO). CaNAC2c silencing was accompanied by the significant down-regulation of the thermotolerance marker genes Heat Shock Protein 24 (CaHSP24) and CaHSP70, as well as Heat Shock Factor B2a (CaHSFB2a; Ashraf et al., 2018; Supplemental Figure S13, A). In contrast, relative transcript levels of thermotolerance-related N. benthamiana genes ASCORBATE PEROXIDASE (NbAPX), NbHSP18, and NbsHSP rose in CaNAC2c-OE in N. benthamiana plants relative to control plants (Supplemental Figure S6, D), and their upregulation was amplified by HTS.
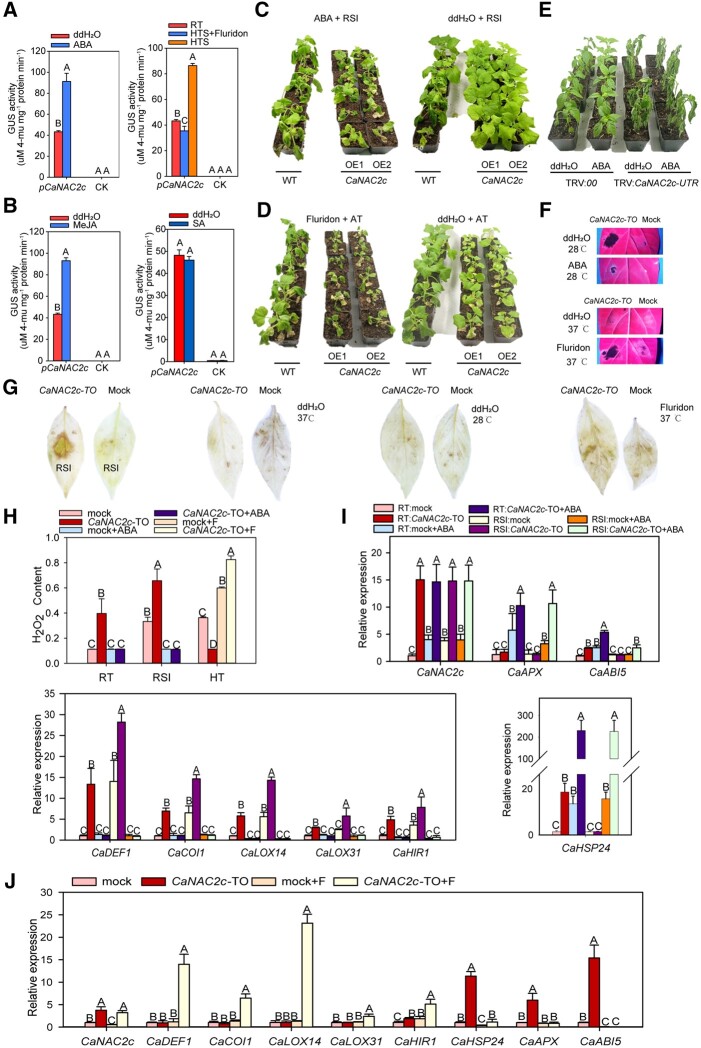
Furthermore, by exogenous application of ABA (or Furidon), SA, or JA, whose success was confirmed by transcript levels of CaABR1 (Choi and Hwang, 2011), CaPR1 (Kim and Hwang, 2014), and CaCOI1 (Hu et al., 2013), respectively (Supplemental Figure S14), the association of CaNAC2c to signaling mediated by ABA, SA, or JA was assayed. We established that transcription of CaNAC2c was upregulated both by an exogenous application of abscisic acid (ABA) and by HTS, based on b-glucuronidase (GUS) activity from a CaNAC2c pro:GUS reporter construct. Notably, the higher transcription rate of CaNAC2c in response to HTS was blocked by treatment of fluridon, an inhibitor of ABA biosynthesis (Figure 2, A). These data suggest that the role of CaNAC2c in promoting thermotolerance is regulated by ABA signaling. In addition, CaNAC2c transcription was upregulated by exogenously applied methyl jasmonate (MeJA) but not by that of salicylic acid (SA; Figure 2, B). To test the possible regulation of ABA signaling in immunity and thermotolerance mediated by CaNAC2c, we detected the effect of exogenously applied ABA and fluridon on response CaNAC2c overexpressing Nicotiana benthamiana plants to RSI and to HTS, respectively, we found that exogenous application of ABA significantly increased the susceptivity of CaNAC2c overexpressing Nicotiana benthamiana plants to RSI (Figure 2, C), and the exogenous application of fluridon significantly reduced the tolerance of Nicotiana benthamiana plants to HTS (Figure 2, D), consistently, the exogenous application of ABA did not restore thermotolerance reduced by CaNAC2c silencing in pepper plants (Figure 2, E) indicating that ABA signaling acts positively in thermotolerance but negatively in immunity against RSI, and CaNAC2c might act downstream ABA signaling.
Figure 2.
CaNAC2c Resistance to RSI is Repressed upon HTS. A and B, Activity of the CaNAC2cpro:GUS reporter to the application of SA (2 mM), ABA (30 μM), MeJA (10 μM), fluridon (10 μM) or HTS. Agrobacterium cells carrying the CaNAC2cpro:GUS reporter were infiltrated into pepper plants leaves, and GUS activity measured after HTS, fluridon, or phytohormones at 24 hpt (CK: pepper plants leaves infiltrated with GV3101 cells carrying pMDC-163). C, Effect of exogenous application of ABA on-resistance of CaNAC2c overexpressing N. benthamiana plants to Ralstonia solanacearum infection. D, Effect of the exogenously applied fluridon on acquired thermotolerance of CaNAC2c overexpressing N. benthamiana plants. E, Effect of exogenous application of ABA on thermotolerance of CaNAC2c-silenced pepper plants. F, HR-like cell death lesions at room temperature or under HTS triggered by transient overexpression of CaNAC2c or exogenous application of fluridon or ABA. G, H2O2 accumulation, displayed by DAB staining in CaNAC2c transiently overexpressing pepper leaves challenged with RSI, HTS, or with fluridon under HTS. H, H2O2 accumulation in CaNAC2c transiently overexpressing pepper leaves challenged with RSI, ABA, HTS, or with fluridon under HTS (F: fluridon). I, Transcript level of CaDEF1, CaCOI1, CaLOX14, CaLOX31, CaHIR1, CaHSP24, CaAPX, or CaABI5 in CaNAC2c transiently overexpressing pepper leaves challenged with RSI, ABA, or with ABA under RSI. J, Transcript level of CaDEF1, CaCOI1, CaLOX14, CaLOX31, CaHIR1, CaHSP24, CaAPX, or CaABI5 in CaNAC2c transiently overexpressing pepper leaves challenged with HTS or with fluridon under HTS (F: fluridon). In C–E, the images were digitally extracted. Data presented are means ± standard error (SE) of four replicates. In A, B, H, I, and J, different capital letters indicate significant differences among means (P < 0.01), as calculated with Fisher’s protected LSD test.
We also discovered that CaNAC2c-TO triggered HR-like cell death (Figure 2, F) at room temperature, resulting in the higher level of H2O2 accumulation upon RSI (Figure 2, G), which was also revealed by DAB staining, but this response was abolished by exogenous ABA treatment (Figure 2, F–H). In contrast, at 37°C, CaNAC2c-TO did not induce any cell death but reduced the accumulation of H2O2, although an exogenous application of fluridon restored cell death (Figure 2, F and G). These results suggest that ABA may prevent the activation of cell death induced by CaNAC2c at 37°C, and CaNAC2c-mediated resistance to RSI is repressed by ABA signaling at high temperature.
Turning to pepper plants transiently overexpressing CaNAC2c, JA signaling dependent gene DEFENSIN 1 (CaDEF1; Choi et al., 2008; Germain et al., 2012; Choi et al., 2015; Kim et al., 2015; Zhang et al., 2018), CaCoI1 (Hu et al., 2013), two lipoxygenase genes CaLOX14 (Sarde et al., 2018), CaLOX31 (Sarde et al., 2019) with the most obvious response to Ralstonia solanacearum infection in pepper by data of our RNA-seq (Supplemental Figure S15) as well as by that of Dang et al. (2013), which encode enzyme crucial for JA biosynthesis, a hypersensitive response (HR)-related gene hypersensitive-induced reaction 1 (CaHIR1; Jung et al., 2008) were all upregulated by RSI as well as by overexpression of CaNAC2c at room temperature, but these upregulations were blocked by the exogenous application of ABA (Figure 2, I), while CaHSP24 that is related to thermotolerance, CaAPX (Wang et al., 2017) that is related to scavenger of ROS including H2O2, and ABA signaling related CaABI5 (Lopez-Molina et al., 2002) were similarly upregulated by overexpression of CaNAC2c in the absence of R. solanacaerum at room temperature, and these upregulations were potentiated by exogenous application of ABA (Figure 2, I). The upregulation of CaLOX14 by overexpression of CaNAC2c upon RSI was much higher than that of CaLOX31, indicating that the biosynthesis of JA upon RSI is mainly determined by CaLOX14. Upon HTS plus exogenous fluridon, the overexpression of CaNAC2c failed to upregulate CaHSP24, CaAPX, or CaABI5, but upregulated all of the tested immunity-related marker genes (Figure 2, J). All these data suggest that ABA signaling positively regulates thermotolerance but negatively regulates immunity against RSI.
Consistently, the thermotolerance related CaHSP24, CaHSP70, and CaHSFB2a were not downregulated by silencing of CaNAC2d, their transcript levels in CaNAC2c silencing were similar to that in CaNAC2c and CaNAC2d simultaneously silenced pepper plants (Supplemental Figure S13, B). In contrast, transcript levels of CaDEF1 were not downregulated by silencing of either CaNAC2c or CaNAC2d, but were significantly decreased by simultaneous silencing of CaNAC2c and CaNAC2d, indicating that CaNAC2c and CaNAC2d function redundancy in pepper immunity against RSI mediated by JA signaling.
CaHSFA5 is directly targeted by CaNAC2c upon HTS but not RSI
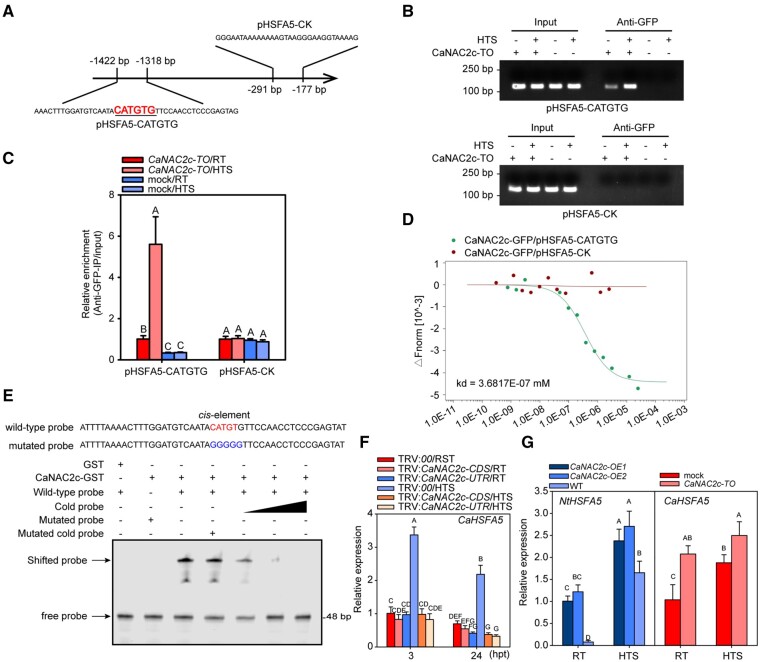
When we performed a co-expression analysis in Pepper Hub (http://www.hnivr.org/; Liu et al., 2017), we noticed that CaHSFA5 was co-expressed with CaNAC2c under HTS (Supplemental Table S4). To test if CaHSFA5 is a target of CaNAC2c, we determined whether CaNAC2c would bind to the CaHSFA5 promoter by chromatin immunoprecipitation followed by PCR or qPCR (ChIP-PCR or ChIP-qPCR; Figure 3, A). Indeed, we amplified a fragment of the CaHSFA5 promoter with a specific primer pair flanking a predicted NAC TF binding site, but not with a control primer pair directed at a region free of NAC TF binding site, indicating that CaNAC2c directly targets CaHSFA5 (Figure 3, B and C). Moreover, HTS significantly enhanced the binding of CaNAC2c to the CaHSFA5 promoter fragment (Figure 3, B and C). We validated the binding of CaNAC2c to its putative binding site in the CaHSFA5 promoter in vitro by microscale thermophoresis (MST) assay (Figure 3, D). Likewise, recombinant CaNAC2c-GST was able to bind the CaHSFA5 promoter in electrophoretic mobility shift assays (EMSA), via the cis-element CATGTG, as mutating it to GGGGGG prevented binding (Figure 3, E). As expected from their co-expression, CaHSFA5 transcript levels decreased when CaNAC2c was silenced in pepper plants challenged with HTS, and was upregulated by the transient overexpression of CaNAC2c. Similarly, NbHSFA5, the presumptive CaHSFA5 ortholog in N. benthamiana, was up-regulated in N. benthamiana CaNAC2c-OE plants (Figure 3, F and G). These results indicate that CaHSFA5 is directly and positively regulated by CaNAC2c upon HTS.
Figure 3.
CaHSFA5 Is a Direct Transcriptional Target of CaNAC2c upon HTS. A, Schematics of the CaHSFA5 promoter, with the CATGTG-containing fragment and a CATGTG-free fragment highlighted for ChIP-PCR of CaHSFA5 by CaNAC2c. B, CaNAC2c directly targets the CaHSFA5 promoter, as shown by ChIP-PCR. Chromatin was isolated from pepper leaves transiently overexpressing CaNAC2c-GFP, sheared into 300–500 bp fragments. The DNA was immunoprecipitated with antibodies against GFP, and the purified DNA was used as a template with specific primer pair of CATGTG-containing promoter region. C, CaNAC2c shows enhanced binding to the CaHSFA5 promoter under HTS, as seen by ChIP-qPCR. D, MST analysis of CaNAC2c binding to the CaHSFA5 promoter, using CaNAC2c-GFP fusion protein transiently overexpressed in pepper leaves and immunoprecipitated with anti-GFP antibody and a CATGTG-containing CaHSFA5 promoter fragment. E, EMSA analysis of CaNAC2x binding to the CaHSFA5 promoter, with recombinant CaNAC2c-GST and a CaHSFA5 promoter fragment containing CATGTG or its mutant version (GGGGGG). F, CaHSFA5 is downregulated by silencing CaNAC2c under HTS. G, CaHSFA5 is upregulated in N. benthamiana plants overexpressing CaNAC2c or by the transient overexpression of CaNAC2c in pepper leaves. In C, F, and G, data presented are means ± standard error (SE) of four replicates, different capital letters indicate significant differences among means (P < 0.01), as calculated with Fisher’s protected LSD test.
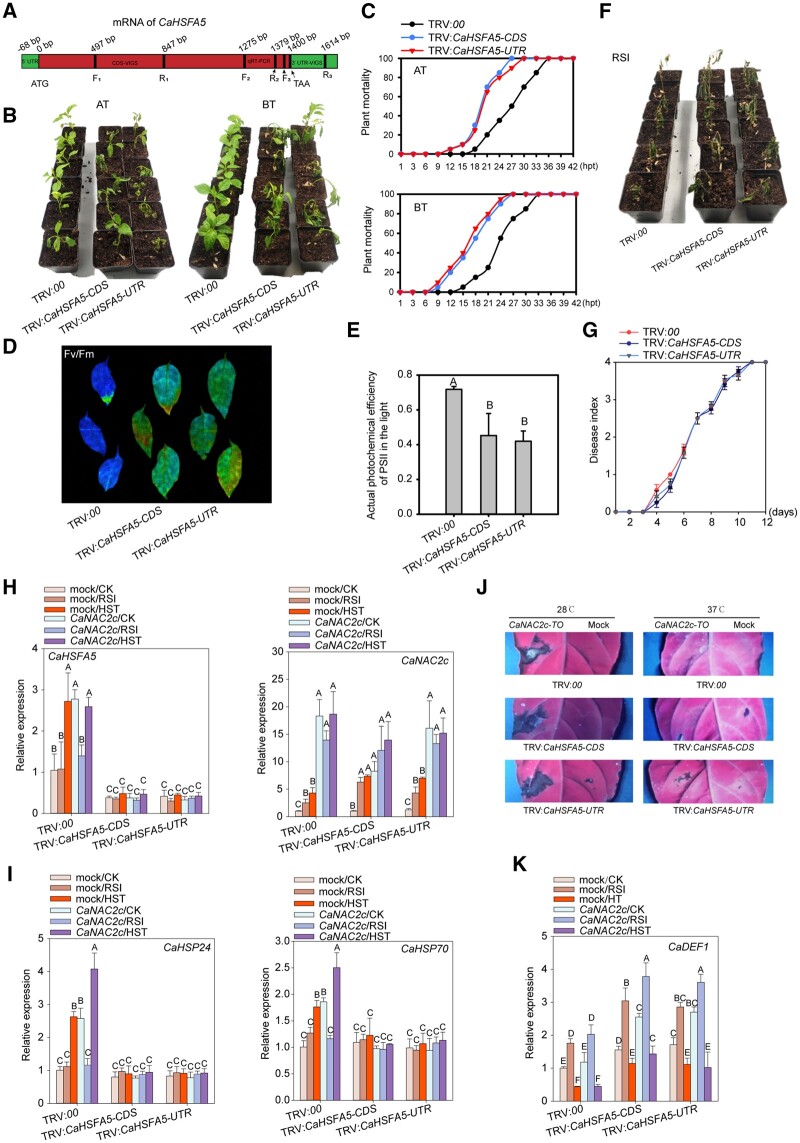
To assay the role of CaHSFA5 in pepper thermotolerance and immunity against RSI, we generated CaHSFA5-silenced pepper plants via VIGS (Figure 4, A). Similar to CaNAC2c, CaHSFA5 acted as a negative regulator of plant growth, as CaHSFA5-silenced plants had larger leaves, as well as longer stems and roots (Supplemental Figure S16). In addition, CaHSFA5-silenced plants were more sensitive to heat stress compared to control plants, with or without pre-treatment with a nonlethal HTS, as evidenced by the higher mortality rates and lower Fv/Fm and △F/Fmʹ (Figure 4, B–E). Upon monitoring the expression of marker genes, we observed that silencing of CaHSFA5 blocked the induction of CaHSP24 and CaHSP70 expression by HTS but did not affect CaDEF1, indicating that CaHSFA5 acts as a positive regulator specifically in thermotolerance (Figure 4, I and K). As might be expected from the normal transcript levels of CaDEF1, the silencing of CaHSFA5 did not change the resistance of pepper plants to RSI (Figure 4, F and G).
Figure 4.
Silencing CaHSFA5 Decreases Thermotolerance in Pepper Plants but does not Affect Resistance to RSI. A, Diagram of the fragments in the coding sequence (CDS) or 3ʹ-untranslated region (UTR) of CaHSFA5 used for VIGS (TRV:CaHSFA5-CDS and TRV:CaHSFA5-3ʹUTR). B and C, CaHSFA5 silencing lowers basal and acquired thermotolerance of pepper plants (24 plants were calculated for mortality). D, CaHSFA5 silencing decreases Fv/Fm of pepper leaves upon HTS. E, CaHSFA5 silencing decreases △F/Fm of pepper leaves upon HTS. F, CaHSFA5 silencing does not affect the resistance of pepper plants to RSI. G, CaHSFA5 silencing does not affect the disease index values of pepper plants upon RSI (24 plants were calculated). H, Successful silencing of CaHSFA5 and overexpression of CaNAC2c. I, CaHSFA5 silencing significantly decreases the expression of thermotolerance-related genes CaHSP24 and CaHSP70 in pepper plants. J, HR-like cell death induced by CaNAC2c-TO at room temperature is not influenced by the silencing of CaHSFA5, and CaNAC2c-TO does not induce cell death at 37°C. K, CaHSFA5 does not affect the induction of CaDEF1 expression by CaNAC2c. In A and F, the images were digitally extracted. Data presented are means ± standard error of four replicates. In C, the data were counted based on 24 pepper plants. In C, E, G, H, I, and K, data presented are means ± standard error (SE) of four replicates, different capital letters indicate significant differences among means (P < 0.01), as calculated with Fisher’s protected LSD test.
To further characterize the role of CaHSFA5 in relation to CaNAC2c, we targeted NbHSFA5, the CaHSFA5 ortholog in N. benthamiana by VIGS. The silencing of NbHSFA5 in N. benthamiana largely rescue the growth retardation (Supplemental Figure S17) displayed by N. benthamiana plants overexpressing CaNAC2c, but at the same time lowered their thermotolerance (Supplemental Figure S18, B and D). However, resistance to R. solanacearum infection was not affected (Supplemental Figure S18, E and F).
CaNAC2c interacts with CaHSP70 under HTS but interacts with CaNAC029 upon RSI
The distinct behavior of CaNAC2c upon HTS or RSI suggested that its function might be modulated by additional regulatory proteins in a context-dependent manner. To isolate these possible interacting partners, we performed a GST pull-down followed by mass spectrometry identification. Accordingly, we incubated recombinant CaNAC2c-GST produced in E. coli with protein extracts from pepper plants challenged with HTS or RSI. All pulled-down proteins were identified by mass spectrometry, revealing a number of proteins that potentially interacted with CaNAC2c under both conditions (Supplemental Table S5). After preliminary screening by Bimolecular Fluorescent Complimentary (BiFC) assay, we selected an HSP70-type protein (XP_016569463.1) which might interact with CaNAC2c specifically under HTS and the NAC-type TF CaNAC029 (XP_016569463.1) which interact with CaNAC2c specifically upon RSI from these potential interacting partners for further analysis. As CaNAC2c regulates thermotolerance in ABA-signaling but mediate pepper immunity against RSI in JA-signaling dependent manner, we tested the response of CaNAC029 and CaHSP70 to the exogenous application of MeJA, SA, or ABA, the result showed that CaNAC029 responded specifically to exogenously applied MeJA, while CaHSP70 responded specifically to exogenous application of ABA(Supplemental Figure S19), indicating that the interactions of CaNAC2c with these two partners might play specific roles in biological processes mediated by JA and ABA signaling, respectively.
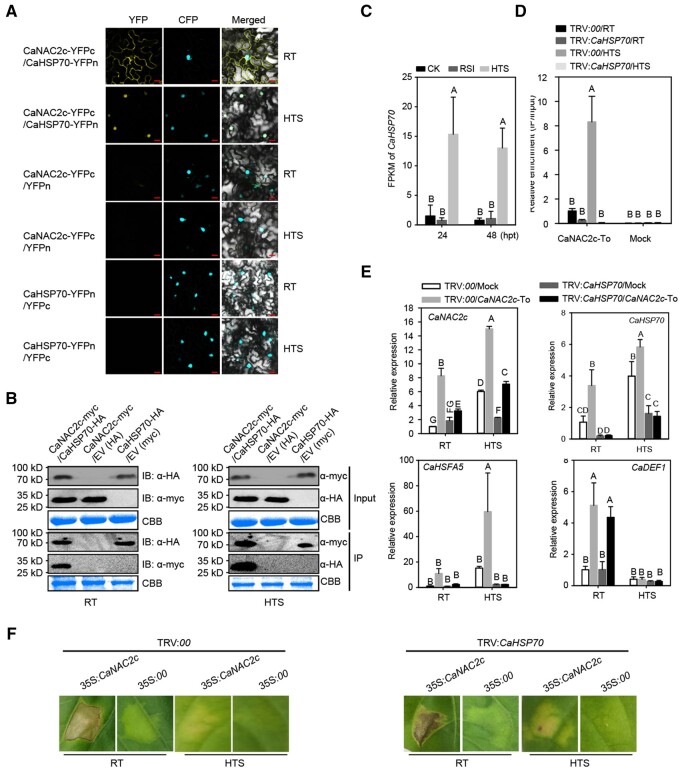
CaHSP70 interacts with CaNAC2c in the nucleus upon HTS to enhance CaNAC2c-mediated thermotolerance but represses immunity to RSI
We established by BiFC assay that CaHSP70 interacts with CaNAC2c in the cytoplasm at room temperature but in the nucleus when exposed to HTS (Figure 5, A). To validate this result, we performed co-immunoprecipitation (co-IP) assays using CaHSP70-HA immunoprecipitated from pepper leaves transiently co-overexpressing CaNAC2c-Myc and CaHSP70-HA with an anti-HA antibody. We tested for the presence of CaNAC2c in the immunoprecipitated by immunoblot analysis with an anti-MYC antibody. CaNAC2c and CaHSP70-HA interacted under both room temperature and high temperature (Figure 5, B). CaHSP70 expression was up-regulated by HTS but not by RSI (Figure 5, C). This result suggested that CaHSP70 may be involved in CaNAC2c-mediated thermotolerance. As mentioned earlier, CaHSFA5 is one of the downstream targets of CaNAC2c for mounting tolerance to high-temperature exposure. We, therefore, examined the effects associated with the overexpression of CaHSP70 on CaNAC2c-mediated transcriptional activation of CaHSFA5 by ChIP-qPCR. We observed that the enrichment of CaNAC2c at the CaHSFA5 promoter increased, as did CaHSFA5 transcript levels, when CaHSP70 was transiently co-overexpressed with CaNAC2c (Supplemental Figure S20), but were drastically reduced when CaHSP70 was silenced in pepper plants (Figure 5, D and E). We conclude that CaHSP70 plays a key role in ameliorating heat tolerance by upregulating CaHSFA5 transcript levels via CaNAC2c binding to the CaHSFA5 promoter.
Figure 5.
The interaction between CaNAC2c and CaHSP70 and its effect on transcription of immunity or thermotolerance-related genes by CaNAC2c. A, BiFC confirmation of the interaction between CaNAC2c and CaHSP70 in N. benthamiana leaves infiltrated with Agrobacterium cells bearing CaNAC2c-YFPC+CaHSP70-YFPN or CaNAC2c-YFPN+CaHSP70-YFPC constructs, NbH3 (histone H3)-CFP was used to indicate the nucleus. Cyan fluorescence and yellow fluorescence, visible light, and merged images were taken on a confocal microscope at 48 hpi. Bars = 25 μm. B, Interaction between CaNAC2c and CaHSP70 in vivo, as determined by Co-IP assay. Proteins were isolated from pepper leaves transiently overexpressing CaNAC2c-Myc and CaHSP70-HA, and CaNAC2c-Flag and its interacting partners were immunoprecipitated with antibody of Flag, the presence of CaHSP70 in the protein complex was assayed by western blotting using antibody of HA. C, Fragments per Kilobase Million (FPKM) of CaHSP70 in pepper plant challenged with HTS or RSI based the RNA-seq data set. D, The effect of CaHSP70 silencing on its deposition on the promoter of CaHSFA5 by ChIP-qPCR. E, The effect of CaHSP70 silencing on the regulation of CaHSFA5 and CaDEF1 by transient overexpression of CaNAC2c under room temperature and upon HTS challenge by RT-qPCR. F, The effect of CaHSP70 silencing on the HR cell death triggered by transient overexpression of CaNAC2c under room temperature and upon HTS challenge. In C, D, and E, data are shown as means ± standard error of four replicates. Different capital letters above the bars indicate significant differences between means (P < 0.01) by Fisher’s protected least-significant-difference (LSD) test.
We then aimed to determine the contribution, if any, of CaHSP70 to immunity against RSI. Notably, the HR-like cell death phenotype triggered by the transient overexpression of CaNAC2c was not affected when silencing CaHSP70 (Figure 5, F). Although CaDEF1 transcript levels were significantly reduced by the transient co-overexpression of CaHSP70 and CaNAC2c at room temperature and high temperature (Supplemental Figure S20, B), they were not affected by the silencing of CaHSP70 when CaNAC2c was transiently overexpressed (Figure 5, E). These data collectively indicate that CaHSP70 interacts with CaNAC2c in the nucleus upon HTS, thereby enhancing CaNAC2c activity toward activating thermotolerance but also repressing its function as a regulator of plant immunity.
CaNAC029 interacts with CaNAC2c to promote the regulation of CaNAC2c in pepper immunity but not thermotolerance
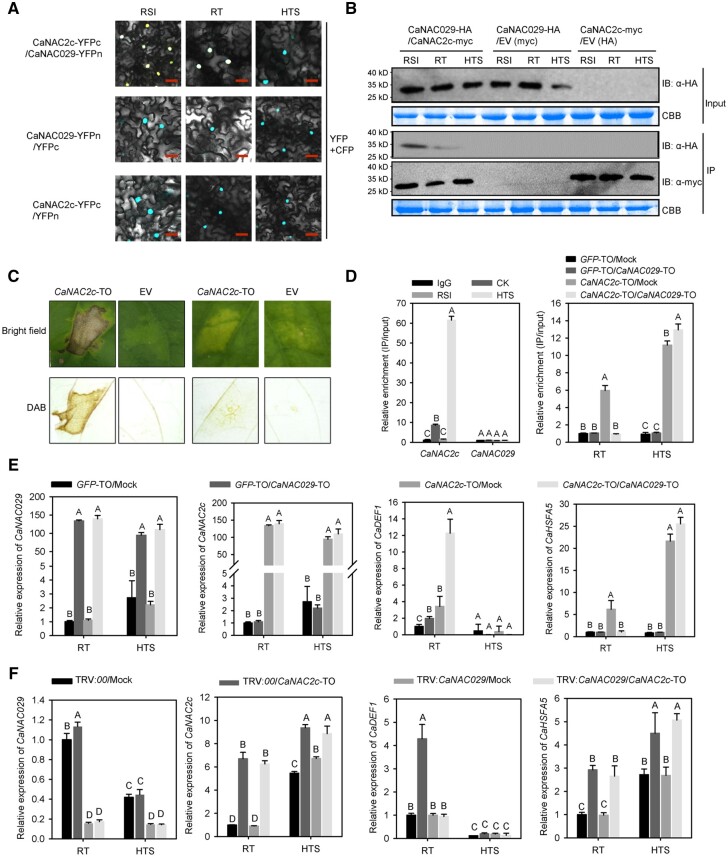
We performed similar BiFC and co-IP assays to validate the interaction between CaNAC2c and CaNAC029: CaNAC2c interacted with CaNAC029 under room temperature and upon RSI in the nucleus, but not upon HTS (Figure 6, A and B). Based on co-IP, CaNAC2c appeared to interact with CaNAC029 quantitatively more upon RSI than at room temperature, while we detected no interaction upon HTS (Figure 6, B).
Figure 6.
The interaction between CaNAC2c and CaNAC029 and its effect on transcription of immunity or thermotolerance related genes by CaNAC2c. A, BiFC confirmation of the interaction between CaNAC2c and CaNAC029 in N. benthamiana leaves infiltrated with Agrobacterium cells bearing CaNAC2c-YFPC+CaNAC029-YFPN or CaNAC2c-YFPN+CaNAC029-YFPC constructs, NbH3 (histone H3)-CFP was used to indicate the nucleus. Cyan fluorescence and yellow fluorescence, visible light, and merged images were taken on a confocal microscope at 48 hpi. Bars = 25 μm. B, Interaction between CaNAC2c and CaHSP70 in vivo, as determined by Co-IP assay. Proteins were isolated from pepper leaves transiently overexpressing CaNAC2c-Myc and CaNAC029-HA, and CaNAC2c-Flag and its interacting partners were immunoprecipitated with antibody of Flag, the presence of CaNAC029 in the protein complex was assayed by western blotting using antibody of HA. C, The effect of CaNAC029 silencing on the HR cell death triggered by transient overexpression of CaNAC2c under room temperature and upon HTS challenge. D, CaNAC029 cannot bind to the promoter of CaHSFA5 upon RSI or HTS, and co-transient overexpression of CaNAC029 and CaNAC2c inhibit the binding of CaNAC2c to CaHSFA5 promoters at room temperature. E, The effect of transient overexpression of CaNAC029 on the regulation of CaHSFA5 and CaDEF1 by CaNAC2c by RT-qPCR. F, The effect of CaNAC029 silencing on the regulation of CaHSFA5 and CaDEF1 by transient overexpression of CaNAC2c under room temperature and upon HTS challenge by RT-qPCR. In D, E, and F, data are shown as means ± standard error of four replicates. Different capital letters above the bars indicate significant differences between means (P < 0.01) by Fisher’s protected least-significant-difference (LSD) test.
We then explored the consequences of the interaction between CaNAC2c and CaNAC029 on the function of CaNAC2c in the induction of HR-like cell death, which play important roles in plant immunity against the pathogen (Jones and Dangl, 2006). To this end, we transient overexpressed CaNAC2c in the leaves of pepper plants with normal levels of CaNAC029 (wild-type controls) or silenced for CaNAC29. The transient overexpression of CaNAC2c triggered a strong HR-like cell death response in wild-type leaves but not when CaNAC029 was silenced (Figure 6, C), indicating that CaNAC2c positively regulates pepper immunity in a CaNAC029-dependent manner. To test the effect of the transient overexpression of CaNAC029 on the targeting and regulation of the immunity-related gene CaDEF1 by CaNAC2c, we performed RT-qPCR by transiently co-overexpressing CaNAC2c and CaNAC029 or transiently overexpressing CaNAC2c alone in pepper leaves. A ChIP-qPCR assay showed that CaNAC029 failed to bind to the CaHSFA5 promoter under RT, RSI, or HTS. In addition, overexpression of CaNAC029 prevented the binding of CaNAC2c to the CaHSFA5 promoter specifically at room temperature but supported full deposition of CaNAC2c at the CaHSFA5 promoter upon HTS (Figure 6, D). The transient co-overexpression of CaNAC029 and CaNAC2c induced CaDEF1 expression to higher levels than CaNAC2c alone at room temperature, but this phenomenon was abolished upon HTS (Figure 6, E). In contrast, CaNAC029 silencing significantly repressed the upregulation of CaDEF1 by transient overexpression of CaNAC2c at room temperature but did not affect the positive regulation of CaHSFA5 by CaNAC2c either at room temperature or upon HTS (Figure 6, F). In addition, transient overexpression of CaNAC029 inhibited the increase of CaHSFA5 transcription induced by CaNAC2c overexpression, but the inhibition was abolished at high temperature. These data indicate that the interaction between CaNAC2c and CaNAC029 promotes the control of gene expression mediated by CaNAC2c in pepper immunity but not in thermotolerance.
The 26S proteasome degrades CaNAC029 to direct CaNAC2c toward thermotolerance under HTS
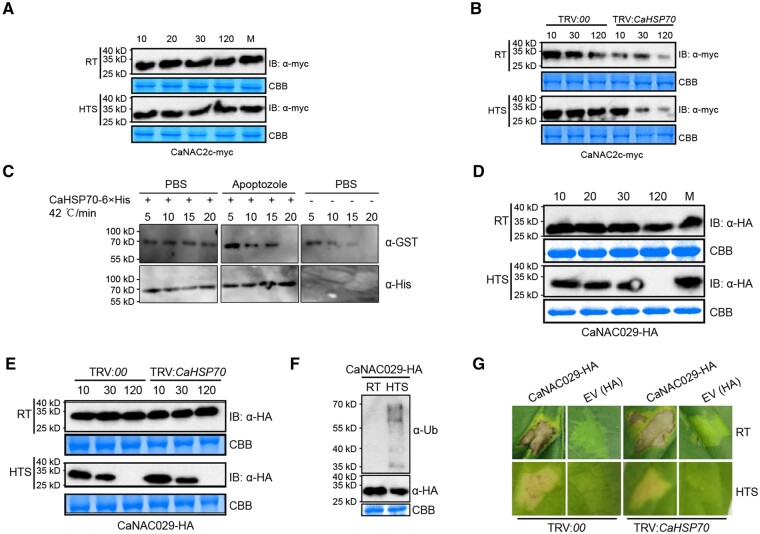
We characterized protein stability for CaNAC2c and CaNAC29 under our experimental conditions. To our surprise, CaNAC2c-Myc remained stable under all conditions tested, including challenge by HTS (Figure 7, A). Unlike CaNAC2c, CaNAC029-HA was stable at room temperature and upon RSI, but was degraded within 2 h upon HTS (Figure 7, D). The 26S proteasome inhibitor MG132 blocked the degradation of CaNAC029 upon HTS, suggesting the involvement of ubiquitin modification and the 26S proteasome (Figure 7, D). Indeed, an immunoblot analysis with an antibody against ubiquitin revealed a ladder-like pattern upon HT, consistent with degradation through ubiquitination (Figure 7, F). Even though CaNAC2c was not degraded in response to HTS in control plants, the silencing of CaHSP70 significantly reduced the stability of CaNAC2c (Figure 7, B). Consistently, we tested the effect of the addition of prokaryotic expressed CaHSP70-6×His on the degradation of CaNAC2c-GST at 42°C, we found that the addition of CaHSP70-6×His significantly blocked the degradation of CaNAC2c-GFP (Figure 7, C). In contrast, silencing CaHSP70 did not affect the degradation of CaNAC029 upon HTS (Figure 7, E), nor did it affect the HR-like cell death caused by transient overexpression of CaNAC029 in leaves (Figure 7, G). These data indicate that CaHSP70 maintains the stability of CaNAC2c upon HTS but not CaNAC029. During heat shock, the rapid elimination of CaNAC029 through the 26S proteasome will quickly target CaNAC2c to the CaHSFA5 promoter and initiate thermotolerance responses.
Figure 7.
Effect of CaHSP70 silencing on HR-like cell death and immunity and expression of thermotolerance-related genes induced by transient overexpression of CaNAC029. A, Stability of CaNAC2c-myc in pepper plants transiently overexpressing CaNAC2c-myc at room temperature or upon HTS. Time since 10 is shown in min. B, Effect of CaHSP70 silencing on the stability of CaNAC2c in pepper plants at room temperature or upon HTS. C, CaNAC2c was protected from degradation by CaHSP70 through in vitro assay using specific inhibitors (200 μM Apoptozole) of CaHSP70. D, Stability of CaNAC029-HA in pepper plants transiently overexpressing CaNAC029-HA at room temperature or upon HTS. Time since 10 is shown in min. E, Effect of CaHSP70 silencing on the stability of CaNAC29-HA in pepper plants transiently overexpressing CaNAC20-HA at room temperature or upon HTS. F, Immunoblot analysis of CaNAC29-HA in pepper plants transiently overexpressing CaNAC29-HA upon HTS with an anti-Ub antibody. G, Effect of CaHSP70 silencing on the HR-like cell death response triggered by the transient overexpression of CaNAC29 at room temperature and upon HTS.
Discussion
To maximize fitness, plants continuously balance their limited resources via diverse mechanisms leading to trade-offs between growth or responses to various stresses. Although TFs are crucial for plant responses to biotic and abiotic stresses, a single TF may be involved in regulating several seemingly disparate processes (Rushton et al., 2010; Nuruzzaman et al., 2013), making it difficult to identify the precise mode of action underlying the balance between plant growth and stress responses. In the present study, we establish that CaNAC2c acts as a negative regulator of pepper growth and as a positive regulator in defense response to HTS and RSI. Notably, whether CaNAC2c directs plant resources toward growth or response to HTS and RSI is dictated by differential and context-specific interactions with the heat shock protein CaHSP70 and the other NAC-type TF CaNAC029.
CaNAC2c acts positively in thermotolerance and in immunity against RSI but negatively in pepper growth
Our data demonstrated that upon HTS or RSI, CaNAC2c was transcriptionally upregulated, leading to enhanced basal and acquired thermotolerance, or enhanced resistance to RSI, depending on the stressor. At the same time, the induction of CaNAC2c also repressed growth, supporting a role for CaNAC2c as a negative regulator of pepper growth but as a positive regulator of thermotolerance and defense against RSI. This result is similar to our previous studies in which we had shown that CaWRKY6 (Cai et al., 2015), CaWRKY40 (Dang et al., 2013), and CabZIP63 (Shen et al., 2016) act positively in pepper responses to both RSI and HTS, supporting the notion that pepper responses to RSI are closely linked to HTS responses. CaNAC2c performed its distinct functions by differential targeting and transcriptional regulation of subsets of genes: for thermotolerance, CaNAC2c directly targeted and induced the expression of thermotolerance-related genes such as CaHSFA5 (Figures 3 and 4 and Supplemental Figure S16), CaHSP24, CaHSP70, and CaHSFB2a (Supplemental Figure S13, A). CaNAC2c also caused reduced H2O2 accumulation, which is associated with the degree of thermotolerance (Yu et al., 2019; Zhuang et al., 2020). In contrast, CaNAC2c acted as a positive regulator for immunity against RSI by upregulating CaDEF1 expression and enhancing the accumulation of H2O2 (Yoshioka et al., 2003), reflecting the different mechanisms behind heat stress response (HSR) and immune responses to pathogens. While CaNAC2c silencing did not affect pepper responses to RSI, this result is due to partial redundancy between CaNAC2c and the highly related CaNAC2d, as the simultaneous silencing of CaNAC2c and CaNAC2d decreased plant resistance to RSI. A comparable level of functional redundancy has been extensively described for immunity-associated genes such as bHLH-type TFs (Xu et al., 2014). That CaNAC2c acts alone in the context of thermotolerance but redundantly with other genes against pathogen attacks may stem from the varying selective pressure imposed by HTS and pathogen attack over the course of evolution. As plants and pathogens attempt to bypass each other’s defense mechanisms, plant immunity has frequently been overcome by new pathogen-derived effectors that target specific plant immune components such as TFs (Canonne et al., 2011). This apparent redundancy may therefore make the immune response more robust in the face of a constant arms race against pathogens, since the removal or inactivation of a subset of redundant components may be functionally compensated for by others.
The immunity-thermotolerance trade-off mediated by CaNAC2c is tuned by differential and context-specific interactions with CaHSP70 and CaNAC029
Our data showed that CaNAC2c targets and regulates CaHSFA5 upon HTS but regulates CaDEF1 when pepper plants are challenged by RSI, indicating that CaNAC2c performs functions in both processes by differential targeting via context-specific interactions with other regulatory proteins (Chi et al., 2013). We tested this hypothesis by isolating possible interacting partners during pepper response to HTS or RSI by pull-down assays followed by mass spectrometry and testing these new interactors by BiFC and co-IP assays against CaNAC2c. We determined that CaNAC2c may interact with either CaHSP70 or CaNAC029 depending on the context: upon HTS, CaNAC2c interacted with CaHSP70 in the nucleus, thereby protecting CaNAC2c from degradation, promoting the targeting of CaNAC2c to the CaHSFA5 promoter and preventing CaNAC2c from activating CaDEF1 (Figures 5 and 7 and Supplemental Figure S18). When pepper plants were challenged by RSI, CaNAC2c interacted with CaNAC029 in the nucleus, leading to enhanced transcript levels of CaDEF1, while the potential activation of HSR by CaNAC2c was blocked (Figure 6), similarly, GmNAC81 in soybean interact with GmNAC30, and this interaction determines the full activation or repression of target promoters (Mendes et al., 2013), we speculate that the heterodimerization of CaNAC2c and CaNAC029 may be required for the full regulation of gene expression by CaNAC2c. To confirm this hypothesis, further study is required to determine the target genes of CaNAC2c and CaNAC029 and to study the precise molecular details behind the regulation of CaNAC2c/CaNAC029 interaction on their transcription. However, we did not observe any interaction between CaNAC2c and CaNAC029, probably due to the degradation of CaNAC029 upon HTS by the 26S proteasome (Figure 6, A, B and 7). Based on these results, we concluded that the balance between pepper responses to HTS and RSI that is mediated by CaNAC2c is modulated by CaHSP70 and CaNAC029, respectively, in a context-specific manner. As HTS and pathogen attacks such as R. solanacearum are two frequently co-occurring stresses in subtropical or tropical climates where pepper originated from, this post-translational regulation might benefit rapid and precise switches between defense responses to different stresses. It is worth pointing out that a subset of regulatory proteins such as MYB (Zhang et al., 2020), xylem NAC domain 1 (XND1; Zhang et al., 2020), CDPKs (Vivek et al., 2016), the phosphatidylinositol 4-kinase PI4Kgamma5 (Yong et al., 2016), radical-induced cell death 1 (RCD1; O'Shea et al., 2015), C-repeat binding factor (CBF; Shan et al., 2014), the protein phosphatase regulator of CBF gene expression 2 (RCF2; Guan et al., 2014), ring-H2 (Greve et al., 2003), RCD1 (O'Shea et al., 2015), and crowded nuclei (CRWN1; Guo et al., 2017) have been shown to interact with NAC TFs, thereby altering their targeting specificities and transcriptional activities. In contrast, to date, neither HSP protein had been demonstrated to interact with a NAC TF, nor one NAC-type TF with another NAC protein to regulate plant immunity.
The growth/defense trade-off mediated by CaNAC2c is regulated at both transcriptional and post-transcriptional levels by interaction with CaHSP70
The different roles of CaNAC2c as negative regulator in pepper growth and positive regulator in thermotolerance and immunity against RSI indicate that CaNAC2c acts as a central regulator of the trade-off between pepper growth and stress responses. Growth-defense tradeoffs are essential for optimizing plant performance and adaptation under stress conditions (de Vries et al., 2017), recent advances in plant physiology and ecology suggest that this mechanism is more complex than just a resource trade-off (de Vries et al., 2017). Signaling mediated by SA (Meldau et al., 2012; Li et al., 2019; Nakagami et al., 2020) and JA (Guo et al., 2018; Howe et al., 2018) have been found in tradeoffs between growth and defense response to biotrophic and to necrotrophic pathogen or herbivore, respectively. CaNAC2c expression remains low but constitutive in the absence of stress, but is upregulated by RSI or HTS, indicating that the lower transcript levels of CaNAC2c are crucial for the evocation of pepper growth under normal conditions. In contrast, upon HTS or RSI, CaNAC2c transcription was upregulated, leading to the activation of distinct defense responses via differential targeting of the encoded CaNAC2c TF. In addition, we saw that CaNAC2c interacts with CaHSP70 in nonstressed pepper plants, but outside the nucleus (Figure 5, A), indicating a possible mechanism to titrate a TF away from its target defense-related genes to prevent their untimely activation (Moore et al., 2011). These data indicate that growth/defense trade-offs mediated by CaNAC2c are regulated at the transcriptional level and the post-translational level by interacting with CaHSP70. Since the overexpression of these TFs generally causes a growth penalty (Liu et al., 2018), it is plausible that plants have adopted reducible expression strategies for TFs involved in stress resistance (Cheng et al., 2018; Sun et al., 2019) to reduce the associated fitness cost. Since CaNAC2c was upregulated by exogenous application of MeJA or ABA and its expression change by overexpression altered the expression of JA- or ABA-signaling dependent marker genes(Figure 2, I and J), CaNAC029 and CaHSP70 were specifically upregulated by exogenous application of MeJA and ABA, respectively, and JA and ABA have been implicated in plant response to necrotrophic stage infection of R. solanacearum, a hemibiotrophic pathogen, and to heat stress, respectively (Kachroo et al., 2003; Hiruma et al., 2013; Huang et al., 2016), it can be speculated that the trade-off between pepper growth and immunity against RSI is mediated by JA signaling, the trade-off between pepper growth and thermotolerance is regulated by ABA signaling, and the balance between pepper immunity against RSI and thermotolerance mediated by CaNAC2c might be regulated by the antagonism between JA and ABA signaling (Robert-Seilaniantz et al., 2011; Kyndt et al., 2017).
Collectively, our data indicate that CaNAC2c acts positively in thermotolerance and immunity against R. solanacearum and negatively in pepper growth. Trade-offs between pepper growth and defense responses are determined by CaNAC2c transcription and CaNAC2c protein via its interaction with CaHSP70, while thermotolerance-immunity trade-off is regulated by CaHSP70 and CaNAC029, respectively, in a CaNAC2c interactor-dependent manner.
Materials and methods
Plant materials and growth conditions
The seeds of HN42, a pepper (Capsicum annuum) inbred line with middle level of thermotolerance and bacterial wilt resistance, and Nicotiana benthamiana were sown on a soil mixture [peat moss: perlite, 2:1 (v/v)] in plastic pots and were placed in a growth room at 28°C, 60–70 µmol photons m−2 s−1, a relative humidity of 70%, and a 16-h light/8-h dark photoperiod.
Construction of vectors
To generate vectors for overexpression, the full-length open reading frames (ORFs) of CaNAC2c, CaNAC029, and CaHSP70 were cloned into the entry vector pDONR207 by BP reaction with appropriate primers (Supplemental Table S1) and then cloned into the destination vectors pEarleyGate101, pDEST-15/17, and pEarleyGate103 by LR reaction, using Gateway cloning techniques (Invitrogen, Carlsbad, CA, USA). For virus-induced gene silencing (VIGS), one or two specific 300–400 bp fragments in the ORFs or 3ʹ-untranslated regions (UTRs) of CaNAC2c/d, CaHSFA5, CaHSP70, and CaNAC029 were PCR-amplified and cloned into the entry vector pDONR207 and then into the pYL279 vector. The specificity of each fragment was confirmed by Basic Local Alignment Search Tool for DNA (BLAST) searches against the pepper Zunla-1 genome (http://peppersequence.genomics.cn/page/species/blast.jsp).
Plant treatment with HTS
Pepper plants were exposed to HTS by transferring 8-leaf stage pepper plants to 42°C with 50% humidity in a growth chamber, while the control plants were kept at 28°C and 50% humidity in another growth chamber until they are harvested for further assay.
Application of plant hormones
Application of plant hormones was carried out as described previously (Dang et al., 2013). Pepper plants at the four-leaf stage were sprayed with 5 mm salicylic acid (SA), 100 mM methyl jasmonate (MeJA; both dissolved in 1:9, v:v ethanol). Mock plants were sprayed with 10% ethanol (1:9, v:v). One-month-old pepper plants were sprayed with 100 mM abscisic acid (ABA) or 10 mM Fluridone and 10 mm ethephon in sterile ddH2O. Control plants were sprayed with sterile ddH2O.
Pathogens and R. solanacearum inoculation
The highly virulent Ralstonia solanacearum strain FJC100301 was used in the present study. The bacterial cell solution used for inoculation was diluted to 108 cfu mL−1 (OD600 = 0.8) or 103 cfu mL−1 (OD600 = 0.3). For root inoculation of pepper or N. benthamiana plants planted in pots, which was well watered before mechanically damaging the roots with scissors and irrigated with 1 mL of R. solanacearum cell suspension (OD600 = 0.8) for each pot. For leaf inoculation with R. solanacearum, we inoculated leaves with 100 μL of R. solanacearum cell suspension (OD600 = 0.3) at each inoculating site using a syringe without a needle. We scored the disease index of more than 24 plants at each time point over the course of the infection cycle based on visual observation (Supplemental Table S3).
DAB and NBT staining and H2O2 content detection
The accumulation of H2O2 and reactive oxygen species (Papageorgiou et al., 2016) was assessed by staining the leaves, roots, or stems from pepper or N. benthamiana plants with 1 mg mL−1 diaminobenzidine (DAB) or Nitrotetrazolium Blue chloride (NBT). After overnight incubation in DAB and NBT, the stained leaves were cleared by boiling in lactic:glycerol:absolute ethanol (1:1:3, v:v:v) and then destained overnight in absolute ethanol. We used Beibo® BBcellProbe® plant hydrogen peroxide H2O2 detection kit to text the level of hydrogen peroxide in tissues and cells. 1:5 ratio of tissue mass (g) and reagent volume (mL; it is recommended to weigh about 0.1 g tissue and add 500 μL Buffer B) were mixed for ice bath homogenization, and then transferred to EP tube and centrifuged at 8,000 g, 4°C 10 min, then took the supernatant and put it on ice for testing. Then 2 μL BBcellProbeTM O11 hydrogen peroxide probe was added to the supernatant of the homogenate and mixed well, then incubated in a 37°C cell incubator in the dark for 20 min. Then, 488 nm excitation wavelength and 525 nm emission wavelength were used to detect the intensity of the sample fluorescence.
Gene silencing by VIGS in pepper plants
To silence CaNAC2c(d), CaHSFA5, CaHSP70, or CaNAC029, we infiltrated the cotyledons of 2-week-old pepper seedlings with Agrobacterium (Agrobacterium tumefaciens) strain GV3101 carrying the vector pTRV-RNA1 (pYL192) and pTRV-RNA2 (pYL279, the empty VIGS vector) or pYL279-CaNAC2c (CaNAC2d, CaHSFA5, CaHSP70, or CaNAC029), mixed and resuspended in induction medium (10 mM MES, 10 mM MgCl2, 200 µM acetosyringone, pH 5.6) at a 1:1 ratio to a final OD600 = 0.6. To simultaneously silence CaNAC2c and CaNAC2d, Agrobacterium cells containing pTRV-RNA1 and pTRV-RNA2-CaNAC2c and cells containing pTRV-RNA1 and pTRV-RNA2-CaNAC2d were mixed at a ratio of 1:1 and resuspended in induction medium to a final OD600 = 0.6. A volume of 100 μL was infiltrated into the cotyledons of 2-week-old pepper seedlings, which were then placed in the dark at 16°C for 56 h, and then moved to a growth room at 28°C, 60–70 µmol photons m−2 s−1, a relative humidity of 70%, and a 16-h light/8-h dark photoperiod. At 15 dpi, the success and specificity of gene silencing were assessed in pepper plants challenged with HTS by measuring the transcript levels of the gene(s) targeted for silencing.
Transient expression of CaNAC2c in pepper leaves
For transient expression analysis, Agrobacterium cells harboring the 35Spro:CaNAC2c-GFP construct (or 35Spro:GFP as control) were grown overnight and then resuspended in induction medium (10 mM MES, 10 mM MgCl2, 200 µM acetosyringone, pH 5.6) to OD600 = 0.8. Approximately 1 mL was infiltrated into the leaves of pepper plants at the eight-leaf stage using a syringe. The infiltrated leaves were collected at the indicated time points for further use.
Generation of transgenic N. benthamiana plants overexpressing CaNAC2c
Nicotiana benthamiana leaf discs were transformed with Agrobacterium strain GV3101 carrying the 35Spro:CaNAC2c-GFP vector according to the method of Regner et al. (1992) and Bardonnet et al. (1994). Nineteen independent T0 transgenic N. benthamiana plants were selected by hygromycin (5 mg L−1) selection and validated by PCR and reverse transcription-quantitative PCR (RT-qPCR). The T0 plants were then allowed to self-pollinate and set seeds. Positive transformants were propagated for two or three generations and subjected to the same selection to obtain T2 and T3 seeds. Two T3 transgenic lines that exhibited moderate levels of CaNAC2c transcripts without phenotypic abnormality were selected for further analysis.
RT-qPCR
We performed RT-qPCR to determine the relative transcript levels of selected genes with specific primers (Supplemental Table S1) and the SYBR Premix Ex Taq II system (TaKaRa) on a BIO-RAD Real-time PCR system (Foster City, CA, USA) according to the manufacturer’s instructions. Total RNA preparation and real-time qPCR were carried out following procedures described in our previous studies from four biological replicates (Cai et al., 2015). Total RNA was isolated from pepper samples using TRIzol Reagent according to the manufacturer’s protocol (Invitrogen, Canada). mRNAs were reverse-transcribed into cDNA using a reverse transcription system and an oligo(dT) primer (Takara Biotechnology, Japan). Data were analyzed by the Livak method (Livak and Schmittgen, 2001; Zhang et al., 2015) and expressed as normalized relative expression level (2−ΔΔCT) of the respective genes. Relative transcript levels were normalized to CaACTIN (GQ339766) and 18S ribosomal RNA (EF564281), respectively.
Chlorophyll fluorescence spectrophotometry
We used a MINI Imaging PAM instrument (Heinz Walz GmbH, Effeltrich, Germany) to measure Fv/Fm and △F/Fm′ values from pepper and N. benthamiana leaves. The plants were adapted to darkness for 15 min before being placed into the instrument for measurements according to the method of Schreiber et al. (2012).
Immunoblot analysis
We extracted total protein from pepper samples by adding extraction buffer (10% glycerol, 25 mM Tris-HCl pH 7.5, 150 mM sodium chloride, 1 mM EDTA, 1% Triton X-100, 10 mM dithiothreitol, 1 × plant protease inhibitor, 2% (w/w) polyvinylpyrrolidone [PVPP]) to samples ground to a fine powder in liquid nitrogen. Protein extracts were incubated at 4°C with anti-hemagglutinin (anti-GFP) agarose beads (Thermo Fisher Scientific, Waltham, MA, USA) overnight. Beads were collected using a magnetic rack and washed 3 times with Tris-buffered saline and Tween-20 (0.05%). Eluted proteins were probed by immunoblotting using anti-GFP–peroxidase antibodies (Abcam, Cambridge, UK).
ChIP assay
ChIP assays were performed according to a previous study (Khan et al., 2018). We inoculated three fully extended pepper leaves at the six-leaf stage with Agrobacterium cells harboring the 35Spro:CaNAC2c-GFP construct. The infiltrated leaves were harvested at 48 h post-inoculation (hpi) and crosslinked in a 1% formaldehyde solution; chromatin was isolated and sheared into 300–500 bp fragments, followed by immunoprecipitation of DNA–protein complexes using anti-GFP antibodies. The crosslinking was then reversed and DNA purified and used as a template for qPCR using primer pairs specific to a CATGTG-box-containing fragment or CATGTG-free fragment in the CaHSFA5 promoter by semi-quantitative PCR using specific primer pairs (Supplemental Table S1).
Production and purification of recombinant CaNAC2c-GST/6×His
We introduced the pDEST-15/17 plasmid harboring T7:CaNAC2c-GST/6×His into Escherichia coli (E. coli) strain BL21 (DE3). Production of the fusion protein was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 20°C for 12 h. To purify the recombinant proteins carrying the GST tag, Beaver BeadsTM GSH (Beaver Biosciences, China) were washed thrice with Buffer A (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) and then mixed with protein extract for 3 h at 4°C. The beads were washed 5 times with Buffer A and the target protein was eluted in Buffer B (50 mM Tris-HCl, 10 mM reduced glutathione, pH 8.0).
Pull-down assays
We immobilized the fusion protein CaNAC2c-6×His (0.1 mg) onto nickel Smart Beads 6FF (Smart-Life Sciences, China) and incubated the mixture under shaking for 3 h at 4°C. Proteins bound to the beads were subsequently washed with wash buffer (20 mM PBS with 0.5 mM imidazole, pH 7.4) and eluted with elution buffer (20 mM PBS with 250 mM imidazole, pH 7.4). Eluted proteins were resolved by SDS-PAGE gel electrophoresis and detected with an antibody against 6×His (PM013; 1:1,000 dilution; MBL International, Woburn, MA, USA).
Liquid chromatography-tandem mass spectrometry analysis
Isolated proteins and potential CaNAC2c interactors were analyzed on an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific), as previously described (Zuo et al., 2001; Wang et al., 2018). The samples were dissolved in 10 μL of a 10% formic acid solution, then analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with an online sodium spray ion source. Peptide samples (5 μL) were loaded onto the trap column (Acclaim PepMapC18, 100 μm × 2 cm; Thermo Fisher Scientific) at a flow rate of 10 μL min−1, and subsequently separated on a 60-min gradient on the analytical column (Acclaim PepMapC18, 75 μm × 15 cm). The column flow was controlled at 300 nL min−1, and the electrospray voltage was 2 kV. Full scan spectra (m/z 350–1,550) were collected at a mass resolution of 60 K, and HCD MS/MS scans were subsequently performed at a resolution of 30 K with a dynamic exclusion for 30 s.
The original mass spectrometry collection files were imported into Proteome Discover 2.1 for retrieval. We searched peptide fragments against the Zunla pepper database and used BLAST searches at the National Center for Biotechnology Information (NCBI) database to annotate the functions of the corresponding proteins.
BiFC assay and subcellular localization
We determined the subcellular localization of CaNAC2c as described previously (Shen et al., 2016). Agrobacterium cells harboring the 35Spro:CaNAC2c-YFP construct were infiltrated into N. benthamiana leaves. The YFP signal was detected 48 hpi. The open reading frames for CaNAC2c or CaNAC029/CaHSP70 in the pDONR vector were directly introduced into the destination vectors pSPYCE or pSPYNE via Gateway cloning to generate 35Spro:CaNAC2c-YFPc-HA and 35Spro:CaNAC029/CaHSP70-YFPn-MYC. Vectors were then introduced into Agrobacterium strain GV3101, and cells harboring 35S:CaNAC2c-YFPc-HA and 35S: CaNAC029/CaHSP70-YFPn-MYC were co-infiltrated into N. benthamiana leaves. BiFC assays were performed as described previously (Choi et al., 2012) and the fluorescent signal from Agrobacterium-infiltrated N. benthamiana leaves was observed at 48 hpi. YFP fluorescence was collected on a confocal microscope (YFP:527 nm/CFP:485 nm/TCS SP8, Leica Microsystems, Germany), Images were obtained at 100 Hz, 42% Exposure Rate, 1.000 Gain, and 5% Offset.
Co-IP assay
Nicotiana benthamiana leaves were infiltrated with 35Spro:CaNAC2c-YFPc-HA and 35Spro:CaNAC029/CaHSP70-YFPn-MYC and harvested at 48 hpi. Total protein extracts were prepared using protein extraction buffer (10% glycerol, 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2% Triton X-100, 10 mM DTT, 1× complete protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA), and 2% (w/v) PVPP). Extracted proteins were incubated with monoclonal anti-HA magnetic beads (Sigma-Aldrich) at 4°C overnight. Beads were then collected with a magnet and washed 3 times with protein extraction buffer. Eluted proteins were separated by SDS-PAGE electrophoresis and immunoblotted using anti-HA-peroxidase antibody or anti-MYC-peroxidase antibody (Sigma-Aldrich)
Electrophoretic mobility shift assay
Cy5-labeled double-stranded DNA fragments containing the CATGTG motif or its mutated version (GGGGGG) were commercially synthesized to use as probes in EMSA. CaNAC2c-GST or GST proteins were incubated with wild-type or mutated probe labeled with a Cy5 fluorochrome in 5× binding buffer (1 M Tris-HCl pH 7.5, 5 M NaCl, 1 M KCl, 1 M MgCl2, 0.5 M EDTA pH 8.0, 10 mg mL−1 BSA). The mixture was separated by PAGE gel and then scanned on an Odyssey@ CLX instrument (LI-COR, USA).
MST assessment of interaction of protein with promoter fragments in solution
The binding of CaNAC2c to the CaHSFA5 promoter was analyzed by microscale thermophoresis (MST) in solution (Zillner et al., 2012). GFP fused to CaNAC2c was used as fluorescent label against a fragment containing the CATGTG motif within the CaHSFA5 promoter, which was amplified by PCR with a specific primer pair, followed by purification. The fragment containing the mutant version of CATGTG motif (GGGGGG) was amplified by PCR by conventional overlapping PCR-based site-directed mutagenesis. The two DNA fragments were used as the nonfluorescent molecules in the assay. Interaction between protein and DNA was measured as described previously (Qiu et al., 2018). We used the Nano Temper Analysis 1.2.20 software to fit the data and determine apparent Kd values (Zillner et al., 2012; Papageorgiou et al., 2016).
Statistical analyses
Statistical analyses were performed with the DPS software package. Data are shown as means ± SD obtained from three or four replicates; different letters indicate significant differences among means (P < 0.01), as calculated with Fisher’s protected least-significant-difference (LSD) test.
Accession numbers
CaNAC2c (XP_016575179.1); CaNAC2d (XP_016569664.1); CaHSFA5 (XM_016695662.1); CaNAC029 (A0A1U8EJR9); CaHSP70 (A0A1U8E6Q9); CaHSFB2a (XP_016564681.1); CaHSP24 (HM132040); CaNPR1 (X61679.1); CaDEF1 (AF442388); CaHIR1 (AAX20040); CaNAC07 (XM_016697660.1); CaNAC08 (XM_016725353.1); CaNAC059 (XM_016723243.1); CaLOX14 (NM_001324652.1); CaLOX31 (NM_001324819.1); CaActin (GQ339766); NbAPX (XP_016432750.1); NbHSFA5 (XM_016600975.1); NbHSP18 (XP_016481364.1); NbsHSP (XP_016463300.1); NbDEF1 (ABU40984.1); NtEF-1a (D63396).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Transcript levels of CaNAC2c and pepper NAC family members to HTS or RSI.
Supplemental Figure S2. Deduced amino acid sequences of pepper NAC2 proteins and their phylogenetic relationship.
Supplemental Figure S3. Nuclear localization of CaNAC2c in N. benthamiana epidermal cells.
Supplemental Figure S4. Silencing of CaNAC2c in pepper enhances thermotolerance.
Supplemental Figure S5. The specificity of CaNAC2c silencing in pepper plants by VIGS.
Supplemental Figure S6. Overexpression of CaNAC2c enhances thermotolerance in N. benthamiana plants.
Supplemental Figure S7. N. benthamiana plants overexpressing CaNAC2c exhibit enhanced Fv/Fm, △F/Fm and decreased ion leakage and lower ROS levels upon HTS.
Supplemental Figure S8. Silencing of CaNAC2c in different pepper germplasms.
Supplemental Figure S9. N. benthamiana plants overexpressing CaNAC2c exhibit enhanced resistance to RSI.
Supplemental Figure S10. Effect of CaNAC2c/2d silencing on the thermotolerance of pepper plants.
Supplemental Figure S11. CaNAC2c silencing promotes the growth of pepper plants.
Supplemental Figure S12. N. benthamiana plants overexpressing CaNAC2c repress growth and delay development.
Supplemental Figure S13. CaNAC2c influences the expression of genes related to thermotolerance and immunity.
Supplemental Figure S14. Relative transcript levels of marker genes by the exogenous application of ABA, Fluridon, SA, or MeJA.
Supplemental Figure S15. FPKMs of members in pepper LOX family in roots of pepper plants challenged with RSI.
Supplemental Figure S16. CaHSFA5 acts as negative regulator of pepper growth.
Supplemental Figure S17. Silencing of CaHSFA5 promotes growth of N. benthamiana plants overexpressing CaNAC2c.
Supplemental Figure S18. Silencing of NbHSFA5 significantly decreases thermotolerance but does not affect resistance of N. benthamiana plants overexpressing CaNAC2c to RSI.
Supplemental Figure S19. CaHSP70 is upregulated by ABA and CaNAC029 is upregulated by MeJA.
Supplemental Figure S20. The effect of transient co-overexpression of CaNAC2c and CaHSP70 on the enrichment of CaNAC2c at the CaHSFA5 promoter and expression of immunity or thermotolerance-related genes at room temperature and upon HTS challenge.
Supplemental Table S1. Primers used in this study.
Supplemental Table S2. Sequence similarity between CaNAC2c predicted protein sequence and its putative orthologs from other plant species.
Supplemental Table S3. Disease index for pepper plants infected with Ralstonia solanacearum.
Supplemental Table S4. Results of CaNAC2c co-expression analysis under HTS in pepperhub.
Supplemental Table S5. Proteins identified as potential interaction candidates with CaNAC2c using LC-MS/MS.
Supplementary Material
Acknowledgments
The authors thank Mark D. Curtis for kindly providing the Gateway destination vectors and Dr. S. P. Dinesh-Kumar of Yale University for the pTRV1 and pTRV2 vectors.
Funding
This work was supported by the National Natural Science Foundation of China (31401890, 31401312), the Scientific Research Foundation of the Graduate School of Fujian Agriculture and Forestry University (324-1122yb068), and Development Fund Project of Fujian Agriculture and Forestry University (CXZX2016158, CXZX2017548).
Conflict of interest statement. The authors have no conflicts of interest to declare.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Shuilin He (shlhe201304@aliyun.com).
References
- Ashraf MF, Yang S, Wu R, Wang Y, Hussain A, Noman A, Khan MI, Liu Z, Qiu A, Guan D. et al. (2018) Capsicum annuum HsfB2a positively regulates the response to Ralstonia solanacearum infection or high temperature and high humidity forming transcriptional cascade with CaWRKY6 and CaWRKY40. Plant Cell Physiol 59:2608–2623 [DOI] [PubMed] [Google Scholar]
- Bardonnet N, Hans F, Serghini MA, Pinck L (1994) Protection against virus infection in tobacco plants expressing the coat protein of grapevine fanleaf nepovirus. Plant Cell Rep 13:357–360 [DOI] [PubMed] [Google Scholar]
- Birkenbihl R, Kracher B, Ross A, Kramer K, Finkemeier I, Somssich I (2018) Principles and characteristics of the Arabidopsis WRKY regulatory network during early MAMP-triggered immunity. Plant J Cell Mol Biol 96:487–502 [DOI] [PubMed] [Google Scholar]
- Breen S, Williams S, Outram M, Kobe B, Solomon P (2017) Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci 22:871–879 [DOI] [PubMed] [Google Scholar]
- Cai H, Yang S, Yan Y, Xiao Z, Cheng J, Wu J, Qiu A, Lai Y, Mou S, Guan D. et al. (2015) CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J Exp Bot 66:3163–3174 [DOI] [PubMed] [Google Scholar]
- Canonne J, Marino D, Jauneau A, Pouzet C, Briere C, Roby D, Rivas S (2011) The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell 23:3498–3511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chandra S, Martin G, Low P (1996) The Pto kinase mediates a signaling pathway leading to the oxidative burst in tomato. Proc Natl Acad Sci USA 93:13393–13397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Yu R, Feng J, Chen H, Eri H, Gao G (2020) NAC transcription factor involves in regulating bacterial wilt resistance in potato. Funct Plant Biol 47:925–936 [DOI] [PubMed] [Google Scholar]
- Chen H, Chu P, Zhou Y, Ding Y, Li Y, Liu J, Jiang L, Huang S (2016) Ectopic expression of NnPER1, a Nelumbo nucifera 1-cysteine peroxiredoxin antioxidant, enhances seed longevity and stress tolerance in Arabidopsis. Plant J Cell Mol Biol 88:608–619 [DOI] [PubMed] [Google Scholar]
- Cheng Q, Dong L, Gao T, Liu T, Li N, Wang L, Chang X, Wu J, Xu P, Zhang S (2018) The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J Exp Bot 69:2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu J, Chen Z (2013) Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant 6:287–300 [DOI] [PubMed] [Google Scholar]
- Choi D, Hwang B (2011) Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23:823–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Hwang IS, Hwang BK (2012) Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24:1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Kim NH, Hwang BK (2015) The pepper phosphoenolpyruvate carboxykinase CaPEPCK1 is involved in plant immunity against bacterial and oomycete pathogens. Plant Mol Biol 89:99–111 [DOI] [PubMed] [Google Scholar]
- Choi HW, Lee BG, Kim NH, Park Y, Lim CW, Song HK, Hwang BK (2008) A role for a menthone reductase in resistance against microbial pathogens in plants. Plant Physiol 148:383–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang FF, Wang YN, Yu L, Eulgem T, Lai Y, Liu ZQ, Wang X, Qiu AL, Zhang TX, Lin J. et al. (2013) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ 36:757–774 [DOI] [PubMed] [Google Scholar]
- Dangl J (1998) Innate immunity. Plants just say NO to pathogens. Nature 394:525–527 [DOI] [PubMed] [Google Scholar]
- Dang FF, Wang YN, Lu Y, Eulgem T, Lai Y, Liu ZQ, Qiu AL (2013) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ 36: 757–774 [DOI] [PubMed] [Google Scholar]
- de Vries J, Evers J, Poelman E (2017) Dynamic plant-plant-herbivore interactions govern plant growth-defence integration. Trends Plant Sci 22:329–337 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98:13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Xu Z, Xu L, Galli M, Gallavotti A, Dooner HK, Chuck G (2020) Necrotic upper tips1 mimics heat and drought stress and encodes a protoxylem-specific transcription factor in maize. Proc Natl Acad Sci USA 117:20908–20919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Zhao J, Tzeng DTW, Liu Y, Deng L, Yang T, Zhai Q, Wu F, Huang Z, Zhou M. et al. (2017) MYC2 orchestrates a hierarchical transcriptional cascade that regulates Jasmonate-mediated plant immunity in tomato. Plant Cell 29:1883–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Liao K, Du H, Xu Y, Song H, Li X, Xiong L (2015) A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot 66:6803–6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Su L, Sun X, Li X, Sun M, Karungo SK, Fang S, Chu J, Li S, Xin H (2016) Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J Exp Bot 67:2829–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, You J, Xie K, Xie W, Xiong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genom 280:547–563 [DOI] [PubMed] [Google Scholar]
- Fobert P, Després C (2005) Redox control of systemic acquired resistance. Curr Opin Plant Biol 8:378–382 [DOI] [PubMed] [Google Scholar]
- Fragkostefanakis S, Roth S, Schleiff E, Scharf KD (2015) Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ 38:1881–1895 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9:436–442 [DOI] [PubMed] [Google Scholar]
- Germain H, Lachance D, Pelletier G, Fossdal CG, Solheim H, Seguin A (2012) The expression pattern of the Picea glauca Defensin 1 promoter is maintained in Arabidopsis thaliana, indicating the conservation of signalling pathways between angiosperms and gymnosperms. J Exp Bot 63:785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve K, La Cour T, Jensen MK, Poulsen FM, Skriver K (2003) Interactions between plant RING-H2 and plant-specific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochemistry 371:97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Yang F, Xia X, Shi Y, Yang S, Cheng W, He S (2018) CaHSL1 acts as a positive regulator of pepper thermotolerance under high humidity and is transcriptionally modulated by CaWRKY40. Front Plant Sci 9:1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q, Yue X, Zeng H, Zhu J (2014) The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26:438–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Major I, Howe G (2018) Resolution of growth-defense conflict: mechanistic insights from jasmonate signaling. Curr Opin Plant Biol 44:72–81 [DOI] [PubMed] [Google Scholar]
- Guo T, Mao X, Zhang H, Zhang Y, Fu M, Sun Z, Kuai P, Lou Y, Fang Y (2017) Lamin-like proteins negatively regulate plant immunity through NAC with transmembrane Motif1-Like9 and nonexpressor of PR GENES1 in Arabidopsis thaliana. Mol Plant 10:1334–1348 [DOI] [PubMed] [Google Scholar]
- Hiruma K, Fukunaga S, Bednarek P, Pislewska-Bednarek M, Watanabe S, Narusaka Y, Shirasu K, Takano Y (2013) Glutathione and tryptophan metabolism are required for Arabidopsis immunity during the hypersensitive response to hemibiotrophs. Proc Natl Acad Sci USA 110:9589–9594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G, Major I, Koo A (2018) Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol 69:387–415 [DOI] [PubMed] [Google Scholar]
- Hu T, Yang Y, Zeng H, Hu Z, Chen Z. (2013) Transgenic pepper plants carrying RNA interference constructs of CaCOI1 gene show severe abnormality. Mol Breed 31:971–979 [Google Scholar]
- Hua J (2009) From freezing to scorching, transcriptional responses to temperature variations in plants. Curr Opin Plant Biol 12:568–573 [DOI] [PubMed] [Google Scholar]
- Huang YC, Niu CY, Yang CR, Jinn TL (2016) The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol 172:1182–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs T, Tschope M, Colling C, Hahlbrock K, Scheel D (1997) Elicitor-stimulated ion fluxes and O2- from the oxidative burst are essential components in triggering defense gene activation and phytoalexin synthesis in parsley. Proc Natl Acad Sci USA 94:4800–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda M, Lamparová L, Zubíková A, Burketová L, Martinec J, Krčková Z (2019) Temporary heat stress suppresses PAMP-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Mol Plant Pathol 20:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Wei Z, Xu J, Chen H, Zhang Y, She X, Macho AP, Ding W, Liao B (2017) Bacterial wilt in China: history, current status, and future perspectives. Front Plant Sci 8:1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J, Dangl J (2006) The plant immune system. Nature 444:323–329 [DOI] [PubMed] [Google Scholar]
- Jung HW, Lim CW, Lee SC, Choi HW, Hwang CH, Hwang BK (2008) Distinct roles of the pepper hypersensitive induced reaction protein gene CaHIR1 in disease and osmotic stress, as determined by comparative transcriptome and proteome analyses. Planta 227:409–425 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Lapchyk L, Fukushige H, Hildebrand D, Klessig D, Kachroo P (2003) Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15:2952–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MI, Zhang Y, Liu Z, Hu J, Liu C, Yang S, Hussain A, Furqan Ashraf M, Noman A, Shen L. et al. (2018) CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int J Mol Sci 19: 1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khedia J, Agarwal P, Agarwal PK (2018) AlNAC4 transcription factor from halophyte aeluropus lagopoides mitigates oxidative stress by maintaining ROS homeostasis in transgenic tobacco. Front Plant Sci 9:1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Hwang B (2014) An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J Exp Bot 65:2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YW, Jung HJ, Park JI, Hur Y, Nou IS (2015) Response of NBS encoding resistance genes linked to both heat and fungal stress in Brassica oleracea. Plant Physiol Biochem 86:130–136 [DOI] [PubMed] [Google Scholar]
- Kim NH, Lee DH, Choi du S, Hwang BK. (2015) The pepper GNA-related lectin and PAN domain protein gene, CaGLP1, is required for plant cell death and defense signaling during bacterial infection. Plant Sci 241:307–315 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf K (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10:310–316 [DOI] [PubMed] [Google Scholar]
- Kyndt T, Nahar K, Haeck A, Verbeek R, Demeestere K, Gheysen G (2017) Interplay between Carotenoids, abscisic acid and jasmonate guides the compatible rice-meloidogyne graminicola interaction. Front Plant Sci 8:951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yun H, Kwon C (2012) Molecular communications between plant heat shock responses and disease resistance. Mol Cells 34:109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fu Q, Chen L, Huang W, Yu D (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233:1237–1252 [DOI] [PubMed] [Google Scholar]
- Li Y, Yang Y, Hu Y, Liu H, He M, Yang Z, Kong F, Liu X, Hou X (2019) DELLA and EDS1 form a feedback regulatory module to fine-tune plant growth-defense tradeoff in arabidopsis. Mol Plant 12:1485–1498 [DOI] [PubMed] [Google Scholar]
- Liu J, Feng L, Li J, He Z (2015a) Genetic and epigenetic control of plant heat responses. Front Plant Sci 6:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kracher B, Ziegler J, Birkenbihl RP, Somssich IE (2015b) Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4:e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Lyu YS, Yang W, Yang ZT, Lu SJ, Liu JX (2020) A membrane-associated NAC transcription factor OsNTL3 is involved in thermotolerance in rice. Plant Biotechnol J 18:1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Wang B, Li Z, Peng Z, Zhang J (2018) TsNAC1 is a key transcription factor in abiotic stress resistance and growth. Plant Physiol 176:742–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Yu H, Deng Y, Zheng J, Liu M, Ou L, Yang B, Dai X, Ma Y, Feng S. et al. (2017) PepperHub, an informatics hub for the chili pepper research community. Mol Plant 10:1129–1132 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32:317–328 [DOI] [PubMed] [Google Scholar]
- Lozano-Duran R, Macho AP, Boutrot F, Segonzac C, Somssich IE, Zipfel C (2013) The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. Elife 2:e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA. et al. (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew I, Das S, Mahto A, Agarwal P (2016) Three rice NAC transcription factors heteromerize and are associated with seed size. Front Plant Sci 7:1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell J, Dangl J (2000) Signal transduction in the plant immune response. Trends Biochem Sci 25:79–82 [DOI] [PubMed] [Google Scholar]
- Meldau S, Ullman-Zeunert L, Govind G, Bartram S, Baldwin IT (2012) MAPK-dependent JA and SA signalling in Nicotiana attenuata affects plant growth and fitness during competition with conspecifics. BMC Plant Biol 12:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes GC, Reis PA, Calil IP, Carvalho HH, Aragao FJ, Fontes EP (2013) GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc Natl Acad Sci USA 110:19627–19632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37:118–125 [DOI] [PubMed] [Google Scholar]
- Moore JW, Loake GJ, Spoel SH (2011) Transcription dynamics in plant immunity. Plant Cell 23:2809–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami S, Saeki K, Toda K, Ishida T, Sawa S (2020) The atypical E2F transcription factor DEL1 modulates growth-defense tradeoffs of host plants during root-knot nematode infection. Sci Rep 10:8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819:97–103 [DOI] [PubMed] [Google Scholar]
- Negi S, Tak H, Ganapathi TR (2018) A banana NAC transcription factor (MusaSNAC1) impart drought tolerance by modulating stomatal closure and H2O2 content. Plant Mol Biol 96:457–471 [DOI] [PubMed] [Google Scholar]
- Nejat N, Mantri N (2017) Plant immune system: crosstalk between responses to biotic and abiotic stresses the missing link in understanding plant defence. Curr Iss Mol Biol 23:1–16 [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea C, Kryger M, Stender EG, Kragelund BB, Willemoes M, Skriver K (2015) Protein intrinsic disorder in Arabidopsis NAC transcription factors: transcriptional activation by ANAC013 and ANAC046 and their interactions with RCD1. Biochem J 465:281–294 [DOI] [PubMed] [Google Scholar]
- Papageorgiou AC, Adam PS, Stavros P, Nounesis G, Meijers R, Petratos K, Vorgias CE (2016) HU histone-like DNA-binding protein from Thermus thermophilus: structural and evolutionary analyses. Extremophiles 20:695–709 [DOI] [PubMed] [Google Scholar]
- Perochon A, Kahla A, Vranić M, Jia J, Malla K, Craze M, Wallington E, Doohan F (2019) A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnol J 17:1892–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci 17:369–381 [DOI] [PubMed] [Google Scholar]
- Qiu A, Lei Y, Yang S, Wu J, Li J, Bao B, Cai Y, Wang S, Lin J, Wang Y. et al. (2018) CaC3H14 encoding a tandem CCCH zinc finger protein is directly targeted by CaWRKY40 and positively regulates the response of pepper to inoculation by Ralstonia solanacearum. Mol Plant Pathol 19:2221–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regner F, Machado AdC, Machado MLdC, Steinkellner H, Katinger H (1992) Coat protein mediated resistance to Plum Pox Virus in Nicotiana clevelandii and N. benthamiana. Plant Cell Rep 11:30–33 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49:317–343 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258 [DOI] [PubMed] [Google Scholar]
- Sarde S, Bouwmeester K, Venegas-Molina J, David A, Boland W, Dicke M (2019) Involvement of sweet pepper CaLOX2 in jasmonate-dependent induced defence against Western flower thrips. J Integr Plant Biol 61:1085–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarde S, Kumar A, Remme R, Dicke M (2018) Genome-wide identification, classification and expression of lipoxygenase gene family in pepper. Plant Mol Biol 98:375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreck R, Baeuerle P (1991) A role for oxygen radicals as second messengers. Trends Cell Biol 1:39–42 [DOI] [PubMed] [Google Scholar]
- Schreiber U, Klughammer C, Kolbowski J (2012) Assessment of wavelength-dependent parameters of photosynthetic electron transport with a new type of multi-color PAM chlorophyll fluorometer. Photosynth Res 113:127–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Kuang JF, Lu WJ, Chen JY (2014) Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ 37:2116–2127 [DOI] [PubMed] [Google Scholar]
- Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC (2013) Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant 6:250–260 [DOI] [PubMed] [Google Scholar]
- Shen L, Liu Z, Yang S, Yang T, Liang J, Wen J, Liu Y, Li J, Shi L, Tang Q. et al. (2016) Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature-high humidity challenge in a positive feedback loop with CaWRKY40. J Exp Bot 67:2439–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Yang S, Yang T, Liang J, Cheng W, Wen J, Liu Y, Li J, Shi L, Tang Q. et al. (2016) CaCDPK15 positively regulates pepper responses to Ralstonia solanacearum inoculation and forms a positive-feedback loop with CaWRKY40 to amplify defense signaling. Sci Rep 6:22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Liu Q, Hu B, Wu W (2017) Photoreceptor PhyB involved in arabidopsis temperature perception and heat-tolerance formation. Int J Mol Sci 18: 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Howell S (2014) Stress sensing in plants by an ER stress sensor/transducer, bZIP28. Front Plant Sci 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Zhang X, Zhang Q, Ji X, Jia Y, Wang H, Niu L, Zhang Y (2019) Comparative transcriptome profiling uncovers a Lilium regale NAC transcription factor, LrNAC35, contributing to defence response against cucumber mosaic virus and tobacco mosaic virus. Mol Plant Pathol 20:1662–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman MG, Rafii MY, Ismail MR, Malek MA, Abdul Latif M (2014) Heritability and genetic advance among chili pepper genotypes for heat tolerance and morphophysiological characteristics. Sci World J 2014:308042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivek PJ, Resmi MS, Sreekumar S, Sivakumar KC, Tuteja N, Soniya EV (2016) Calcium-dependent protein kinase in ginger binds with importin-alpha through its junction domain for nuclear localization, and further interacts with NAC transcription factor. Front Plant Sci 7:1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wu B, Yin H, Fan Z, Li J (2017) Overexpression of CaAPX induces orchestrated reactive oxygen scavenging and enhances cold and heat tolerances in tobacco. BioMed Res Int 2017:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang X, Goatley M, Ervin E (2014) Heat shock proteins in relation to heat stress tolerance of creeping bentgrass at different N levels. PLoS One 9:e102914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao Y, Li Z, Hsu CC, Liu X, Fu L, Hou YJ, Du Y, Xie S, Zhang C. et al. (2018) Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol Cell 69:100–112.e106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Kapos P, Cheng YT, Li M, Zhang Y, Li X (2014) NLR-associating transcription factor bHLH84 and its paralogs function redundantly in plant immunity. PLoS Pathog 10:e1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue GP, Drenth J, McIntyre CL (2015) TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J Exp Bot 66:1025–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Chen N, Qu YY, Dong XC, Meng QW, Zhao SJ (2008) Overexpression of sweet pepper glycerol-3-phosphate acyltransferase gene enhanced thermotolerance of photosynthetic apparatus in transgenic tobacco. J Integr Plant Biol 50:613–621 [DOI] [PubMed] [Google Scholar]
- Yong T, Zhao CY, Tan ST, Xue HW, Qu LJ (2016) Arabidopsis type II phosphatidylinositol 4-kinase PI4Kγ5 regulates auxin biosynthesis and leaf margin development through interacting with membrane-bound transcription factor ANAC078. PLoS Genet 12:e1006252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones J, Doke N (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15:706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Wang L, Zhao R, Sheng J, Zhang S, Li R, Shen L (2019) Knockout of SlMAPK3 enhances tolerance to heat stress involving ROS homeostasis in tomato plants. BMC Plant Biol 19:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HX, Ali M, Feng XH, Jin JH, Huang LJ, Khan A, Lv JG, Gao SY, Luo DX, Gong ZH. (2018) CaSBP12A novel transcription factor gene negatively regulates the defense response against in Pepper (L.). Int J Mol Sci 20: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen Y, Zhao L, Li C, Yu J, Li T, Yang W, Su H, Wang L (2020) A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol 40:413–423 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Kuang H, Chen C, Yan J, Do-Umehara H, Liu X, Dada L, Ridge K, Chandel N, Liu J (2015) The kinase Jnk2 promotes stress-induced mitophagy by targeting the small mitochondrial form of the tumor suppressor ARF for degradation. Nat Immunol 16:458–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Luo F, Zhong Y, He J, Li L (2020) Modulation of NAC transcription factor NST1 activity by XYLEM NAC DOMAIN1 regulates secondary cell wall formation in Arabidopsis. J Exp Bot 71:1449–1458 [DOI] [PubMed] [Google Scholar]
- Zhuang K, Gao Y, Liu Z, Diao P, Sui N, Meng Q, Meng C, Kong F (2020) WHIRLY1 regulates HSP21.5A expression to promote thermotolerance in tomato. Plant Cell Physiol 61:169–177 [DOI] [PubMed] [Google Scholar]
- Zillner K, Jerabek-Willemsen M, Duhr S, Braun D, Langst G, Baaske P (2012) Microscale thermophoresis as a sensitive method to quantify protein: nucleic acid interactions in solution. Methods Mol Biol 815:241–252 [DOI] [PubMed] [Google Scholar]
- Zuo X, Echan L, Hembach P, Tang HY, Speicher KD, Santoli D, Speicher DW (2001) Towards global analysis of mammalian proteomes using sample prefractionation prior to narrow pH range two-dimensional gels and using one-dimensional gels for insoluble and large proteins. Electrophoresis 22:1603–1615 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.